Abstract
Adverse environmental conditions have a detrimental impact on crop growth and development, and cause protein denaturation or misfolding. The binding protein (BiP) plays an important protective role by alleviating endoplasmic reticulum (ER) stress induced by misfolded proteins. In this study, we characterized three BiP genes (CaBiP1, CaBiP2, and CaBiP3) in pepper, an economically important vegetable and spice species. The role of CaBiP1 in plant tolerance to ER stress and adverse environmental conditions (including heat, salinity, osmotic and drought stress) were investigated. All the expected functional and signaling domains were detected in three BiP proteins, but the motifs and exon-intron distribution differed slightly in CaBiP3. CaBiP1 and CaBiP2 were constitutively expressed in all the tested tissues under both normal and stressed conditions, whereas CaBiP3 was mainly expressed following stress. Silencing of CaBiP1 reduced pepper tolerance to ER stress and various environment stresses, and was accompanied by increased H2O2 accumulation, MDA content, relative electric leakage (REL), water loss rate, and a reduction in soluble protein content and relative water content (RWC) in the leaves. Conversely, overexpression of CaBiP1 in Arabidopsis enhanced tolerance to ER stress and multiple environment stresses, as demonstrated by an increase in germination rate, root length, survival rate, RWC, the unfolded protein response (UPR) pathway, and a decrease in water loss rate. Our results suggest that CaBiP1 may contribute to plant tolerance to abiotic stresses by reducing ROS accumulation, increasing the water-retention ability, and stimulating UPR pathways and expression of stress-related genes.
Keywords: BiP, pepper, Arabidopsis, ER stress, abiotic stresses, UPR pathway
Introduction
With advancing global warming, extreme weather events, especially high temperatures and droughts in arid and semiarid areas, pose an increasing threat to crop productivity. To develop crop varieties that better tolerate these adverse conditions, a greater understanding of the mechanisms involved in environment stresses is crucial (Thiry et al., 2016). In general, the growth and development of plants requires proteins to function in a normal manner, but the elaborate folding of proteins is easily disturbed by adverse environmental conditions, whereas it is well known that the accumulation of misfolded proteins is harmful to plant health (Howell, 2013).
The ER is the site of the protein secretory pathway in plant cells, and it is responsible for the folding and assembly of about one third of all cellular proteins (Deng et al., 2013). When misfolded proteins accumulate in the ER, the balance between folding pressure and folding capacity is broken, resulting ER stress (Wan and Jiang, 2016). During evolution, plants have developed a comprehensive mechanism to mitigate ER stress induced by adverse environmental conditions. One important strategy is up-regulating the transcription of ER chaperones to enhance the capacity of the protein folding machinery (Fanata et al., 2013).
Binding protein is a member of the Hsp70 family that include an HDEL or KDEL ER retention factor motif at the C-terminus (Denecke et al., 1991). BiP is one of the most abundant chaperones in ER lumen, and it has an ATP-binding domain and a protein- binding domain at the N- and C-terminus, respectively, the latter of which allows it bind to the hydrophobic surfaces of nascent proteins to protect them from aggregation via an ATP-dependent mechanism (Howell, 2013).
Under normal growth conditions, BiP genes are highly expressed in plant tissues and during plant developmental events with high cellular secretory activity and/or a high proportions of rapidly dividing cells, such as male and female gametogenesis (Maruyama et al., 2014, 2015). When plants are subjected to the ER stress induced by adverse environmental factors or ER stressors, such as heat in Arabidopsis (Deng et al., 2011), rice (Jung et al., 2013), and soybean (Zhang et al., 2015), drought in wheat (Zhu et al., 2014), tunicamycin (TM) exposure in Arabidopsis (Noh et al., 2003), dithiothreitol (DTT) exposure in rice (Wakasa et al., 2012), BiP genes are up-regulated via the UPR (Wan and Jiang, 2016). Once misfolded proteins accumulate in the ER, the bZIP28 and IRE1 branches of UPR are activated, and the transcription factors of bZIP28 and bZIP60 are released and progressively relocate to the nucleus to up-regulate UPR-related genes including BiP (Howell, 2013). Therefore, the up-regulation of BiP gene is a marker of the UPR (Cheng et al., 2015).
Overexpression of BiP genes also enhances plant tolerance to environment stresses. Yang et al. (2016) found that overexpression of BiP genes and the exogenous chemical chaperones sodium 4-phenylbutyrate (PBA) alleviated the ER stress induced by DTT and high temperatures. Overexpression of soybean BiP in tobacco conferred tolerance to water deficit during plant growth by preventing endogenous oxidative stress (Alvim et al., 2001), and similar results were observed in soybean (Valente et al., 2009). Leborgne-Castel et al. (1999) also found that overexpressing BiP in transgenic plants mitigated ER stress and reduced the UPR. In addition, silencing of BiP genes in tomato compromised Ve1-mediated resistance to Verticillium dahlia (Liebrand et al., 2014), and plants overexpressing BiP line displayed hypersensitivity to Pseudomonas syringae pv tomato in soybean and tobacco (Carvalho et al., 2014b).
Unlike mammals and yeast that possess only one copy in their genomes, plant genomes contain multiple highly conserved BiP genes, with six in rice (Sarkar et al., 2013) and tobacco (Denecke et al., 1991), three in Arabidopsis (Noh et al., 2003) and wheat (Zhu et al., 2014), two in soybean (Zhang et al., 2015), and 22 BiP-like genes are present in maize (Li et al., 2012). However, different BiP members exhibit different expression patterns. For instance, while AtBiP-1 and AtBiP-2 in Arabidopsis show significant basal expression in unstressed cells, AtBiP3 is normally expressed at much lower levels but is highly induced under stress conditions (Noh et al., 2003). OsBiP1 is constitutively expressed in various tissues in rice, whereas OsBiP4 and OsBiP5 appear not to be expressed in any tissue under normal conditions, but they are highly up-regulated following exposure to DTT (Wakasa et al., 2012). Therefore, characterizing the expression patterns and functions of BiP genes in response to abiotic stresses will greatly contribute to our understanding of plant tolerance to adverse environmental conditions.
In our previous study, we identified three BiP genes, CaBiP1 (CaHsp70-8, CA01g00570), CaBiP2 (CaHsp70-7, CA03g20120), and CaBiP3 (CaHsp70-10, Capana08g001522), in the genome of pepper (Capsicum annuum L.), an economically important vegetable and spice crop (Guo et al., 2016). In the present study, we further analyzed the structure and expression pattern of these genes under normal and stressed conditions. Virus-induced gene silencing (VIGS) and overexpression analyses were performed to investigate the functions of CaBiP1 in response to multiple abiotic stresses (including heat, salinity, osmotic, and drought stress) in both pepper and Arabidopsis. Our results provide insight into the function of BiP in the plant response to environment stresses.
Materials and Methods
Plant Materials and Growth Conditions
The R9 thermotolerant pepper line (introduced from the World-Asia Vegetable Research and Development Center, PP0042-51) and the Arabidopsis ecotype Col-0 variety were used in this study. Pepper seedlings were grown under normal conditions (26°C/20°C day/night, 200 μmol⋅m-2⋅s-1 illumination intensity, thermo- and photoperiod of 16 h light /8 h dark cycle, and 70 % relative humidity) in a controlled climate chamber, and Arabidopsis seedlings were grown under 22°C/18°C day/night conditions.
Sequence Analysis of CaBiP Proteins
The amino acid sequences of CaBiP1, 2, and 3 were downloaded from the PGD1 (Qin et al., 2014) and PGP2 (Kim et al., 2014) pepper genome databases. Arabidopsis AtBiP amino acid sequences were obtained from Genbank3 (Noh et al., 2003) and rice OsBiP sequences were downloaded from Rice Genome Annotation Project4 (RGAP) (Sarkar et al., 2013).
Alignment of full-length BiP amino acid sequences from pepper, rice and Arabidopsis was performed using the online program of Clustal Omega5, and the phylogenetic tree was constructed using MEGA 6 with the neighbor-joining method, p-distance substitution model and 1000 bootstrap replicates (Tamura et al., 2013). Functional and signaling domains in CaBiPs were identified based on published Arabidopsis and rice literatures (Noh et al., 2003; Sarkar et al., 2013). The identification of conserved motifs in CaBiP proteins was carried out using the MEME program6 with the following parameters: normal motif discovery mode, maximum number of motifs = 9, a motif site distribution in which each gene has none or only one motif, and a motif width between four and 200 amino acids. Structural diagrams for exon-intron analysis were generated using the online program GSDS7.
Subcellular Localization of CaBiP1 Protein
The CaBiP-1 ORF without a termination codon was amplified using specific primer pair GFP-CaBiP1-F and GFP-CaBiP1-R (Supplementary Table S1) from a cDNA template isolated from R9 leaf material grown under normal growth conditions, and then the PCR product was cloned into the pBI221 expression vector containing green GFP. An empty vector without CaBiP-1 was used as the control. Particle bombardment was performed to introduce recombinant plasmid into onion epidermal cells. ER-Tracker Red (Beyotime, C1041, China), a specific fluorescent probe for ER, was used to highlight this cellular component. Details of the methods can be found in our previous study (Guo et al., 2014).
Virus-Induced Gene Silencing (VIGS) of CaBiP1
A 346 bp fragment of the CaBiP1 ORF was amplified by gene-specific primer pair TRV2-CaBiP1-F and TRV2-CaBiP1-R (Supplementary Table S1) from a cDNA template isolated from R9 leaf material grown under normal growth conditions, and the PCR product was cloned into the pMD19T vector (Takara, Dalian, China). After digestion with restriction enzymes of Xba I and Kpn I, the CaBiP1 fragment was cloned into the pTRV2 virus expression vector to generate the TRV2:CaBiP1 silencing construct. The empty TRV2:00 vector without CaBiP1 was used as the control, and TRV2:CaPDS (phytoene desaturase gene) was used as a marker for gene silencing. Agrobacterium tumefaciens strain GV3101 cells containing TRV2:CaBiP1, TRV2:00 or TRV2:CaPDS were separately injected into the leaves of the R9 thermotolerant pepper line as described by Wang et al. (2013). When the photo-bleaching phenotype was evident in pepper seedlings carrying TRV2:CaPDS, the silencing efficiency of pTRV2:CaBiP1 was assessed by qRT-PCR with the primer pair of qCaBiP1-F and qCaBiP1-R (Supplementary Table S1).
Generation of Arabidopsis Lines Overexpressing CaBiP1
The full-length CaBiP1 ORF was amplified from cDNA isolated from R9 leaf material grown under normal growth conditions with the gene-specific primer pair of CaBiP1-F and CaBiP1-R (Supplementary Table S1). The amplification product was inserted into the plant transformation binary vector pBI121 between the CaMV-35S promoter and the nos (nopaline synthase) terminator. The resultant pBI121 vector was transformed into Arabidopsis ecotype Col-0 using the floral dip method intermediated by Agrobacterium GV3101 (Clough and Bent, 1998). Transgenic plants were obtained by screening successive generations for kanamycin resistance, and T3 seeds were used for subsequent experiments.
Experimental Treatments and Samples Collection
For tissue-specific expression analysis of CaBiP genes, young leaves, flower buds, fruits (about 1 cm in length), stems and roots were collected from pepper plants grown under normal conditions. Seedlings at the six-leaf stage were used for abiotic stress treatments. For abiotic stress treatments involving abscisic acid (ABA), H2O2, DTT, the plants were sprayed with 0.1 mM ABA, 1 mM H2O2, or 15 mM DTT until leaves were thoroughly wetted, and leaves were collected at 0, 1, 3, 6, 12, and 24 h post treatment. For salt and osmotic stress experiments, the roots of the seedling were soaked in 200 mM NaCl and 200 g⋅L-1 PEG6000, respectively, and leaves and roots were sampled at 0, 1, 3, 6, 12, and 24 h post treatment. For heat treatment, pepper seedlings were incubated at 45°C, and leaves were collected at 0, 0.5, 1, 2, 4, and 6 h post treatment. All samples were immediately frozen in liquid nitrogen and kept at -80°C for RNA extraction.
Pepper seedlings of TRV2:CaBiP1 and TRV2:00 were used for abiotic stress treatments. For ER stress, pepper seedlings were sprayed with 30 mM DTT until leaves were thoroughly wetted and incubated for 24 h. For heat stress, pepper seedlings were exposed to 45°C for 24 h then allowed to recover for 5 days under normal conditions. For osmotic stress, the seedlings were soaked in 300 g⋅L-1 PEG for 24 h. For drought stress, pepper seedlings were deprived of water for 10 days, after which the RWC was determined. For dehydration tests, detached leaves were placed on the bench and weighted from 0 to 240 min at intervals of 30 min. For salt stress, pepper seedlings were divided into two groups, and one group was irrigated with 300 mM NaCl for 14 days, while the root of the other group was soaked in 300 mM NaCl for 24 h. After treatment, pepper leaves were sampled immediately used for determination of H2O2, REL, MDA, and soluble protein.
Seeds from Arabidopsis overexpressing CaBiP1 and wild-type Col-0 were germinated on MS plates with 3 mM DTT for ER stress, and 5-day-old seedlings grown on normal medium were transferred to medium with 2 mM DTT for 15 days. For heat stress, the MS plates with 7-day-old transgenic Arabidopsis seedlings were immersed in a water bath at 45°C for 50 min, then recovered at 22°C for 5 days, while 2-week-old seedlings in pots were heat-treated at 45°C for 6 h and recovered at 22°C for 7 days in a controlled temperature chamber. For salt and osmotic stress tests, Arabidopsis seeds were grown on MS medium with 100 mM NaCl or 200 mM mannitol for 4 days to determine the germination rate, and on medium with 75 mM NaCl or 150 mM mannitol for 7 days to determine root length. For salt stress experiments, 2-week-old seedlings were watered with 300 mM NaCl for 15 days to determine the survival rate. For dehydration tests, the detached Arabidopsis leaves were placed on the bench and weighted from 0 to 240 min at intervals of 30 min. Arabidopsis seedlings were deprived of water for 12 days to measure the RWC, root length, and re-watered for 2 days to determine the survival rate. In addition, 3-week-old Arabidopsis seedlings were soaked with root in 300 mM NaCl or 300 mM mannitol solutions, or sprayed with 30 mM DTT, or exposed to high temperatures of 45°C, and leaves were collected at 0, 3, and 9 h post treatments for genes expression analysis. For drought stress, 2-week-old Arabidopsis seedlings were withheld water for 10 days for gene expression analysis. All experiments were performed with three biological replicates.
Measurement of REL, H2O2, MDA, RWC, and Soluble Protein
Pepper leaf discs were used to measure the REL according to the method of Yin et al. (2014), and the MDA content was determined using the thiobarbituric acid reaction according to Dhindsa and Thorpe (1981). H2O2 levels were assessed by the DAB staining method (Dang et al., 2013), and the soluble protein content was measured by the protein dye-binding method with Coomassie light blue (Bradford, 1976). RWC was measured as described by Chakraborty et al. (2012). All experiments were performed with three biological replicates.
Total RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
Total RNA was extracted from sampled plant tissues using the Trizol (Invitrogen, Carlsbad, CA, United States) method, and the residual genomic DNA was digested by RNase-free DNase I (Promega, Madison, WI, United States). The first-strand cDNA was synthesized using the PrimeScriptTM Kit according to the manufacturer’s instructions (TaKaRa, Tokyo, Japan). Primer pairs (Supplementary Table S1) were designed by NCBI Primer-BLAST8, and qRT-PCR was performed using SYBR® Premix Ex TaqTM II (TaKaRa) as described previously by Wang et al. (2013). Relative gene expression levels were analyzed according to the 2-ΔΔCT method (Livak and Schmittgen, 2001), and CaUBI3 and AtActin2 were used as internal controls in pepper and Arabidopsis, respectively. Significance tests for differences in gene expression levels between control and stress treatments were performed using the Student’s t-test method at the α = 0.05 and 0.01 levels.
Results
Sequence Analysis of CaBiP Proteins
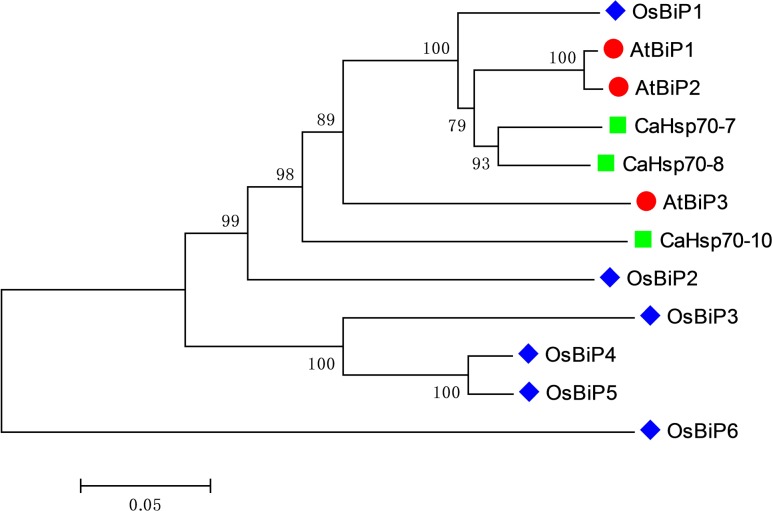
The phylogenetic tree of BiP proteins from pepper, Arabidopsis and rice showed that CaHsp70-8 and CaHsp70-7 share a close evolutionary relationship with AtBiP1 and AtBiP2, respectively, while CaHsp70-10 is more closely related to AtBiP3 and OsBiP2 (Figure 1). Therefore, CaHsp70-8, CaHsp70-7, and CaHsp70-10 were renamed as CaBiP1, CaBiP2, and CaBiP3, respectively.
FIGURE 1.
Phylogenetic tree of BiP family members from representative plant species. Ca, Capsicum annuum L.; At, Arabidopsis thaliana; Os, Oryza sativa.
Alignment of CaBiP protein sequences with AtBiPs (AtBiP1, NP_198206; AtBiP2, NP_851119; AtBiP3, NP_172382) and OsBiPs (OsBiP1, XP_015625618; OsBiP2, XP_015629631; OsBiP3, XP_015638605; OsBiP4, XP_015638801; OsBiP5, BAT04227; OsBiP6, LOC_Os01g33360) revealed high conservation in the signaling and functional domains. Seven domains and nine motifs were identified in CaBiPs based on published Arabidopsis and rice literatures (Noh et al., 2003; Sarkar et al., 2013) (Figures 2A,B). A signal peptide sequence (SP) for membrane transport was detected at the beginning of the N-terminus in CaBiPs, and the cut-off point GI (labeled by black arrow), a conserved site separating ATPase domain from the peptide-binding domain, was also found. In the N-terminal ATPase region, all the essential domains for BiP ATPase activity, including Domain 1 (motif 5) and Domain 2 (N-terminus of motif 1) for phosphate-binding, and Domain 4 (motif 2) for adenosine binding, are conserved in the predicted CaBiP proteins. Meanwhile, Domain 3 (C-terminus of motif 1) for calmodulin binding was also identified in CaBiPs. The C-terminal protein binding region has a highly conserved five residue core (shown by red arrows) that mediates hydrogen bonding with peptide substrates. However, Domain 5 (motif 6), which is essential for binding to peptide substrates, differs between CaBiP3 and CaBiP1/CaBiP2. Although CaBiP3 has the same pattern in motif distribution pattern found in CaBiP1 and CaBiP2, motif 7 containing the SP and motif 9 containing the ER retention signal are absent in CaBiP3 (Figure 2B and Supplementary Table S2). Analysis of gene structure also revealed that CaBiP3 lacks an intron that is present in CaBiP1 and CaBiP2 (Figure 2C).
FIGURE 2.
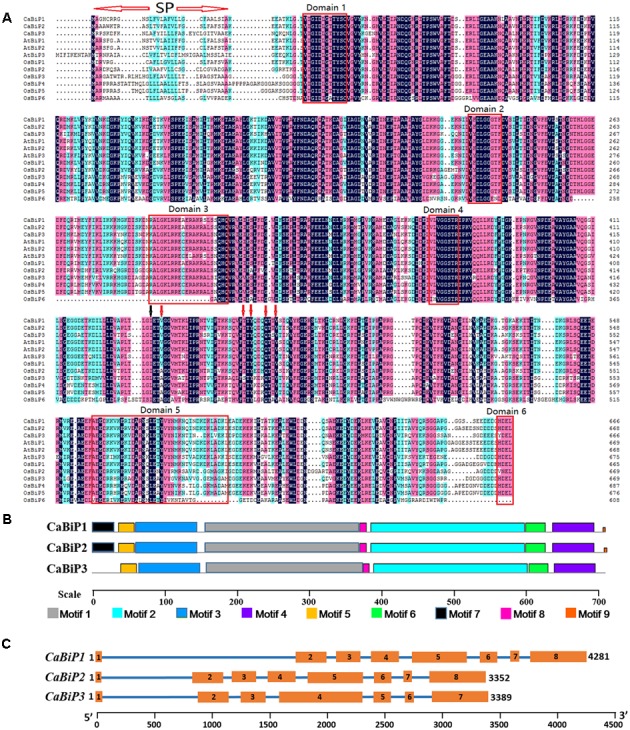
Sequence analysis of CaBiP genes. (A) Conserved amino acid domains in CaBiPs shown by amino acid sequence alignment with orthologs in Arabidopsis and rice. SP, the signal peptide. Red arrows indicate hydrogen bonds, black arrow indicates the the cut-off point GI. (B) Motifs of CaBiP proteins identified by MEME tools. Different motifs are indicated by different colors. (C) Distribution of introns and extrons in CaBiP genes. Blue lines = introns, orange rectangles = extrons.
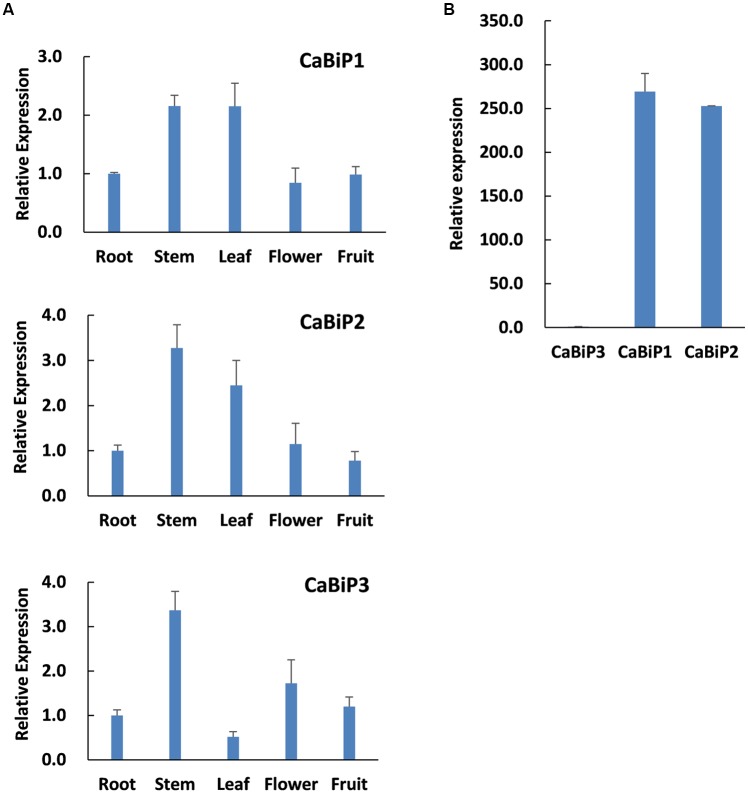
Expression Profiles of CaBiP Genes in Pepper Tissues
Expression profiles of the three CaBiP genes in pepper root, stem, leaf, flower and fruit tissues were determined by qRT-PCR with transcript-specific primers (Supplementary Table S1). The results indicated that the three CaBiP genes were diversely expressed in roots, stems, leaves, flowers, and fruits (Figure 3A), and the overall expression level of CaBiP3 was much lower than CaBiP1 and CaBiP2 (Figure 3B). In addition, the expression levels of CaBiP1 and CaBiP2 genes were obviously higher in stem and leaf tissue than in other organs, while CaBiP3 expression was higher in stem and flower (Figure 3A).
FIGURE 3.
Tissue-specific expression CaBiP genes. (A) Expression of CaBiP1, CaBiP2 and CaBiP3 in pepper root, stem, leaf, flower and fruit. (B) Relative expression of CaBiP genes in pepper root. The expression level of CaUBI3 was used as the internal control. Bars shows the standard deviation of expression levels from three biological replications.
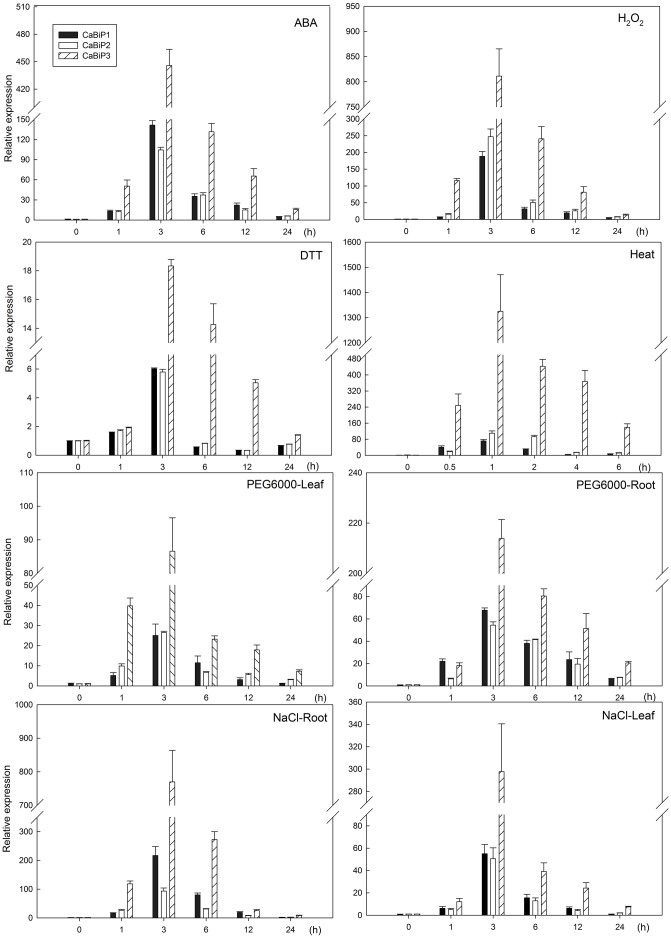
Expression of CaBiP Genes in Pepper Under Abiotic Stresses in Pepper
Expression of all CaBiP genes was induced under abiotic stress conditions involving ABA, H2O2, DTT, heat, salt, and osmotic stresses (Figure 4). Peak expression occurred earlier in heat stress (1 h) than in other treatments (3 h), and the fold change in CaBiP3 expression was significantly higher than that of CaBiP1 and CaBiP2 in all treatments. In addition, CaBiP was expressed more highly in roots than in leaves under osmotic stress and salt stress.
FIGURE 4.
Expression of CaBiP genes following salt, osmotic, heat, ER, ABA, and H2O2 stress treatments. Bars show the standard deviation of expression levels from three biological replications.
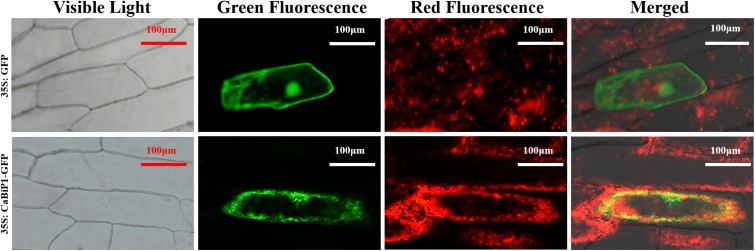
Subcellular Localization of CaBiP1 in the ER
Under the control of CaMV-35S promoter, the 35S:CaBiP1-GFP construct was transiently expressed in onion epidermal cells. While both the green fluorescence from fusion protein of CaBiP1 and GFP and the red fluorescence from ER-Tracker Red were detected by laser confocal microscopy, their merged yellow fluorescence was also observed (Figure 5). The results indicate CaBiP1 is located in the ER.
FIGURE 5.
Subcellular localization of the CaBiP1-GFP fusion protein in onion epidermal cells. CaMV35S:GFP was used as the control, and ER-Tracker Red was recruited to mark the ER. CaMV35S, constitutive promoter from the cauliflower mosaic virus. GFP, Bars = 100 μm. More than 10 transgenic cells were observed, and all of them had the similar result. One of them was presented here.
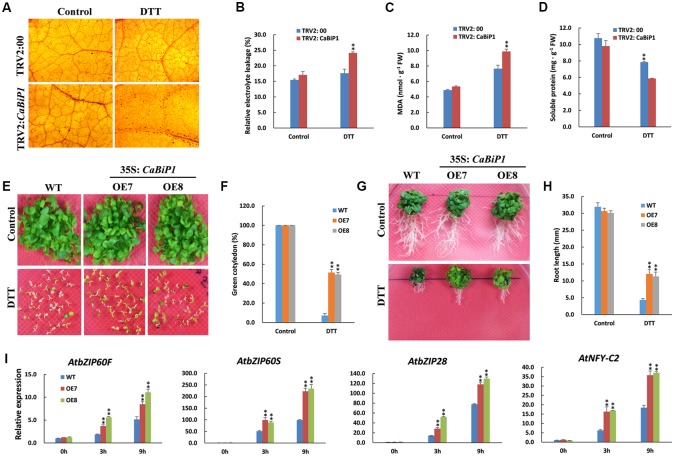
ER Stress Tolerance Is Altered in CaBiP1-Silenced and CaBiP1-Overexpressing Plants
Pepper seedlings harboring TRV2:CaBiP1 demonstrated a silencing efficiency of over 70%, and Arabidopsis transgenic lines OE7 and OE8 were used to perform abiotic stress treatments. No visible difference was observed between either TRV2:CaBiP1 and TRV2:00 pepper lines, and the same was true for CaBiP1-OE lines and WT Arabidopsis plants (Supplementary Figure S1).
Following ER stress induced by 30 mM DTT, the H2O2 content in pepper leaves, as revealed by dark brown dots after DAB staining, was increased in both TRV2:CaBiP1 and TRV2:00 lines, albeit to a greater extent in the former (Figure 6A). Similar results were also observed for REL levels and MDA content (Figures 6B,C). By contrast, the soluble protein content was decreased in the leaves of both TRV2:CaBiP1 and TRV2:00 plants following ER stress, and the decrease was again more significant in the former (Figure 6D).
FIGURE 6.
Altered ER stress tolerance in CaBiP1-silenced and CaBiP1-overexpressing plants. (A–D) H2O2 accumulation, REL, MDA content, and soluble protein content in leaves of TRV2:00 and TRV2:CaBiP1 pepper seedlings grown under ER stress induced by spraying 30 mM DTT for 24 h. Spraying with distilled water was performed on control plants. (E,F) Establishment of WT and CaBiP1-OE Arabidopsis seedling under ER stress conditions induced by 3 mM DTT. The percentage of seedlings with green cotyledons was calculated at the14th day after sowing. (G,H) Root growth of 5-day-old WT and transgenic Arabidopsis seedlings exposed to 2 mM DTT for 15 days. (I) Expression analysis of ER stress-responsive genes in WT and CaBiP1-OE lines sprayed with 30 mM DTT for the designed time duration. AtActin2 was used as the internal control. Data are the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) and a double asterisk (p < 0.01) based on the results of the Student’s t-test.
Under normal growth conditions, no difference was observed in the germination rate between CaBiP1-OE lines and WT Arabidopsis lines. However, under ER stress induced by 3 mM DTT, the germination rate was significantly higher in CaBiP1-OE plants (Supplementary Figure S2A). Furthermore, when the treatment was extended to 14 days, chlorosis of cotyledons was more severe in WT than in CaBiP1-OE lines (Figures 6E,F). After Arabidopsis seedlings at 5-day-old were transferred to MS medium containing 2 mM DTT, the growth was inhibited in both CaBiP1-OE and WT lines, but fresh weight and root length were obviously higher in the CaBiP1-OE plants (Figures 6G,H and Supplementary Figure S2B). Additionally, expression of some genes in the UPR pathway, such as AtbZIP60F, AtbZIP60S, AtbZIP28, and AtNF-YC2 (encoding a component of the AtbZIP28 transcription complex; Liu and Howell, 2010), were up-regulated when the Arabidopsis seedlings were exposed to DTT, and the fold change was greater in transgenic lines than in WT plants (Figure 6I).
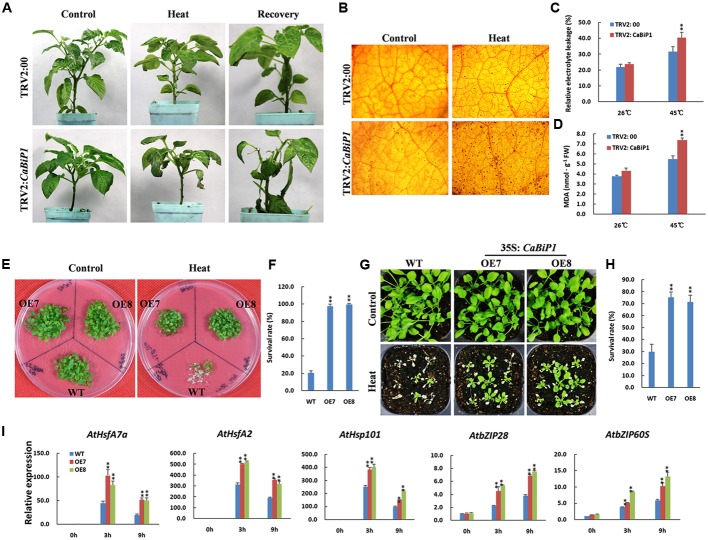
Heat Tolerance Is Altered in CaBiP1-Silenced and CaBiP1-Overexpressing Plants
When TRV2:CaBiP1 and TRV2:00 pepper seedlings were subjected to 45°C for 24 h, sunburn-like symptoms of heat stress was observed in TRV2:CaBiP1 leaves but not in those of TRV2:00 plants. After recovery under normal conditions for 5 days, the injured leaves in TRV2:CaBiP1 fell off, while TRV2:00 leaves resumed growth (Figure 7A). Under heat stress, H2O2, MDA content, and REL levels were increased in both TRV2:CaBiP1 and TRV2:00 lines, and all three indicators were markedly higher in the former (Figures 7B–D).
FIGURE 7.
Altered heat tolerance in CaBiP1-silenced and CaBiP1-overexpressing plants. (A) TRV2:CaBiP1 and TRV2:00 pepper seedlings following heat stress at 45°C for 24 h and recovery for 5 days under 25°C normal conditions. Plants continually grown at 25°C were used as controls. (B–D) H2O2 accumulation,REL and MDA content in pepper seedlings leaves treated with 45°C heat stress for 24 h. (E,F) Survival rate of 7-day-old WT and CaBiP1-OE Arabidopsis seedlings submerged in a 45°C water bath for 50 min and recovered at 22°C for 5 days. (G,H) Survival rate of 2-week-old Arabidopsis seedlings in soil after treatment at 45°C for 6 h then recovered for 5 days. (I) Expression of ER stress- and heat stress-related genes in 3-week-old WT and CaBiP1-OE Arabidopsis seedlings treated at 45°C for the designed time duration. AtActin2 was used as the internal control. Data are the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) and a double asterisk (p < 0.01) based on the results of the student’s t-test.
Arabidopsis seedlings of 1-week-old were treated at 45°C for 50 min, then recovered for 5 days under normal conditions, and only 20% of WT plants survived, compared with more than 90% of CaBiP1-OE plants (Figures 7E,F). When 2-week-old seedlings of WT and CaBiP1-OE lines in pots were subjected to 45°C for 6 h then recovered for 5 days, about 30% of WT plants survived, compared with 74.1% for OE7 and 70.4% for OE8 plants (Figures 7G,H). During heat stress treatments, both UPR-related genes (AtbZIP60S and AtbZIP28) and heat responsive genes (AtHsfA2, AtHsfA7a, and AtHsp101) were obviously induced in both WT and transgenic Arabidopsis lines. However, the induction of gene expression in transgenic lines was more intense than in WT plants. Furthermore, the expression levels of three heat response genes peaked at 3 h, while those of two UPR genes increased continuously during the treatments (Figure 7I).
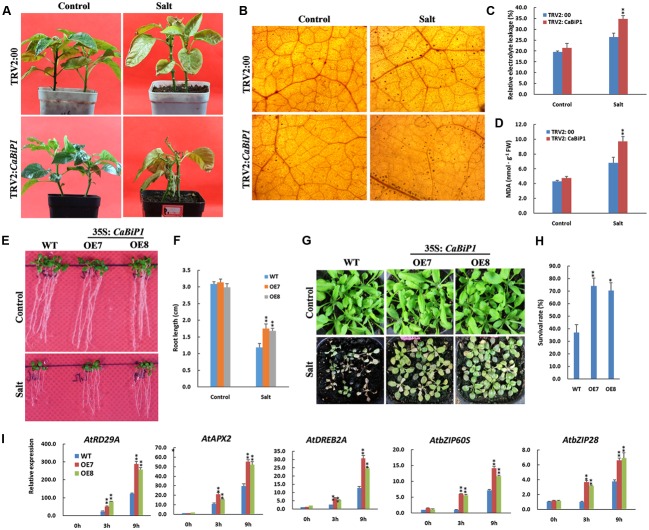
Salt Tolerance Is Altered in CaBiP1-Silenced and CaBiP1-Overexpressing Plants
After irrigation with 300 mM NaCl solution for 14 days, the leaves of CaBiP1-silenced pepper seedlings became yellowish and wilted and eventually fell off, while in TRV2:00 plants, only yellowness was observed (Figure 8A). When the root of TRV2:CaBiP1 and TRV2:00 seedlings were soaked in 300 mM salt solution for 24 h, the accumulation of H2O2 in TRV2:CaBiP1 leaves was higher than in TRV2:00 leaves (Figure 8B), and the MDA content was also 1.4-fold higher in TRV2:CaBiP1 leaves (Figure 8C). Similarly, REL levels were higher in TRV2:CaBiP1 (36.8%) than TRV2:00 (26.4%) (Figure 8D).
FIGURE 8.
Altered salt tolerance in CaBiP1-silenced and CaBiP1–overexpressing plants. (A) TRV2:00 and TRV2:CaBiP1 peppers seedlings irrigated with 300 mM NaCl solution for 14 days. (B–D) H2O2 accumulation, REL and MDA content in pepper seedlings leaves treated with 300 mM NaCl solution for 24 h. (E,F) Root growth of WT and transgenic lines grown under 75 mM NaCl for 7 days. (G,H) Survival rate of 2-week-old Arabidopsis seedlings in soil treated with 300 mM NaCl for 15 days. (I) Expression of salt stress- and ER stress-related genes in leaves of 3-week-old WT and CaBiP1-OE hydroponic seedlings treated with 300 mM NaCl s for the designed time duration. AtActin2 was used as the internal control gene. Data are the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) and a double asterisk (p < 0.01) based on the results of student’s t-test.
When Arabidopsis seedlings were grown on MS medium with 100 mM NaCl for 4 days, the germination rate of CaBiP1-OE lines was obviously higher than that of WT plants (Supplementary Figure S3). Additionally, in Arabidopsis seedlings grown on MS plates containing 75 mM NaCl for 7 days, the roots of CaBiP1-OE lines were significantly longer than those of WT plants (Figures 8E,F). Following watering of 2-week-old seedlings with a high concentration of NaCl (300 mM) for 15 days, CaBiP1-OE seedlings showed a higher salt tolerance than WT plants; the survival rates of CaBiP1-OE7 and OE8 plants were 2.0- and 1.9-fold higher than WT, respectively (Figures 8G,H). In terms of gene expression, salt stress-related genes (AtAPX2, AtDREB2A, and AtRD29A) and UPR-related genes (AtbZIP60S and AtbZIP28) displayed higher levels of transcription in CaBiP1-OE plants than in WT plants (Figure 8I). Interestingly, the expression of all the tested genes revealed continuous enhancement throughout the duration of salt stress experiments (Figure 8I).
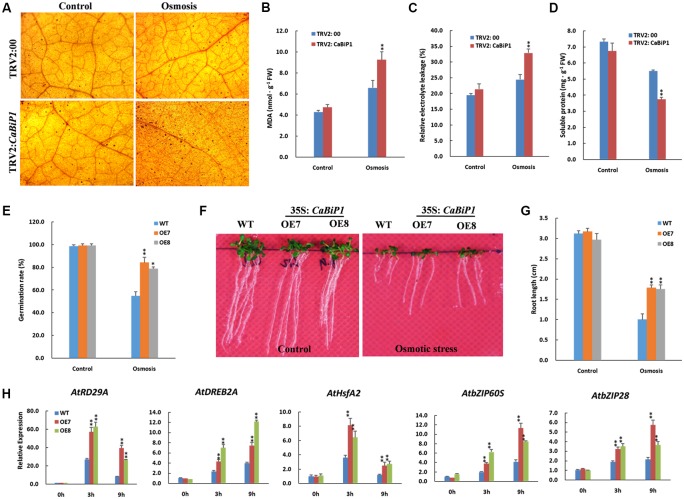
Osmotic Stress Tolerance Is Altered in CaBiP1-Silenced and CaBiP1-Overexpressing Plants
After soaking in 300 g⋅L-1 PEG6000 solution for 24 h, TRV2:CaBiP1 pepper seedlings accumulated more H2O2 than did TRV2:00 plants, as shown by the dark brown dots in leaves after DAB staining (Figure 9A). Similarly, the MDA content and REL levels in TRV2:CaBiP1 seedlings were obviously higher than those of TRV2:00 lines following osmotic stress (Figures 9B,C). By contrast, the soluble protein content in TRV2:CaBiP1 leaves was significantly lower than in TRV2:00 leaves (Figure 9D). Following osmotic stress, Arabidopsis seeds from CaBiP1-OE plants grown on MS medium containing 200 mM mannitol displayed a remarkably high germination rate compared with that of WT plants; seed germination in OE7 and OE8 lines reached to 84 and 78% at the 4th day, respectively, whereas that of WT was only 54% (Figure 9E). On the 7th day after germination on MS medium containing 150 mM mannitol, CaBiP1-OE Arabidopsis seedlings exhibited stronger tolerance to osmotic stress, as evidenced by a 1.8-fold increase in root length compared with WT plants (Figures 9F,G). The qRT-PCR analysis indicated that osmotic stress-related genes (AtDREB2A, AtRD29A, and AtHsfA2) were highly induced in stressed leaves (Figure 9H). Notably, AtRD29A, a marker for osmotic stress, as up-regulated by 5.0- and 3.3-fold at 9 h in OE7 and OE8 lines compared with WT plants. Similarly, CaBiP1-OE lines showed higher expression levels of ER stress-related genes (AtbZIP60S and AtbZIP28) at 3 h and 9 h following exposure to 300 mM mannitol. Additionally, expression of AtRD29A and AtHsfA2 peaked at 3 h, while expression of the other three genes increased continuously throughout the treatment (Figure 9H).
FIGURE 9.
Altered osmotic tolerance in CaBiP1-silenced and CaBiP1-overexpressing plants. (A–D) H2O2 accumulation, REL, MDA content, and soluble protein content in TRV2:00 and TRV2:CaBiP1 pepper seedlings leaves grown under osmotic stress simulated by soaking in 300 g/L PEG6000 for 24 h. Treatment with distilled water was used as the control. (E) Seed germination rate of WT and CaBiP1-OE lines grown under osmotic stress by adding 200 mM mannitol in the medium for 4 days. (F,G) Root growth of WT and transgenic Arabidopsis lines germinated under 150 mM mannitol for 7 days. (H) Expression of osmotic stress- and ER stress-related genes in the leaves of 3-week-old WT and CaBiP1-OE hydroponic seedling grown under 300 mM mannitol for designed time duration. AtActin2 was used as the internal control gene. Data are the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) and a double asterisk (p < 0.01) based on the results of the Student’s t-test.
Drought/Dehydration Tolerance Is Altered in CaBiP1-Silenced and CaBiP1-Overexpressing Plants
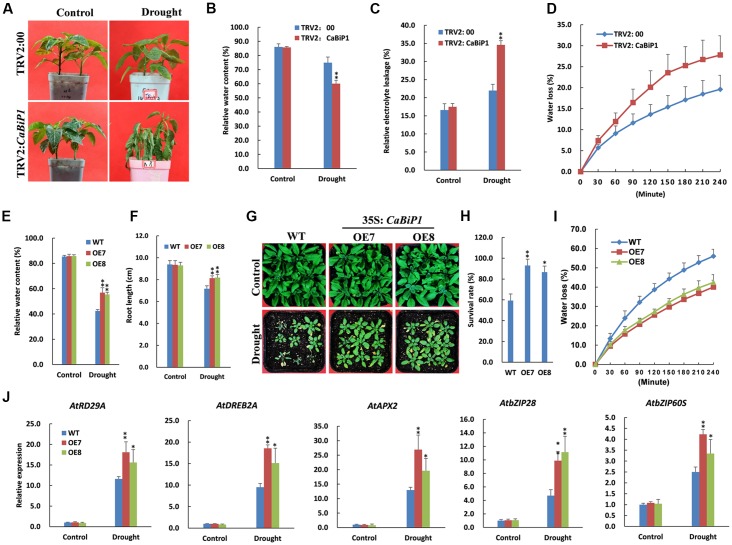
At the end of the drought stress treatment that involved withholding water for 10 days, wilting symptoms were observed in pepper leaves of TRV2:CaBiP1 lines but not TRV2:00 (Figure 10A). Furthermore, TRV2:CaBiP1 plants exhibited higher REL level and lower RWC than TRV2:00 (Figures 10B,C). Similarly, dehydration of pepper leaves placed on a bench for 4 h resulted in a rate of water loss that was higher in gene-silenced pepper seedlings compared with controls (Figure 10D).
FIGURE 10.
Altered drought tolerance in CaBiP1-silenced and CaBiP1-overexpressing plants. (A–C) Symptoms, RWC and REL in the leaves of TRV2:CaBiP1 and TRV2:00 pepper seedlings grown under drought stress performed by withholding water for 10 days. (D) Water loss rate of detached leaves from pepper TRV2:CaBiP1 and TRV2:00 seedlings. (E,F) RWC and root length of WT and CaBiP1-OE lines grown under drought stress by withholding water for 12 days. (G,H) Survival rate of WT and CaBiP1-OE lines grown under drought stress by withholding water for 12 days then rehydrated for 2 days. (I) Water loss rate of detached leaves from WT and CaBiP1-OE seedlings. (J) Expression of drought stress- and ER stress-related genes in the leaves of WT and CaBiP1-OE lines grown under drought stress by withholding water for 10 days. AtActin2 was used as the internal control gene. Data are the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) and a double asterisk (p < 0.01) based on the Student’s t-test.
Following the withholding of water for 12 days, Arabidopsis seedlings became severely withered, but CaBiP1-OE seedlings exhibited higher RWC and longer root than WT plants (Figures 10E,F). After re-watering for 2 days, more than 90% of CaBiP1-OE Arabidopsis seedlings survived, whereas just 59% WT plants remained alive (Figures 10G,H). In addition, under dehydration conditions, the rate of water loss for detached rosette leaves from transgenic seedlings was markedly lower than that in WT leaves (Figure 10I). Additionally, CaBiP1-OE lines showed a more obvious induction in the drought stress-related genes (AtDREB2A, AtRD29A, and AtAPX2) and ER stress-related genes (AtbZIP60S and AtbZIP28) under drought stress than did WT plants (Figure 10J).
Discussion
A major negative effect of abiotic stress on plant cells is the accumulation of misfolded and denatured proteins, which further induces ER stress. The alleviation of ER stress is therefore key to plant survival under adverse environmental conditions (Wan and Jiang, 2016). BiP, an Hsp70 family member located in the ER, binds to nascent proteins to protect them from aggregation (Howell, 2013). Characterization and functional analysis of BiPs will therefore contribute to our understanding of plant tolerance to abiotic stress.
In our previous study, three BiP proteins (CaBiP1, CaBiP2, and CaBiP3) were identified in the genome of pepper (Guo et al., 2016). Amino acid sequence alignment revealed the conserved functional and signaling domains in CaBiPs, including an ATP-binding domain at the N-terminus (Domain 1, 2, and 4), a protein-binding domain at the C-terminus (Domain 5), and an ER retention factor (Domain 6) (Figure 2A) (Noh et al., 2003; Sarkar et al., 2013), indicating the importance of the different domains and ER localization of CaBiP1 (Figure 5).
CaBiP1 and CaBiP2 share high sequence similarity, whereas CaBiP3 is more distantly related, with diversity particularly high in Domain 5 (motif 6), Domain 6, motif 7, and motif 9 (Figures 2A,B), and difference in the exon-intron structure (Figure 2C). Correspondingly, differences in expression pattern were also observed between CaBiP3 and CaBiP1/CaBiP2. While CaBiP1 and CaBiP2 were constitutively expressed in all tested tissues (Figure 3), transcription of CaBiP3 was barely detectable under normal growth conditions, but expression was strongly induced by ER stress and various environmental stresses such as heat, osmotic and salt stress, and so by exposure to the signaling molecules such as ABA and H2O2 (Figure 4). Similar phenomena were also found in Arabidopsis and rice, in which AtBiP1, AtBiP2 and OsBiP1 were constitutively expressed in various tissues, whereas mRNA levels of AtBiP3, OsBiP4, and OsBiP5 remained low under normal conditions but were strongly up-regulated by ER stress agents of TM or DTT (Noh et al., 2003; Wakasa et al., 2012).
In agreement with our results, Noh et al. (2003) found that AtBiP3 was distinguishable from AtBiP1 and AtBiP2 in several ways, including the regulatory elements in the promoter region, genomic organization, and sequence homology. Furthermore, Wakasa et al. (2012) also suggested that OsBiP4 and OsBiP5 may have a function that is distinct from that of OsBiP1. Guo et al. (2016) revealed that the cis-elements in CaBiP3 were significantly different from those in CaBiP1 and CaBiP2. These results indicate a relationship between the gene structures of CaBiPs and their expression patterns, and even biological functions, but further research is needed.
The contribution of CaBiP1 in plant tolerance to environmental stress was revealed by VIGS experiments in pepper and overexpression experiments in Arabidopsis. Our results suggest that the silencing of CaBiP1 decreased the tolerance of pepper to multiple abiotic stresses, including heat, osmotic, salt, drought/dehydration and ER stress, and conversely, overexpression of CaBiP1 increased the tolerance of Arabidopsis to these stresses. Under stress conditions, the CaBiP1-silenced pepper line presented more severe injury symptoms based on various physiological indicators, while CaBiP1-overexpressing Arabidopsis lines displayed higher tolerance to adverse environments in terms of seed germination rate, seedling survival rate, and root length (Figures 6–10). Similar results were also found following ER and heat stresses in Arabidopsis (Yang et al., 2016), and following water deficit in tobacco (Alvim et al., 2001) and pathogen infection experiments in tomato (Liebrand et al., 2014). These results suggest that CaBiP1 confers plant enhanced tolerance to multiple environmental stress factors.
Abiotic stress induces the production of ROS such as H2O2, a secondary messenger that regulates protective mechanism of plant cells. However, excess ROS causes irreversible damage leading to cell death (Ozgur et al., 2014). After treatment with DTT and following abiotic stress, the accumulation of ROS was higher in CaBiP1-silenced pepper leaves than in WT plants, and the MDA content and REL were consequently elevated (Figures 6–10). These results suggest that down-regulation of the CaBiP1 gene worsens the cell injuries raised by adverse environmental conditions that cause accumulation of ROS. In accordance with our results, Alvim et al. (2001) also found that antisense silencing of the BiP gene diminishes water deficit-induced oxidative stress in tobacco, which is characterized by increased superoxide dismutase activity.
In addition, under ER and osmotic stress conditions, the soluble protein content in CaBiP1-silenced pepper leaves was significantly lower than in the WT line (Figures 6, 9), consistent with the study by Yang et al. (2016), in which overexpression of AtBiP1 alleviated autophagy pressure in Arabidopsis and decreased protein degradation following heat stress. These results indicate that CaBiP1 may protect plant cells from environmental stress by preventing protein aggregation. CaBiP1-OE Arabidopsis plants exhibited slower water loss in detached rosette leaves, but higher RWC than WT plants under water deficit conditions (Figures 10C,D). Conversely, CaBiP1-silenced pepper leaves showed faster water loss, but lower RWC than control plants under water deficit conditions (Figures 10E,I). Similarly, when the soybean BiP gene was overexpressed in soybean and tobacco, transgenic lines were better able to maintain cellular homeostasis under water stress conditions, and displayed higher leaf water content and reduced withering (Alvim et al., 2001; Valente et al., 2009; Carvalho et al., 2014a). Since water deficit can be caused by other abiotic stress such as heat, salt and osmotic stress, these results indicate that CaBiP1 confers plant tolerance to adverse environmental conditions by increasing water retention capacity of the cells.
When Arabidopsis seedlings were exposed to ER stress induced by DTT, transcription of UPR genes bZIP28 and bZIP60S was continuously increased throughout the treatment (Figure 6). Similarly, expression of UPR genes was also continuously enhanced under the duration of various abiotic stresses (Figures 7–10). Based on these combined results in Arabidopsis, it appears ER stress is always concomitant with abiotic stress, and alleviation of ER stress is therefore conducive to improving plant tolerance to adverse environments. Furthermore, under abiotic stress treatments, expression of AtbZIP28 and AtbZIP60S in CaBiP1-overexpressing Arabidopsis lines was markedly higher than in WT plants (Figures 7–10). Since AtbZIP28 and AtbZIP60 are transcription factors involved in up-regulating protective genes in different branches of the UPR pathway (Howell, 2013), these results suggest overexpression of CaBiP1 confers higher efficiency of the protein folding machinery under stress conditions. However, Leborgne-Castel et al. (1999) reported that overexpression of BiP in tobacco decreased the UPR pathway, and up-regulation of BiP in the transgenic line was sufficiently elevated to assist protein folding during ER stress. Based on the role of BiP as a sensor in ER stress signaling (Bertolotti et al., 2000; Shen et al., 2002), we believe CaBiP1 performs protective functions as both a molecular chaperone and UPR regulator, consistent with research on Arabidopsis by Srivastava et al. (2013).
Similarly, under abiotic stress conditions, expression of stress-related genes was elevated in both CaBiP1-overexpressing and WT Arabidopsis lines, but levels were higher in the former (Figures 7–10). Transcription of DREB2A (a major transcription factor functioning under stress conditions, Nakashima et al., 2000) and APX2 (an important H2O2-scavenging enzyme, Suzuki et al., 2013) increased continuously throughout the treatment of salt, osmotic and drought stress (Figures 8–10), which suggests overexpression of CaBiP1 promote plant responsivity to water deficit and the ability to remove ROS. Expression level of HsfA2, a regulatory amplifier of heat-response genes (Schramm et al., 2006), peaked at 3 h then fell down under the treatment of heat and osmotic stress (Figures 7–9), which implies that overexpression of CaBiP1 enhances the function of HsfA2 by up-regulating its expression level but not changing its expression pattern. Expression of RD29A, a marker for plant response to water deficit stress (Msanne et al., 2011), kept increasing throughout the treatment of salt and drought stress, while peaked at 3 h then fell down under osmotic stress (Figures 8–10), which hints that RD29A is more responsive to salt and drought stress, and overexpression of CaBiP1 advancing this responsivity of RD29A. Based on the discussion above, it can be assumed CaBiP1 improves plant tolerance to environmental stresses by the up-regulating stress-responsive genes, but elucidating the exact mechanisms and signaling pathways involved requires need further study.
Conclusion
BiP plays important roles in helping plants to cope with ER stress induced by adverse environmental conditions. In this study, we characterized the sequence and expression patterns of three BiP genes in pepper. While CaBiP1 and CaBiP2 were constitutively expressed in all tissues under normal and stressed conditions, CaBiP3 was mainly transcribed under stress treatments. The silencing of CaBiP1 lowered the tolerance of pepper plants to ER and environmental stresses, whereas overexpression of CaBiP1 in Arabidopsis enhanced of tolerance to these stresses. Our study suggests that CaBiP1 may contribute to tolerance to abiotic stress in pepper by reducing ROS accumulation, increasing the water retention ability, and enhancing the UPR pathways and expression of stress-related genes.
Author Contributions
HW and ML designed the experiments. HW, HN, and YZ performed the research. HW drafted the manuscript. LM revised the paper and contributed reagents/materials/analysis tools. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31572114, U1603102), the Shaanxi Agriculture Science and Technology Projects (Grant No. 2014K01114101), and the Opening Fund of Key Laboratory for Crop Biotechnology of Xinjiang Uygur Autonomous Region (Grant No. XJYS030212014103).
Abbreviations
- BiP
binding proteins
- DAB
diaminobenzidine
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- Hsp
heat shock proteins
- MDA
malondialdehyde
- ORF
open reading frame
- qRT-PCR
quantitative real-time PCR
- REL
relative electric leakage
- RWC
relative water content
- UPR
unfolding protein response.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01122/full#supplementary-material
Confirmation of CaBiP1-silenced pepper seedlings and CaBiP1-overexpressing Arabidopsis lines. (A) Phenotypes of CaBiP1-silenced pepper seedlings. CaPDS, phytoene desaturase gene for chlorophyll synthesis; TRV2:00, control plants harboring the empty TRV2 vector. (B) Measurement of the silencing efficiency for CaBiP1 expression in silenced pepper seedlings at the 30th day after inoculation. (C) Screening of CaBiP1-overexpressing Arabidopsis lines in MS medium containing kanamycin. (D) Measurement of CaBiP1 expression levels in CaBiP1-overexpressing Arabidopsis lines by semi-quantitative PCR. The AtActin2 gene was used as the internal control. Error bars represent standard deviation from three biological replicates.
Enhanced tolerance to ER stress induced by DTT in CaBiP1-overexpressing Arabidsopsis lines. (A) Germination rate on MS medium with 3 mM DTT. (B) Fresh weight of 5-day-old CaBiP1-OE Arabidsopsis seedlings grown on MS medium with 2 mM DTT for 15 days. Data represent the mean ± standard deviation from three biological replicates. Statistical significance is indicated as by a single asterisk (p < 0.05) and double asterisk (p < 0.01) based on the results of the Student’s t-test.
Seed germination rate in WT and CaBiP1-OE (OE7 and OE8) Arabidsopsis lines grown on MS medium with 100 mM NaCl. Data represent the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) based on the results of the Student’s t-test.
References
- Alvim F. C., Carolino S. M. B., Cascardo J. C. M., Nunes C. C., Martinez C. A., Otoni W. C., et al. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126 1042–1054. 10.1104/pp.126.3.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell. Biol. 2 326–332. 10.1038/35014014 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Carvalho H. H., Brustolini O. J. B., Pimenta M. R., Mendes G. C., Gouveia B. C., Silva P. A., et al. (2014a). The molecular chaperone binding protein BiP prevents leaf dehydration-induced cellular homeostasis disruption. PLoS ONE 9:e86661 10.1371/journal.pone.0086661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H. H., Silva P. A., Mendes G. C., Brustolini O. J. B., Pimenta M. R., Gouveia B. C., et al. (2014b). The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiol. 164 654–670. 10.1104/pp.113.231928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K., Sairam R. K., Bhattacharya R. C. (2012). Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol. Biochem. 51 90–101. 10.1016/j.plaphy.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Cheng Q., Zhou Y., Liu Z., Zhang L., Song G., Guo Z., et al. (2015). An alternatively spliced heat shock transcription factor, OsHSFA2dl, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 17 419–429. 10.1111/plb.12267 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dang F. F., Wang Y. N., Yu L., Eulgem T., Lai Y., Liu Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36 757–774. 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- Denecke J., Goldman M. H. S., Demolder J., Seurinck J., Botterman J. (1991). The tobacco luminal binding protein is encoded by a multigene family. Plant Cell 3 1025–1035. 10.1105/tpc.3.9.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7247–7252. 10.1073/pnas.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Srivastava R., Howell S. H. (2013). Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int. J. Mol. Sci. 14 8188–8212. 10.3390/ijms14048188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa R. S., Thorpe T. A. (1981). Leaf Senescence: Correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 32 93–101. 10.1093/jxb/32.1.93 [DOI] [Google Scholar]
- Fanata D. W. I., Lee S. Y., Lee K. O. (2013). The unfolded protein response in plants: a fundamental adaptive cellular response to internal and external stresses. J. Proteomics 93 356–368. 10.1016/j.jprot.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Guo M., Liu J. H., Ma X., Zhai Y. F., Gong Z. H., Lu M. H. (2016). Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 252 246–256. 10.1016/j.plantsci [DOI] [PubMed] [Google Scholar]
- Guo M., Zhai Y. F., Lu J. P., Chai L., Chai W. G., Gong Z. H., et al. (2014). Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. Int. J. Mol. Sci. 15 19741–19759. 10.3390/ijms151119741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant. Biol. 64 477–499. 10.1146/annurev-arplant-050312-120053 [DOI] [PubMed] [Google Scholar]
- Jung K. H., Gho H. J., Nguyen M. X., Kim S. R., An G. (2013). Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Gen. 13 391–402. 10.1007/s10142-013-0331-6 [DOI] [PubMed] [Google Scholar]
- Kim S., Park M., Yeom S. I., Kim Y. M., Lee J. M., Lee H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet 46 270–278. 10.1038/ng.2877 [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N., Jelitto-Van Dooren E. P., Crofts A. J., Denecke J. (1999). Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11 459–470. 10.1105/tpc.11.3.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Humbert S., Howell S. H. (2012). ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes 5:144 10.1186/1756-0500-5-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand T. W., Kombrink A., Zhang Z., Sklenar J., Jones A. M., Robatzek S., et al. (2014). Chaperones of the endoplasmic reticulum are required for ve1-mediated resistance to verticillium. Mol. Plant Pathol. 15 109–117. 10.1111/mpp.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. X., Howell S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 782–796. 10.1105/tpc.109.072173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maruyama D., Endo T., Nishikawa S. (2015). BiP3 supports the early stages of female gametogenesis in the absence of BiP1 and BiP2 in Arabidopsis thaliana. Plant. Signal. Behav. 10:e1035853 10.1080/15592324.2015.1035853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D., Sugiyama T., Endo T., Nishikawa S. (2014). Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant Cell Physiol. 55 801–810. 10.1093/pcp/pcu018 [DOI] [PubMed] [Google Scholar]
- Msanne J., Lin J., Stone J. M., Awada T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234 97–107. 10.1007/s00425-011-1387-y [DOI] [PubMed] [Google Scholar]
- Nakashima K., Shinwari Z. K., Sakuma Y., Seki M., Miura S., Shinozaki K., et al. (2000). Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 42 657–665. 10.1023/A:1006321900483 [DOI] [PubMed] [Google Scholar]
- Noh S. J., Kwon C. S., Oh D. H., Moon J. S., Chung W. I. (2003). Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311 81–91. 10.1016/S0378-1119(03)00559-6 [DOI] [PubMed] [Google Scholar]
- Ozgur R., Turkan I., Uzilday B., Sekmen A. H. (2014). Endoplasmic reticulum stress triggers ROS signalling, changes the redox state, and regulates the antioxidant defence of Arabidopsis thaliana. J. Exp. Bot. 65 1377–1390. 10.1093/jxb/eru034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Yu C. S., Shen Y. O., Fang X. D., Chen L., Min J. M., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A 111 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar N. K., Kundnani P., Grover A. (2013). Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18 427–437. 10.1007/s12192-012-0395-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F., Ganguli A., Kiehlmann E., Englich G., Walch D., von Koskull-Döring P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60 759–772. 10.1007/s11103-005-5750-x [DOI] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3 99–111. 10.1016/S1534-5807(02)00203-4 [DOI] [PubMed] [Google Scholar]
- Srivastava R., Deng Y., Shah S., Rao A. G., Howell S. H. (2013). Binding protein is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25 1416–1429. 10.1105/tpc.113.110684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Sejima H., Harper J., Mittler R. (2013). Enhanced seed production under prolonged heat stress conditions in Arabidopsis thaliana plants deficient in cytosolic ascorbate peroxidase 2. J. Exp. Bot. 64 253–263. 10.1093/jxb/ers335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry A. A., Dulanto P. N. C., Reynolds M. P., Davies W. J. (2016). How can we improve crop genotypes to increase stress resilience and productivity in a future climate? a new crop screening method based on productivity and resistance to abiotic stress. J. Exp.Bot. 67 5593–5603. 10.1093/jxb/erw330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente M. A. S., Faria J. A. Q. A., Soares-Ramos J. R. L., Reis P. A. B., Pinheiro G. L., Piovesan N. D., et al. (2009). The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 60 533–546. 10.1093/jxb/ern296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y., Hayashi S., Takaiwa F. (2012). Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236 1519–1527. 10.1007/s00425-012-1714-y [DOI] [PubMed] [Google Scholar]
- Wan S., Jiang L. (2016). Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253 753–764. 10.1007/s00709-015-0842-1 [DOI] [PubMed] [Google Scholar]
- Wang J. E., Liu K. K., Li D. W., Zhang Y. L., Zhao Q., He Y. M., et al. (2013). A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophthora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14 3158–3177. 10.3390/ijms14023158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Srivastava R., Howell S. H., Bassham D. C. (2016). Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 85 83–95. 10.1111/tpj.13091 [DOI] [PubMed] [Google Scholar]
- Yin Y. X., Guo W. L., Zhang Y. L., Ji J. J., Xiao H. J., Yan F., et al. (2014). Cloning and characterisation of a pepper aquaporin, CaAQP, which reduces chilling stress in transgenic tobacco plants. Plant Cell Tissue Organ. Cult. 118 431–444. 10.1007/s11240-014-0495-3 [DOI] [Google Scholar]
- Zhang L., Zhao H. K., Dong Q. L., Zhang Y. Y., Wang Y. M., Li H. Y., et al. (2015). Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 6:773 10.3339/fpls.2015.00773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Hao P., Chen G., Han C., Li X., Zeller F. J., et al. (2014). Molecular cloning, phylogenetic analysis, and expression profiling of endoplasmic reticulum molecular chaperone BiP genes from bread wheat (Triticum aestivum L.). BMC Plant Biol. 14:260 10.1186/s12870-014-0260-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of CaBiP1-silenced pepper seedlings and CaBiP1-overexpressing Arabidopsis lines. (A) Phenotypes of CaBiP1-silenced pepper seedlings. CaPDS, phytoene desaturase gene for chlorophyll synthesis; TRV2:00, control plants harboring the empty TRV2 vector. (B) Measurement of the silencing efficiency for CaBiP1 expression in silenced pepper seedlings at the 30th day after inoculation. (C) Screening of CaBiP1-overexpressing Arabidopsis lines in MS medium containing kanamycin. (D) Measurement of CaBiP1 expression levels in CaBiP1-overexpressing Arabidopsis lines by semi-quantitative PCR. The AtActin2 gene was used as the internal control. Error bars represent standard deviation from three biological replicates.
Enhanced tolerance to ER stress induced by DTT in CaBiP1-overexpressing Arabidsopsis lines. (A) Germination rate on MS medium with 3 mM DTT. (B) Fresh weight of 5-day-old CaBiP1-OE Arabidsopsis seedlings grown on MS medium with 2 mM DTT for 15 days. Data represent the mean ± standard deviation from three biological replicates. Statistical significance is indicated as by a single asterisk (p < 0.05) and double asterisk (p < 0.01) based on the results of the Student’s t-test.
Seed germination rate in WT and CaBiP1-OE (OE7 and OE8) Arabidsopsis lines grown on MS medium with 100 mM NaCl. Data represent the mean ± standard deviation from three biological replicates. Statistical significance is indicated by a single asterisk (p < 0.05) based on the results of the Student’s t-test.