ABSTRACT
Despite the introduction of effective drugs to treat patients with chronic hepatitis C virus (HCV) infection, a vaccine would be the only means to substantially reduce the worldwide disease burden. An incomplete understanding of how HCV interacts with its human host and evades immune surveillance has hampered vaccine development. It is generally accepted that in infected individuals, a narrow repertoire of exhausted T cells is a hallmark of persistent infection, whereas broad, vigorous CD4+ and CD8+ T cell responses are associated with control of acute hepatitis C. We employed a vaccine approach based on a mixture of peptides (pepmix) spanning the entire sequence of HCV nonstructural protein 3 (NS3) in cross-priming cationic liposomes (CAF09) to facilitate a versatile presentation of all possible T cell epitopes, regardless of the HLA background of the vaccine recipient. Here, we demonstrate that vaccination of mice with NS3 pepmix broadens the repertoire of epitope-specific T cells compared to the corresponding recombinant protein (rNS3). Moreover, vaccination with rNS3 induced only CD4+ T cells, whereas the NS3 pepmix induced a far more vigorous CD4+ T cell response and was as potent a CD8+ T cell inducer as an adenovirus-vectored vaccine expressing NS3. Importantly, the cellular responses are dominated by multifunctional T cells, such as gamma interferon-positive (IFN-γ+) tumor necrosis factor alpha-positive (TNF-α+) coproducers, and displayed cytotoxic capacity in mice. In conclusion, we present a novel vaccine approach against HCV, inducing a broadened T cell response targeting both immunodominant and potential subdominant epitopes, which may be key elements to counter T cell exhaustion and prevent chronicity.
IMPORTANCE With at least 700,000 annual deaths, development of a vaccine against hepatitis C virus (HCV) has high priority, but the tremendous ability of the virus to dodge the human immune system poses great challenges. Furthermore, many chronic infections, including HCV infection, have a remarkable ability to drive initially strong CD4+ and CD8+ T cell responses against dominant epitopes toward an exhausted, dysfunctional state. Thus, new and innovative vaccine approaches to control HCV should be evaluated. Here, we report on a novel peptide-based nanoparticle vaccine strategy (NS3 pepmix) aimed at generating T cell immunity against potential subdominant T cell epitopes that are not efficiently targeted by vaccination with full-length recombinant protein (rNS3) or infection with HCV. As proof of concept, we found that NS3 pepmix excels in broadening the repertoire of epitope-specific, multifunctional, and cytotoxic CD4+ and CD8+ T cells compared to vaccination with rNS3, which generated only CD4+ T cell responses.
KEYWORDS: hepaciviruses, HCV, hepatitis, vaccine, cellular immunity, hepatitis C virus
INTRODUCTION
Hepatitis C virus (HCV) is a major health problem worldwide, with more than 170 million persistently infected people. Persistent HCV infection can lead to chronic inflammation of the liver, cirrhosis, liver failure, and liver cancer, ultimately causing 700,000 annual HCV-related deaths (1). Despite the recent breakthrough in HCV therapy with the introduction of direct-acting antivirals (DAAs) resulting in viral clearance rates above 90%, the impact of these drugs on the global disease burden remains unknown, as high prices and development of viral resistance represent important challenges to current treatment strategies (2–4). Furthermore, successful treatment with DAAs does not prevent HCV reinfection, which is an issue of concern in highly exposed patient groups, such as intravenous drug users (5, 6). Thus, development of a vaccine to prevent or potentially help resolve a chronic infection would have a tremendous impact on the HCV epidemic worldwide.
HCV is spontaneously controlled in a proportion of those infected, and several studies have demonstrated an important role for T cells in protection against HCV, fueling the hope that development of an effective vaccine is an achievable goal. A strong and broad CD8+ T cell response targeting multiple epitopes within the nonstructural proteins can be detected in the early phase of infection in patients who manage to combat the virus and control infection (7–10). Moreover, a vigorous, broad, and sustained CD4+ T cell response accounts for the induction of a strong gamma interferon (IFN-γ) response with a direct antiviral effect, as well as the capacity to help induce and maintain multifunctional CD8+ T cells. This results in rapid control of HCV and potentially a self-limiting course of infection (11–14). In addition, the importance of a sustained CD4+ T cell response is highlighted by studies of experimentally infected chimpanzees showing that CD4+ T cell depletion immediately before reinfection results in incomplete HCV resolution, even though HCV-specific CD8+ T cells have been developed during the primary infection (15). Thus, the induction of both CD8+ and CD4+ T cell responses is very important for effective control of HCV.
Most peptide-based vaccines aimed at inducing T cell responses target one or a few epitopes strongly recognized during natural infection or in silico-predicted HLA-restricted epitopes. However, this strategy has proven inadequate in chronic viral infections, where, in particular, broader T cell responses targeting multiple epitopes confer better protection against viral replication (16–19). The superior efficacy of a broad T cell response could be partially ascribed to blocking of viral immune evasion, as mutational escape from T cells targeting a particular epitope can still involve susceptibility to other epitope-specific T cells (20–22). By mechanisms still poorly understood, a hierarchy of T cell responses targeting various epitopes is established after natural processing and presentation of antigens on antigen-presenting cells (APCs). Immunodominant epitopes are epitopes targeted following the processing of whole protein derived from vaccination or natural infection (23). Responses against subdominant epitopes, on the other hand, are not promoted after vaccination with whole protein or during infection but can be induced by vaccination with, e.g., peptides, truncated proteins, or types of antigens from which dominant epitopes have been removed (23–25). Nevertheless, responses against these pathogen-derived subdominant epitopes can be pursued to exert functional responses that target infectious agents or cancer cells expressing the protein in question.
Narrow T cell repertoires targeting relatively few immunodominant viral epitopes are observed in persistently infected individuals (26, 27), and even strong initial responses against such epitopes tend to wane over time, as continued antigen stimulation may drive polyfunctional T cells toward an exhausted, dysfunctional state (28–31). Lately, it has been speculated that a vaccine-induced response against subdominant epitopes can offset T cell exhaustion by refocusing the T cell response against epitopes that are not heavily targeted during a chronic infection. Recent studies have shown that it is indeed possible to induce functional responses against subdominant epitopes in several systems (25, 32–34).
In an attempt to broaden the CD4+ and CD8+ T cell repertoire to include responses against multiple HCV epitopes, including both dominant and subdominant epitopes, we utilized a mixture of 62 overlapping 20-mer peptides (pepmix) covering the amino acid sequence of the relatively conserved nonstructural protein 3 (NS3). To promote uptake by APCs and presentation on both major histocompatibility complex class I (MHC-I) and MHC-II, a peptide-based vaccine should be formulated in a suitable adjuvant. Extracellular antigens taken up by APCs require cross presentation on MHC-I to induce CD8+ T cell responses. This process, which directs antigens from the endosomal to the cytosolic compartment, can be enhanced in a type I IFN-dependent manner by stimulation with the Toll-like receptor 3 (TLR-3) ligand polyinosinic-poly(C) [poly(I·C)] (35–37). The CAF01 adjuvant, with a good safety record and proven to induce potent T helper type 1 (TH1)/interleukin 17 (IL-17)-inducing responses in humans (38, 39), has recently been refined to enhance both CD4+ and CD8+ T cell responses in the cationic liposomal formulation CAF09, consisting of dimethyldioctadecylammonium bromide (DDA), monomycolyl glycerol (MMG), and poly(I·C) (40).
Here, we find that HCV NS3 pepmix formulated in CAF09 is able to broaden the T cell repertoire by inducing responses against several epitopes not induced by immunization with recombinant NS3 protein (rNS3). Furthermore, NS3 pepmix excels in its ability to generate both robust CD4+ and CD8+ T cell responses, whereas rNS3 and an adenovirus expressing NS3 (AdNS3) are capable of generating solely CD4+ or CD8+ T cell responses, respectively. Importantly, we find that NS3 pepmix induces robust, polyfunctional T cell responses where not only CD8+ but also CD4+ T cells are capable of killing antigen-expressing target cells in vivo.
RESULTS
Vaccination with HCV NS3 pepmix induces multifunctional CD4+ and CD8+ T cells.
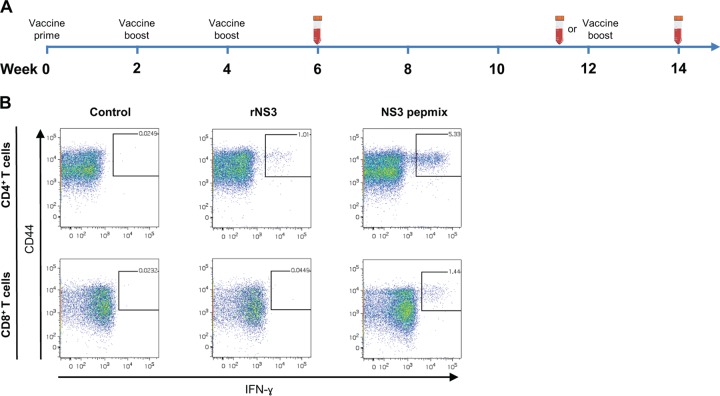
To compare vaccination with recombinant HCV protein to a strategy based on vaccination with a mixture of overlapping peptides spanning the corresponding identical amino acid sequence, CB6F1 mice were vaccinated three times at 2-week intervals with HCV rNS3 or NS3 pepmix, both formulated in the cationic adjuvant CAF09. We performed a series of experiments to assess the vaccine-induced T cell responses at various time points, as indicated in Fig. 1A. At week 6, splenocytes were isolated and restimulated with a pool of NS3 peptides (the 62 peptides spanning the sequence of the HCV NS3 protein), and the magnitude of the T cell response was assessed by intracellular (IC) flow cytometry. Figure 1B shows representative CD4+ (top row) and CD8+ (bottom row) T cell responses in control mice and mice vaccinated with rNS3 or NS3 pepmix. We found that vaccination with both rNS3 and NS3 pepmix induced robust antigen-specific CD4+ T cell responses (Fig. 1B, top row, and 2A, bottom). However, the frequencies of IFN-γ-, tumor necrosis factor alpha (TNF-α)-, and IL-2-producing CD4+ T cells were significantly higher in mice vaccinated with NS3 pepmix (3.7% ± 0.6%, 4.3% ± 0.5%, and 0.8% ± 0.1%, respectively) than in mice vaccinated with rNS3 (0.8% ± 0.1%, 1.6% ± 0.1%, and 0.3% ± 0.02%, respectively) (Fig. 2A, bottom). Vaccination with rNS3 did not generate CD8+ T cell responses, whereas NS3 pepmix induced a significant number of antigen-specific CD8+ T cells capable of producing IFN-γ (0.9% ± 0.3%), TNF-α (0.7% ± 0.2%), and IL-2 (0.2% ± 0.02%) (Fig. 1B, bottom row, and 2B, bottom). It is noteworthy that even a 10-fold reduction of the antigen amount in the NS3 pepmix did not alter the immunological outcome (data not shown). Thus, vaccination with NS3 pepmix not only yields a more vigorous cellular response than vaccination with rNS3, but also facilitates priming of both CD4+ and CD8+ T cells, which is critical for effective control of HCV.
FIG 1.
(A) Overview of vaccinations and gating strategy for flow cytometry data. The diagram indicates the time points for vaccination and/or bleeding of mice. (B) Vaccines were administered three times at 2-week intervals (weeks 0, 2, and 4), and immune responses were assessed at week 6, 12, or 14. In the last case, mice received a third booster vaccination at week 12. Representative FACS plots indicate gating on the IFN-γ-producing population of CD44+ CD4+ T cells (top) or CD44+ CD8+ T cells (bottom) from mice at week 6 after the priming vaccination with rNS3 or NS3 pepmix.
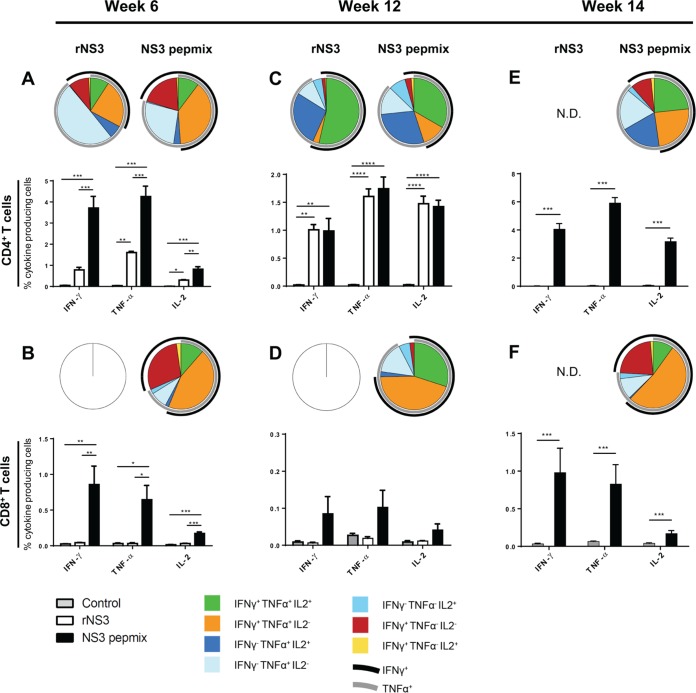
FIG 2.
Frequency and functional characterization of antigen-specific cytokine-producing T cells after vaccination with HCV rNS3 and NS3 pepmix. CB6F1 mice were vaccinated three times at 2-week intervals with rNS3 or NS3 pepmix formulated in CAF09, as indicated. Splenocytes isolated from individual mice were restimulated with a pool of 62 NS3 peptides and analyzed by multiparameter flow cytometry at various time points. The bar charts show frequencies of NS3-specific IFN-γ-, TNF-α-, and IL-2-producing CD44+ CD4+ T cells (A, C, and E) or CD44+ CD8+ T cells (B, D, and F) out of the total CD4+ or CD8+ T cell population at week 6 (A and B) (n = 4) and week 12 (C and D) (n = 4) after the priming vaccination. (E and F) In a parallel experiment, a third boost was administered to NS3 pepmix-vaccinated mice at week 12, and T cell responses were assessed 14 weeks after the priming vaccination (rNS3 was not done [N.D.]; n = 8). The data are shown as means and standard errors of the mean (SEM). The pie charts show cytokine-producing CD44+ CD4+ T cells or CD44+ CD8+ T cells divided into seven distinct subpopulations based on their ability to coproduce IFN-γ, TNF-α, and IL-2 in any combination. The black arcs indicate IFN-γ-producing T cell subsets, and the gray arcs indicate TNF-α-producing T cell subsets. The data shown are representative of two independent experiments. *, P < 0.05; **, P, < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Protective responses against HCV depend not only on the magnitude of the response, but also on the quality of the T cell response. In particular, effector memory T cells (TEM) with the ability to coproduce IFN-γ and TNF-α show enhanced killing of intracellular pathogens (7, 28, 41). We therefore analyzed the composition of multifunctional T cells contributing to the overall immune response based on their ability to produce IFN-γ, TNF-α, and IL-2. Vaccination with NS3 pepmix clearly induced an effector-like response dominated by CD4+ and CD8+ T cells capable of producing IFN-γ and TNF-α simultaneously (Fig. 2A and B, indicated with overlapping black/gray arcs on the pie charts). In contrast, vaccination with rNS3 skewed CD4+ T cells toward a cytokine profile associated with a less differentiated response dominated by TNF-α+ single positives and with a smaller proportion of IFN-γ+ TNF-α+ double positives (41).
A vaccine targeting HCV and other agents causing chronic infections should not only be capable of inducing a vigorous effector T cell response, but also avoid loss of functionality over time. We therefore evaluated the immune responses at week 12 and found highly significant CD4+ T cell responses at comparable levels in rNS3- and NS3 pepmix-vaccinated mice at this later stage (Fig. 2C, bottom). With the lack of continued antigen exposure, the surviving CD4+ T cells after the contraction phase were mainly found to be IL-2+ producers, either as triple-positive or as TNF-α+ IL-2+ double-positive T cells (Fig. 2C, top). At this time point, the rather minute CD8+ T cell response mainly consisted of triple-positive or IFN-γ+ TNF-α+ double-positive T cells (Fig. 2D).
Finally, to examine the ability of the NS3 pepmix-induced response to expand and to reestablish its initial effector-like profile upon antigen stimulation, we boosted NS3 pepmix-vaccinated mice in a parallel experiment at week 12 and evaluated the immune response at week 14. We found that the composition of multifunctional CD4+ and CD8+ T cells indeed resembled that observed at week 6 (Fig. 2E and F, top) and that the magnitudes of cytokine responses were at least as high as or higher than those observed at earlier time points (Fig. 2E and F, bottom).
HCV NS3 pepmix immunization induces broad cellular responses.
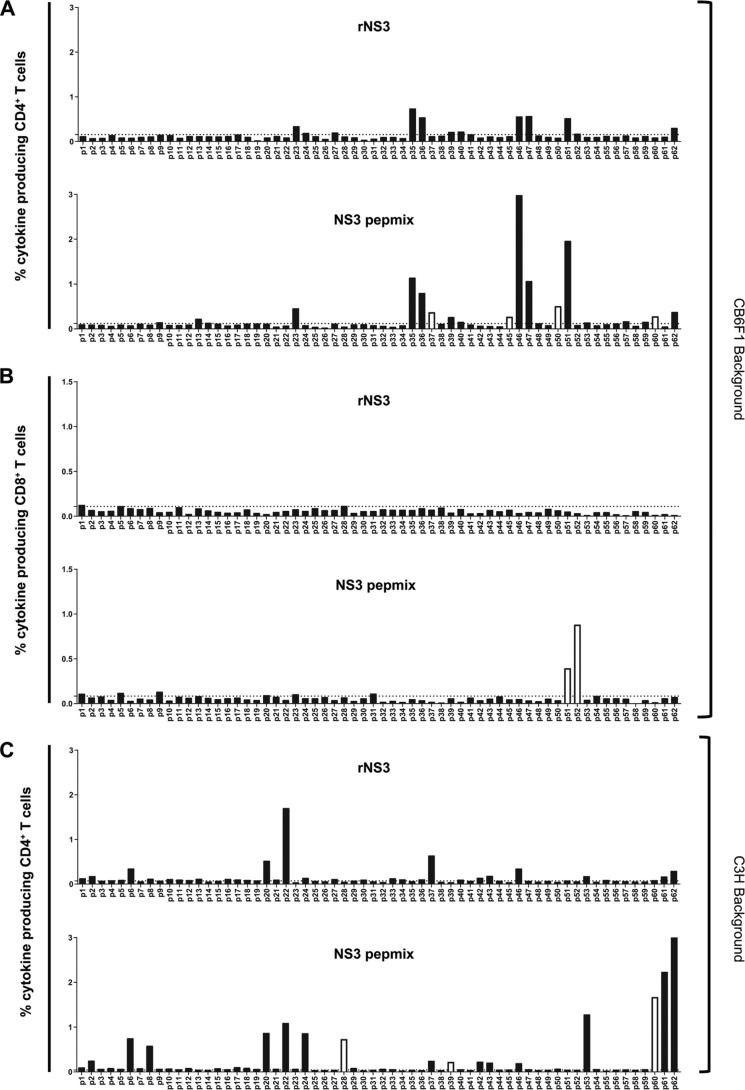
Protein and peptides may be processed very differently once the vaccine is taken up by APCs (42, 43), and we therefore speculated that NS3 pepmix could induce T cell responses with distinct epitope specificity or with the capacity to broaden the response relative to that with rNS3 vaccination. To investigate the epitope patterns induced by rNS3 and NS3 pepmix, splenocytes isolated from vaccinated CB6F1 mice at week 6 were pooled in their respective groups and subsequently restimulated ex vivo in separate wells with 62 individual peptides spanning the complete HCV NS3 sequence. The vaccine-induced cellular response was measured by IC flow cytometry. We found that both rNS3 (Fig. 3A, top) and NS3 pepmix (Fig. 3A, bottom) induced CD4+ T cell responses against epitopes within peptides p23, p35, p36, p39, p40, p46, p47, p51, and p62. However, NS3 pepmix induced responses against additional epitopes not targeted following rNS3 vaccination. These epitopes were located in peptides p37, p45, p50, and p60. In parallel with IC flow cytometry, IFN-γ enzyme-linked immunosorbent assay (ELISA) confirmed these findings and, in addition, revealed minute IFN-γ responses against peptides p29 and p32 in both vaccine groups and a p58-specific response in NS3 pepmix-vaccinated mice (data not shown; summarized in Table 1). Furthermore, we found CD8+ T cells induced by NS3 pepmix to be specific for epitopes present in peptides p51 and p52 (Fig. 3B, bottom).
FIG 3.
Epitope mapping of the antigen-specific T cell repertoire after vaccination with rNS3 or NS3 pepmix. CB6F1 and C3H mice were vaccinated three times at 2-week intervals with rNS3 or NS3 pepmix formulated in CAF09, as indicated. Splenocytes from four mice from each vaccine group were isolated and pooled 6 weeks after the priming vaccination. The cells were restimulated with each of the 62 individual peptides spanning the entire NS3 sequence. The peptide identifiers (ID) are indicated on the x axis. The bars indicate the total frequency of epitope-specific CD44+ CD4+ T cells (A) or CD44+ CD8+ T cells (B) in CB6F1 mice with the ability to produce IFN-γ, TNF-α, or IL-2 in any combination. (C) Similarly, the frequencies of cytokine-producing CD44+ CD4+ T cells are shown in a C3H background (no epitope-specific cytokine-producing CD44+ CD8+ T cells were detected). Open bars, cytokine responses observed only in mice vaccinated with NS3 pepmix; dotted lines, medium background. The data show responses from mice pooled in groups (n = 4) and are representative of two independent experiments.
TABLE 1.
Overview of the epitope-specific T cell repertoire in CB6F1 mice assessed by IFN-γ ELISA and IC flow cytometrya
| Peptide no. | Amino acid sequence | rNS3 |
NS3 pepmix |
||||
|---|---|---|---|---|---|---|---|
| IFN-γ ELISA | CD4+ | CD8+ | IFN-γ ELISA | CD4+ | CD8+ | ||
| p23 | QGYKVLVLNPSVAATLGFGA | + | + | − | + | + | − |
| p29 | GGAYDIIICDECHSTDSTTI | + | − | − | + | − | − |
| p32 | ETAGARLVVLATATPPGSVT | + | − | − | + | − | − |
| p35 | LSNNGEIPFYGKAIPIEAIK | + | + | − | + | + | − |
| p36 | GKAIPIEAIKGGRHLIFCHS | + | + | − | + | + | − |
| p37b | GGRHLIFCHSKKKCDELAAK | − | − | − | + | + | − |
| p39 | LTGLGLNAVAYYRGLDVSVI | − | + | − | + | + | − |
| p40 | YYRGLDVSVIPPIGDVVVVA | − | + | − | + | + | − |
| p45b | DPTFTIETTTVPQDAVSRSQ | − | − | − | − | + | − |
| p46 | VPQDAVSRSQRRGRTGRGRS | + | + | − | + | + | − |
| p47 | RRGRTGRGRSGIYRFVTPGE | + | + | − | + | + | − |
| p50b | LCECYDAGCAWYELTPAETS | − | − | − | + | + | − |
| p51c | WYELTPAETSVRLRAYLNTP | + | + | − | + | + | + |
| p52c | VRLRAYLNTPGLPVCQDHLE | − | − | − | + | − | + |
| p58b | AQAPPPSWDQMWKCLIRLKP | − | − | − | + | − | − |
| p60b | TLHGPTPLLYRLGAVQNEVI | − | − | − | + | + | − |
| p62 | LTHPITKYIMACMSADLEVVT | + | + | − | + | + | − |
| Total no. positive | 9 | 9 | 0 | 16 | 13 | 2 | |
Peptide-specific recall responses were determined by IFN-γ ELISA or intracellular expression of IFN-γ, TNF-α, or IL-2 by CD4+ and CD8+ T cells assessed by flow cytometry. +, positive; −, negative.
Peptide sequence with epitopes recognized only in NS3 pepmix-vaccinated mice.
Peptide sequence with CD8+ T cell epitopes recognized only after NS3 pepmix vaccination.
In silico prediction indicated the presence of distinct CD8+ T cell epitopes not shared by the peptide p51 and p52 amino acid sequences (Table 2). To assess any possible cross-reactivity between shared amino acid sequences within these peptides experimentally, mice were vaccinated with individual p51 or p52 peptides formulated in CAF09. Vaccination with p51 alone induced a p51-specific CD8+ T cell response that could not be obtained by p52 restimulation (and vice versa), suggesting that there is no cross-reactivity between p51 and p52 (data not shown).
TABLE 2.
In silico prediction of CD8+ T cell epitopes in peptides p51 and p52 in CB6F1 micea
| Peptideb | Epitope sequence | Allele | MHC IC50 (nM) |
|---|---|---|---|
| p51 | SVRLRAYL | H-2b | 344.7 |
| p51 | WYELTPAETSV | H-2d | 537.7 |
| p52 | RAYLNTPGL | H-2b | 68.8 |
| p52 | AYLNTPGL | H-2d | 122.0 |
In silico prediction of strong CD8+ T cell epitopes defined by their 50% inhibitory concentration (IC50) values were computed for H-2b and H-2d backgrounds by use of the IEDB Analysis Resource NetMHCpan algorithm.
Peptide sequences were as follows (identical residues are in italics): p51, WYELTPAETSVRLRAYLNTP; p52, VRLRAYLNTPGLPVCQDHLE.
The expanded repertoire of epitope-specific T cells obtained by vaccination with NS3 pepmix demonstrates the existence of additional CD4+ and CD8+ T cell responses (summarized in Table 1). As the patterns of epitope-specific T cells induced by vaccination differ depending on the genetic background of the recipient, we assessed whether NS3 pepmix would also induce a broadened response in a mouse strain different from CB6F1. In an experiment similar to that described above, immunization of C3H mice confirmed that NS3 pepmix induced a broadened CD4+ T cell response compared to rNS3 vaccination (Fig. 3C). Neither rNS3 nor NS3 pepmix induced any CD8+ T cell response in this mouse strain (data not shown). However, it should be noted that T cell epitopes are restricted to the unique haplotype of inbred homozygous CB6F1 and C3H mice. Thus, the genetic diversity of MHC molecules in humans allowing additional dominant, as well as subdominant, T cell responses is not reflected in the inbred mouse strains used here, which could explain the absence of CD8+ T cell responses. Importantly, the specific epitopes recognized differed in the two mouse strains, which underlines the strength of employing a panel of overlapping peptides to ensure a broader presentation of all possible T cell epitopes in any given genetic background rather than vaccination with preselected peptides or full-length protein.
NS3 pepmix induces CD8+ T cell responses comparable to those induced by an adenoviral vectored NS3 vaccine.
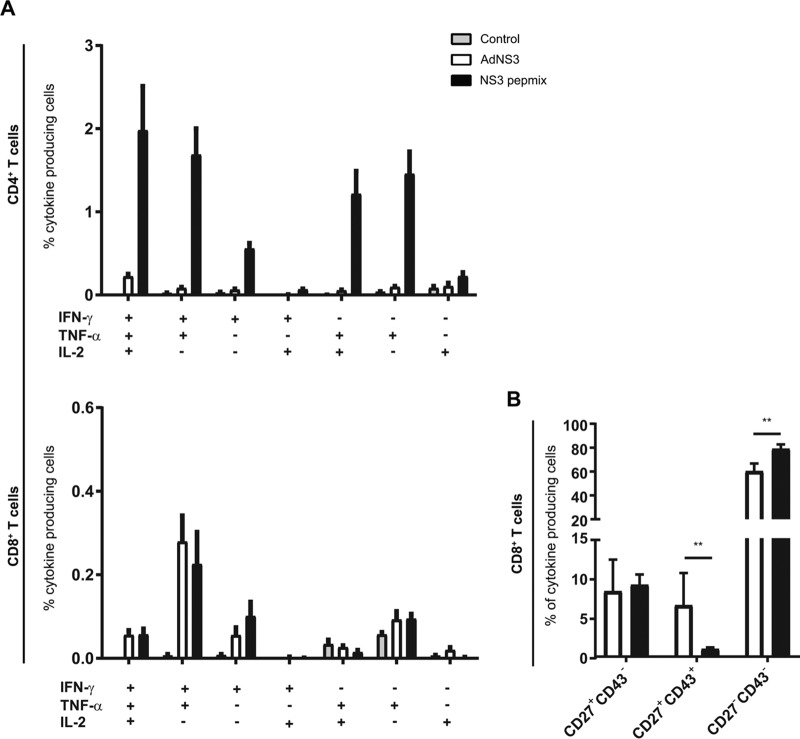
Generation of cytotoxic T lymphocyte (CTL) responses is regarded as particularly crucial for controlling cancer and viral and other intracellular infections. Adenoviral vectors are generally less proficient CD4+ T cell inducers but generate superior CD8+ T cell responses and have been widely used as platforms for vaccine development targeting cancer and viral infections (44). To assess the potential of NS3 pepmix to generate CD8+ T cell responses by cross-presentation compared to adenoviral vectors, we employed an adenoviral-vector-based vaccine, AdNS3, expressing NS3 with a sequence identical to that of NS3 pepmix and administered it in its optimal dose and regimen to CB6F1 mice (45). Vaccination with AdNS3 did not induce any significant CD4+ T cell response, while NS3 pepmix induced robust CD4+ T cell responses producing IFN-γ, TNF-α, or IL-2 at week 6 (Fig. 4A, top). The two vaccines induced significant CD8+ T cell responses at comparable levels, mainly consisting of T cells with the capacity to coproduce IFN-γ and TNF-α (Fig. 4A, bottom left). Based on the expression of CD27 and CD43, we divided the cytokine-producing CD8+ T cell population into three subpopulations to assess their activation and differentiation status (46, 47) (Fig. 4B). The proportions of CD27hi CD43lo long-lived central memory phenotypes were similar in AdNS3- and NS3 pepmix-vaccinated mice (8.3% ± 1.7% versus 9.1% ± 0.5%). Conversely, AdNS3 induced a higher proportion of CD27hi CD43hi CD8+ T cells (6.5% ± 1.7% versus 1.0% ± 0.1%), which are reported to display higher proliferative capacity (47) but in turn do not induce as strong recall responses as the long-lived effector-like CD27lo CD43lo phenotypes, which are favored by NS3 pepmix vaccination (78.9% ± 1.6%) compared to AdNS3-vaccinated mice (60.3% ± 2.6%).
FIG 4.
Vaccine-induced antigen-specific cytokine profile after vaccination with AdNS3 or NS3 pepmix. (A) CB6F1 mice were vaccinated three times at 2-week intervals with NS3 pepmix formulated in CAF09 (black bars) or once with AdNS3 at week 4 (white bars). Splenocytes were isolated 12 days after the final vaccination and restimulated with a pool of all 62 NS3 peptides. Surface markers and intracellular-cytokine production by antigen-specific T cells were determined by flow cytometry. The bars indicate the frequencies of seven distinct subpopulations of CD44+ CD4+ T cells (top) and CD44+ CD8+ T cells (bottom) based on their ability to produce IFN-γ, TNF-α, or IL-2 in any combination. (B) Cytokine-producing CD8+ T cells were further divided into three populations based on their expression of CD27 and CD43. The data are shown as means and SEM (n = 8 to 10). The plots are based on data from two independent experiments. **, P < 0.01.
Specific killing by cytotoxic CD8+ and CD4+ T cells.
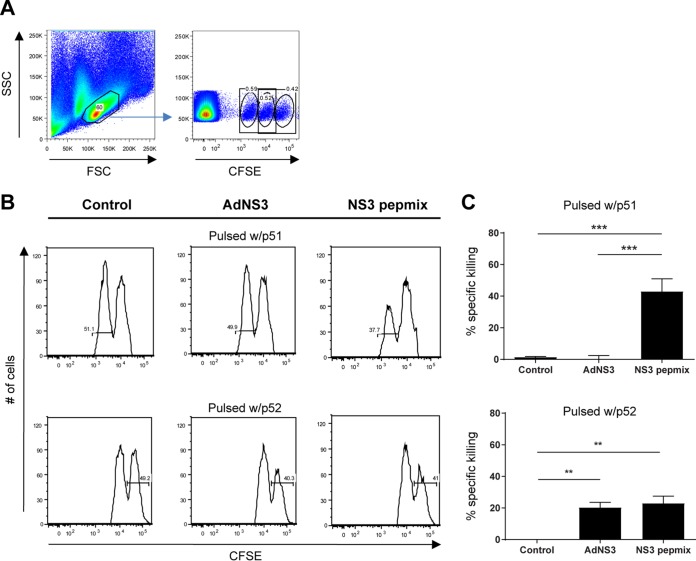
Prompt destruction of infected cells is a key trait associated with antiviral CD8+ T cells. As no robust small-animal HCV challenge model currently exists (48, 49), we employed an in vivo cytotoxicity assay to assess the killing capacity of the vaccine-induced T cells. It is conceivable that CD8+ T cell responses in AdNS3-immunized mice may be underestimated, as AdNS3-induced CD8+ T cells might target epitopes not facilitated by restimulation and processing of 20-mer peptides. This concern was addressed in our prior study, where peptides of various lengths based on in silico prediction of CD8+ T cell epitopes were used in restimulation assays (45). In line with the findings here, where we restimulated with peptides p51 and p52 (data not shown), only restimulation with peptide NS3514–531 (RAYLNTPGLPVCQDHLEF, contained in the sequence of p52), and not NS3500–510 (WYELTPAETSV, contained in the sequence of p51), resulted in a recall response in AdNS3-vaccinated C57BL/6 mice. Reactivity against NS3500–510 was, however, found in an in vivo cytotoxicity assay. Based on these data, we chose to pulse carboxyfluorescein succinimidyl ester (CFSE)-labeled target cells with the peptides containing these predicted CD8+ T cell epitopes (p51 or p52). The CFSE-labeled cells were injected into vaccinated CB6F1 mice at week 6 after initial priming. Splenocytes were retrieved 18 h postinjection, and p51- or p52-specific lysis was determined by measuring the frequency of the remaining target cells (Fig. 5A). Vaccination with AdNS3 and NS3 pepmix displayed comparable killing of target cells loaded with p52 (20.3% ± 3.3% and 23.0% ± 4.5%, respectively) (Fig. 5B and C, bottom). In contrast, killing of cells loaded with p51 was observed only in mice vaccinated with NS3 pepmix (42.9% ± 8.0%) (Fig. 5B and C, top). The lack of AdNS3-induced cytotoxicity against p51 could be explained by the different mouse strains used (CB6F1 versus C57BL/6).
FIG 5.
Cytotoxic potential of vaccine-induced CD8+ T cells. CB6F1 mice were vaccinated three times at 2-week intervals with NS3 pepmix formulated in CAF09 or once with AdNS3 at week 4. Ten days after the final vaccination, CFSE-labeled target cells from naive mice were pulsed with the single peptide p51 or p52 or left untouched and injected i.v. into vaccinated mice. Eighteen hours postinjection, splenocytes were isolated and analyzed by flow cytometry to determine killing of target cells. (A) Lymphocytes were gated based on their FSC versus SSC profiles (left), and the three target cell populations were identified based on their CFSE fluorescence (right). (B) Representative histograms showing specific lysis of CFSElow cells pulsed with p51 (top) and CFSEhigh cells pulsed with p52 (bottom) in vaccinated mice. (C) Specific percent killing of target cells pulsed with p51 (top) and p52 (bottom) within the two vaccination groups. The data show means and SEM (n = 5). The plots are representative of two independent experiments. **, P < 0.01; ***, P < 0.001.
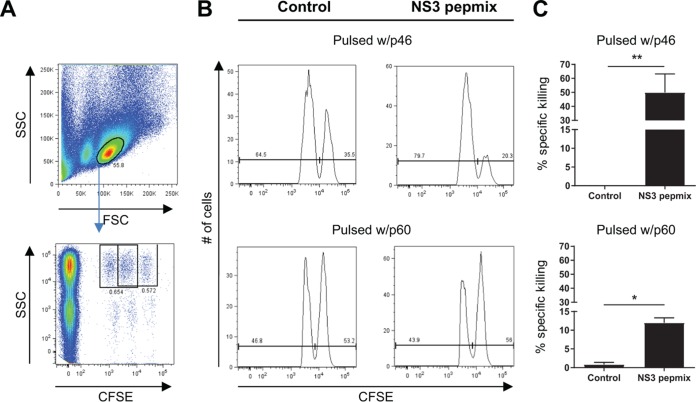
In addition to aiding the CD8+ T cell response and secreting antiviral cytokines, such as IFN-γ and TNF-α, recent studies suggest that granzyme B- and perforin-expressing CD4+ T cells with direct cytolytic activity play an important role in control of viral infections, such as human immunodeficiency virus (HIV), influenza virus, and HCV infections (50–53). We therefore investigated the cytotoxic potential of CD4+ T cells induced by NS3 pepmix by performing an in vivo cytotoxicity assay with target cells pulsed with selected peptides known to comprise CD4+, but not CD8+, T cell epitopes (Fig. 6). We found significant cytolytic activity against target cells displaying p46 (50.0% ± 13.2%) (Fig. 6B and C, top) or p60 (12.0% ± 1.3%) (Fig. 6B and C, bottom).
FIG 6.
CD4+ T cells contribute to vaccine-induced cytotoxicity. CB6F1 mice were vaccinated three times at 2-week intervals with NS3 pepmix formulated in CAF09, as indicated. Ten days after the final vaccination, CFSE-labeled target cells from naive mice were pulsed with the single peptide p46 or p60 or left untouched and injected i.v. into vaccinated mice. Eighteen hours postinjection, splenocytes were isolated and analyzed by flow cytometry to determine killing of target cells. (A) Lymphocytes were gated based on their FSC versus SSC profiles (top), and the target cell populations were identified based on their CFSE fluorescence and MHC-II expression (bottom). (B) Histograms showing specific lysis of CFSEhigh cells pulsed with p46 (top) and CFSElow cells pulsed with p60 (bottom) in vaccinated mice. (C) Specific percent killing of target cells pulsed with p46 (top) and p60 (bottom) within the two vaccination groups. The data show means and SEM (n = 6). *, P < 0.05; **, P < 0.01.
In summary, we have shown that vaccination with NS3 pepmix generates not only CD8+, but also CD4+ T cell responses with the capacity to eliminate cells by targeting multiple HCV epitopes.
DISCUSSION
The numerous ways in which HCV dodges the immune system have made vaccine development extremely challenging (54–57). Thus, the design of an effective vaccine should reflect the need for both CD4+ and CD8+ T cell responses for effective control of HCV replication observed in patients (11, 58). Here, we report on a novel HCV vaccine platform, NS3 pepmix, based on a library of overlapping 20-mer peptides spanning the entire sequence of the NS3 protein and delivered in a novel liposomal adjuvant (CAF09) with the capacity to induce both robust CD4+ and CD8+ T cell responses in mice. In contrast, vaccination with the corresponding full-length protein induced a CD4+ T cell response but no CD8+ T cells, whereas an adenoviral construct expressing NS3 induced only CD8+ T cell responses. Although adenovirus-based vaccines can be significantly improved, e.g., by fusing NS3 to the MHC class II chaperone protein invariant chain to obtain a broadened and enhanced cellular response, the CD4+ T cell response is still relatively limited (45, 59). Recently, the induction of broad and multifunctional cellular responses targeting NS3-NS5B after immunization with the adenoviral vectors AdCh3NSmut and Ad6NSmut in healthy volunteers has been reported (60, 61). The protective efficacy of this promising vaccine candidate is currently being assessed in phase I/II trials.
Indeed, broad antigen-specific CD4+ and CD8+ T cell responses targeting multiple NS3-NS5 epitopes correlate with control of HCV infection in patients, whereas a narrow response is distinctive of chronic disease (13, 16, 62, 63). This is also seen during chronic HIV infection, where the repertoire of responding T cells has been found to condense into a few dominant epitopes (64). Adding to this, a strong initial response against dominant epitopes tends to wane over time, as continued antigen stimulation may drive polyfunctional T cells toward an exhausted, dysfunctional state (65, 66). Recently, it has been speculated that an ideal vaccine against viruses resulting in chronic infections should not only generate robust cellular responses against strongly recognized epitopes, but should also aim to induce responses against subdominant epitopes, which may offset potential T cell exhaustion by inducing new multifunctional responses against viral epitopes that are not heavily targeted during infection (67).
Concordant with this idea, neither therapeutic approaches, including dendritic cells loaded with peptides, nor peptide vaccination based on immunodominant epitopes successfully reestablished a functional T cell response resulting in a decreased viral load in chronically infected HCV patients (68, 69). Likewise, AdCh3NSmut and Ad6NSmut, which were found to induce broad and multifunctional responses in healthy individuals, failed to enhance or broaden the exhausted and narrower T cell responses when employed in a therapeutic setting, indicating the benefit of inducing de novo T cell responses against epitopes that are not under high antigenic pressure and therefore not driven to a dysfunctional state during natural infection (70). Thus, the concept of inducing new T cell immunity by targeting subdominant epitopes could be a promising approach, not limited to a therapeutic context but also to prevent (re)infection or development of chronicity in patients, e.g., intravenous drug users, who have cleared the virus after successful treatment but nevertheless display dysfunctional HCV-specific T cells. Induction of new CD4+ and CD8+ T cell responses against subdominant epitopes to subvert T cell exhaustion is currently being pursued in HIV/simian immunodeficiency virus (SIV) and tuberculosis vaccinology (17–19, 71–74). Here, we demonstrate that HCV NS3 pepmix broadens the repertoire of epitope-specific T cells compared to vaccination with the corresponding full-length protein. Currently, no small-animal model that faithfully resembles HCV pathogenesis allows the study of adaptive immunity, i.e., responses against dominant and subdominant epitopes and their protective efficacy (48, 49). However, the results from this study support the idea that the NS3 pepmix approach may facilitate induction of T cell responses against not only dominant, but also subdominant, HCV epitopes, which could expose an Achilles heel of persisting HCV. Furthermore, a broadened antigen-specific response recognizing multiple epitopes within the conserved NS3 region is not only key to minimizing the risk of mutational escape by formation of quasispecies in infected individuals, but may also overcome the genetic heterogeneity of HCV within and across distinct genotypes (20, 21, 75–77). Moreover, selection of vaccine antigens does not have to rely on prediction algorithms or restimulation assays to identify responses against subdominant epitopes, which tend to be below or close to the detection level.
The primary structure of the antigen plays a pivotal role in processing of T cell epitopes. It is indeed well established that single amino acid residues or sequences flanking CD8+ T cell epitopes in proteins can impact proteasomal cleavage dramatically and thus be detrimental to induction of CD8+ T cell responses (78–80). It has been shown that very strong CD8+ T cell responses could be obtained by vaccination with 5 μg of TB10.3 protein (80). In our study, we found that a 4-fold dose difference in rNS3 (5 μg versus 20 μg) did not alter the frequency of CD4+ T cells or facilitate priming of CD8+ T cells (data not shown). Similarly, a 10-fold reduction of antigens in the NS3 pepmix did not result in significantly lower CD4+ and CD8+ T cell responses. It seems plausible that the primary structure, rather than the dose, of rNS3 is suboptimal for processing of the CD8+ T cell epitopes and that a strategy utilizing overlapping peptides may bypass a potential detrimental influence on antigen processing and presentation imposed by flanking regions in order to prime CD8+ T cells and broaden the CD4+ T cell repertoire.
Once an infection has been established, it is desirable to induce effector T cells with the ability to coproduce IFN-γ and TNF-α, as these polyfunctional subsets exhibit enhanced killing of intracellular pathogens and malignant cells (28, 81, 82). Recently, we reported that CAF09 enhanced and skewed CD8+ and CD4+ T cells toward a TEM response that was able to control tumors in a therapeutic human papillomavirus 16 (HPV-16) E7-expressing model (40). In concordance with these findings, another group reported that a vigorous vaccine-induced TEM response dominated by double-positive IFN-γ+ TNF-α+ CD8+ T cells controlled tumors more efficiently than a central memory T cell (TCM) phenotypic response (82). Here, we found that NS3 pepmix induced an effector-like response, which predominantly consisted of polyfunctional CD4+ and CD8+ T cells capable of producing IFN-γ and TNF-α simultaneously. Compared with AdNS3, we found similar levels of CD27hi CD43lo long-lived memory CD8+ T cells within the cytokine-producing population. Both vaccine groups were dominated by CD27lo CD43lo long-lived effector CD8+ T cells, which have been associated with swift protective immunity against challenge with vaccinia virus and Listeria monocytogenes (46, 47). The high frequency of CD27lo CD43lo subpopulations is mirrored in IFN-γ+ TNF-α+ double positives comprising the majority of the cytokine-producing CD8+ T cells. Due to the reduced expression of CD62L compared to IFN-γ+ TNF-α+ IL-2+ triple positives (data not shown), these cells are able to migrate to the periphery, where they can promptly engage invading pathogens.
Although the importance of broad CD4+ T cell responses for control of HCV is recognized, the overall trend in HCV vaccine development tends to favor CD8+ T cell responses at the expense of CD4+ T cells (60, 83, 84). In addition to aiding the CD8+ T cell response, recent evidence suggests that granzyme B- and perforin-expressing CD4+ T cells can exert direct cytolytic activity on cells in influenza virus, HIV, hepatitis B virus, and HCV infections (50–53). Here, we demonstrate the in vivo cytotoxic capacity of CD4+ T cells induced by HCV NS3 pepmix vaccination. This adds to the multifaceted performance of NS3 pepmix vaccination, given that the broadened and vigorous CD4+ T cell response not only is vital for induction and maintenance of CD8+ T cells, but could also have a direct role in elimination of potentially HCV-infected cells. The ability of the vaccine-induced immune response to eliminate potential target cells by release of cytotoxic compounds was also demonstrated in vivo for CD8+ T cells. Although this assay clearly does not recapitulate the mechanisms by which endogenous antigens expressed by HCV are processed and how the virus manipulates the processing pathway and MHC expression, the use of 20-mers rather than minimal epitopes most likely ensures that antigen is internalized and processed in order to facilitate presentation on and stabilization of MHC-I and -II molecules (85). The fact that we can induce comparable levels of p52-specific CD8+ T cells through two very distinct pathways for antigen processing (AdNS3 and peptide-based vaccines), however, does indicate an overlap in the final presentation on MHC. Thus, we consider that in vivo cytotoxicity mediated by vaccinated mice represents T cells that recognize internalized and processed antigens presented on MHC molecules.
Overall, the data presented suggest that the NS3 pepmix vaccine can induce a highly antiviral T cell response with a phenotypic profile resembling that of HCV controllers. This vaccine strategy seems to support multiple indispensable components of T cell immunity, and in particular, the engagement of sustained, broadened, and cytotoxic CD4+ T cell responses could fill a gap in the established paradigm of HCV vaccinology, where the main emphasis is on induction of CD8+ T cell responses. It would be of interest in future studies to evaluate this novel vaccine approach prophylactically or therapeutically and also to assess its ability to counter HCV-driven T cell exhaustion by targeting subdominant epitopes and preventing persistent infection. A better understanding of how “new” T cell immunity can be induced by refocusing cellular responses toward subdominant epitopes may also contribute to the development of immunotherapies against cancer or human pathogens, such as HIV or Mycobacterium tuberculosis, which can establish chronic infections and drive the immune response to exhaustion.
MATERIALS AND METHODS
Animals.
Female CB6F1 (C57BL/6 × BALB/c) and C3H mice aged 6 to 8 weeks were purchased from Harlan Scandinavia, Allerød, Denmark. The animals were kept at the experimental-animal facilities at Statens Serum Institut and handled by authorized personnel. All experiments were performed according to the guidance of the Danish Ministry of Justice and Animal Protection Committees and in compliance with European Community Directive 86/609.
Antigens and adjuvants.
NS3 from the cloned HCV strain J4, genotype 1b, was used as the antigen (86, 87). rNS3 was produced in Escherichia coli by ProMab Biotechnologies (California, USA). A library of 62 20-mer peptides overlapping by 10 amino acids and spanning the entire sequence of NS3 was synthesized by standard Fmoc chemistry procedures on Tenta Gel S RAM resin in house (88) or purchased (GeneCust, Luxemburg). Peptides were isolated by preparative high-pressure liquid chromatography (Waters, Massachusetts, USA) to purities greater than 95% and characterized by matrix-assisted laser desorption ionization–time of flight mass spectrometry (Bruker Daltonics, Bremen, Germany). The peptides were then lyophilized and reconstituted in dimethyl sulfoxide (DMSO) (Sigma-Aldrich) at 20 mg/ml. The vaccine antigens were tested in combination with the cationic adjuvant formulation CAF09 at a dose of 250 μg DDA-50 μg MMG-50 μg poly(I·C). The adjuvant was prepared as described previously (40, 89). Mice were injected with 20 μg rNS3 or with a mixture of the 62 overlapping peptides (NS3 pepmix) consisting of 10 μg of each peptide. Altered rNS3 antigen doses ranging from 5 to 20 μg did not affect the immunological outcome, nor did NS3 pepmix consisting of 1 μg of each peptide (data not shown). Control mice were injected with CAF09 alone. The vaccines were administered intraperitoneally at a total volume of 0.25 ml. The mice were vaccinated three times at 2-week intervals. If indicated, the mice received a 3rd booster 12 weeks after the priming vaccination. AdNS3 vaccination was administered in its optimal regimen (data not shown): once in a volume of 30 μl in the footpad (2 × 107 infectious units). The AdNS3 vaccine was produced as described previously and is based on the same HCV NS3 sequence used for rNS3 and NS3 pepmix (45). Compared with NS3 pepmix, AdNS3 vaccination was administered at week 4.
Isolation of cells from mice.
Blood was drawn via the facial vein or by periorbital puncture, and peripheral blood mononuclear cells (PBMCs) were purified using Lympholyte (Cederlane, Burlington, NC) according to the manufacturer's protocol. Splenocytes were obtained by homogenization through a 100-μm cell strainer (BD Pharmingen) followed by red blood cell lysis in 0.84% NH4Cl and two washes with RPMI 1640 (Gibco, Invitrogen, Taastrup, Denmark). All cell cultures were performed in Nucleon microtiter plates (Nunc, Roskilde, Denmark) with RPMI 1640 supplemented with 1 mM glutamine, 1% pyruvate, 1% penicillin-streptomycin, 1% HEPES, and 10% fetal bovine serum (FBS) (all from Gibco, Taastrup, Denmark).
Cytokine secretion assay.
PBMCs or splenocytes cultured in 96-well microtiter plates in a volume of 200 μl containing 2 × 105 cells/well were stimulated with NS3 peptides (individually or as a pool of 62 peptides [2 μg/ml of each]). Cell supernatants were harvested after 5 days of incubation at 37°C and 5% CO2 in a humidified incubator. Levels of antigen-specific secreted IFN-γ were measured using ELISA, as previously described (90).
Intracellular flow cytometry.
Splenocytes were stimulated with 2 μg/ml NS3 peptides (individually or as a pool of 62 peptides [2 μg/ml of each]) in 96-well plates for 1 h in the presence of 1 μg/ml anti-CD28 (clone 37.51) and anti-CD49d (clone 9C10; MFR4.B) (both from BD Pharmingen) and subsequently incubated for 5 to 6 h at 37°C with 10 μg/ml brefeldin A (Sigma-Aldrich). Following overnight storage at 4°C, the cells were washed in fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline [PBS] containing 1% FBS) and subsequently stained for 30 min at 4°C for surface markers with monoclonal antibodies (MAbs) as indicated. Anti-CD4−allophycocyanin-Cy7 (clone GK1.5), anti-CD8-peridinin chlorophyll protein (PerCP)-Cy5.5 (clone 53-6.7), and anti-CD44-fluorescein isothiocyanate (FITC) (clone IM7) (all from BD Pharmingen) were diluted 1:600. The cells were then washed in FACS buffer, permeabilized using a Cytofix/Cytoperm kit (BD Pharmingen) according to the manufacturer's instructions, and stained intracellularly for 30 min at 4°C in dilutions of 1:200 using anti-IFN-γ–phycoerythrin (PE)–Cy7 (eBioscience), anti-TNF-α–PE (BD Pharmingen), or anti-IL-2–allophycocyanin (BD Pharmingen). The activation and differentiation status of CD8+ T cells was assessed as described above with anti-CD8-PerCP-Cy5.5 diluted 1:600, anti-CD43-Alexa Fluor 488 (clone 1B11; BioLegend), anti-CD27-allophycocyanin (clone LG.7F9; eBioscience), anti-IFN-γ–PE–Cy7, anti-TNF-α–PE, and anti-IL-2–allophycocyanin–Cy7 (clone Jes6-5H4; BD Pharmingen) diluted 1:200. The cells were subsequently washed, resuspended in FACS buffer, and then analyzed using a six-color BD FACSCanto flow cytometer (BD Biosciences). Finally, the flow cytometry data were analyzed using FlowJo software v8.8.6 (Tree star Ashland). Lymphocytes were gated based on their forward scatter (FSC) versus side scatter (SSC) profile and were gated into CD4+ or CD8+ populations. The CD4+ CD44+ and CD8+ CD44+ T cells were further divided into seven distinct subpopulations based on their ability to produce IFN-γ, TNF-α, or IL-2 in any combination.
Specific cell lysis in vivo.
Splenocytes from naive CB6F1 mice were divided into three fractions and labeled for 10 min at room temperature with carboxyfluorescein succinimidyl ester (Invitrogen) diluted in PBS at concentrations of 0.4 μM (CFSElow), 2 μM (CFSEmid), and 10 μM (CFSEhigh). The reaction was stopped by adding FBS to a final concentration of 30%, and the reaction mixture was placed on ice for 2 min. The two fractions, CFSElow and CFSEhigh, were pulsed with10 μg/ml of individual NS3 peptides as indicated in the figures for 30 min to 2 h at 37°C. CFSEmid-labeled cells were left unpulsed. Here, the concentrations of cells in all the fractions were adjusted to equal numbers. The three fractions were subsequently mixed at a 1:1:1 ratio, followed by three washes in RPMI plus 10% FBS. Finally, the cell number was adjusted to 3.3 × 107 cells/ml, and 300 μl of the cell suspension was injected intravenously (i.v.) into naive or vaccinated recipient mice 10 days after the final vaccination. Eighteen hours postinjection, the mice were sacrificed and splenocytes were isolated. Additionally, for the in vivo cytotoxicity assay based on CD4+ T cell epitopes (p46 and p60), splenocytes were labeled with anti-MHC-II–PerCP–Cy5.5 (BioLegend; cloneM5/114.15.2). CFSE-labeled lymphocytes were identified by flow cytometry based on their FSC versus SSC profiles, and the cells pulsed with CD4+ epitopes were further selected for expression of MHC-II. Epitope-specific lysis of cells pulsed with the indicated peptides was calculated using the following equation: 1 − [(percent pulsed vaccinated/percent unpulsed vaccinated) − (percent pulsed control/percent control)] (91).
In silico prediction of CD8+ T cell epitopes.
In silico prediction of CD8+ T cell epitopes within the sequences of NS3 peptides p51 and p52 was performed by using the MHC-I processing tool version 2009-09-01 NetMHCpan algorithm provided by the Immune Epitope Database (IEDB) Analysis Resource (http://tools.immuneepitope.org/analyze/html/mhc_processing.html).
Statistical analysis.
The statistical analysis of differences in cytokine expression and cytotoxicity between multiple vaccination groups was tested using analysis of variance (ANOVA), followed by Tukey's posttest. The Mann-Whitney test was used for comparison of cytokine responses and cytotoxicity between two groups only. P values of <0.05 were considered significant and are marked with asterisks in the figures. All statistical analyses were performed using Prism 6 software (GraphPad, San Diego, CA).
ACKNOWLEDGMENTS
We thank Jurgita Nørup and Sabaheta Babajic (Department of Drug Design and Pharmacology, University of Copenhagen, Copenhagen, Denmark) and Rune Fledelius Jensen and Camilla Haumann Rasmussen (Department of Infectious Disease Immunology, Statens Serum Institut, Copenhagen, Denmark) for excellent technical assistance. We especially thank Joshua Woodworth (Department of Infectious Disease Immunology, Statens Serum Institut, Copenhagen, Denmark) for discussions on subdominant epitopes and support in setting up in vivo cytotoxicity experiments.
This project was funded by the Innovation Fund Denmark project 060-2009-3, the Danish Cancer Society, the Amager and Hvidovre Hospital Research Fund, the Aase og Ejnar Danielsens Fond, the Novo Nordisk Foundation, and a Ph.D. stipend from the Faculty of Health and Medical Sciences, University of Copenhagen.
The content is solely our responsibility and does not necessarily reflect the official views of the funding sources listed above.
P.A. is a coinventor of patents relating to cationic liposomes as adjuvants and the use of overlapping peptide panels targeting dominant and subdominant epitopes by vaccination. All rights have been assigned to the Statens Serum Institut, a nonprofit government research facility. We have no other disclosures or conflicts of interest with respect to the manuscript.
REFERENCES
- 1.GBD 2013 Mortality and Causes of Death Collaborators. 2015. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385:117–171. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sølund C, Krarup H, Ramirez S, Thielsen P, Roge BT, Lunding S, Barfod TS, Madsen LG, Tarp B, Christensen PB, Gerstoft J., Laursen AL, Bukh J, Weis N. 2014. Nationwide experience of treatment with protease inhibitors in chronic hepatitis C patients in Denmark: identification of viral resistance mutations. PLoS One 9:e113034. doi: 10.1371/journal.pone.0113034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez S, Mikkelsen LS, Gottwein JM, Bukh J. 2016. Robust HCV genotype 3a infectious cell culture system permits identification of escape variants with resistance to sofosbuvir. Gastroenterology 151:973–985.e2. doi: 10.1053/j.gastro.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Pawlotsky JM. 2016. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 151:70–86. doi: 10.1053/j.gastro.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Sulkowski MS, Eron JJ, Wyles D, Trinh R, Lalezari J, Wang C, Slim J, Bhatti L, Gathe J, Ruane PJ, Elion R, Bredeek F, Brennan R, Blick G, Khatri A, Gibbons K, Hu YB, Fredrick L, Schnell G, Pilot-Matias T, Tripathi R, Da Silva-Tillmann B, McGovern B, Campbell AL, Podsadecki T. 2015. Ombitasvir, paritaprevir co-dosed with ritonavir, dasabuvir, and ribavirin for hepatitis C in patients co-infected with HIV-1: a randomized trial. JAMA 313:1223–1231. doi: 10.1001/jama.2015.1328. [DOI] [PubMed] [Google Scholar]
- 6.Sarrazin C, Isakov V, Svarovskaia ES, Hedskog C, Martin R, Chodavarapu K, Brainard DM, Miller MD, Mo H, Molina JM, Sulkowski MS. 2017. Late relapse versus hepatitis C virus reinfection in patients with sustained virologic response after sofosbuvir-based therapies. Clin Infect Dis 64:44–52. doi: 10.1093/cid/ciw676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciuffreda D, Comte D, Cavassini M, Giostra E, Buhler L, Perruchoud M, Heim MH, Battegay M, Genne D, Mulhaupt B, Malinverni R, Oneta C, Bernasconi E, Monnat M, Cerny A, Chuard C, Borovicka J, Mentha G, Pascual M, Gonvers JJ, Pantaleo G, Dutoit V. 2008. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. Eur J Immunol 38:2665–2677. doi: 10.1002/eji.200838336. [DOI] [PubMed] [Google Scholar]
- 8.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med 194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callendret B, Bukh J, Eccleston HB, Heksch R, Hasselschwert DL, Purcell RH, Hughes AL, Walker CM. 2011. Transmission of clonal hepatitis C virus genomes reveals the dominant but transitory role of CD8(+) T cells in early viral evolution. J Virol 85:11833–11845. doi: 10.1128/JVI.02654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin EC, Park SH, Nascimbeni M, Major M, Caggiari L, de Re V, Feinstone SM, Rice CM, Rehermann B. 2013. The frequency of CD127(+) hepatitis C virus (HCV)-specific T cells but not the expression of exhaustion markers predicts the outcome of acute HCV infection. J Virol 87:4772–4777. doi: 10.1128/JVI.03122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aberle JH, Formann E, Steindl-Munda P, Weseslindtner L, Gurguta C, Perstinger G, Grilnberger E, Laferl H, Dienes HP, Popow-Kraupp T, Ferenci P, Holzmann H. 2006. Prospective study of viral clearance and CD4(+) T-cell response in acute hepatitis C primary infection and reinfection. J Clin Virol 36:24–31. doi: 10.1016/j.jcv.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Diepolder HM, Zachoval R, Hoffmann RM, Wierenga EA, Santantonio T, Jung MC, Eichenlaub D, Pape GR. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 346:1006–1007. doi: 10.1016/S0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 13.Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, Wurcel AG, Timm J, Jones AM, Mothe B, Allen TM, McGovern B, Lewis-Ximenez L, Sidney J, Sette A, Chung RT, Walker BD. 2005. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol 175:3603–3613. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 14.Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, Thomas DL, Cox AL. 2010. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology 138:315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 16.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med 191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frahm N, Kiepiela P, Adams S, Linde CH, Hewitt HS, Sango K, Feeney ME, Addo MM, Lichterfeld M, Lahaie MP, Pae E, Wurcel AG, Roach T, St John MA, Altfeld M, Marincola FM, Moore C, Mallal S, Carrington M, Heckerman D, Allen TM, Mullins JI, Korber BT, Goulder PJ, Walker BD, Brander C. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol 7:173–178. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 18.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones NA, Wei X, Flower DR, Wong M, Michor F, Saag MS, Hahn BH, Nowak MA, Shaw GM, Borrow P. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med 200:1243–1256. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, Hahn BH, Korber BT. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 22.Diamond MS. 2003. Evasion of innate and adaptive immunity by flaviviruses. Immunol Cell Biol 81:196–206. doi: 10.1046/j.1440-1711.2003.01157.x. [DOI] [PubMed] [Google Scholar]
- 23.Melief CJ, van der Burg SH. 2008. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat Rev Cancer 8:351–360. doi: 10.1038/nrc2373. [DOI] [PubMed] [Google Scholar]
- 24.Olsen AW, Hansen PR, Holm A, Andersen P. 2000. Efficient protection against Mycobacterium tuberculosis by vaccination with a single subdominant epitope from the ESAT-6 antigen. Eur J Immunol 30:1724–1732. doi:. [DOI] [PubMed] [Google Scholar]
- 25.Woodworth JS, Aagaard CS, Hansen PR, Cassidy JP, Agger EM, Andersen P. 2014. Protective CD4 T cells targeting cryptic epitopes of Mycobacterium tuberculosis resist infection-driven terminal differentiation. J Immunol 192:3247–3258. doi: 10.4049/jimmunol.1300283. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Anton LC, Bennink JR, Yewdell JW. 2000. Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity 12:83–93. doi: 10.1016/S1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- 27.La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC. 2006. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc Natl Acad Sci U S A 103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med 13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 29.Golden-Mason L, Palmer B, Klarquist J, Mengshol JA, Castelblanco N, Rosen HR. 2007. Upregulation of PD-1 expression on circulating and intrahepatic hepatitis C virus-specific CD8+ T cells associated with reversible immune dysfunction. J Virol 81:9249–9258. doi: 10.1128/JVI.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin B, Hennecke N, Lohmann V, Kayser A, Neumann-Haefelin C, Kukolj G, Bocher WO, Thimme R. 2014. Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol 61:538–543. doi: 10.1016/j.jhep.2014.05.043. [DOI] [PubMed] [Google Scholar]
- 31.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. 2005. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology 42:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman VR, Jensen KJ, Jensen SS, Leo-Hansen C, Jespersen S, da Silva Te D, Rodrigues CM, Janitzek CM, Vinner L, Katzenstein TL, Andersen P, Kromann I, Andreasen LV, Karlsson I, Fomsgaard A. 2013. Therapeutic vaccination using cationic liposome-adjuvanted HIV type 1 peptides representing HLA-supertype-restricted subdominant T cell epitopes: safety, immunogenicity, and feasibility in Guinea-Bissau. AIDS Res Hum Retroviruses 29:1504–1512. doi: 10.1089/aid.2013.0076. [DOI] [PubMed] [Google Scholar]
- 33.Steffensen MA, Pedersen LH, Jahn ML, Nielsen KN, Christensen JP, Thomsen AR. 2016. Vaccine targeting of subdominant CD8+ T cell epitopes increases the breadth of the T cell response upon viral challenge, but may impair immediate virus control. J Immunol 196:2666–2676. doi: 10.4049/jimmunol.1502018. [DOI] [PubMed] [Google Scholar]
- 34.Holst PJ, Jensen BA, Ragonnaud E, Thomsen AR, Christensen JP. 2015. Targeting of non-dominant antigens as a vaccine strategy to broaden T-cell responses during chronic viral infection. PLoS One 10:e0117242. doi: 10.1371/journal.pone.0117242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durand V, Wong SY, Tough DF, Le Bon A. 2004. Shaping of adaptive immune responses to soluble proteins by TLR agonists: a role for IFN-alpha/beta. Immunol Cell Biol 82:596–602. doi: 10.1111/j.0818-9641.2004.01285.x. [DOI] [PubMed] [Google Scholar]
- 36.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 37.Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D, Borrow P, Tough DF. 2003. Cross-priming of CD8+ T cells stimulated by virus-induced type I interferon. Nat Immunol 4:1009–1015. doi: 10.1038/ni978. [DOI] [PubMed] [Google Scholar]
- 38.van Dissel JT, Joosten SA, Hoff ST, Soonawala D, Prins C, Hokey DA, O'Dee DM, Graves A, Thierry-Carstensen B, Andreasen LV, Ruhwald M, de Visser AW, Agger EM, Ottenhoff TH, Kromann I, Andersen P. 2014. A novel liposomal adjuvant system, CAF01, promotes long-lived Mycobacterium tuberculosis-specific T-cell responses in human. Vaccine 32:7098–7107. doi: 10.1016/j.vaccine.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 39.Nordly P, Rose F, Christensen D, Nielsen HM, Andersen P, Agger EM, Foged C. 2011. Immunity by formulation design: induction of high CD8+ T-cell responses by poly(I:C) incorporated into the CAF01 adjuvant via a double emulsion method. J Control Release 150:307–317. doi: 10.1016/j.jconrel.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Korsholm KS, Hansen J, Karlsen K, Filskov J, Mikkelsen M, Lindenstrom T, Schmidt ST, Andersen P, Christensen D. 2014. Induction of CD8+ T-cell responses against subunit antigens by the novel cationic liposomal CAF09 adjuvant. Vaccine 32:3927–3935. doi: 10.1016/j.vaccine.2014.05.050. [DOI] [PubMed] [Google Scholar]
- 41.Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 42.Billeskov R, Grandal MV, Poulsen C, Christensen JP, Winther N, Vingsbo-Lundberg C, Hoang TT, van Deurs B, Song YH, Aagaard C, Andersen P, Dietrich J. 2010. Difference in TB10.4 T-cell epitope recognition following immunization with recombinant TB10.4, BCG or infection with Mycobacterium tuberculosis. Eur J Immunol 40:1342–1354. doi: 10.1002/eji.200939830. [DOI] [PubMed] [Google Scholar]
- 43.Aagaard C, Dietrich J, Andersen P. January 2012. Expanding the T cell repertoire to include subdominant epitopes by vaccination with antigens delivered as protein fragments or peptide cocktails. US patent 8105614 B2.
- 44.Steffensen MA, Holst PJ, Steengaard SS, Jensen BA, Bartholdy C, Stryhn A, Christensen JP, Thomsen AR. 2013. Qualitative and quantitative analysis of adenovirus type 5 vector-induced memory CD8 T cells: not as bad as their reputation. J Virol 87:6283–6295. doi: 10.1128/JVI.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mikkelsen M, Holst PJ, Bukh J, Thomsen AR, Christensen JP. 2011. Enhanced and sustained CD8+ T cell responses with an adenoviral vector-based hepatitis C virus vaccine encoding NS3 linked to the MHC class II chaperone protein invariant chain. J Immunol 186:2355–2364. doi: 10.4049/jimmunol.1001877. [DOI] [PubMed] [Google Scholar]
- 46.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med 204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. 2013. Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity 38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bukh J. 2012. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 142:1279–1287.e3. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 49.von Schaewen M, Gaska JM, Ploss A. 2016. New animal models for hepatitis C, p 275–297. In Miyamura T, Lemon SM, Walker CM, Wakita T (ed), Hepatitis C virus. I. Cellular and molecular virology. Springer, Tokyo, Japan. [Google Scholar]
- 50.Jellison ER, Kim SK, Welsh RM. 2005. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol 174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 51.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, Rychert J, Rosenberg ES, Piechocka-Trocha A, Brass AL, Brenchley JM, Walker BD, Streeck H. 2012. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med 4:123ra25. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aslan N, Yurdaydin C, Wiegand J, Greten T, Ciner A, Meyer MF, Heiken H, Kuhlmann B, Kaiser T, Bozkaya H, Tillmann HL, Bozdayi AM, Manns MP, Wedemeyer H. 2006. Cytotoxic CD4 T cells in viral hepatitis. J Viral Hepat 13:505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 53.Brown DM, Lee S, Garcia-Hernandez MDL, Swain SL. 2012. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol 86:6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forns X, Bukh J, Purcell RH. 2002. The challenge of developing a vaccine against hepatitis C virus. J Hepatol 37:684–695. doi: 10.1016/S0168-8278(02)00308-2. [DOI] [PubMed] [Google Scholar]
- 55.Mikkelsen M, Bukh J. 2007. Current status of a hepatitis C vaccine: encouraging results but significant challenges ahead. Curr Infect Dis Rep 9:94–101. doi: 10.1007/s11908-007-0003-6. [DOI] [PubMed] [Google Scholar]
- 56.Park SH, Rehermann B. 2014. Immune responses to HCV and other hepatitis viruses. Immunity 40:13–24. doi: 10.1016/j.immuni.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Man John Law L, Landi A, Magee WC, Lorne Tyrrell D, Houghton M. 2013. Progress towards a hepatitis C virus vaccine. Emerg Microbes Infect 2:e79. doi: 10.1038/emi.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauer GM, Barnes E, Lucas M, Timm J, Ouchi K, Kim AY, Day CL, Robbins GK, Casson DR, Reiser M, Dusheiko G, Allen TM, Chung RT, Walker BD, Klenerman P. 2004. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Holst PJ, Sorensen MR, Mandrup Jensen CM, Orskov C, Thomsen AR, Christensen JP. 2008. MHC class II-associated invariant chain linkage of antigen dramatically improves cell-mediated immunity induced by adenovirus vaccines. J Immunol 180:3339–3346. doi: 10.4049/jimmunol.180.5.3339. [DOI] [PubMed] [Google Scholar]
- 60.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, Meyer J, Huddart R, Smith K, Townsend R, Brown A, Antrobus R, Ammendola V, Naddeo M, O'Hara G, Willberg C, Harrison A, Grazioli F, Esposito ML, Siani L, Traboni C, Oo Y, Adams D, Hill A, Colloca S, Nicosia A, Cortese R, Klenerman P. 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med 4:115ra1. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swadling L, Capone S, Antrobus RD, Brown A, Richardson R, Newell EW, Halliday J, Kelly C, Bowen D, Fergusson J, Kurioka A, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Siani L, Traboni C, Hill A, Colloca S, Davis M, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. 2014. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci Transl Med 6:261ra153. doi: 10.1126/scitranslmed.3009185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schulze Zur Wiesch J, Ciuffreda D, Lewis-Ximenez L, Kasprowicz V, Nolan BE, Streeck H, Aneja J, Reyor LL, Allen TM, Lohse AW, McGovern B, Chung RT, Kwok WW, Kim AY, Lauer GM. 2012. Broadly directed virus-specific CD4+ T cell responses are primed during acute hepatitis C infection, but rapidly disappear from human blood with viral persistence. J Exp Med 209:61–75. doi: 10.1084/jem.20100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grakoui A, Crispe IN. 2016. Presentation of hepatocellular antigens. Cell Mol Immunol 13:293–300. doi: 10.1038/cmi.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto H, Matano T. 2008. Anti-HIV adaptive immunity: determinants for viral persistence. Rev Med Virol 18:293–303. doi: 10.1002/rmv.577. [DOI] [PubMed] [Google Scholar]
- 65.Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature 362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 66.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol 77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aubert RD, Kamphorst AO, Sarkar S, Vezys V, Ha SJ, Barber DL, Ye L, Sharpe AH, Freeman GJ, Ahmed R. 2011. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc Natl Acad Sci U S A 108:21182–21187. doi: 10.1073/pnas.1118450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gowans EJ, Roberts S, Jones K, Dinatale I, Latour PA, Chua B, Eriksson EM, Chin R, Li S, Wall DM, Sparrow RL, Moloney J, Loudovaris M, Ffrench R, Prince HM, Hart D, Zeng W, Torresi J, Brown LE, Jackson DC. 2010. A phase I clinical trial of dendritic cell immunotherapy in HCV-infected individuals. J Hepatol 53:599–607. doi: 10.1016/j.jhep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klade CS, Schuller E, Boehm T, von Gabain A, Manns MP. 2012. Sustained viral load reduction in treatment-naive HCV genotype 1 infected patients after therapeutic peptide vaccination. Vaccine 30:2943–2950. doi: 10.1016/j.vaccine.2012.02.070. [DOI] [PubMed] [Google Scholar]
- 70.Kelly C, Swadling L, Capone S, Brown A, Richardson R, Halliday J, von Delft A, Oo Y, Mutimer D, Kurioka A, Hartnell F, Collier J, Ammendola V, Del Sorbo M, Grazioli F, Esposito ML, Di Marco S, Siani L, Traboni C, Hill AV, Colloca S, Nicosia A, Cortese R, Folgori A, Klenerman P, Barnes E. 2016. Chronic hepatitis C viral infection subverts vaccine-induced T-cell immunity in humans. Hepatology 63:1455–1470. doi: 10.1002/hep.28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korsholm KS, Karlsson I, Tang ST, Brandt L, Agger EM, Aagaard C, Andersen P, Fomsgaard A. 2013. Broadening of the T-cell repertoire to HIV-1 Gag p24 by vaccination of HLA-A2/DR transgenic mice with overlapping peptides in the CAF05 adjuvant. PLoS One 8:e63575. doi: 10.1371/journal.pone.0063575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedrich TC, Valentine LE, Yant LJ, Rakasz EG, Piaskowski SM, Furlott JR, Weisgrau KL, Burwitz B, May GE, Leon EJ, Soma T, Napoe G, Capuano SV III, Wilson NA, Watkins DI. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol 81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karlsson I, Brandt L, Vinner L, Kromann I, Andreasen LV, Andersen P, Gerstoft J, Kronborg G, Fomsgaard A. 2013. Adjuvanted HLA-supertype restricted subdominant peptides induce new T-cell immunity during untreated HIV-1-infection. Clin Immunol 146:120–130. doi: 10.1016/j.clim.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Aagaard CS, Hoang TT, Vingsbo-Lundberg C, Dietrich J, Andersen P. 2009. Quality and vaccine efficacy of CD4+ T cell responses directed to dominant and subdominant epitopes in ESAT-6 from Mycobacterium tuberculosis. J Immunol 183:2659–2668. doi: 10.4049/jimmunol.0900947. [DOI] [PubMed] [Google Scholar]
- 75.Farci P, Bukh J, Purcell RH. 1997. The quasispecies of hepatitis C virus and the host immune response. Springer Semin Immunopathol 19:5–26. doi: 10.1007/BF00945022. [DOI] [PubMed] [Google Scholar]
- 76.Bukh J. 2016. The history of hepatitis C virus (HCV): basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control. J Hepatol 65:S2–S21. doi: 10.1016/j.jhep.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 77.Major ME. 2016. Hepatitis C: new clues to better vaccines? Gut 65:4–5. doi: 10.1136/gutjnl-2015-309829. [DOI] [PubMed] [Google Scholar]
- 78.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. 1997. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol 158:3227–3234. [PubMed] [Google Scholar]
- 79.Shastri N, Serwold T, Gonzalez F. 1995. Presentation of endogenous peptide/MHC class I complexes is profoundly influenced by specific C-terminal flanking residues. J Immunol 155:4339–4346. [PubMed] [Google Scholar]
- 80.Lindenstrom T, Aagaard C, Christensen D, Agger EM, Andersen P. 2014. High-frequency vaccine-induced CD8(+) T cells specific for an epitope naturally processed during infection with Mycobacterium tuberculosis do not confer protection. Eur J Immunol 44:1699–1709. doi: 10.1002/eji.201344358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lichterfeld M, Yu XG, Waring MT, Mui SK, Johnston MN, Cohen D, Addo MM, Zaunders J, Alter G, Pae E, Strick D, Allen TM, Rosenberg ES, Walker BD, Altfeld M. 2004. HIV-1-specific cytotoxicity is preferentially mediated by a subset of CD8(+) T cells producing both interferon-gamma and tumor necrosis factor-alpha. Blood 104:487–494. doi: 10.1182/blood-2003-12-4341. [DOI] [PubMed] [Google Scholar]
- 82.van Duikeren S, Fransen MF, Redeker A, Wieles B, Platenburg G, Krebber WJ, Ossendorp F, Melief CJ, Arens R. 2012. Vaccine-induced effector-memory CD8+ T cell responses predict therapeutic efficacy against tumors. J Immunol 189:3397–3403. doi: 10.4049/jimmunol.1201540. [DOI] [PubMed] [Google Scholar]
- 83.Wedemeyer H, Schuller E, Schlaphoff V, Stauber RE, Wiegand J, Schiefke I, Firbas C, Jilma B, Thursz M, Zeuzem S, Hofmann WP, Hinrichsen H, Tauber E, Manns MP, Klade CS. 2009. Therapeutic vaccine IC41 as late add-on to standard treatment in patients with chronic hepatitis C. Vaccine 27:5142–5151. doi: 10.1016/j.vaccine.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 84.Fournillier A, Frelin L, Jacquier E, Ahlen G, Brass A, Gerossier E, Holmstrom F, Broderick KE, Sardesai NY, Bonnefoy JY, Inchauspe G, Sallberg M. 2013. A heterologous prime/boost vaccination strategy enhances the immunogenicity of therapeutic vaccines for hepatitis C virus. J Infect Dis 208:1008–1019. doi: 10.1093/infdis/jit267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Germain RN, Rinker AG Jr. 1993. Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature 363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 86.Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 87.Bukh J, Meuleman P, Tellier R, Engle RE, Feinstone SM, Eder G, Satterfield WC, Govindarajan S, Krawczynski K, Miller RH, Leroux-Roels G, Purcell RH. 2010. Challenge pools of hepatitis C virus genotypes 1-6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis 201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nielsen SL, Frimodt-Moller N, Kragelund BB, Hansen PR. 2007. Structure–activity study of the antibacterial peptide fallaxin. Protein Sci 16:1969–1976. doi: 10.1110/ps.072966007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davidsen J, Rosenkrands I, Christensen D, Vangala A, Kirby D, Perrie Y, Agger EM, Andersen P. 2005. Characterization of cationic liposomes based on dimethyldioctadecylammonium and synthetic cord factor from M. tuberculosis (trehalose 6,6′-dibehenate)—a novel adjuvant inducing both strong CMI and antibody responses. Biochim Biophys Acta 1718:22–31. doi: 10.1016/j.bbamem.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 90.Dietrich J, Billeskov R, Doherty TM, Andersen P. 2007. Synergistic effect of bacillus calmette guerin and a tuberculosis subunit vaccine in cationic liposomes: increased immunogenicity and protection. J Immunol 178:3721–3730. doi: 10.4049/jimmunol.178.6.3721. [DOI] [PubMed] [Google Scholar]
- 91.Hermans IF, Silk JD, Yang J, Palmowski MJ, Gileadi U, McCarthy C, Salio M, Ronchese F, Cerundolo V. 2004. The VITAL assay: a versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J Immunol Methods 285:25–40. doi: 10.1016/j.jim.2003.10.017. [DOI] [PubMed] [Google Scholar]