Abstract
The ozone-depleting and greenhouse gas, nitrous oxide (N2O), is mainly consumed by the microbially mediated anaerobic process, denitrification. N2O consumption is the last step in canonical denitrification, and is also the least O2 tolerant step. Community composition of total and active N2O consuming bacteria was analyzed based on total (DNA) and transcriptionally active (RNA) nitrous oxide reductase (nosZ) genes using a functional gene microarray. The total and active nosZ communities were dominated by a limited number of nosZ archetypes, affiliated with bacteria from marine, soil and marsh environments. In addition to nosZ genes related to those of known marine denitrifiers, atypical nosZ genes, related to those of soil bacteria that do not possess a complete denitrification pathway, were also detected, especially in surface waters. The community composition of the total nosZ assemblage was significantly different from the active assemblage. The community composition of the total nosZ assemblage was significantly different between coastal and off-shore stations. The low oxygen assemblages from both stations were similar to each other, while the higher oxygen assemblages were more variable. Community composition of the active nosZ assemblage was also significantly different between stations, and varied with N2O concentration but not O2. Notably, nosZ assemblages were not only present but also active in oxygenated seawater: the abundance of total and active nosZ bacteria from oxygenated surface water (indicated by nosZ gene copy number) was similar to or even larger than in anoxic waters, implying the potential for N2O consumption even in the oxygenated surface water.
Keywords: N2O consuming bacteria, nosZ gene, microarray, oxygen minimum zone, Eastern Tropical South Pacific
Introduction
N2O is a major ozone-depleting substance and a greenhouse gas whose radiative forcing per mole is 298 times that of carbon dioxide (IPCC, 2007; Ravishankara et al., 2009). Oxygen minimum zones (OMZs) are the most intense marine sources of N2O and are hot spots of rapid N2O cycling (Martinez-Rey et al., 2015). OMZs are marine regions with a strong O2 gradient (oxycline) overlying an oxygen deficient zone (ODZ) where O2 concentration is low enough to induce anaerobic processes. The global expansion and intensification of OMZs, which are predicted to result from global warming, further stress the importance of understanding N2O cycling in these regions (Codispoti, 2010). N2O production and consumption are driven by marine bacteria (Naqvi et al., 2000). The dominant microbial process for N2O cycling is denitrification, the sequential reduction of NO3- to NO2-, NO, N2O and finally to N2 (Zumft, 1997; Naqvi et al., 2000). Denitrification could stop at an intermediate step before N2 if O2 concentration exceeds the threshold for the latter step or if electron donors are depleted (Ward et al., 2008; Dalsgaard et al., 2012; Babbin et al., 2014). Thus O2 concentration or electron donor availability could also control the N2O budget. N2O concentrations and net N2O production rates (N2O production minus N2O consumption) were found to peak at the oxic-suboxic interface in OMZs, due to excess production from nitrification and incomplete denitrification (Nicholls et al., 2007; Ji et al., 2015; Trimmer et al., 2016). However, while multiple processes can produce N2O, reduction by N2O consuming bacteria is the only known biological N2O sink.
N2O consumption is the final step of denitrification, and is the least O2 tolerant step (Bonin et al., 1989; Körner and Zumft, 1989). N2O consumption rates have been measured in ODZs at depths where O2 concentration ranged from very low to below the detection limit (Wyman et al., 2013; Babbin et al., 2015). N2O consumption by denitrification and genes involved in N2O reduction have also been detected in oxygenated seawater (Farías et al., 2009; Wyman et al., 2013). Characterizing the distribution and environmental regulation of this step is necessary for a complete quantification of the oceanic N2O budget and will improve our ability to predict oceanic N2O emissions under global climate change.
N2O consumption is catalyzed by the enzyme nitrous oxide reductase, encoded by the nosZ gene. A recent study of nosZ genes found a lower diversity of nosZ genes in ODZ waters than in the upper oxycline of the OMZ (Castro-González et al., 2015). The distribution of nosZ genes was related to O2 concentration, which suggested that the quantity and composition of nosZ genes and the diversity of denitrifying bacteria might influence the microbial potential for N2O consumption.
We aimed to determine the distribution and community composition of total and transcriptionally active (abbreviated as ‘active’ hereafter) nosZ assemblages based on the presence (DNA) and expression (RNA) of nosZ genes in the OMZ of the Eastern Tropical South Pacific (ETSP), one of the three major OMZs in the world ocean. Three hypotheses were tested in the study: (1) the community compositions of total and active nosZ assemblages differ between coastal and off-shore stations because the quantity and quality of the nutrients at the two stations differ due to different contributions from land and sediment; (2) quantities and composition of total and active nosZ assemblages are related to O2 concentration because N2O consumption is the least O2 tolerant step in conventional denitrification; and (3) the distribution of the active nosZ assemblage is more related to N2O concentration than that of total nosZ assemblage because the former indicates live and active organisms.
Materials and Methods
Experimental Sites and Sampling
Samples were collected on the R/V Nathaniel B. Palmer during June to July 2013 (cruise NBP 1305) in the OMZ of the ETSP at the off-shore station (BB1; 14.0°S, 81.2°W) and the coastal station (BB2; 20.50°S, 70.70°W) (Supplementary Figure 1). Particulate material was collected in Niskin bottles mounted on the standard conductivity-temperature-depth (CTD) rosette system (Seabird Electronics, Seattle, WA, United States) at four depths at each station (BB1: 60, 130, 300, and 1000 m; BB2: 60, 115, 300, and 1000 m) and concentrated by filtration (up to 4 L) through Sterivex filters (0.22 μm). Filters were flash frozen in liquid nitrogen onboard and stored at -80°C until DNA and RNA extraction was performed.
Temperature, salinity, sigma theta, bottom depth and pressure at each station were measured on the SBE 911+ CTD system. Fluorescence, representing chlorophyll a, was measured using a single channel fluorometer (Wet labs, Philomath, OR, United States) mounted on the CTD. Oxygen distributions were determined using the STOX sensor (detection limit = 10 nM) mounted on the CTD rosette (Revsbech et al., 2009). Ammonium, nitrite and nitrate concentrations were measured using standard colorimetric protocols (UNESCO, 1994). N2O concentration was determined using mass spectrometry (Ji et al., 2015). N∗ is the deviation of measured dissolved inorganic nitrogen (DIN = nitrate + nitrite + ammonia) from predicted DIN by Redfield ratio and the world-ocean nitrogen to phosphate regression relationship (Deutsch et al., 2001). Environmental data were reported by Ji et al. (2015) and are provided in Supplementary Table 1.
DNA and RNA Extractions
Both DNA and RNA were extracted from eight Sterivex filters using the plant tissue protocol of the All Prep DNA/RNA Mini Kit (50) using a QIAcube (Qiagen). Reverse transcription from RNA to cDNA was performed using SuperScript® III First-Strand Synthesis System for RT-PCR (InvitrogenTM by Life TechnologiesTM). Excess RNA was removed by RNase at the end of the synthesis.
Quantitative PCR Assays
The abundance of total and active nosZ assemblages were estimated by quantitative PCR (qPCR) using SYBR® Green based assays using protocols described previously (Jayakumar et al., 2013). Primers nosZ1F and nosZ1R (Henry et al., 2006) were used to amplify a 259-bp conserved fragment of the nosZ gene. Known quantities (∼20–25 ng) of DNA and cDNA samples were assayed along with a minimum of five serial dilutions of plasmids containing nosZ gene, no template controls and no primer controls, all in triplicate on the same plate. To maintain continuity and consistency among qPCR assays, a subset of samples from the first qPCR assay was run with subsequent assays and fresh standard dilutions were prepared for each assay. DNA, cDNA and the concentrated standards were quantified prior to every assay using PicoGreen fluorescence (Molecular Probes, Eugene, OR, United States) calibrated with several dilutions of phage lambda standards, to account for DNA loss due to freeze thaw cycles. qPCR assays were run on a Stratagene MX3000P (Agilent Technologies, La Jolla, CA, United States). Automatic analysis settings were used to determine the threshold cycle (Ct) values. The copy numbers (number of copies of the gene sequence detected in the sample) were calculated according to: Copy number = (ng ∗ number/mole)/(bp ∗ ng/g ∗ g/mole of bp) and then converted to copy number per ml seawater filtered, assuming 100% extraction efficiency.
Microarray Experiments
DNA and cDNA qPCR products were used as targets for microarray experiments to characterize the community composition of total and active nosZ assemblages, respectively. Triplicate qPCR products from each depth were pooled. nosZ gene targets were purified and extracted from agarose gels using the QIAquick gel extraction kit (Qiagen). Purified DNA qPCR products from eight depths and cDNA qPCR products from seven depths were used to prepare targets for microarray analysis.
Microarray targets were prepared from the qPCR products following the protocol of Ward and Bouskill (2011). Briefly, dUaa was incorporated into purified DNA and cDNA during linear amplification using the BioPrime kit (InvitrogenTM). The dUaa-Klenow product was labeled with Cy3 (dissolved in dimethyl sulfoxide), purified using QIAquick columns (Qiagen) and quantified by Nanodrop 2000 (Thermo Scientific). Duplicate Cy3 products for each sample were hybridized at 65°C overnight (16 h) onto replicate microarrays under ozone free conditions. Hybridized microarrays were washed and scanned with an Axon 4300 laser scanner.
nosZ Microarray
The microarray (BC016) contains 114 nosZ archetype probes. Each probe is a 90-bp sequence comprised of a 70-bp nosZ gene fragment and a 20-bp control region. Each archetype probe represents, and hybridizes with, all nosZ sequences with >85% identity, based on published sequences available in 2013. There are 71 NosZ archetypes, which represent typical or Clade I nosZ genes, and 43 WNZ archetypes, which represent the atypical or Clade II nosZ genes. The development of the microarray is described in Jayakumar et al. (in preparation) and the sequences are shown in Supplementary Table 2.
Data Analyses
Fluorescence signal intensities for nosZ probes hybridized to the microarrays were obtained using GenePix Pro 7 software. The fluorescence ratio (FR) of each feature is defined as the ratio Cy3/Cy5 (70-mer probe/20-mer standard for each feature). The FR for each nosZ archetype was calculated as the average of probe signal intensities for duplicate features on the same microarray. Normalized fluorescence ratio (FRn) was calculated by dividing the FR of each nosZ probe by the maximum nosZ FR on the same microarray. FRn is the proxy of the relative abundance of each archetype and was used for further analyses.
Detrended correspondence analysis (DCA) was performed to analyze the overall microbial community composition. A dissimilarity test was performed using Permutational Multivariate Analysis of Variance (adonis). α-diversities (Shannon diversity indices) of total and active nosZ assemblages were calculated. β-diversities (Bray–Curtis dissimilarities) of total and active nosZ assemblages between different sites (i.e., depths) were calculated to perform a Mantel test. The Mantel test was used to determine significant environmental variables correlated with microbial community composition. These analyses were carried out using the vegan package in R (version 3.3.1). A maximum likelihood phylogenetic tree was built from aligned archetype sequences with MEGA 7 software. FRn values for each archetype at different depths from both stations were visualized on the phylogenetic tree by iTOL1. The copy number of nosZ genes at each depth is given as mean (± standard error) of the qPCR triplicates.
Results
Abundance and Depth Distribution of Total and Active nosZ Assemblages
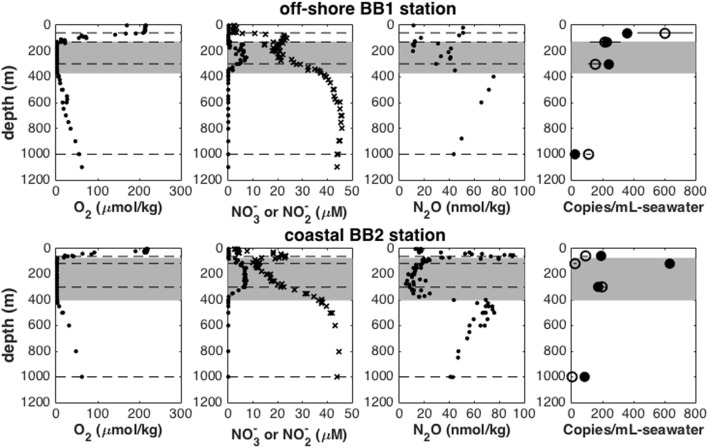
At stations BB1 and BB2, the continuously undetectable O2 concentration, the local nitrite maximum and the nitrate deficit at intermediate depths (130–370 m at BB1; 75–400 m at BB2) all indicated the presence of ODZs (gray areas in Figure 1). Sampling depths were chosen to represent water column features defined by oxygen concentration, as measured with the in situ STOX sensor: oxygenated surface water, upper oxycline [characterized by sharp O2 concentration gradient ranging from saturation to below detection limit (<10 nM)], top of the ODZ (O2 concentration <10 nM), core of the ODZ (O2 concentration <10 nM) and lower oxycline (O2 concentration >10 nM). The abundance of the total nosZ genes ranged from 24.1 (±1.4) copies mL-1 in a sample from the lower oxycline to 636.4 (±28.3) copies mL-1 in a sample from the ODZ (Figure 1). As for the active nosZ assemblage, the lowest abundance of active nosZ genes was 5.1 (±0.5) copies mL-1 in a sample from the lower oxycline and the highest abundance was 604.6 (±103.7) copies mL-1 in a sample from the surface water.
FIGURE 1.
O2, NO3- (x), NO2-, N2O and copy number of total (DNA, filled circles) and active (RNA, open circles) profiles at off-shore station BB1 and coastal station BB2. Dashed lines indicate depths where samples were collected. Gray areas indicate ODZs.
The abundance of total and active nosZ assemblages showed different distribution patterns at the two stations (Figure 1). At station BB1, the abundance of both total and active nosZ genes was highest in a sample from the surface water, and decreased with depth. At station BB2, the abundance of both total and active nosZ genes peaked in samples from the ODZ and was lowest in samples from the lower oxycline. The active nosZ genes were most abundant in the sample from 300 m. However, the total nosZ genes were most abundant in the sample from 115 m, where the abundance of the active nosZ genes was only 3% of the total.
Diversity and Dominant Archetypes of Total and Active nosZ Assemblages
The distribution of FRn of the total or active archetypes was similar across all depths within the same station (Figure 2). The average α-diversity was not significantly different (student’s t-test, P = 0.102) between the total assemblages (3.21) and the active assemblages (2.60) (Table 1). The least diverse total assemblage was from the lower oxycline (1000 m of station BB2), but the two least diverse active assemblages were from the ODZs (130 m of BB1 and 300 m of BB2).
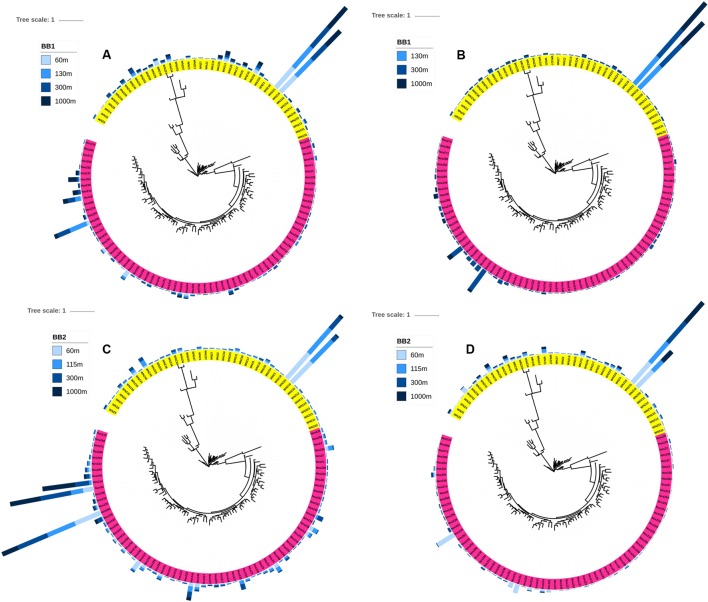
FIGURE 2.
Maximum likelihood trees for (A,C) total and (B,D) active nosZ archetypes at off-shore station BB1 (A,B) and coastal station BB2 (C,D). Pink = typical nosZ clade, yellow = atypical nosZ clade. Bars in outer cycles stand for mean FRn of duplicates of each archetype.
Table 1.
α-diversities of total (DNA) and active (RNA) nosZ genes at off-shore station BB1 and coastal station BB2.
| Depth(m) | DNA | RNA | Depth(m) | DNA | RNA | ||
|---|---|---|---|---|---|---|---|
| Off-shore station BB1 | 60 | 3.18 | Coastal station BB2 | 60 | 3.33 | 3.39 | |
| 130 | 3.09 | 1.29 | 115 | 3.56 | 2.75 | ||
| 300 | 3.57 | 3.37 | 300 | 3.57 | 1.63 | ||
| 1000 | 3.29 | 2.56 | 1000 | 2.10 | 3.19 | ||
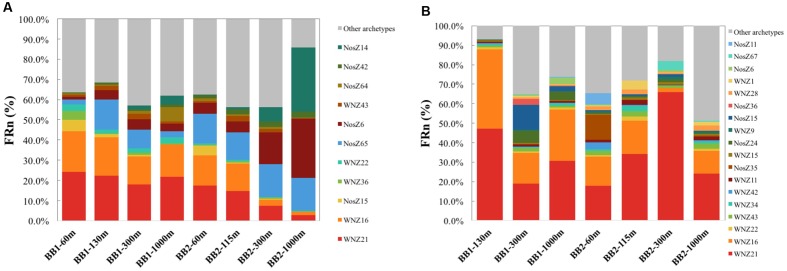
The FRn distribution of nosZ archetypes showed that a very limited number of archetypes dominated the total or the active nosZ assemblages (Figure 2). Dominant archetypes were affiliated with bacteria from various environments, including salt marsh, soil, marine sediment, marine hot spring and activated sludge of a wastewater treatment plant (Supplementary Tables 3, 4). The FRn of the top five most abundant archetypes accounted for 48.9 to 83.3% of the total nosZ hybridization signal (Figure 3A and Supplementary Table 3). Notably, the highest percentage (83.3%) was from the sample from the lower oxycline (1000 m) at station BB2 and the most abundant archetype (NosZ42, an uncultured clone of nosZ gene derived from salt marsh sediments; Kearns et al., 2015) accounted for 31.6% of total FRn. However, this archetype was not among the top five archetypes of the active nosZ assemblage in the same sample (Figure 3B and Supplementary Table 4). The two most dominant typical nosZ archetypes in the total assemblage were NosZ6, derived from an uncultured clone from salt marsh sediments (Kearns et al., 2015), which is closely related to Marinobacter, and NosZ65, derived from Marinobacter sp. BSs20148 from marine sediment (Song et al., 2013) (Figure 3A and Supplementary Table 3). In contrast, NosZ6 and NosZ65 were not dominant in the active assemblage (Figure 3B and Supplementary Table 4).
FIGURE 3.
(A) total (DNA) and (B) active (RNA) nosZ archetypes whose FRn signals are among the top five highest in any of the depths at off-shore station BB1 and coastal station BB2.
The sample from the ODZ (130 m) at station BB1 illustrates the contrasts observed between total and active nosZ assemblages. The FRns of the top three dominant archetypes (WNZ21, WNZ16 and NosZ65) in the total nosZ assemblage were comparable to each other, and they constituted 56.1% of the total community (Supplementary Table 3). However, NosZ65 was nearly undetectable in the active assemblage and the top two dominant archetypes (WNZ21 and WNZ16) accounted for 87.8% of the active assemblage in the same sample (Supplementary Table 4). The total nosZ assemblage was much more diverse than the active assemblage at this depth (Table 1). The representative sequences of WNZ21 and WNZ16 archetypes were derived from nosZ gene sequences of Anaeromyxobacter dehalogenans strain DCP18 (Chee-Sanford et al., unpublished) and an uncultured bacterium clone obtained from agricultural soils (Sanford et al., 2012), respectively. WNZ21 and WNZ16 archetypes were not only dominant in the active assemblage in one sample from the ODZ, but were among the top five abundant archetypes of both total and active assemblages in almost all samples (Figure 3).
Community Composition of Total and Active nosZ Assemblages
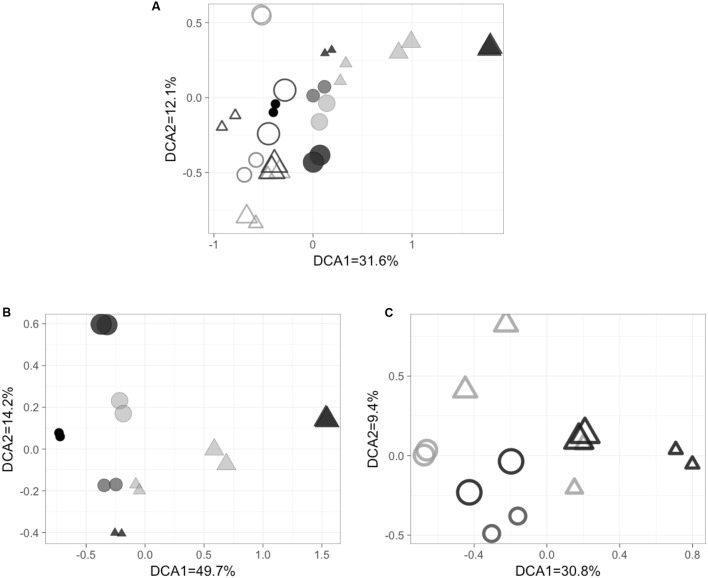
Functional gene microarrays were used to describe the community composition of nosZ assemblages. FRn values from duplicate microarrays replicated well (r2 = 0.802–0.997) (Supplementary Figure 2) and each pair of duplicates clustered together in the DCA plots (Figure 4).
FIGURE 4.
Detrended correspondence analysis (DCA) results of (A) total and active nosZ genes, (B) the total genes only and (C) the active genes only. Filled symbol stands for total nosZ genes, open symbol stands for active nosZ genes. Circles = off-shore station BB1 and triangles = coastal station BB2. The symbol size represents depths (the larger, the deeper). Symbol color represents O2 concentration (the darker, the higher). Symbols with the same shape, color and size were duplicates. Three pairs of symbols in lightest gray in (B) indicates samples from the ODZ with O2 concentration under detection limit.
The two-dimensional DCA model including both DNA and RNA microarray results explained 43.7% of the community composition of nosZ assemblages with 31.6% explained by the first axis and 12.1% explained by the second axis (Figure 4A). The clearest pattern was the clear separation of total (filled symbols) and active (open symbols) nosZ assemblages, indicating that they were different from each other. The significance (P < 0.001) of the difference between total and active assemblages was confirmed by the dissimilarity test (Table 2). Therefore, the community composition of total and active nosZ assemblages was further analyzed by two DCA models separately to better examine other patterns.
Table 2.
P-values of dissimilarity test (adonis).
| DNA and RNA samples | DNA samples only | RNA samples only | |
|---|---|---|---|
| DNA versus RNA | <0.001 | — | — |
| Site dependence | 0.034 | <0.001 | 0.039 |
The DCA model of DNA microarray results explained 63.9% of the composition of the total nosZ assemblage (Figure 4B). The total nosZ community composition revealed site difference and O2 dependence. Samples from the same station clustered together in the DCA model, indicating community composition was different between the off-shore station (BB1) and the coastal station (BB2). The site difference was statistically significant (P < 0.001) based on the dissimilarity test (Table 2). Besides the geographical pattern, composition of the total nosZ assemblage was also affected by O2 concentration (Figure 4B). Samples from the ODZs clustered together, while samples with higher O2 concentrations were distinct from the ODZ samples and different from each other. O2 concentration of the seawater might not be the most important driver of microbial community composition, however, since the O2 pattern was not captured by either axis of the DCA model.
The DCA model of RNA microarray results explained 40.2% of the community composition of the active nosZ assemblage (Figure 4C). Significant site difference (P = 0.039) of the active community composition was also revealed by the DCA model (Figure 4C) and the dissimilarity test (Table 2). However, the clustering based on O2 concentration that was observed in the total nosZ assemblage was not observed for the active nosZ assemblage.
Environmental Variables Correlated with the Community Composition
The composition of total and active nosZ assemblages was correlated with different environmental variables based on a Mantel test (Table 3). Relative depth, nitrate concentration, temperature, density (sigma theta) and pressure were significantly related to the β-diversity of the total nosZ assemblage. However, the β-diversity of the active nosZ assemblage was significantly related to N2O concentration, nitrite concentration and fluorescence. Bottom depth, which was dramatically different between two stations and is a proxy for important ecological differences between the two sites, was a significant factor for both total and active nosZ assemblages.
Table 3.
Mantel tests between total (DNA) or active (RNA) nosZ genes and environmental factors.
| Total nosZ | Active nosZ | |||
|---|---|---|---|---|
| r | P | r | P | |
| Oxygen | 0.169 | 0.190 | -0.046 | 0.636 |
| Relative Depth1 | 0.848 | 0.001 | -0.170 | 0.886 |
| Nitrate | 0.355 | 0.009 | -0.149 | 0.913 |
| Nitrite | 0.090 | 0.178 | 0.300 | 0.019 |
| Nitrous Oxide | -0.089 | 0.684 | 0.415 | 0.014 |
| Temperature | 0.483 | 0.002 | -0.153 | 0.937 |
| Ammonium | 0.155 | 0.234 | -0.008 | 0.458 |
| Salinity | 0.258 | 0.069 | -0.165 | 0.941 |
| Sigma Theta2 | 0.439 | 0.007 | -0.088 | 0.774 |
| Bottom Depth | 0.320 | 0.001 | 0.255 | 0.012 |
| Pressure | 0.492 | 0.003 | -0.168 | 0.949 |
| Fluorescence | 0.059 | 0.290 | 0.320 | 0.015 |
1Relative depth was calculated by dividing measured depth by the bottom depth of each station. 2Sigma Theta was density calculated with in situ salinity and potential temperature at zero pressure. Bolded P values indicate significant correlation (P < 0.05).
Discussion
Abundance and Diversity of nosZ Assemblages
Oxygen minimum zones are sites of high N2O flux to the atmosphere (Law and Owens, 1990; Arévalo-Martínez et al., 2015). N2O consuming organisms are the only biological sink for N2O. Hence their abundance and community composition in the OMZ may be important in understanding the N2O flux. The abundance of total and active N2O consuming bacteria in the OMZ of the ETSP was estimated by measuring nosZ gene copy number (Figure 1). The relationship between abundance of N2O consuming bacteria and depth in this study differed from that of denitrifiers indicated by nirS gene copy number at the same stations (Ji et al., 2015): the abundances of the total and active N2O consuming bacteria in the surface water were similar to or higher than those in the ODZs (Figure 1), but the abundance of denitrifiers in the surface water was two orders of magnitude smaller than that in the ODZs. This difference suggests that the two genes represent functionally different groups.
nirS and nosZ also differed in their absolute abundance. The highest abundance of N2O consuming bacteria was only a few hundred copies mL-1, which was three orders of magnitude smaller than the highest abundance of denitrifiers measured at the same stations (Ji et al., 2015). It is assumed that both genes are present in the genome as single copy genes, although there are exceptions for nosZ (Sanford et al., 2012). One possible explanation for the differences in both distribution and abundance of the nosZ assemblage and the nirS assemblage is that not all N2O consuming bacteria contain the complete denitrification gene sequence (Sanford et al., 2012). The atypical nosZ genes are associated with bacteria that lack the other steps in the conventional denitrification pathway. Notably, bacteria with only nosZ genes but no other denitrification genes were overrepresented in the genomes of marine bacteria compared to other ecosystems (Graf et al., 2014). nirS, however, was preferentially associated with bacteria that contained a complete denitrification pathway (Graf et al., 2014).
Another contributing factor may be the specificity or bias of the PCR primers. The nosZ primers used in this study were optimized to amplify all known nosZ sequences as of 2006, and should therefore represent the large database of both terrestrial and marine sequences available at the time. However, it is clear that they might underrepresent the atypical N2O consuming bacteria, which were not known at the time. The nirS primers used in the previous analysis of these samples (Ji et al., 2015) are potentially biased toward marine sequences (Braker et al., 1998) and may underrepresent more diverse sequences now available from other environments. One way to improve the nosZ coverage is to use multiple primer sets targeting different groups of nosZ archetypes.
The N2O consuming bacteria are a small component of the total microbial assemblage, but are still quite diverse (Jones et al., 2013), so they are difficult to characterize by pure culture or metagenomics. The microarray, which was designed specifically to target N2O consuming bacteria using more than 100 nosZ gene probes (Figure 2), may be a better tool to capture these underrepresented organisms without cultivation or detection of rare sequences in complex metagenomic datasets. The high reproducibility of microarrays reported previously (Bulow et al., 2008) was confirmed in this study in that duplicates for each sample run on two different microarrays clustered together in DCA (Figure 4) and had high r2 of linear regressions (Supplementary Figure 2).
Based on the FRn values of diverse nosZ archetypes determined by microarray hybridization (Figure 2), a very limited number of archetypes dominated the total or the active assemblages. Moreover, the top five active archetypes accounted for larger percentage of the assemblage than that of the total archetypes (Figure 3), consistent with the less diverse active assemblage compared to the total assemblage (Table 1). These findings imply that although the total nosZ assemblage is very diverse, a few nosZ archetypes might be the major contributors of N2O consumption at the study sites. The relative abundance of active archetypes, however, might uncouple that of enzymes of different archetypes and/or the contribution of different archetypes to the N2O consumption rate due to different stabilities of enzymes from different archetypes.
Total and Active Community Compositions and Their Controlling Environmental Variables
The active nosZ assemblage was different from the total assemblage in both abundance profiles and community composition as detected by qPCR and nosZ microarray hybridization analysis, respectively. The highest abundance of the total N2O consuming bacteria indicated by nosZ DNA copy number was 636.4 (±28.3) copies mL-1 in the sample from the ODZ (115 m) at station BB2, but the abundance of the active bacteria in the same sample was only 21.1 (±5.2) copies mL-1 (Figure 1). In the sample from oxygenated surface seawater (60 m) at station BB1, the abundance of the active N2O consuming bacteria was 604.6 (±103.7) copies mL-1, but the abundance of the total bacteria was only 357.2 (±12.5) copies mL-1 (Figure 1).
Significant differences between the community composition of total and active N2O consuming bacteria were indicated by the results of DCA and dissimilarity test (Figure 4A; Table 2). The different community composition was attributed to the differences in the distribution of FRn of more than 100 archetypes, especially the dominant ones. NosZ6 and NosZ65 were dominant in the total nosZ assemblage but were minor components in the active assemblage. NosZ42 was the most abundant archetype in the total nosZ assemblage from the lower oxycline (1000 m) at station BB2, but was not among the top five archetypes of the active assemblage. These differences between active and total nosZ communities are consistent with observations from soil and salt marsh sediments in which the active component of the microbial assemblage was apparently more responsive to environmental conditions (Barnard et al., 2013; Kearns et al., 2016).
Despite the differences between the total and the active N2O consuming assemblages, their distributions both depended on geographic location. The composition of total and active nosZ assemblages was significantly different between the coastal and the off-shore stations as indicated by the results of DCA and dissimilarity test. Since the two stations shared similar dominant nosZ archetypes, the geographical divergence mainly reflected differences among the large number of rare archetypes between the two stations. The more negative N∗ at the coastal station BB2 (Supplementary Table 1) indicates more intense nitrogen-loss fueled by more organic matter. The different amount of organic matter, which supports the metabolism of heterotrophic nosZ bacteria, might partially contribute to the geographical differences of nosZ assemblages. Geographical differences might also result from different nutrient sources at the two stations, since their distance to the sediment (bottom depth) and to the shore were dramatically different. The dependence of geographic location was also observed for ammonia oxidizing archaea (Peng et al., 2013) and for nirS denitrifiers (Jayakumar et al., 2013) in the ETSP and Arabian Sea OMZs.
In addition to geographical patterns, the total and active assemblages were correlated with different environmental variables (Table 3). Depth and environmental parameters that co-varied with depth (including temperature, density and pressure) were major drivers of the β-diversity of the total N2O consuming assemblages, implying different organisms coexist in the water column by occupying different ecological niches. However, N2O concentration difference was a major driver of the β-diversity of the active nosZ assemblages, implying the active nosZ community was a better indicator for N2O consumption potential.
nosZ Assemblage in Oxygenated Seawater
The role of the nosZ assemblage in oxygenated seawater has been ignored because N2O consumption is considered the least oxygen tolerant anaerobic step in the conventional denitrification pathway (Zumft, 1997). However, nosZ genes were abundant in oxygenated surface water in the Southern Indian Ocean (Raes et al., 2016) and nosZ mRNAs were detected in the oxic regions in the Arabian Sea (Wyman et al., 2013). Our study confirmed that a nosZ assemblage was not only present but also active in oxygenated surface water in the OMZ of the ETSP. In particular, atypical nosZ archetypes, usually associated with N2O consuming bacteria lacking a complete denitrification pathway, were present and active in surface waters. In addition, the most abundant archetypes of total and active nosZ communities were both atypical nosZ archetypes (WNZ21 and WNZ16), implying the significant contribution of atypical archetypes to the nosZ communities and the necessity to consider atypical archetypes while analyzing the potential of N2O consumption.
N2O reductase enzymes from denitrifiers had very low O2 tolerance (Bonin et al., 1989; Körner and Zumft, 1989); on the contrary, nosZ assemblages were detected in the oxygenated surface waters and O2 concentration was not significantly correlated with the active microbial community, as indicated by DCA and Mantel test. The survival of N2O consuming bacteria in oxic layers and their O2-independence might be attributed to anoxic micro-environments created by phytoplankton microaggregates or particles. Free-living and particle-associated microbes from the same seawater sample can have different community compositions (Delong et al., 1993). More specifically, a recent study in the OMZ of the ETSP showed that nosZ mRNAs were 28-fold more abundant on particles (>1.6 μm) compared to free-living microbes (0.2–1.6 μm) (Ganesh et al., 2015). Additionally, nosZ mRNA co-occurred with the cyanobacterium Trichodesmium in oxic water in the Arabian Sea (Wyman et al., 2013). Consistently, fluorescence, a proxy for chlorophyll a, was significantly correlated with the β-diversity of the active nosZ assemblages in this study (Table 3).
The active nosZ community in oxygenated surface water might capture N2O produced in deeper seawater and thus reduce the flux into the atmosphere. Thus, evaluating the nosZ community is essential to the prediction of the oceanic N2O emissions. Moreover, the oceanic N2O emissions represent net fluxes, which are controlled by both N2O production and N2O consumption. Some N2O flux models (Suntharalingam and Sarmiento, 2000; Martinez-Rey et al., 2015; Trimmer et al., 2016) do not parameterize N2O consumption, and other models either consider N2O consumption only in suboxic or anoxic waters (Cornejo and Farías, 2012; Babbin et al., 2015) or estimate N2O consumption assuming it is constrained by O2 concentration (Zamora et al., 2012). Failing to consider the O2-independent, non-denitrification N2O consumption potential in these O2 forcing models might contribute to their uncertainty and the variation among different models. Additionally, the N2O consuming organisms have not been fully investigated. Besides denitrifiers and atypical N2O consuming bacteria analyzed in this study, other organisms (Trichodesmium and Crocosphaera) also exhibited N2O consuming capacity under laboratory conditions (Farías et al., 2013), suggesting that their significance in the environment warrants further investigation.
Conclusion
The results described above support two (1 and 3) of the initial hypotheses. (1) Compositions of total and active nosZ assemblages were different between the coastal station and the off-shore station mainly due to their dramatic differences of distance to the sediment and to the shore, which are very likely to result in different environmental conditions (i.e., different phytoplankton assemblages, different nutrients and organic matter). (2) The abundances of total and active nosZ assemblages in oxygenated seawater were similar to or larger than those in the ODZs, implying the potential for N2O consumption even in oxygenated surface water. Atypical nosZ archetypes, which may lack a complete denitrification pathway, dominated both total and active nosZ assemblages. (3) The total and active nosZ assemblages were significantly different from each other. The community composition of the total nosZ assemblage showed O2 dependence and shifted along depth gradients and environmental gradients associated with depth, but fluorescence, N2O and nitrite concentration were significantly correlated with the composition of the transcriptionally active community. We conclude that the difference between active and total nosZ assemblages may be related to differential response to environmental conditions by different components of the diverse natural assemblage and that the presence of nosZ assemblage in surface waters should be investigated to determine their actual N2O reduction capabilities.
Author Contributions
XS and BW designed the experiments. AJ and BW collected samples. XS and AJ performed experiments. XS analyzed the data. XS and BW wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge all scientists and the crew of the R/V Nathaniel B. Palmer for assistance in sample collection.
Funding. This paper was supported by an NSF grant to BW and AJ (OCE-1029951).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01183/full#supplementary-material
References
- Arévalo-Martínez D. L., Kock A., Löscher C. R., Schmitz R. A., Bange H. W. (2015). Massive nitrous oxide emissions from the tropical South Pacific Ocean. Nat. Geosci. 8 530–533. 10.1038/ngeo2469 [DOI] [Google Scholar]
- Babbin A. R., Bianchi D., Jayakumar A., Ward B. B. (2015). Rapid nitrous oxide cycling in the suboxic ocean. Science 348 1127–1129. 10.1126/science.aaa8380 [DOI] [PubMed] [Google Scholar]
- Babbin A. R., Keil R. G., Devol A. H., Ward B. B. (2014). Oxygen control nitrogen loss in the ocean. Science 344 406–408. 10.1126/science.1248364 [DOI] [PubMed] [Google Scholar]
- Barnard R. L., Osborne C. A., Firestone M. K. (2013). Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7 2229–2241. 10.1038/ismej.2013.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin P., Gilewicz M., Bertrand J. C. (1989). Effects of oxygen on each step of denitrification on Pseudomonas nautica. Can. J. Microbiol. 35 1061–1064. 10.1139/m89-177 [DOI] [Google Scholar]
- Braker G., Fesefeldt A., Witzel K.-P. (1998). Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64 3769–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow S. E., Francis C. A., Jackson G. A., Ward B. B. (2008). Sediment denitrifier community composition and nirS gene expression investigated with functional gene microarrays. Environ. Microbiol. 10 3057–3069. 10.1111/j.1462-2920.2008.01765.x [DOI] [PubMed] [Google Scholar]
- Castro-González M., Ulloa O., Farías L. (2015). Structure of denitrifying communities reducing N2O at suboxic waters off northern Chile and Perú. Rev. biol. Mar. Ocean. 50 95–110. 10.4067/S0718-19572015000100008 [DOI] [Google Scholar]
- Codispoti L. A. (2010). Interesting times for marine N2O. Science 327 1339–1340. 10.1126/science.1184945 [DOI] [PubMed] [Google Scholar]
- Cornejo M., Farías L. (2012). Following the N2O consumption in the oxygen minimum zone of the eastern South Pacific. Biogeosciences 9 3205–3212. 10.5194/bg-9-3205-2012 [DOI] [Google Scholar]
- Dalsgaard T., Thamdrup B., Farías L., Peter Revsbech N. (2012). Anammox and denitrification in the oxygen minimum zone of the eastern South Pacific. Limnol. Oceanogr. 57 1331–1346. 10.4319/lo.2012.57.5.1331 [DOI] [Google Scholar]
- Delong E. F., Franks D. G., Alldredge A. L. (1993). Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38 924–934. 10.4319/lo.1993.38.5.0924 [DOI] [Google Scholar]
- Deutsch C., Key R. M., Sarmiento J. L., Ganachaud A. (2001). Denitrification and N2 fixation in the Pacific Ocean. Glob. Biogeochem. Cycles 15 483–506. 10.1029/2000GB001291 [DOI] [Google Scholar]
- Farías L., Castro-González M., Cornejo M., Charpentier J., Faúndez J., Boontanon N., et al. (2009). Denitrification and nitrous oxide cycling within the upper oxycline of the eastern tropical South Pacific oxygen minimum zone. Limnol. Oceanogr. 54 132–144. 10.4319/lo.2009.54.1.0132 [DOI] [Google Scholar]
- Farías L., Faúndez J., Fernández C., Cornejo M., Sanhueza S., Carrasco C. (2013). Biological N2O fixation in the Eastern South Pacific Ocean and marine cyanobacterial cultures. PLoS ONE 8:e63956 10.1371/journal.pone.0063956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh S., Bristow L. A., Larsen M., Sarode N., Thamdrup B., Stewart F. J. (2015). Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J. 9 2682–2696. 10.1038/ismej.2015.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf D. R. H., Jones C. M., Hallin S. (2014). Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE 9:e114118 10.1371/journal.pone.0114118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S., Bru D., Stres B., Hallet S., Philippot L., Burgundy I., et al. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72 5181–5189. 10.1128/AEM.00231-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2007). “Changes in atmospheric constituents and in radiative forcing,” in Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (Cambridge: Cambridge University Press; ). 10.1103/PhysRevB.77.220407 [DOI] [Google Scholar]
- Jayakumar A., Peng X., Ward B. B. (2013). Community composition of bacteria involved in fixed nitrogen loss in the water column of two major oxygen minimum zones in the ocean. Aquat. Microb. Ecol. 70 245–259. 10.3354/ame01654 [DOI] [Google Scholar]
- Ji Q., Babbin A., Jayakumar A., Oleynik S., Ward B. B. (2015). Nitrous oxide production by nitrification and denitrification in the Eastern Tropical South Pacific oxygen minimum zone. Geophys. Res. Lett. 42 755–764. 10.1002/2015GL065934 [DOI] [Google Scholar]
- Jones C. M., Graf D. R., Bru D., Philippot L., Hallin S. (2013). The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 7 417–426. 10.1038/ismej.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns P. J., Angell J. H., Feinman S. G., Bowen J. L. (2015). Long-term nutrient addition differentially alters community composition and diversity of genes that control nitrous oxide flux from salt marsh sediments. Estuar. Coast. Shelf Sci. 154 39–47. 10.1016/j.ecss.2014.12.014 [DOI] [Google Scholar]
- Kearns P. J., Angell J. H., Howard E. M., Deegan L. A., Stanley R. H. R., Bowen J. L., et al. (2016). Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nat. Commun. 7:12881 10.1038/ncomms12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner H., Zumft W. G. (1989). Expression of denitrification enzymes in response to the dissolved oxygen level and respiratory substrate in continuous culture of Pseudomonas stutzeri. Appl. Environ. Microbiol. 55 1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. S., Owens N. J. P. (1990). Significant flux of atmospheric nitrous oxide from the northwest Indian Ocean. Nature 346 826–828. 10.1038/346826a0 [DOI] [Google Scholar]
- Martinez-Rey J., Bopp L., Gehlen M., Tagliabue A., Gruber N. (2015). Projections of oceanic N2O emissions in the 21st century using the IPSL Earth system model. Biogeosciences 12 4133–4148. 10.5194/bg-12-4133-2015 [DOI] [Google Scholar]
- Naqvi S. W., Jayakumar D. A., Narvekar P. V., Naik H., Sarma V. V., D’Souza W., et al. (2000). Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408 346–349. 10.1038/35042551 [DOI] [PubMed] [Google Scholar]
- Nicholls J. C., Davies C. A., Trimmer M. (2007). High-resolution profiles and nitrogen isotope tracing reveal a dominant source of nitrous oxide and multiple pathways of nitrogen gas formation in the central Arabian Sea. Limnol. Oceanogr. 52 156–168. 10.4319/lo.2007.52.1.0156 [DOI] [Google Scholar]
- Peng X., Jayakumar A., Ward B. B. (2013). Community composition of ammonia-oxidizing archaea from surface and anoxic depths of oceanic oxygen minimum zones. Front. Microbiol. 4:177 10.3389/fmicb.2013.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes E. J., Bodrossy L., Van De Kamp J., Holmes B., Hardman-Mountford N., Thompson P. A., et al. (2016). Reduction of the powerful greenhouse gas N2O in the south-eastern Indian Ocean. PLoS ONE 11:e0145996 10.1371/journal.pone.0145996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravishankara A. R., Daniel J. S., Portmann R. W. (2009). Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326 123–125. 10.1126/science.1176985 [DOI] [PubMed] [Google Scholar]
- Revsbech N. P., Larsen L. H., Gundersen J., Dalsgaard T., Ulloa O., Thamdrup B. (2009). Determination of ultra-low oxygen concentrations in oxygen minimum zones by the STOX sensor. Limnol. Oceanogr. Methods 7 371–381. 10.4319/lom.2009.7.371 [DOI] [Google Scholar]
- Sanford R. A., Wagner D. D., Wu Q., Chee-Sanford J. C., Thomas S. H., Cruz-Garcia C., et al. (2012). Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc. Natl. Acad. Sci. U.S.A. 109 19709–19714. 10.1073/pnas.1211238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Ren L., Li X., Yu D., Yu Y., Wang X., et al. (2013). Complete genome sequence of Marinobacter sp. BSs20148. Genome Announc. 1:e00236–13 10.1128/genomeA.00236-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam P., Sarmiento J. L. (2000). Factors governing the oceanic nitrous oxide distribution: simulations with an ocean genera circulation model. Glob. Biogeochem. Cycles 14 429–454. 10.1029/1999GB900032 [DOI] [Google Scholar]
- Trimmer M., Chronopoulou P.-M., Maanoja S. T., Upstill-Goddard R. C., Kitidis V., Purdy K. J. (2016). Nitrous oxide as a function of oxygen and archaeal gene abundance in the North Pacific. Nat. Commun. 7:13451 10.1038/ncomms13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNESCO (1994). Protocols for the Joint Global Ocean Flux Study (JGOFS) Core Measurements, ed. Intergovernmental Oceanographic Commission, Educational United Nations, Scientific and Cultural Organization. Paris: UNESCO. [Google Scholar]
- Ward B. B., Bouskill N. J. (2011). “The utility of functional gene arrays for assessing community composition, relative abundance, and distribution of ammonia- oxidizing bacteria and archaea,” in Methods in Enzymology, 1st Edn Vol. 496 eds Klotz M. G., Stein L. Y. (Burlington, NJ: Elsevier Inc.), 373–396. 10.1016/B978-0-12-386489-5.00015-4 [DOI] [PubMed] [Google Scholar]
- Ward B. B., Tuit C. B., Jayakumar A., Rich J. J., Moffett J., Naqvi S. W. A. (2008). Organic carbon, and not copper, controls denitrification in oxygen minimum zones of the ocean. Deep. Res. I Oceanogr. Res. Pap. 55 1672–1683. 10.1016/j.dsr.2008.07.005 [DOI] [Google Scholar]
- Wyman M., Hodgson S., Bird C. (2013). Denitrifying alphaproteobacteria from the Arabian Sea that express nosZ, the gene encoding nitrous oxide reductase, in oxic and suboxic waters. Appl. Environ. Microbiol. 79 2670–2681. 10.1128/AEM.03705-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora L. M., Oschlies A., Bange H. W., Huebert K. B., Craig J. D., Kock A., et al. (2012). Nitrous oxide dynamics in low oxygen regions of the Pacific: insights from the MEMENTO database. Biogeosciences 9 5007–5022. 10.5194/bg-9-5007-2012 [DOI] [Google Scholar]
- Zumft W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.