Abstract
In the process of profiling the secondary metabolites of actinobacteria isolated from the Saudi coastal habitats for production of antibiotics and anti-cancer drugs, the cultures of strain WH1 that was identified as Streptomyces heliomycini exhibited strong antibacterial activity against Staphylococcus aureus. By means of MS and NMR techniques, the active compounds were characterized as actinomycins X0β, X2, and D, respectively. The research on the productivity of this strain for actinomycins revealed that the highest production of actinomycins X0β, X2, and D was reached in the medium MII within 5% salinity and pH 8.5. In this optimized condition, the fermentation titers of actinomycins X0β, X2, and D were 107.6 ± 4.2, 283.4 ± 75.3, and 458.0 ± 76.3 mg/L, respectively. All the three actinomycins X0β, X2, and D showed potent cytotoxicities against the MCF-7, K562, and A549 tumor cell lines, in which actinomycin X2 was the most active against the three tumor cell lines with the IC50 values of 0.8–1.8 nM. Both actinomycins X2 and D showed potent antibacterial activities against S. aureus and the methicillin-resistant S. aureus, Bacillus subtilis, and B. cereus and the actinomycin X2 was more potent.
Keywords: marine-derived actinobacteria, Streptomyces heliomycini WH1, actinomycins, cytotoxicity, antimicrobial activity
Introduction
The actinobacteria have been reported as the producers of two-thirds of the microbially-derived antibiotics known today (Newman et al., 2003). However, the rate for identification of novel compounds has decreased significantly from the widely explored normally terrestrial strains (Lam, 2006; Tiwari et al., 2015). Therefore, the discovery of new strains of actinobacteria may be the first and the key step to obtain novel compounds with bioactivity and subsequently to discover the natural product-based drugs. Increasing number of studies show that unusual and underexplored habitats, such as desert and marine ecosystems, are a rich source of novel actinobacteria with the capacity to produce new compounds with bioactivities (Bister et al., 2004; Bull and Stach, 2007; Fu et al., 2011, 2012, 2014; Wang et al., 2013).
Actinomycins are a class of chromopeptide antibiotics produced by Streptomyces sp., most of which share the same phenoxazinone chromophore. Actinomycin D (Act-D) is the most extensively studied example and is widely used as an anti-tumor drug for treatment of childhood rhabdomyosarcoma and Wilms' tumor, etc. The binding of Act-D to DNA is the basis for the antitumor activity (Koba and Konopa, 2005). This characteristic also makes Act D and 7-aminoactinomycin D as the useful tools for biological investigation (Chen Chiao et al., 1979). Act D also exhibit antiviral activity against coxsackievirus B3 (Saijets et al., 2003) and human immunodeficiency virus HIV-1 (Rill and Hecker, 1996), as well as the enzyme inhibitors against sereine proteinases (Betzel et al., 1993), acid phosphatase (Kapp and Okada, 1972), and tryptophan 2,3-dioxygenase (Killewich et al., 1975). However, its structurally related actinomycins (Acts), Act-X2 and Act-X0β, have not been well investigated for their medicinal properties due to the limits of the available amounts (Kurosawa et al., 2006). To discover new actinobacterial strains and optimize their cultural conditions to produce Acts, we carried out screenings of the marine-derived actinobacterial strains from the coastal habitats of Saudi Arabia. A producing strain of Acts, designated WH1, was identified as Streptomyces heliomycini whose products showed significant inhibition on the growth of Staphyloccocus aureus. A chemical study on the ethyl acetate (EtOAc) extract of the fermentation broth of S. heliomycini WH1 resulted in the isolation and identification of three Acts, Act-X0β (1), Act-X2 (2), and Act-D (3) (Figure 1). All three Acts exhibited more potent cytotoxicities on the A549, MCF-7 and K562 tumor cell lines than adriamycin in which Act-X2 is the most active with the IC50 values of 0.8–1.8 nM and the lowest toxicity against the human embryo liver cell strand (L02 cells) with the values of the selective index (SI) of 5.2–12.2. Moreover, all the three Acts displayed more active or comparable antibacterial to ciprofloxacin hydrochloride against Staphylococcus aureus and the methicillin-resistant S. aureus (MRSA), Bacillus subtilis, and Bacillus cereus with MICs of 0.04–2.48 μM. In addition, the productivity on actinomycins of S. heliomycini WH1 under different cultural conditions were investigated.
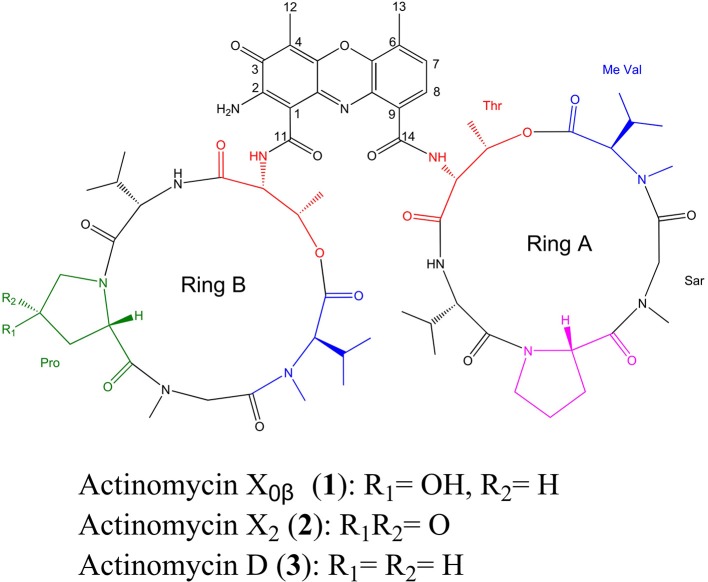
Figure 1.
Chemical structures of Acts X0β (1), X2 (2), and D (3) from Streptomyces heliomycini WH1.
Materials and methods
General experimental procedures
Silica gel (200–300 mesh) and on plates pre-coated with silica gel GF254 (10–40 μm) (Qingdao Marine Chemical Factory, Qingdao, China) were used in vacuum liquid chromatography (VLC) and thin layer chromatography (TLC), respectively. The optical rotation was measured by a Jasco P-1020 digital polarimeter. IR and UV spectra were recorded on a Nicolet Nexus 470 spectrophotometer using KBr discs and on a Hitachi UH5300 UV-Visible spectrophotometer, respectively. NMR spectra of Acts X2 and D were measured by a JEOL JNM-ECP 600 spectrometer while Act-X0β was recorded on a Bruker Avance III 600 spectrometer and the chemical shifts were recorded as δ values using TMS as internal standard. Compounds were separately injected into the Q-TOF Ultima Global GAA076 LC mass spectrometer to obtained mass spectra. The cultures were analyzed over an analyzing YMC-ODS-A chromatographic column (4.6 × 250 mm, 5 μm) eluted with 80% H2O-MeOH (v/v) at a flow rate of 1 mL/min by a Shimadzu LC-6AD HPLC equipment and detected at λmax 443 nm. The actinomycins were purified over a semi-preparative YMC-ODS-A chromatographic column (10 × 250 mm, 5 μm) eluted with 80% H2O-MeOH (v/v) at a flow of 4 mL/min by a Waters 1525 HPLC equipment. The RPMI 1640 powder with L-glutamine (Gln) and without NaHCO3 was from Life Technologies Corporation (USA). The ciprofloxacin hydrochloride was from J&K Scientific Ltd. (Beijing, China), and ketoconazole and itraconazole were from Energy Chemical (Shanghai, China). Phosphate Buffer Solution (PBS, 0.01 M, pH 7.2–7.4) and RPMI 1640 liquid medium for cell culture were from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China).
Bacterial material
The actinobacterial strain WH1 was isolated from a sandy soil sample collected at the coastal region of the Arabian Gulf at Jobail industrial city, in the Eastern Province of Saudi Arabia, and identified as S. heliomycini WH1 according to its phenotypic and phylogenetic characters (Figures S1, S2). The strain was deposited in our laboratories in 20% glycerol at −80°C. The working strain was prepared on ISP2 agar slants and stored at 4°C. The human pathogenic bacteria, B. subtilis (ATCC 6051), Escherichia coli (ATCC 11775), Pseudomonas aeruginosa (ATCC 10145), Staphyloccocus aureus (ATCC 6538) and MRSA (ATCC 43300), and the pathogenic fungi, Candida albicans (ATCC 10231) and Candida glabrata (ATCC 2001) were purchased from the Institute of microbiology, Chinese Academy of Sciences. The aquatic pathogenic bacteria, B. cereus (ATCC 14579), Vibrio vulnificus (ATCC 27562), and Vibrio parahaemolyticus (ATCC 17802) were purchased from Guangdong Institute of Microbiology (GIM). Aspergillus fumigates AF293 was given by Prof. Ling Lu, Nanjing Normal University, and Vibrio alginolyticus, Vibrio splendidus, and Aeromonas hydrophila were given by Prof. Xiangli Tian, Fisheries College of Ocean University of China.
Isolation and identification of strain WH1
The collected sandy soil sample was air dried at room temperature (rt) for 7 days and then serially diluted up to 10−4 and inoculated in triplicates onto two selective media recommended for the isolation of actinobacteria, M1 (Mincer et al., 2002) and MM (Hozzein et al., 2008). After incubation at 28°C for 3 weeks, the isolate under study was picked and purified by streaking on the isolation medium twice each for 14 days at 28°C. The pure culture was maintained on slants at 4°C and preserved as a mixture of hyphae and spores in 20% glycerol at −80°C.
The purified isolate was characterized by its morphological characteristics (mycelia, cell morphology, and spore surface) by examining coverslip cultures on ISP2 agar plates grown at 28°C for 14 days by light and scanning electron (JEOL M6060) microscopes as described in the International Streptomyces Project (ISP) (Shirling and Gottlieb, 1966). The isomer type of the diaminopimelic acid in the cell wall and the whole-organism sugars were determined according to the standard methods of Hasegawa et al. (1983) and Staneck and Roberts (1974), respectively. The genomic DNA extraction, PCR amplification of the 16S rRNA gene, and sequencing of the PCR product were carried out as described before (Hozzein and Goodfellow, 2007). The obtained sequence was deposited in Genbank (accession No. AB184712) and compared with available 16S rRNA gene sequences of validly published species from the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/; Kim et al., 2012).
Cultural media
The isolation medium was MM agar medium containing 0.05% glucose, 0.05% yeast extract, 0.05% MgSO4·7H2O, 0.05% NaCl, 0.1% K2HPO4, 1.8% agar, and 1 L seawater, pH 7.5. The working strain was prepared on ISP2 agar slants composed of 0.4% glucose, 1% malt extract, 0.4% yeast extract, 1.8% agar, and 1 L 50% seawater, pH 7.5. The MM liquid medium, soybean meal medium (2% soybean meal and 1 L seawater, pH 8.0), and the MI–MIV media were used to investigate the productivity of WH1 for actinomycins. MI–MIV media contained 2.0% yeast extract, 0.15% KH2PO4, 0.05% MgSO4·7H2O and 1 L seawater (pH 8.0), 2.0% soybean meal, 0.15% KH2PO4, 0.05% MgSO4·7H2O and 1 L seawater (pH 8.0), 2.25% soluble starch, 0.5% yeast extract, 0.15% KH2PO4, 0.05% MgSO4·7H2O and 1 L seawater (pH 8.0), and 2.25% glucose, 0.5% yeast extract, 0.15% KH2PO4, 0.05% MgSO4·7H2O and 1 L seawater (pH 8.0), respectively. The LB agar medium consisted of 1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.8% agar, and 1 L tap water (pH 7.4), while the YPD agar medium consisted of 1% yeast extract, 2% peptone, 2% glucose, 1.8% agar, and 1 L tap water (pH 7.0). The 2216E agar medium was prepared by 1% peptone, 0.5% yeast extract powder, 1.8% agar, and 1 L seawater (pH 7.8). The PDA agar medium contained 20% potato, 2% glucose, 1.8% agar, and 1 L tap water.
Fermentation and extraction
S. heliomycini WH1 was fermented in ten 500-mL Erlenmeyer flasks each containing 150 mL MM liquid medium and was shaken for 10 days at 28°C and 180 rpm. The fermentation broth was extracted three times each with 1,500 mL EtOAc. The EtOAc phase was combined and evaporated to dryness under reduced pressure by a rotary evaporator to give the EtOAc extracts (0.5 g). S. heliomycini WH1 was also cultured in different media under different pH and salinity. The chemo-diversity of the EtOAc extracts was investigated by high performance liquid chromatography (HPLC, Figure S3).
Purification and identification of the acts
The EtOAc extract (0.5 g) was separated into five fractions on a VLC silica gel column using a step gradient elution with 100:0, 50:1, 30:1, 10:1, and 0:100 (v/v) of CH2Cl2-MeOH. Then the fractions 2–4 containing actinomycins were combined and purified by semi-preparative HPLC using YMC-pack semi-preparative chromatographic column (ODS-A) eluted with 80% H2O-MeOH (v/v) at a flow rate of 4 mL/min to give Act-X0β (1) (1.0 mg, tR 9.6 min), Act-X2 (2) (3.7 mg, tR 11.2 min) and Act-D (3) (6.6 mg, tR 13.0 min). The isolation yields of Acts X0β, X2, and D were 0.7, 2.5, and 4.4 mg/L, respectively.
Sample preparation for analysis of the acts production
The strain WH1 was fermented with three parallels for 10 days at 180 rpm and 28°C in a 500 mL Erlenmeyer flask containing 150 ml of MM liquid, soybean meal, and MI–MIV media, and the pH value was adjusted to a certain value before sterilization. The initial pH values were adjusted by 20% hydrochloric acid (HCl) or 4% sodium hydroxide (NaOH). The 0, 3, 5, and 7% salinity were prepared by tap water, seawater, and sea water supplemented with NaCl, respectively. Each experiment was carried out in three parallel. The fermentation broths were extracted thrice with EtOAc (each 250 mL), and concentrated to dryness in vacuo to give the extracts for HPLC analysis.
Analysis of the acts production
The production of Acts was estimated by establishing the standard curve between the HPLC peak areas and the concentrations of Acts. The standard curve was established using standard solutions from 0.1 to 10 μg/mL on a YMC-pack C18 analytical column with 1 mL/min of flow rate and detection at λmax 443 nm. Linear curves and their fitting equations were established using Origin 9.0. The production of Acts was calculated according to the fitting equations.
Cytotoxic assay of the acts
The cytotoxicities on three human cancer cell lines, nonsmall cell lung cancer cell line (A549), breast cancer cell line (MCF-7), and myelogenous leukemia cell line (K562), along with human embryo liver L02 cell line were assayed. The method for A549, MCF-7 and L02 was the 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Mosmann, 1983), while the one for K562 was the cell counting Kit-8 (CCK-8) (Tominaga et al., 1999). A549, MCF-7, K562, and L02 cell lines were cultured for 3–5 d in the RPMI-1640 liquid medium supplemented with 10% FBS under a humidified atmosphere with 5% CO2 and 95% air at 37°C. Then 100 μL of the cell suspensions with a density of 3 × 105 cells mL−1 was plated in the 96-well microtiter plates and incubated for 12 h. The 200 μM testing DMSO solutions of the samples were diluted into 12.5 and then to 0.012 μM by the continuous 2-fold dilution method with RPMI-1640 medium. Then, the obtained test solutions (100 μL) were added into above wells each containing 100 μL cell suspension and further incubated for 72 h. The 20 μL 0.5% MTT solution (in PBS) was added to each well containing A549, MCF-7, and L02 cell lines and further incubated for 4 h. The culture broth was then gently pipetted and the DMSO (150 μL) was added to dissolve the formed formazan crystals. Absorbance of the solution was determined on a Spectra Max Plus plate reader at 570 nm. The CCK-8 solution was added to each well containing K562 cell and further incubated for 6 h, absorbance was determined on a Spectra Max Plus plate reader at 450 nm. The inhibition rates were calculated as ((Ablankcontrol − Asample)/Ablankcontrol × 100%). The half-maximal inhibitory concentration (IC50) is defined as the concentration within 50% inhibition. Adriamycin was used as the positive control with the IC50 values of 1.30, 0.30, and 1.00 μM for A549, K562, and MCF-7, and the CC50 value of 0.40 μM for L02 cell lines, respectively. The selective index (SI) is defined as the value of CC50/IC50.
Antimicrobial assay of the acts
The antimicrobial activities against human pathogenic bacteria (B. subtilis, E. coli, P. aeruginosa, S. aureus, and MRSA) and pathogenic fungi (C. albicans, C. glabrata, Aspergillus fumigatus AF293) and aquatic pathogenic bacteria (A. hydrophila, B. cereus, V. alginolyticus, V. parahaemolyticus, V. splendidus, and V. vulnificus) were evaluated using the filter paper disc method. The pathogenic strains were cultivated in LB agar plates at 37°C for bacteria and in YPD agar plates at 37°C for fungi. The testing methanol (MeOH) solutions (1 mg/mL) of the samples and positive control (ciprofloxacin hydrochloride for bacteria, and ketoconazole for C. albicans and C. glabrata) were diluted into 500–1.95 μg/mL by the continuous 2-fold dilution method with MeOH. Then 10 μL of the testing solutions were separately added to the paper disc (5 mm diameter). After evaporation to dryness, the drug paper discs were added into the cultural plates of the pathogenic microorganisms and incubated at 28°C for 12 h. Inhibition zones were then recorded as mm in diameter. The samples were first tested for their inhibitory zone diameters (IZDs) at the concentration of 1 mg/mL. Only those active samples with IZDs ≥14 mm were tested for their minimum inhibitory concentration (MIC) by 2-fold dilution method (Fu et al., 2013). The drug solutions of the three actinomycins, extracts and ciprofloxacin hydrochloride (positive control) were respectively prepared by a serial 2-fold dilution method from 100 to 0.049 μg/mL with the LB liquid medium for S. aureus and B. subtilis and 2216E liquid medium for B. cereus. The pathogenic bacterial colony with 24 h old grown on the LB (S. aureus and B. subtilis) or 2216E (B. cereus) agar plates were transferred into a 50-mL tube containing 30 mL fresh corresponding liquid media and incubated for 12 h at 28°C and 180 rpm. The final bacterial suspension was adjusted to the density of 5 × 105 CFU/mL with fresh corresponding liquid media and was added into the 96-well plates. Each well contains 100 μL bacterial suspension and 100 μL of the testing solution. The medium (100 μL) equipped with 100 μL bacterial suspension was used as the corresponding negative controls and the medium (200 μL) was used as blank controls. Each experiment was carried out in three parallel wells. All plates were stationary incubated for 15 h at 37°C. The minimal inhibitory concentration (MIC) was the lowest drug concentration at which no bacteria were grown, that is, the wells were more transparent than the negative control examined by eyes. In addition, the antifungal activity against A. fumigatus AF293 was also assayed. 3-(N-morpholino) propansulfonic acid (MOPS, 6.906 g) and glucose (4 g) were dissolved in 65 mL of deionized water at 60°C and then cooled to rt. And then 2.08 g RPMI 1640 powder with L-Gln and without NaHCO3 was added to the solution. The pH was adjusted to 7.0 with 5.0% NaOH and the solution volume was adjusted to 90 mL by adding deionized water. The A. fumigatus was grown on PDA at 28°C for a week and the mature spores were suspended in 0.9% saline and the density was adjusted to 2 × 104 CFU/mL with above fresh-prepared RPMI 1640 medium. The drug solution was prepared by dissolving extracts (50 mg/mL) or compounds (10 mg/mL) in DMSO and then diluted to 100-fold with the sterile water so that the final concentration of DMSO was less than 1%. One hundred microliters of the drug solution was added into 96-well plates (Costar 3599) that each well contains 100 μL A. fumigatus AF293 suspension within the density of 2 × 104 CFU/mL. The 96-well plates were incubated in a wet box at 28°C for 4–7 days. Itraconazole, A. fumigatus AF293 suspension without drugs and the fresh-prepared RPMI 1640 medium were used as the positive control, growth control, and the negative control, respectively. Each experiment was set in three parallels. The MIC was the lowest drug concentration at which no fungal growth was observed compared to the growth control.
Results
Identification of strain WH1
The observed morphological features of strain WH1 showed that it produced extensively branched hyphae bearing long spore chains with smooth surfaces (Figure S1). The cell walls of the strain WH1 contained LL-DAP as the characteristic amino acid of the peptidoglycan and its whole-organism sugar patter have glucose, galactose, and mannose as the characteristic sugars. These characters indicated that WH1 belongs to genus Streptomyces. The taxonomic position of WH1 and its affiliation to genus Streptomyces was confirmed by the analysis of the 16S rRNA gene sequence against the available validly published species. The sequence analysis using the EzTaxon-e server showed that strain WH1 had 100% similarity to S. heliomycini NCBR 15899(T). These results indicated that the strain WH1 under study is a strain of S. heliomycini.
The identification of acts X0β, X2, and D
HPLC analysis revealed that there were three peaks with the typical UV absorption of Acts at λmax 203, 225, and 443 nm in the cultures of S. heliomycini WH1 (Wang et al., 2014) (Figure S3). Chemical investigations resulted in the isolation of the three Acts, compounds 1–3, by VLC and semi-preparative HPLC. By means of specific rotation, MS and NMR data, their structures were identified. The 13C NMR spectra of compounds 1–3 showed the characteristic skeleton resonances of Acts at δ 34–40 for N-CH3 (× 4) and δ 165–175 for N-C=O (× 12), further indicating the nature of the Acts of compounds 1–3. All the 13C NMR spectra of compounds 1–3 showed 62 carbon signals (Figures S5, S8, S11) and the ESIMS of compounds 1–3 showed molecular peaks at m/z 1272.06 [M+H]+ (Figure S6), 1270.03 [M+H]+ (Figure S9), and 1256.05[M+H]+ (Figure S12). Their molecular formulas were further determined as C62H86O17N12, C62H84N12O17, and C62H86N12O16 from the HRESIMS peaks at m/z 1271.6305[M+H]+ (Figure S6), 1269.6155[M+H]+ (Figure S9), and 1255.6365[M+H]+ (Figure S12), respectively. The differences of 13C NMR spectra of compounds 1–3 are that a methylene carbon signal (δC 23.0) in 3 is replaced by an oxygenated methine carbon signal (δC 70.0) in 1 and a carbonyl signal (δC208.8) in 2, respectively. These data suggested that compounds 1–3 might be corresponding to Acts X0β, X2, and D. The consistence of NMR (Figures S4, S5, S7, S8, S10, S11, Tables 1–3) and [α]D with those reported further supported Acts 1–3 were Act-X0β (1) (Lifferth et al., 1999), Act-X2 (2) (Lifferth et al., 1999) and Act-D (3) (Wang et al., 2014), respectively (Figure 1).
Table 1.
1H (600 MHz) and 13C (150 MHz) NMR Date of Act-X0β (1) in CDCl3a.
| Ring-A | Ring-B | |||||
|---|---|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | Position | δC, type | δH (J in Hz) | |
| Thr | 1 | 168.8, C | 1 | 169.0, C | ||
| 2 | 55.5, CH | 4.81 (dd, 7.0, 2.5) | 2 | 54.9, CH | 4.49 (dd, 6.5, 2.8) | |
| 3 | 74.7, CH | 5.24 (m) | 3 | 75.4, CH | 5.24 (m) | |
| 4 | 17.7, CH3 | 1.28 (d, 6.3) | 4 | 17.8, CH3 | 1.24 (d, 6.0) | |
| NH | 7.44 (d, 6.9) | NH | 7.50 (d, 6.4) | |||
| Val | 1 | 173.5, C | 1 | 173.3, C | ||
| 2 | 58.9, CH | 3.57 (dd, 10.2, 3.5) | 2 | 58.1, CH | 3.55 (dd, 8.9, 4.3) | |
| 3 | 32.0, CH | 2.16 (m) | 3 | 31.9, CH | 2.12 (m) | |
| 4 | 19.2, CH3 | 0.75 (d, 2.7) | 4 | 19.2, CH3 | 0.74 (d, 2.7) | |
| 5 | 19.2, CH3 | 0.96 (d, 7.4) | 5 | 19.0, CH3 | 0.94 (d, 1.6) | |
| NH | 8.17 (d, 6.2) | NH | 7.91 (d, 5.5) | |||
| Pro | 1 | 173.2, C | 1 | 173.0, C | ||
| 2 | 57.0, CH | 5.97 (d, 9.2) | 2 | 56.5, CH | 6.05 (d, 9.6) | |
| 3 | 31.2, CH2 | 1.84 (m), 2.78 (m) | 3 | 31.4, CH2 | 4.13 (m), 3.95 (m) | |
| 4 | 23.0, CH2 | 2.06 (m), 2.26 (m) | 4 | 70.0, CH | 4.70 (m) | |
| 5 | 47.6, CH2 | 3.85 (m), 3.73 (m) | 5 | 54.7, CH2 | 3.56 (m), 3.08 (m) | |
| Sar | 1 | 166.5, C | 1 | 166.5, C | ||
| 2 | 51.3, CH2 | 4.72 (d, 17.4), 3.63 (d, 17.4) | 2 | 51.5, CH2 | 4.55 (d, 17.4), 3.59 (d, 17.4) | |
| MeVal | NMe | 35.0, CH3 | 2.87 (s) | NMe | 35.1, CH3 | 2.87 (s) |
| 1 | 167.6, C | 1 | 167.7, C | |||
| 2 | 71.3, CH | 2.71 (d, 9.3) | 2 | 71.4, CH | 2.67 (d, 9.4) | |
| 3 | 27.0, CH | 2.78 (m) | 3 | 27.0, CH | 2.64 (m) | |
| 4 | 21.7, CH3 | 0.96 (m) | 4 | 21.7, CH3 | 0.94 (m) | |
| 5 | 19.4, CH3 | 0.75 (d, 2.7) | 5 | 19.2, CH3 | 0.74 (d, 3.1) | |
| NMe | 39.5, CH3 | 2.95 (s) | NMe | 39.3, CH3 | 2.94 (s) | |
| chromophore | 1 | 102.7, C | ||||
| 2 | 147.1, C | |||||
| 3 | 179.1, C | |||||
| 4 | 113.6, C | |||||
| 4a | 145.0, C | |||||
| 5a | 140.7, C | |||||
| 6 | 128.5, C | |||||
| 7 | 130.4, CH | 7.35 (d, 7.8) | ||||
| 8 | 126.2, CH | 7.64 (d, 7.8) | ||||
| 9 | 131.3, C | |||||
| 9a | 129.6, C | |||||
| 10a | 146.1, C | |||||
| 11 | 166.2, C | |||||
| 12 | 7.9, CH3 | 2.22 (s) | ||||
| 13 | 15.3, CH3 | 2.54 (s) | ||||
| 14 | 173.0, C | |||||
Thr, Threonine; Val, Valine; Pro, Proline; Sar, Sarcosine.
Table 3.
1H (500 MHz) and 13C (125 MHz) NMR Date of Act-D (3) in CDCl3a.
| Ring-A | Ring-B | |||||
|---|---|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | Position | δC, type | δH (J in Hz) | |
| Thr | 1 | 169.0, C | 1 | 168.5, C | ||
| 2 | 55.4, CH | 4.60 (d, 5.1) | 2 | 55.0, CH | 4.48 (d, 5.2) | |
| 3 | 75.2, CH | 5.20 (d, 5.1) | 3 | 75.1, CH | 5.16 (d, 5.6) | |
| 4 | 17.5, CH3 | 1.26 (s) | 4 | 18.0, CH3 | 1.26 (s) | |
| NH | 7.19 (d, 6.4) | NH | 7.81 (d, 5.8) | |||
| Val | 1 | 173.8, C | 1 | 173.5, C | ||
| 2 | 59.0 CH | 3.54 (m) | 2 | 58.8, CH | 3.55 (m) | |
| 3 | 31.9, CH | 2.16 (m) | 3 | 31.6, CH | 2.08 (m) | |
| 4 | 19.4, CH3 | 1.12 (d, 5.7) | 4 | 19.2, CH3 | 1.12 (d, 5.7) | |
| 5 | 19.2, CH3 | 0.9 (d, 6.7) | 5 | 19.1, CH3 | 0.89 (d, 6.7) | |
| NH | 8.09 (d, 5.3) | NH | 7.94 (d, 5.7) | |||
| Pro | 1 | 173.4 C | 1 | 173.4, C | ||
| 2 | 56.4, CH | 6.02 (d, 9.0) | 2 | 56.3, CH | 5.98 (d, 9.0) | |
| 3 | 31.1, CH2 | 1.88 (m), 2.67 (m) | 3 | 31.4, CH2 | 1.87 (m), 2.67 (m) | |
| 4 | 23.1, CH | 2.17 (m), 2.25 (m) | 4 | 23.0, CH2 | 2.15 (m), 2.25 (m) | |
| 5 | 47.7, CH2 | 3.72 (m) | 5 | 47.4, CH2 | 3.82 (m) | |
| Sar | 1 | 166.4, C | 1 | 166.1, C | ||
| 2 | 51.5, CH2 | 4.76 (d, 17.6), 3.61 (d, 17.6) | 2 | 51.5, CH2 | 4.70 (d, 17.4), 3.64 (d, 17.5) | |
| MeVal | NMe | 35.0, CH3 | 2.88 (s) | NMe | 35.0, CH3 | 2.88 (s) |
| 1 | 167.8, C | 1 | 167.7, C | |||
| 2 | 71.6, CH | 2.67 (m) | 2 | 71.4, CH | 2.67 (m) | |
| 3 | 27.0, CH | 2.67 (m) | 3 | 27.0, CH | 2.67 (m) | |
| 4 | 21.8, CH3 | 0.96 (d, 7.4) | 4 | 21.7, CH3 | 0.95 (d, 6.6) | |
| 5 | 19.4, CH3 | 0.75 (s) | 5 | 19.2, CH3 | 0.75 (s) | |
| NMe | 39.4, CH3 | 2.90 (s) | NMe | 39.3, CH3 | 2.94 (s) | |
| chromophore | 1 | 101.8, C | ||||
| 2 | 147.8, C | |||||
| 3 | 179.2, C | |||||
| 4 | 113.6, C | |||||
| 4a | 145.2, C | |||||
| 5a | 140.6, C | |||||
| 6 | 127.7, C | |||||
| 7 | 130.4, CH | 7.37 (d, 7.7) | ||||
| 8 | 126.0, CH | 7.64 (d, 7.7) | ||||
| 9 | 132.7, C | |||||
| 9a | 129.2, C | |||||
| 10a | 146.0, C | |||||
| 11 | 169.0, C | |||||
| 12 | 7.9, CH3 | 2.25 (s) | ||||
| 13 | 15.2, CH3 | 2.56 (s) | ||||
| 14 | 166.6.0, C | |||||
Thr, Threonine; Val, Valine; Pro, Proline; Sar, Sarcosine.
Table 2.
1H (500 MHz) and 13C (125 MHz) NMR Date of Act-X2 (2) in CDCl3a.
| Ring-A | Ring-B | |||||
|---|---|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | Position | δC, type | δH (J in Hz) | |
| Thr | 1 | 168.7, C | 1 | 168.8, C | ||
| 2 | 55.0, CH | 4.55 (m) | 2 | 54.8, CH | 4.48 (m) | |
| 3 | 74.7, CH | 5.15 (m) | 3 | 74.6, CH | 5.24 (m) | |
| 4 | 17.2, CH3 | 1.14 (d, 6.4) | 4 | 17.7, CH3 | 1.26 (d, 7.3) | |
| NH | 7.17 (d, 7.2) | NH | 7.67 (d, 6.1) | |||
| Val | 1 | 173.5, C | 1 | 174.0, C | ||
| 2 | 58.5, CH | 3.57 (dd, 9.3, 6.3) | 2 | 57.2, CH | 3.70 (dd, 10.4, 6.3) | |
| 3 | 31.9, CH | 2.10 (m) | 3 | 31.7, CH | 2.09 (m) | |
| 4 | 18.9, CH3 | 0.90 (d, 6.8) | 4 | 18.8, CH3 | 0.89 (d, 6.8) | |
| 5 | 19.2, CH3 | 1.12 (d,6.8) | 5 | 19.2, CH3 | 1.15 (d, 6.8) | |
| NH | 7.68 (d, 6.2) | NH | 8.19 (d, 5.9) | |||
| Pro | 1 | 173.1, C | 1 | 172.7, C | ||
| 2 | 56.4, CH | 5.95 (d, 8.9) | 2 | 54.3, CH | 6.55 (d, 11.0) | |
| 3 | 31.0, CH2 | 1.84 (m), 2.75 (m) | 3 | 41.9, CH2 | 3.85 (m), 2.33 (m) | |
| 4 | 23.0, CH | 2.24 (m) | 4 | 208.8, C | ||
| 5 | 47.4, CH2 | 3.91 (m), 3.74 (m) | 5 | 52.9, CH2 | 4.55 (m), 3.89 (m) | |
| Sar | 1 | 166.0, C | 1 | 165.9, C | ||
| 2 | 51.3, CH2 | 4.72 (d, 17.3), 3.62 (d, 17.3) | 2 | 51.3, CH2 | 4.57 (d, 17.3), 3.62 (d, 17.3) | |
| MeVal | NMe | 34.8, CH3 | 2.88 (s) | NMe | 34.9, CH3 | 2.89 (s) |
| 1 | 166.5, C | 1 | 166.3, C | |||
| 2 | 71.3, CH | 2.68 (m) | 2 | 71.5 CH | 2.68 (m) | |
| 3 | 26.9, CH | 2.68 (m) | 3 | 27.0, CH | 2.68 (m) | |
| 4 | 21.6, CH3 | 0.94 (d, 5.8) | 4 | 21.7, CH3 | 0.97 (d, 5.5) | |
| 5 | 19.1, CH3 | 0.74 (d, 5.8) | 5 | 19.1, CH3 | 0.73 (d, 5.5) | |
| NMe | 31.7, CH3 | 2.92 (s) | NMe | 31.9, CH3 | 2.93 (s) | |
| chromophore | 1 | 101.7, C | ||||
| 2 | 147.4, C | |||||
| 3 | 179.0, C | |||||
| 4 | 113.6, C | |||||
| 4a | 145.0, C | |||||
| 5a | 140.5, C | |||||
| 6 | 127.8, C | |||||
| 7 | 130.3, CH | 7.35 (d, 7.8) | ||||
| 8 | 126.2, CH | 7.60 (d, 7.8) | ||||
| 9 | 132.1, C | |||||
| 9a | 129.1, C | |||||
| 10a | 145.9, C | |||||
| 11 | 167.4, C | |||||
| 12 | 7.8, CH3 | 2.24 (s) | ||||
| 13 | 15.1, CH3 | 2.55 (s) | ||||
| 14 | 172.7, C | |||||
Thr, Threonine; Val, Valine; Pro, Proline; Sar, Sarcosine.
The standard curves for analysis of the productions of acts
The standard curves of Acts X0β, X2, and D were established by means of HPLC-UV. The liner regression equations for ActsX0β, X2, and D were respectively obtained as X = 1.90E–6Y – 0.107 (R2 = 0.9992) (Figure 2), X = 1.14E–6Y – 0.072 (R2 = 0.9994) (Figure 3), and X = 1.01E–6Y – 0.028 (R2 = 0.9993) (Figure 4), where Y is the weight of Acts (μg) and X is the peak area. All curves showed good linear relationships that could be used to estimate the production of the Acts from the corresponding HPLC peaks' areas.
Figure 2.
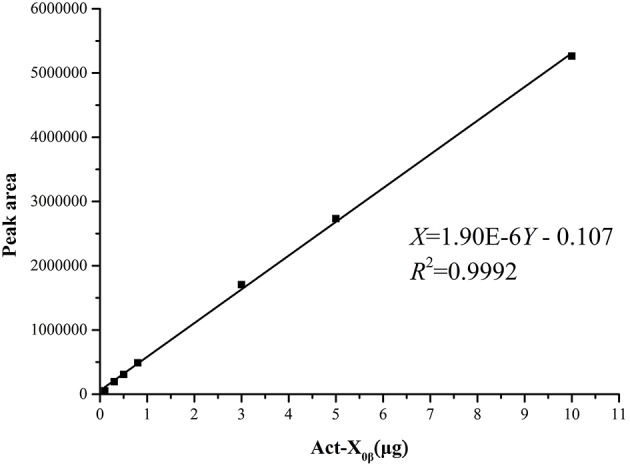
Standard carve of Act-X0β.
Figure 3.
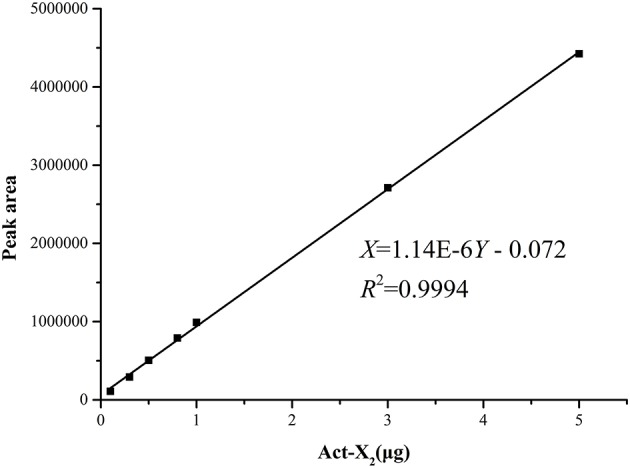
Standard carve of Act-X2.
Figure 4.
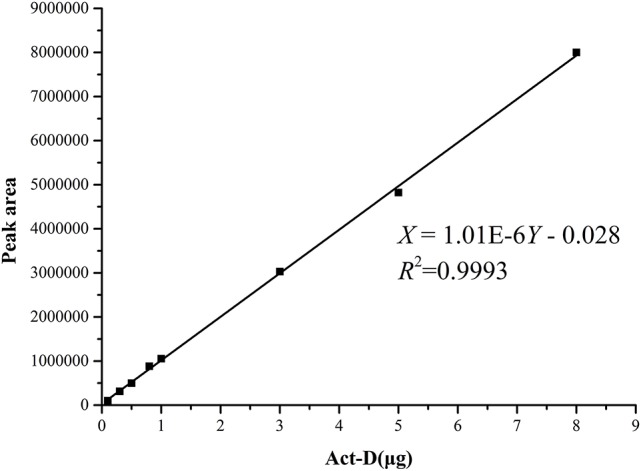
Standard carve of Act-D.
The effects of pH, salinity, and media on productivity of acts from S. heliomycini WH1
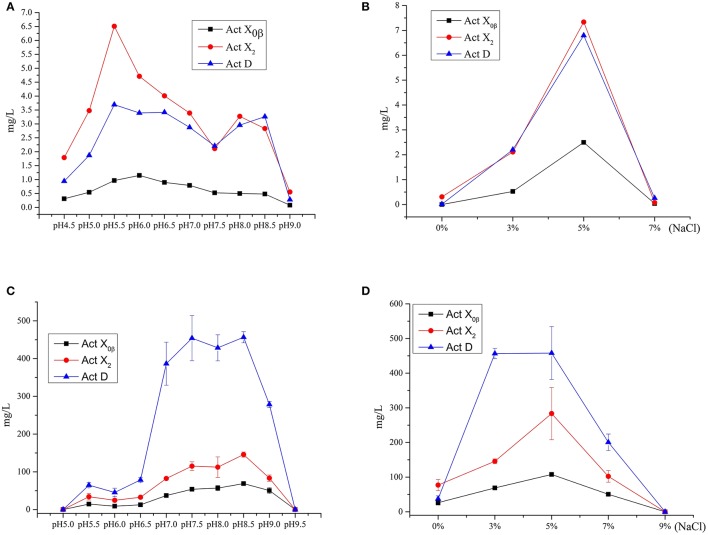
The effect of the initial pH values on the production was studied in the MM liquid medium with 3% salinity (natural seawater), whose initial pH was increased to 9.0 from 4.5 at an interval of 0.5. The results (Figure 5A, Table S1) showed that the production of Acts X0β, X2, and D reached the highest at pH 6.0, 5.5, and 5.5 in the MM medium, whose yields were 1.2, 6.5 and 3.7 mg/L, respectively. This indicated that 5.5 is the most suitable initial pH value for producing both Acts D and X2 in MM medium, and is also suitable for producing Act-X0β that was 1.0 mg/L only after the production at pH 6.0. The reason is that all the components in the MM liquid medium were completely dissolved at pH 5.5. Only under this pH value, the MM liquid medium is clear and transparent that is easy to be used by microorganisms.
Figure 5.
The effect of the initial pH values and salinity on the productions of Acts X0β, X2 and D. (A) The effect of the initial pH values on the productions of Acts X0β, X2 and D in the liquid medium MM. (B) The effect of the salinity on the productions of Acts X0β, X2 and D in the liquid medium MM. (C) The effect of the initial pH values on the productions of Acts X0β, X2 and D in the liquid medium MII. (D) The effect of the salinity on the productions of Acts X0β, X2 and D in the liquid medium MII.
The effect of the salinity on the Acts production was studied in the MM liquid medium at pH 5.5. The salinity of the medium was designed as 0, 3, 5, and 7%, respectively. The results (Figure 5B, Table S2) showed that the highest production of Acts X0β, X2, and D was under 5% salinity with the yields of 2.5, 7.3, and 6.8 mg/L, respectively, while the corresponding production under 3% salinity was 0.5, 2.1, and 2.2 mg/L, respectively. However, the Acts fermentation titers under these conditions in MM liquid medium are too low to be satisfactory. Therefore, the other media were adopted to improve the productions of the Acts.
The study on the productivity of Acts in the soybean meal and MI–MIV liquid media (Table 4) showed that the highest production of all the Acts X0β, X2, and D was in the MII liquid medium with the yields of 56.8 ± 6.8, 112.4 ± 27.2, and 428.5 ± 34.5 mg/L, respectively. The results indicated that the soybean meal supplemented with the minor elements Mg, K, P, and S, that is MII liquid medium, is favorable for the production performance of actinomycins by strain S. heliomycini WH1. Therefore, the productivity of Acts was further optimized in the liquid medium MII by investigating the effects of pH values and salinity. The results showed that initial pH 8.5 was the most suitable pH value for production of all the Acts X0β, X2, and D in the medium MII with the yields of 68.8 ± 1.2, 145.7 ± 6.8, and 456.5 ± 14.7 mg/L (Figure 5C, Table S3) at 3% salinity, respectively. On the basis of initial pH 8.5, the highest productivity of all the Acts X0β, X2, and D was obtained in the liquid medium MII with 5% salinity which reached to 107.6 ± 4.2, 283.4 ± 75.3, and 458.0 ± 76.3 mg/L (Figure 5D, Table S4), respectively.
Table 4.
The productions of Acts X0β, X2, and D in different media (mg/L).
| Medium | Act-X0β | Act-X2 | Act-D |
|---|---|---|---|
| MM | 0.7 ± 0.02 | 2.5 ± 0.2 | 4.4 ± 0.2 |
| MI | 36.0 ± 3.4 | 34.2 ± 3.5 | 173.4 ± 20.5 |
| MII | 56.8 ± 6.8 | 112.4 ± 27.2 | 428.5 ± 34.5 |
| MIII | 38.7 ± 1.7 | 90.7 ± 14.1 | 162.0 ± 10.3 |
| MIV | 53.8 ± 5.4 | 219.7 ± 23.8 | 269.7 ± 40.3 |
| Soybean meal | 61.5 ± 15.7 | 121.6 ± 32.0 | 330.8 ± 64.2 |
The bioactivities of acts from S. heliomycini WH1
The cytotoxicities of Acts X0β, X2, and D on the A549, MCF-7, and K562 and L02 cell lines were examined. The results indicated that all the three Acts exhibited strong cytotoxicities on the three tumor cell lines and the one normal cell line with the IC50 and CC50 values of 0.8–157.4 nM (Table 5). Among them, Act-X2 displayed the strongest activities on the three human tumor cell lines, A549, MCF-7 and K562, and the lowest toxicity on the human normal embryo liver L02 cell line. Thus, Act-X2 showed the highest selective index (SI) for the three tested tumor cell lines (10.3, 12.2, and 5.2, respectively), indicating a potential of Act-X2 as a drug candidate for treatment of human cancers. As far as we known, there were no reports on the cytotoxicity of Acts X0β and X2 on the A549, MCF-7 and K562 tumor cells.
Table 5.
The cytotoxicity and selective index (SI) of Acts X0β, X2, and D.
| Cell lines | Act-X0β | Act-X2 | Act-D | Adriamycin | ||||
|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | SI | IC50 (nM) | SI | IC50 (nM) | SI | IC50 (nM) | SI | |
| A549 | 63.8 ± 2.9 | 2.5 ± 0.3 | 0.9 ± 0.1 | 10.4 ± 1.9 | 3.4 ± 0.1 | 3.9 ± 0.3 | 1,300 | 0.3 |
| MCF-7 | 85.2 ± 12.2 | 1.9 ± 0.4 | 0.8 ± 0.1 | 12.2 ± 1.8 | 2.6 ± 0.1 | 4.9 ± 0.4 | 1,000 | 0.4 |
| K562 | 128.3 ± 6.1 | 1.2 ± 0.2 | 1.8 ± 0.1 | 5.2 ± 0.5 | 3.3 ± 0.3 | 4.0 ± 0.5 | 300 | 1.3 |
| L02 (CC50) | 157.4 ± 14.7 | 9.3 ± 0.7 | 13.0 ± 0.5 | 400 | ||||
The antimicrobial activities against the human and aquatic pathogenic microbes, A. hydrophila, B. subtilis, B. cereus, E. coli, P. aeruginosa, S. aureus, MRSA, V. vulnificus, V. alginolyticus, V. parahaemolyticus, V. splendidus, C. albicans, C. glabrata, A. fumigatus AF293, and were evaluated. The results indicated that both Act-X2 and Act-D showed comparable or stronger antimicrobial activities against S. aureus, MRSA, B. subtilis, and B. cereus to ciprofloxacin hydrochloride (a positive control, MIC 0.1–12.5 μM) with MIC values of 0.04–0.15 μM (Table 6), while Act-X0β displayed very weak inhibitions on S. aureus and B. subtilis (MIC 0.3–2.5 μM). All the three Acts were not active against the tested pathogenic fungi and other bacteria at the concentration of 1 mg/mL. Except for the antibacterial activities of Act-X2 on the S. aureus and B. cereus (Xiong et al., 2008) and Act-D on the S. aureus (Bian et al., 2003), there were no other reports on the antibacterial activities of Acts D, X2 and X0β against MRSA and B. subtilis, the Act-X0β against S. aureus and B. cereus, as well as the Act-D against B. cereus. And the Acts D and X2 were more active. These results revealed the potential use of Act-X2 and Act-D in the treatment of infectious diseases caused by S. aureus, B. subtilis, and B. cereus, especially by MRSA.
Table 6.
The MIC values (μM) of Acts X0β, X2, and D on the pathogenic bacteria.
| Bacteria | Act-X0β | Act-X2 | Act-D | Ciprofloxacin hydrochloride |
|---|---|---|---|---|
| S. aureus | 0.31 | 0.15 | 0.31 | 0.53 |
| MRSA | 2.48 | 0.15 | 0.31 | 12.48 |
| B. subtilis | 0.31 | 0.08 | 0.04 | 0.13 |
| B. cereus | 1.23 | 0.04 | 0.08 | 8.50 |
Discussion
Actinomycins were firstly reported in 1940 from Actinomyces antibioticus (Waksman and Woodruff, 1940). Since then, more than 30 actinomycins have been discovered from the natural sources including Acts AI (BI, XI), AII, AIV (BIV, D, XIV), AV (BV, XV), and Acts C1–C3 (Roussos and Vining, 1956), Acts E1, E2, and F1–F4 (Brockmann, 1961), Acts G1–G6 (Lackner et al., 2000a; Bitzer et al., 2006), X0α–X0δ (Brockmann, 1961), Acts Y1–Y5 (Bitzer et al., 2009), and Acts Y6–Y9 (Cai et al., 2016), Acts Z1–Z5 (Lackner et al., 2000b), and Act Zp(Cai et al., 2016). To date, there are many Streptomyces species capable of producing actinomycins, but few of them were reported to produce relatively large quantities of one or two major actinomycins. Streptomyces parvulus (Foster and Katz, 1981), S. halstedii, S. anulatus (Praveen et al., 2008a), S. sindenensis (Praveen et al., 2008b), and S. griseoruber (Praveen and Tripathi, 2009) are examples to produce Act D, among which the highest production is 620 mg/L (Praveen et al., 2008a). Streptomyces nasri YG62 (Elnaggar et al., 1998) and S. triostinicus (Singh et al., 2009) are examples to produce Act X2 whose production reached to 443 mg/L (Singh et al., 2009). Streptomyces sp. MITKK-103 (Kurosawa et al., 2006), Streptomyces sp. JAU4234 (Xiong et al., 2008) and Streptomyces sp. MS449 (Chen et al., 2012) can simultaneously produce Acts D, X0β and X2 and strain MS449 produced the highest production of Acts D and X2 with the yields of 1,770 and 1,920 mg/L, respectively. No examples were found to produce Act X0β solely. This study identified Acts X0β, X2, and D by comparison of 13C NMR data to those reported ones (Lifferth et al., 1999; Wang et al., 2014) with the errors less than 0.5 ppm. And the productions of Acts D, X0β, and X2 by the marine-derived S. heliomycini WH1 in the optimized fermentation conditions were significantly improved by 100, 110, and 150 folds, respectively, relative to those in the isolation medium (MM medium). Among these actinomycins, Act-D has been extensively studied and widely used in the treatment of malignant tumors, such as Wilms' tumor and childhood rhabdomyosarcoma (Womer, 1997). However, the cytotoxicity of Acts X2 and X0β against the tumor cells and antibacterial activities of Acts D, X2, and X0β against pathogenic bacteria were received very little attention. Compared to Act D, Act X2 showed stronger cytotoxicity toward HL-60 cells (Kurosawa et al., 2006) and better antibacterial activity against MTB H37Rv (Chen et al., 2012) and S. aureus and B. cereus (Xiong et al., 2008). Our investigation firstly demonstrated that Act-X2 displayed the strongest activities against MCF-7, A549, and K562 human tumor cell lines, and the lowest toxicity on L02 human normal embryo liver cell line, indicating the potential use of Act-X2 as a cancer drug candidate. In addition, our fresh results on the strong antibacterial activity of Acts D and X2 against MRSA and B. subtilis along with Act-D against B. cereus indicated the potential use of Act-D and Act-X2 in the treatment of the infections caused by those human and aquatic pathogenic bacteria, especially by MRSA. Therefore, the present study revealed that actinobacteria from newly-explored, special or extreme environments could be a potential pool for drug discovery.
Author contributions
The authors from China contribute to the isolation and identification of Acts, the optimization of fermentation conditions, as well as the assays of the cytotoxic and antimicrobial activities, and prepared the paper. The authors from Saudi Arabia are responsible for the isolation and identification of the marine-derived actinobacterial strain, Streptomyces heliomycini WH1.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81561148012) and the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award number (12-BIO2630-02).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01147/full#supplementary-material
Data sheet
The colony and micrograph of Streptomyces heliomycini WH1 and the MS and NMR spectra of compounds 1–3.
References
- Betzel C., Rachev R., Dolashka P., Genov N. (1993). Actinomycins as proteinase inhibitors. Biochim. Biophys. Acta 1161, 47–51. 10.1016/0167-4838(93)90194-V [DOI] [PubMed] [Google Scholar]
- Bian Z.-L., Xiong P.-Y., Shen D.-S. (2003). Determination of bactericidal effect of six antitumor antibiotics on five antibiotic-resistant bacteria. Pharm. Care Res. 3, 253–255. 10.3969/j.issn.1671-2838.2003.04.014 [DOI] [Google Scholar]
- Bister B., Bischoff D., Strobele M., Riedlinger J., Reicke A., Wolter F., et al. (2004). Abyssomicin C–a polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed Engl. 43, 2574–2576. 10.1002/anie.200353160 [DOI] [PubMed] [Google Scholar]
- Bitzer J., Gesheva V., Zeeck A. (2006). Actinomycins with altered threonine units in the β-peptidolactone. J. Nat. Prod. 69, 1153–1157. 10.1021/np060063g [DOI] [PubMed] [Google Scholar]
- Bitzer J., Streibel M., Langer H., Grond S. (2009). First Y-type actinomycins from Streptomyces with divergent structure-activity relationships for antibacterial and cytotoxic properties. Org. Biomol. Chem. 7, 444–450. 10.1039/B815689A [DOI] [PubMed] [Google Scholar]
- Brockmann H. (1961). Chemistry of the actinomycins. Pure Appl. Chem. 2, 405–424. 10.1351/pac196102030405 [DOI] [Google Scholar]
- Bull A. T., Stach J. E. M. (2007). Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15, 491–499. 10.1016/j.tim.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Cai W., Wang X., Elshahawi S. I., Ponomareva L. V., Liu X., McErlean M. R., et al. (2016). Antibacterial and cytotoxic actinomycins Y6–Y9 and Zp from Streptomyces sp. Strain Gö-GS12. J. Nat. Prod. 79, 2731–2739. 10.1021/acs.jnatprod.6b00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Chiao Y. C., Gurudath Rao K., Hook J. W., III., Krugh T. R., Sengupta S. K. (1979). 7-Amino-actinomycin D complexes with deoxynucleotides as model for binding of the drug to DNA. Biopolymers 18, 1749–1762. 10.1002/bip.1979.360180712 [DOI] [PubMed] [Google Scholar]
- Chen C., Song F., Wang Q., Abdel-Mageed W. M., Guo H., Fu C., et al. (2012). A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X2 and actinomycin D with potent anti-tuberculosis activity. Appl. Microbiol. Biot. 95, 919–927. 10.1007/s00253-012-4079-z [DOI] [PubMed] [Google Scholar]
- Elnaggar M. Y. M., Elaassar S. A., Hashem M. A., Stoodley R. J., Raynor C. M., Sigee D. C. (1998). Production of actinomycin X2 by immobilized Streptomyces nasri YG62 mycelia. Microbios 95, 165–179. [Google Scholar]
- Foster J. W., Katz E. (1981). Control of actinomycin D biosynthesis in Streptomyces parvulus: regulation of tryptophan oxygenase activity. J. Bacteriol. 148, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P., Kong F., Li X., Wang Y., Zhu W. (2014). Cyanogramide with a new spiro[indolinone-pyrroloimidazole] skeleton from Actinoalloteichus cyanogriseus. Org. Lett. 16, 3708–3711. 10.1021/ol501523d [DOI] [PubMed] [Google Scholar]
- Fu P., Kong F., Wang Y., Wang Y., Liu P., Zuo G., et al. (2013). Antibiotic metabolites from the coral-associated actinomycete Streptomyces sp. OUCMDZ-1703. Chinese J. Chem. 31, 100–104. 10.1002/cjoc.201201062 [DOI] [Google Scholar]
- Fu P., Liu P., Li X., Wang Y., Wang S., Hong K., et al. (2011). Cyclic bipyridine glycosides from the marine-derived actinomycete Actinoalloteichus cyanogriseus WH1-2216-6. Org. Lett. 13, 5948–5951. 10.1021/ol202245s [DOI] [PubMed] [Google Scholar]
- Fu P., Zhuang Y., Wang Y., Liu P., Qi X., Gu K., et al. (2012). New indolocarbazoles from a mutant strain of the marine-derived actinomycete Streptomyces fradiae 007M135. Org. Lett. 14, 6194–6197. 10.1021/ol302940y [DOI] [PubMed] [Google Scholar]
- Hasegawa T., Takizawa M., Tanida S. (1983). A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322. 10.2323/jgam.29.319 [DOI] [Google Scholar]
- Hozzein W. N., Ali M. I. A., Rabei W. (2008). A new preferential medium for enumeration and isolation of desert actinomycetes. World J. Microbiol. Biotechnol. 24, 1547–1552. 10.1007/s11274-007-9641-y [DOI] [Google Scholar]
- Hozzein W. N., Goodfellow M. (2007). Nonomuraea aegyptia sp. nov., a novel actinomycete isolated from a sand dune. Antonie van Leeuwenhoek 92, 165–171. 10.1007/s10482-007-9145-7 [DOI] [PubMed] [Google Scholar]
- Kapp L. N., Okada S. (1972). Actinomycin D induction of acid phosphatase in synchronized L5178Y mouse leukemia cells. Exp. Cell Res. 72, 473–479. 10.1016/0014-4827(72)90016-X [DOI] [PubMed] [Google Scholar]
- Killewich L., Schutz G., Feigelson P. (1975). Functional level of rat liver tryptophan 2,3-dixoygenase messenger RNA during superinduction of enzyme with actinomycin D. Proc. Natl. Acad. Sci. U.S.A. 72, 4285–4287. 10.1073/pnas.72.11.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Koba M., Konopa J. (2005). Actinomycin D and its mechanisms of action. Postepy Hig. Med. Dosw. 59, 290–298. Available online at: http://www.phmd.pl/fulltxt.php?ICID=16586 [PubMed] [Google Scholar]
- Kurosawa K., Bui V. P., Van Essendelft J. L., Willis L. B., Lessard P. A., Ghiviriga I., et al. (2006). Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2-producer. Appl. Microbiol. Biot. 72, 145–154. 10.1007/s00253-005-0240-2 [DOI] [PubMed] [Google Scholar]
- Lackner H., Bahner I., Shigematsu N., Pannell L. K., Mauger A. B. (2000b). Structures of five components of the actinomycin Z complex from Streptomyces fradiae, two of which contain 4-chlorothreonine. J. Nat. Prod. 63, 352–356. 10.1021/np990416u [DOI] [PubMed] [Google Scholar]
- Lackner H., Hulsmann H., Heinze S., Simon H., Bar H., Zimmer C., et al. (2000a). A new actinomycin-type chromopeptide from Streptomyces sp. HKI-0155. J. Antibiot. 53, 84–87. 10.7164/antibiotics.53.84 [DOI] [PubMed] [Google Scholar]
- Lam K. S. (2006). Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9, 245–251. 10.1016/j.mib.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Lifferth A., Bahner I., Lackner H., Schäfer M. (1999). Synthesis and structure of proline ring modified actinomycins of the X-type. Z. Naturforsch. B 54, 681–691. 10.1515/znb-1999-0518 [DOI] [Google Scholar]
- Mincer T. J., Gensen P. R., Kauffman C. A., Fenical W. (2002). Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microbiol. 68, 5005–5011. 10.1128/AEM.68.10.5005-5011.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. (1983). Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63. 10.1016/0022-1759(83)90303-4 [DOI] [PubMed] [Google Scholar]
- Newman D. J., Cragg G. M., Snader K. M. (2003). Natural products as sources of new drugs over the period 1981−2002. J. Nat. Prod. 66, 1022–1037. 10.1021/np030096l [DOI] [PubMed] [Google Scholar]
- Praveen V., Tripathi C. K. M. (2009). Studies on the production of actinomycins D by Streptomyces griseoruber—a novel source. Lett. Appl. Microbiol. 49, 450–455. 10.1111/j.1472-765X.2009.02689.x [DOI] [PubMed] [Google Scholar]
- Praveen V., Tripathi C. K. M., Bihari V. (2008a). Studies on optimum fermentation conditions for actinomycin D production by two new strains of Streptomyces spp. Med. Chem. Res. 17, 114–112. 10.1007/s00044-007-9042-7 [DOI] [Google Scholar]
- Praveen V., Tripathi C. K. M., Bihari V., Srivastava S. C. (2008b). Production of actinomycins D by a new isolate, Streptomyces sindenensis. Ann. Microbiol. 58, 109–114. 10.1007/BF03179453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rill R. L., Hecker K. H. (1996). Sequence-specific actinomycins D binding to single-stranded DNA inhibits HIV reverse transcriptase and other polymerases. Biochemistry 35, 3525–3533. 10.1021/bi9530797 [DOI] [PubMed] [Google Scholar]
- Roussos G., Vining C. (1956). Isolation and properties of pure actinomycins. J. Chem. Soc. 2, 2469–2474. 10.1039/jr9560002469 [DOI] [Google Scholar]
- Saijets S., Ylipaasto P., Vaarala O., Hovi T., Roivainen M. (2003). Enterovirus infection and activation of human umbilical vein endothelial cells. J. Med. Virol. 70, 430–439. 10.1002/jmv.10413 [DOI] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Singh V., Khan M., Khan S. A., Tripathi C. K. M. (2009). Optimization of actinomycin V production by Streptomyces triostinicus using artificial neural network and genetic algorithm. Appl. Microbiol. Biotechnol. 82, 379–385. 10.1007/s00253-008-1828-0 [DOI] [PubMed] [Google Scholar]
- Staneck J. L., Roberts G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari K., Upadhyay D. J., Mösker E., Süssmuth R., Gupta R. K. (2015). Culturable bioactive actinomycetes from the Great Indian Thar Desert. Ann. Microbiol. 65, 1901–1914. 10.1007/s13213-014-1028-3 [DOI] [Google Scholar]
- Tominaga H., Ishiyama M., Ohseto F., Sasamoto K., Hamamoto T., Suzuki K., et al. (1999). A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal. Commun. 36, 47–50. 10.1039/a809656b [DOI] [Google Scholar]
- Waksman S. A., Woodruff H. B. (1940). Bacteriostatic and bactericidal substances produced by a soil actinomyces. Exp. Biol. Med. 45, 609–614. 10.3181/00379727-45-11768 [DOI] [Google Scholar]
- Wang C., Liu P.-P., Wang Y., Sun K.-L., Jia H.-J., Zhu W.-M. (2014). Optimization of fermentation conditions for production of actinomycin D by marine-derived Strptomyces parvulus OUCMDZ-2554. Chinese J. Mar. Drugs 33, 34–42. 10.13400/j.cnki.cjmd.2014.03.006 [DOI] [Google Scholar]
- Wang Z., Fu P., Liu P., Wang P., Hou J., Li W., et al. (2013). New pyran-2-ones from alkalophilic actinomycete, Nocardiopsis alkaliphila sp. Nov. YIM-80379. Chem. Biodivers. 10, 281–287. 10.1002/cbdv.201200086 [DOI] [PubMed] [Google Scholar]
- Womer B. R. (1997). Soft tissue sarcomas. Eur. J. Cancer 33, 2230–2236. 10.1016/S0959-8049(97)00334-1 [DOI] [PubMed] [Google Scholar]
- Xiong Z., Tu X., Tu G. (2008). Optimization of medium composition for actinomycin X2 production by Streptomyces spp JAU4234 using response surface methodology. J. Ind. Microbiol. Biotechnol. 35, 729–734. 10.1007/s10295-008-0339-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.