ABSTRACT
HIV-1 is rare among viruses for having a low number of envelope glycoprotein (Env) spikes per virion, i.e., ∼7 to 14. This exceptional feature has been associated with avoidance of humoral immunity, i.e., B cell activation and antibody neutralization. Virus-like particles (VLPs) with increased density of Env are being pursued for vaccine development; however, these typically require protein engineering that alters Env structure. Here, we used instead a strategy that targets the producer cell. We employed fluorescence-activated cell sorting (FACS) to sort for cells that are recognized by trimer cross-reactive broadly neutralizing antibody (bnAb) and not by nonneutralizing antibodies. Following multiple iterations of FACS, cells and progeny virions were shown to display higher levels of antigenically correct Env in a manner that correlated between cells and cognate virions (P = 0.027). High-Env VLPs, or hVLPs, were shown to be monodisperse and to display more than a 10-fold increase in spikes per particle by electron microscopy (average, 127 spikes; range, 90 to 214 spikes). Sequencing revealed a partial truncation in the C-terminal tail of Env that had emerged in the sort; however, iterative rounds of “cell factory” selection were required for the high-Env phenotype. hVLPs showed greater infectivity than standard pseudovirions but largely similar neutralization sensitivity. Importantly, hVLPs also showed superior activation of Env-specific B cells. Hence, high-Env HIV-1 virions, obtained through selection of producer cells, represent an adaptable platform for vaccine design and should aid in the study of native Env.
IMPORTANCE The paucity of spikes on HIV is a unique feature that has been associated with evasion of the immune system, while increasing spike density has been a goal of vaccine design. Increasing the density of Env by modifying it in various ways has met with limited success. Here, we focused instead on the producer cell. Cells that stably express HIV spikes were screened on the basis of high binding by bnAbs and low binding by nonneutralizing antibodies. Levels of spikes on cells correlated well with those on progeny virions. Importantly, high-Env virus-like particles (hVLPs) were produced with a manifest array of well-defined spikes, and these were shown to be superior in activating desirable B cells. Our study describes HIV particles that are densely coated with functional spikes, which should facilitate the study of HIV spikes and their development as immunogens.
KEYWORDS: antigenicity, broadly neutralizing antibodies, envelope glycoprotein, fluorescence-activated cell sorting, HIV-1, vaccine design, virus-like particles, cellPACK, electron microscopy, viral infectivity
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) displays around 7 to 14 envelope (Env) spikes per virion (1–3). This low number of spikes is unusual for enveloped viruses in comparison to numbers for influenza virus (∼400 to 500 spikes), vesicular stomatitis virus ([VSV] ∼1,200 spikes), Rous sarcoma virus (up to 118 spikes) (4), murine leukemia virus ([MuLV] ∼100 spikes), and simian immunodeficiency virus ([SIV] ∼70 spikes); measles and respiratory syncytial virus also have a dense coat of spikes in electron micrographs (5). The consequences and underlying basis for the low copy number of HIV-1 Env are incompletely understood. However, the scarcity of spikes on HIV-1 may be an adaptation to evade humoral immunity by (i) limiting multivalent engagement with B cell receptors (BCRs) and hence the B cell response (6, 7) and (ii) preventing bivalent binding by antibody that might otherwise enhance neutralization (5). Whereas HIV-1 Env copy number has been studied at subnormal levels, increasing the levels of functional Env on virions has been met with limited success.
Because spikes are sparse across different subtypes of HIV-1, elements that regulate Env abundance are likely to be conserved. There are conserved regions in the surface subunit, gp120, and transmembrane (TM) subunit, gp41; however, these regions of Env are typically important for folding or targets of broadly neutralizing antibodies (bnAbs). Modifications to the C-terminal tail (CTT) of gp41 have been shown to enhance Env copy number with SIV (8–10). With HIV-1, however, manipulating the CTT has either enhanced Env modestly (i.e., 3-fold) or decreased its levels (11). CTT modification also affects the antigenicity of HIV-1 Env (12) and can alter neutralization sensitivity in an Env-dependent manner (13–17). Stable cell line production of Env (18), and substitutions with a foreign TM domain have been shown to enhance Env on virus-like particles (VLPs); but the effects are either modest, or the reports lack details about the integrity, function, and antigenicity of trimeric Env (19–22).
Efforts to enhance immune responses to Env have involved multimerization of soluble native-like trimers (gp140s) on nanoparticles by fusion with self-assembling proteins or by conjugation to liposomes (23–25). These soluble trimers mimic the structure and antigenicity of native spikes (26); however, in many cases truncations to the CTT, TM domain, and the membrane-proximal external region (MPER) perturb Env structure, albeit to various degrees (8, 12, 14, 17, 27), expose nonneutralizing epitopes at the base of the spike (28), and/or eliminate epitopes of MPER bnAbs. Full-length Env spikes have also been multimerized on proteoliposomes, but immobilization and chemical fixation procedures could have distorted Env display (29). In sum, these artificial particles have the potential to create unwanted “neo-epitopes” or lose bnAb epitopes on Env. To our knowledge, a high-density display on HIV-1 of native spikes (n > 100) has not been clearly demonstrated but could circumvent some of the above issues and be useful for vaccine design.
Here, we asked whether the host cell limits the number of spikes on HIV-1. We transduced a population of human cells to express native Env and sorted these by multiple iterations of fluorescence-activated cell sorting (FACS) for a phenotype featuring high levels of bnAbs (bnAbhigh phenotype) and low levels of non-nAbs (non-nAblow phenotype). Resulting cells were stained efficiently by trimer-specific bnAbs and not by non-nAbs. Remarkably, VLPs generated from these cells present an average of >120 Env spikes per virion by electron microscopy (EM), as supported by biochemical methods. We designate these high-Env VLPs, or hVLPs. Despite differences in average Env copy numbers of over 1 order of magnitude between hVLPs and normal pseudotyped virus, there was surprisingly no strong or consistent difference in global neutralization sensitivities. Sequencing of Env from sorted cells revealed the presence of a spontaneous stop codon in the CTT from gp41; the partial CTT truncation, however, did not disturb Env antigenicity and was alone insufficient for the high density of Env. The selection of the producer cell crucially contributed to the high-Env phenotype. Notably, hVLPs show superior activation of Env-specific B cells. The augmentation of Env trimers on cells and progeny hVLPs thus provides opportunities for vaccine design that includes native Env in a membrane environment.
RESULTS
Cell sorting enhances Env display.
We showed previously that transfection of human cells using a molecular clone of HIV-1 yields characteristically low levels of Env spikes on cognate virions (30). Our attempts to increase Env content using DNA transfection, including env codon optimization, use of a constitutive cytomegalovirus (CMV) promoter, optimized leader sequence, and truncation of the CTT did not significantly increase the number of mature Env trimers on virions but did produce an excess of immature or misfolded Env debris (30). We considered that impediments to a dense display of spikes on the membrane may be intrinsic to the producer cell.
We designed a screen to augment the Env display on the cell surface. We chose to display a relatively stable Env, Comb-mut, which was identified previously for its ability to withstand harsh conditions and thus may be relatively well behaved prior to and following incorporation into virions (31). We also combined codon optimization of env and a strong CMV promoter to eliminate Rev dependence of the transcript and to support constitutive transcription of env, respectively (32, 33). Thus, rather than use an inducible promoter that limits the duration of exposure of Env to the cell (34), we had the intention of selecting cells that tolerate high-level, continuous production of Env.
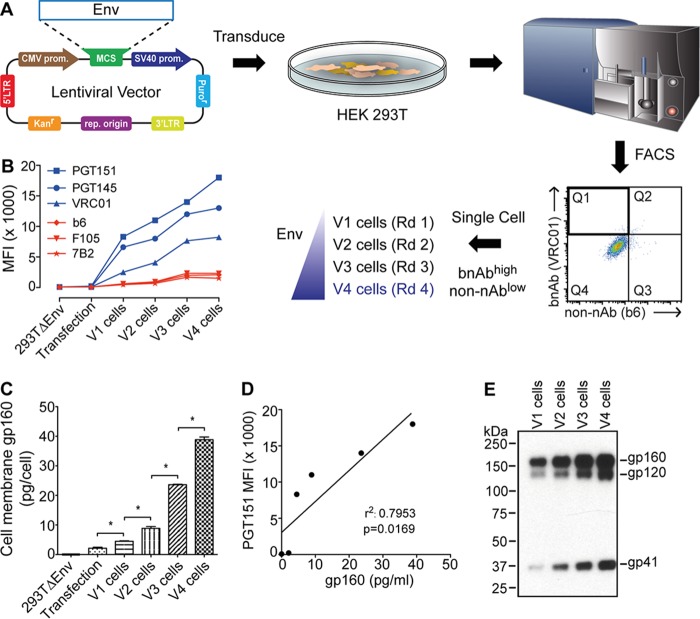
A lentiviral vector facilitated transduction and chromosomal integration of the env transgene into human (HEK293T) cells. Following transduction, cells were expanded and stained in bulk using two antibodies, VRC01 and b6, to the CD4 binding site (CD4bs) of the subunit gp120. VRC01 is a bnAb that recognizes both mature trimeric spikes and other forms of gp120 (35), whereas b6 is a non-nAb that cannot bind the CD4bs when gp120 is assembled on mature trimeric spikes (31). We used FACS to sort for cells with high levels of VRC01 and low levels of b6 binding (VRC01high b6low phenotype), anticipating that these would be enriched for trimeric spikes. A single cell with this phenotype was expanded into a stable cell line, named V1 cells, which flow cytometry consistently showed to have a bnAbhigh non-bnAblow phenotype.
V1 cells were subjected to three more rounds of FACS, each time gating on the VRC01high b6low phenotype to obtain a single cell for subsequent expansion and resorting (Fig. 1A). Cell lines from each consecutive round of selection, named V1, V2, V3, and V4, were assessed for bnAb binding by flow cytometry. Cells from each round of selection showed increased levels of staining with bnAbs, including PGT151 and PGT145 that are specific for trimeric Env, and binding remained low with non-nAbs b6, F105, and 7B2 (Fig. 1B). The observed increases in binding to quaternary bnAbs unrelated to VRC01 used in FACS strongly suggest that membrane Env was increasing in amount rather than in affinity for VRC01. We also prepared total membrane protein from cell lines V1 to V4 and probed these using Env-specific antibodies in an enzyme-linked immunosorbent assay (ELISA). Membrane Env increased with each round of sorting (Fig. 1C), which correlated further with bnAb binding to sorted cells using flow cytometry (Fig. 1D) (P = 0.0169). We conclude that sorting cells for a VRC01high b6low phenotype led to a progressive increase in Env trimer content on the surface of sorted cells.
FIG 1.
Sorting cells using bnAb and FACS to express high levels of HIV-1 Env trimers. (A) Human cells (HEK293T) were transduced with a lentiviral vector bearing env, and FACS was used to acquire single cells having a bnAbhigh non-nAblow phenotype (i.e., VRC01high b6low). A total of four rounds of sorting were performed. (B) Env-bearing cells expanded from each round (V1 to V4) were probed using a panel of bnAbs and non-nAbs by flow cytometry. The antibody staining profile of Env-bearing cells is diagnostic of mature Env trimers and increases with each round of sorting. (C) Env content (gp160 equivalents) in membrane fractions prepared from cell lines V1 to V4 as determined by ELISA. (D) Correlation between cell surface staining by bnAb as shown in panel B and the quantity of membrane Env as shown in panel C. (E) V1 to V4 cells were normalized to 300 cells per sample and solubilized, and Env was separated and probed using SDS-PAGE and Western blotting; the blot is representative of at least two independent experiments. LTR, long terminal repeat; SV40, simian virus 40; prom, promoter; MCS, multiple cloning site; MFI, mean fluorescence intensity; rep origin, replication of origin. *, P < 0.05.
To investigate whether the increase in cell surface Env on cell lines V1 to V4 was due to an increase in total Env expression in the cells or to a redistribution of mature Env to cell surfaces, we solubilized the same number of cells from each cell line V1 to V4 and probed the total Env content by SDS-PAGE and Western blotting using a cocktail of antibodies to gp120 and gp41. The Western blot revealed a steady increase of not only mature gp120 and gp41 subunits but also immature gp160, with apparently equal increases in expression levels among all three glycoprotein species (Fig. 1E). These results contradict a model in which there was a disproportionate increase in processing and surface display of mature Env and, rather, support a model in which total expression of Env increased as the cells were selected and in which the processing of gp160 into gp120 and gp41 kept pace with the increase in gp160.
Presence of CTT truncation.
Sequencing of genomic DNA of both the V1 and V4 cell lines revealed the presence of a premature stop codon in the env transgene at position 755 (755*) in the CTT of gp41 (Fig. 2A). No other mutations were observed, and the stop codon 755* was not in the original transduction vector and so most likely arose spontaneously following integration into the host chromosome. Stop codon 755* truncates 102 amino acid residues from the CTT, preserving the Kennedy epitope but removing the lentiviral lytic peptide (LLP) motifs LLP-2, LLP-3, and LLP-1 (Fig. 2A). Notably, stop codons proximal to this position have been observed in lentiviruses that have been passaged in cell culture (36, 37).
FIG 2.
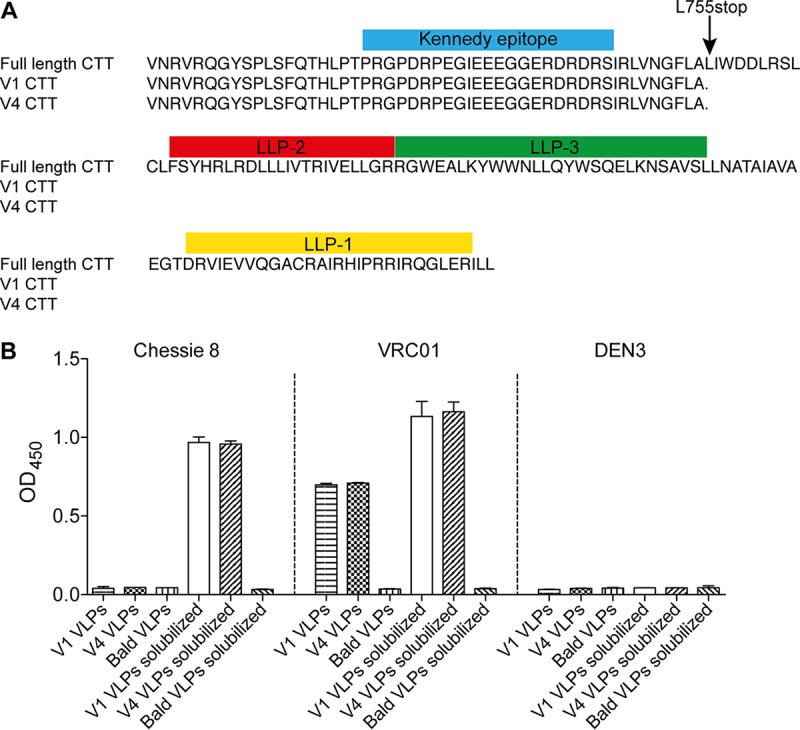
The CTT of gp41 is partially truncated and is not exposed on hVLPs. (A) Sequence alignment of the Env CTTs from V1 and V4 cells. The env transgenes from V1 and V4 cell lines were sequenced and aligned to full-length Comb-mut. Only the C-terminal tail (CTT) of gp41 is shown where the only mutation was found, i.e., L555*. Described sequence motifs in the CTT are marked with colored boxes above the corresponding sequence. LLP, lentivirus lytic peptide. (B) Kennedy epitope on the CTT of gp41 is occluded on intact hVLPs. Purified V1 and V4 VLPs were immobilized and probed in an ELISA using the anti-Kennedy epitope antibody Chessie 8. Bald VLPs and anti-dengue virus DEN3 antibody were included as negative virus controls and negative antibody controls, respectively. Solubilized VLPs are treated with detergent to liberate gp41 from virions and were used as positive controls.
Truncations in the CTT have been shown to alter conformation or exposure of epitopes surrounding the receptor binding sites on gp120 or the MPER of gp41 (12, 14). However, as an anticipated consequence of our FACS strategy, V4 cells show favorable antigenicity with high binding to quaternary bnAbs and minimal binding by non-nAbs b6 and F105 to the CD4bs. MPER bnAbs 2F5, 4E10, and 10E8 also show minimal binding to V4 cells, as expected of mature, native Env trimers (38). We will return to the effect of the CTT truncation on membrane Env in the following section.
Generation of HIV-1 virus-like particles with enhanced levels of Env trimer.
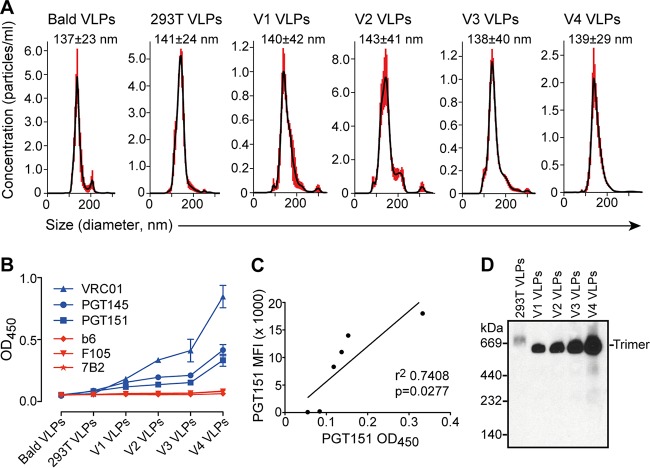
Next, we asked whether observed increases in Env on sorted cells results in an increase of Env on virus-like particles (VLPs) released from these cells. We transfected cell lines V1 to V4 with DNA of an Env-deficient molecular clone, pSG3ΔEnv, to provide components for pseudotyped VLP formation (e.g., Gag, Pol, and accessory elements). VLPs were purified from cell culture supernatants using iodixanol density gradient centrifugation (30). We assessed particle size (dispersity) of the VLPs using nanoparticle tracking analysis (NTA), which simultaneously evaluates light scattering and Brownian motion (39). VLPs produced from V1, V2, V3, and V4 cells were found to be monodisperse, with nearly identical average diameters ranging from 138 to 143 nm, indicating pure, intact, and homogeneous particles of a size expected of HIV-1 virions (Fig. 3A). Purified, intact VLPs were normalized for Gag (p24) content, immobilized, and then probed in an ELISA using a panel of bnAbs and non-nAbs. Binding of bnAbs VRC01, PGT145, and PGT151 increased incrementally from V1 to V4 VLPs (Fig. 3B). Notably, these ELISA binding data on VLPs correlate with the above binding data obtained from flow cytometry of the cognate VLP producer cells (Fig. 3C).
FIG 3.
hVLPs are homogeneous and display trimeric spikes. (A) Nanoparticle tracking analysis (NTA) of purified VLPs, including bald VLPs, normal pseudotyped virus (293T VLPs), and V1 to V4 hVLPs shows that particles are monodisperse with an average diameter of 137 to 143 nm. (B) Virus ELISA was used to probe V1 to V4 hVLPs immobilized on microwells with a panel of bnAbs and non-nAbs. (C) Correlation between bnAb staining of producer cells by flow cytometry and bnAb binding to hVLPs by virus ELISA. (D) V1 to V4 hVLPs were solubilized in nonionic detergent, and Env was separated and probed using BN-block and Western blotting. OD450, optical density at 450 nm.
To determine the oligomeric state of Env on VLPs, we turned to blue native (BN)-PAGE. VLPs from V1, V2, V3, and V4 cells were solubilized in nonionic detergent, and then proteins were separated by BN-PAGE and stained by Western blotting using Env-specific antibodies. A clear band is seen at the predicted position of an Env trimer, with staining increasing incrementally from V1 to V4 hVLPs (Fig. 3D), again in agreement with antigenicity data from ELISAs and the flow cytometry experiments described above. Taken together, the results suggest the presence of well-folded, trimeric Env on both VLPs and producer cells.
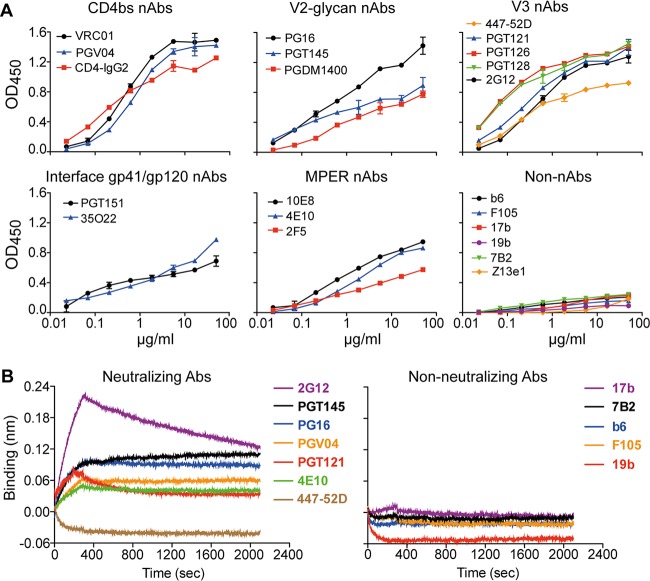
VLPs produced from V4 cells showed the highest abundance of Env and hence will be referred to as V4 hVLPs or hVLPs. We probed V4 hVLPs in a virus ELISA using a panel of antibodies. The virus ELISA revealed an antigenicity profile expected for well-ordered spikes (Fig. 4A). Thus, hVLPs were recognized by a variety of bnAbs as follows: binding of VRC01, PGV04, and CD4-IgG2 to the CD4bs; PG16, PGT145, and PGDM1400 to V2; PGT121, PGT126, PGT128, and 2G12 to the N332 glycan supersite; PGT151, 35O22, to the gp120-gp41 interface; 10E8, 2F5, and 4E10 to the MPER. We note that the V3 crown antibody, 447-52D, bound to V4 hVLPs; however, this is not entirely unexpected since 447-52D shows some weak neutralizing activity against Comb-mut (see below), and binding by 447-52D was less than that of bnAbs against the N332 supersite. Antibodies b6, F105, 17b, 7B2, and Z13e1 also showed little to no binding to V4 hVLPs in the ELISA (Fig. 4A), consistent with the expected antigenicity of Comb-mut native trimers since Comb-mut is not neutralized by these antibodies (see below). hVLPs also showed no binding with an antibody to the Kennedy epitope of the CTT, Chessie 8, in agreement with a prior report that this epitope is generally not exposed on virions (Fig. 2B) (40, 41).
FIG 4.
V4 hVLPs display antigenically correct trimeric spikes. (A) V4 hVLPs were immobilized and probed by ELISA using a panel of bnAbs and non-nAbs. (B) V4 hVLPs were captured on wheat germ agglutinin-coated biosensors and probed using a subset of bnAbs and non-nAbs by biolayer light interferometry (BLI).
To assess antigenicity of VLPs using a different method, purified V4 hVLPs were captured onto wheat germ agglutinin (WGA)-coated biosensors and then probed with antibodies using biolayer light interferometry (BLI). The BLI results clearly show that V4 hVLPs are recognized by bnAbs 2G12, PGV04, PG16, PGT145, PGT121, and 4E10 and not by several non-nAbs, including b6, F105, 17b, 7B2, and 19b (Fig. 4B). One discrepancy was with 447-52D, which showed no significant binding in this BLI assay but did show binding in the virus ELISA. The reason for this discrepancy is unclear but might be due to differences in immobilization conditions that expose V3 in the ELISA but not the BLI assay, or other factors might be involved. Overall, the antigenicity profile of membrane Env on V4 hVLPs is consistent with that of well-folded, trimeric spikes, with little or no evidence of gp41 stumps or misfolded Env debris.
EM of HIV-1 VLPs.
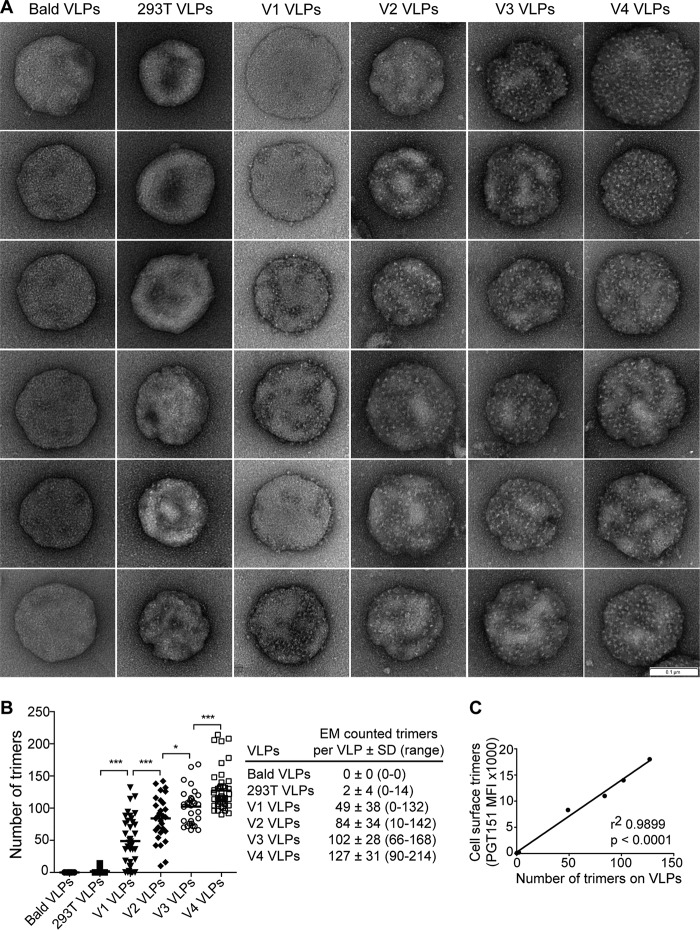
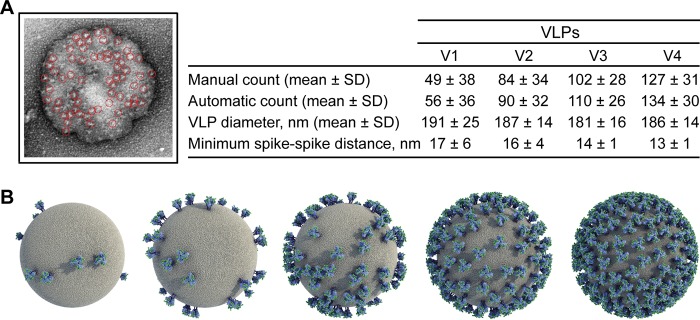
Next, we used electron microscopy (EM) to visualize intact VLPs. We negatively stained particles using uranyl formate, which we found suitable for visualizing opaque virions without significant disruption or exposure of condensed Gag cores (Fig. 5A). VLPs were roughly 181 to 191 nm in diameter, around the anticipated size of HIV-1 virions though slightly larger than that determined by NTA (3). The enlarged diameter in EM is consistent with a slight flattening of virions due to immobilization and staining on the carbon grid. Importantly, distinct puncta or projections equal in size to that predicted of a trimeric spike, i.e., ∼10 to 15 nm, were clearly visible against the darker VLPs and frequently appeared in a trimeric shape.
FIG 5.
Electron microscopy (EM) of hVLPs. (A) Representative VLPs, including bald particles, VLPs generated by transient transfection (293T), and (h)VLPs V1 to V4 visualized using negative stain EM. Bar, 100 nm. (B) Visible spikes or puncta were counted on individual virions (bald VLPs, n = 24; 293T VLPs, n = 19; V1 VLPs, n = 37; V2 VLPs, n = 29; V3 VLPs, n = 31; V4 VLPs, n = 46), and the total number of estimated trimers is plotted per virion. Each dot in the graph represents an individual virion. (C) Relative staining of producer cells by trimer-specific bnAb correlates with the average number of spikes per virion (P < 0.0001). *, P < 0.05; ***, P < 0.001.
We manually counted visible spikes on numerous virions. To estimate the number of spikes per hVLP, we doubled the number of observed spikes since negatively stained VLPs are opaque, and the opposite face is occluded (Fig. 5B). Notably, the average number of spikes per hVLP from V1, V2, V3, and V4 cells correlated with staining of Env trimers on producer cells as determined by flow cytometry (Fig. 5C) (P value of <0.0001). We also counted the number of spikes using a semiautomated image-processing approach (42). The resulting count and spike position were in good agreement with the manual count (Fig. 6A). Minimum spike-spike distances per particle were also determined; the distance decreased with increasing copy number, as expected, and reached a minimum of 13 nm with the hVLP V4 particles (Fig. 6A). We conclude that the number of spikes on hVLPs was enhanced through the iterative FACS of producer cells from an average of 49 to 56 spikes per particle for V1 hVLPs to 127 to 134 spikes per particle for V4 hVLPs. With V4 hVLPs, this average count is as much as 10-fold above that typical for HIV-1 (n = 7 to 14) (2, 3).
FIG 6.
Computational analyses and modeling of Env on VLPs. (A) Env spikes from EM images were counted using a semiautomated procedure. The hVLP spike count, diameter, and minimum spike-spike distances are tabulated. (B) Computational models of virions were generated using cellPACK. From left to right, the five models illustrate a random distribution of an increasing number of spikes: 10 (average number of spikes for HIV-1 virions), 49 (average number of spikes for V1 hVLPs), 127 (average number of spikes for V4 hVLPs), 214 (highest number of spikes estimated from an EM image of a single V4 hVLP), and 330 (theoretical maximum number of spikes determined to fit on the same size of particle).
We noted that there was some heterogeneity in the number of spikes per particle in the V1 image set, where some particles showed few, if any, spikes whereas others had as many as 132 (Fig. 5B). We did not, however, observe VLPs without spikes in subsequent selection rounds. The spikes appear to be distributed more or less randomly on all particles. Whereas nonrandom distributions of spikes have been reported with immature HIV-1 due to interaction between the gp41 CTT and Pr55Gag (43, 44), the random distribution with our hVLPs is largely consistent with these being mature particles and having a partial truncation in the CTT. Using a computational approach, cellPACK (45, 46) (Fig. 6B), we modeled virions with a random distribution of 10, 49, 127, 214, and 330 spikes. These spike counts correspond with, respectively, (i) regular HIV-1 virions (2, 3), (ii) V1 hVLPs, (iii) V4 hVLPs, (iv) the highest number of spikes on a single V4 hVLP estimated from EM images, and (v) the theoretical maximum number of spikes calculated using cellPACK. On visual inspection, the models compare favorably to cognate representative hVLPs in the EM images, both in terms of density and distribution of Env on the particles. We note that the highest number of spikes determined for V4 hVLPs, which was 214, is close to, albeit still below, a theoretical maximum number of 330 spikes, suggesting perhaps that the number of spikes might have been further increased with additional rounds of sorting.
Infectivity and neutralization properties of hVLPs.
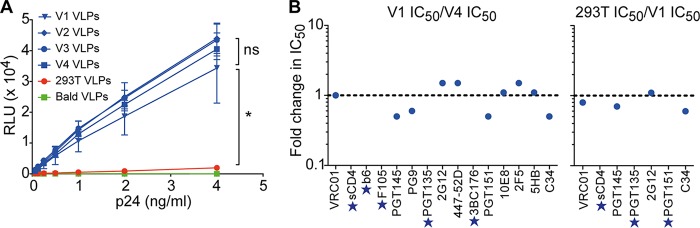
Wide separation between Env spikes on HIV-1 presumably retards clustering of receptor-engaged spikes, so increased density of Env is anticipated to enhance infectivity of HIV-1 (5, 9, 13, 47–51). To address this possibility, hVLPs were produced from V1, V2, V3, and V4 cells and then assayed for infection of TZM-bl CD4+ CCR5+ target cells. Infectivity of VLPs was determined both from cell culture supernatant and following purification by iodixanol density gradient centrifugation using normal Comb-mut (gp160) pseudovirus that was produced by transient transfection as a control. hVLPs from V1, V2, V3, and V4 cells were found to be ∼20-fold more infectious than normal pseudovirus, whereas hVLPs were equally infectious with each other (Fig. 7A). Thus, an apparent maximum in infectivity is reached when the average number of spikes is between that of normal pseudovirus (∼0.5) and V1 VLPs (∼49) and does not increase further with V4 VLPs (∼127). Wild-type HIV-1 notably falls within this range, with an average of ∼7 to 14 spikes (1, 3).
FIG 7.
Infectivity and neutralization characteristics of hVLPs. (A) hVLPs are more infectious than standard pseudovirions generated in 293T cells but show no significant difference in infectivity levels between hVLPs V1, V2, V3, and V4. (B) A comparison of the neutralization sensitivities of V1 and V4 hVLPs (left) as well as of standard (gp160) pseudovirions produced by transient transfection and V1 VLPs against a panel of bnAbs (right). Results are plotted as the fold change in IC50s in each panel. A star indicates that antibodies did not reach an IC50 at the highest or lowest concentration tested, as shown in Fig. 8. *, P < 0.05.
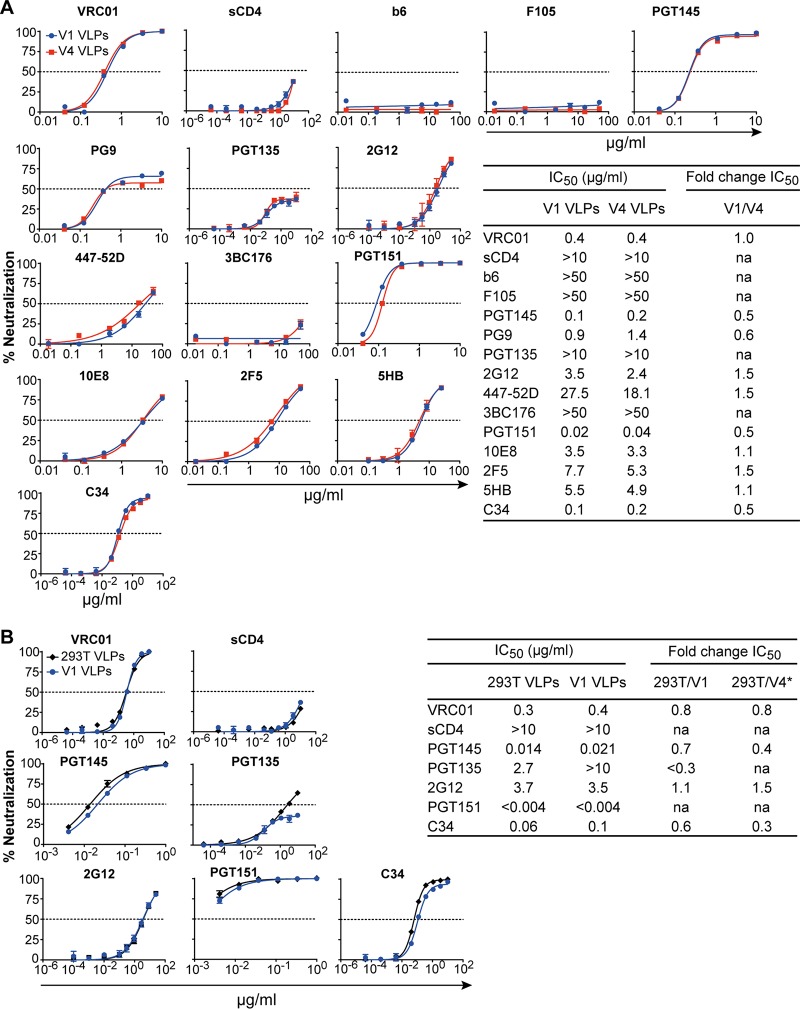
An increase in spike density conceivably might affect the sensitivity of HIV-1 to bnAbs, either due to changes in viral infectivity (9), alteration of epitope exposure from CTT truncation (12, 14), or an increased chance of bivalent binding of IgG between spikes (5). We therefore performed neutralization assays using a panel of bnAbs against V1 and V4 VLPs to determine whether the difference in Env displays between these two VLP preparations affected neutralization sensitivity. The V1 and V4 VLPs showed broadly similar neutralization sensitivities against this diverse panel of bnAbs (i.e., 50% inhibitory concentrations [IC50s] were generally within 2-fold), despite a 3-fold difference in apparent copy numbers of Env (Fig. 7B and 8A). However, the dose-response curve with PG9 plateaued with different maximum percent neutralization (MPN) values against V4 hVLPs and V1 hVLPs, i.e., at 66% and 57%, respectively (Fig. 8A). We also performed neutralization assays comparing Comb-mut gp160 full-length pseudotyped virus prepared by transient transfection (293T VLPs) side by side with V1 VLPs against several bnAbs. The differences in IC50s between 293T VLPs and V1 VLPs were generally less than 2-fold (Fig. 8B). We note that IC50s with V1 VLPs in this experiment were lower than those in the earlier experiment, at least with respect to PGT145, PGT135, and PGT151, most likely reflecting assay sensitivity variation between the two experiments. To attempt to compare neutralization sensitivities between 293T VLPs and V4 VLPs, we used the fact that V1 VLPs were included in both experiments and thus calculated the following ratio: (IC50 293T VLPs/IC50 V1 VLPsexperiment B)/(IC50 V4 VLPs/IC50 V1 VLPsexperiment A). The ratios of IC50s of 293T VLPs and V4 VLPs calculated in this way were largely similar; however, PGT145 and C34 were ∼3-fold less potent against V4 VLPs. It was also notable from these two experiments that PGT135 neutralization activity against V1 and V4 cells appeared to plateau at ∼40%, while the neutralization curve against 293T VLPs trended further upwards. Taken together, the neutralization data indicate that neutralization sensitivities between 293T VLPs, V1 VLPs, and V4 VLPs were largely similar for most bnAbs; however, modest epitope-specific differences were also observed that might reflect differences in spike densities, presence of the partial truncation in the CTT, or perhaps as yet undetermined factors.
FIG 8.
Neutralization of hVLPs by a panel of bnAbs. Results are shown for a comparison of neutralization sensitivities to bnAbs between V1 and V4 VLPs (A) as well as between V1 and standard pseudovirions generated in 293T cells (293T VLPs) (B). The estimated fold change in the IC50 between 293T VLPs and V4 VLPs shown in panel B (*) was calculated using IC50s from two independent experiments, both of which included V1 VLPs, as follows: fold change in IC50 = (IC50 293T VLPs/IC50 V1 VLPsexperiment B)/(IC50 V4 VLPs/IC50 V1 VLPsexperiment A). na, not applicable (the fold change in IC50 could not be calculated because the antibodies did not reach an IC50 at the highest or lowest concentration tested); sCD4, soluble CD4.
That neutralization of VLPs was largely similar between full-length and CTT-truncated Env 755* agrees with the flow cytometry experiments of cognate producer cells in which the partial CTT truncation of Env did not grossly alter major bnAb epitopes. Taken together, the results from flow cytometry, virus ELISA, and neutralization assays show that the partial CTT truncation 755* has limited effects on the Env ectodomain and its antigenic profile for most bnAbs.
To determine whether the L755* CTT truncation might increase infectivity and Env display of hVLPs, we tested the infectivity of Comb-mut 755* produced by transient transfection of 293T cells and found that it was equal to that of normal (full-length gp160) pseudotyped virus (Fig. 9A). Similar to the infectivity results, when the virions were normalized by particle content and then probed on an ELISA with anti-gp120 antibodies b12, 2G12, and 447-52D, the 755* virions produced by transient transfection showed levels of Env (gp120 equivalents) similar to those of virus pseudotyped with full-length gp160 even when a 10-fold excess of gp160 plasmid DNA was used (Fig. 9B). We conclude that in the context of transient transfection at least the CTT truncation 755* does not improve Env display or virus infectivity.
FIG 9.
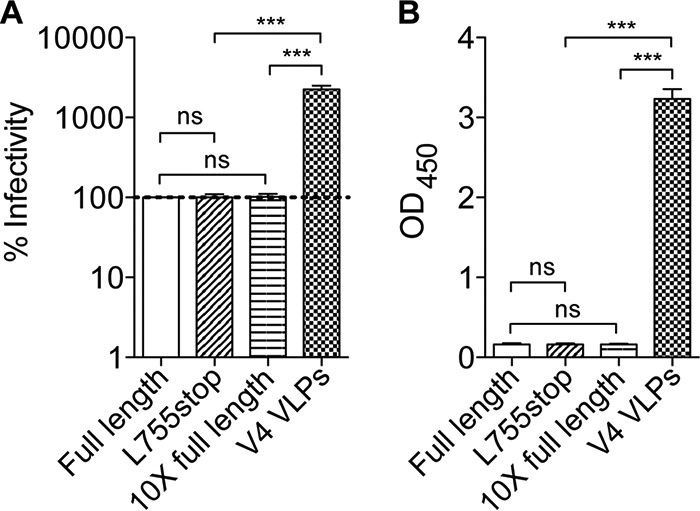
CTT truncation (755*) does not increase infectivity or Env content of VLPs produced by transient transfection of 293T cells. Virions were pseudotyped with Comb-mut gp160 (full-length), CTT-truncated Comb-mut (L755stop), and a 10-fold excess of plasmid DNA of full-length gp160 and were compared to V4 hVLPs. Virions were normalized for particle concentration and then assayed for percent infectivity in TZM-bl reporter cells (A) as well as for Env incorporation (B). For the latter, virions were probed with anti-gp120 antibodies b12, 2G12, and 447-52D in an ELISA, and the optical density (OD) at 450 nm is shown. ns, not significant. ***, P < 0.001.
hVLPs effectively activate VRC01 B cells.
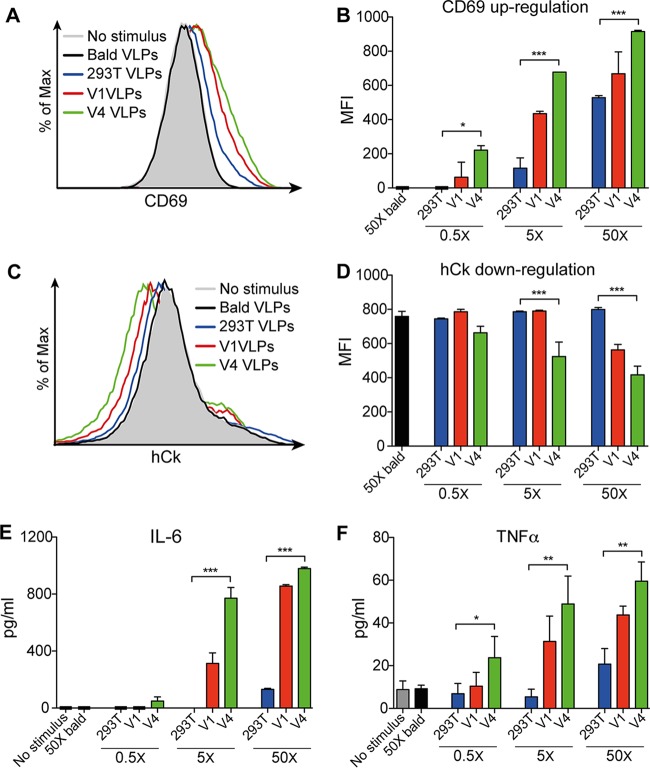
Clustering of B cell receptors (BCRs) is a crucial step in antigen-driven B cell clonal expansion and maturation that leads to high-affinity antibody responses (52). Whereas normal pseudovirions fail to activate Env-specific B cells (6), multivalent display of Env on hVLPs should, in principle at least, augment activation of BCRs. To address this, V1 and V4 hVLPs, along with wild-type pseudovirions, were purified, normalized for p24 content, and coincubated with a B cell line expressing the BCR of bnAb VRC01 (human Ig kappa light chain [hCk]) (6). Activation of VRC01 B cells was quantified by probing (i) upregulation of the activation marker CD69 (6), (ii) downregulation of the BCR (6), and (iii) release of the proinflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) (53). Notably, a rank order of B cell activation was observed that correlates with the content of membrane Env on hVLPs (Fig. 10A). This is best seen with staining of CD69 that increases incrementally not only according to the dose of particle (p24 equivalents) but also according to Env content: i.e., V4 hVLP > V1 hVLP > pseudovirions (Fig. 10B). A similar trend is observed with BCR downregulation (Fig. 10C). Downregulation of VRC01 (hCk) was not detected for normal pseudovirions or with the lowest dose of any particle but is apparent with higher doses of hVLPs and was strongest with V4 hVLPs at the highest dose (50× concentrated) (Fig. 10D). The superior capacity of hVLPs to activate VRC01-expressing B cells was further confirmed by detection of increased levels of proinflammatory cytokines IL-6 (Fig. 10E) and TNF-α (Fig. 10F) in the cell culture supernatant. We conclude from the above results that hVLPs activate B cells of a bnAb in vitro with an efficiency that increases with the number of spikes per virion and that continues to increase well above the Env density found in nature, i.e., at least up to 127 spikes per particle.
FIG 10.
hVLPs efficiently activate Env-specific B cells. B cells expressing BCRs of the anti-gp120 CD4bs bnAb VRC01 were cultured with three different concentrations of bald VLPs, normal pseudotyped virus (293T VLPs), V1 VLPs, or V4 VLPs. Following 24 h, flow cytometry of B cells was used to determine levels of the activation marker CD69 (A and B) and the B cell receptor hCk, which was downregulated upon activation (C and D), while levels of proinflammatory cytokines IL-6 (E) and TNF-α (F) in cell culture supernatants were determined by ELISA. Data shown are an average of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
The low number of spikes on HIV-1 is atypical for viruses and is a feature that has been suggested to help it evade humoral immunity (7). Increasing Env density is an attractive way to augment B cell responses, but to date a dense array of functional, membrane-anchored, trimeric spikes on HIV-1 has not been clearly demonstrated. Here, we describe the generation of hVLPs with high numbers of spikes per virion, >10-fold beyond the amount of typical HIV-1. This abundance of membrane Env was verified in concert with a comprehensive assessment of its molecular properties that confirmed the predominance of well-ordered trimeric spikes using various techniques. This high-Env phenotype was acquired using a FACS strategy that selected for antigenicity of native trimers (i.e., cells recognized by bnAbs but not non-nAbs) and overcame an inherent resistance to high-Env display in the producer cell.
Our FACS experiment resulted in the selection of an Env that contained a stop codon in CTT, 755*, that removes 102 amino acid residues from Env. The deleted region contains functional motifs, including membrane-lytic peptide motifs LLP-1, LLP-2, and LLP-3, an NF-κB activation motif (762CLFSYHRLRDLL773) (54), and an endocytosis dileucine motif (853LL854) (27), whereas at least one endocytosis motif (709GYSPL713) is retained in V4 Env 755*. Mutations in LLPs to calmodulin binding motifs or to dileucine motifs in the CTT have been shown to mitigate cytopathic effects of Env (55) or to slow endosome recycling from the cell surface (27, 56, 57), respectively. Hence, the abundance of Env observed with V4 cells might have been facilitated at least in part by removing one or more of these motifs. However, it must be emphasized that additional factors were required since (i) Env content from the selection of cell lines V1 to V4 increased and both V1 and V4 cell lines had identical Env sequences and CTT partial truncations, and (ii) transient transfections with Env proteins having the same CTT truncations produced similar amounts of Env as full-length Env.
Alterations to the CTT can disrupt Env conformation; however, any disruption by the partial 755* truncation would have to be limited since V4 cells and hVLPs were strongly recognized by quaternary bnAbs and not by non-nAbs. Moreover, we saw no consistent global differences in neutralization sensitivities with different bnAbs between normal pseudovirus and V1 or V4 hVLPs. The CTT can interact with Pr55Gag to affect incorporation and distribution of Env on immature virions (43, 58). Whether a partial CTT truncation at 755* affects distribution of Env on immature virions in a Pr55Gag-dependent manner remains to be determined (59).
Our results support a role of the producer cell in limiting spikes on HIV-1. Several host factors have been shown to regulate Env either positively, for example, Rab11-FIP1C and Rab14 (60), or negatively, for example, IFITMs (61) and MARCH-8 (62). However, the effects of these proteins on Env were reportedly modest (e.g., 3-fold or less). Their impacts in different cell types and Env backgrounds will be important to establish. Env content in V4 hVLPs might additionally relate to the following: genetic copy number, chromosome accessibility, or epigenetic regulation associated with the integrated transgene; chaperones or other factors that affect Env folding or the unfolded protein response (UPR), Env maturation, Env trafficking, endocytic recycling of Env, or suppression of cell death; or some combination of these factors. Whether these or other unknown factors contributed to the high-Env phenotype will be worth investigating in future studies. It would also be interesting to determine if high-Env display occurs in infected individuals, considering the observed high infectivity of hVLPs and their enhanced visibility to B cells.
A previous study has shown that increasing Env from less than one gp120 molecule per virion to an average of nine increased the infectivity of HIV-1 (49). We find that V1, V2, V3, and V4 hVLPs (ranging from an average of 49 to 127 spikes per virion) are equally infectious and ∼20-fold more infectious than normal pseudovirus that averages ∼0.5 spikes per virion. It is tempting to speculate that HIV-1 will reach maximum infectivity near to its natural number of spikes of 7 to 14 spikes per virion and that a dense array of Env is unnecessary for maximum infectivity. A caveat, however, is that infectivity requirements can differ with the type of target cell, particularly with cells having low levels of CD4 (e.g., macrophages) (63), or in the case of Envs of low intrinsic infectivity (48, 50, 51). Additional studies to determine the infectivity of hVLPs in which target cells and envelopes are varied will therefore be informative.
That the density of Env had a limited effect on the neutralization sensitivity of virions using diverse bnAbs was unexpected as a prior study had shown that SIV was more resistant to neutralizing antibodies with an increase in Env copy number (13). However, in that study truncations in the CTT of SIV gp41 had strong effects on infectivity and so might have altered Env conformation and/or stability. Nevertheless, a dense array of HIV-1 spikes affords a possibility for bivalent binding of IgG between spikes and the potential to increase neutralization potency by the avidity effect (5). The minimum distance between Env trimers at 127 spikes per (V4) particle was determined to be ∼13 nm, while that spanned by two Fab arms of an IgG is ∼15 nm (5). Hence, many spikes per particle will have a neighbor within the span of a single IgG molecule. However, many spikes will be outside this reach distance. Moreover, spike-spike distance may or may not predict bivalent straddling of neighboring spikes by an IgG molecule. We conjecture that HIV-1 may have evolved to limit bivalent engagement of bnAb IgGs through restricting allowable angles of approach in addition to increasing the interspike distance (5) so that spikes will tend to bind 1:1 with IgG. Further studies are warranted to investigate this hypothesis. Overall, however, bnAb affinity for trimeric Env seems to be the major determinant of neutralization for both hVLPs and HIV-1, in broad agreement with prior studies (64).
Limited epitope-specific differences in neutralization sensitivities were observed between 293T, V1, and V4 VLPs. Such differences might be due to variations in Env copy number, CTT length, glycosylation, membrane lipid content of the VLPs, or perhaps other undetermined factors; future studies are needed to better understand how these factors affect the neutralization sensitivity of HIV-1. PGT135, for example, binds to the N332 glycan supersite and showed maximum neutralization at ∼40% with V1 and V4 VLPs but not 293T VLPs. A previous study showed that microheterogeneity in N-linked glycosylation at N332, N386, and N392 can affect PGT135 neutralization and that the presence of Man9GlcNAc2 at N392 likely blocks PGT135 binding to create the observed partial neutralization phenotype (65). It is possible that 293T VLPs, V1 VLPs, and V4 VLPs differ somewhat in glycosylation; however, we note that the neutralization abilities of other glycan-specific bnAbs (PGT145, PGT151, and 2G12) were relatively similar among these VLPs, so such differences are likely to be limited.
Densely studded HIV-1 hVLPs are reminiscent of other viruses such as human papilloma virus (HPV), influenza virus, and hepatitis B virus (HBV), which are coated with spikes and are associated with potent long-lasting neutralizing antibody responses as a correlate of protection (52, 66). However, HIV-1 Env shows greater genetic and antigenic diversity than most other viruses and is more densely shielded by glycan, and the fact remains that no reported vaccine, multivalent or otherwise, has elicited bnAbs against HIV-1 (25, 64, 67). Encouragingly, arrays of well-ordered gp140 trimers with an SOS disulfide (68) or a native flexible linker (NFL) (69) and mutation I559P on liposomes and self-assembling nanoparticles have shown some capacity to increase humoral responses to Env (23–25). Specific immunization strategies with hVLPs or other Env arrays combined with rational design might benefit humoral responses as well. Notably, our hVLPs display well-ordered trimers that are not well ordered when produced as a soluble gp140 (31), so hVLPs can increase the Env sequence space available for trimerization. Moreover, whereas gp140s expose non-nAb epitopes on the N/C termini at the base of Env (28), hVLPs preserve native packing of subunits that includes the MPER epitopes (e.g., 10E8). Finally, hVLPs can equally be made with natural and designed Env molecules, which can be further screened according to purpose, such as for enhanced exposure of conserved epitopes or recognition by desirable germ line BCRs (70).
HIV-1 pseudovirions fail to activate Env-specific B cells (6), but we have shown that hVLPs are highly efficient in this respect. B cell activation improved as a function of both hVLP dose and Env copy number, the latter increasing up to the highest density of Env tested. Maximizing Env density would seem to be helpful for vaccine design. The potential for greater B cell activation, however, has to be weighed against the possibility for enhanced off-target responses. Such issues can be addressed only through immunization experiments. Assessment of B cell activation across different BCR idiotypes may also provide insight into whether and how hVLPs activate a particular response (7, 52). Lastly, B cell responses might be improved further by equipping hVLPs with immunomodulatory molecules. Future studies will focus on using hVLPs as immunogens and evaluating B cell and antibody responses.
MATERIALS AND METHODS
Antibodies and cells.
Monoclonal antibodies (human IgG) were obtained from the following sources (target epitope and subunit in parentheses): VRC01 (71) (CD4bs, gp120) from J. Mascola (Vaccine Research Center, NIH); b6 (72), PGV04 (CD4bs, gp120) (73), PGT145 (V2 glycan, gp120) (74), and PGT151 (glycan, gp41) (75) a kind gift from D. Burton (The Scripps Research Institute [TSRI]); F105 (CD4bs, gp120) (76); 2G12 (mannose patch, gp120) (77), 4E10 (78) (NWFDIT, gp41), 2F5 (ELDKWA, gp41) (79), and 447-52D (V3 crown) (80) from H. Katinger (Polymun); PGT121, PGT126, PGT128 (N332 supersite, gp120) (74) PG16 (N160 supersite, gp120) (74), and C34 peptide (NHR, gp41) from the NIH AIDS Research and Reference Reagent Program [ARRRP]; 35O22 (gp41-gp120 interface) (81) from M. Connors (NIH); and 7B2 (disulfide loop, gp41) (82) and 17b (CD4i, gp120) (83) from J. Robinson (Tulane). PG9 (N160 supersite, gp120), PGT135 (N332 supersite, gp120) (74), 3BC176 (gp41-gp120 interface), 5HB (CHR, gp41), 10E8 (MPER, gp41) (84), and Z13e1 (MPER, gp41) (85) were produced in house; soluble CD4 was purchased from Progenics; and anti-human Fc-allophycocyanin (APC) and anti-mouse Fc-APC were purchased from SouthernBiotech. For FACS, VRC01 and b6 were conjugated to APC and fluorescein isothiocyanate (FITC), respectively, using BioPAINT Antibody Labeling (Imgenex). 293T cells were purchased from the American Type Culture Collection. TZM-bl cells (86) were obtained from the NIH ARRRP and contributed by J. Kappes and X. Wu. VRC01-expressing B cells (6) were a kind gift from D. Nemazee (TSRI). All cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 20 mM l-glutamine, 100 U of penicillin/ml, and 100 μg/ml of streptomycin. Medium for Env cells and VRC01-expressing B cells was supplemented with 2 μg/ml puromycin (Life Technologies).
Generation of lentiviral particles for transduction.
Comb-mut, a hyperstable mutant of the clade B ADA gp160 (31), was codon optimized and subcloned as an XhoI-NotI fragment into the lentiviral expression vector pLenti-III-HA (where HA is hemagglutinin) (Applied Biological Materials), which contains a puromycin gene for selection of stable cell lines. pLenti-Comb-mut DNA was purified using a Plasmid Plus Maxikit (Qiagen). Lentiviral particles were generated in 293T cells by cotransfection with three plasmids: (i) pLenti-Comb-mut, (ii) the packaging plasmid psPAX2 (Addgene), and (iii) the vesicular stomatitis virus G envelope (VSV-G)-expressing plasmid pMD2.G (Addgene). Lentiviral particles were collected at 2 days posttransfection; culture supernatant was cleared of debris by centrifugation at 2,000 × g for 15 min, passed through a 0.45-μm-pore-size filter, and centrifuged at 60,000 × g for 1 h. Lentiviral particles were pelleted, resuspended at 100× concentration in phosphate-buffered saline (PBS), and titrated using a quantitative PCR (qPCR) Lentiviral Titration kit (Applied Biological Materials).
FACS.
For each round of sorting, 107 cells were washed with PBS and labeled with Fixable Aqua Dead Cell Stain (Life Technologies) for the detection of dead cells. Cells were washed with FACS buffer (PBS supplemented with 2% heat-inactivated FBS) and stained with 4 μg/ml APC-conjugated VRC01 and FITC-conjugated b6 for 15 min at 4°C. Single cells were sorted using a BD FACSAria II cell sorter (BD Biosciences) into microwells containing 200 μl of DMEM containing 20% FBS, 20 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 μg/ml puromycin.
Flow cytometry.
Cells, labeled as above, were washed with FACS buffer and aliquoted into a round-bottom 96-well plate at 2 × 105 cells per well. Cells were resuspended in 50 μl of FACS buffer containing primary Ab and incubated for 15 min at 4°C. Cells were washed with FACS buffer, resuspended in 50 μl of APC-conjugated anti-Fc secondary Ab, and incubated for 15 min at 4°C. Samples were washed, and cells were acquired on a BD LSR II flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (Tree Star). Dead cells and doublets were excluded from the analysis.
Generation and purification of hVLPs and pseudovirus.
VLPs were generated by transfecting Env cell lines with an Env-deficient HIV-1 backbone plasmid, pSG3ΔEnv (NIH ARRRP; contributed by J. Kappes and X. Wu). Pseudotyped virus was generated by cotransfection of normal 293T cells with pLenti-III-Comb-mut and pSG3ΔEnv. Negative-control (bald) VLPs were generated by transfecting 293T cells with pSG3ΔEnv. Supernatants containing VLPs were collected at 2 days posttransfection and clarified by centrifugation at 3,000 × g for 15 min, and then VLPs were pelleted at 60,000 × g for 1 h. VLPs were resuspended 100× concentrated in PBS. VLPs were purified as previously described (87). Briefly, 100× concentrated VLPs were centrifuged through a 6 to 18% iodixanol (Optiprep; Axis-Shield) density gradient (formed by layering iodixanol in 1.2% increments) at 200,000 × g for 1.5 h at 4°C in an SW41 Ti rotor (Beckman). One-milliliter gradient fractions were collected starting from the top.
ELISAs. (i) Env ELISA.
Microwells (Corning) were coated with Galanthus nivalis lectin (10 μg/ml in PBS; Sigma-Aldrich), blocked with 4% nonfat dry milk (NFDM), and washed using PBS–0.05% Tween 20. Samples were added to the wells for 1 h at 37°C. Following a washing step, wells were probed with primary Abs (5 μg/ml) for 1 h at 37°C. Wells were washed and probed with secondary anti-mouse IgG-horseradish peroxidase (HRP) (Jackson), washed again, and developed using TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Thermo Scientific). Absorbance was measured at 450 nm using a VersaMax Absorbance Microplate Reader (Molecular Devices).
(ii) Virus ELISA.
Virus ELISA was performed as described above except that 0.05% Tween was excluded from the washing steps to preserve native Env.
(iii) p24 ELISA.
For the p24 ELISA, microwells were coated with 5 μg/ml of sheep anti-p24 (Aalto), blocked with 4% NFDM, and washed using PBS–0.05% Tween 20. VLPs were lysed by adding 0.1% Empigen and added to the anti-p24-coated wells. Following 2 h of incubation at 37°C, p24 was probed with sheep anti-p24-AP (alkaline phosphatase conjugate; Aalto) and detected using an AMPAK amplification kit (Argene) according to the manufacturer's instructions.
SDS-PAGE and Western blotting.
Cells were incubated in Laemmli buffer (Bio-Rad) containing 50 mM dithiothreitol (DTT) for 5 min at 100°C and then were separated on 8 to 16% Novex Tris-glycine gels (Life Technologies) at 120 V for 1 h. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) and probed using a cocktail of antibodies to gp120 (b12, 2G12, and F425-B4e8; 2 μg/ml each) and gp41 (10E8, Z13e1, and A1; 1 μg/ml each), and the blot was developed using Pierce ECL Plus Western blotting substrate (Life Technologies).
BN-PAGE and Western blotting.
Virus was solubilized with 1% n-dodecyl-β-d-maltopyranoside (DDM), loaded onto a 3 to 12% gradient Native Page BisTris gel (Invitrogen), and run according to the manufacturer's directions. Proteins were transferred onto a PVDF membrane and blotted as for SDS-PAGE.
NTA.
Size distribution and concentration of hVLPs were determined by nanoparticle tracking analysis (NTA) on a NanoSight NS300 instrument (Malvern, Ltd., United Kingdom). An average of three 30-s measurements per sample were taken under a syringe pump flow rate of ∼10 μl/min. At least 109 particles were analyzed in total during each measurement. The hydrodynamic diameter of the particles is determined by tracking the individual trajectory of particles moving under Brownian motion (39). The software NTA, version 3.1, was used for the analysis.
BLI.
Biolayer light interferometry (BLI) was performed using an Octet RED system (ForteBio) according to the manufacturer's protocol. Briefly, as previously described (25), biotinylated wheat germ agglutinin (WGA; Vector Laboratories) was captured on streptavidin biosensors (ForteBio), followed by blocking with 0.1% nonfat dry milk (NFDM) in PBS for 180 s and then a wash for 180 s in blocking buffer. Next, VLPs were captured onto WGA sensors for 30 min, followed by a wash for 1 h. Biosensors were immersed in 30 μg/ml monoclonal antibodies (MAbs) for 300 s to allow association of trimers with antibody, followed by dissociation for 30 min. A constant temperature of 30°C was maintained inside the instrument during all reactions.
Electron microscopy.
Purified VLPs were applied for 2 min onto glow-discharged, carbon-coated grids (Electron Microscopy Sciences). Excess sample was removed by capillary action, and the grids were immediately placed on a droplet of 2% uranyl formate for 2 min. Excess stain was removed, and grids were examined on a Philips CM100 electron microscope (FEI) at 80 kV. Images were acquired with a Megaview III charge-coupled-device (CCD) camera (Olympus Soft Imaging Solutions). Spikes were counted manually from EM images. In addition, a semiautomated approach was taken according to the procedure of Audrey et al. (42). Briefly, local contrast normalization was applied to the EM images, followed by contour extraction. The spikes were identified by a local-maximum approach and circled. Spike count, particle diameter, and minimal spike-spike distances were calculated from these processed images. The different scripts and images can be provided on request. The resulting images for all VLPs with contour and spikes detected are available at http://autopack.org/home/hiv_stained.
Computational modeling of virion particles.
Virion particles were modeled using cellPACK, an open-source project (http://cellPACK.org) designed to assemble large-scale models from molecular building blocks, as previously described (45, 46). With the number of spikes (10, 49, 127, 214, and 330) and particle size (140 nm) provided as input, cellPACK randomly distributed spikes on the surface of the spherical particle.
B cell activation assay.
WEHI-231 mouse B cells, engineered to express human VRC01 B cell receptor (BCR) on the cell surface in the presence of doxycycline (6), were cocultured (5 × 105 cells per well) with purified VLPs in the presence of 1 μg/ml doxycycline (Sigma). After 24 h, cells were processed for flow cytometry, as described above, and probed with FITC-conjugated anti-mouse CD69 and APC-conjugated anti-human Ig kappa light chain (hCk) antibodies (Biolegend). A Ready-SET-go ELISA (eBioscience) was used for detection of cytokines IL-6 and TNF-α in the supernatants of cocultured cells.
Neutralization assay.
Mixtures of pseudotyped virus or hVLPs and antibody were incubated for 1 h at 37°C, prior to addition to TZM-bl target cells. After 72 h at 37°C, cells were lysed, Bright-Glo luciferase reagent (Promega) was added, and luminescence in relative light units (RLU) was measured using an Orion luminometer (Berthold).
Statistics.
Experimental groups were compared using a nonparametric Mann-Whitney U test or analysis of variance and Bonferroni posttest using GraphPad Prism. Data are represented as means with standard deviations (±SD).
ACKNOWLEDGMENTS
We thank Malcolm Wood and Theresa Fassel in TSRI's Center for Cell Ultrastructure and Morphology for assistance with negative staining electron microscopy (transmission electron microscopy). We thank David Nemazee for the VRC01 B cell line, as well as Clayton Deighan and Yu Feng for technical advice.
We gratefully acknowledge funding from National Institutes of Health (NIAID) R01s AI098602 and AI114401 (M.B.Z.), P01 AI104722 (R.T.W.), and P41GM103426-23 (A.J.O.) and from the James B. Pendleton Charitable Trust (M.B.Z.).
REFERENCES
- 1.Chertova E, Bess JW Jr, Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, Arthur LO. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu P, Chertova E, Bess J Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 4.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. 2008. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J Mol Biol 376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein JS, Bjorkman PJ. 2010. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog 6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, Hangartner L, Le K, Sok D, Jardine J, Lifson J, Wu X, Mascola JR, Poignard P, Binley JM, Chakrabarti BK, Schief WR, Wyatt RT, Burton DR, Nemazee D. 2012. Anti-HIV B cell lines as candidate vaccine biosensors. J Immunol 189:4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiller J, Chackerian B. 2014. Why HIV virions have low numbers of envelope spikes: implications for vaccine development. PLoS Pathog 10:e1004254. doi: 10.1371/journal.ppat.1004254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manrique JM, Celma CC, Affranchino JL, Hunter E, Gonzalez SA. 2001. Small variations in the length of the cytoplasmic domain of the simian immunodeficiency virus transmembrane protein drastically affect envelope incorporation and virus entry. AIDS Res Hum Retroviruses 17:1615–1624. doi: 10.1089/088922201753342022. [DOI] [PubMed] [Google Scholar]
- 9.Yuste E, Reeves JD, Doms RW, Desrosiers RC. 2004. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J Virol 78:6775–6785. doi: 10.1128/JVI.78.13.6775-6785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaBranche CC, Sauter MM, Haggarty BS, Vance PJ, Romano J, Hart TK, Bugelski PJ, Marsh M, Hoxie JA. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol 69:5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murakami T, Freed EO. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci U S A 97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. 2015. HIV-1 envelope. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuste E, Johnson W, Pavlakis GN, Desrosiers RC. 2005. Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J Virol 79:12455–12463. doi: 10.1128/JVI.79.19.12455-12463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards TG, Wyss S, Reeves JD, Zolla-Pazner S, Hoxie JA, Doms RW, Baribaud F. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J Virol 76:2683–2691. doi: 10.1128/JVI.76.6.2683-2691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durham ND, Yewdall AW, Chen P, Lee R, Zony C, Robinson JE, Chen BK. 2012. Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol 86:7484–7495. doi: 10.1128/JVI.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyner AS, Willis JR, Crowe JE Jr, Aiken C. 2011. Maturation-induced cloaking of neutralization epitopes on HIV-1 particles. PLoS Pathog 7:e1002234. doi: 10.1371/journal.ppat.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwata T, Kaori T, Enomoto I, Yoshimura K, Matsushita S. 2013. Increased infectivity in human cells and resistance to antibody-mediated neutralization by truncation of the SIV gp41 cytoplasmic tail. Front Microbiol 4:117. doi: 10.3389/fmicb.2013.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammonds J, Chen X, Fouts T, DeVico A, Montefiori D, Spearman P. 2005. Induction of neutralizing antibodies against human immunodeficiency virus type 1 primary isolates by Gag-Env pseudovirion immunization. J Virol 79:14804–14814. doi: 10.1128/JVI.79.23.14804-14814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deml L, Kratochwil G, Osterrieder N, Knuchel R, Wolf H, Wagner R. 1997. Increased incorporation of chimeric human immunodeficiency virus type 1 gp120 proteins into Pr55gag virus-like particles by an Epstein-Barr virus gp220/350-derived transmembrane domain. Virology 235:10–25. doi: 10.1006/viro.1997.8669. [DOI] [PubMed] [Google Scholar]
- 20.Guo L, Lu X, Kang SM, Chen C, Compans RW, Yao Q. 2003. Enhancement of mucosal immune responses by chimeric influenza HA/SHIV virus-like particles. Virology 313:502–513. doi: 10.1016/S0042-6822(03)00372-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang BZ, Liu W, Kang SM, Alam M, Huang C, Ye L, Sun Y, Li Y, Kothe DL, Pushko P, Dokland T, Haynes BF, Smith G, Hahn BH, Compans RW. 2007. Incorporation of high levels of chimeric human immunodeficiency virus envelope glycoproteins into virus-like particles. J Virol 81:10869–10878. doi: 10.1128/JVI.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vzorov AN, Wang L, Chen J, Wang BZ, Compans RW. 2016. Effects of modification of the HIV-1 Env cytoplasmic tail on immunogenicity of VLP vaccines. Virology 489:141–150. doi: 10.1016/j.virol.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Sliepen K, Ozorowski G, Burger JA, van Montfort T, Stunnenberg M, LaBranche C, Montefiori DC, Moore JP, Ward AB, Sanders RW. 2015. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 12:82. doi: 10.1186/s12977-015-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He L, de Val N, Morris CD, Vora N, Thinnes TC, Kong L, Azadnia P, Sok D, Zhou B, Burton DR, Wilson IA, Nemazee D, Ward AB, Zhu J. 2016. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat Commun 7:12041. doi: 10.1038/ncomms12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, Wyatt RT. 2016. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep 15:1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postler TS, Desrosiers RC. 2013. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J Virol 87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu JK, Crampton JC, Cupo A, Ketas T, van Gils MJ, Sliepen K, de Taeye SW, Sok D, Ozorowski G, Deresa I, Stanfield R, Ward AB, Burton DR, Klasse PJ, Sanders RW, Moore JP, Crotty S. 2015. Murine antibody responses to cleaved soluble HIV-1 envelope trimers are highly restricted in specificity. J Virol 89:10383–10398. doi: 10.1128/JVI.01653-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leaman DP, Lee JH, Ward AB, Zwick MB. 2015. Immunogenic display of purified chemically cross-linked HIV-1 spikes. J Virol 89:6725–6745. doi: 10.1128/JVI.03738-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leaman DP, Kinkead H, Zwick MB. 2010. In-solution virus capture assay helps deconstruct heterogeneous antibody recognition of human immunodeficiency virus type 1. J Virol 84:3382–3395. doi: 10.1128/JVI.02363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leaman DP, Zwick MB. 2013. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog 9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golden A, Austen DA, van Schravendijk MR, Sullivan BJ, Kawasaki ES, Osburne MS. 1998. Effect of promoters and signal sequences on the production of secreted HIV-1 gp120 protein in the baculovirus system. Protein Expr Purif 14:8–12. doi: 10.1006/prep.1998.0926. [DOI] [PubMed] [Google Scholar]
- 33.Haas J, Park EC, Seed B. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol 6:315–324. doi: 10.1016/S0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 34.Herschhorn A, Finzi A, Jones DM, Courter JR, Sugawara A, Smith AB III, Sodroski JG. 2011. An inducible cell-cell fusion system with integrated ability to measure the efficiency and specificity of HIV-1 entry inhibitors. PLoS One 6:e26731. doi: 10.1371/journal.pone.0026731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtkotte D, Pfeiffer T, Pisch T, Bosch V. 2006. Selection and characterization of a replication-competent human immunodeficiency virus type 1 variant encoding C-terminally truncated Env. AIDS Res Hum Retroviruses 22:57–65. doi: 10.1089/aid.2006.22.57. [DOI] [PubMed] [Google Scholar]
- 37.Jones DR, Suzuki K, Piller SC. 2002. A 100-amino acid truncation in the cytoplasmic tail of glycoprotein 41 in the reference HIV type 1 strain RF. AIDS Res Hum Retroviruses 18:513–517. doi: 10.1089/088922202317406664. [DOI] [PubMed] [Google Scholar]
- 38.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. 2008. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright M. 2012. Nanoparticle tracking analysis for the multiparameter characterization and counting of nanoparticle suspensions. Methods Mol Biol 906:511–524. doi: 10.1007/978-1-61779-953-2_41. [DOI] [PubMed] [Google Scholar]
- 40.Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. 2013. Detailed topology mapping reveals substantial exposure of the “cytoplasmic” C-terminal tail (CTT) sequences in HIV-1 Env proteins at the cell surface. PLoS One 8:e65220. doi: 10.1371/journal.pone.0065220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postler TS, Martinez-Navio JM, Yuste E, Desrosiers RC. 2012. Evidence against extracellular exposure of a highly immunogenic region in the C-terminal domain of the simian immunodeficiency virus gp41 transmembrane protein. J Virol 86:1145–1157. doi: 10.1128/JVI.06463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrey P, Kieu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot PG, Hue-Beauvais C, Houba-Herin N, Duranthon V, Devinoy E, Beaujean N, Gaudin V, Maurin Y, Debey P. 2010. Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Comput Biol 6:e1000853. doi: 10.1371/journal.pcbi.1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyma DJ, Kotov A, Aiken C. 2000. Evidence for a stable interaction of gp41 with Pr55Gag in immature human immunodeficiency virus type 1 particles. J Virol 74:9381–9387. doi: 10.1128/JVI.74.20.9381-9387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bugelski PJ, Maleeff BE, Klinkner AM, Ventre J, Hart TK. 1995. Ultrastructural evidence of an interaction between Env and Gag proteins during assembly of HIV type 1. AIDS Res Hum Retroviruses 11:55–64. doi: 10.1089/aid.1995.11.55. [DOI] [PubMed] [Google Scholar]
- 45.Johnson GT, Autin L, Al-Alusi M, Goodsell DS, Sanner MF, Olson AJ. 2015. cellPACK: a virtual mesoscope to model and visualize structural systems biology. Nat Methods 12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 2014. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discuss 169:23–44. doi: 10.1039/C4FD00017J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magnus C, Regoes RR. 2010. Estimating the stoichiometry of HIV neutralization. PLoS Comput Biol 6:e1000713. doi: 10.1371/journal.pcbi.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brandenberg OF, Magnus C, Rusert P, Regoes RR, Trkola A. 2015. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog 11:e1004595. doi: 10.1371/journal.ppat.1004595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeSantis MC, Kim JH, Song H, Klasse PJ, Cheng W. 2016. Quantitative correlation between infectivity and Gp120 density on HIV-1 virions revealed by optical trapping virometry. J Biol Chem 291:13088–13097. doi: 10.1074/jbc.M116.729210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klasse PJ. 2007. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology 369:245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klasse PJ, Moore JP. 1996. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J Virol 70:3668–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachmann MF, Jennings GT. 2010. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat Rev Immunol 10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 53.Yi AK, Krieg AM. 1998. Rapid induction of mitogen-activated protein kinases by immune stimulatory CpG DNA. J Immunol 161:4493–4497. [PubMed] [Google Scholar]
- 54.Postler TS, Desrosiers RC. 2012. The cytoplasmic domain of the HIV-1 glycoprotein gp41 induces NF-κB activation through TGF-β-activated kinase 1. Cell Host Microbe 11:181–193. doi: 10.1016/j.chom.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller MA, Mietzner TA, Cloyd MW, Robey WG, Montelaro RC. 1993. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res Hum Retroviruses 9:1057–1066. doi: 10.1089/aid.1993.9.1057. [DOI] [PubMed] [Google Scholar]
- 56.Sauter MM, Pelchen-Matthews A, Bron R, Marsh M, LaBranche CC, Vance PJ, Romano J, Haggarty BS, Hart TK, Lee WM, Hoxie JA. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol 132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol 73:1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, Muller B, Hell SW, Krausslich HG. 2012. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 59.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, Spearman P. 2013. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog 9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J, Li M, Wilkins J, Ding S, Swartz TH, Esposito AM, Zheng YM, Freed EO, Liang C, Chen BK, Liu SL. 2015. IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep 13:145–156. doi: 10.1016/j.celrep.2015.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, Fujita H, Tokunaga K. 2015. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med 21:1502–1507. doi: 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
- 63.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, Swanstrom R. 2014. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol 88:1858–1869. doi: 10.1128/JVI.02477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwick MB, Burton DR. 2007. HIV-1 neutralization: mechanisms and relevance to vaccine design. Curr HIV Res 5:608–624. doi: 10.2174/157016207782418443. [DOI] [PubMed] [Google Scholar]
- 65.Pritchard LK, Spencer DI, Royle L, Vasiljevic S, Krumm SA, Doores KJ, Crispin M. 2015. Glycan microheterogeneity at the PGT135 antibody recognition site on HIV-1 gp120 reveals a molecular mechanism for neutralization resistance. J Virol 89:6952–6959. doi: 10.1128/JVI.00230-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plotkin SA. 2010. Correlates of protection induced by vaccination. Clin Vaccine Immunol 17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Taeye SW, Moore JP, Sanders RW. 2016. HIV-1 envelope trimer design and immunization strategies to induce broadly neutralizing antibodies. Trends Immunol 37:221–232. doi: 10.1016/j.it.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roben P, Moore JP, Thali M, Sodroski J, Barbas CF III, Burton DR. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol 68:4821–4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Falkowska E, Ramos A, Feng Y, Zhou T, Moquin S, Walker LM, Wu X, Seaman MS, Wrin T, Kwong PD, Wyatt RT, Mascola JR, Poignard P, Burton DR. 2012. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J Virol 86:4394–4403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol G Principal Investigators, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr 6:7–14. [PubMed] [Google Scholar]
- 77.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 79.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67:6642–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol 66:7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pincus SH, Fang H, Wilkinson RA, Marcotte TK, Robinson JE, Olson WC. 2003. In vivo efficacy of anti-glycoprotein 41, but not anti-glycoprotein 120, immunotoxins in a mouse model of HIV infection. J Immunol 170:2236–2241. doi: 10.4049/jimmunol.170.4.2236. [DOI] [PubMed] [Google Scholar]
- 83.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nelson JD, Brunel FM, Jensen R, Crooks ET, Cardoso RM, Wang M, Hessell A, Wilson IA, Binley JM, Dawson PE, Burton DR, Zwick MB. 2007. An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10. J Virol 81:4033–4043. doi: 10.1128/JVI.02588-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dettenhofer M, Yu XF. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol 73:1460–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]