Abstract
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disease characterized by a reduced platelet count and an increased risk of bleeding. Although immense research has improved our understanding of ITP, the pathogenesis remains unclear. Here, we investigated the involvement of 25 single-nucleotide polymorphisms (SNPs) of the inflammation-related genes, including CD24, CD226, FCRL3, IL2, IRF5, ITGAM, NLRP3, CARD8, PTPN22, SH2B2, STAT4, TNFAIP3, and TRAF1, in the pathogenesis and treatment response of ITP. We recruited 312 ITP inpatients and 154 healthy participants in this case–control study. Inflammation-related SNP genotyping was performed on the Sequenom MassARRAY iPLEX platform. The expression of TNFAIP3 mRNA was determined by quantitative real-time RT-PCR. All SNPs in healthy controls were consistent with Hardy–Weinberg equilibrium. Statistical analysis revealed that rs10499194 in TNFAIP3 was significantly associated with a decreased risk of ITP after Bonferroni multiple correction (codominant, CT vs. CC, OR = 0.431, 95% CI = 0.262–0.711, p = 0.001; dominant, TT/CT vs. CC, OR = 0.249, 95% CI = 0.141–0.440, p = 0.000). Besides, TNFAIP3 expression was significantly higher in patients with CT and pooled CT/TT genotypes compared with CC genotype (p = 0.001; p = 0.001). Interestingly, rs10499194 was also associated with corticosteroid-sensitivity (codominant, CT vs. CC, OR = 0.092, 95% CI = 0.021–0.398, p = 0.001; dominant, TT/CT vs. CC, OR = 0.086, 95% CI = 0.020–0.369, p = 0.001; allelic, T vs. C, OR = 0.088, 95% CI = 0.021–0.372, p = 0.001). Furthermore, no significant association was found between inflammation-related SNPs and the severity or refractoriness of ITP after Bonferroni multiple correction. Our findings suggest that rs10499194 may be a protective factor for susceptibility and corticosteroid sensitivity in ITP patients.
Keywords: primary immune thrombocytopenia, inflammation, single-nucleotide polymorphism, susceptibility, treatment
Introduction
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disease characterized by a reduced platelet count and an increased risk of bleeding (1). The pathogenesis of ITP includes enhanced platelet clearance and impaired platelet production, which is related to T cell-mediated effects, B cell-mediated effects, tolerance checkpoint defects, and more (2, 3). Although immense research has improved our understanding of ITP, the etiology is still not completely clear. However, it has been widely accepted that both genetic and environmental factors play a significant role in the pathogenesis of ITP (4).
Autoimmune disease is characterized by self-antigen-induced chronic activation of immune system and eventually leads to tissue inflammation in genetically predisposed individuals (5). ITP is an autoimmune disease and closely related to inflammation (6, 7). Besides, inflammation can trigger ITP. For example, serum uric acid, an inflammatory mediator, played a pathophysiological role in the occurrence of ITP (8). However, ITP can also induce inflammation through platelet dysfunction. Recent research revealed that the acute phase of platelet destruction in severe ITP may trigger inflammation in the lung, which may induce interstitial lung disease (9). Furthermore, platelets mediate inflammation and immune-mediated disorders through multiple mechanisms, such as release of pro-inflammatory mediators, surface inflammation-related molecules, and interaction with leukocytes and endothelial cells (10, 11). Even after being phagocytosed by the mononuclear phagocyte system, platelets modulated survival surface molecules in monocytes (12). In others words, platelets regulated inflammation even after being eliminated from the circulation.
Single-nucleotide polymorphisms (SNPs), the most common pattern of genetic variants, have drawn attention because of their universal distribution and gradually uncovered biological functions (13). Many SNPs of inflammatory cytokine genes, including IL-17F, IL-10, TNFA, TNFB, TGF-beta 1, IL-6, INF-gamma, and IL-1A, influence patients’ susceptibility to ITP (14–18). Among them, IL-17F rs763780 G allele has been identified as a protective factor in ITP, while TNFB + 252G/A A allele was a risk factor. There were also some SNPs associated with the severity of ITP. IL-10 rs1800872 AA genotype presented a significant association with severity of ITP in a cohort of Egyptian population (p = 0.008) (14). The IL-10 gene polymorphism was also identified to be associated with the severity in Japanese population (p = 0.01) (19). When it comes to corticosteroid sensitivity of ITP patients, an earlier study reported that stromal cell-derived factor-1 (SDF-1) rs2297630 GA and AA genotypes significantly increased the corticosteroid treatment sensitivity compared with GG genotype (p = 0.007) (20). Interestingly, a recent study showed that this SDF-1 polymorphism was associated with corticosteroid dependence in pediatric ITP patients (21). Furthermore, researchers identified that ATP-binding cassette gene B1 gene (ANCB1) G2677T/A polymorphism was significantly associated with corticosteroid sensitivity (22). Besides, there were several researches on the association between SNPs and the refractoriness of ITP. For example, it was identified that TNFA-308G/A A allele was a risk factor for refractory ITP (17). In addition, genetic variants of a cluster of inflammation-related genes, including CD24, CD226, FCRL3, IL2, IRF5, ITGAM, NLRP3, CARD8, PTPN22, SH2B3, STAT4, TNFAIP3, and TRAF1, are associated with diverse autoimmune diseases. For example, the rs8734 polymorphism in CD24 was associated with susceptibility to diverse autoimmune diseases including rheumatoid arthritis (RA), autoimmune thyroid disease (AITD), systemic lupus erythematosus (SLE), multiple sclerosis (MS), and inflammatory bowel disease (IBD) (23–29). In addition, four SNP candidates in FCRL3, including rs11264799, rs7528684, rs945635, and rs3761959, have been identified as potential risk factors in multiple autoimmune diseases, including RA, AITD, SLE, MS, autoimmune Addison’s disease, and autoimmune pancreatitis (30–36). Finally, rs6822844, located between the IL2 gene and the nearby IL21 gene, was associated with RA, IBD, type 1 diabetes, and psoriasis (37–40). Therefore, there are multiple inflammation-related SNPs that are robustly associated with multiple autoimmune diseases. The above studies demonstrated that multiple genetic factors involved in the pathogenesis of ITP and other autoimmune diseases. However, most previous researches only verified the associations between SNPs and ITP susceptibility or ITP treatment. None of them associated the polymorphisms with complete clinical data including susceptibility, severity, corticosensitivity, and refractoriness.
Given that ITP is an inflammatory and autoimmune disease and given the above associations between inflammation-related SNPs and other autoimmune diseases, we hypothesized that these SNPs were also associated with primary ITP. The aim of our study was to investigate the association between inflammation-related gene polymorphisms and the pathogenesis of primary ITP in the Chinese Han population.
Materials and Methods
Study Participants
In this case–control study, 312 ITP inpatients were recruited between January 2007 and April 2016 from the Department of Hematology, Qilu Hospital, Shandong University, Jinan, China.
Patients were diagnosed with primary ITP according to the International Working Group guidelines (41). And other autoimmune diseases or underlying immune dysregulation were excluded by medical history, clinical manifestations, physical examinations, radiologic findings and laboratory findings such as HIV and HCV testing, direct antiglobulin test, antiphospholipid antibody testing, antinuclear antibody testing, antithyroid antibody, and thyroid function testing, and testing for other acute and persistent infections. Cases were stratified by severity, corticosteroid sensitivity, and refractoriness based on the standardized definitions (1). Severe ITP is defined by the presence of bleeding symptoms at presentation sufficient to mandate treatment, or by the occurrence of new bleeding symptoms requiring additional therapeutic intervention with a different platelet-enhancing agent or an increased dose to relieve symptoms caused by low platelet count (platelet count may even <10 × 109/L). The corticosteroid regimen is dexamethasone 40 mg p.o. daily for four consecutive days (non-responders received an additional 4-day course of dexamethasone) or prednisone 1.0 mg/kg body weight p.o. daily for four consecutive weeks. Corticosteroid sensitivity is defined as a platelet count ≥30 × 109/L with at least a twofold increase from the baseline count and without bleeding after corticosteroid management. Requirements for additional interventions were considered as corticosteroid resistance. Refractory ITP patients satisfy two criteria. First, they should fail splenectomy. Second, they either manifest severe ITP or have a risk of bleeding that required therapy in the opinion of the attending physician.
For the control group, 205 healthy participants were enrolled. Controls were randomly selected from healthy volunteers with no symptoms of ITP and no history of other autoimmune diseases. All participants were Han Chinese and no genetic associations were found between any participants.
The study was approved by the Medical Ethical Committee of Qilu Hospital, Shandong University. Written informed consent was obtained from each participant in accordance with the Declaration of Helsinki.
DNA Extraction and Genotyping
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) that had been isolated from whole blood samples (5 mL) from participants by a standard protocol. DNA concentration and purity were accessed at 260/280 absorbance by a NanoDrop spectrophotometer. The DNA extraction was stored at −20°C until genotyping. Inflammation-related SNPs (summarized in Table 1) genotyping was performed on a MassArray system (Sequenom iPLEXassay, BGI Tech., Beijing, China), which is based on a multiplex PCR reaction, a locus-specific single-base extension reaction, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
Table 1.
Selected genes and SNPs.
| Genes | SNPs |
|---|---|
| CD24 | rs8734 |
| FCRL3 | rs11264799, rs7528684, rs945635, rs3761959 |
| CD226 | rs763361 |
| IL2 | rs6822844 |
| IRF5 | rs2004640, rs2280714, rs10954213 |
| ITGAM | rs1143679 |
| NLRP3 | rs35829419, rs4353135, rs10754558 |
| CARD8 | rs2043211 |
| PTPN22 | rs33996649, rs1310182 |
| SH2B3 | rs3184504 |
| STAT4 | rs7574865, rs10181656 |
| TNFAIP3 | rs6920220, rs10499194, rs2230926, rs5029939 |
| TRAF1 | rs10818488 |
SNP, single-nucleotide polymorphism.
Statistical Analysis
The p value of the Hardy–Weinberg equilibrium (HWE) was calculated using the calculator available at the Helmholtz Center Munich website. In addition to allelic frequencies, we analyzed genotypic frequencies under three genetic models, specifically, the codominant, dominant, and recessive models. Associations between the SNPs and ITP susceptibility, severity, corticosteroid sensitivity, and refractoriness were calculated by a chi-squared (χ2) test or a Fisher’s exact test. Univariate and multivariate binary logistic regression analyses were used to analyze adjusted p values and odds ratios (ORs) with a corresponding 95% confidence interval (95% CI). A two-tailed p < 0.05 (or adjusted p value by Bonferroni multiple testing) was considered statistically significant. All statistical analyzes were performed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA).
RNA Extraction and Real-time RT-PCR of TNFAIP3
Total RNA of PBMCs was isolated by TRIzol reagent (Invitrogen). RNA was converted into cDNA using the PrimeScript RT reagent kit (Perfect Real Time; Takara) according to the manufacturer’s instructions. Multiplex real-time RT-PCR was performed for TNFAIP3 and the endogenous control β-actin on an ABI PRISM_7500 Sequence Detection System (Applied Biosystems) using SYBR Green (Toyobo) as a double-strand DNA-specific binding dye. The primers for all mRNA assays were intron spanning. The PCR reactions were cycled 40 times after initial denaturation (95°C, 10 min) at 95°C for 15 s and at 60°C for 30 s. The primers for TNFAIP3 and β-actin are as follows: TNFAIP3 forward: GTGTATTTTGGGACTCCAGA, TNFAIP3 reverse: ACTTCTGGCAGTATCCTTCA; β-actin forward: CACCAACTGGGACGACAT, β-actin reverse: GCACAGCCTGGATAGCAAC. We used the comparative threshold cycle (Ct) method for relative quantification of TNFAIP3 mRNA according to relative expression software tool (Michael) (42). The amplification efficiency between the target (TNFAIP3) and the reference control (β-actin) were compared to use the delta Ct (ΔΔCt) calculation.
Results
Study Population
Demographic and clinical characteristics of controls and ITP patients are summarized in Table 2. All inflammation-related SNPs in the control group were in accordance with HWE. No significant deviations were observed after Bonferroni multiple correction (p > 0.002, Table S1 in Supplementary Material).
Table 2.
Demographic and clinical characteristics.
| Controls | ITP patients | |
|---|---|---|
| No. | 205 | 312 |
| Age, mean ± SD | 45.72 ± 13.41 | 39.96 ± 13.69 |
| Gender (M/F) | 75/130 | 116/196 |
| ITP severity, n (%) | ||
| Severe ITP | NA | 189 (60.6) |
| Non-severe ITP | NA | 123 (39.4) |
| Treatment, n (%) | ||
| No use of corticosteroid | NA | 50 (16.0) |
| Corticosteroid-sensitive | NA | 156 (50.0) |
| Corticosteroid-resistant | NA | 106 (34.0) |
| Refractory ITP | NA | 21 (6.7) |
| Non-refractory ITP | NA | 291 (93.3) |
M, male; F, female; NA, not applicable; ITP, immune thrombocytopenia.
Association between Inflammation-Related SNPs and ITP Susceptibility
Four genetic models were used to analyze the association between the 25 inflammation-related SNPs and ITP. We analyzed the relationship between every single locus and the susceptibility to ITP by a chi-squared (χ2) test or a Fisher’s exact test (Table S1 in Supplementary Material). Preliminary screening showed that allelic frequencies of rs8734 in CD24 and genotypic frequencies of rs11264799 in FCRL3 under the codominant model were significantly associated with susceptibility to ITP (p < 0.05). In addition, both the allelic and genotypic frequencies of rs10499194 in TNFAIP3 under the codominant and dominant models were significantly associated with the susceptibility to ITP (p < 0.05). Among the above SNPs, only rs10499194 was associated with susceptibility of ITP after Bonferroni multiple correction.
Univariate logistic regression analysis revealed that for rs8734 in CD24, allele A in place of G was significantly associated with susceptibility to ITP after adjusting for age and gender (p = 0.035, Table 3). For rs11264799 in FCRL3, the CT rather than CC genotype was significantly associated with susceptibility to ITP under the codominant model (p = 0.037, Table 3). For rs10499194 in TNFAIP3, CT and CC/CT genotypes were both statistically significant compared to CC (p = 0.001 and p = 0.000, respectively, Table 3). rs10499194 allelic distribution also showed a statistically significant difference (p = 0.003; Table 3). Interestingly, all three inflammation-related polymorphisms demonstrated a protective effect. Among these polymorphisms, only rs10499194 presented a significant association with susceptibility to ITP with univariate logistic regression analysis after Bonferroni multiple correction.
Table 3.
Association between selected SNPs and ITP risk.
| Gene | SNP | Model/allele | Genotype/allele | Controls |
ITP patients |
OR (95% CI) | Adjusted p value | ||
|---|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | ||||||
| CD24 | rs52812045 | Allele | G | 244 | 59.5 | 411 | 65.9 | 1.000 | |
| A | 166 | 40.5 | 213 | 34.1 | 0.754 (0.580–0.981) | 0.035 | |||
| FCRL3 | rs11264799 | Codominant | CC | 119 | 58.0 | 205 | 65.7 | 1.000 | |
| TT | 3 | 1.5 | 11 | 3.5 | 0.544 (0.678–9.547) | 0.166 | |||
| CT | 83 | 40.5 | 96 | 30.8 | 0.668 (0.458–0.975) | 0.037 | |||
| TNFAIP3 | rs10499194 | Codominant | CC | 160 | 78.0 | 277 | 88.8 | 1.000 | |
| TT | 1 | 0.5 | 2 | 0.6 | 1.235 (0.106–14.385) | 0.866 | |||
| CT | 44 | 21.5 | 33 | 10.6 | 0.431 (0.262–0.711) | 0.001 | |||
| Dominant | CC | 160 | 78.0 | 292 | 93.6 | 1.000 | |||
| TT/CT | 45 | 22.0 | 20 | 6.4 | 0.249 (0.141–0.440) | 0.000 | |||
| Allele | C | 364 | 88.8 | 587 | 94.1 | 1.000 | |||
| T | 46 | 11.2 | 37 | 5.9 | 0.499 (0.315–0.791) | 0.003 | |||
SNP, single-nucleotide polymorphism; CI, confidence interval; OR, odds ratio; ITP, immune thrombocytopenia.
Adjusted p value calculated with univariate logistic regression.
Bold highlights statistical significance (p < 0.05).
Next, we performed a multivariate logistic regression analysis under the codominant model. We found that the heterozygous genotypes of FCRL3 rs11264799 and TNFAIP3 rs10499194 significantly decreased the risk of ITP compared with homozygous major alleles (p = 0.029 and p = 0.001, respectively, Table 4). When we analyzed the combined influence of allelic distribution of both CD24 rs52812045 and TNFAIP3 rs10499194, we found a statistical difference in TNFAIP3 rs10499194 between ITP patients and controls (p = 0.006, Table 5).
Table 4.
Association between selected SNPs and ITP risk under codominant model.
| Genes | SNP | Genotype | Controls |
ITP patients |
OR (95% CI) | Adjusted p value | ||
|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | |||||
| FCRL3 | rs11264799 | CC | 119 | 58.0 | 205 | 65.7 | 1.000 | |
| TT | 3 | 1.5 | 11 | 3.5 | 2.703 (0.719–10.161) | 0.141 | ||
| CT | 83 | 40.5 | 96 | 30.8 | 0.653 (0.446–0.958) | 0.029 | ||
| TNFAIP3 | rs10499194 | CC | 160 | 78.0 | 277 | 88.8 | 1.000 | |
| TT | 1 | 0.5 | 2 | 0.6 | 1.285 (0.104–15.869) | 0.845 | ||
| CT | 44 | 21.5 | 33 | 10.6 | 0.415 (0.250–0.689) | 0.001 | ||
SNP, single-nucleotide polymorphism; CI, confidence interval; OR, odds ratio; ITP, immune thrombocytopenia.
Adjusted p value calculated with multivariate logistic regression.
Bold highlights statistical significance (p < 0.05).
Table 5.
Association between selected SNPs and ITP risk in allelic analysis.
| Genes | SNP | Allele | Controls |
ITP patients |
OR (95% CI) | Adjusted p value | ||
|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | |||||
| CD24 | rs52812045 | G | 244 | 59.5 | 411 | 65.9 | 1 | |
| A | 166 | 40.5 | 213 | 34.1 | 0.785 (0.0620–1.023) | 0.073 | ||
| TNFAIP3 | rs10499194 | C | 364 | 88.8 | 587 | 94.1 | 1.000 | |
| T | 46 | 11.2 | 37 | 5.9 | 0.522 (0.329–0.829) | 0.006 | ||
SNP, single-nucleotide polymorphism; CI, confidence interval; OR, odds ratio; ITP, immune thrombocytopenia.
Adjusted p value calculated with multivariate logistic regression.
Bold highlights statistical significance (p < 0.05).
Association between Inflammation-Related SNPs and ITP Severity
We next evaluated the association of the inflammation-related SNPs with ITP disease severity. CARD8 rs2043211 and TRAF1 rs10818488 genotypes were statistically different between severe and non-severe ITP patients according to chi-squared analyses (p < 0.05, Table S2 in Supplementary Material). However, neither difference persisted after Bonferroni multiple correction. CARD8 rs2043211 lost the statistical difference after adjusting for age and gender factors by univariate binary logistic regression under dominant model (AA + AT vs. TT: OR = 0.589, 95% CI = 0.343–1.013, p = 0.056). After adjusting for age and gender, ITP patients carrying the AG genotype of TRAF1 rs10818488 showed a 1.713-fold increased risk of developing severe ITP compared with patients carrying major genotype GG under codominant model (OR = 1.713, 95% CI = 1.008–2.911, p = 0.047). In summary, only the AG genotype of TRAF1 rs10818488 increased the risk of severe ITP among patients.
Association between Inflammation-Related SNPs and Corticosteroid Sensitivity
After studying associations with the pathogenesis of ITP, we explored the association between the inflammation-related polymorphisms and the treatment of ITP. Specifically, we studied corticosteroid sensitivity and refractoriness among patients with different genotypes or alleles. To study corticosteroid sensitivity, we divided the patients who received corticosteroid treatment into two groups, the corticosteroid-sensitive group (n = 156) and the corticosteroid-resistant group (n = 106).
We found statistically significant associations between inflammation-related polymorphisms and corticosteroid sensitivity. rs945635, rs7528684, and rs3761959 of FCRL3 and rs2004640 of IRF5 under the dominant model were significantly associated with corticosteroid-sensitivity (p < 0.05, Table S3 in Supplementary Material). rs4353135 of NLRP3 under both the codominant and recessive models showed significant association with corticosteroid sensitivity (p < 0.05, Table S3 in Supplementary Material). Allelic frequencies and genotypic frequencies under the codominant and dominant models of TNFAIP3 rs10499194 revealed statistically significant differences between the corticosteroid-sensitive and corticosteroid-resistant groups (p < 0.05, Table S3 in Supplementary Material). TNFAIP3 rs10499194 remained significant after Bonferroni multiple correction.
Allelic and genotypic frequencies were analyzed by logistic regression analysis. After univariate logistic regression analysis, compared with major allele homozygotes, minor allele homozygotes, and heterozygotes of FCRL3 rs945635, rs7528684, and rs3761959 and IRF rs2004640 were still significantly associated with corticosteroid-sensitivity under the dominant model after adjusting for age and gender (p = 0.029, p = 0.029, p = 0.029, p = 0.024, respectively, Table 6). Genotypic frequencies of NLRP3 rs4353135 were significantly different under the recessive model between the corticosteroid-sensitive and -resistant groups (p = 0.028, Table 6). Genotypic and allelic frequencies of TNFAIP3 rs10499194 were significantly different under the codominant and dominant models (all p = 0.001, Table 6). Importantly, the observed differences in TNFAIP3 rs10499194 were still statistically significant after Bonferroni multiple correction (p < 0.002).
Table 6.
Association between selected SNPs and corticosteroid-sensitivity of ITP patients.
| Gene | SNP | Model/allele | Genotype/allele | Sensitive |
Resistant |
OR (95% CI)* | p Value* | OR (95% CI)# | p Value# | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | % | Count | % | ||||||||
| TNFAIP3 | rs10499194 | Codominant | CC | 128 | 82.1 | 104 | 98.1 | 1 | |||
| TT | 2 | 1.3 | 0 | 0.0 | – | 0.999 | |||||
| CT | 26 | 16.7 | 2 | 1.9 | 0.092 (0.021–0.398) | 0.001 | |||||
| Dominant | CC | 128 | 82.1 | 104 | 98.1 | 1.000 | 1.000 | ||||
| TT/CT | 28 | 17.9 | 2 | 1.9 | 0.086 (0.020–0.369) | 0.001 | 0.069 (0.016–0.302) | 0.000 | |||
| Allele | C | 282 | 90.4 | 210 | 99.1 | 1.000 | |||||
| T | 30 | 9.6 | 2 | 0.9 | 0.088 (0.021–0.372) | 0.001 | |||||
| IRF5 | rs2004640 | Dominant | GG | 96 | 61.5 | 51 | 48.1 | 1.000 | 1.000 | ||
| TT/GT | 60 | 38.5 | 55 | 51.9 | 1.793 (1.079–2.980) | 0.024 | 1.996 (1.162–3.429) | 0.012 | |||
| FCRL3 | rs3761959 | Dominant | CC | 65 | 41.7 | 30 | 28.3 | 1.000 | 1.000 | ||
| TT/CT | 91 | 58.3 | 76 | 71.7 | 1.807 (1.062–3.076) | 0.029 | 2.107 (1.208–3.674) | 0.009 | |||
| FCRL3 | rs7528684 | Dominant | AA | 65 | 41.7 | 30 | 28.3 | 1.000 | 1.000 | ||
| GG/AG | 91 | 58.3 | 76 | 71.7 | 1.807 (1.062–3.076) | 0.029 | 2.107 (1.208–3.674) | 0.009 | |||
| FCRL3 | rs945635 | Dominant | CC | 65 | 41.7 | 30 | 28.3 | 1.000 | 1.000 | ||
| GG/CG | 91 | 58.3 | 76 | 71.7 | 1.807 (1.062–3.076) | 0.029 | 2.107 (1.208–3.674) | 0.009 | |||
| NLRP3 | rs4353135 | Codominant | TT | 49 | 31.4 | 36 | 34.0 | 1.000 | |||
| GG | 24 | 15.4 | 29 | 27.4 | 1.599 (0.798–3.205) | 0.186 | |||||
| GT | 83 | 53.2 | 41 | 38.7 | 0.684 (0.385–1.212) | 0.193 | |||||
| Recessive | TT/GT | 132 | 84.6 | 77 | 72.6 | 1.000 | |||||
| GG | 24 | 15.4 | 29 | 27.4 | 1.991 (1.077–3.681) | 0.028 | |||||
SNP, single-nucleotide polymorphism; CI, confidence interval; OR, odds ratio; ITP, immune thrombocytopenia.
*Calculated by univariate logistic regression.
#Calculated by multivariate logistic regression analysis under dominant model.
Bold highlights statistical significance (p < 0.05).
Multivariate logistic regression showed significant associations between FCRL3 rs945635, rs7528684, and rs3761959, IRF rs2004640, and TNFAIP3 rs10499194 and corticosteroid sensitivity (p = 0.009, p = 0.009, p = 0.009, p = 0.012, and p = 0.000, respectively, Table 6). TNFAIP3 rs10499194 was significantly associated with corticosteroid-sensitivity under both univariate and multivariate logistic regression analyses after Bonferroni multiple correction (p < 0.002).
Association between Inflammation-Related SNPs and ITP Refractoriness
To study refractoriness, we divided the enrolled patients into two subgroups: those refractory to splenectomy treatment and those who responded to medical treatment. We analyzed the association between the inflammation-related SNPs and refractoriness of ITP using chi-squared tests for preliminary screening. The genotypic distribution of STAT4 rs7574869 was significantly associated with refractoriness under the codominant model (p < 0.05, Table S4 in Supplementary Material). No statistical differences were found between other SNPs and refractoriness (p > 0.05). However, the association between STAT4 rs7574869 and refractoriness was not statistically significant after Bonferroni multiple correction. When the GG genotype was used as reference, neither the TT nor GT genotypes were significantly associated with refractoriness after adjusting for the age and gender (p = 0.998 and p = 0.108, respectively).
Association of TNFAIP3 rs10499194 Polymorphism and Expression Levels of TNFAIP3
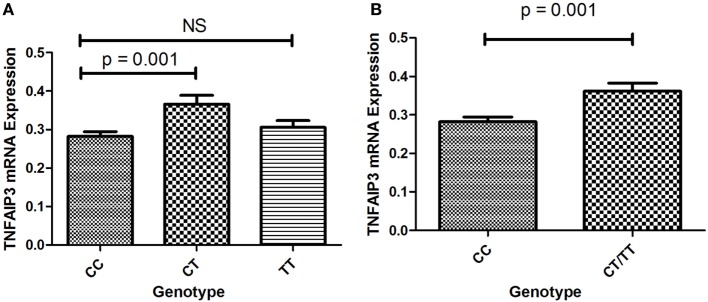
To determine whether TNFAIP3 rs10499194 was a protective factor for ITP, we further explored the effects of this polymorphism on TNFAIP3 expression. The number of ITP patients with TT genotype was small (n = 2) in this study, which may lead to an underpowered assessment. The expression of TNFAIP3 was assessed in 85 ITP patients (60 cases of CC, 2 cases of TT, and 23 cases of CT) by quantitative real-time RT-PCR. As shown in Figure 1A, patients with CT genotype showed higher levels of TNFAIP3 mRNA expression than those with CC genotype (p = 0.001). We found that TNFAIP3 expression was significantly higher in samples with pooled CT/TT genotype compared with the CC genotype (p = 0.001; Figure 1B). The result showed that CC genotype on TNFAIP3 rs10499194 was a protective factor for ITP.
Figure 1.
(A) Expression of TNFAIP3 mRNA in ITP patients with the CC, CT and TT genotypes. (B) Expression of TNFAIP3 mRNA in ITP patients with the CC and pooled CT/TT genotypes.
Discussion
Immune thrombocytopenia is an autoimmune and inflammatory disease characterized by immune-mediated platelet destruction. Although ITP has been associated with inflammation, only a few studies have identified the association between inflammation-related SNPs and the pathogenesis of ITP (43). By contrast, inflammation-related gene polymorphisms have been associated with multiple autoimmune diseases.
In this study, we assessed the associations between 25 inflammation-related SNPs and the occurrence and treatment of ITP in the Chinese Han population. Of these SNPs, the frequencies of the homozygous minor allele of FCRL3 rs11264799 and TNFAIP3 rs10499194 were significantly decreased in ITP patients. The A allele of CD24 rs8734 was associated with a decreased risk of ITP. In addition, compared with the CC genotype, individuals carrying pooled TT/CT genotypes of TNFAIP3 rs10499194 had a decreased risk of ITP under the dominant model. These three SNPs were all protective factors.
Our findings contrast evidence found regarding other autoimmune diseases (26, 44–48). For example, in the Spanish population, no association was found between FCRL3 rs11264799 and MS (44). Interestingly, however, the C allele of FCRL3 rs7528684 was a protective factor for MS. This suggests that FCRL3 polymorphisms may protect individuals from autoimmune diseases, which is somewhat consistent with our findings. In the Israeli population, individuals with CD24 rs8734 polymorphisms had an increased risk of IBD, ulcerative colitis, and Crohn’s disease (26). In the Chinese population, a significant association was not found between TNFAIP3 rs10499194 and SLE (45); however, carriers of the CT genotype and combined TT/CT genotype had an increased risk for RA in the Chinese Han population (46). A recent meta-analysis revealed that there was no association between TNFAIP3 rs10499194 and RA in Europeans, but a significant association in Asians (OR = 1.254, 95% CI = 1.101–1.429, p = 6.7 × 10−4) (48). The discrepancies between our results and previous research may be attributed to differences in the populations and autoimmune diseases studied.
When it comes to TNFAIP3 rs10499194, there were several researches identified that it was significantly associated with the susceptibility of autoimmune diseases. Strong associations were observed between TNFAIP3 rs10499194 and juvenile idiopathic arthritis (OR = 0.74, 95% CI = 0.61–0.91, p < 0.004) (49). Prahalad et al demonstrated that TNFAIP3 rs10499194 had a significant protective effect against childhood onset RA (OR = 0.60, 95% CI = 0.44–0.83, p = 0.002) (50). Consistent with our findings, TNFAIP3 rs2230926 and rs5029939 were significantly different between chronic ITP and control groups (43), suggesting that TNFAIP3 polymorphisms may affect the susceptibility to ITP. We found that TNFAIP3 rs10499194 was significantly associated with the susceptibility of ITP even after Bonferroni multiple correction. Therefore, TNFAIP3 rs10499194 may be an important susceptibility-related SNP for ITP.
To investigate whether TNFAIP3 rs10499194 was a functional polymorphism, quantitative real-time RT-PCR was performed to evaluate TNFAIP3 expression. The data showed that individuals with CT genotype on TNFAIP3 rs10499194 locus showed higher levels of TNFAIP3 mRNA expression compared with the CC genotype, which might play a role in the susceptibility of ITP. It is widely accepted that TNFAIP3 is a deubiquitinating protein which can deregulate pro-inflammatory signal pathways including NF-κB- and IRF3-dependent gene expression by deubiquitinating specific signaling molecules (51, 52). Some researchers revealed that TNFAIP3 was a central gatekeeper in inflammation and immunity (53). A recent study demonstrated that lack of TNFAIP3 in B cells resulted in overexpression of pro-inflammatory cytokines, which caused inflammation and autoimmunity in aged mice (54). TNFAIP3-deficient B cells displayed enhanced proliferation and development of autoantibodies (55). In our functional analysis, individuals with CT genotype on TNFAIP3 rs10499194 locus showed higher levels of TNFAIP3 mRNA expression compared with the wild genotype CC, which is in agreement with our genotyping analysis that CT genotype is a protective factor for ITP. However, the mechanism of polymorphism on TNFAIP3 expression awaits further investigations.
Furthermore, we found that the AG genotype of TRAF1 rs10818488 was significantly associated with increased risk of severe ITP after adjusting for age and gender. These novel findings will be studied further in order to understand the molecular mechanism of these genetic polymorphisms.
In addition to the pathogenesis of ITP, the mechanism of corticosteroid resistance remains poorly understood. In acute lymphoblastic leukemia, the NLRP3-CASP1 inflammasome induced glucocorticoid resistance (56). Overexpression of CASP1 may promote cleavage of the glucocorticoid receptor, which decreased glucocorticoid sensitivity. We identified that NLRP3 rs4353135 was significantly associated with increased risk of corticosteroid resistance of ITP under the recessive model; however, this observation was not significant after Bonferroni multiple correction. Thus, we propose that the NLRP3 rs4353135 polymorphism may enhance expression of the NLRP3-CASP1 inflammasome and lead to corticosteroid resistance. Future studies will examine this hypothesis.
Importantly, TNFAIP3 rs10499194 was associated with corticosteroid-sensitivity after Bonferroni correction. Future work will examine the potential of TNFAIP3 rs10499194 as a biomarker for corticosteroid sensitivity.
There were some limitations to our study. On the one hand, the participants enrolled in our study were solely from Chinese Han population. Larger trials investigating multi-racial populations are needed to examine whether the associations between the inflammation-related SNPs and ITP exist in the general population. Second, as mentioned earlier, the biological functions and signaling pathways of these SNPs are not yet known and future research with global collaborations are required. Third, there was a potential selection bias in our study. Only inpatients were recruited in our study, as the corticosteroid-sensitivity and refractoriness of outpatients were difficult to follow-up in our hospital. The inpatients have more severe thrombocytopenia and bleeding symptoms. The bleeding symptoms may include gastrointestinal hemorrhage, extensive skin and mucosal hemorrhage, or intracranial hemorrhage. Besides, the 25 susceptibility loci, we tested were selected basing on the pervious reported loci associated with other autoimmune diseases. The candidate gene approach cannot cover all susceptible loci, which may lead to the selection bias.
Conclusion
Our investigation of SNPs and ITP provides interesting results. We found that inflammation-related SNPs, especially TNFAIP3 rs10499194, may be genetic risk factors associated with the development and treatment of ITP. Our findings may lead to clinicians screening for these SNPs in order to predict the prognosis and guide the treatment of ITP.
Statement of Prior Presentation
Part of this study was presented as a poster with a title of “Inflammation-related Gene Polymorphisms Associated with Susceptibility to Primary Immune Thrombocytopenia” (Abstract Code: 3737) and won ASH Abstract Achievement Award at the 58th ASH Annual Meeting and Exposition in San Diego, CA, USA, December 3–6, 2016.
Ethics Statement
This study was carried out in accordance with the recommendations of Medical Ethical Committee of Qilu Hospital, Shandong University with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Medical Ethical Committee of Qilu Hospital, Shandong University.
Author Contributions
JP, MH, and X-hZ designed research, analyzed data, and wrote the paper; JL performed research, analyzed data, and wrote the paper; SM, LS, CM, and CG performed research and analyzed data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Alexandra Marshall (Marshall Medical Communications) for her editorial assistance. This work was supported by grants from the Major Research plan of the National Natural Science Foundation of China (91442204), National Natural Science Foundation for Distinguished Young Scholars of China (81125002), National Natural Science Foundation of China (81370623, 81470284 and 81500094), State Program of National Natural Science Foundation of China for Innovative Research Group (81321061), State Key Clinical Specialty of China for Blood Disorders, and Tai Shan Scholar Foundation.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00744/full#supplementary-material.
References
- 1.Rodeghiero F, Stasi R, Gernsheimer T, Michel M, Provan D, Arnold DM, et al. Standardization of terminology, definitions and outcome criteria in immune thrombocytopenic purpura of adults and children: report from an International Working Group. Blood (2009) 113(11):2386–93. 10.1182/blood-2008-07-162503 [DOI] [PubMed] [Google Scholar]
- 2.Olsson B, Andersson PO, Jernås M, Jacobsson S, Carlsson B, Carlsson LM, et al. T-cell-mediated cytotoxicity toward platelets in chronic idiopathic thrombocytopenic purpura. Nat Med (2003) 9(9):1123–4. 10.1038/nm921 [DOI] [PubMed] [Google Scholar]
- 3.McMillan R, Wang L, Tomer A, Nichol J, Pistillo J. Suppression of in vitro megakaryocyte production by antiplatelet autoantibodies from adult patients with chronic ITP. Blood (2004) 103(4):1364–9. 10.1182/blood-2003-08-2672 [DOI] [PubMed] [Google Scholar]
- 4.Cines DB, Bussel JB, Liebman HA, Luning Prak ET. The ITP syndrome: pathogenic and clinical diversity. Blood (2009) 113(26):6511–21. 10.1182/blood-2009-01-129155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doria A, Zen M, Bettio S, Gatto M, Bassi N, Nalotto L, et al. Autoinflammation and autoimmunity: bridging the divide. Autoimmun Rev (2012) 12(1):22–30. 10.1016/j.autrev.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 6.Kazatchkine MD, Kaveri SV. Immunomodulation of autoimmune and inflammatory diseases with intravenous immune globulin. N Engl J Med (2001) 345(10):747–55. 10.1056/NEJMra993360 [DOI] [PubMed] [Google Scholar]
- 7.Imbach P. Development and research in idiopathic thrombocytopenic purpura: an inflammatory and autoimmune disorder. Pediatr Blood Cancer (2006) 47(5 Suppl):685–6. 10.1002/pbc.20969 [DOI] [PubMed] [Google Scholar]
- 8.Dal MS, Karakus A, Aydin BB, Ekmen MO, Ulas T, Ayyildiz O. Serum uric acid and inflammation in patients with immune thrombocytopenic purpura: preliminary results. Eur Rev Med Pharmacol Sci (2015) 19(22):4385–9. [PubMed] [Google Scholar]
- 9.Fontana V, Horstman LL, Donna E, Dudkiewicz P, Ahn ER, Ahn YS. Interstitial lung disease (ILD) and severe ITP. Hematology (2007) 12(1):75–80. [DOI] [PubMed] [Google Scholar]
- 10.Boudreau LH, Duchez AC, Cloutier N, Soulet D, Martin N, Bollinger J, et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood (2014) 124(14):2173–83. 10.1182/blood-2014-05-573543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weyrich AS, Lindemann S, Zimmerman GA. The evolving role of platelets in inflammation. J Thromb Haemost (2003) 1(9):1897–905. 10.1046/j.1538-7836.2003.00304.x [DOI] [PubMed] [Google Scholar]
- 12.Lang D, Dohle F, Terstesse M, Bangen P, August C, Pauels HG, et al. Down-regulation of monocyte apoptosis by phagocytosis of platelets: involvement of a caspase-9, caspase-3, and heat shock protein 70-dependent pathway. J Immunol (2002) 168(12):6152–8. 10.4049/jimmunol.168.12.6152 [DOI] [PubMed] [Google Scholar]
- 13.Zhang N, Huo Q, Wang X, Chen X, Long L, Guan X, et al. A genetic variant in p63 (rs17506395) is associated with breast cancer susceptibility and prognosis. Gene (2014) 535(2):170–6. 10.1016/j.gene.2013.11.038 [DOI] [PubMed] [Google Scholar]
- 14.El Ghannam D, Fawzy IM, Azmy E, Hakim H, Eid I. Relation of interleukin-10 promoter polymorphisms to adult chronic immune thrombocytopenic purpura in a cohort of Egyptian population. Immunol Invest (2015) 44(7):616–26. 10.3109/08820139.2015.1064948 [DOI] [PubMed] [Google Scholar]
- 15.Li H, Zhou Z, Tai W, Feng W, Zhang D, Gu X, et al. Decreased frequency of IL-17F rs763780 site allele G is associated with genetic susceptibility to immune thrombocytopenia in a Chinese population. Clin Appl Thromb Hemost (2017). 23(5):466–71. 10.1177/1076029615618022 [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Xiong YZ, Li T, Li Y, Gu SQ, Wang YM, et al. Interleukin-17A and -17F gene polymorphisms in Chinese population with chronic immune thrombocytopenia. Ann Clin Lab Sci (2016) 46(3):291–7. [PubMed] [Google Scholar]
- 17.Pavkovic M, Angelovic R, Popova-Simjanovska M, Karanfilski O, Trpkovska-Terzieva S, Sotirova T, et al. Single nucleotide polymorphisms of the inflammatory cytokine genes: interleukin-1B, tumor necrosis factors-A and tumor necrosis factor-B in adult patients with immune thrombocytopenia. Pril (Makedon Akad Nauk Umet Odd Med Nauki) (2015) 36(1):109–15. [DOI] [PubMed] [Google Scholar]
- 18.Pehlivan M, Okan V, Sever T, Balci SO, Yilmaz M, Babacan T, et al. Investigation of TNF-alpha, TGF-beta 1, IL-10, IL-6, IFN-gamma, MBL, GPIA, and IL1A gene polymorphisms in patients with idiopathic thrombocytopenic purpura. Platelets (2011) 22(8):588–95. 10.3109/09537104.2011.577255 [DOI] [PubMed] [Google Scholar]
- 19.Saitoh T, Kasamatsu T, Inoue M, Mitsui T, Koiso H, Yokohama A, et al. Interleukin-10 gene polymorphism reflects the severity of chronic immune thrombocytopenia in Japanese patients. Int J Lab Hematol (2011) 33(5):526–32. 10.1111/j.1751-553X.2011.01320.x [DOI] [PubMed] [Google Scholar]
- 20.Lyu M, Li Y, Hao Y, Sun T, Liu W, Lyu C, et al. Stromal cell-derived factor-1 rs2297630 polymorphism associated with platelet production and treatment response in Chinese patients with chronic immune thrombocytopenia. Platelets (2016) 27(4):338–43. 10.3109/09537104.2015.1103368 [DOI] [PubMed] [Google Scholar]
- 21.Ku FC, Tsai CR, Der Wang J, Wang CH, Chang TK, Hwang WL. Stromal-derived factor-1 gene variations in pediatric patients with primary immune thrombocytopenia. Eur J Haematol (2013) 90(1):25–30. 10.1111/ejh.12025 [DOI] [PubMed] [Google Scholar]
- 22.Xuan M, Li H, Fu R, Yang Y, Zhang D, Zhang X, et al. Association of ABCB1 gene polymorphisms and haplotypes with therapeutic efficacy of glucocorticoids in Chinese patients with immune thrombocytopenia. Hum Immunol (2014) 75(4):317–21. 10.1016/j.humimm.2014.01.013 [DOI] [PubMed] [Google Scholar]
- 23.Goris A, Maranian M, Walton A, Yeo TW, Ban M, Gray J, et al. CD24 Ala/Val polymorphism and multiple sclerosis. J Neuroimmunol (2006) 175(1–2):200–2. 10.1016/j.jneuroim.2006.03.009 [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Yang Y, Liang Z, Kang M, Kuang Y, Li F. Association between the CD24 Ala57Val polymorphism and risk for multiple sclerosis and systemic lupus erythematosus: a meta-analysis. Sci Rep (2015) 5:9557. 10.1038/srep09557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue N, Watanabe M, Hayashi F, Hidaka Y, Iwatani Y. The association between a functional polymorphism in the CD24 gene and the development of autoimmune thyroid diseases. Tissue Antigens (2013) 81(3):161–3. 10.1111/tan.12070 [DOI] [PubMed] [Google Scholar]
- 26.Lisiansky V, Kraus S, Naumov I, Kazanov D, Nabiochtchikov I, Toledano O, et al. Role of CD24 polymorphisms in the susceptibility to inflammatory bowel disease. Int J Biol Markers (2014) 29(1):e62–8. 10.5301/jbm.5000072 [DOI] [PubMed] [Google Scholar]
- 27.Piotrowski P, Lianeri M, Wudarski M, Lacki JK, Jagodzinski PP. CD24 Ala57Val gene polymorphism and the risk of systemic lupus erythematosus. Tissue Antigens (2010) 75(6):696–700. 10.1111/j.1399-0039.2010.01447.x [DOI] [PubMed] [Google Scholar]
- 28.Sánchez E, Abelson AK, Sabio JM, González-Gay MA, Ortego-Centeno N, Jiménez-Alonso J, et al. Association of a CD24 gene polymorphism with susceptibility to systemic lupus erythematosus. Arthritis Rheum (2007) 56(9):3080–6. 10.1002/art.22871 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez E, Fernandez-Gutierrez B, Gonzalez-Gay MA, Balsa A, García A, Rodríguez L, et al. Investigating the role of CD24 gene polymorphisms in rheumatoid arthritis. Ann Rheum Dis (2008) 67(8):1197–8. 10.1136/ard.2007.084475 [DOI] [PubMed] [Google Scholar]
- 30.Martínez A, Mas A, de Las Heras V, Bartolomé M, Arroyo R, Fernández-Arquero M, et al. FcRL3 and multiple sclerosis pathogenesis: role in autoimmunity? J Neuroimmunol (2007) 189(1–2):132–6. 10.1016/j.jneuroim.2007.06.018 [DOI] [PubMed] [Google Scholar]
- 31.Owen CJ, Kelly H, Eden JA, Merriman ME, Pearce SH, Merriman TR. Analysis of the Fc receptor-like-3 (FCRL3) locus in Caucasians with autoimmune disorders suggests a complex pattern of disease association. J Clin Endocrinol Metab (2007) 92(3):1106–11. 10.1210/jc.2006-2183 [DOI] [PubMed] [Google Scholar]
- 32.Bajpai UD, Swainson LA, Mold JE, Graf JD, Imboden JB, McCune JM. A functional variant in FCRL3 is associated with higher Fc receptor-like 3 expression on T cell subsets and rheumatoid arthritis disease activity. Arthritis Rheum (2012) 64(8):2451–9. 10.1002/art.34457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, Sawada T, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet (2005) 37(5):478–85. 10.1038/ng0605-652c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez-Bello J, Jiménez-Morales S, Espinosa-Rosales F, Gómez-Vera J, Gutiérrez A, Velázquez Cruz R, et al. Juvenile rheumatoid arthritis and asthma, but not childhood-onset systemic lupus erythematosus are associated with FCRL3 polymorphisms in Mexicans. Mol Immunol (2013) 53(4):374–8. 10.1016/j.molimm.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 35.Sanchez E, Callejas JL, Sabio JM, de Haro M, Camps M, de Ramón E, et al. Polymorphisms of the FCRL3 gene in a Spanish population of systemic lupus erythematosus patients. Rheumatol (2006) 45(8):1044–6. 10.1093/rheumatology/kel160 [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Yang LH, Zuo J, Liang YL, Li PQ, Liu W, et al. Fc receptor-like 3 gene polymorphisms confer susceptibility to rheumatoid arthritis in a Chinese population. Hum Immunol (2010) 71(12):1203–8. 10.1016/j.humimm.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 37.Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, et al. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut (2009) 58(6):799–804. 10.1136/gut.2008.166918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fichna M, Zurawek M, Fichna P, Ziolkowska-Suchanek I, Januszkiewicz D, Nowak J. Polymorphic variant at the IL2 region is associated with type 1 diabetes and may affect serum levels of interleukin-2. Mol Biol Rep (2013) 40(12):6957–63. 10.1007/s11033-013-2815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren RB, Smith RL, Flynn E, Bowes J, UKRAG Consortium. Eyre S, et al. A systematic investigation of confirmed autoimmune loci in early-onset psoriasis reveals an association with IL2/IL21. Br J Dermatol (2011) 164(3):660–4. 10.1111/j.1365-2133.2011.10237.x [DOI] [PubMed] [Google Scholar]
- 40.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet (2007) 81(6):1284–8. 10.1086/522037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Provan D, Stasi R, Newland AC, Blanchette VS, Bolton-Maggs P, Bussel JB, et al. International consensus report on the investigation and management of primary immune thrombocytopenia. Blood (2010) 115(2):168–86. 10.1182/blood-2009-06-225565 [DOI] [PubMed] [Google Scholar]
- 42.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res (2002) 30(9):e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Yang J, Liu L, Zhang D, Zhou K, Li H, et al. The polymorphisms of tumor necrosis factor-induced protein 3 gene may contribute to the susceptibility of chronic primary immune thrombocytopenia in Chinese population. Platelets (2016) 27(1):26–31. 10.3109/09537104.2015.1022142 [DOI] [PubMed] [Google Scholar]
- 44.Matesanz F, Fernández O, Milne RL, Fedetz M, Leyva L, Guerrero M, et al. The high producer variant of the Fc-receptor like-3 (FCRL3) gene is involved in protection against multiple sclerosis. J Neuroimmunol (2008) 195(1–2):146–50. 10.1016/j.jneuroim.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 45.You Y, Wang Z, Deng G, Hao F. Lack of association between Fc receptor-like 3 gene polymorphisms and systemic lupus erythematosus in Chinese population. J Dermatol Sci (2008) 52(2):118–22. 10.1016/j.jdermsci.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Li W, Zhang X, Zhao L, Zhang X, Jiang L, et al. Single nucleotide polymorphisms in TNFAIP3 were associated with the risks of rheumatoid arthritis in northern Chinese Han population. BMC Med Genet (2014) 15:56. 10.1186/1471-2350-15-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Su X, Zhang K, Zhou R. The Fc receptor-like 3 gene polymorphisms and susceptibility to autoimmune diseases: an updated meta-analysis. Autoimmunity (2013) 46(8):547–58. 10.3109/08916934.2013.835804 [DOI] [PubMed] [Google Scholar]
- 48.Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Associations between TNFAIP3 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm Res (2012) 61(6):635–41. 10.1007/s00011-011-0397-3 [DOI] [PubMed] [Google Scholar]
- 49.Prahalad S, Hansen S, Whiting A, Guthery SL, Clifford B, McNally B, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum (2009) 60(7):2124–30. 10.1002/art.24618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prahalad S, Conneely KN, Jiang Y, Sudman M, Wallace CA, Brown MR, et al. Susceptibility to childhood-onset rheumatoid arthritis: investigation of a weighted genetic risk score that integrates cumulative effects of variants at five genetic loci. Arthritis Rheum (2013) 65(6):1663–7. 10.1002/art.37913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shembade N, Harhaj EW. Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cell Mol Immunol (2012) 9(2):123–30. 10.1038/cmi.2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verstrepen L, Verhelst K, van Loo G, Carpentier I, Ley SC, Beyaert R. Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem Pharmacol (2010) 80(12):2009–20. 10.1016/j.bcp.2010.06.044 [DOI] [PubMed] [Google Scholar]
- 53.Coornaert B, Carpentier I, Beyaert R. A20: central gatekeeper in inflammation and immunity. J Biol Chem (2009) 284(13):8217–21. 10.1074/jbc.R800032200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu Y, Vahl JC, Kumar D, Heger K, Bertossi A, Wójtowicz E, et al. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood (2011) 117(7):2227–36. 10.1182/blood-2010-09-306019 [DOI] [PubMed] [Google Scholar]
- 55.Hövelmeyer N, Reissig S, Xuan NT, Adams-Quack P, Lukas D, Nikolaev A, et al. A20 deficiency in B cells enhances B-cell proliferation and results in the development of autoantibodies. Eur J Immunol (2011) 41(3):595–601. 10.1002/eji.201041313 [DOI] [PubMed] [Google Scholar]
- 56.The NLRP3-CASP1 inflammasome induces glucocorticoid resistance in ALL. Cancer Discov (2015) 5(7):OF18. 10.1158/2159-8290.CD-RW2015-090 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.