ABSTRACT
Chrysochromulina ericina virus CeV-01B (CeV) was isolated from Norwegian coastal waters in 1998. Its icosahedral particle is 160 nm in diameter and encloses a 474-kb double-stranded DNA (dsDNA) genome. This virus, although infecting a microalga (the haptophyceae Haptolina ericina, formerly Chrysochromulina ericina), is phylogenetically related to members of the Mimiviridae family, initially established with the acanthamoeba-infecting mimivirus and megavirus as prototypes. This family was later split into two genera (Mimivirus and Cafeteriavirus) following the characterization of a virus infecting the heterotrophic stramenopile Cafeteria roenbergensis (CroV). CeV, as well as two of its close relatives, which infect the unicellular photosynthetic eukaryotes Phaeocystis globosa (Phaeocystis globosa virus [PgV]) and Aureococcus anophagefferens (Aureococcus anophagefferens virus [AaV]), are currently unclassified by the International Committee on Viral Taxonomy (ICTV). The detailed comparative analysis of the CeV genome presented here confirms the phylogenetic affinity of this emerging group of microalga-infecting viruses with the Mimiviridae but argues in favor of their classification inside a distinct clade within the family. Although CeV, PgV, and AaV share more common features among them than with the larger Mimiviridae, they also exhibit a large complement of unique genes, attesting to their complex evolutionary history. We identified several gene fusion events and cases of convergent evolution involving independent lateral gene acquisitions. Finally, CeV possesses an unusual number of inteins, some of which are closely related despite being inserted in nonhomologous genes. This appears to contradict the paradigm of allele-specific inteins and suggests that the Mimiviridae are especially efficient in spreading inteins while enlarging their repertoire of homing genes.
IMPORTANCE Although it infects the microalga Chrysochromulina ericina, CeV is more closely related to acanthamoeba-infecting viruses of the Mimiviridae family than to any member of the Phycodnaviridae, the ICTV-approved family historically including all alga-infecting large dsDNA viruses. CeV, as well as its relatives that infect the microalgae Phaeocystic globosa (PgV) and Aureococcus anophagefferens (AaV), remains officially unclassified and a source of confusion in the literature. Our comparative analysis of the CeV genome in the context of this emerging group of alga-infecting viruses suggests that they belong to a distinct clade within the established Mimiviridae family. The presence of a large number of unique genes as well as specific gene fusion events, evolutionary convergences, and inteins integrated at unusual locations document the complex evolutionary history of the CeV lineage.
KEYWORDS: Aureococcus anophagefferens virus, Chrysochromulina ericina virus, Haptolina ericina virus, Megamimivirinae, Mesomimivirinae, Mimiviridae, nucleocytoplasmic virus, Phaeocystis globosa virus
INTRODUCTION
Several new viral families have been recently created (or proposed) following the discovery of highly diverse double-stranded DNA (dsDNA) giant viruses (initially defined as those with particles visible under a light microscope), all of which exhibit large genomes (>300 kb) and infect unicellular eukaryotes (reviewed in references 1–3). Among those new families, the most populated is the Mimiviridae, and it is the only one officially recognized by the International Committee on Viral Taxonomy (ICTV). The Mimiviridae comprises two registered genera: the Mimivirus and the Cafeteriavirus. The Mimivirus genus includes several dozen members distributed among three clades (A, B, and C), all of which infect Acanthamoeba and have pseudoicosahedral particles approximately 700 nm in diameter and genomes of about one megabase in length (4). The genus Cafeteriavirus contains a single member, Cafeteria roenbergensis virus (CroV). This virus is markedly different from the mimiviruses, having smaller particles (300 nm in diameter) and a smaller, 730-kb genome, with its host being the heterotrophic stramenopile C. roenbergensis (5). Prior to defining these two genera, metagenomic studies had already hinted at the presence of mimivirus relatives in marine environments (6). The successful isolation and characterization of several of these viruses showed that they correspond to smaller icosahedral particles (140 to 180 nm in diameter) packing smaller dsDNA genomes (370 to 475 kb in length) (7–9). In core gene phylogenies, these viruses clearly cluster with the Mimiviridae, although they appear to constitute a distinct clade (Fig. 1) (1, 2, 7, 8). This clade comprises only viruses infecting photosynthetic hosts (i.e., microalgae) from different taxa: haptophyceae for Phaeocystis globosa virus (PgV) and Haptolina ericina virus and stramenopile for Aureococcus anophagefferens virus (AaV). This emerging subgroup within the Mimiviridae appears to include other lesser characterized members, such as Phaeocystis pouchetii virus (PpV), Prymnesium kappa virus (PkV) (10), and Pyramimonas orientalis virus (PoV). It also includes a number of other nonisolated candidates predicted solely from the assembly of metagenomics sequence data (11), such as the Organic Lake phycodnaviruses (OLPV1 and -2) (12). Since these viruses infect algae, a paraphyletic group of organisms, they were originally classified as members of the Phycodnaviridae, although this historical family increasingly encompasses viruses with little phylogenetic relationship (13, 14). One of the goals of the present study is to clarify this issue.
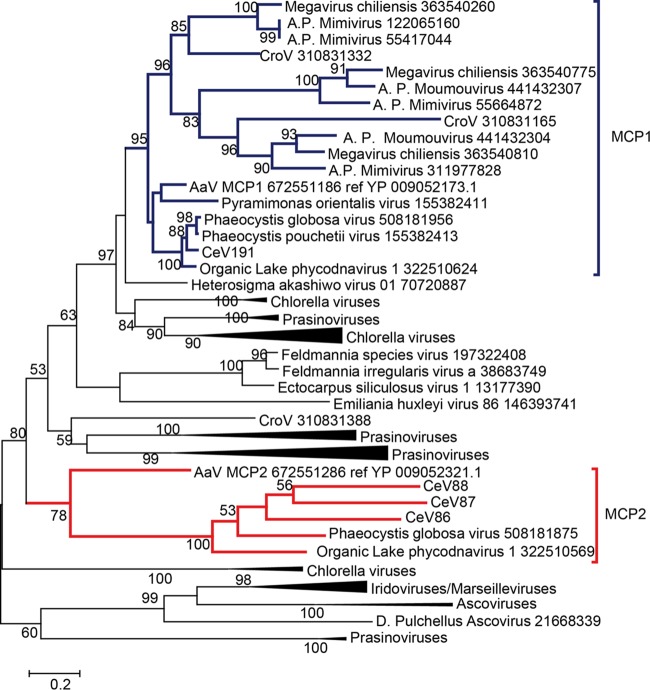
FIG 1.
Phylogeny of the concatenation of MCP, DNAPol B, and DNA packaging ATPase. The phylogenetic tree was built using PhyML (58) based on multiple alignments generated using Expresso (60). The tree was rooted with Ectocarpus siliculosus virus (Phaeovirus). Representatives of the genera Mimivirus (blue) and Cafeteriavirus (red) have been included, as well as the three fully sequenced alga-infecting Mimiviridae relatives (green). The tree was drawn using MEGA7 (61).
Aside from their gene-based phylogenetic clustering within the Mimiviridae, the members of this emerging clade exhibit additional features, such as AT-rich genomes encoding full DNA transcription and replication machinery (needed for their intracytoplasmic replication) (2) and a special version of the MutS mismatch DNA repair protein strangely related to octocorals (15). Most of them also encode an asparagine synthase (AsnS) (11). The Mimiviridae family also harbors the only viruses known to allow the replication of virophages (7, 12, 16–18).
Chrysochromulina ericina virus (CeV) was isolated from Norwegian coastal waters in 1998 but was only recently fully sequenced (9). It replicates within the cytoplasm of H. ericina (formerly Chrysochromulina ericina) with a lytic cycle lasting 14 to 19 h, resulting in the release of thousands of icosahedral particles 160 nm in diameter (19). Its host is distributed worldwide. Here, we performed a detailed comparative analysis of the genome of CeV and of its closest fully sequenced relatives (PgV and AaV) to provide support for their classification within a new clade in the Mimiviridae, as well as to investigate their evolutionary relationship with the rest of the family.
RESULTS
CeV genome global analysis.
In line with its virion size, the CeV genome (473,558 bp) is larger than those of PgV and AaV (Table 1) and is the largest among those of alga-infecting viruses from any other family. We identified 512 putative protein-coding genes with an average length of 280 codons (ranging from 41 to 2,317 codons) and 12 tRNA genes (two tRNALeu, two tRNASer, one tRNAAla, one tRNAIle, two tRNALys, one tRNAGln, one tRNAAsn, one tRNAArg, and one tRNAGly). Intergenic sequences are very short (82.5 nucleotides [nt] on average) and exhibit higher A+T contents than average (85% versus 73.7%), suggesting that the loss of C+G nucleotides is an ongoing evolutionary process only slowed down by the negative selection pressure applied on protein-coding regions (Table 1). Compared to the NCBI nonredundant sequence database NR (including Mimiviridae), 43% (218) of the predicted proteins did not exhibit a significant match (E value of <10−5 by BLASTP). This proportion of ORFans (i.e., without recognizable homologs within the whole NR database) is similar to that of other alga-infecting Mimiviridae (AaV, 45%; OLPV1, 44%; PgV, 43%). Among the 293 predicted proteins with a database homolog, 221 (75.4%) had their best match in eukaryote-infecting large dsDNA viruses, most of which (214/221, i.e., 96.8%) were members of the Mimiviridae family, mainly PgV (with 144 best matches) and the two OLPVs (with 30 best matches). The 72 nonviral best matches were distributed between bacteria (30) and eukaryotes (43), including 7 open reading frames (ORFs) in haptophytes (i.e., the taxon of the CeV host), pointing out potential horizontal gene transfers (HGT). A comparison (dot plot) of the orthologous gene positions in the genomes of CeV's closest relatives indicates numerous rearrangements (data not shown).
TABLE 1.
Genomic features of the Mimiviridae
| Virus | Genome size (kbp) | No. of ORFs | Avg ORF size (bp) | Genome GC% | Coding density | GC% |
|
|---|---|---|---|---|---|---|---|
| ORF | Inter-ORF | ||||||
| Megavirus | 1,259 | 1,120 | 1,015 | 25.3 | 0.90 | 26.5 | 14.1 |
| Mimivirus | 1,181 | 979 | 1,080 | 28.0 | 0.90 | 29.1 | 18.3 |
| CroV | 730 | 544 | 1,025 | 23.3 | 0.76 | 24.3 | 20.1 |
| AaV | 371 | 377 | 870 | 28.7 | 0.88 | 29.9 | 19.8 |
| OLPV1a | 697 | 29.5 | 0.86 | 30.2 | 25.0 | ||
| PgV | 460 | 434 | 960 | 32.0 | 0.91 | 33.7 | 15.3 |
| CeV | 474 | 512 | 840 | 25.4 | 0.91 | 26.3 | 15.7 |
OLPV1 statistics are based on a 344,723-bp-long contig (12).
Phylogenetic analyses.
To reconstruct the relationship between the viruses composing the Mimiviridae alga-infecting clade, we used a concatenation of the DNA polymerases, ATP DNA-packaging enzymes, and closest orthologs of the major capsid protein (MCP1). Consistent with the distribution of CeV best hits in the NR database, PgV appears to be its closest relative among fully sequenced viruses (Fig. 1). However, when we included OLPV1 in the analysis, no consistent pattern was found for the branching order of CeV, PgV, and OLPV after they diverged from their common ancestor with AaV. Moreover, applying the Shimodaira and Hasegawa (SH) test (see Materials and Methods) to each of the 39 groups of orthologous proteins shared by the 4 viruses (CeV, PgV, OLPV, and AaV) did not produce a conclusive answer. Such ambiguous results could be due to divergence times too close to be resolved, orthologous gene exchanges among these viruses, and/or compositional constraint similarities blurring the phylogenetic signal.
Gene content.
Using OrthoMCL (20), we analyzed the groups of orthologues shared by mimivirus, CroV, and the three fully sequenced alga-infecting viruses, CeV, PgV, and AaV (Fig. 2). A striking result is that among the hundreds of proteins encoded by these clearly related viruses, only 30 are shared by all of them (a number dropping down to 19 when including Moumouvirus and Megavirus). Thus, including the new clade of alga-infecting viruses in the Mimiviridae causes the extended family to rest on an amazingly small proportion of common core genes. On the other hand, each viral genome exhibits a high number of unique genes (i.e., without recognizable homologs in the other Mimiviridae) (305 for CeV, 274 for AaV, 232 for PgV, 387 for CroV, and 845 for mimivirus) (Fig. 2). As 68% of CeV-unique genes correspond to ORFans, postulating a high frequency of horizontal exchanges with known viruses or cellular organisms is clearly not sufficient to explain their origin. Finally, some paralogs are sporadically present in the various viruses. Altogether, these large differences in the gene contents of these various viruses, which nevertheless share a strong phylogenetic signal of common ancestry, are rather puzzling and at least suggest a complex evolutionary history of the family.
FIG 2.
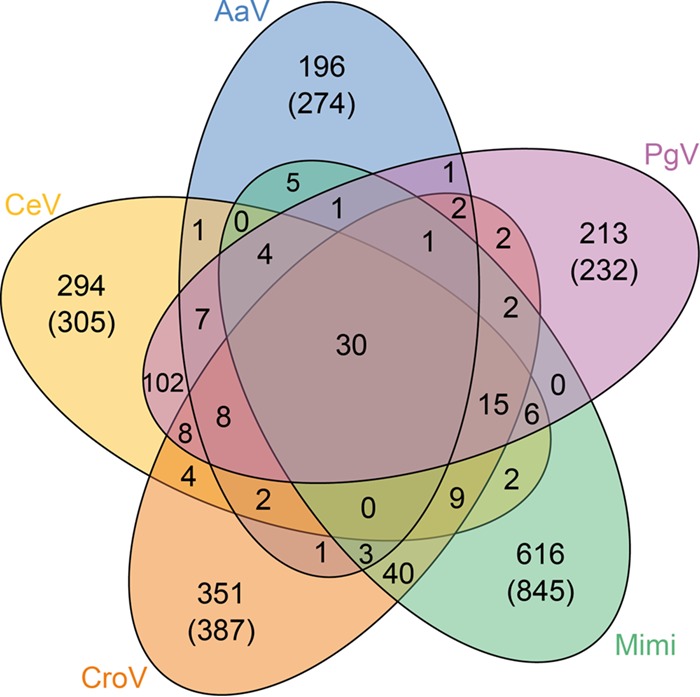
Venn diagram indicating the global proximity in gene content of CeV, its two closest relatives, PgV and AaV, and one member of each genus of the family Mimiviridae (Cafeteriavirus genus, CroV; Mimivirus genus, Mimi). The numbers in parentheses correspond to the raw number of encoded proteins without a homolog in the four other viruses. The numbers without parentheses indicate how many distinct clusters they constitute. The analysis was driven using OrthoMCL software (20), with a 10−5 E-value threshold and 1.5-mcl inflation parameter.
Unique features common to the alga-infecting Mimiviridae.
We further investigated if the alga-infecting viruses possessed common genes/functions not shared with the other Mimiviridae. As PgV, CeV, and AaV infect photosynthetic protozoans (at variance with the other Mimiviridae), we postulated that genomic features exclusive to them could be linked to this specific lifestyle. Only 7 of such shared genes were identified, among which a single one corresponds to a predicted function: an ERCC4-type DNA repair nuclease (YP_009173624.1, or CeV_369). Such enzymes are usually part of the cellular response against UV-induced DNA damage (21, 22), also known to occur in eukaryotic viruses (23). While infecting photosynthetic hosts, alga-infecting Mimiviridae might be exposed to sunlight-induced genome damages, making these nucleases useful for repair. Such an ERCC4-dependent process extends the known diversity of viral responses to light-induced DNA damage. Other marine viruses possess a variety of mechanisms to repair DNA damage, which are either host dependent or host independent (23–25). These might be essential for the maintenance of viral communities (24), perhaps indirectly by protecting their host, as suggested by the reduced sensitivity to UV-B stress of microalgae cocultured with viruses compared to those in virus-free cultures (23).
Unrelated to the above-described function, another common feature of the alga-infecting Mimiviridae is the presence of two paralogs of the second-largest subunit of the DNA-directed RNA polymerase II (RPB2) (CeV_276 and CeV_288). As none of these copies appears defective and their sequences are quite divergent (only 34.2% identical), two distinct forms of transcriptional complexes might be formed following their interaction with the single RPB1 subunit encoded by these viruses. Combinations of different subunits leading to different versions of RNA polymerases IV and V (pol IV and pol V, respectively) are known in plants (26). These complexes evolved specialized roles (e.g., nonredundant gene silencing) (27, 28). Similarly, alternative RNA pol II complexes formed in alga-infecting Mimiviridae could play different roles. The duplicated paralogs present longer branches (Fig. 3), consistent with an accelerated rate of evolution. The presence of a C-terminal extension on the sequences of this group of paralogs would also be consistent with a functional modification: such additional residues could cause a change in substrate specificity. Phylogenetic reconstruction (Fig. 3) suggests that the RPB2 paralogs originated from a single duplication that occurred after the divergence from the rest of the Mimiviridae. In addition, AaV exhibits two copies of RNA polymerase II large subunits (RPB1) (AaV_242 and AaV_320) (8).
FIG 3.
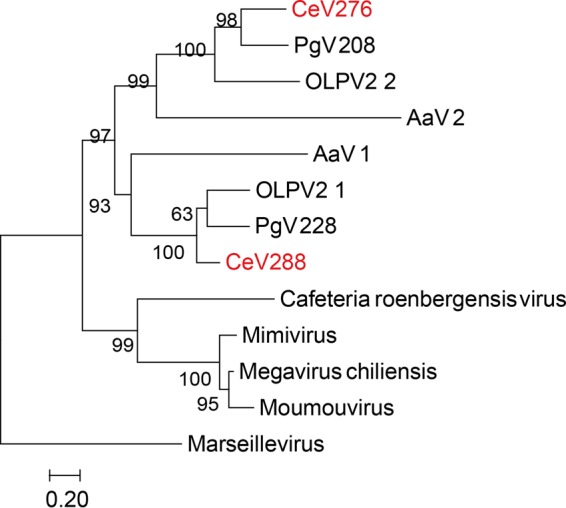
Duplication of the second largest subunit of the DNA-directed RNA polymerase II (RPB2) in CeV, PgV, and AaV. A maximum likelihood phylogenetic tree (58) of RPB2, aligned using Tcoffee (62) (Mcoffee mode), was constructed. Statistical branch supports (in percentages) for SH-like local support tests are given beside nodes. This phylogeny strongly supports a separate lineage leading to CeV, AaV, and PgV.
The alga-infecting Mimiviridae also share two distinct versions of major capsid proteins. The MCP1 paralogs exhibit a phylogeny consistent with the “species” tree and correspond to the least-divergent version of the major capsid protein common to all Mimiviridae (Fig. 4, top). The Mimivirus and Cafeteriavirus genera possess a second copy of MCP1, the duplication of which clearly predates their divergence (Fig. 4). In contrast, another type of MCP2 paralog (Fig. 4, bottom) is uniquely found in CeV (CeV_86, CeV_87, and CeV_88), PgV, and AaV. It probably results from an exchange with other alga-infecting large DNA viruses.
FIG 4.
Relationship of the two major capsid protein homologs (MCP) found in CeV, PgV, and AaV. A maximum likelihood phylogenetic tree (58) of MCP, aligned using Tcoffee (62) (Expresso mode), was constructed. Statistical branch supports (in percentages) are given beside nodes.
Altogether, these phyletic patterns specific to AaV, PgV, and CeV further support grouping them in a clade distinct from the other Mimiviridae.
Specific features shared only by CeV and PgV.
CeV, PgV, and OLPV share homologs to the cold shock protein, which is known to act as an RNA chaperone (29). This protein, not found in other eukaryotic large dsDNA viruses, either was acquired by the alga-infecting lineage and later lost by AaV or was acquired after its divergence from AaV. We noticed that some dsDNA phages also encode similar cold shock proteins, suggesting that proper RNA folding is a recurrent constraint among unrelated viruses.
The analysis of PgV's genomic repeats led to the identification of an ORFan present in 12 copies, designated PgV_MIGE (major interspersed genomic element) (7). CeV possesses six copies of MIGE homologs. Interestingly, phylogenetic reconstruction grouped the PgV's MIGE and CeV's MIGE in separate clusters (Fig. 5). This suggests that MIGE was initially a single-copy gene in each virus before undergoing multiple duplications after their divergence. Analysis of CeV-MIGE did not hint at the mechanism by which this genetic element is duplicated and/or moved around. At variance with PgV, MIGE homologs in CeV lack an associated noncoding highly conserved region (7).
FIG 5.
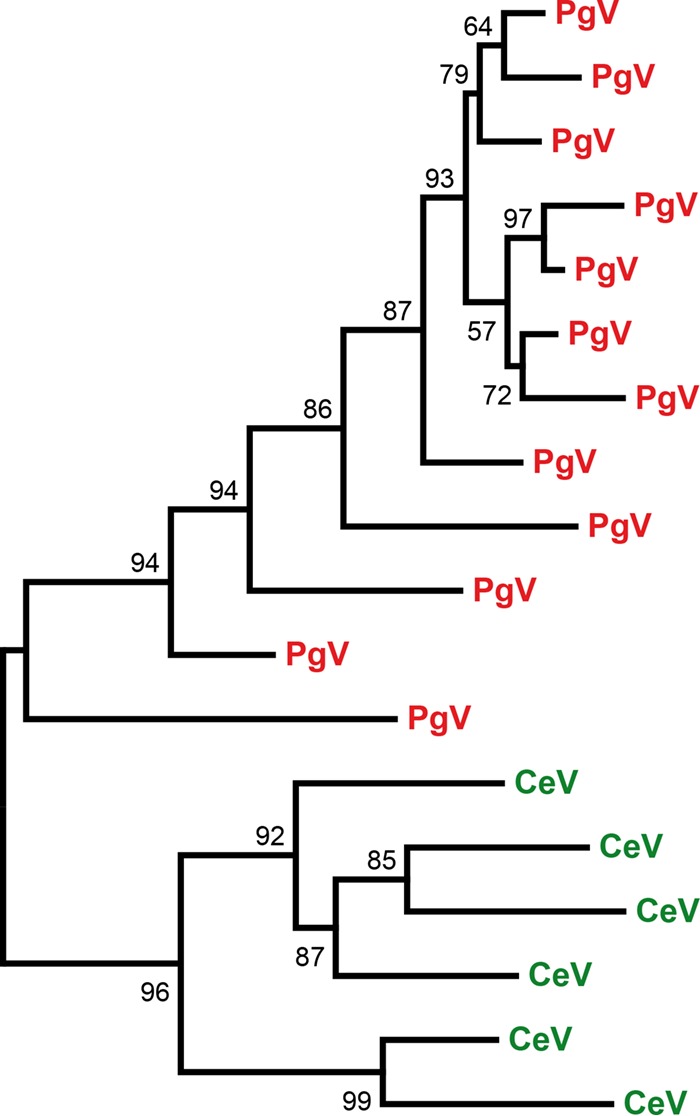
Independent MIGE spreading in CeV and PgV. A maximum likelihood phylogenetic tree (58) of MIGE protein sequences, aligned using Muscle (57), was constructed. Statistical branch supports (in percentages) for SH-like local support tests are given beside nodes.
CeV and PgV also share a fusion event between a DNA polymerase X (DNAPolX) and a DNA ligase that is discussed in a later section.
Predicted functions unique to CeV.
Among the 305 proteins unique to CeV, only 106 (35%) have a database homolog, among which only 52 are associated with functional attributes (listed in Table 2). Except for one light-harvesting complex protein (CeV_128) that will be discussed in a later section, none of these predicted functions have previously been shown to be specifically linked to the viral infection of algae.
TABLE 2.
Protein-coding genes unique to CeV and associated with functional attributes
| ORF no. | Predicted function or detected domain |
|---|---|
| CeV_002 | Multiple glycosyltransferase domain |
| CeV_003 | Methyltransferase |
| CeV_008 | Ubox domain |
| CeV_009 | Alpha-1,2-fucosyltransferase |
| CeV_023 | Alkylated (methylated) DNA repair protein |
| CeV_033 | Papain-like cysteine peptidase |
| CeV_053 | RING-finger domain |
| CeV_096 | Putative patatin-like phospholipase |
| CeV_113 | Intein containing GDP-mannose 4,6-dehydratase |
| CeV_128 | Light-harvesting complex protein |
| CeV_137 | Superoxide dismutase Cu-Zn |
| CeV_139 | Arginase |
| CeV_146 | Trans-2-enoyl-coenzyme A reductase (TER) and 2,4-dienoyl-coenzyme A reductase (DECR) |
| CeV_149 | Class V aminotransferase |
| CeV_151 | Putative phosphotransferase |
| CeV_152 | Quaternary ammonium transporter |
| CeV_154 | Fe-S cluster assembly scaffold protein |
| CeV_155 | Zinc finger, C3HC4 type domain-containing protein, RING superfamily |
| CeV_161 | Putative prenyltransferase |
| CeV_171 | Phospholipase/carboxylesterase |
| CeV_176 | Collagen and repeat-containing protein |
| CeV_179 | Multiple type acyltransferase domains |
| CeV_180 | Glucosyltransferase TPR |
| CeV_183 | Glycosyltransferase family 2 |
| CeV_184 | Collagen triple helix |
| CeV_194 | PAN/APPLE-like domain |
| CeV_195 | Repeat containing Hsp70-like protein |
| CeV_196 | Ubox/RING superfamily domain |
| CeV_201 | RING-finger domain |
| CeV_213 | Protein disulfide isomerase (PDIa) |
| CeV_218 | Glycosyl hydrolase family 16 |
| CeV_233 | Putative AHH-like nuclease |
| CeV_252 | Proline-rich repeats |
| CeV_256 | Toll-like receptor |
| CeV_265 | Partial perforin domain-like |
| CeV_267 | Putative AAA+ family ATPase |
| CeV_311 | Galactose binding lectin domain |
| CeV_323 | Putative permease |
| CeV_324 | N-acyltransferase/N-myristoyltransferase |
| CeV_327 | Putative AAA+ family ATPase |
| CeV_359 | Link (hyaluronan-binding) domain |
| CeV_361 | Ring finger domain |
| CeV_366 | Protein disulfide-isomerase domain |
| CeV_372 | Hsp70-like protein |
| CeV_373 | Acetylpolyamide aminohydrolase (histone deacetylase) |
| CeV_404 | ATP-dependent Clp protease, proteolytic subunit |
| CeV_415 | Putative syntaxin, SNARE domain |
| CeV_433 | Class II HMG-box domain |
| CeV_440 | HMG box domain |
| CeV_463 | RING domain |
| CeV_464 | YABBY domain |
| CeV_467 | Catalytic core of Asn/Asp-ARNt synthetase |
CeV, a privileged host for spreading inteins?
Inteins are mobile genetic elements encoding protein introns that remove themselves from host proteins through an autocatalytic excision and protein-splicing process (30). Fully functional inteins encode a homing endonuclease that mediates their integration at specific genomic sites. Different classes of inteins are found associated with the same, usually highly conserved, proteins (and thus genomic sequences), most of which are essential enzymes involved in DNA processing, replication, or synthesis. Nonallelic inteins (i.e., inteins not associated with the same insertion sites and/or protein-coding genes) do not exhibit significant sequence similarity, although they all presumably share an ancestor (30).
CeV encodes 8 inteins, to date the largest number among viruses. These inteins are inserted in 7 different ORFs: two in the ribonucleotide reductase large subunit (CeV_219), one in a DEAD-like RNA helicase (CeV_416), one in the DNA polymerase (CeV_365), one in an ATP-dependent Lon peptidase (CeV_043), one in an RPB2 paralog (CeV_288), one in a GDP-mannose 4,6-dehydratase (CeV_113), and one at the C terminus of a protein (CeV_451), concatenating an N-terminal U-box domain and a von Willebrand factor (vWA) domain. In general, intein insertions are strongly biased toward DNA-processing enzymes (30, 31). Among other explanations (30, 31), this bias might result from the fact that viruses are the main vectors of inteins within eukaryotes (32) and that viral metabolisms are mostly limited to DNA processing. In that context, the inteins hosted in a CeV's GDP-mannose 4,6-dehydratase and Lon peptidase are noticeable exceptions, to our knowledge the first such cases reported in eukaryotic viruses. With their extended metabolisms, viruses with larger genomes, such as members of the Mimiviridae, might thus expand the range of homing genes/enzymes for inteins. According to the current paradigm, inteins can be maintained only within essential genes. This is likely true for 5 of the 7 genes cited above (that belong to the Mimiviridae core genes) (3), the exception being the GDP-mannose 4,6-dehydratase that has no homolog in other Mimiviridae. Such a function might nevertheless be important, as it was independently acquired by a number of other large dsDNA viruses that infect various algae (Chlorella and Prasinophyceae) (33). The CeV homolog might thus be necessary in an alga-infecting context. The closest homologues to CeV GDP-mannose 4,6-dehydratase are intein-free bacterial enzymes (although intein-containing bacterial enzymes exist that may not have been sequenced yet).
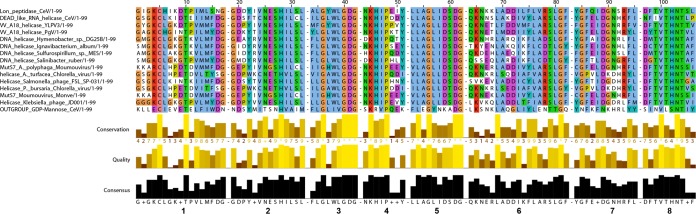
Inteins normally insert in highly conserved regions of essential proteins. As the paradigm goes, the strong conservative constraints exerted on these regions ensures the correct excision (from the protein) or homing (into the DNA) processes. Surprisingly, we found that two similar inteins of the same prototype (standard class 1 with a homing endonuclease of the LAGLIDADG type) have been inserted in two unrelated CeV genes/proteins: a Lon protease and a DEAD box helicase (Fig. 6 and Table 3). Moreover, a similar intein helicase-targeted allele is inserted in different enzymes in other large DNA viruses: MutS7 in Moumouvirus, the VVA18 helicase in PgV and Yellowstone Lake phycodnavirus (YLP) (percent identity with CeV Lon protease intein ranging from 42.5% to 54%), and, besides the Mimiviridae, DNA helicase B of 2 chloroviruses (identity with CeV Lon protease intein, 40% and 38%). Other related inteins are hosted by DNA helicases from phages or bacteria (identity with CeV Lon protease intein ranging from 39% to 43%). The 8 blocks specific to this intein prototype (34) are well conserved (Fig. 6). Although unrelated, these enzymes share a P-loop NTPase domain. All inteins but those in MutS7 are inserted in the ATP/GTP binding site (i.e., the Walker A or P-loop domain), precisely at the GK/T site. This might be sufficient for this prototype of intein to properly excise. More puzzling is the way by which this intein might have jumped from one enzyme to another. Indeed, after being cut by the intein-encoded endonuclease, the free intein gene should proceed with homing by homologous base pairing with the intein-containing allele, which serves as the template for the polymerase. This scenario appears unlikely given the limited similarity of the extein DNA sequences (Fig. 7). Another intriguing fact is that the intein is not inserted in the P-loop domain of MutS7 but is inserted at an AR/S site, 30 amino acids upstream. Furthermore, these amino acids are not conserved in the intein-free MutS7 found in other Mimiviridae. Altogether, these cases constitute violations of the current allele-specific intein paradigm. They suggest a mechanism specific to the Mimiviridae spreading inteins while widening their homing range.
FIG 6.
Helicase-type intein inserted in the CeV Lon peptidase. Multiple alignment, computed with Muscle (57), of the 8 blocks (numbered below the alignment) characteristic of inteins (Table 3). The intein hosted by the CeV GDP-mannose 4,6-dehydratase is included for comparison.
TABLE 3.
High similarity of the CeV Lon peptidase intein to those usually found in various DNA helicases
| Intein (no.) | % Similarity to intein no.a: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Lon peptidase, CeV (1) | 100 | 44 | 44 | 54 | 42 | 40 | 41 | 39 | 43 | 40 | 42 | 38 | 42 | 43 |
| DEAD-like RNA helicase, CeV (2) | 43 | 100 | 40 | 44 | 36 | 35 | 37 | 33 | 42 | 36 | 40 | 36 | 41 | 37 |
| Putative VV A18 helicase, YLPV3 (3) | 44 | 40 | 100 | 49 | 42 | 40 | 44 | 41 | 42 | 37 | 44 | 37 | 42 | 47 |
| VV A18-like helicase, PgV (4) | 54 | 43 | 50 | 100 | 43 | 44 | 46 | 41 | 42 | 42 | 46 | 42 | 42 | 45 |
| DNA helicase, Hymenobacter sp. (5) | 42 | 35 | 42 | 43 | 100 | 59 | 62 | 58 | 36 | 33 | 41 | 33 | 35 | 43 |
| DNA helicase, Ignavibacterium album (6) | 40 | 36 | 40 | 43 | 59 | 100 | 60 | 62 | 37 | 32 | 41 | 32 | 36 | 45 |
| DNA helicase, Sulfurospirillum sp. (7) | 41 | 36 | 44 | 46 | 62 | 60 | 100 | 55 | 34 | 32 | 38 | 32 | 34 | 45 |
| DNA helicase, Salinibacter ruber (8) | 39 | 33 | 41 | 41 | 58 | 62 | 55 | 100 | 34 | 33 | 37 | 33 | 34 | 42 |
| MutS7, A. P. Moumouvirus (9) | 43 | 42 | 42 | 42 | 36 | 36 | 34 | 34 | 100 | 37 | 37 | 36 | 94 | 40 |
| Helicase, ATCV NTS-1 (10) | 40 | 35 | 37 | 42 | 33 | 31 | 32 | 33 | 37 | 100 | 35 | 75 | 37 | 51 |
| Helicase, Salmonella phage (11) | 42 | 40 | 44 | 46 | 41 | 40 | 38 | 37 | 37 | 35 | 100 | 36 | 38 | 36 |
| Hypothetical protein B508R, PBCV (12) | 38 | 35 | 37 | 42 | 33 | 32 | 32 | 33 | 36 | 75 | 36 | 100 | 35 | 41 |
| MutS7, Moumouvirus Monve (13) | 42 | 41 | 42 | 42 | 35 | 36 | 34 | 34 | 94 | 37 | 38 | 35 | 100 | 25 |
| Helicase, Klebsiella phage (14) | 43 | 37 | 47 | 45 | 43 | 44 | 45 | 42 | 40 | 37 | 51 | 36 | 41 | 100 |
| Outgroup GDP-mannose CeV | 24 | 26 | 22 | 20 | 25 | 20 | ||||||||
The pairwise percentages of identity between these homologous inteins is given. The values for the nonallelic intein hosted by CeV GDP-mannose 4,6 dehydratase is included for comparison when pairwise alignment was possible. YLPV3, Yellowstone Lake phycodnavirus 3; ATCV, Acanthocystis turfacea chlorella virus; PBCV, Paramecium bursaria chlorella virus.
FIG 7.
Extein DNA sequences in different genes hosting similar intein alleles (Fig. 6) are not similar.
Convergent acquisition of host genes.
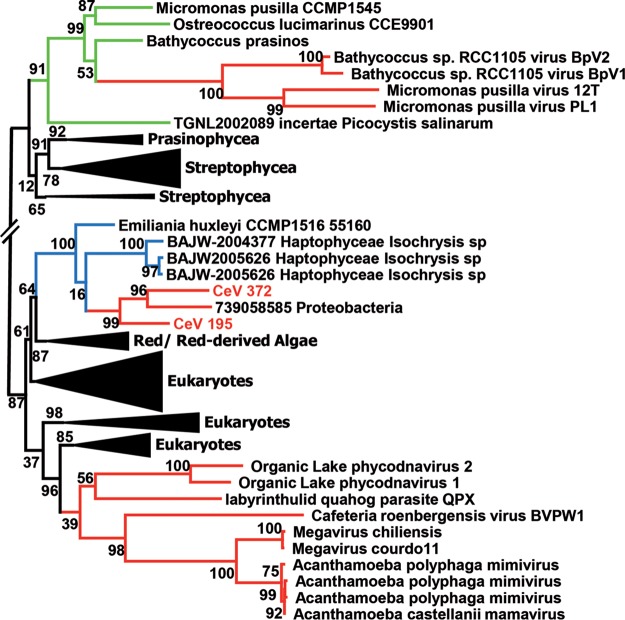
Detailed attention was paid to the CeV-encoded light-harvesting complex protein (LHC) (CeV_128) of the LIL (light-harvesting light) family. The presence of such a gene coding for a component of the photosynthesis apparatus was rather unexpected. Further analyses led to the discovery that other alga-infecting viruses encode proteins of the LIL family (Fig. 8). Their phylogenies clearly suggest that they were acquired from their hosts through three independent events (one for CeV and two for the prasinoviruses) (Fig. 8). Besides the expected chlorophyll binding (CB) motifs and 3 predicted transmembrane helices (35), all viral LIL proteins contain an N-terminal transit peptide targeting them to their respective host chloroplast type (36) (secondary red for H. ericina and primary green for the prasinophytes). This suggests that these proteins are functional. Rather than collecting incoming light for photosynthesis, some nucleus-encoded eukaryotic LIL proteins are involved in photoprotection, such as nonphotochemical quenching (NPQ) (35, 37). This might be the role of the viral LIL homologs.
FIG 8.
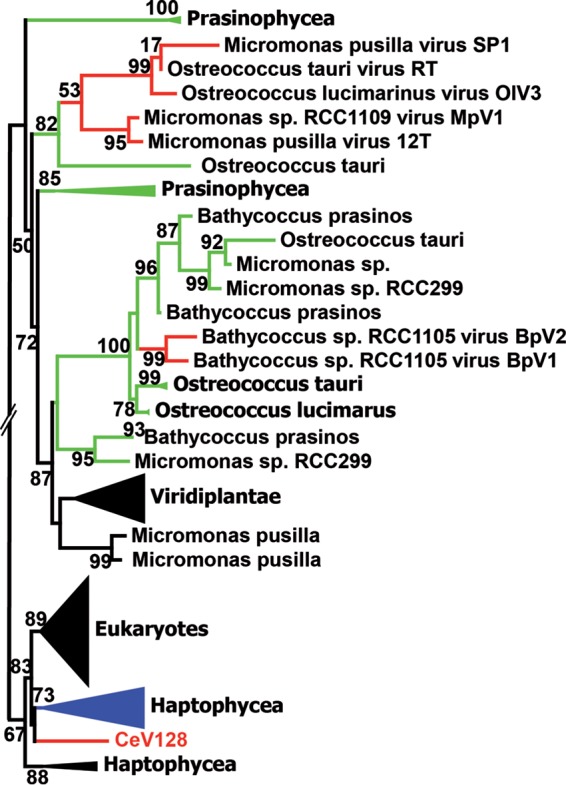
Convergent acquisitions of host LIL proteins. A maximum likelihood phylogenetic tree (58) of LIL proteins, aligned using Muscle (57), was constructed. Statistical branch supports (in percentages) for the S-H-like local support tests are given beside nodes. Branches corresponding to viruses are colored red, with blue for haptophyceae and green for prasinophyceae. Sequences used for the analysis were selected using BLAST Explorer as implemented in Phylogeny.fr (63).
We then systematically looked for additional cases of convergent acquisition involving other genes. We identified 2 more, concerning the DNAK (HSP70) chaperone (CeV_372 and CeV_195) (Fig. 9) and a U-box domain at the C terminus of the CeV_008-encoded protein (data not shown). CeV and two prasinoviruses have independently acquired HSP70, likely from their hosts (Fig. 9). Interestingly, other Mimiviridae also encode homologs of this chaperone. However, these proteins appear to derive from an ancestral version of HSP70 already present at the root of the family tree. Thus, the HSP70 proteins found today in CeV might result from a nonhomologous replacement of the ancestral Mimiviridae version of the protein. The U-box domain mediates the ubiquitin conjugation of protein targeted for degradation in the proteasome (38). CeV (CeV_008) and EhV (EMVG_00184) appear to have independently acquired host-derived U-box domains.
FIG 9.
Convergent acquisitions of host HSP70 proteins. A maximum likelihood phylogenetic tree (58) of HSP70, aligned using Muscle (57), was constructed. Statistical branch supports (in percentages) for S-H-like local support test are given beside nodes. Viral branches are colored red, with blue for haptophyceae and green for prasinophyceae.
Gene fusions in CeV.
Gene fusions are genomic rearrangements that are thought to facilitate the coexpression and/or assembly of proteins initially encoded separately. The involved proteins could physically interact or be functionally associated (39, 40).
We systematically screened for potential gene fusions in the CeV genome by comparing the topology of homologous genes in other Mimiviridae. Two clear cases were identified: one (CeV_489) is a fusion between the DNAPolX and the NAD-dependent DNA ligase (also found in PgV), and the other (CeV_007) is a fusion between the uridyltransferase and the UDP-glucose 4,6-dehydratase (UDG). We did not detect such fusions in other viruses or cellular organisms.
The DNAPolX/NAD-dependent DNA ligase fusion makes functional sense, as these enzymes normally work in succession when participating in DNA repair: DNAPolX fills single-nucleotide gaps before their ends are sealed by the ligase (41, 42). CroV homologs of these two proteins are encapsidated, suggesting their role in prereplicative DNA repair (43). We noticed that the CeV and PgV fusion proteins exhibit very different linkers that could have resulted from 2 independent events. However, the analysis of similar fusions in the environmental database showed that such linkers are not conserved. The CeV and PgV proteins thus likely resulted from a unique fusion event prior to their divergence and that of their linkers. Interestingly, some bacteria also exhibit a fusion between an ATP-dependent DNA ligase and a DNA polymerase domain (44). This might constitute a case of functional evolutionary convergence between bacteria and eukaryotic viruses resulting in more efficient DNA repair enzymes.
The unique fusion between a uridyltransferase domain and UDG identified in CeV (CeV_007) might optimize the synthesis of l-rhamnose, known to be involved in the glycosylation of structural proteins in mimivirus and certain chloroviruses (45). This fusion occurred after the duplication of the uridyltransferase gene (CeV_479), originally involved in the three-component UDP-N-acetylglucosamine biosynthetic pathway (46) found in large Mimiviridae. This duplication might have opened the way to the fusion event (47).
DISCUSSION
Convergent evolution in CeV and other large dsDNA viruses.
This study identified several cases where homologs of the same gene were independently acquired by CeV and other viruses, most likely from their hosts. The best example of such convergent acquisition is that of the light-harvesting complex protein (LHC) that is found in CeV and seven species of prasinoviruses (Fig. 8). Such gene transfers are reminiscent of the acquisition of core photosystem components (including a remote member of the LIL family, HLIP) by cyanophages from their bacterial hosts (35, 37, 48). Thus, the manipulation of host photosynthesis by viruses might be a recurrent theme in evolution, most likely to meet the increased energy burden of the production of virions by the infected cell.
DNA repair is another general function that has been the object of convergent innovations, this time through gene fusions. The DNAPolX-NAD-dependent DNA ligase fusion identified in CeV (and shared with PgV) is echoed by that of a DNAPolX-AP-endonuclease found in poxviruses (49) and that of an Mre11 and Rad50 domain in mimivirus (50). These proteins are likely parts of an optimized dsDNA strand break repair machinery.
The unique fusion between a uridyltransferase domain and UDG identified in CeV (CeV_479), the presence of several enzymes for the synthesis of rhamnose in mimivirus (45), and the fused sequential enzymes found in OtV5, OmV1, and OlV1 (YP_001648294.1, YP_009172960.1, YP_004061822.1) are other examples of convergent innovations targeting the same biosynthetic pathway, probably central to the glycosylation of viral structural proteins (45).
CeV also acquired copies of the HSP70 chaperone (CeV_195 and CeV_372) that clearly originated from a different source than the one encoded by its close relative, OLPV, as well as CroV and acanthamoeba-infecting Mimiviridae. It is also different from the one acquired by several prasinoviruses from their hosts (Fig. 9).
Finally, CeV and EhV have independently acquired U-box domain-encoding sequences, suggesting the active involvement of the host proteasome in the infectious process, perhaps as a source of recycled amino acids (51).
The puzzling origin of CeV's many unique genes.
We previously pointed out the paradox of CeV exhibiting so many unique protein-coding genes (294/512, or 57.4%), while the rest of them overwhelmingly had their closest relative in PgV or other Mimiviridae. This paradox is amplified by the small number of core genes, i.e., genes common to all of these obviously related viruses (Fig. 2). This paradox would be partially explained if most of these so-called unique genes were not real but were false-positive calls from an overestimating (albeit standard) (9) bioinformatic annotation procedure. Such ambiguities are best solved by validating gene predictions using transcriptomic data that unfortunately is not available for CeV. However, we can estimate the actual proportion of real genes among the unique ones using CroV, for which such data exist. According to the original publication (5), CroV has 544 protein-coding genes, of which 438 genes were tested for transcription and 274 (63%) found positive. We then examined how this proportion changed when separately considering unique versus shared CroV genes. Out of the 438 tested genes, 299 are unique, of which 177 (59.2%) fall in the expressed category. On the other hand, 97 of the 139 shared genes were found to be expressed (69.7%). Even though these numbers denote a significant difference in transcript detection in favor of shared genes (P value of <0.05 for the table [177, 122; 97, 42] by Fisher's exact test), they nevertheless suggest that a large proportion (59.2/69.7, or 84.9%) of CroV unique genes are real (postulating that their transcripts should be detected in the same proportion as those of shared genes). Extending the same reasoning to CeV and using the same ratio would reduce the number of unique true genes from 294 to 250, i.e., still half of its total gene complement.
The evolutionary scenario susceptible to leading to the presence of so many CeV-unique genes while preserving the strong phylogenetic affinity globally exhibited by the genes shared among the Mimiviridae remains to be elucidated. As most of the unique CeV genes are ORFans, a scenario involving a huge number of gene acquisitions from cellular organisms (or known viruses) is unlikely, short of postulating an equally huge mutation rate erasing their phylogenetic origins. There is no evidence of such genomic instability in these large dsDNA viruses. On the contrary, they are well equipped with high-fidelity DNA replication and repair machineries (our results here and reference 52). The converse model, postulating a reductive evolution from a common Mimiviridae ancestor (2) with a genome large enough to accommodate the number of unique genes indicated in Fig. 2, appears increasingly unlikely (although viruses with thousands of genes are known to exist [53]). An alternative to the opposite and equally unlikely accretion and reduction evolutionary scenarios would be to postulate that these DNA viruses de novo generate new protein-coding genes (and functions) by a totally unknown mechanism. Demonstrating such a capacity would definitely put viruses on the center stage of biological evolution.
Proposed taxonomy of an extended Mimiviridae family.
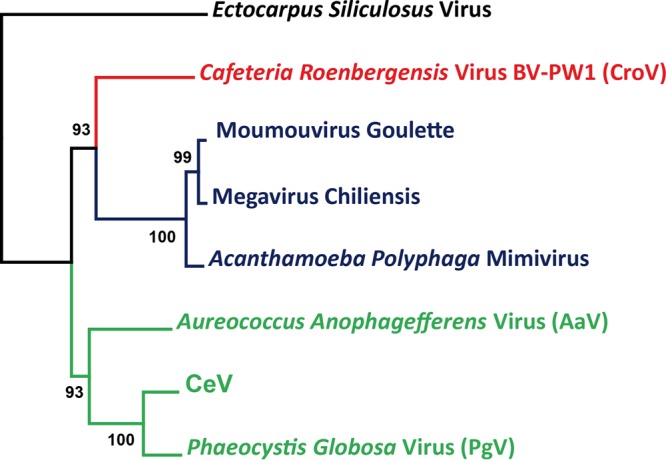
The present analysis of the CeV genome adds to the mounting consensus that despite their large differences in gene content, particle size, host range, and ecology, a group of alga-infecting large dsDNA viruses (CeV, PgV, OLPV, and AaV) and the acanthamoeba-infecting Mimiviridae (genus, Mimivirus) belong to the same family and share an ancestor (3, 7, 8, 13). This family also includes CroV (5), a virus that infects the heterotrophic stramenopile Cafeteria roenbergensis, as the sole member of the genus Cafeteriavirus. In addition to the sharing of unique genes (such as the mismatch DNA repair protein MutS7 and the puzzling asparagine synthase), large A+T rich genomes, and a full DNA transcription and replication apparatus, divergent members of this virus group (mimivirus, CroV, Pgv, and OLPV) exhibit a unique association with virophages, small dsDNA viruses replicating as parasites of their intracytoplasmic virion factories. Since CeV, PgV, and their relatives (such as Phaeocystis pouchettii virus or the Organic Lake phycodnaviruses) infect unicellular algae, they are referred to as unclassified new members of the family Phycodnaviridae in the literature as well as in sequence databases. As presently recognized by the ICTV, the family Phycodnaviridae includes six genera: Raphidovirus, Coccolithovirus, Phaeovirus, Chlorovirus, Prasinovirus, and Prymnesiovirus. As more alga-infecting viruses are characterized, it is clear that an increasing number of them do not fit within this established family, the name of which (“phyco” means “algae”) has become a source of confusion. This highlights the danger in classifying viruses within clades named after their hosts, as there is increasing evidence that the same host can be infected by phylogenetically distinct viruses (such as Acanthamoeba being infected by five different types of giant dsDNA viruses: mimivirus, marseillevirus, pandoravirus, pithovirus, and mollivirus) (2). Conversely, the Mimiviridae family (described here) (but also the Asfarviridae family [54]) shows that viruses with strong phylogenetic relationships can infect hosts belonging to branches that diverged at the earliest time of eukaryote history.
To help clarify the classification of alga-infecting viruses and acknowledge the phylogenetic affinity of CeV, PgV, and AaV with CroV and the mimivirus group (Fig. 1), we propose to divide the family Mimiviridae into two subfamilies. One, tentatively named the “Megamimivirinae” (i.e., the largest Mimiviridae), should include the 3 clades (A, B, and C) of the existing Mimivirus genus (4) and CroV as the prototype of the existing Cafeteriavirus genus. A new subfamily, named the “Mesomimivirinae” (i.e., the still large but smaller Mimiviridae), should include CeV and PgV (as well as the partially sequenced OLPV1 and OLPV2) as a new genus and the outlier AaV as the prototype of yet another distinct genus. Redefined as proposed, the new family Mimiviridae would clearly separate the above-described large alga-infecting viruses from the Phycodnaviridae family while acknowledging their relationship with the acanthamoeba-infecting mimiviruses. At the same time, the range of sequence divergence exhibited by the core proteins (such as DNA pol B, MutS7, or the packaging ATPase) within the Mimiviridae will remain comparable to that observed within other large dsDNA virus families, such as the Poxviridae (also divided into two subfamilies, Chordopoxvirinae and Entomopoxvirinae). As this paper was in review, findings from metagenomic studies suggested that yet another lineage of large Mimiviridae remains to be characterized (55).
MATERIALS AND METHODS
Genome sequencing, assembly, and annotation.
The procedures for genome sequencing, assembly, and annotation were described previously in reference 9.
Identification of gene fusions.
We mapped the CeV predicted proteins onto other Mimiviridae genomes using TBLASTN. When different segments of a CeV protein were found to best match at two distant locations in the target genome, the corresponding ORFs were submitted to further phylogenetic analyses to confirm their orthologous relationship with the different parts of the candidate CeV fusion protein. BLAST best-scoring sequences belonging to bacteria, archaea, eukaryotes, and viruses were included in the analysis to establish that the fusion occurred within the Mimiviridae lineage.
S-H test.
To discriminate between the three possibilities, (i) OLPV emerged first after AaV, (ii) PgV emerged first, or (iii) CeV emerged first, we performed multiple Shimodaira and Hasegawa (S-H) tests (56) as follows. First, we identified the 39 clusters of orthologs present in the proteomes of AaV, OLPV, PgV, and CeV using OrthoMCL. Second, 4 proteins of each of the 39 clusters were aligned using MUSCLE (57), the resulting 39 multiple alignments were visually validated, their gapped positions removed, and the corresponding likelihood matrices for the three tree topology were computed with PhyML (58).
Third, the CONSEL procedure (59) (www.sigmath.es.osaka-u.ac.jp/shimo-lab/prog/consel/) was applied to perform the S-H test itself (i.e., a computation of the P value for each of the possible topologies by comparison of the matrix of the log likelihoods).
The test was not conclusive. Among the 117 (3 × 39) trees, we could reject only three of them at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Matthieu Legendre, Sebastien Santini, Adrien Villain, and Olivier Poirot for their helpful discussions and for help with the sequence analysis software used for this work. We also thank Chantal Abergel for her help with the final versions of the figures.
The IGS laboratory is supported by the Centre National de la Recherche Scientifique and Aix-Marseille University. We acknowledge the use of the PACA-Bioinfo Platform, supported by France-Génomique (ANR-10-INBS-0009) and Institut Français de Bioinformatique (ANR-11-INSB-0013). L. Gallot-Lavallée is supported by a PhD award from Aix-Marseille University.
REFERENCES
- 1.Fischer MG. 2016. Giant viruses come of age. Curr Opin Microbiol 31:50–57. doi: 10.1016/j.mib.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Abergel C, Legendre M, Claverie J-M. 2015. The rapidly expanding universe of giant viruses: mimivirus, pandoravirus, pithovirus and mollivirus. FEMS Microbiol Rev 39:779–796. doi: 10.1093/femsre/fuv037. [DOI] [PubMed] [Google Scholar]
- 3.Yutin N, Colson P, Raoult D, Koonin EV. 2013. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J 10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol 4:1324–1330. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer MG, Allen MJ, Wilson WH, Suttle CA. 2010. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A 107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monier A, Larsen JB, Sandaa R-A, Bratbak G, Claverie J-M, Ogata H. 2008. Marine mimivirus relatives are probably large algal viruses. Virol J 5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santini S, Jeudy S, Bartoli J, Poirot O, Lescot M, Abergel C, Barbe V, Wommack KE, Noordeloos AAM, Brussaard CPD, Claverie J-M. 2013. Genome of Phaeocystis globosa virus PgV-16T highlights the common ancestry of the largest known DNA viruses infecting eukaryotes. Proc Natl Acad Sci U S A 110:10800–10805. doi: 10.1073/pnas.1303251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moniruzzaman M, LeCleir GR, Brown CM, Gobler CJ, Bidle KD, Wilson WH, Wilhelm SW. 2014. Genome of brown tide virus (AaV), the little giant of the Megaviridae, elucidates NCLDV genome expansion and host–virus coevolution. Virology 466-467:60–70. [DOI] [PubMed] [Google Scholar]
- 9.Gallot-Lavallée L, Pagarete A, Legendre M, Santini S, Sandaa R-A, Himmelbauer H, Ogata H, Bratbak G, Claverie J-M. 2015. The 474-kilobase-pair complete genome sequence of CeV-01B, a virus infecting Haptolina (Chrysochromulina) ericina (Prymnesiophyceae). Genome Announc 3:e01413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johannessen TV, Bratbak G, Larsen A, Ogata H, Egge ES, Edvardsen B, Eikrem W, Sandaa R-A. 2015. Characterisation of three novel giant viruses reveals huge diversity among viruses infecting Prymnesiales (Haptophyta). Virology 476:180–188. doi: 10.1016/j.virol.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Mozar M, Claverie J-M. 2014. Expanding the Mimiviridae family using asparagine synthase as a sequence bait. Virology 466-467:112–122. [DOI] [PubMed] [Google Scholar]
- 12.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Gibson JA, Cavicchioli R. 2011. Virophage control of Antarctic algal host-virus dynamics. Proc Natl Acad Sci U S A 108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson WH, Van Etten JL, Allen MJ. 2009. The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol 328:1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama F, Ueki S. 2016. Evolution and phylogeny of large DNA viruses, Mimiviridae and Phycodnaviridae including newly characterized Heterosigma akashiwo virus. Front Microbiol 7:1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogata H, Ray J, Toyoda K, Sandaa R-A, Nagasaki K, Bratbak G, Claverie J-M. 2011. Two new subfamilies of DNA mismatch repair proteins (MutS) specifically abundant in the marine environment. ISME J 5:1143–1151. doi: 10.1038/ismej.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villain A, Gallot-Lavallée L, Blanc G, Maumus F. 2016. Giant viruses at the core of microscopic wars with global impacts. Curr Opin Virol 17:130–137. doi: 10.1016/j.coviro.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV, Raoult D. 2012. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A 109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer MG, Hackl T. 2016. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 540:288–291. doi: 10.1038/nature20593. [DOI] [PubMed] [Google Scholar]
- 19.Sandaa R-A, Heldal M, Castberg T, Thyrhaug R, Bratbak G. 2001. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology 290:272–280. doi: 10.1006/viro.2001.1161. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerveld A, Hoeijmakers JHJ, van Duin M, de Wit J, Odijk H, Pastink A, Wood RD, Bootsma D. 1984. Molecular cloning of a human DNA repair gene. Nature 310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 22.Busch D, Greiner C, Lewis K, Ford R, Adair G, Thompson L. 1989. Summary of complementation groups of UV-sensitive CHO cell mutants isolated by large-scale screening. Mutagenesis 4:349–354. doi: 10.1093/mutage/4.5.349. [DOI] [PubMed] [Google Scholar]
- 23.Jacquet S, Bratbak G. 2003. Effects of ultraviolet radiation on marine virus-phytoplankton interactions. FEMS Microbiol Ecol 44:279–289. doi: 10.1016/S0168-6496(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 24.Weinbauer MG, Wilhelm SW, Suttle CA, Garza DR. 1997. Photoreactivation compensates for UV damage and restores infectivity to natural marine virus communities. Appl Environ Microbiol 63:2200–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuta M, Schrader JO, Schrader HS, Kokjohn TA, Nyaga S, McCullough AK, Lloyd RS, Burbank DE, Landstein D, Lane L, Van Etten JL. 1997. Chlorella virus PBCV-1 encodes a homolog of the bacteriophage T4 UV damage repair gene denV. Appl Environ Microbiol 63:1551–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tucker SL, Reece J, Ream TS, Pikaard CS. 2010. Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harbor Symp Quant Biol 75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- 27.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu J-K, Hagen G, Guilfoyle TJ, Pasa-Tolić L, Pikaard CS. 2009. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haag JR, Pikaard CS. 2011. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 29.Rudan M, Schneider D, Warnecke T, Krisko A. 2015. RNA chaperones buffer deleterious mutations in E. coli. eLife 4:e04745. doi: 10.7554/eLife.04745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. 2002. Inteins: structure, function, and evolution. Annu Rev Microbiol 56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 31.Novikova O, Jayachandran P, Kelley DS, Morton Z, Merwin S, Topilina NI, Belfort M. 2016. Intein clustering suggests functional importance in different domains of life. Mol Biol Evol 33:783–799. doi: 10.1093/molbev/msv271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novikova O, Topilina N, Belfort M. 2014. Enigmatic distribution, evolution, and function of inteins. J Biol Chem 289:14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piacente F, Gaglianone M, Laugieri ME, Tonetti MG. 2015. The autonomous glycosylation of large DNA viruses. Int J Mol Sci 16:29315–29328. doi: 10.3390/ijms161226169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perler FB, Olsen GJ, Adam E. 1997. Compilation and analysis of intein sequences. Nucleic Acids Res 25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelken J, Brinkmann H, Adamska I. 2010. Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patron NJ, Waller RF. 2007. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays News Rev Mol Cell Dev Biol 29:1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- 37.Büchel C. 2015. Evolution and function of light harvesting proteins. J Plant Physiol 172:62–75. doi: 10.1016/j.jplph.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. 2001. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 39.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. 1999. Detecting protein function and protein-protein interactions from genome sequences. Science 285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 40.Yanai I, Wolf YI, Koonin EV. 2002. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol 3:research0024.1-research0024.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. 1996. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J Biol Chem 271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 42.Yamtich J, Sweasy JB. 2010. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta 1804:1136–1150. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer MG, Kelly I, Foster LJ, Suttle CA. 2014. The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription and DNA repair. Virology 466-467:82–94. [DOI] [PubMed] [Google Scholar]
- 44.Della M, Palmbos PL, Tseng H-M, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. 2004. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 45.Parakkottil Chothi M, Duncan GA, Armirotti A, Abergel C, Gurnon JR, Van Etten JL, Bernardi C, Damonte G, Tonetti M. 2010. Identification of an l-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J Virol 84:8829–8838. doi: 10.1128/JVI.00770-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piacente F, Bernardi C, Marin M, Blanc G, Abergel C, Tonetti MG. 2014. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology 24:51–61. doi: 10.1093/glycob/cwt089. [DOI] [PubMed] [Google Scholar]
- 47.Ohno S. 1970. Evolution by gene duplication. Springer-Verlag, New York, NY. [Google Scholar]
- 48.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- 49.Afonso CL, Tulman ER, Lu Z, Oma E, Kutish GF, Rock DL. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol 73:533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida T, Claverie J-M, Ogata H. 2011. Mimivirus reveals Mre11/Rad50 fusion proteins with a sporadic distribution in eukaryotes, bacteria, viruses and plasmids. Virol J 8:427. doi: 10.1186/1743-422X-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. 2011. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 52.Doutre G, Philippe N, Abergel C, Claverie J-M. 2014. Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to virus fitness. J Virol 88:14340–14349. doi: 10.1128/JVI.02414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie J-M, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 54.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, Klose T, Rossmann M, Colson P, Raoult D, La Scola B. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol 89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, Kyrpides NC, Koonin EV, Woyke T. 2017. Giant viruses with an expanded complement of translation system components. Science 356:82–85. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- 56.Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1116. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 57.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 59.Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 60.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. 2006. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res 34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 63.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]