ABSTRACT
African swine fever virus, a double-stranded DNA virus that infects pigs, is the only known member of the Asfarviridae family. Nevertheless, during our isolation and sequencing of the complete genome of faustovirus, followed by the description of kaumoebavirus, carried out over the past 2 years, we observed the emergence of previously unknown related viruses within this group of viruses. Here we describe the isolation of pacmanvirus, a fourth member in this group, which is capable of infecting Acanthamoeba castellanii. Pacmanvirus A23 has a linear compact genome of 395,405 bp, with a 33.62% G+C content. The pacmanvirus genome harbors 465 genes, with a high coding density. An analysis of reciprocal best hits shows that 31 genes are conserved between African swine fever virus, pacmanvirus, faustovirus, and kaumoebavirus. Moreover, the major capsid protein locus of pacmanvirus appears to be different from those of kaumoebavirus and faustovirus. Overall, comparative and genomic analyses reveal the emergence of a new group or cluster of viruses encompassing African swine fever virus, faustovirus, pacmanvirus, and kaumoebavirus.
IMPORTANCE Pacmanvirus is a newly discovered icosahedral double-stranded DNA virus that was isolated from an environmental sample by amoeba coculture. We describe herein its structure and replicative cycle, along with genomic analysis and genomic comparisons with previously known viruses. This virus represents the third virus, after faustovirus and kaumoebavirus, that is most closely related to classical representatives of the Asfarviridae family. These results highlight the emergence of previously unknown double-stranded DNA viruses which delineate and extend the diversity of a group around the asfarvirus members.
KEYWORDS: giant viruses, pacmanvirus, Asfarviridae, Acanthamoeba castellanii, NCLDV, faustovirus, kaumoebavirus, African swine fever virus
INTRODUCTION
The discovery impacts of “giant viruses,” such as Acanthamoeba polyphaga mimivirus (1), pandoraviruses (2), and pithoviruses (3), and their interactions with the world of protists are very common and do not need a long introduction. A large community of previously unknown viruses is emerging among the former nucleocytoplasmic large DNA viruses (NCLDV). We are witnessing a growing number of new isolates with extraordinary properties in gene content, particle resistance, and/or morphological appearance (3–7). Genome sequencing and advances in coculture isolation strategies have led to an increase in the discovery of new viruses (8, 9). Some NCLDV are well studied because of their role in infecting algae, in the case of Phycodnaviridae (10), or in swine and wild boar infections, in the case of members of the Asfarviridae (11). The latter, viruses of the species African swine fever virus (ASFV), are double-stranded DNA (dsDNA) arboviruses that were described for the first time in 1921 by Montgomery (12). They are responsible for hemorrhagic fever in pigs, with very high mortality rates. Until 2 years ago, ASFV was the only known complete virus of this family (13). In 2009, partial sequencing of Heterocapsa circularisquama DNA virus (HcDNAV) showed a close link with Asfarviridae (14), and this virus represents a cooccurrence of marine and specific mammalian viruses in the same family. In 2015, we reported the isolation of a new virus, faustovirus (15), using the protist Vermamoeba vermiformis as a cell support in our coculture process. Another virus, kaumoebavirus, was isolated using the same strategy (16). These two viruses represent a new group which is most closely related to asfarviruses, raising the question of whether they are distant relatives of asfarviruses or comprise putative new families.
In the present study, we describe the isolation of a new giant virus on Acanthamoeba castellanii. This virus branched within the cluster encompassing asfarviruses, faustoviruses, and kaumoebavirus. We named it pacmanvirus because of the broken capsid that we sometimes observed in our first transmission electron micrographs.
RESULTS
Virus isolation, replicative cycle, and host specificity.
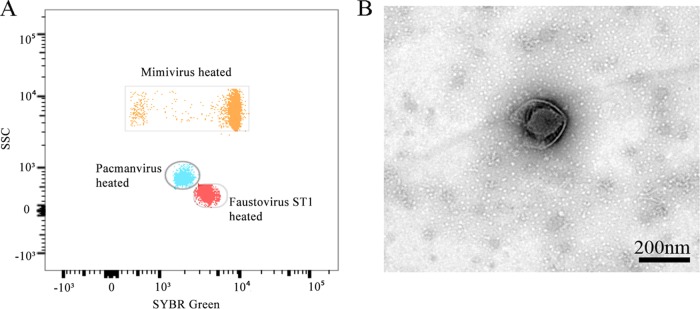
Following the procedure described in our recent work (7), using an inverted microscope and the same series of 24 different samples from Algeria, we observed the presence of a new unknown lysis agent in the sample well designated A23. We tested the potential host specificity of the lytic agent on two other amoebae: V. vermiformis and Dictyostelium discoideum. We did not observe any cytopathic effect on these two amoebae. Next, using flow cytometry and our previous data collected on known giant viruses through our gating strategy, we detected a different population (Fig. 1A) that was less fluorescent on fluorescein isothiocyanate (FITC) than the faustovirus gate. Negative staining performed on the supernatant showed a broken capsid (Fig. 1B). This virus was named pacmanvirus A23.
FIG 1.
Initial data on pacmanvirus. (A) Flow cytometry dot plot showing the different viral profiles, in SSC (side scatter) versus FITC (SYBR green DNA contents), of mimivirus and faustovirus strain ST1 (unpublished data) as controls and of pacmanvirus, distinguishable at the levels of capsid shape (optical properties) and DNA components. (B) Negative staining of a culture supernatant of pacmanvirus with its particular capsid aspect.
Pacmanvirus has extremely fast replication in A. castellanii compared to that of mimivirus. The burst of amoeba begins after only 6 h, and complete lysis occurs after 8 h. Pacmanvirus seems to be extremely well adapted to phagocytosis by its amoebal host. After 15 min of contact, we observed the presence of some virus in the vacuole (Fig. 2B) and then in the cytoplasm (Fig. 2C) of the amoeba. The opening of the capsid was not observed, but the virus was able to escape the phagosomal process. Surprisingly, at a very early time of infection, some viruses were observed close to the mitochondria, where a probable interaction takes place (Fig. 2D), but without any membrane fusion (see Fig. S1 in the supplemental material at http://www.mediterranee-infection.com/article.php?laref=873&titer=pacmanvirus). The viral particles presented an irregular icosahedral form with an average size of 175 nm. This form corresponds to the inner membrane of the virus also observed in cryo-sections (see the sections on global ultrastructure below). DNA leakage was never clearly visualized in our ultrathin sections, even though some viral particles in the cytoplasm appeared to be empty. This step was followed by a traditional eclipse phase known to take place in the replication cycle of most common giant viruses. At 3 h postinfection, the remodeling of the amoebal cytoplasm could clearly be observed with the establishment of an early viral factory (Fig. 2E). New virions were synthesized in the cytoplasm at a virus assembly site constituting a typical virus factory, visible only at 4 h postinfection (Fig. 2F). At 6 h postinfection, we noticed that mature viruses were organized in a well-aligned and -distributed geometric order, sometimes occupying the whole amoeba and sometimes in different areas of the amoeba at the periphery of the virus factory (Fig. 2G to I). Finally, after 8 h postinfection, total lysis of the cell was observed.
FIG 2.
Pacmanvirus infectious cycle realized on Acanthamoeba castellanii. (A) Pacmanvirus entry by typical phagocytosis and internalization into A. castellanii. (B and C) Viral particles with capsids could be detected inside phagocytotic vacuoles. (D) Two viruses without capsids clearly observed close to amoebal mitochondria. (E) At 3 h postinfection, primary virus factories (marked by asterisks) started to appear at a distinct location in the cytoplasm facing the nucleus of the amoeba. (F) At 4 h postinfection, early synthesized virions could be visualized in the area of the viral factory. (G to I) At 6 h postinfection, mature viruses occupied a large part of the amoebal cytoplasm, especially at the periphery of the virus factory.
Genome analysis.
A complete genome was obtained by assembly and through Sanger sequencing of 6 gap regions obtained in the first genome draft version. Pacmanvirus A23 is a double-stranded DNA virus of 395,405 bp, with an estimated G+C content of 33.6%, which is lower than those of ASFV, kaumoebavirus, and faustoviruses (Table 1). In addition, the genome is ≈2.3 times longer than that of ASFV BA71V, ≈44,600 bp longer than that of kaumoebavirus, and 60,400 to 95,600 bp shorter than those of faustoviruses. The pacmanvirus genome could not be closed by use of different primer sets, which, together with genome assembly analyses, suggests that it is linear. A dot plot analysis of the pacmanvirus genome against itself revealed an exceptionally small number of points not following the straight line (see Fig. S2 in the supplemental material at http://www.mediterranee-infection.com/article.php?laref=873&titer=pacmanvirus), suggesting the total absence of large repeats or inverted regions and other kinds of paralogous regions. Clearly, in a manner congruent with the results obtained with the EMBOSS palindrome and CRISPRFinder programs, these findings indicate that pacmanvirus possesses a linear compact genome without large repeated regions. Finally, this is similar to what was observed for kaumoebavirus and ASFV (16), but not for faustoviruses (17). Even if kaumoebavirus and pacmanvirus seem to have similar genome structure organizations, multiple-genome alignment (see Fig. S3 in the supplemental material at http://www.mediterranee-infection.com/article.php?laref=873&titer=pacmanvirus) with MAUVE showed that 7 regions of the pacmanvirus genome are more similar to the faustovirus genomes.
TABLE 1.
Main genomic comparative characteristics of Asfarviridae and other new viruses
| Parameter | Value or description |
|||
|---|---|---|---|---|
| Pacmanvirus | Kaumoebavirus | Faustovirus | ASFV BA71V | |
| Genome length (bp) | 395,405 | 350,731 | 455,803–491,024 | 170,101 |
| GC content (%) | 33.62 | 43.7 | 37.14a | 38.9 |
| Genome structure | Linear | Circular | Circularb | Linear |
| Coding density (%) | 89.3 | 86 | 85c | 88.6 |
| No. of tRNAs | 1 (Ile) | 0 | 0 | 0 |
Average value.
Expected for faustovirus strain Liban.
Score for faustovirus strain E12.
One tRNA, an isoleucine-tRNA, was detected. This was the first time that a tRNA was detected in a virus from the viral cluster encompassing ASFV, kaumoebavirus, and faustoviruses.
For gene prediction, we decided to keep all the common genes predicted by two different software packages: GeneMarkS and Prodigal. Indeed, with faustovirus and kaumoebavirus analyses (15, 16), we observed that small coding sequences were conserved and may encode elements constituting the viral capsid. Finally, in the absence of transcriptomic and proteomic analyses, we decided to keep 465 genes in the pacmanvirus gene repertoire, which represents a coding density of about 89.3% along the 353,121 bp (mean GC% of the coding sequences ≈ 34.7%).
BLASTp searches with the 465 pacmanvirus genes found matches for 221 genes in the nr database, including 135 best hits with viruses, 45 with eukaryotes, and 41 with prokaryotes (including 38 with bacteria). Regarding virus category, a large majority of the best hits (n = 84) were obtained with faustoviruses, whereas 9 were obtained with kaumoebavirus, 11 with marseilleviruses, 9 with mimiviruses, and only 2 with African swine fever virus. Regarding eukaryotes, only 3 best hits were obtained with Acanthamoeba castellanii strain Neff, and 4 were obtained with Dictyostelium spp. We observed that 244 protein sequences (≈52.5%) had no hit in the nr database, and the corresponding genes were therefore classified as ORFan genes (i.e., open reading frames [ORFs] with no detectable homology to other ORFs in a database). A total of 79 proteins (17.0% of all predicted proteins) presented a size smaller than 100 amino acids, and 13 of them (16.5%) had a hit in the nr database. Only 155 proteins (33.3%) had a functional annotation. Paralogous gene searches by use of BLASTClust showed that the most represented family of paralogous genes encode zinc finger C2H2/integrases (6 copies). Three copies of a gene encoding a protein containing a bacteriophage T5 ORF172 domain are present in the pacmanvirus genome, and restriction endonuclease genes are also present as 3 copies. Other paralogous genes are all annotated as hypothetical. These findings indicate that the paralogous genes present in pacmanvirus are a very limited set of genes and are similar to what was observed for kaumoebavirus. They confirm the idea of a compact genome, in contrast to those of pandoraviruses, for example, whose genomes exhibit a high level of gene duplication (2). This may be explained for the pandoraviruses by the accumulation of miniature inverted-repeat transposable element (MITE) transposons in their genomes (18).
In contrast to faustovirus E12, whose genome carries genes for a large number of MORN repeat-containing proteins (15; 3.05% of the 492 faustovirus E12 proteins), pacmanvirus A23 shows only 6 such proteins (1.3% of the 465 proteins), whereas kaumoebavirus presents 3 such proteins (≈0.65% of the 465 proteins). Some pacmanvirus proteins were not found in kaumoebavirus, asfarviruses, or faustoviruses. For example, we observed 3 copies of genes encoding proteins containing PAN/apple domains, as well as a gene predicted to encode a tRNA histidine guanylyltransferase (Thg1).
A phylogenetic reconstruction based on the DNA polymerase B gene family (Fig. 3) points out extreme distances between the four viral groups represented by asfarviruses, faustoviruses, kaumoebavirus, and pacmanvirus. Whereas asfarvirus strains are strongly similar, faustovirus strains exhibit more diversity between and within 4 lineages. Maximum likelihood reconstruction based on the DNA polymerase gene supports the ideas that kaumoebavirus, ASFV, pacmanvirus, and faustoviruses have a common ancestor and that kaumoebavirus divergence preceded those of the other three viral groups. Taken together, these results highlight that pacmanvirus is the prototype isolate of a new viral group.
FIG 3.
Maximum likelihood tree reconstruction based on the DNA polymerase B gene. iTol visualization was used with the deletion branch option for values inferior to 0.5. We colored the Mimiviridae and extended family in blue, the pithoviruses and cedratvirus in red, some Phycodnaviridae, pandoraviruses, and Mollivirus sibericum in green, some Poxviridae in purple, the Asco-Iridoviridae in orange, and the Marseilleviridae in gray.
Global ultrastructure characterization of pacmanvirus A23.
Bioinformatic analyses highlight a difference in the organization of the capsid regions of the ASFV, pacmanvirus, kaumoebavirus, and faustovirus genomes. In addition, the pacmanvirus major capsid protein (MCP) locus seems to resemble that of ASFV more than those of faustoviruses and kaumoebavirus. Indeed, the MCP is encoded by a single open reading frame, as is the case for ASFV. But even if the MCP gene is a unique ORF, a BLASTp homology search revealed a more important alignment score with faustovirus (92% coverage and 63% identity). Moreover, a BLASTp search with the pacmanvirus MCP sequence against environmental metagenome proteins (env_nr database) detected 3 homologous proteins (with an alignment score of up to 200) in a marine metagenome. These data confirm the idea of an extremely diverse undescribed family and support the hypothesis of a group encompassing the traditional Asfarviridae.
Cryo-electron microscopy (cryo-EM) micrographs of purified virus showed icosahedral virus particles with a spiky outer surface and a distinct inner membrane (Fig. 4A). A three-dimensional (3D) cryo-EM reconstruction assuming icosahedral symmetry to an average resolution of 15 Å gave an overall external structure comparable to that of faustovirus (19), measuring about 2,500 Å in diameter, clearly indicating the presence of an inner membrane. No distinct inner capsid shell similar to that of faustovirus has been resolved at this resolution. The outer protein shell is arranged in icosahedral fashion, with an h value of 7 and a k value of 13, giving a triangulation number of 309. Similar to that of faustovirus, the MCP forming the pseudo-hexagonal capsomeres around the 5-fold vertices of the icosahedral virion shows a double jelly-roll fold, with the exception of the capsomere at the icosahedral 5-fold axis, which has a 5-fold symmetry and is different from the MCP. The double jelly-roll fold of pacmanvirus MCP could be confirmed by fitting the faustovirus MCP trimer (PDB ID 5J7O) into a 15-Å cryo-EM map of pacmanvirus, with a map correlation of 65.76%. We also identified the protein (PACV_341) at the icosahedral 5-fold axis as the putative minor capsid protein, with a single jelly-roll fold similar to those of all known dsDNA viruses. PACV_341 is half the size of the MCP. Secondary structure predictions using the programs HHpred (https://toolkit.tuebingen.mpg.de/hhpred) and Quick2D (https://toolkit.tuebingen.mpg.de/quick2_d) suggested a beta-strand-rich fold.
FIG 4.
Cryo-EM 3D reconstruction of pacmanvirus A23. (A) Cryo-EM micrograph of pacmanvirus showing a distinct inner membrane (arrows). (B) Cryo-EM reconstruction of pacmanvirus at 15 Å. The capsomeres near the 5-fold axes are colored dark blue for clarity. (C) Cross section showing the distinct inner membrane in the cryo-EM density map.
Deeper analysis of gene partitioning and pangenomes.
Analysis of the best reciprocal hits by use of Proteinortho software revealed a core gene group containing 31 genes (Fig. 5) shared by ASFV, faustoviruses, kaumoebavirus, and pacmanvirus genomes. These core genes include a functional set of genes involved in replication (i.e., RNA polymerase 1, 2, 3, and 5 subunit genes) or DNA damage repair (e.g., ERCC4 domain-containing protein gene) (see Table S1 in the supplemental material at http://www.mediterranee-infection.com/article.php?laref=873&titer=pacmanvirus). Moreover, ASFV, faustoviruses, and pacmanvirus share 7 genes also involved in replication or capsid formation. Of 7,730 proteins, based on reciprocal best hits, we determined that there are 1,936 clusters comprising at least one known protein itself coming from at least one of the 25 different genomes. Of 465 pacmanvirus proteins, 371 (79.8%) represent unique protein clusters. In addition, pacmanvirus proteins are shared in 42 clusters with proteins from faustovirus strains. Overall, faustoviruses and pacmanvirus share proteins within 83 clusters, and faustoviruses therefore represent the most closely related viruses to pacmanvirus, to date. ASFV and kaumoebavirus share 40 and 55 clusters with faustovirus, respectively, whereas these numbers are 43 and 47, respectively, with pacmanvirus. Moreover, the largest pangenome was observed for the faustoviruses (801 clusters), which is explained by the four distinct faustovirus lineages and the availability of 9 strains (17) (Fig. 1). In conclusion, the newcomer pacmanvirus dropped the precedent core genome between faustovirus, kaumoebavirus, and ASFV from 33 to 31 genes. This is only a small reduction compared to the 371 unique proteins reported above.
FIG 5.
Venn diagram representing coorthologous genes from asfarviruses, faustoviruses, kaumoebavirus, and pacmanvirus. *, same number or cluster of analysis for the group of faustovirus and asfarvirus; **, same number or cluster of analysis for the group of kaumoebavirus, faustovirus, and ASFV.
Regarding ASFVs, their analysis has allowed us to better understand the newly identified viruses faustovirus, kaumoebavirus, and pacmanvirus. Based on a previous analysis of asfarvirus genomes (20), we explored the expansion of this cluster by detecting orthologous genes and searching for reciprocal best hits with BLASTp. In the case of nucleotide metabolism, transcription, replication, repair, and some other structural proteins, we detected that the structural gene complex associated in ASFV with the capsid maturation process, as well as RNA polymerase subunits, has conserved homologs in kaumoebavirus, faustoviruses, and pacmanvirus. However, ASFV genomes possess at least 8 particular genes, i.e., the IAP apoptosis inhibitor A224L gene (21), the Bcl-2 apoptosis inhibitor A179L gene (22), the inhibitor of host gene transcription A238L gene (23), the C-type lectin-like EP153R gene (24), the CD2-like hemadsorption to infected cells EP402R gene (25), the similar neurovirulence factor DP71L gene (26), the NifS-like QP383R gene, and the phosphoprotein binding ribonucleoprotein-K CP204L gene. All of these are involved in host cell interactions, but only one gene, QP383R, seems to have orthologs in faustoviruses and pacmanvirus. QP383R and its orthologs possess a NifS domain whose presumed function is similar to that of cysteine desulfurase. All of these data can be explained by the progressive genome reduction undergone by ASFV in order to adapt to its mammalian host, resulting in a long-term progressive adaptation like that described for the families Iridoviridae and Ascoviridae (27).
DISCUSSION
Our analysis shows that pacmanvirus strain A23 constitutes a bona fide new giant virus. The detection of 310 genes encoding hypothetical proteins, including 244 ORFan genes, highlights that a considerable part of this virus's components is still completely unknown. The genome length of pacmanvirus ranges between that of kaumoebavirus and that of faustovirus. Nevertheless, pacmanvirus showed the lowest G+C content among the four analyzed viruses, and its genome content shared a core of genes encoding only 31 essential proteins with the other members of this viral cluster. The addition of a new prototype virus does not affect the core genome of this cluster very much. Indeed, beyond the presence of these relatively steady core genes, we observed that genes associated with virus replication are conserved between ASFV, kaumoebavirus, faustovirus, and pacmanvirus. In contrast, we did not observe the conservation in the last three viruses of described proteins which may modulate host cell interaction as seen in ASFVs.
In fact, ASFVs multiply in pig monocytes and can be transmitted by ticks (28), and they may have other unexplored reservoirs. In contrast, faustoviruses and kaumoebavirus grow in V. vermiformis, while pacmanvirus multiplies in A. castellanii. The major capsid protein is encoded in a single ORF in pacmanvirus and ASFV, while it is located in different loci in the case of kaumoebavirus and faustoviruses. We do not know if this process is related to host range as for the double protein capsid studied for faustovirus (15, 17, 19) and the predicted capsid protein of kaumoebavirus (16). Despite all this, the ASFV virion structure includes an outer lipid membrane, an icosahedral protein capsid, and an inner lipid membrane (29) that is strongly involved in cell infection. Nevertheless, an outer lipid membrane seems to be absent in the pacmanvirus, faustovirus, and kaumoebavirus structures.
Another point raised by this study lies in the earliest steps of pacmanvirus infection, where a particularly close interaction of the viral capsid with mitochondria can be observed prior to the eclipse phase, without membrane fusion. Unfortunately, we were not able to observe the way in which the viral DNA was delivered into the amoeba. This process needs further observation and more experiments.
The isolation of pacmanvirus confirms that the coculture strategy on protists in association with flow cytometry (8, 30) detection from culture supernatant is an extremely fast, sensitive, and simple way to detect new viruses.
Considering the recent expansion of newly described giant amoeba viruses, it seems difficult to define and apply the concept of a new family for pacmanvirus. The concept of family needs to be clarified, and we prefer to use the term “group” or “cluster” of viruses, pending further viral descriptions. Pacmanvirus is the closest known virus to faustoviruses. Finally, our knowledge of pacmanvirus and future isolates will probably deeply reshape the current knowledge of Asfarviridae in many ways.
MATERIALS AND METHODS
Culture procedure.
A classic coculture protocol (9) was followed, with Acanthamoeba castellanii strain Neff as the cell support. The cell suspension was centrifuged at 700 × g for 10 min, and then the pellet was resuspended in starvation medium (15). The quantity of cells in the suspension was controlled to obtain a final concentration of 5 × 105 amoebae/ml. Concentrations of antibiotic and antifungal mixtures similar to those reported recently were used (7). Next, 250 μl of amoebal suspension per well was inoculated into the wells of a 48-well plate. Twenty-four environmental samples from Algeria (50-μl aliquots) were inoculated by direct inoculation into wells numbered A1 to A24. The sample in well A23 was collected from El Taref City on the road to El Kala (GPS localization, 36.764787, 8.361114).
Flow cytometry detection.
In order to obtain more information about the DNA content and the nature of the lytic agent, a gating strategy (8) was developed using flow cytometry based on side scatter and DNA content. For this strategy, after cytopathic effect and lysis detection, a fraction of the virus-lysed supernatant was centrifuged at 700 × g for 10 min to exclude large debris in the pellet. The supernatant was stained by use of SYBR green dye (SYBR green I nucleic acid gel stain; Molecular Probes, Life Technologies) at a dilution of 1:100, with heating at 80°C for 3 min. Finally, the sample was diluted to 1:1,000 in distilled water before collection of data on a BD LSR Fortessa (BD Biosciences) cytometer. The data were compared with those for previously known gated viruses by use of FlowJo software.
Transmission electron microscopy.
For pacmanvirus, we used exactly the same protocols as in the case of cedratvirus (7) for negative staining, embedding, and transmission electron microscopy. The only difference was the infection time points at which the samples were collected. The early time points were 0, 15, 30, and 45 min and 1 h postinfection, followed by the late time points of 2, 3, 4, 6, 8, and 10 h postinfection. Micrographs were collected on a Tecnai G20 electron microscope (FEI, Germany) operating at 200 keV, and the size of the particles was measured using ImageJ (https://imagej.nih.gov/ij/).
Cryo-EM data collection and processing.
Purified pacmanvirus particles were plunged frozen into liquid ethane by applying 2-μl samples onto C-flat holey carbon grids (CF-2/2-4C; 2.0-μm hole by 2.0-μm space) (Electron Microscopy Sciences) by use of a CryoPlunge 3 device (Gatan), with a blot time of 6 s. Micrographs were collected on an FEI Titan Krios electron microscope equipped with a Gatan K2 Summit detector at a magnification of ×18,000 in “superresolution mode,” resulting in a pixel size of 0.81 Å. A total of 1,044 micrographs were collected, with an exposure time of 10 s/micrograph and a dose of ∼7e−/Å2/s, using Leginon software (31). About 1,956 particles were boxed manually by using the e2boxer.py module of the EMAN2 software package (32). The data set was not split into two subsets due to the availability of very few particles. Initial model generation, orientation refinement, and selected particle centering were done using the jspr program (33). After 9 rounds of 3D classification (and the application of an outer mask for initial model generation), a total of 1,746 particles were used to generate a cryo-EM map at an average resolution of 15 Å, calculated using the 0.5 Fourier shell correlation (FSC) criterion. Figures were prepared using CHIMERA (34).
Genome sequencing and genome assembly.
The genomic DNA (gDNA) of pacmanvirus A23 was sequenced on a MiSeq sequencer (Illumina Inc., San Diego, CA) with 2 applications: paired-end and mate pair sequencing. Both strategies were barcoded in order to be combined, respectively, with 11 other genomic projects prepared according to the instructions of a Nextera XT DNA sample prep kit (Illumina) and 11 projects prepared according to the instructions of a Nextera Mate Pair sample prep kit (Illumina). The gDNA was quantified to 343.3 ng/μl by a Qubit assay with a high-sensitivity kit (Life Technologies, Carlsbad, CA).
To prepare the paired-end library, 1 ng of gDNA was fragmented and amplified by limited PCR (12 cycles), introducing dual-index barcodes and sequencing adapters. After purification on AMPure XP beads (Beckman Coulter Inc., Fullerton, CA), the libraries were then normalized and pooled for sequencing on the MiSeq sequencer. Automated cluster generation and paired-end sequencing with dually indexed 2- by 250-bp reads were performed in a 9-h run.
A total information set of 9.0 Gb was obtained, with a 1,019,000/mm2 cluster density and with a cluster-passing quality control filter of 90.2% (17,374,744 passed filtered reads). Within this run, the index representation for pacmanvirus A23 was determined to be 3.42%. The 594,320 paired-end reads were trimmed and filtered according to read quality.
The mate pair library was prepared with 1.5 μg of genomic DNA, using the Illumina Nextera mate pair guide. The genomic DNA sample was simultaneously fragmented and tagged with a mate pair junction adapter. The profile of the fragmentation was validated on an Agilent 2100 BioAnalyzer instrument (Agilent Technologies Inc., Santa Clara, CA) with a DNA 7500 LabChip. The obtained fragments presented a size range of 1 to 10 kb, with an optimal mean size distribution of 6.7 kb. No size selection was performed, and 600 ng of tagged fragments was circularized. The circularized DNA was mechanically sheared to small fragments with an optimal size of 1,136 bp in T6 tubes on a Covaris S2 device (Covaris, Woburn, MA). The library profile was visualized on a high-sensitivity BioAnalyzer LabChip (Agilent Technologies Inc., Santa Clara, CA), and the final concentration of the library was measured at 7.53 nmol/liter. The libraries were normalized to 2 nM, pooled with 11 other projects, denatured, and diluted to 15 pM. The automated cluster generation and 2- by 250-bp sequencing run were performed in a 39-h run.
A total information set of 8.3 Gb was obtained, with a 910,000/mm2 cluster density and with a cluster-passing quality control filter of 92.8% (16,316,457 passed filter clusters). Within this run, the index representation for pacmanvirus A23 was determined to be 3.44%. The 561,759 paired-end reads were filtered according to read quality.
The pacmanvirus genome assembly was obtained using the Abyss assembler (35) and SSPACE software v1.0 (36). Primer-BLAST (37) was used to design specific primers in order to close the gaps in six regions of the initial draft genome.
Genome organization.
Repeats were studied by using the CRISPRfinder program with standard defaults (38) in association with the EMBOSS explorer palindrome program (http://emboss.bioinformatics.nl/cgi-bin/emboss/palindrome), with a maximum length of 200 nucleotides and a minimum of 100 nucleotides. A dot plot was created based on the results of BLAST nucleotide searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genome annotation and comparative analysis.
We computed an analysis of tRNA by using the ARAGORN (39) and tRNAscan-SE 1.21 (40) software programs. Protein and gene predictions were successively launched on GeneMarkS (41) and Prodigal (42). Finally, the complete sets of ORFs in the genome predicted by both methods were conserved. A BLASTp search was done against the NCBI GenBank nonredundant sequence (nr) database and against all the ORFomes (constituted by all predicted proteins) of specific new viruses still unavailable in the GenBank database, as is the case with the 2 faustoviruses Liban and E9 and with kaumoebavirus, with an E value threshold of 10−3. We then performed a specific BLASTp search with the predicted proteins of our virus against the NCVOGs (COGs defined for NCLDV and giant amoeba viruses) (43), with similar parameters. BLASTClust (44) was used to detect paralogous genes with 70% coverage and 30% identity.
Global genomic alignment of nucleotides was done by computing with the MAUVE program (45), using the genome fasta files for each genome, with the default parameters. Proteinortho v4 (46) was used to detect true coorthologous genes between pacmanvirus, asfarviruses, faustoviruses, and kaumoebavirus, with 60% coverage, 20% identity, and an E value of 10−2 as thresholds. For that purpose, we built a collection of predicted proteins for each of the following viruses: faustovirus E9 (501 proteins), faustovirus Liban (518 proteins), faustovirus E23 (495 proteins), faustovirus E24 (496 proteins), faustovirus E12 (492 proteins), faustovirus D5a (490 proteins), faustovirus D5b (491 proteins), faustovirus D6 (495 proteins), faustovirus D3 (485 proteins), kaumoebavirus (465 proteins), ASFV Warthog/Namibia/WART80:1980 (160 proteins), ASFV tick/South Africa/Pretoriuskop Pr4/1996 (159 proteins), ASFV pig/Kenya/KEN-05/1950 (160 proteins), ASFV OURT 88/3 (157 proteins), ASFV Georgia 2007/1 (188 proteins), ASFV E75 (163 proteins), ASFV Benin 97/1 (156 proteins), ASFV 47/Ss/2008 (235 proteins), ASFV Ken05/Tk1 (168 proteins), ASFV BA71V nonvirulent (152 proteins), ASFV BA71 (161 proteins), ASFV NHV (158 proteins), and ASFV Lisboa L60 (163 proteins).
Phylogenetic analysis.
Alignments were performed in the MUSCLE program (47). The FastTree program (48) was used to compute the maximum likelihood tree, with 1,000 bootstrap replications, using JTT model substitution (standard default).
Accession number.
The viral genome is available in the EMBL-EBI database under accession number LT706986.
ACKNOWLEDGMENTS
We are thankful to Claire Andreani for her help with manuscript correction and to Emeline Baptiste for the genome submission.
Part of this work was supported by NIH grant R01AI011219 to Michael G. Rossmann.
REFERENCES
- 1.La Scola B. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie J-M, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 3.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Couté Y, Rivkina E, Abergel C, Claverie J-M. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A 111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic J-M, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie J-M. 2015. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci U S A 112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levasseur A, Andreani J, Delerce J, Bou Khalil J, Robert C, La Scola B, Raoult D. 2016. Comparison of a modern and fossil pithovirus reveals its genetic conservation and evolution. Genome Biol Evol 8:2333–2339. doi: 10.1093/gbe/evw153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreani J, Aherfi S, Bou Khalil JY, Di Pinto F, Bitam I, Raoult D, Colson P, La Scola B. 2016. Cedratvirus, a double-cork structured giant virus, is a distant relative of pithoviruses. Viruses 8:300. doi: 10.3390/v8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalil JYB, Robert S, Reteno DG, Andreani J, Raoult D, La Scola B. 2016. High-throughput isolation of giant viruses in liquid medium using automated flow cytometry and fluorescence staining. Front Microbiol 7:26. doi: 10.3389/fmicb.2016.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou Khalil JY, Andreani J, Raoult D, La Scola B. 2016. A rapid strategy for the isolation of new faustoviruses from environmental samples using Vermamoeba vermiformis. J Vis Exp 2016:e54104. doi: 10.3791/54104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 11.Wardley RC, de Andrade CM, Black DN, de Castro Portugal FL, Enjuanes L, Hess WR, Mebus C, Ordas A, Rutili D, Sanchez Vizcaino J, Vigario JD, Wilkinson PJ, Moura Nunes JF, Thomson G. 1983. African swine fever virus. Arch Virol 76:73–90. doi: 10.1007/BF01311692. [DOI] [PubMed] [Google Scholar]
- 12.Montgomery RE. 1921. On a form of swine fever occurring in British East Africa (Kenya Colony). J Comp Pathol Ther 34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- 13.Dixon LK, Escribano JM, Martins C, Rock DL, Salas ML, Wilkinson PJ. 2005. Asfarviridae, p 135–143. In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, UK. [Google Scholar]
- 14.Ogata H, Toyoda K, Tomaru Y, Nakayama N, Shirai Y, Claverie J-M, Nagasaki K. 2009. Remarkable sequence similarity between the dinoflagellate-infecting marine girus and the terrestrial pathogen African swine fever virus. Virol J 6:178. doi: 10.1186/1743-422X-6-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajrai L, Benamar S, Azhar E, Robert C, Levasseur A, Raoult D, La Scola B. 2016. Kaumoebavirus, a new virus that clusters with faustoviruses and Asfarviridae. Viruses 8:278. doi: 10.3390/v8110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benamar S, Reteno DGI, Bandaly V, Labas N, Raoult D, La Scola B. 2016. Faustoviruses: comparative genomics of new Megavirales family members. Front Microbiol 7:3. doi: 10.3389/fmicb.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, Feschotte C, Wu Z, Mueller RL. 2015. DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol 13:38. doi: 10.1186/s12915-015-0145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose T, Reteno DG, Benamar S, Hollerbach A, Colson P, La Scola B, Rossmann MG. 2016. Structure of faustovirus, a large dsDNA virus. Proc Natl Acad Sci U S A 113:6206–6211. doi: 10.1073/pnas.1523999113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon LK, Chapman DAG, Netherton CL, Upton C. 2013. African swine fever virus replication and genomics. Virus Res 173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Nogal ML, González de Buitrago G, Rodríguez C, Cubelos B, Carrascosa AL, Salas ML, Revilla Y. 2001. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J Virol 75:2535–2543. doi: 10.1128/JVI.75.6.2535-2543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banjara S, Caria S, Dixon LK, Hinds MG, Kvansakul M. 2017. Structural insight into African swine fever virus A179L-mediated inhibition of apoptosis. J Virol 91:e02228-16. doi: 10.1128/JVI.02228-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granja AG, Perkins ND, Revilla Y. 2008. A238L inhibits NF-ATc2, NF-kappa B, and c-Jun activation through a novel mechanism involving protein kinase C-theta-mediated up-regulation of the amino-terminal transactivation domain of p300. J Immunol 180:2429–2442. doi: 10.4049/jimmunol.180.4.2429. [DOI] [PubMed] [Google Scholar]
- 24.Hurtado C, Bustos MJ, Granja AG, de León P, Sabina P, López-Viñas E, Gómez-Puertas P, Revilla Y, Carrascosa AL. 2011. The African swine fever virus lectin EP153R modulates the surface membrane expression of MHC class I antigens. Arch Virol 156:219–234. doi: 10.1007/s00705-010-0846-2. [DOI] [PubMed] [Google Scholar]
- 25.Borca MV, Carrillo C, Zsak L, Laegreid WW, Kutish GF, Neilan JG, Burrage TG, Rock DL. 1998. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J Virol 72:2881–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Moon A, Childs K, Goodbourn S, Dixon LK. 2010. The African swine fever virus DP71L protein recruits the protein phosphatase 1 catalytic subunit to dephosphorylate eIF2alpha and inhibits CHOP induction but is dispensable for these activities during virus infection. J Virol 84:10681–10689. doi: 10.1128/JVI.01027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piégu B, Asgari S, Bideshi D, Federici BA, Bigot Y. 2015. Evolutionary relationships of iridoviruses and divergence of ascoviruses from invertebrate iridoviruses in the superfamily Megavirales. Mol Phylogenet Evol 84:44–52. doi: 10.1016/j.ympev.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Guinat C, Gogin A, Blome S, Keil G, Pollin R, Pfeiffer DU, Dixon L. 2016. Transmission routes of African swine fever virus to domestic pigs: current knowledge and future research directions. Vet Rec 178:262–267. doi: 10.1136/vr.103593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernáez B, Guerra M, Salas ML, Andrés G. 2016. African swine fever virus undergoes outer envelope disruption, capsid disassembly and inner envelope fusion before core release from multivesicular endosomes. PLoS Pathog 12:e1005595. doi: 10.1371/journal.ppat.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalil JYB, Langlois T, Andreani J, Sorraing J-M, Raoult D, Camoin L, La Scola B. 2017. Flow cytometry sorting to separate viable giant viruses from amoeba co-culture supernatants. Front Cell Infect Microbiol 6:e17722. doi: 10.3389/fcimb.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Guo F, Jiang W. 2013. Single particle cryo-electron microscopy and 3-D reconstruction of viruses, p 401–443. In Kuo J. (ed), Electron microscopy: methods and protocols. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 35.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boetzer M, Pirovano W. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics 15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alva V, Nam S-Z, Söding J, Lupas AN. 2016. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]