Abstract
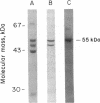
A critical step in the import of nuclear-encoded precursor proteins into mitochondria involves proteolytic cleavage of their amino-terminal leader peptides by processing proteases found in the mitochondrial matrix. We report here the characterization of the general matrix processing protease from rat liver mitochondria. The final enzyme preparation consisted of two polypeptides, a catalytically active 55-kDa subunit and a 52-kDa one. To deduce the complete primary structure of the 55-kDa subunit, we first sequenced its mature amino terminus and several tryptic peptides derived from the pure protein. Next, using mixed oligonucleotide primers that had sequences based on two of these peptides, we synthesized a partial cDNA probe by selective amplification of liver RNA with the polymerase chain reaction. The amplified probe was then used to obtain a nearly full-length clone from a rat liver cDNA library. This cDNA codes for 508 amino acid residues, including 16 residues of an amino-terminal leader peptide, the cleavage site of which is located two polypeptide bonds downstream from an arginine residue. The mature portion has a predicted molecular mass of 55.2 kDa; it shows 36% identity with the mitochondrial processing peptidases of Saccharomyces cerevisiae and Neurospora crassa. A conserved structural feature is a putative, negatively charged alpha-helix, located in the amino-terminal half of the subunit; this element might be important for the recognition of positively charged leader peptides characteristic of mitochondrial precursor proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Attardi G., Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Böhni P. C., Daum G., Schatz G. Import of proteins into mitochondria. Partial purification of a matrix-located protease involved in cleavage of mitochondrial precursor polypeptides. J Biol Chem. 1983 Apr 25;258(8):4937–4943. [PubMed] [Google Scholar]
- Cheng M. Y., Hartl F. U., Martin J., Pollock R. A., Kalousek F., Neupert W., Hallberg E. M., Hallberg R. L., Horwich A. L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 1989 Feb 16;337(6208):620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cornette J. L., Cease K. B., Margalit H., Spouge J. L., Berzofsky J. A., DeLisi C. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987 Jun 5;195(3):659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schatz G. Protein unfolding and the energetics of protein translocation across biological membranes. Cell. 1988 Feb 26;52(4):481–483. doi: 10.1016/0092-8674(88)90458-8. [DOI] [PubMed] [Google Scholar]
- Evans E. A., Gilmore R., Blobel G. Purification of microsomal signal peptidase as a complex. Proc Natl Acad Sci U S A. 1986 Feb;83(3):581–585. doi: 10.1073/pnas.83.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Daum G., Schatz G. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J Biol Chem. 1982 Nov 10;257(21):13034–13041. [PubMed] [Google Scholar]
- Hartl F. U., Pfanner N., Nicholson D. W., Neupert W. Mitochondrial protein import. Biochim Biophys Acta. 1989 Jan 18;988(1):1–45. doi: 10.1016/0304-4157(89)90002-6. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Schmidt B., Wachter E., Weiss H., Neupert W. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 1986 Dec 26;47(6):939–951. doi: 10.1016/0092-8674(86)90809-3. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G., Schneider H., Schmidt B., Tropschug M., Hartl F. U., Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988 Jun 3;53(5):795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hodges P. E., Rosenberg L. E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich A. L., Kalousek F., Mellman I., Rosenberg L. E. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J. 1985 May;4(5):1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E. C., Pesold-Hurt B., Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984 Dec 10;178(2):306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- Isaya G., Fenton W. A., Hendrick J. P., Furtak K., Kalousek F., Rosenberg L. E. Mitochondrial import and processing of mutant human ornithine transcarbamylase precursors in cultured cells. Mol Cell Biol. 1988 Dec;8(12):5150–5158. doi: 10.1128/mcb.8.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. E., Yaffe M. P. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988 Dec 1;7(12):3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek F., Darigo M. D., Rosenberg L. E. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem. 1980 Jan 10;255(1):60–65. [PubMed] [Google Scholar]
- Kalousek F., Hendrick J. P., Rosenberg L. E. Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7536–7540. doi: 10.1073/pnas.85.20.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaput J., Goltz S., Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor. Functional implications of the pre sequence for protein transport into mitochondria. J Biol Chem. 1982 Dec 25;257(24):15054–15058. [PubMed] [Google Scholar]
- Kolansky D. M., Conboy J. G., Fenton W. A., Rosenberg L. E. Energy-dependent translocation of the precursor of ornithine transcarbamylase by isolated rat liver mitochondria. J Biol Chem. 1982 Jul 25;257(14):8467–8471. [PubMed] [Google Scholar]
- Lee C. C., Wu X. W., Gibbs R. A., Cook R. G., Muzny D. M., Caskey C. T. Generation of cDNA probes directed by amino acid sequence: cloning of urate oxidase. Science. 1988 Mar 11;239(4845):1288–1291. doi: 10.1126/science.3344434. [DOI] [PubMed] [Google Scholar]
- Ou W. J., Ito A., Okazaki H., Omura T. Purification and characterization of a processing protease from rat liver mitochondria. EMBO J. 1989 Sep;8(9):2605–2612. doi: 10.1002/j.1460-2075.1989.tb08400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R. A., Hartl F. U., Cheng M. Y., Ostermann J., Horwich A., Neupert W. The processing peptidase of yeast mitochondria: the two co-operating components MPP and PEP are structurally related. EMBO J. 1988 Nov;7(11):3493–3500. doi: 10.1002/j.1460-2075.1988.tb03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn O. B. Statistical analysis of the distribution of amino acid residues among helical and non-helical regions in globular proteins. J Mol Biol. 1969 Jun 28;42(3):501–510. doi: 10.1016/0022-2836(69)90238-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer M., Schmidt B., Neupert W. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. Eur J Biochem. 1982 Jun 15;125(1):109–116. doi: 10.1111/j.1432-1033.1982.tb06657.x. [DOI] [PubMed] [Google Scholar]
- Schneider C., Newman R. A., Sutherland D. R., Asser U., Greaves M. F. A one-step purification of membrane proteins using a high efficiency immunomatrix. J Biol Chem. 1982 Sep 25;257(18):10766–10769. [PubMed] [Google Scholar]
- Schneider H., Arretz M., Wachter E., Neupert W. Matrix processing peptidase of mitochondria. Structure-function relationships. J Biol Chem. 1990 Jun 15;265(17):9881–9887. [PubMed] [Google Scholar]
- Sztul E. S., Hendrick J. P., Kraus J. P., Wall D., Kalousek F., Rosenberg L. E. Import of rat ornithine transcarbamylase precursor into mitochondria: two-step processing of the leader peptide. J Cell Biol. 1987 Dec;105(6 Pt 1):2631–2639. doi: 10.1083/jcb.105.6.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989 Dec 22;59(6):1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Perz A., Sebald W. The imported preprotein of the proteolipid subunit of the mitochondrial ATP synthase from Neurospora crassa. Molecular cloning and sequencing of the mRNA. EMBO J. 1982;1(5):565–571. doi: 10.1002/j.1460-2075.1982.tb01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M. P., Ohta S., Schatz G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 1985 Aug;4(8):2069–2074. doi: 10.1002/j.1460-2075.1985.tb03893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Jensen R. E., Yaffe M. P., Oppliger W., Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988 Dec 1;7(12):3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwizinski C., Neupert W. Precursor proteins are transported into mitochondria in the absence of proteolytic cleavage of the additional sequences. J Biol Chem. 1983 Nov 10;258(21):13340–13346. [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]