ABSTRACT
Human herpesviruses 6A and 6B (HHV-6A/B) can integrate their genomes into the telomeres of human chromosomes using a mechanism that remains poorly understood. To achieve a better understanding of the HHV-6A/B integration mechanism, we made use of BRACO-19, a compound that stabilizes G-quadruplex secondary structures and prevents telomere elongation by the telomerase complex. First, we analyzed the folding of telomeric sequences into G-quadruplex structures and their binding to BRACO-19 using G-quadruplex-specific antibodies and surface plasmon resonance. Circular dichroism studies indicate that BRACO-19 modifies the conformation and greatly stabilizes the G-quadruplexes formed in G-rich telomeric DNA. Subsequently we assessed the effects of BRACO-19 on the HHV-6A initial phase of infection. Our results indicate that BRACO-19 does not affect entry of HHV-6A DNA into cells. We next investigated if stabilization of G-quadruplexes by BRACO-19 affected HHV-6A's ability to integrate its genome into host chromosomes. Incubation of telomerase-expressing cells with BRACO-19, such as HeLa and MCF-7, caused a significant reduction in the HHV-6A integration frequency (P < 0.002); in contrast, BRACO-19 had no effect on HHV-6 integration frequency in U2OS cells that lack telomerase activity and elongate their telomeres through alternative lengthening mechanisms. Our data suggest that the fluidity of telomeres is important for efficient chromosomal integration of HHV-6A and that interference with telomerase activity negatively affects the generation of cellular clones containing integrated HHV-6A.
IMPORTANCE HHV-6A/B can integrate their genomes into the telomeres of infected cells. Telomeres consist of repeated hexanucleotides (TTAGGG) of various lengths (up to several kilobases) and end with a single-stranded 3′ extension. To avoid recognition and induce a DNA damage response, the single-stranded overhang folds back on itself and forms a telomeric loop (T-loop) or adopts a tertiary structure, referred to as a G-quadruplex. In the current study, we have examined the effects of a G-quadruplex binding and stabilizing agent, BRACO-19, on HHV-6A chromosomal integration. By stabilizing G-quadruplex structures, BRACO-19 affects the ability of the telomerase complex to elongate telomeres. Our results indicate that BRACO-19 reduces the number of clones harboring integrated HHV-6A. This study is the first of its kind and suggests that telomerase activity is essential to restore a functional telomere of adequate length following HHV-6A integration.
KEYWORDS: BRACO-19, chromosomal integration, G-quadruplex, HHV-6, telomere
INTRODUCTION
Human herpesvirus 6A (HHV-6A) and HHV-6B are two distinct DNA viruses that belong to the subfamily Betaherpesvirinae. Despite their high genome sequence similarities, these viruses possess different biological and epidemiological properties (1). HHV-6B is a ubiquitous virus that infects almost 100% of the human population. It is the etiological agent of the febrile illness roseola infantum, also known as the sixth childhood eruptive disease (2, 3). Reactivation of HHV-6B in immunosuppressed individuals is associated with adverse clinical outcomes, including life-threatening encephalitis or graft rejection in transplant patients (4). The diseases associated with HHV-6A infection are not clearly established. During latency, human herpesviruses typically maintain their genomes as extrachromosomal nuclear episomes. How HHV-6A/B achieve latency is still unclear, and since these viruses can integrate their genomes into host chromosomes, integration is considered one possible mode of latency (5). HHV-6 integration was first described by Luppi et al., who observed the presence of the integrated HHV-6 genome in the chromosomes of freshly isolated peripheral blood mononuclear cells (PBMC) (6, 7). HHV-6A/B integration can occur in germinal cells and be transmitted vertically to descendants (8, 9). This condition is termed inherited chromosomally integrated HHV-6 (iciHHV-6) and is present in about 0.2% to 1% of the human population across the world (reviewed in references 10 and 11). In 2015, Gravel et al. analyzed DNA samples from 20,000 Quebecers (Canada) for iciHHV-6 and concluded that iciHHV-6A/B represents a predisposing risk factor for angina pectoris (12).
HHV-6A/B integration can occur in various chromosomes, with viral genomes consistently detected in telomeric regions that are located at chromosome termini (5, 10, 13–15). Telomeres are composed of double-stranded TTAGGG repeats (8 to 13 kbp long) followed by a 30- to 200-bp single-stranded 3′ overhang (16–19). Telomeres protect the chromosome against information loss and instability (20, 21) and are shortened every time a cell divides.
The HHV-6A/B genome consists of a unique sequence (U) that is flanked by G-rich direct repeat regions (DR) that harbor the packaging sequences (pac1 and pac2) and two arrays of either perfect or imperfect telomeric repeats (TMR) at the genome termini (22–26). The presence of these repeats at the ends of the viral genomes and the fact that chromosomal integration occurs in telomeres led to the hypothesis that HHV-6A/B integrates the human genome by homologous recombination (HR) between viral TMRs and chromosomal telomeric sequences (10, 27). Wallaschek et al. recently demonstrated that the viral telomeric repeats are required for efficient HHV-6A integration into host telomeres (28). Analysis of the integrated virus genome revealed that the perfect TMR of the DRR region are fused to the host chromosome accompanied by a loss of the pac2 sequence, consistent with an integration mediated by HR between the viral TMR and the host telomere (5). At the other end of the viral genome, the pac1 region is also lost and additional telomeric repeats are added, suggesting that the TMR in DRL is a substrate for the generation of a new telomere (29–31).
It has been shown that nucleic acid G-rich sequences, such as those at telomeres, can fold into peculiar secondary structures called G-quadruplexes (32). Four Gs bind via Hoogsteen-type hydrogen bond base pairing to yield G-quartets. The stacking of at least two G-quartets leads to formation of the G-quadruplex through π-π interactions between aromatic systems of G-quartets. K+ cations in the central cavity relieve repulsion among oxygen atoms and specifically support G-quadruplex formation and stability (33). Biologically relevant G-quadruplexes normally form intramolecular structures that, based on the strand orientation, can adopt three main topologies: parallel, antiparallel, and hybrid-type (34).
In this work, we set out to determine whether a loss in telomere fluidity would affect the ability of HHV-6A to integrate its genome into host DNA. To achieve this, we used BRACO-19, a 3,6,9-trisubstituted acridine derivative designed by molecular modeling to interact with and stabilize the G-quadruplex DNA structures formed in human telomeres while displaying very low affinity toward duplex DNA (35). BRACO-19 binds and stabilizes G-quadruplexes present in the single-stranded region of telomeres. BRACO-19 also inhibits telomerase activity and provokes the displacement of the telomerase complex (36, 37).
In the current study, we first confirmed the specificity of BRACO-19 and next assessed its effect on HHV-6A integration. Our results demonstrate that BRACO-19 does not affect cell viability or HHV-6A entry, as determined by measuring the amount of intracellular viral DNA. BRACO-19 did, however, affect our ability to generate HeLa and MCF-7 clones with integrated HHV-6A. Taken together, our data provide evidence that a fluid interaction between the HHV-6A genome and the telomeres is needed for chromosomal integration.
RESULTS
Biophysical characterization of BRACO-19 binding to G-quadruplex telomere structures.
To ensure the correct folding of oligonucleotides into G-quadruplex structures, we initially performed an enzyme-linked immunosorbent assay (ELISA) using the BG4 and 1H6 antibodies that specifically bind to G-quadruplex structures. We demonstrated that the BG4 antibodies preferentially bind G-rich oligonucleotides, such as the telomeric [(TTAGGG)4] and myc oligonucleotides. The latter corresponds to a DNA sequence present in the promoter of the MYC gene and was previously reported to fold into a G-quadruplex structure (38–40) (Fig. 1). Binding of 1H6 antibodies to G-rich oligonucleotides [(TTAGGG)4] was also observed. Marginal binding to the single-stranded DNA (ssDNA) was recorded, as previously reported (41, 42). These results confirm that under our conditions, both the telomeric and myc oligonucleotides fold into G-quadruplexes.
FIG 1.
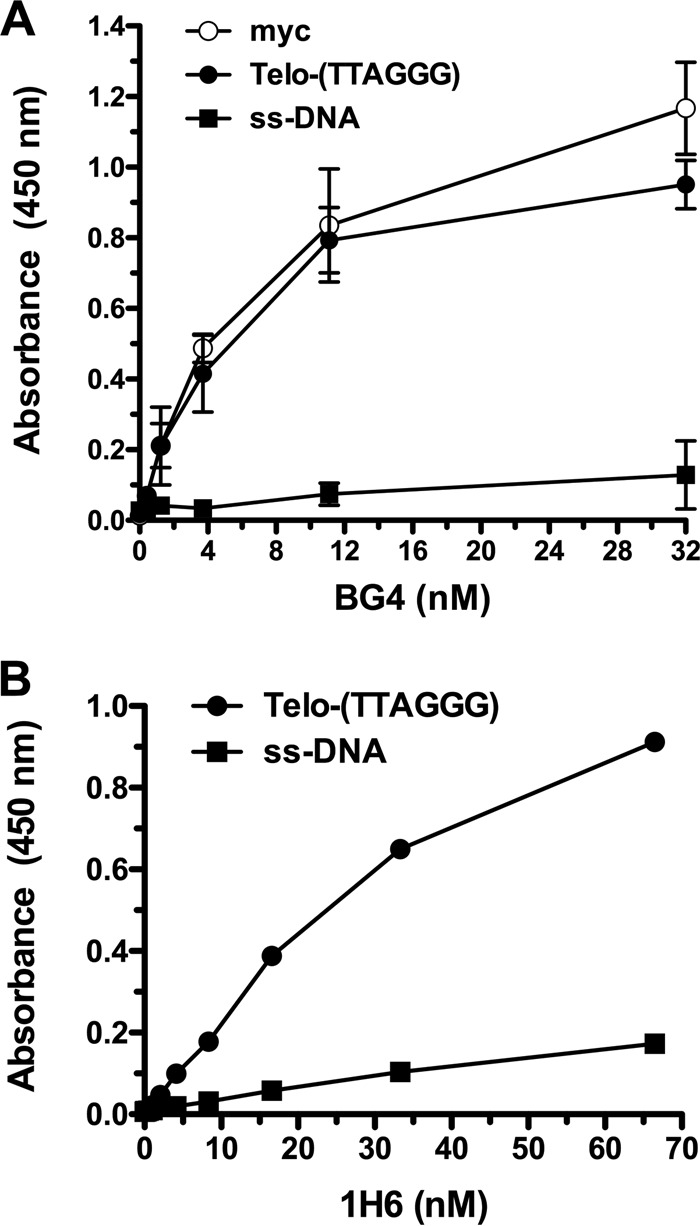
Specificity of the BG4 and 1H6 antibodies for G-quadruplex structures. Binding curves, as determined by ELISA, showing that the BG4 (A) and 1H6 (B) antibodies have high affinity for intramolecular DNA G-quadruplex structures [myc and Telo-(TTAGGG)] relative to non-G-quadruplex-containing DNA (ssDNA). Results (absorbance at 450 nm) are expressed as means ± standard deviations from 4 independent experiments.
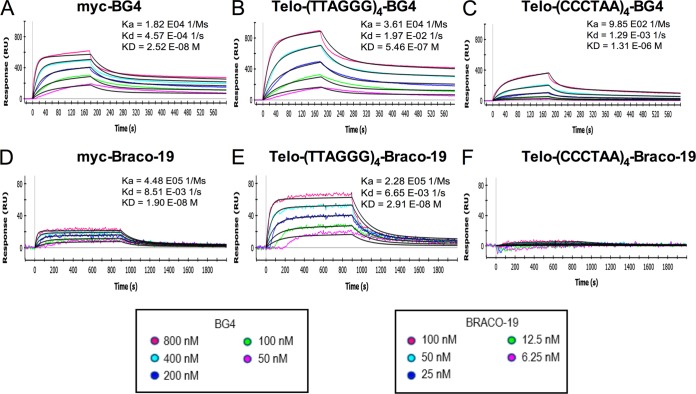
We next analyzed the binding of BRACO-19 to the telomeric and myc oligonucleotides by surface plasmon resonance. BRACO-19 was designed to bind specifically to G-quadruplexes with the assumption that each of its chains would occupy a groove of its ligand, as it stacks between two G-quadruplexes: its aromatic planar core stacks between two G-quadruplexes, and each of the two positively charged chains occupies the groove of each bound G-quadruplex (35, 36, 43). As a positive control, the BG4 antibody was used. BG4 antibody efficiently bound the G-quadruplex-folded telomeric sequence (KD [equilibrium constant] = 5.46 × 10−7 M) and the myc promoter sequence (KD = 2.52 × 10−8 M), confirming the ELISA results. Much weaker (100× less than that of myc) binding of the BG4 antibody was observed against the C-rich telomeric sequence (KD = 1.31 × 10−6 M). The binding of BRACO-19 was studied next. As shown, BRACO-19 efficiently binds to both G-rich telomeric and myc G-quadruplex oligonucleotides, with KDs in the range of 10−8 M. In contrast, minimal binding to the C-rich telomeric sequence was observed, and no constant could be calculated (Fig. 2F). These results confirm that BRACO-19 specifically binds to oligonucleotides forming G-quadruplex structures.
FIG 2.
Association and dissociation curves of ligands (BG4 and BRACO-19) to the DNA oligonucleotides by surface plasmon resonance. Curves of the association and dissociation over time between the ligands (BG4 antibody and BRACO-19) and the DNA sequences [myc promoter, Telo-G-rich telomere (TTAGGG)4, and C-rich telomere (CCCTAA)4], corresponding to association (Ka), dissociation (Kd), and equilibrium (KD) constants. Curves with BG4 (0 nM to 800 nM) are shown in panels A to C, and curves with BRACO-19 (0 nM to 100 nM) are in panels D to F.
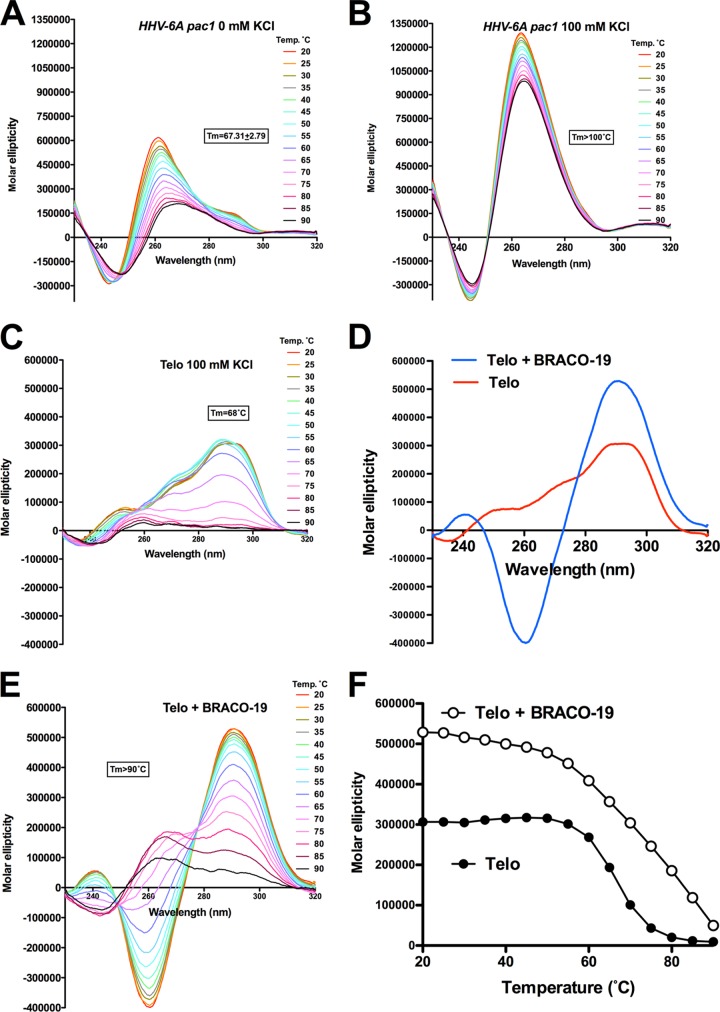
Folding of HHV-6A pac1 and telomeric DNA into G-quadruplexes and binding of BRACO-19 to the telomeric DNA were confirmed by circular dichroism (CD) analysis. HHV-6A pac1 sequence exhibited a parallel G-quadruplex topology (minimum near 240 nm and maximum near 260 nm), characterized by very high molar ellipticities (Fig. 3A and B). Stability of the telomeric G-quadruplex was measured by thermal unfolding, or Tm, i.e., the melting temperature obtained at half unfolding. The pac1 sequence was very stable in the absence of K+, with a Tm of 67°C, whereas the addition of K+ (100 mM) further stabilized the G-quadruplex structure (Tm of >100°C). The telomeric G-quadruplex folded oligonucleotide displays a CD spectrum characteristic of a hybrid type conformation (maximum near 290 nm and shoulder near 260 nm) (Fig. 3C) (34). Upon addition of BRACO-19, the CD spectrum drastically changed, with an increase in the two positive peaks at 290 and 240 nm and formation of a negative peak at 260 nm, signals that have been empirically shown to be signatures of the antiparallel G-quadruplex topology (Fig. 3D) (34). The Tm of the telomeric sequence in the absence of BRACO-19 was 66.8°C, and it increased to values of >90°C in the presence of the compound, with a net variation in Tm of >23.2°C (Fig. 3E and F). These results indicate HHV-6A pac1 and G-rich telomeric sequences can fold into G-quadruplex and that BRACO-19 specifically binds and greatly stabilizes G-quadruplex structures.
FIG 3.
CD spectra and thermal unfolding of HHV-6A pac1 and telomeric (Telo) AGGG(TTAGGG)3 oligonucleotides. (A and B) CD spectra and thermal unfolding of HHV-6A pac1 oligonucleotide in the absence (A) or in the presence (B) of 100 mM K+. (C) CD spectra and thermal unfolding of telomeric oligonucleotides in the presence of 100 mM K+. (D) CD spectra of telomeric oligonucleotide in the absence or in the presence of BRACO-19 (16 μM). (E) CD spectra and thermal unfolding of telomeric oligonucleotide in the presence of 100 mM KCl and BRACO-19 (16 μM). (F) Relative melting curves of the telomeric oligonucleotides in the absence or in the presence of BRACO-19 (16 μM) plotted at the wavelength corresponding to the maximum CD signal (291 nm).
Toxicity analyses of BRACO-19.
To assess BRACO-19 concentrations to be used without induction of cytotoxic effects, we performed MTT cell proliferation assays in U2OS, HeLa, and MCF-7 cells. The cells were incubated for 3 days with various concentrations of BRACO-19, ranging from 0 to 2 μM. No toxicity was observed with BRACO-19 concentrations up to 1 μM (Fig. 4). A concentration of 2 μM reduced the survival and proliferation of all cell types. All experiments were thus performed with concentrations of BRACO-19 of ≤1 μM.
FIG 4.
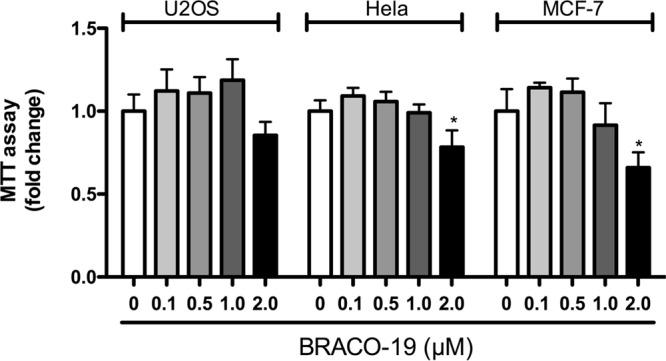
Determination of BRACO-19 toxicity. U2OS, HeLa, and MCF-7 cells were incubated with the indicated concentrations of BRACO-19 for 3 days, after which cell number and viability were determined using the MTT assay. Results are expressed as mean ratio ± SD relative to the control in the absence of BRACO-19. *, P < 0.05.
Effects of BRACO-19 on HHV-6A DNA levels.
The HHV-6A genome contains G-rich sequences, including telomeric repeats that might interact with BRACO-19 and affect viral genome replication. We determined the amount of HHV-6A DNA at different times postinfection in BRACO-19-treated or untreated cells. U2OS, HeLa, and MCF-7 cells were infected with HHV-6A (U1102) for 4 h, 48 h, or 72 h in the absence or in the presence of BRACO-19 (1 μM). Intracellular DNA was extracted and the amount of HHV-6A DNA was assessed by quantitative PCR (qPCR). We could demonstrate that for most time points tested (except for 4 h in U2OS cells), BRACO-19 had no impact on intracellular viral DNA levels (Fig. 5A), indicating that the compound had limited effects on virus entry or the fate of the HHV-6A genome. The fact that we observed a significant decrease in viral DNA at 48 h and 72 h suggests that the cellular environment of these 3 cell lines does not allow efficient HHV-6A DNA replication. As suggested previously (44), semipermissive or nonpermissive cells are preferred cell types for the study of viral integration, since permissive cells are, for the most part, destroyed during productive infection. In support of this, even though immediate-early (IE1) and early (P41) proteins could be detected in 10 to 20% of U2OS cells (Fig. 5B), expression of late proteins and release of mature virions in the culture supernatant was not detected (data not shown).
FIG 5.
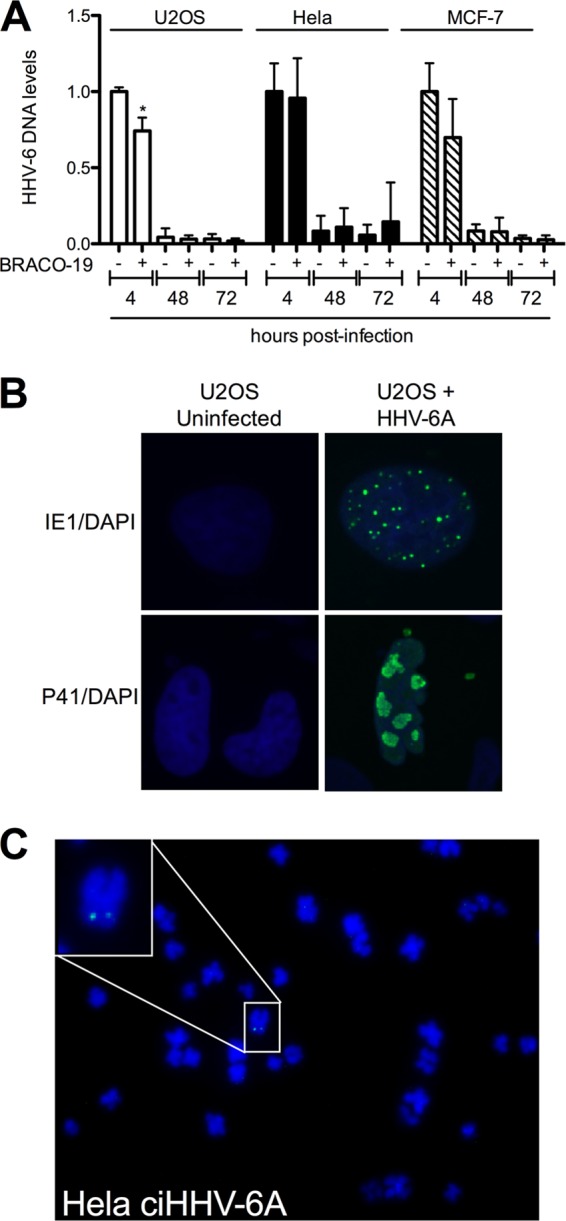
Effects of BRACO-19 on intracellular HHV-6A DNA levels. (A) U2OS, HeLa, and MCF-7 cells were infected with HHV-6A and incubated with 1 μM BRACO-19 for 4 h, 48 h, and 72 h, after which the intracellular HHV-6 DNA content was estimated by qPCR. Results are expressed as mean ± SD (n = 4) HHV-6 DNA levels relative to cellular DNA (GAPDH gene) following normalization to the 4-h BRACO-19-untreated time point. *, P < 0.05. (B) U2OS cells were infected with HHV-6A, and 48 h later the cells were processed for immunofluorescence analysis using anti-immediate early 1 antibodies or anti-P41 (early) antibodies (green). Nuclei were visualized using 4′,6-diamidino-2-phelylindole (DAPI) (blue). (C) Metaphase spread of a HeLa ciHHV-6A+ clone. The HHV-6A genome was detected using a digoxigenin-labeled probe (green) by FISH. Chromosomes were visualized using DAPI (blue).
Effects of BRACO-19 on HHV-6 chromosomal integration.
We next examined the effects of BRACO-19 on HHV-6A chromosomal integration. The frequency of HHV-6A integration was determined using two recently described integration assays, one based on the isolation of cell clones and the other using bulk cultures, that provide equivalent estimates (44). Using these assays, most clones analyzed contained approximately 1 copy of HHV-6A/B genome/cell, as determined by in situ hybridization and droplet digital PCR (ddPCR) (44). For the first three experiments, HeLa and U2OS cells were either left untreated or were treated with 1 μM BRACO-19 prior to and during infection. After 48 h, the medium was changed and cells were seeded in 96-well plates and left without BRACO-19 for the rest of the experiment. For the fourth assay, cells were left with BRACO-19 for 72 h, and the concentration was subsequently reduced to 0.1 μM for the rest of the experiment to determine whether a longer exposure to BRACO-19 would affect the outcome of the experiment. Single-cell cloning was performed and the frequency of clones containing chromosomally integrated HHV-6A (ciHHV-6A+) was determined by qPCR from the four independent experiments (Table 1). In HeLa cells, BRACO-19 treatment resulted in a 45% reduction in HHV-6A integration frequency relative to the untreated control cells (P = 0.0017). In contrast, no statistically significant difference (P = 1.00) was observed in the HHV-6A integration frequency of U2OS cells treated with BRACO-19 compared to untreated cells. Prolonging the exposition to BRACO-19 had no additional effects on the proportion of ciHHV-6A+ clones obtained. To ensure that clones contained integrated HHV-6A, we performed FISH analyses on several clones. A representative result is presented in Fig. 5C.
TABLE 1.
Effects of BRACO-19 on HHV-6A integration frequency in HeLa and U2OS cells
| Cell line and treatment | No. of clones |
Integration frequencya (%) | |
|---|---|---|---|
| ciHHV-6− | ciHHV-6+ | ||
| HeLa | 413 | 82 | 16.6 |
| HeLa + BRACO-19 | 338 | 34 | 9.1* |
| U2OS | 202 | 17 | 7.8 |
| U2OS + BRACO-19 | 137 | 11 | 7.4 |
*, P = 0.0017.
To confirm these results, we included an additional cell line, MCF-7, and used the ddPCR analysis platform that allowed the detection of ciHHV-6A+ cells in polyclonal populations without the need to produce individual clones. BRACO-19-treated and untreated HeLa, MCF-7, and U2OS cells were infected with HHV-6A and allowed to grow for approximately 1 month. Cellular DNA was isolated and the percentage of cells containing integrated HHV-6A was determined by ddPCR, as described previously (44). ddPCR analyses confirmed that the integration frequency was significantly reduced in HeLa (52.66%) (P = 0.010) and MCF-7 (51.05%) (P = 0.009) cells treated with BRACO-19 (Table 2). A reduction of 20% of the integration frequency was observed in U2OS cells in the presence of BRACO-19, but it did not reach statistical significance (P = 0.056). Taken together, our data suggest that integration and/or maintenance of the HHV-6A viral genome is significantly reduced in HeLa and MCF-7 cells treated with BRACO-19 but not in telomerase-negative U2OS.
TABLE 2.
Effects of BRACO-19 on chromosomal integration of HHV-6A into HeLa, MCF-7, and U2OS cells
| Cell line and treatment | No. of copies of U65-U66/no. of copies of RPP30 | % ciHHV-6A+ cellsa |
|---|---|---|
| HeLa | 42.2/1,070 | 7.9 |
| HeLa + BRACO-19 | 16.7/893 | 3.74* |
| MCF-7 | 40.1/701 | 11.4 |
| MCF-7 + BRACO-19 | 18.6/665 | 5.58** |
| U2OS | 151/1,683 | 18 |
| U2OS + BRACO-19 | 67.6/980 | 13.8 |
*, P = 0.010; **, P = 0.009.
DISCUSSION
Although HHV-6A/B chromosomal integration was first thought to be an evolutionary dead-end, it is now thought to be a way HHV-6A/B, and several other herpesviruses that harbor telomeres at their genome ends, maintain their virus genome during latency (45). The integrated virus genome can be excised from latently infected cells and reinitiate lytic replication (5, 46–49). Certain HHV-6A/B proteins, such as U94, possess many characteristics, such as exonuclease and helicase activities, a preferential DNA binding to telomeric sequences, and the ability to hydrolyze ATP, all of which are compatible with a role in viral integration (50). However, recent results indicate that U94 is dispensable for HHV-6A integration in vitro (51). HHV-6A/B chromosomal integration is thought to occur via homologous recombination. Viral integration always occurs in telomeres, and it has been shown that the viral telomeric sequences are essential for efficient integration (28). Even though the formal integration mechanism has yet to be demonstrated experimentally, sequencing of ciHHV-6A/B indicates that the TMR of the DRR of the genome is fused to the host chromosome telomeric repeats with loss of pac2 sequence from DRR. Such a structure is compatible with HR-mediated viral integration (5, 30, 31).
To better understand the HHV-6A integration process, we made use of BRACO-19, a compound that stabilizes G-quadruplex structures (36, 37). The binding of BRACO-19 to telomeric G-quadruplexes was confirmed by surface plasmon resonance, while circular dichroism spectra coupled with melting analyses indicated that BRACO-19 stabilizes the G-quadruplex structure. Considering that HR is a dynamic process that requires that two sequences bind together and elongate with the intervention of a polymerase, we expected that the rigidity of the telomeres, and possibly the viral TMRs as well, in the presence of BRACO-19 would affect chromosomal integration. Indeed, we observed that in HeLa and MCF-7 cells, the frequency of chromosomal integration was reduced by about 50% when exposed to the drug. The reduction in integration frequency is not due to negative effects of BRACO-19 on HHV-6A entry, as no difference in viral genome copy numbers was observed between mock- and BRACO-19-treated cells (Fig. 4). Furthermore, considering that HR operates before cells enter mitosis, during and shortly after the S and G2 phases of the cell cycle (52), we have confirmed that BRACO-19 did not influence cell cycle progression under the experimental conditions tested (data not shown). Thus, reduced fluidity of telomeric sequences negatively affected the ability to generate clones of cells with integrated virus. In contrast to HeLa and MCF-7 cells, no statistically significant effects of BRACO-19 on the frequency of HHV-6A chromosomal integration in U2OS cells were noted. Several hypotheses may be formulated to explain this difference. First, HeLa and MCF-7 cells express telomerase and use this enzyme to elongate their telomeres. Upon viral integration, the viral TMR on the left end (from the DRL) of the genome is short and must be elongated to prevent cells from entering senescence. Inhibition of telomerase activity by stabilization of the G-quadruplex viral telomere likely prevents telomere elongation early after integration and may cause cells containing the integrated HHV-6A genome to die prematurely. In contrast, U2OS cells do not express telomerase and use an alternative telomere elongation mechanism (ALT). ALT is mostly based on HR (53), and repression of HR appears to be loosened in ALT+ cells to allow telomere maintenance, which might be the cause of the more frequent homologous recombination and telomere sister chromatid exchange observed in these cells (54, 55). In ALT+ cells, the viral telomere likely gets elongated by HR in a manner similar to host cell telomeres. Second, it was recently reported that ALT+ cells have a much longer telomeric overhang (40 to 400 nucleotides) (56) than telomerase-positive cells (65 to 140 nucleotides) (57). This difference in telomeric overhang length may influence the folding of the telomeric DNA into a G-quadruplex and affect BRACO-19 binding. Third, another important difference between ALT+ and telomerase-expressing cells is the frequent association of the promyelocytic leukemia protein (PML) nuclear bodies at telomeres in ALT+ cells (58, 59). In addition to PML, these ALT-associated PML nuclear bodies (APBs) contain many other proteins that form ring-like structures around the telomeric DNA that may negatively influence DNA folding or affect BRACO-19 binding. Furthermore, some of these proteins present at telomeres of ALT+ cells include the BLM and WRN RecQ helicases that bind telomeric DNA (60–62) and can unwind G-quadruplex structures (63–65), thereby limiting the potential number of G-quadruplex structures. Considering that ligands that stabilize G-structures are also reported to inhibit the processivity of these enzymes (66), the net in vivo impact of G-quadruplex unfolding by helicases and stabilization by BRACO-19 remains to be determined.
The presence of G-quadruplexes in viral genomes was previously reported (reviewed in reference 67). Interestingly, BRACO-19 negatively affects genome-processing activities, such as replication and transcription of herpes simplex virus 1 (68), Epstein-Barr virus (69), and human immunodeficiency virus type 1 (70). Unlike these viruses, however, in silico analyses of the HHV-6A/B genomes indicate that potential G-quadruplex structures are located exclusively in the viral TMR and not within coding regions (70). BRACO-19 therefore is not expected to affect the expression of HHV-6A/B genes. Whether BRACO-19 affects viral DNA replication is yet to be studied in detail. The cell lines used in this study are not very permissive to HHV-6A infection/replication and thus are not adequate to address the effects of BRACO-19 on viral DNA replication. However, considering that HHV-6A TMRs, located at the extremities of the viral genome, contain telomeric and pac1 sequences that can fold into G-quadruplex (Fig. 3) and likely form similar structures during viral DNA unwinding and replication, one might expect BRACO-19 to affect viral replication or DNA packaging. Analysis of the HHV-6B pac1 sequence also indicates folding into a parallel G-quadruplex structure (data not shown). Thermal unfolding studies indicate that relative to HHV-6A pac1 (63% GC), HHV-6B pac1 (59% GC) was less stable, with a Tm of 57°C and a Tm of 67°C in the absence or in the presence of 100 mM KCl, respectively. How and if this variation in G-quadruplex stability of pac1 sequences translates into biological differences during HHV-6A and HHV-6B infection or integration remains to be determined.
In summary, we presented data indicating that the G-quadruplex stabilizing agent BRACO-19 negatively affects HHV-6A integration in telomerase-expressing cells. Our results suggest that following the recombination between the host and viral telomeres at one end of the virus genome, the viral telomere at the other DRL end needs to be rapidly elongated. If this event is prevented, through the action of BRACO-19 that inhibits telomerase activity, expansion of clones with integrated virus is compromised. It is well established that the self-renewal potential of cells is directly proportional to telomere lengths and telomerase activity (71, 72). It is also known that the shortest telomere, not average telomere length, is critical for cell viability and chromosome stability (73). When the number of telomeric repeats falls below 13, chromosomal instability is observed (74). At the time of integration, the TMR in DRL contains approximately 50 to 60 repeats (23, 75). Considering that with each cell division 10 to 20 telomeric repeats are lost (76), in the absence of elongation, chromosomal instability would occur within 2 to 3 cell divisions. The study of DNA damage responses at the extremity of the viral genome should help in understanding the importance of telomere elongation processes following integration and its importance in preventing premature cellular senescence. Lastly, based on these results we also surmise that the long-term maintenance of integrated HHV-6A/B following infection of primary somatic cells that lack telomerase expression and do not have telomere elongation mechanisms is improbable. To be successful, HHV-6A/B integration likely needs to occur in primary cells expressing telomerase, such as gametes, stem cells, or activated T lymphocytes, the latter being primary HHV-6 target cells (77). In fact, T and B lymphocytes are unique among differentiated somatic cell types in terms of their ability to respond to specific or nonspecific stimuli by proliferation and continued expansion, accompanied by TERT upregulation (78, 79). However, the factors/conditions favoring integration over productive infection in highly permissive cells such as T lymphocytes remain to be elucidated.
MATERIALS AND METHODS
Cell lines and viruses.
U2OS (osteosarcoma) cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM; Corning Cellgro, Manassas, VA, USA) supplemented with 10% Nu serum (Corning Cellgro), nonessential amino acids (Corning Cellgro), HEPES, sodium pyruvate (Wisent Inc., St-Bruno, Québec, Canada), and 5 μg/ml plasmocin (Invitrogen, San Diego, CA, USA). HeLa (cervix epithelial) and MCF-7 (mammary epithelial) cells (ATCC) were cultured in the same medium but supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) instead of Nu serum. HSB-2 (ATCC CCL-120.1), human T lymphoblastic cells, were cultured in RPMI 1640 (Corning Cellgro) supplemented with 10% Nu serum (Corning Cellgro), HEPES, and 5 μg/ml plasmocin (InvivoGen, San Diego, CA, USA). HHV-6A was propagated in HSB-2 as previously described (80).
BRACO-19 cytotoxicity.
BRACO-19 (N,N′-(9-((4-(dimethylamino)phenyl)amino) acridine-3,6-diyl)bis(3-(pyrrolidin-1-yl) propanamide) was purchased from Sigma-Aldrich Canada. To evaluate the cytotoxic effect of BRACO-19, a 3-(4-5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed. U2OS, HeLa, and MCF-7 cells were plated at 5,000 cells/well in 96-well plates (6 wells/condition). Cells were treated with increasing concentrations of BRACO-19 (0 to 2 μM) and incubated at 37°C. After 72 h, cell survival was evaluated by adding 10 μl/well MTT solution (TACS MTT cell proliferation assay; R&D Systems). Cells were incubated for 4 h at 37°C, and 110 μl/well of SDS-HCl solution (10% sodium dodecyl sulfate, 0.01 M HCl) subsequently was added. Cells were incubated overnight at 37°C, and the absorbance was determined at 620 nm (Infinite M200 microplate reader; Tecan).
Effect of BRACO-19 on HHV-6 DNA levels.
To investigate the effect of BRACO-19 on the initial phase of HHV-6 infection, U2OS, HeLa, and MCF-7 cells were seeded in 12-well culture plates at 8 × 104 (U2OS) or 4 × 104 (HeLa and MCF-7) cells/well and treated before (2 h) and during HHV-6A infection (multiplicity of infection [MOI] of 1) for 4 h at 37°C. Cells were then washed three times with phosphate-buffered saline (PBS) and cultured in complete medium with or without BRACO-19 (1 μM). After 4 h, 2 days, and 3 days of infection, cells were washed extensively, treated with trypsin to remove attached virions and detached cells, and washed again before DNA isolation using a QIAamp DNA blood minikit (Qiagen Inc., Toronto, ON, Canada) by following the manufacturer's instructions. One hundred nanograms of DNA was analyzed by qPCR with a Rotor-Gene Q (Qiagen) using a Rotor-Gene multiplex PCR kit (Qiagen) and HHV-6-specific primers as previously described (81). As a cellular control, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific probe and primers (IDT) were used with the following sequences: FWD, 5′-GTCCCTCAATATGGTCCTGTC-3′; REV 5′-TTCTCCATGGTGGTGAAGAC-3′; probe, 5′-/5HEX/CGACGTACT/ZEN/CAGCGCCAGCATC/3IABkFQ/-3′.
G-quadruplex-specific antibodies.
The pSANG10-3F-BG4 plasmid was a gift from the laboratories of S. Balasubramanian and J. McCafferty (Addgene plasmid number 55756) (41). The single-chain Flag-BG4 antibody was expressed in Escherichia coli and purified by affinity chromatography as described previously (41). The 1H6 monoclonal antibody (42) was a gift from P. Lansdorp.
ELISA.
The ELISA was performed with the BG4 or 1H6 antibodies that bind specifically to the G-quadruplex structures (41, 42). Biotinylated oligonucleotides of the Myc promoter (5′-Biosg/ACTACTACTGGGGAGGGTGGGGAGGGTGGGGAAGG-3′) and telomeric repeats (5′-Biosg/ACTACTACTGGTTAGGGTTAGGGTTAGGGTTAGGGTTAG-3′), which fold in G-quadruplex, and ssDNA (5′-Biosg/ACTACTACTGGCATAGTGCGTGGGCG-3′), which supports formation of G-quadruplexes, were purchased from IDT (Coralville, Iowa, USA). Ninety-six-well plates were coated with an avidin solution at 5 μg/ml in water at 37°C overnight. The oligonucleotides were folded by incubation at 95°C for 10 min in buffer (10 mM Tris-HCl, pH 7.5, 100 mM KCl) and were left to cool down at room temperature overnight. The oligonucleotides were added to the wells at a concentration of 2 pmol/well. The G-quadruplex-specific antibody was added at different concentrations (BG4 from 0 to 32 nM and 1H6 from 0 to 266 nM). After 3 washes with PBS–0.1% Tween (PBS-T), 1 μg/ml of anti-FLAG antibody (Applied Biological Materials, Richmond, BC, Canada) was added (for BG4 only) and the plate was incubated for 1 h at room temperature. After 3 additional washes, 0.1 ml of horseradish peroxidase-conjugated goat anti-mouse antibody (1:20,000 dilution) (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) was added to each well for 1 h at room temperature. After washing, the tetramethylbenzidine substrate (BD Biosciences, Mississauga, ON, Canada) was added and the reaction allowed to proceed for 15 min, after which 0.05 ml of sulfuric acid was added. The absorbance of each well was measured at 450 nm with an Infinite M200 microplate reader.
Surface plasmon resonance.
All surface plasmon resonance experiments were conducted using the ProteOn XPR36 apparatus (Bio-Rad, Mississauga, Canada). Biotin-labeled DNA oligonucleotides were attached to the surface of a NeutrAvidin-coated NLC chip (Bio-Rad). The NLC chip was preconditioned by injecting, two times sequentially, 50 mM NaOH and 1 M NaCl in the two directions (horizontally and vertically). The same biotinylated oligonucleotides used for the ELISA (Myc, human Telo G-rich, and human Telo C-rich [5′-Biosg/ACTACTACTTAACCCTAACCCTAACCCTAACCCTAACCCT-3′]) were diluted to 25 nM in running buffer (10 mM HEPES-KOH, pH 7.4, 0.2 M KCl, 3 mM EDTA, and 0.005% Tween 20). The equivalent of 60 response units (RU) of biotinylated oligonucleotides was attached to the NLC chip. The chip then was ready for protein binding analyses. Between each of these injections, a three-step regeneration (2 M NaCl, 5 mM NaOH plus 0.5 M NaCl, 0.1% SDS) program was performed to remove residual binding. BG4 was injected at 50 μl/min over 3 min, followed by a dissociation time of 10 min. BRACO-19 was injected at 25 μl/min over 15 min, followed by a dissociation time of 30 min.
Spectroscopic analysis.
HHV-6A pac1 sequence (5′-CCCCCGGGGGGGCTAAAAAAAGGGGGGTAA-3′) from U1102 strain (26) and the telomeric DNA oligonucleotide (5′-AGGGTTAGGGTTAGGGTTAGGG-3′) were diluted from stock to a final concentration of 4 μM in lithium cacodylate buffer (10 mM, pH 7.4, with or without 100 mM KCl). All samples were annealed by heating at 95°C for 5 min, gradually cooled to room temperature, and measured after 24 h. BRACO-19 was added after DNA annealing at a final concentration of 16 μM. CD spectra were recorded on a Chirascan-Plus (Applied Photophysics, Leatherhead, United Kingdom) equipped with a Peltier temperature controller using a quartz cell of 5-mm optical path length and a scanning speed of 50 nm/min with a response time of 4 s over a wavelength range of 230 to 320 nm. The reported spectrum of each sample represents the average of 2 scans at 20°C and is baseline corrected for signal contributions due to the buffer. Observed ellipticities were converted to mean residue ellipticity (θ) with the following equation: θ = deg × cm2 × dmol−1. For the determination of Tm, spectra were recorded over a temperature range of 20 to 90°C, with a temperature increase of 5°C/min. Tm values were calculated by the van't Hoff equation applied for a two-state transition from a folded to unfolded state, assuming that the heat capacity of the folded and unfolded states are equal.
HHV-6 integration assay.
HeLa and U2OS cells were seeded in 48-well plates at 1 × 104 cells per well and cultured overnight. BRACO-19 (1 μM) was added to half of the wells 3 h before infection. All cells were infected at an MOI of 5 for 4 h, washed with PBS, and cultured in media with or without BRACO-19 (1 μM) for 48 h for the first three assays. Cells were then detached with trypsin, counted, and seeded at 2 cells per well in 96-well plates. Wells containing more than one clone were excluded. Clones were harvested at confluence (approximately 30 days) and then detached with trypsin and grown in 24-well plates. When confluent, DNA was isolated from cells and tested for the presence of HHV-6 DNA by qPCR. In a fourth assay, cells were infected and cultured for 72 h with BRACO-19 (1 μM) and then seeded in 96-well plates with or without BRACO-19 (100 nM). Medium and BRACO-19 were changed every 3 days for the rest of the experiment.
To confirm the above-described results, we measured the frequency of integrated HHV-6 in bulk cultures using droplet digital PCR (ddPCR) instead of isolating individual clones (44). Briefly, HeLa, MCF-7, and U2OS cells were infected using the same conditions described above. Three days after infection, cells were transferred into a 25-cm2 flask and cultured with or without BRACO-19 (100 nM) for 21 days. At that point in time, cells were detached using trypsin and DNA was isolated and tested by ddPCR for the presence of HHV-6 and the cellular reference gene RPP30, as described previously (82). U65-U66-specific primers were validated previously (12, 83) and used for the detection of the integrated HHV-6 genome.
Statistical analyses.
Clone integration frequencies were analyzed using Fisher's exact test, and BRACO-19 cytotoxicity and DNA levels were analyzed using t test. A P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by a grant from the Canadian Institutes of Health awarded to L.F. (MOP_123214). S.G.G. is supported by a studentship from the Fonds de Recherche Québec-Santé.
S.G.-G., A.G., S.A., and N.W. performed experiments. L.F., B.B.K., and S.N.R. made contributions to the design and interpretation of experiments, in addition to providing key reagents. S.G.-G. and L.F. wrote the paper.
REFERENCES
- 1.Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067. [DOI] [PubMed] [Google Scholar]
- 3.Tesini BL, Epstein LG, Caserta MT. 2014. Clinical impact of primary infection with roseoloviruses. Curr Opin Virol 9:91–96. doi: 10.1016/j.coviro.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill JA, Zerr DM. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol 9:53–60. doi: 10.1016/j.coviro.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A 107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. 1993. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol 40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 7.Luppi M, Barozzi P, Marasca R, Torelli G. 1994. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia 8(Suppl 1):S41–S45. [PubMed] [Google Scholar]
- 8.Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I. 1999. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 94:1545–1549. [PubMed] [Google Scholar]
- 9.Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, Kondo K, Okada S, Yamanishi K. 2004. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol 73:465–473. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- 10.Morissette G, Flamand L. 2010. Herpesviruses and chromosomal integration. J Virol 84:12100–12109. doi: 10.1128/JVI.01169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, Flamand L, Gautheret-Dejean A, Hall CB, Kamble RT, Kuehl U, Lassner D, Lautenschlager I, Loomis KS, Luppi M, Lusso P, Medveczky PG, Montoya JG, Mori Y, Ogata M, Pritchett JC, Rogez S, Seto E, Ward KN, Yoshikawa T, Razonable RR. 2012. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol 22:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravel A, Dubuc I, Morissette G, Sedlak RH, Jerome KR, Flamand L. 2015. Inherited chromosomally integrated human herpesvirus 6 as a predisposing risk factor for the development of angina pectoris. Proc Natl Acad Sci U S A 112:8058–8063. doi: 10.1073/pnas.1502741112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. 1995. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol 46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 14.Daibata M, Taguchi T, Taguchi H, Miyoshi I. 1998. Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br J Haematol 102:1307–1313. doi: 10.1046/j.1365-2141.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 15.Nacheva EP, Ward KN, Brazma D, Virgili A, Howard J, Leong HN, Clark DA. 2008. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J Med Virol 80:1952–1958. doi: 10.1002/jmv.21299. [DOI] [PubMed] [Google Scholar]
- 16.Takubo K, Aida J, Izumiyama-Shimomura N, Ishikawa N, Sawabe M, Kurabayashi R, Shiraishi H, Arai T, Nakamura K. 2010. Changes of telomere length with aging. Geriatr Gerontol Int 10(Suppl 1):S197–S206. doi: 10.1111/j.1447-0594.2010.00605.x. [DOI] [PubMed] [Google Scholar]
- 17.Makarov VL, Hirose Y, Langmore JP. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88:657–666. doi: 10.1016/S0092-8674(00)81908-X. [DOI] [PubMed] [Google Scholar]
- 18.McElligott R, Wellinger RJ. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J 16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai W, Shay JW, Wright WE. 2005. Human telomeres maintain their overhang length at senescence. Mol Cell Biol 25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Rane G, Dai X, Shanmugam MK, Arfuso F, Samy RP, Lai MK, Kappei D, Kumar AP, Sethi G. 2016. Ageing and the telomere connection: An intimate relationship with inflammation. Ageing Res Rev 25:55–69. doi: 10.1016/j.arr.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 21.de Lange T. 2009. How telomeres solve the end-protection problem. Science 326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin ME, Efstathiou S, Craxton M, Macaulay HA. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol 73:8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol 73:8053–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng H, Dewhurst S. 1998. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J Virol 72:320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson BJ, Dewhurst S, Gray D. 1994. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol 68:3007–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufer BB, Flamand L. 2014. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol 9C:111–118. doi: 10.1016/j.coviro.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Wallaschek N, Sanyal A, Pirzer F, Gravel A, Mori Y, Flamand L, Kaufer BB. 2016. The telomeric repeats of human herpesvirus 6A (HHV-6A) are required for efficient virus integration. PLoS Pathog 12:e1005666. doi: 10.1371/journal.ppat.1005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arbuckle JH, Pantry SN, Medveczky MM, Prichett J, Loomis KS, Ablashi D, Medveczky PG. 2013. Mapping the telomere integrated genome of human herpesvirus 6A and 6B. Virology 442:3–11. doi: 10.1016/j.virol.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, Hidalgo-Bravo A, Zhang E, Cotton VE, Mendez-Bermudez A, Wig G, Medina-Calzada Z, Neumann R, Jeffreys AJ, Winney B, Wilson JF, Clark DA, Dyer MJ, Royle NJ. 2014. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res 42:315–327. doi: 10.1093/nar/gkt840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohye T, Inagaki H, Ihira M, Higashimoto Y, Kato K, Oikawa J, Yagasaki H, Niizuma T, Takahashi Y, Kojima S, Yoshikawa T, Kurahashi H. 2014. Dual roles for the telomeric repeats in chromosomally integrated human herpesvirus-6. Sci Rep 4:4559. doi: 10.1038/srep04559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipps HJ, Rhodes D. 2009. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Sen D, Gilbert W. 1990. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 34.Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, Webba da Silva M. 2011. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew Chem Int Ed Engl 50:10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 35.Read M, Harrison RJ, Romagnoli B, Tanious FA, Gowan SH, Reszka AP, Wilson WD, Kelland LR, Neidle S. 2001. Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc Natl Acad Sci U S A 98:4844–4849. doi: 10.1073/pnas.081560598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell NH, Parkinson GN, Reszka AP, Neidle S. 2008. Structural basis of DNA quadruplex recognition by an acridine drug. J Am Chem Soc 130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 37.Zhou G, Liu X, Li Y, Xu S, Ma C, Wu X, Cheng Y, Yu Z, Zhao G, Chen Y. 2016. Telomere targeting with a novel G-quadruplex-interactive ligand BRACO-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget 7:14925–14939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oganesian L, Bryan TM. 2007. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays 29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 40.Simonsson T, Pecinka P, Kubista M. 1998. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res 26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. 2013. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM Jr, Sen D, Lansdorp PM. 2014. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res 42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haider SM, Neidle S, Parkinson GN. 2011. A structural analysis of G-quadruplex/ligand interactions. Biochimie 93:1239–1251. doi: 10.1016/j.biochi.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Gravel A, Dubuc I, Wallaschek N, Gilbert-Girard S, Collin V, Hall-Sedlak R, Jerome KR, Mori Y, Carbonneau J, Boivin G, Kaufer BB, Flamand L. 2017. Cell culture systems to study human herpesvirus 6A/B chromosomal integration. J Virol 91:e00437-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osterrieder N, Wallaschek N, Kaufer BB. 2014. Herpesvirus genome integration into telomeric repeats of host cell chromosomes. Annu Rev Virology 1:215–235. doi: 10.1146/annurev-virology-031413-085422. [DOI] [PubMed] [Google Scholar]
- 46.Endo A, Watanabe K, Ohye T, Suzuki K, Matsubara T, Shimizu N, Kurahashi H, Yoshikawa T, Katano H, Inoue N, Imai K, Takagi M, Morio T, Mizutani S. 2014. Molecular and virological evidence of viral activation from chromosomally integrated human herpesvirus 6A in a patient with X-linked severe combined immunodeficiency. Clin Infect Dis 59:545–548. doi: 10.1093/cid/ciu323. [DOI] [PubMed] [Google Scholar]
- 47.Gravel A, Hall CB, Flamand L. 2013. Sequence analysis of transplacentally acquired human herpesvirus 6 DNA is consistent with transmission of a chromosomally integrated reactivated virus. J Infect Dis 207:1585–1589. doi: 10.1093/infdis/jit060. [DOI] [PubMed] [Google Scholar]
- 48.Hill JA, Sedlak RH, Zerr DM, Huang ML, Yeung C, Myerson D, Jerome KR, Boeckh MJ. 2015. Prevalence of chromosomally integrated human herpesvirus 6 in patients with human herpesvirus 6-central nervous system dysfunction. Biol Blood Marrow Transplant 21:371–373. doi: 10.1016/j.bbmt.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prusty BK, Krohne G, Rudel T. 2013. Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation. PLoS Genet 9:e1004033. doi: 10.1371/journal.pgen.1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trempe F, Gravel A, Dubuc I, Wallaschek N, Collin V, Gilbert-Girard S, Morissette G, Kaufer BB, Flamand L. 2015. Characterization of human herpesvirus 6A/B U94 as ATPase, helicase, exonuclease and DNA-binding proteins. Nucleic Acids Res 43:6084–6098. doi: 10.1093/nar/gkv503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallaschek N, Gravel A, Flamand L, Kaufer BB. 2016. The putative U94 integrase is dispensable for human herpesvirus 6 (HHV-6) chromosomal integration. J Gen Virol 97:1899–1903. doi: 10.1099/jgv.0.000502. [DOI] [PubMed] [Google Scholar]
- 52.Branzei D, Foiani M. 2008. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 53.Cesare AJ, Reddel RR. 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 54.de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 55.Wang RC, Smogorzewska A, de Lange T. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Min J, Wright WE, Shay JW. 2017. Alternative lengthening of telomeres can be maintained by preferential elongation of lagging strands. Nucleic Acids Res 45:2615–2628. doi: 10.1093/nar/gkw1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chai W, Du Q, Shay JW, Wright WE. 2006. Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Wu G, Lee WH, Chen PL. 2000. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J Biol Chem 275:30618–30622. [DOI] [PubMed] [Google Scholar]
- 59.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59:4175–4179. [PubMed] [Google Scholar]
- 60.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. 2004. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell 14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 61.Schawalder J, Paric E, Neff NF. 2003. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol 4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. 2002. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 63.Chatterjee S, Zagelbaum J, Savitsky P, Sturzenegger A, Huttner D, Janscak P, Hickson ID, Gileadi O, Rothenberg E. 2014. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun 5:5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fry M, Loeb LA. 1999. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem 274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 65.Sun H, Karow JK, Hickson ID, Maizels N. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 66.Li JL, Harrison RJ, Reszka AP, Brosh RM Jr, Bohr VA, Neidle S, Hickson ID. 2001. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry 40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- 67.Metifiot M, Amrane S, Litvak S, Andreola ML. 2014. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res 42:12352–12366. doi: 10.1093/nar/gku999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Artusi S, Nadai M, Perrone R, Biasolo MA, Palu G, Flamand L, Calistri A, Richter SN. 2015. The herpes simplex virus-1 genome contains multiple clusters of repeated G-quadruplex: implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res 118:123–131. doi: 10.1016/j.antiviral.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norseen J, Johnson FB, Lieberman PM. 2009. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol 83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biswas B, Kandpal M, Jauhari UK, Vivekanandan P. 2016. Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genomics 17:949. doi: 10.1186/s12864-016-3282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 72.Harley CB, Futcher AB, Greider CW. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 73.Hemann MT, Strong MA, Hao LY, Greider CW. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67–77. doi: 10.1016/S0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 74.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. 2007. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev 21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gravel A, Ablashi D, Flamand L. 2013. Complete genome sequence of early passaged human herpesvirus 6A (GS strain) isolated from North America. Genome Announc 1:00012-13. doi: 10.1128/genomeA.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. 2000. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem 275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 77.Lusso P, Markham PD, Tschachler E, di Marzo Veronese F, Salahuddin SZ, Ablashi DV, Pahwa S, Krohn K, Gallo RC. 1988. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6). J Exp Med 167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hiyama K, Hirai Y, Kyoizumi S, Akiyama M, Hiyama E, Piatyszek MA, Shay JW, Ishioka S, Yamakido M. 1995. Activation of telomerase in human lymphocytes and hematopoietic progenitor cells. J Immunol 155:3711–3715. [PubMed] [Google Scholar]
- 79.Weng NP, Levine BL, June CH, Hodes RJ. 1996. Regulated expression of telomerase activity in human T lymphocyte development and activation. J Exp Med 183:2471–2479. doi: 10.1084/jem.183.6.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flamand L, Gosselin J, D'Addario M, Hiscott J, Ablashi DV, Gallo RC, Menezes J. 1991. Human herpesvirus 6 induces interleukin-1 beta and tumor necrosis factor alpha, but not interleukin-6, in peripheral blood mononuclear cell cultures. J Virol 65:5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaworska J, Gravel A, Fink K, Grandvaux N, Flamand L. 2007. Inhibition of transcription of the beta interferon gene by the human herpesvirus 6 immediate-early 1 protein. J Virol 81:5737–5748. doi: 10.1128/JVI.02443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sedlak RH, Cook L, Huang ML, Magaret A, Zerr DM, Boeckh M, Jerome KR. 2014. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem 60:765–772. doi: 10.1373/clinchem.2013.217240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flamand L, Gravel A, Boutolleau D, Alvarez-Lafuente R, Jacobson S, Malnati MS, Kohn D, Tang YW, Yoshikawa T, Ablashi D. 2008. Multicenter comparison of PCR assays for detection of human herpesvirus 6 DNA in serum. J Clin Microbiol 46:2700–2706. doi: 10.1128/JCM.00370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]