ABSTRACT
Mammarenaviruses are enveloped viruses with a bisegmented negative-stranded RNA genome that encodes the nucleocapsid protein (NP), the envelope glycoprotein precursor (GPC), the RNA polymerase (L), and a RING matrix protein (Z). Viral proteins are synthesized from subgenomic mRNAs bearing a capped 5′ untranslated region (UTR) and lacking 3′ poly(A) tail. We analyzed the translation strategy of Tacaribe virus (TCRV), a prototype of the New World mammarenaviruses. A virus-like transcript that carries a reporter gene in place of the NP open reading frame and transcripts bearing modified 5′ and/or 3′ UTR were evaluated in a cell-based translation assay. We found that the presence of the cap structure at the 5′ end dramatically increases translation efficiency and that the viral 5′ UTR comprises stimulatory signals while the 3′ UTR,specifically the presence of a terminal C+G-rich sequence and/or a stem-loop structure, down-modulates translation. Additionally, translation was profoundly reduced in eukaryotic initiation factor (eIF) 4G-inactivated cells, whereas depletion of intracellular levels of eIF4E had less impact on virus-like mRNA translation than on a cell-like transcript. Translation efficiency was independent of NP expression or TCRV infection. Our results indicate that TCRV mRNAs are translated using a cap-dependent mechanism, whose efficiency relies on the interplay between stimulatory signals in the 5′ UTR and a negative modulatory element in the 3′ UTR. The low dependence on eIF4E suggests that viral mRNAs may engage yet-unknown noncanonical host factors for a cap-dependent initiation mechanism.
IMPORTANCE Several members of the Arenaviridae family cause serious hemorrhagic fevers in humans. In the present report, we describe the mechanism by which Tacaribe virus, a prototypic nonpathogenic New World mammarenavirus, regulates viral mRNA translation. Our results highlight the impact of untranslated sequences and key host translation factors on this process. We propose a model that explains how viral mRNAs outcompete cellular mRNAs for the translation machinery. A better understanding of the mechanism of translation regulation of this virus can provide the bases for the rational design of new antiviral tools directed to pathogenic arenaviruses.
KEYWORDS: arenavirus, mRNA, translation
INTRODUCTION
The Arenaviridae family comprises the genera Reptarenavirus and Mammarenavirus; the latter, which includes important human pathogens, is further subdivided based on phylogeny, serological properties, and geographical distribution into two main groups: the Old World (OW) and New World (NW) viruses (1). Tacaribe mammarenavirus (TCRV), a prototype of NW viruses, is nonpathogenic and closely related to Junín virus (JUNV), the etiological agent of Argentinian hemorrhagic fever (2). Like all arenaviruses, TCRV is enveloped, with a genome composed of two negative-stranded RNA segments (S and L), each encoding two proteins with an ambisense strategy. The S segment encodes the nucleocapsid protein (NP) and the precursor (GPC) of the envelope glycoprotein complex (GP), which mediates receptor binding and cell entry. The L segment encodes the viral RNA-dependent RNA polymerase (L) and a RING matrix protein (Z) that is involved in virus morphogenesis and egress (3, 4). NP and L protein are produced early during infection; NP associates with the viral genomic and antigenomic RNAs and with L protein to form nucleocapsids that promote viral genome transcription and replication (5–7). Coding sequences in both RNA segments are arranged in opposite orientations and separated by a noncoding intergenic region (IGR), predicted to fold into 1 or 2 strong RNA stem-loop (hairpin) structures (8–10). In particular, the predicted conformation of the S-segment IGR from NW mammarenaviruses is well conserved, consisting of two stem-loop structures, each of which comprise an uninterrupted 10- to 16-bp stem with high G+C content and a 3- to 4-nucleotide (nt) loop (4).
Arenavirus coding sequences are expressed from subgenomic mRNAs transcribed from the 3′ region of genomes and antigenomes. The NP and L mRNAs are transcribed from the genomic RNAs, whereas the GPC and Z mRNAs are synthesized from the antigenomic RNAs. The 5′ untranslated regions (UTRs) of arenaviral mRNAs bear a 5′ cap structure followed by a short, host-derived sequence (3 to 7 nt) acquired by a cap-snatching mechanism (11). In contrast to most eukaryotic mRNAs, viral mRNAs lack a 3′ poly(A) tail. We have previously demonstrated that the 3′ ends of the four TCRV mRNAs map within the corresponding IGR and that the four mRNA 3′ UTRs can theoretically fold into a strong hairpin configuration (12). Characterization of the 3′ termini of JUNV S-derived mRNAs revealed similar results (13). Moreover, using a TCRV S-minigenome system, we have shown that the presence of a predicted single-hairpin structure at the IGR is required for transcription termination, supporting the notion that IGRs contain transcription termination signals (14). Although the predicted structures have also been proposed to play a role in viral translation (8, 15), their specific involvement in viral gene expression has remained unclear.
Translation initiation is a tightly regulated and rate-limiting process during which a number of translation factors contribute to assemble a complete (80S) ribosome at the start codon of the mRNA. Eukaryotic initiation factor 4E (eIF4E) associates with the scaffolding protein eIF4G and the RNA helicase eIF4A to form the eIF4F complex and binds to the cap structure present at the 5′ terminus of eukaryotic mRNAs. eIF4F recruits the 43S preinitiation complex (40S ribosomal subunit, eIF1, eIF1A, eIF2, tRNAi Met, and eIF3) that scans the 5′ UTR in the 5′ to 3′ direction until a favorably positioned AUG initiation codon is encountered. The interaction between eIF4G and poly(A)-binding protein (PABP), bound at the terminal 3′ poly(A) tail, stimulates translation by bringing the mRNA 5′ and 3′ ends together in a “closed loop” topology. Start codon recognition is followed by recruitment of the 60S ribosomal subunit to form an elongation-competent 80S ribosome (16, 17).
Viruses have developed various strategies to take control of the host translation machinery at the initiation, elongation, or termination step. Picornaviruses, retroviruses, and caliciviruses, for example, code for proteases that cleave host translation factors, including eIF4G, thus preventing host cap-dependent translation. Viral mRNAs translation remains unaffected because of the usage of internal ribosome entry sites (IRES) to initiate the process via a cap-independent mechanism (18–20). Differently, rotavirus nonstructural protein NSP3 binds to a consensus sequence at the 3′ UTR of the capped and nonpolyadenylated viral mRNAs and interacts with eIF4G at the PABP-binding site, impairing the translation of cellular mRNAs (21). Prior studies demonstrated that infection of Vero cells with TCRV induces the inhibition of cellular protein synthesis (22). Alteration of host gene expression in cells infected with the Old World lymphocytic choriomeningitis mammarenavirus (LCMV) has also been documented (9). Recent evidence suggests that virus replication can take place in the absence of functional eIF4E. Indeed, silencing of eIF4E did not reduce viral nucleoprotein synthesis or virus yield in JUNV-infected cells; similar results were obtained for TCRV and Pichinde, another NW mammarenavirus (23). In addition, based on the observation that JUNV NP associates with eIF4A and eIF4G but not with eIF4E in infected cells, and that NP can be pulled down by 7 methyl-cap Sepharose beads from infected-cell lysates, it has been proposed that NP may bind to the cap structure and eventually act as a surrogate of eIF4E (23). However, the molecular mechanism by which arenavirus mRNAs outcompete cellular mRNAs has not been yet elucidated.
In this study, using synthetic transcripts and a cell-based translation assay, we directly analyzed the contribution of 5′ and 3′ untranslated sequences in TCRV mRNA translation. We found that the viral 5′ UTR displays a stimulatory effect, whereas the 3′ UTR may negatively modulate translation. By employing RNA interference (RNAi)-mediated silencing and factor depletion, we evaluated the requirement of key host initiation factors in this process. Interestingly, our results suggest that viral translation depends on eIF4G but displays a low dependence on the cap-binding initiation factor 4E. Additionally, translation of a synthetic virus-like mRNA was not stimulated upon NP overexpression or in the context of viral infection. Our results suggest that TCRV mRNAs may engage yet-unidentified noncanonical host factors to initiate translation.
RESULTS
Role of TCRV untranslated sequences: downregulatory effect of the 3′ UTR in viral translation.
To analyze the role of viral mRNA UTR sequences in translation, we employed a translation assay based on capped synthetic RNAs obtained by in vitro transcription from T7 promoter-controlled constructs. A virus-like mRNA (5′wt/3′wt_1), mimicking the TCRV NP mRNA, was synthesized using plasmid p5′wt/3′wt_1 as the template (Fig. 1A). Transcript 5′wt/3′wt_1 comprises a 5-nt nonviral sequence preceding the viral 5′ UTR, which is fused to the reporter firefly luciferase (FLUC) open reading frame (ORF), followed by the viral 3′ UTR. A variant of the viral transcript, harboring a 2-nt mutation that creates a BsaAI site to facilitate engineering (5′wt/3′wt_2), was obtained from plasmid p5′wt/3′wt_2 (Materials and Methods). Translation from these mRNAs was examined after transfection into mammalian cells. The synthetic mRNA RLUC, which expresses the Renilla reniformis luciferase enzyme in a cap-independent manner, was included as an internal control in the transfection mix. As illustrated in Fig. 1C, the mRNA 5′wt/3′wt_2 (henceforth called 5′wt/3′wt) mediated levels of FLUC activity comparable to those exhibited by 5′wt/3′wt_1, indicating that generation of the BsaAI sequence had no effect on translation. An uncapped version of the virus-like transcript was also synthesized and tested for translation (5′Δcap/3′wt). Importantly, the absence of the cap structure at the 5′ end decreased translation about 250-fold compared with the capped transcript, suggesting that viral mRNA translation is cap dependent.
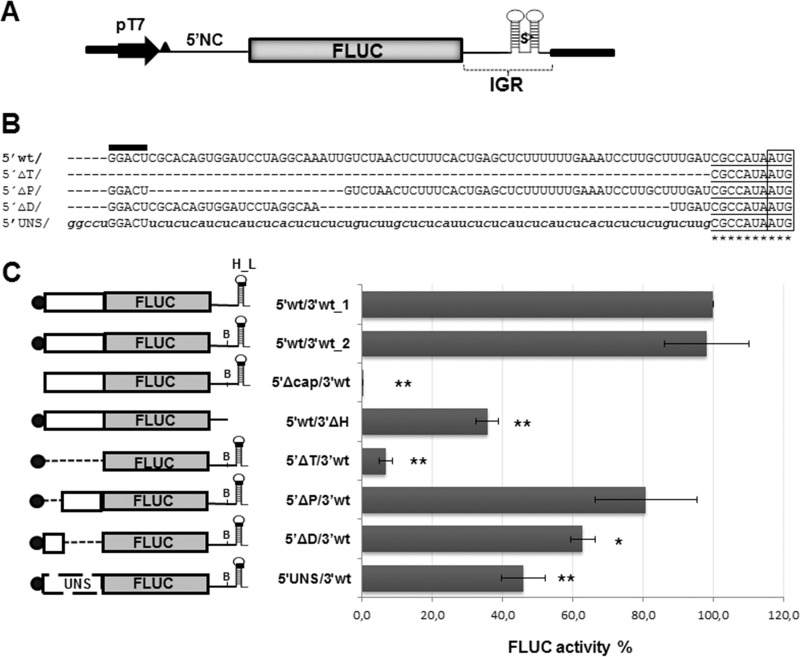
FIG 1.
(A) Schematic representation of plasmid p5′wt/3′wt_1. This plasmid allows the synthesis of mRNA 5′wt/3′wt_1, which mimics the TCRV NP mRNA. The construct, comprising the TCRV S antigenome 5′ noncoding sequence (5′NC), fused to the FLUC ORF followed by the TCRV S antigenome intergenic region (IGR), is located downstream of the T7 RNA polymerase promoter (pT7). Nonviral nucleotides preceding the 5′NC are indicated by a black triangle. The position of the SmaI (S) restriction site used for plasmid linearization is shown. (B) Alignment of the viral 5′ UTR sequence to the corresponding region of mutant mRNAs. The wild-type sequence is indicated with capital letters (5′wt/); 5′ nonviral nucleotides are marked with a black bar on top. The AUG codon is boxed; the neighboring sequence is underlined. Substitutions are indicated with lowercase letters. Unmodified viral residues are marked with asterisks. (C) Translation activity of the synthetic virus-like mRNAs. (Left) Schematic representation of synthetic transcripts. The 5′ cap structure is represented with a black circle; the H_L sequence and the position of the BsaAI sequence (B) are indicated. UNS, unstructured sequence. (Right) BSR cells were transfected with each of the indicated mRNAs, along with the RLUC transcript as an internal control. FLUC and RLUC activities were determined in cell lysates 8 h later, as indicated in Materials and Methods. Mean normalized FLUC values (FLUC/RLUC; means ± SD) from three independent experiments (each performed in triplicate) are shown as a percentage of data corresponding to 5′wt/3′wt_1 mRNA, taken as 100% (*, P < 0.05; **, P < 0.005).
A set of mutant plasmids carrying deletions or substitution of the 5′ UTR was generated based on plasmid p5′wt/3′wt_2. Following linearization with SmaI, these constructs were utilized for in vitro synthesis of mutant capped transcripts, which were assayed for translation upon transfection into BSR cells (Fig. 1C). FLUC synthesis from 5′ΔT/3′wt, which retains the AUG context but lacks 74 out of 81 bases of the 5′ UTR (Fig. 1B), was substantially reduced compared with its counterpart carrying the complete viral 5′ UTR sequence. Deletion of 24 residues proximal to the 5′ end (transcript 5′ΔP/3′wt) resulted in levels of FLUC activity 81% of those mediated by the wild-type mRNA, while deletion of 43 nt distal to the 5′ end (transcript 5′ΔD/3′wt) decreased reporter activity to levels 63% with respect to those of the control. These results are consistent with previous reports demonstrating that a short (<20 nt) UTR sequence strongly diminishes translation efficiency (24) and suggested that translation inhibition could correlate with the deletion size. To address the question of whether the length of the 5′ UTR is important for translation regulation, almost the entire 5′ UTR sequence (69 out of 81 nt) was replaced with a sequence consisting in pyrimidine tracts interrupted by A or G residues that would not be expected to adopt a stable configuration. Indeed, it would be stabilized by free energy values substantially higher than those corresponding to the wild-type structure, as determined by mfold (ΔG: −0.80 kcal/mol and −14.30 kcal/mol, respectively [http://unafold.rna.albany.edu/?q=mfold/rna-folding-form]). The resulting mutant 5′UNS/3′wt, in which the context of the AUG codon was preserved (Fig. 1B), showed levels of FLUC activity reduced to 46% of those determined for the control transcript (Fig. 1C). These results indicated that restoring the 5′ UTR to the wild-type length, but with an unstructured sequence, was not sufficient to achieve wild-type translation levels, suggesting that the viral 5′ UTR contains specific signals that stimulate translation.
On the other hand, as a first step to evaluate the role of the 3′ UTR, plasmid p5′wt/3′wt_2 was linearized with BsaAI to generate a capped mutant mRNA lacking 51 out of 74 nt of the 3′ UTR (mutant 5′wt/3′ΔH). The deleted sequence includes a predicted highly stable hairpin structure (H_L) which is conserved across mammarenaviruses. A decrease of 65% in FLUC activity was observed when translation of mutant 5′wt/3′ΔH was examined in transfected BSR cells (Fig. 1C), indicating that deletion of the H_L sequence is detrimental for viral translation.
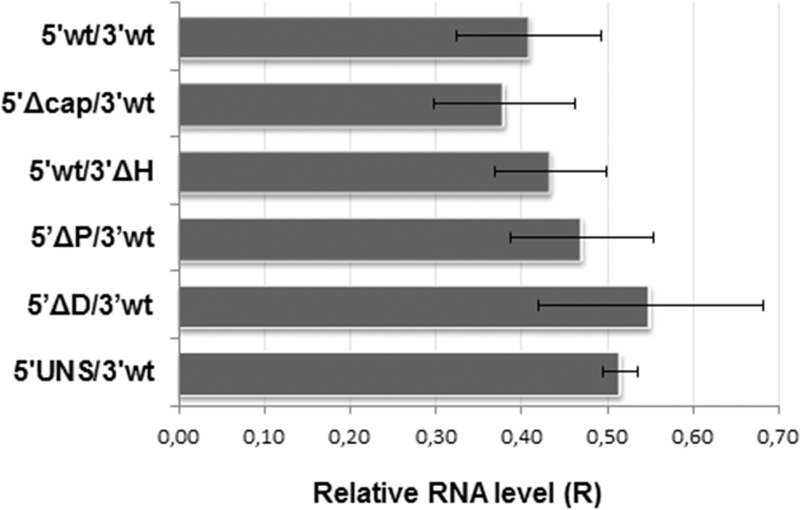
Since modifications in the UTR sequences could affect intracellular stability of synthetic RNAs, we employed reverse transcription followed by quantitative PCR (RT-qPCR) to measure the relative levels of wild-type or mutant mRNAs within cells at the experimental endpoint, as an estimation of mRNA decay (Fig. 2). The results showed estimated levels of wild-type mRNA at 8 h posttransfection (hpt) about 40% of those at 4 hpt (R = 0.41). Comparable remaining levels were observed for mutant mRNAs, indicating that changes affecting the integrity of the 5′ or 3′ UTR did not reduce the stability of transcripts throughout the course of the experiment. These results are particularly relevant in the case of mutants 5′wt/3′ΔH and 5′UNS/3′wt, which displayed significantly decreased levels of translation, and support the notion that regulation of TCRV mRNA translation may involve specific structural elements and/or sequence motifs residing in both the 5′ and 3′ UTRs.
FIG 2.
Stability of synthetic transcripts. BSR cells were transfected in duplicates with each of the indicated mRNAs. Total cellular RNA was extracted at 4 and 8 hpt and used to amplify a fragment of the FLUC ORF, and as a control, a fragment of the GAPDH ORF, by RT-qPCR. For each transcript, the mRNA level at 8 hpt relative to that at 4 hpt was calculated using the 2−ΔΔCT equation. Data correspond to the averages and SD from at least 2 independent experiments.
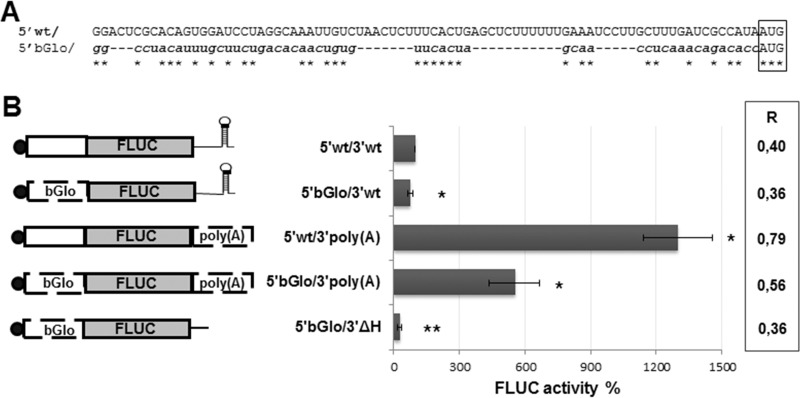
To further investigate the role of viral 5′ and 3′ UTRs, the translation ability of the virus-like mRNA was compared to that of chimeric mRNAs (Fig. 3). A synthetic transcript was created which bears the 5′ UTR from human β-globin (βGlo) mRNA, and a 53-nt 3′ poly(A) tail flanking the FLUC ORF. In addition, either the βGlo 5′ UTR or the poly(A) tail was swapped for the viral 5′ or 3′ UTR in 5′wt/3′wt, to generate transcript 5′βGlo/3′wt or 5′wt/3′poly(A), respectively. As a control, a 3′ truncated version of 5′βGlo/3′wt was also synthesized (5′βGlo/3′ΔH). Translation from each of these mRNAs was monitored in transfected BSR cells (Fig. 3B). Substitution of the viral 5′ UTR with the 5′βGlo sequence, which differs in length and sequence (Fig. 3A), resulted in a 20% inhibition of FLUC activity. In contrast, replacement of the viral 3′ UTR with the poly(A) tract caused a 14-fold increase in translation levels compared with those with 5′wt/3′wt. Translation of the 5′βGlo/3′poly(A) transcript, which carries substitutions of both 5′ and 3′ sequences, reached levels only 5-fold higher than those observed for the virus-like transcript, consistent with the inhibitory effect of replacing the 5′ UTR. In line with previous observations, deletion of the H_L sequence caused a 3-fold reduction in translation from 5′βGlo/3′ΔH compared with 5′βGlo/3′wt (Fig. 3B). Determination of the relative mRNA levels at the experimental endpoint by RT-qPCR showed that replacement of the 5′ UTR had no substantial impact, while change of the viral 3′ UTR with the poly(A) tail caused a nonsignificant increase, on transcript stability (Fig. 3B, right). These results suggest that the viral 5′ UTR comprises stimulatory signal/s which are absent in the 5′ UTR βGlo and that the viral 3′ UTR modulates translation through a mechanism differing from that mediated by the poly(A) tail of cellular mRNAs.
FIG 3.
(A) Alignment of the viral 5′ UTR sequence (5′ wt/) to the 5′ UTR from human β-globin (5′βGlo/) mRNA, comprised in chimeric transcripts. The AUG codon is boxed. Identical residues are marked with asterisks. βGlo mRNA has GenBank accession number NM_000518.4. (B) Translation activities of chimeric mRNAs. BSR cells were transfected with the indicated mRNAs, and normalized FLUC activity (FLUC/RLUC) was determined in cell lysates at 8 hpt. Values correspond to the means (±SD) from at least two independent experiments, each performed in triplicate. Data are presented as a percentage of those corresponding to 5′wt/3′wt mRNA, taken as 100% (*, P < 0.05; **, P < 0.001). A schematic of each transcript is depicted on the left. The 5′ cap structure is represented with a black circle; the poly(A) tract and the 5′ UTR of βGlo mRNA are indicated. The mRNA level at 8 hpt relative to that at 4 hpt was calculated using the 2−ΔΔCT equation (chart on the right). Data correspond to the averages from at least 2 independent experiments. Standard deviations ranged from 5.3% [5′bGlo/3′poly(A)] to 20% [5′wt/3′wt; 5′wt/3′poly(A)].
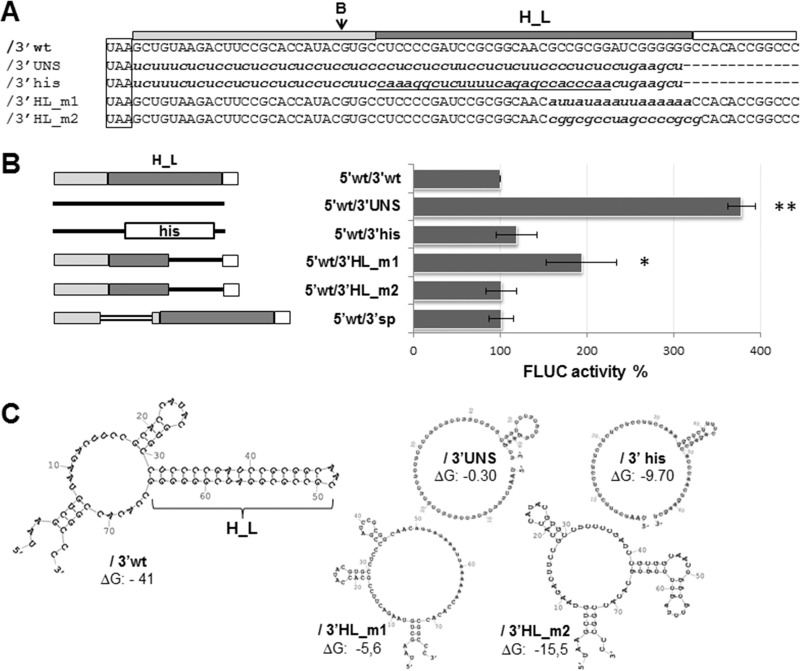
According to previous reports (9, 12, 13), the 3′ UTR of arenavirus mRNAs could fold into a major stem-loop structure (H_L) stabilized by 10 to 12 GC base pairs. In order to further investigate the role of this putative structural element in viral translation, mutant transcripts with partial or total replacements of the 3′ UTR sequence were generated and assayed for their translation abilities (Fig. 4). The results showed that the transcript 5′wt/3′UNS, whose 3′ UTR consists in a pyrimidine-rich sequence predicted to destabilize RNA folding, directed translation levels almost 4-fold higher than 5′wt/3′wt. In contrast, when half of the 3′ UTR in mutant 5′wt/3′UNS was replaced with the 3′-terminal stem-loop (His) of mammalian histone mRNA, which is involved in mRNA processing and stability (25), translation activity was restored to wild-type levels (5′wt/3′his), demonstrating that a nonviral stem-loop structure might functionally replace H_L. In addition, to address the question of whether the position of H_L affects translation, we generated the transcript 5′wt/3′sp, which contains a 48-nt insertion at the BsaAI site that distances H_L from the stop codon (Materials and Methods). Translation of mutant 5′wt/3′sp reached levels comparable with those of the wild-type transcript (Fig. 4). Similar results were obtained when alternative spacers differing in sequence and length (up to 192 nt) were inserted at the same position (data not shown), showing that the distance between the 3′-terminal stem-loop and the stop codon has no detectable impact on translation.
FIG 4.
(A) Alignment of the 5′wt/3′wt 3′ UTR sequence to the corresponding region of mutant mRNAs. The wild-type sequence (/3′wt) is indicated with capital letters, and substitutions are indicated with lowercase letters; the histone 3′-terminal stem-loop sequence is underlined. The position of the BsaAI (B) enzyme recognition site is indicated with an arrow; the H_L sequence is marked with a dark gray box (on top), the stop codon is boxed. (B) (Left) Schematics of the 3′ UTR of wild-type and mutant mRNAs. The 27-nt sequence proximal to the stop codon (light gray box), H_L (dark gray box), and the terminal 12-nt sequence (empty box) are indicated. Substitution sequences are represented with solid lines, except for the histone 3′ terminal stem-loop (his box). The inserted sequence in 5′wt/3′sp is depicted with a double line. (Right) Translation of mutant mRNAs. BSR cells were transfected with the indicated capped mRNAs; FLUC and RLUC activities were determined in cell lysates 8 h later. Mean normalized FLUC values (±SD) from three independent experiments (each conducted in triplicate) are shown as a percentage of those corresponding to 5′wt/3′wt mRNA, taken as 100% (*, P < 0.05; **, P < 0.005). (C) Wild-type and mutant mRNA 3′ UTRs. Predicted secondary structures were obtained by using mfold (46). Free energies (ΔG) are indicated in kilocalories per mole.
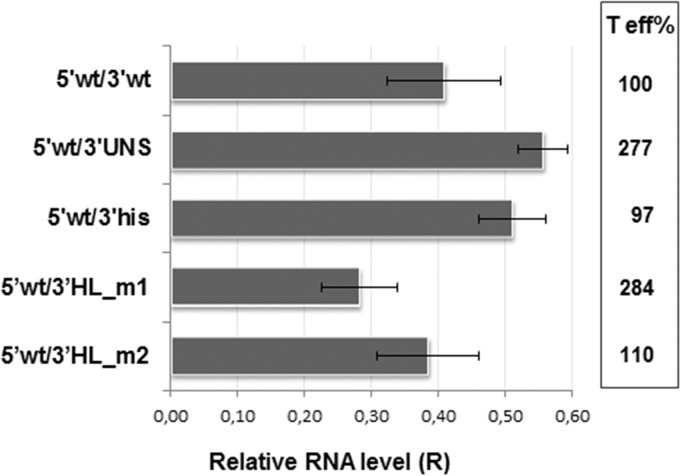
Further analysis of mutant mRNAs bearing partial substitutions intended to destabilize the predicted H_L structure revealed that replacement of the 3′ half of H_L by a 16-nt AU tract (mutant 5′wt/3′HL_m1) caused a 2-fold increase in FLUC activity compared with mRNA 5′wt/3′wt. In contrast, change of the 3′ half of H_L with a C+G-rich sequence (mutant 5′wt/3′HL_m2) resulted in wild-type levels of translation (Fig. 4). Quantification of the endpoint relative amounts of FLUC RNA showed that mutant mRNAs decayed at similar or slightly reduced levels compared to those of the wild-type mRNA (Fig. 5). In order to relate both parameters, we defined translation efficiency as the ratio between mean FLUC values (Fig. 4B) and average relative endpoint FLUC mRNA levels. Accordingly, transcripts 5′wt/3′UNS and 5′wt/3′HL_m1 showed a nearly 3-fold increase in translation efficiency, compared with 5′wt/3′wt (Fig. 5, right). Overall, these results support the notion that a terminal sequence with high C+G content and/or a terminal stem-loop structure at the 3′ UTR could negatively modulate viral translation.
FIG 5.
Stability of synthetic transcripts. BSR cells were transfected in duplicates with each of the indicated RNAs. Total cellular RNA was extracted at 4 and 8 hpt and used to amplify a fragment of the FLUC and GAPDH ORFs by RT-qPCR (Materials and Methods). The mRNA level at 8 hpt relative to that at 4 hpt was calculated as for Fig. 2. Data correspond to the averages (±SD) from at least 2 independent experiments. Translation efficiency (T eff%) was estimated as the ratio between mean FLUC values (from Fig. 4) and the corresponding average of relative endpoint RNA levels (R). T eff for 5′wt/3′wt was set as 100%.
Participation of cellular factors in TCRV translation.
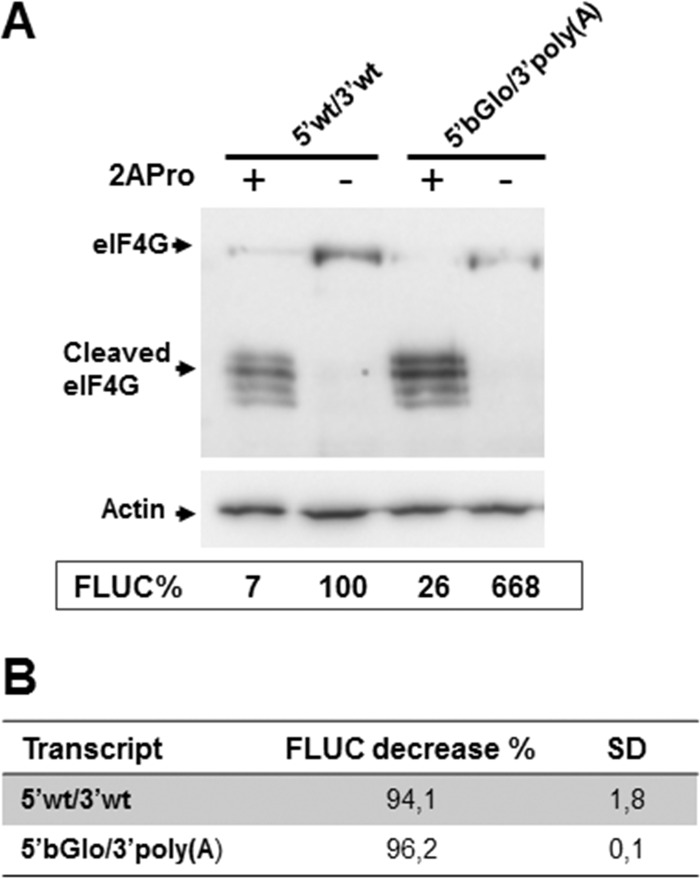
We next examined the extent to which viral translation may depend on key host translation initiation factors, eIF4G and eIF4E. First, to investigate the participation of eIF4G, BSR cells were transfected with the synthetic mRNAs, along with a plasmid expressing poliovirus 2A protease (2APro), which cleaves both eIF4G isoforms (eIF4GI and eIF4GII), thus abrogating eIF4G activity. Upon incubation for 8 h, cells were lysed and assayed for FLUC activity (Fig. 6). Western blotting of cell lysates confirmed the cleavage of eIF4G in cells transfected with the 2APro-expressing plasmid (2APro+). As expected, translation of the cell-like 5′βGlo/3′poly(A) transcript was strongly inhibited in eIF4G-depleted cells, decreasing to 4% of those detected in control cells. Comparable reduction levels of FLUC activity were determined for transcript 5′wt/3′wt upon eIF4G cleavage (Fig. 6). Similar levels of inhibition were observed in eIF4G-inactivated human HEK293T cells (data not shown). Together, the results strongly suggest that eIF4G is required for viral mRNA translation in host cells.
FIG 6.
Requirement of eIF4G for viral mRNA translation. (A) BSR cells were transfected with each of the indicated capped mRNAs, along with either a plasmid expressing poliovirus 2A protease (+) or pTM1 empty vector as a control (−), as indicated at the top. Cell lysates, harvested 8 h later, were subjected to Western blotting to detect eIF4G; actin was probed as a control for gel loading (upper blots). Cell lysates were also analyzed for FLUC activity, as described in legend to Fig. 1. Mean FLUC activity data (FLUC%, chart) from a representative experiment, carried out in triplicate, are presented as a percentage of the mean value determined for 5′wt/3′wt mRNA in the absence of 2APro, taken as 100%. (B) For each transcript, FLUC activity decrease (percent) was calculated by subtracting mean FLUC values in depleted cells from those determined in control cells (set as 100%). Data correspond to the means (±SD) from two independent experiments performed in triplicate.
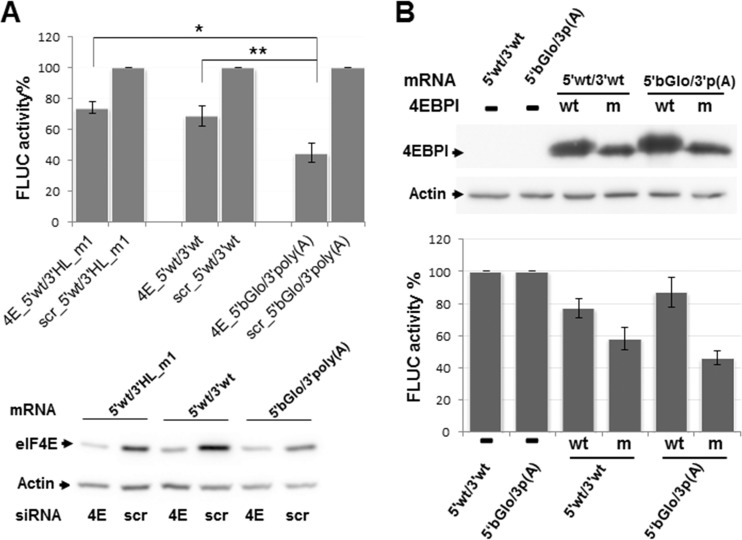
Next, to investigate the requirement of the cap-binding initiation factor 4E for viral translation, HEK293T cells were transfected with a pool of four small interfering RNAs (siRNAs) directed against the eIF4E gene, and the translation efficiency of synthetic mRNAs was analyzed under conditions of low intracellular levels of eIF4E. Indeed, analysis of the cell lysates confirmed that cellular eIF4E levels were substantially diminished (75 to 90%) in cells transfected with the targeting siRNAs compared with those transfected with control (scrambled) oligonucleotides (Fig. 7A, bottom). As expected, FLUC activity directed by 5′βGlo/3′poly(A) in silenced cells was reduced to levels nearly 45% of those determined in control cells. In contrast, comparable levels of eIF4E depletion had significantly less impact on translation of 5′wt/3′wt; mean values of FLUC activity in eIF4E-silenced cells were 70% of those in control cells. Translation of the mutant 5′wt/3′HL_m1 was similarly affected, suggesting that the 3′ H_L structure is not responsible for the relative low eIF4E-dependence exhibited by the viral mRNA (Fig. 7A, top).
FIG 7.
Effect of eIF4E depletion. (A) HEK293T cells were transfected with siRNAs against the host factor eIF4E (4E), or with nontargeted siRNAs (scr) and 42 h later were transfected with the indicated capped mRNAs. Following 6 h of incubation, FLUC activity (top) was determined in cell lysates, as indicated in Materials and Methods. For each transcript, mean FLUC values (±SD) determined in depleted cells are shown as a percentage of those in control cells, taken as 100%. Data correspond to four independent experiments, each conducted in triplicate (**, P < 0.005; *, P < 0.05). Aliquots of the cell lysates were analyzed by Western blotting to detect eIF4E and actin, using specific antibodies (bottom). (B) HEK293T cells were transfected in triplicate with plasmids expressing HA-tagged versions of wild-type (wt) or mutant (m) 4EBPI, or left untransfected (−). At 40 hpt, cells were transfected with the indicated synthetic transcripts, and cell lysates were obtained 8 h later. Expression of wild-type and mutant 4EBPI was evidenced by Western blotting using an anti-HA antibody as indicated in Materials and Methods; representative images are shown (top). Samples were also assayed for FLUC activity (bottom). For each transcript, mean FLUC activities in 4EBPI-expressing cells are shown as a percentage of those determined in nonoverexpressing control cells (100%). Data correspond to the means (±SD) from two independent experiments performed in triplicate. The cell-like mRNA 5′βGlo/3′poly(A) is indicated as 5′bGlo/3′p(A).
In order to further evaluate the translation efficiency of 5′wt/3′wt and 5′βGlo/3′poly(A) under low-eIF4E-availability conditions, we used a mutant version of the eIF4E-binding protein (4E-BP1), bearing alanine substitutions at four key phosphorylation sites (26). As previously reported, it was expected that unphosphorylated 4E-BP1 sequesters the eIF4E factor impairing eIF4F complex assembly (26). Thus, mRNA expression was evaluated in cells overexpressing either wild-type or mutant 4E-BP1. Nonoverexpressing cells were included as a control (Fig. 7B). In agreement with published data (26), the results showed moderately reduced FLUC levels for both virus- and cell-like transcripts upon overexpression of wild-type 4E-BP1. Additionally, in cells expressing mutant 4E-BP1, translation of the cell-like mRNA was inhibited to 46%, while the virus-like transcript was translated at levels near to 60% of those observed in control cells (Fig. 7B). Overall, these results were consistent with our finding that translation of the virus-like transcript is less sensitive to eIF4E availability than the cell-like mRNA and suggested that viral mRNAs may engage an alternative factor(s) for in vivo translation initiation.
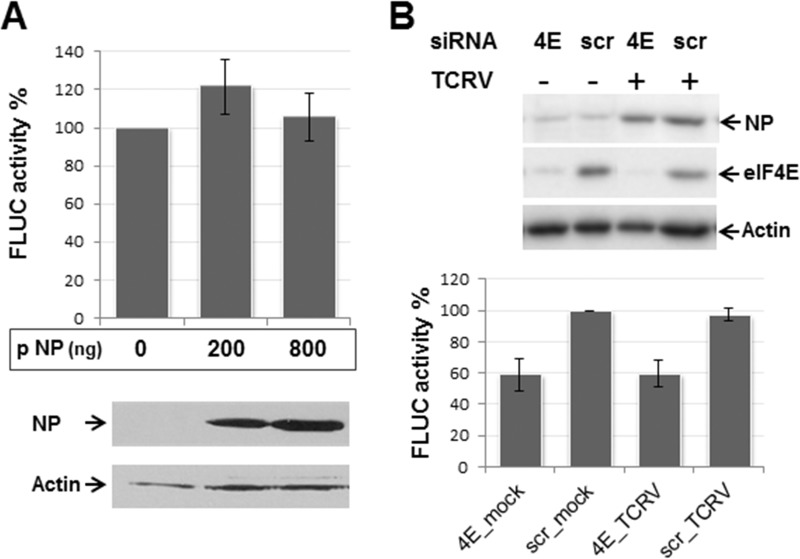
Taking into consideration previous studies, which have proposed that NP might replace eIF4E in JUNV translation (23), we then asked whether NP could modulate virus-like mRNA translation efficiency. To address this question, TCRV NP was overexpressed in BSR cells for 24 h, followed by synthetic mRNA transfection and FLUC activity determination. The results showed that 5′wt/3′wt was translated in the absence of NP at levels comparable to those determined in cells expressing increasing amounts of NP (Fig. 8A). To further substantiate these results, the effect of viral proteins on 5′wt/3′wt translation was examined in TCRV-infected human cells, depleted or not of eIF4E. As expected, expression of the eIF4E-targeted siRNAs reduced levels of eIF4E to 10% of those observed for control siRNAs (Fig. 8B, top). Consistent with our previous results, in the absence of all four viral proteins, silencing of eIF4E resulted in translation of the virus-like mRNA at levels about 60 to 70% of those detected in nonsilenced control cells. Importantly, the results revealed that TCRV infection had no effect on FLUC activity, in either eIF4E-depleted or nondepleted cells (Fig. 8B, bottom). Likewise, FLUC translation levels were unaffected when BHK cells were infected with TCRV prior to the transfection with the 5′wt/3′wt mRNA (data not shown). Taken together, these results suggest that viral translation does not require, and is in fact unaffected by, NP expression, either when transiently overexpressed or in the context of the viral infection.
FIG 8.
Analysis of the effect of NP on viral translation. (A) BSR cells were transfected in triplicate with the indicated amounts (in nanograms per well) of the TCRV NP-expressing plasmid (indicated as pNP). Control cells were transfected with 800 ng of empty pTM1 vector (0 pNP). At 4 hpt, cells were washed and transfected with capped 5′wt/3′wt transcript along with control mRNA RLUC. Cell lysates obtained 8 h later were assayed for FLUC activity (graph). Data correspond to the average (±SD) of at least three independent experiments, shown as percentage of mean values from control cells, taken as 100%. Samples were also analyzed for NP expression by Western blotting; images correspond to a representative experiment. (B) HEK293T cells were transfected with the eIF4E-targeted (4E), or scrambled (scr) siRNAs, and infected 24 h later with TCRV at a multiplicity of infection of 0.1 PFU/ml (+) or mock infected (−). At 18 h postinfection, cells were transfected with the virus-like transcript along with control mRNA RLUC. Cell lysates, obtained 6 h later, were analyzed by Western blotting (top), to detect eIF4E and NP. Actin was probed as gel loading control. Representative images are presented. Samples were also assayed for FLUC activity (bottom). Mean FLUC values are shown as a percentage of those in nondepleted, noninfected cells (scr_mock), taken as 100%. Data correspond to the mean (±SD) from two independent experiments performed in triplicate.
DISCUSSION
The mechanism used by arenaviruses to regulate translation of viral mRNAs has remained largely unknown. This study provides, for the first time, direct evidence on the molecular determinants involved in translation of TCRV, the prototypic New World mammarenavirus. As a first approach, we focused on elucidating the contribution of viral 5′ and 3′ mRNA nontranslated sequences. Using a synthetic transcript that mimics the TCRV NP mRNA and a cell-based translation assay, we found that the presence of a cap structure at the transcript 5′ terminus highly stimulated translation (Fig. 1), strongly suggesting a cap-dependent strategy. Further analysis revealed that mutant transcripts carrying truncated 5′ UTRs exhibited translation efficiencies that seemed to inversely correlate with the length of the deletion. However, when the entire viral 5′ UTR sequence was replaced with an unrelated sequence, likely unstructured, translation levels were further decreased, with no detectable change in transcript stability (Fig. 1 and 2). Despite the possibility that the qPCR-based assay may have technical limitations to finely measure subtle differences in mRNA decay, our data suggest that viral translation efficiency is influenced by specific sequence motifs and/or structure elements contained in the 5′ noncoding region. The fact that replacement of the viral 5′ UTR with the 5′ UTR from the human β-globin mRNA moderately reduced translation efficiency of an mRNA carrying the viral 3′ UTR, but caused a pronounced decrease in translation of a 3′poly(A)+ transcript (Fig. 3), strengthens the idea that the viral 5′ UTR bears specific signal(s) that stimulates translation.
Our findings raise the question of whether the stimulatory effect on translation exerted by the NP mRNA 5′ UTR might extend to the remainder viral mRNAs. A bioinformatics analysis (data not shown) revealed common features on the 5′ UTR sequences across the four viral mRNAs from clade B NW mammarenaviruses, which include TCRV and JUNV as well as other South American pathogenic viruses. A cap-proximal region can be defined by a conserved motif that spans the terminal 27- to 36-nt sequence (MEME [http://meme-suite.org/tools/meme]). Within this region, a stem-loop structure comprising the highly conserved 19-nt terminal sequence is predicted in all four viral mRNAs (Clustal [http://www.ebi.ac.uk/Tools/msa/clustalo/] and LocaRNA [http://rna.informatik.uni-freiburg.de/LocARNA/Input.jsp]). A second, internal region displaying no sequence or structural conservation can be identified within the 5′ UTR of NP, GPC, and Z mRNAs but is absent in the L mRNA. This region contains potential recognition sites for RNA binding proteins (RBP), such as heterogeneous nuclear ribonucleoproteins (hnRNPs) (RBPmap [http://rbpmap.technion.ac.il/]). Interestingly, it has been reported that silencing of a number of hnRNPs, known to regulate translation, reduces virus yield in JUNV-infected cells, suggesting that hnRNPs might be required during infection (27, 28). We speculate that the 5′ conserved terminal motif (or eventually the terminal stem-loop) in concert with the internal UTR sequence harboring RBP recognition site(s) might facilitate the recruitment of the 43S preinitiation complex to viral mRNA. Based on this assumption, it is possible to envisage that the stimulatory effect on translation observed for the 5′ UTR of mRNA NP may not extend to the 5′ UTR of the L mRNA, which might contribute to the low expression levels of L protein, as manifested throughout viral infection.
The 3′ UTR of the TCRV NP mRNA can potentially fold into a very stable hairpin structure (H_L [Fig. 4C]), similar to that predicted in the 3′ UTR of the other TCRV mRNAs (4). Interestingly, we observed a drastic reduction in translation efficiency upon deletion of the entire H_L, indicating that this region plays an important role in this process (Fig. 1 to 3). Notably, translation levels were not affected when the position of H_L was changed by introducing a spacer sequence (Fig. 4). A deeper analysis on 3′ UTR mutant mRNAs appeared to indicate that the stability of predicted secondary structures in the region may influence the mRNA translation efficiency. Indeed, we found that the 3′ UTR from mutant transcripts displaying wild-type levels of translation efficiency (5′wt/3′his and 5′wt/3′HL_m2) may theoretically fold into a nearly terminal hairpin comprising a stem stabilized by at least four GC base pairs. In contrast, the 3′ UTR from mRNAs showing higher levels of translation efficiency show less stable secondary structures (5′wt/3′UNS and 5′wt/3′HL_m1 [Fig. 4C]). In this regard, it is worth noting that circular-dichroism analysis of synthetic 35-nt RNA oligonucleotides spanning the 3′ H_L region revealed a remarkable higher thermal stability for wild-type H_L than for the corresponding sequence of mutant 5′wt/3′HL_m1 (data not shown).
Taken together, our data suggest that the secondary structure of the 3′-terminal region and/or its G+C content may play a downregulatory role in translation. In agreement with our observations, studies employing an LCMV S-minigenome system have shown that replacement of the S-IGR with the L-IGR, which displays a higher degree of complexity and stability than the former, diminished reporter gene expression from the NP locus (15).
Translation initiation is tightly regulated by the availability of host initiation factors. Particularly, host eIF4E is expressed at low levels in most cell types (29), and several viruses have evolved mechanisms to control eIF4E availability or assembly of the eIF4F complex (18, 30). Previous studies on JUNV and TCRV have reported that while the functional activity of eIF4GI and eIF4A is required, eIF4E is unessential for virus multiplication (23). In agreement with these precedents, here we show that while viral mRNA translation depends on eIF4G, it appears to be less dependent on eIF4E than a chimeric mRNA carrying a cellular 5′ UTR and a poly(A) tail (Fig. 6 and 7). Based on these results, it is tempting to speculate that in vivo, arenaviruses may engage additional host factors to operate a noncanonical cap-dependent mechanism for translation initiation. The observation that mutant 5′wt/3′HL_m1 behaved in eIF4E-depleted cells similarly to the wild-type virus-like transcript suggests that the viral reduced dependence on eIF4E might not be linked to the integrity of the 3′ H_L hairpin structure. Nevertheless, these results do not discard the possibility that binding of yet-undefined host factors to the G+C-rich 3′ UTR sequence may contribute to translation initiation. Notably, we found that replacement of the 3′ UTR by a poly(A) tail caused a dramatic increase in translation (Fig. 3), further supporting that the viral translation strategy differs from that used by most cellular mRNAs. Why arenaviruses have evolved a strategy that relies in an apparently disadvantageous 3′ UTR may be explained by a possible participation of this region in an eIF4E-independent translation mechanism, together with the relevant role of noncoding intergenic sequences in arenavirus transcription and replication (10, 14).
Several viruses, such as hantavirus and influenza virus, use viral proteins as noncanonical factors to drive cap-dependent translation initiation of viral mRNAs (26, 31, 32). Even though JUNV NP has been hypothesized to have a role in viral translation (23), our findings that neither overexpression of TCRV NP nor infection with TCRV impacted the virus-like mRNA translation efficiency (Fig. 8) support the notion that NP would not be a functional surrogate of eIF4E for arenavirus translation initiation. In line with this, it is worth noting that a putative cap-binding pocket initially predicted by crystallographic studies in the amino-terminal domain of Lassa mammarenavirus NP has been later ascribed to the RNA binding site and that in vitro experiments have shown that NP does not bind to cap-conjugated agarose beads (33, 34). Additionally, it has been observed that viral mRNAs are excluded from replication-transcription complexes where NP colocalizes with host translation initiation factors eIF4G and eIF4A, supporting the idea that the presence of NP within these assemblies would be unrelated to viral translation (35, 36).
In summary, our results indicate that TCRV mRNAs are translated using a cap-dependent mechanism, whose efficiency relies on the interplay between stimulatory signal(s) in the 5′ UTR and modulatory element/s in the 3′ UTR, likely including a 3′-terminal G+C-rich secondary structure. Data also show that viral translation displays a low dependence on eIF4E, which suggests that viral mRNAs may engage noncanonical host factors for translation initiation. As suggested for dengue virus (37), mammarenavirus translation initiation might alternate from a canonical to a noncanonical cap-dependent mechanism, in which NP becomes dispensable. Further studies oriented to identify alternative initiation factors required for viral translation would provide future targets for development of antiviral strategies.
MATERIALS AND METHODS
Cells and virus.
BSR (a clone of BHK-21) and HEK293T cells were grown in a 5% CO2 atmosphere at 37°C, using Glasgow minimum essential medium (GMEM; Invitrogen) and Dulbecco's modified Eagle's medium (DMEM; Invitrogen), respectively. Growth media were supplemented with glutamine (2 mM), 5 to 10% fetal calf serum (FCS) (Sigma-Aldrich), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Thermo Fisher Scientific). HEK293T cells were also supplemented with sodium 1 mM pyruvate and 0.1 mM nonessential amino acids (Gibco). TCRV stocks were prepared as indicated previously (22).
Plasmids.
Plasmid pTCRV-NP, which expresses the TCRV NP protein, and plasmid pMG-FLUC, expressing a TCRV S-minigenome that includes the firefly luciferase (FLUC) reporter gene, were generated previously (38, 39). Plasmid pMG-FLUC was used as the template for PCR amplification, using a forward primer (Fw_T75′UTR) including the T7 promoter sequence and a reverse primer (Rv_3′IGR) covering positions 1518 to 1538 of the TCRV S genome. The PCR product, comprising (5′ to 3′) the T7 promoter sequence followed by 5 nonviral nucleotides (randomly selected), the TCRV S antigenomic segment 5′ noncoding sequence fused to the FLUC ORF, and the complete TCRV S antigenomic IGR, was cloned into the PvuII site of plasmid pAT153/PvuII/8 (40) to generate plasmid p5′wt/3′wt_2. Both pMG-FLUC and p5′wt/3′wt_2 contain two previously introduced nucleotide changes within the IGR sequence that create a BsaAI restriction site (39). Two point substitutions that change back the BsaAI sequence to the TCRV wild-type sequence were introduced into p5′wt/3′wt_2 by site-directed mutagenesis, using a QuikChange site-directed mutagenesis kit (Stratagene) and primers containing the mutation. The resulting plasmid, designated p5′wt/3′wt_1, expresses a transcript that mimics the wild-type TCRV NP mRNA.
Plasmids p5′ΔT/3′wt, p5′ΔP/3′wt and p5′ΔD/3′wt, encoding transcripts with 5′ UTR deletions, and plasmids p5′wt/3′UNS, p5′wt/3′HL_m1, p5′wt/3′HL_m2, and p5′wt/3′his, which encode mRNAs with substitutions at the 3′ UTR, were generated by standard PCR and cloning strategies. To construct p5′UNS/3′wt and p5′βGlo/3′wt, a StuI restriction site was first introduced into p5′ΔT/3′wt between the T7 promoter sequence and the FLUC ORF by site-directed mutagenesis, as indicated above. The resultant plasmid was designated p5′ΔT_StuI/3′wt. Then two DNA fragments were generated by PCR, each comprising a StuI site and either the 5′ UTR sequence from human β-globin (βGlo) mRNA or a pyrimidine-rich UTR sequence (UNS). After digestion with StuI and BclI, each fragment was inserted between the StuI and BclI sites into p5′ΔT_StuI/3′wt. Similarly, plasmids p5′βGlo/3′wt and p5′wt/3′wt_2 were modified to replace the viral 3′ noncoding sequence with a poly(A) sequence. The resulting plasmids were named p5′βGlo/3′poly(A) and p5′wt/3′poly(A), respectively. Plasmid p5′wt/3′sp was generated by inserting a synthetic 48-bp DNA fragment into the BsaAI site of p5′wt/3′wt_2. The inserted sequence corresponds to two copies of the 24-nt sequence neighboring the stop codon within the viral 3′ UTR.
To construct plasmid pRLUC, used to generate a synthetic mRNA expressing Renilla reniformis luciferase enzyme (RLUC), a DNA fragment carrying the RLUC ORF was synthesized by PCR using specific primers and the pGL4.70hRluc plasmid (Promega) as the template. The PCR product was purified and inserted under the control of the T7 promoter between the NcoI and SmaI sites of pTM1 vector (41).
Oligonucleotides were provided by Macrogen, Inc. (Korea). All constructs were checked by dideoxynucleotide double-stranded DNA sequencing (Macrogen, Inc.). All primer sequences and vector maps are available upon request.
Plasmid pCMV-T7pol expresses the bacteriophage T7 RNA polymerase under the control of cytomegalovirus promoter (42) and was kindly provided by Martin A. Billeter (University of Zurich, Irchel, Switzerland). Plasmid pTM1-2A, expressing poliovirus 2A protease (43) was provided by Luis Carrasco (Centro de Biología Molecular Severo Ochoa, CSIC-UAM, Madrid, Spain). Plasmids pcDNA3-HA-4E-BP1 wt and pcDNA3-HA-4E-BP1 4A, which express hemagglutinin (HA)-tagged versions of the wild-type eIF4E-binding protein I (4E-BPI) and a nonphosphorylatable 4E-BP1 mutant, respectively (26), were kindly provided by Amelia Nieto (Centro Nacional de Biotecnología, Madrid, Spain).
In vitro RNA synthesis.
Plasmids p5′wt/3′UNS and p5′wt/3′his were linearized at the HindIII site of of pAT153/PvuII/8 vector. Unless otherwise indicated, pRLUC, p5′wt/3′wt_1, p5′wt/3′wt_2, and the rest of p5′wt/3′wt_2-derived mutant plasmids were digested with SmaI. To obtain transcript 5′βGlo/3′ΔH (Fig. 3B), plasmid p5′βGlo/3′wt was linearized at the BsaAI restriction site. Linearized plasmids were purified by phenol-chloroform extraction and ethanol precipitation. In vitro transcription was performed in a 20-μl reaction volume using 200 ng of purified linearized DNA, 0.4 mM GTP, 1 mM (each) ATP, CTP, and UTP, 1 mM cap analog [m7G(5′)ppp(5′)G; New England BioLabs Inc.], and T7 enzyme mix (MEGAScript T7 kit; Ambion). The reaction mixture was incubated for 3 h at 37°C. Then the DNA template was removed by treatment with RNase-free DNase (Ambion), the RNA product was purified through Sephadex G-50 columns (illustra MicroSpin, GE Healthcare), and its integrity was verified by electrophoresis on agarose gels.
In vitro synthesis of RLUC mRNA, as well as synthesis of the uncapped version of 5′wt/3′wt, was carried out by following the protocol described above, with the omission of the cap analog from the reaction mixture.
Transfection of RNA and siRNA.
Mammalian cell monolayers, grown in 24-well plates, were transfected with the desired mRNAs (100 to 200 ng/well) along with the RLUC mRNA (50 ng/well) using Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to the manufacturer's specifications, except that FCS was omitted from transfection medium. At 4 hpt, the transfection mixture was removed, cell monolayers were washed with serum-free medium and then covered with 0.5 ml per well of medium containing 2% FCS, and incubation proceeded for 4 h at 37°C. When indicated (see legend to Fig. 6), mRNA and plasmid DNA were cotransfected, as described above.
For silencing experiments, HEK293T cells grown in 24-well plates were transfected with 12.5 pmol per well of either targeted or scrambled siRNA (Dharmacon-GE) using Lipofectamine 2000, as indicated above.
DNA transfections.
Mammalian cell monolayers grown in 24-well plates were transfected with the desired plasmids using Lipofectamine 2000, according to the manufacturer's instructions. Expression of NP from pTCRV-NP, or of poliovirus 2A protease from plasmid pTM1-2A, was driven by the T7 RNA polymerase supplied in trans by cotransfection with 0.5 μg per well of pCMV-T7pol. The total amount of transfected DNA was kept constant by the addition of empty pTM1 vector DNA.
Western blotting.
Proteins were resolved by SDS-PAGE in gels containing 12% (for NP and eIF4E) or, in a two-step gel 8 to 12% (for eIF4G), polyacrylamide and then transferred to a nitrocellulose membrane (Hybond-ECL; GE Healthcare). Blots were stained with Ponceau-S (Sigma-Aldrich) before immunodetection, which was performed according to a protocol previously described (38). Primary antibodies were rabbit anti-TCRV NP polyclonal serum for NP (44) or commercially available monoclonal anti-actin (Thermo Fisher Scientific) or polyclonal anti-eIF4E, anti-eIF4G, or anti-HA tag (Cell Signaling) antibodies. Horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used according to the supplier's specifications. Protein bands were visualized with SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) on GBox equipment (Syngene; Sinoptics) and quantified by densitometry using ImageJ software (45).
RNA purification and RT-qPCR.
Intracellular total RNA was purified using TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer's instructions, and quantified using a NanoDrop spectrophotometer. Aliquots of 100 ng of purified RNA were treated with RQ1 DNase, RNase free (Promega), and then retrotranscribed with SuperScript II reverse transcriptase (Thermo Fisher Scientific) using primers specific for FLUC (LUC 633, ACGCAGGCAGTTCTATGAG) and the glyceraldehyde-3-phosphate dehydrogenase (GAPDHqrv, CAGAAGGTGCGGAGATGATGA) housekeeping gene. The primers for amplification of a FLUC or GAPDH 110-bp DNA fragment were designed by Primer Express software v3.0.1 (Applied Biosystems). RT-qPCRs were run in triplicate, using 1/20 diluted cDNA, 300 nmol of each primer (LUC1362fw, 5′ GGAATCCATCTTGCTCCAACA 3′, and LUC1471Rv, 5′ TTCCGTGCTCCAAAACAAC 3′, or GAPDHqfw, 5′ TGCTGGTGCCGAGTATGTTG 3′, and GAPDHqrv), and 10 μl of 2× Power SYBR green (Thermo Fisher Scientific) in a final volume of 20 μl. PCR cycle conditions were set as follows: preincubation for 10 min at 95°C followed by 40 cycles, each including 15 s at 95°C and 60 s at 60°C. Dissociation curves were generated at the end of the run to verify the specificity of the reaction product. Relative quantification was performed by Applied Biosystems 7500 real-time PCR software v2.0.6 (Thermo Fisher Scientific) based on cycle threshold (CT) values. Average FLUC CT values were normalized to the average CT values for GAPDH, and the relative level of FLUC (R) was estimated as ΔΔCT-based fold change using the 2−ΔΔCT equation.
FLUC and RLUC activity determination.
Lysis of transfected cells and quantification of FLUC and RLUC activities on a Biotek FLx800 luminometer were performed using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. For each sample, FLUC activity was normalized against the corresponding value of RLUC activity.
Statistics.
Statistical analyses were performed using the SPSS 17.0 statistical software package (SPSS, Inc., Chicago, IL). The statistical significance between normalized luciferase activity values (FLUC/RLUC) was analyzed using one-way analysis of variance (ANOVA), with Bonferroni post hoc test. Data from RT-qPCR assays were analyzed with one-way ANOVA.
ACKNOWLEDGMENTS
We are grateful to Andrea Gamarnik (Fundación Instituto Leloir, Argentina) and Maria Eugenia Loureiro (ICT Milstein, Argentina) for helpful discussions and critical readings of the manuscript. We thank Amelia Nieto (Centro Nacional de Biotecnología, Madrid, Spain), Martin A. Billeter (University of Zurich, Irchel, Switzerland), and Luis Carrasco (Centro de Biología Molecular Severo Ochoa, CSIC-UAM, Madrid, Spain) for generously providing reagents. We also appreciate the contributions of Florencia Linero (University of Buenos Aires, Argentina), Rodrigo Jacamo (MD Anderson Cancer Center, Houston, TX), Carlos Palacios (ICT Milstein, Argentina), and Viviana Castilla (University of Buenos Aires, Argentina). The technical assistance of J. Acevedo and S. Rojana is acknowledged.
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and CONICET. A.D., G.P.G., L.S., and N.L. are research investigators of CONICET. S.F. and M.G.N. received a fellowship from CONICET.
REFERENCES
- 1.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Peralta LA, Coto CE, Weissenbacher MC. 1993. The Tacaribe complex: the close relationship between a pathogenic (Junin) and a nonpathogenic (Tacaribe) arenavirus, p 281–296. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY. [Google Scholar]
- 3.Buchmeier MJ, de La Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1828. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Franze-Fernández MT, Iapalucci S, López N, Rossi C. 1993. Subgenomic RNAs of Tacaribe virus, p 113–132. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY. [Google Scholar]
- 5.Hass M, Golnitz U, Muller S, Becker-Ziaja B, Gunther S. 2004. Replicon system for Lassa virus. J Virol 78:13793–13803. doi: 10.1128/JVI.78.24.13793-13803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López N, Jacamo R, Franze-Fernandez MT. 2001. Transcription and RNA replication of Tacaribe virus genome and antigenome analogs require N and L proteins: Z protein is an inhibitor of these processes. J Virol 75:12241–12251. doi: 10.1128/JVI.75.24.12241-12251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergeron E, Chakrabarti AK, Bird BH, Dodd KA, McMullan LK, Spiropoulou CF, Nichol ST, Albarino CG. 2012. Reverse genetics recovery of Lujo virus and role of virus RNA secondary structures in efficient virus growth. J Virol 86:10759–10765. doi: 10.1128/JVI.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer BJ, de la Torre JC, Southern PJ. 2002. Arenaviruses: genomic RNAs, transcription, and replication. Curr Top Microbiol Immunol 262:139–157. [DOI] [PubMed] [Google Scholar]
- 10.Pinschewer DD, Perez M, de la Torre JC. 2005. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol 79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolakofsky D. 1993. The unusual mechanism of arenavirus RNA synthesis, p 103–112. In Salvato MS. (ed), The Arenaviridae. Plenum Press, New York, NY. [Google Scholar]
- 12.Iapalucci S, Lopez N, Franze-Fernandez MT. 1991. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology 182:269–278. doi: 10.1016/0042-6822(91)90670-7. [DOI] [PubMed] [Google Scholar]
- 13.Tortorici MA, Albarino CG, Posik DM, Ghiringhelli PD, Lozano ME, Rivera Pomar R, Romanowski V. 2001. Arenavirus nucleocapsid protein displays a transcriptional antitermination activity in vivo. Virus Res 73:41–55. doi: 10.1016/S0168-1702(00)00222-7. [DOI] [PubMed] [Google Scholar]
- 14.López N, Franze-Fernandez MT. 2007. A single stem-loop structure in Tacaribe arenavirus intergenic region is essential for transcription termination but is not required for a correct initiation of transcription and replication. Virus Res 124:237–244. doi: 10.1016/j.virusres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki M, Ngo N, Cubitt B, Teijaro JR, de la Torre JC. 2015. General molecular strategy for development of arenavirus live-attenuated vaccines. J Virol 89:12166–12177. doi: 10.1128/JVI.02075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Au HH, Jan E. 2014. Novel viral translation strategies. Wiley Interdiscip Rev RNA 5:779–801. doi: 10.1002/wrna.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrero S, Batisse J, Libre C, Bernacchi S, Marquet R, Paillart JC. 2015. HIV-1 replication and the cellular eukaryotic translation apparatus. Viruses 7:199–218. doi: 10.3390/v7010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh D, Mohr I. 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez S, Oceguera A, Sandoval-Jaime C. 2016. Stress response and translation control in rotavirus infection. Viruses 8:162. doi: 10.3390/v8060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López R, Franze-Fernandez MT. 1985. Effect of Tacaribe virus infection on host cell protein and nucleic acid synthesis. J Gen Virol 66(Part 8):1753–1761. [DOI] [PubMed] [Google Scholar]
- 23.Linero F, Welnowska E, Carrasco L, Scolaro L. 2013. Participation of eIF4F complex in Junin virus infection: blockage of eIF4E does not impair virus replication. Cell Microbiol 15:1766–1782. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 25.Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. 2002. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol 22:7853–7867. doi: 10.1128/MCB.22.22.7853-7867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgui I, Yanguez E, Sonenberg N, Nieto A. 2007. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J Virol 81:12427–12438. doi: 10.1128/JVI.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunetti JE, Scolaro LA, Castilla V. 2015. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a host factor required for dengue virus and Junín virus multiplication. Virus Res 203:84–91. doi: 10.1016/j.virusres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Maeto CA, Knott ME, Linero FN, Ellenberg PC, Scolaro LA, Castilla V. 2011. Differential effect of acute and persistent Junin virus infections on the nucleo-cytoplasmic trafficking and expression of heterogeneous nuclear ribonucleoproteins type A and B. J Gen Virol 92:2181–2190. doi: 10.1099/vir.0.030163-0. [DOI] [PubMed] [Google Scholar]
- 29.Gingras AC, Raught B, Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 30.Jan E, Mohr I, Walsh D. 2016. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu Rev Virol 3:283–307. doi: 10.1146/annurev-virology-100114-055014. [DOI] [PubMed] [Google Scholar]
- 31.Yángüez E, Rodriguez P, Goodfellow I, Nieto A. 2012. Influenza virus polymerase confers independence of the cellular cap-binding factor eIF4E for viral mRNA translation. Virology 422:297–307. doi: 10.1016/j.virol.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mir MA, Panganiban AT. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J 27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL, de la Torre JC Jr, Saphire EO. 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A 108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird NL, York J, Nunberg JH. 2012. Arenavirus infection induces discrete cytosolic structures for RNA replication. J Virol 86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knopp KA, Ngo T, Gershon PD, Buchmeier MJ. 2015. Single nucleoprotein residue modulates arenavirus replication complex formation. mBio 6:e00524-15. doi: 10.1128/mBio.00524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgil D, Polacek C, Harris E. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol 80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Casabona JC, Levingston Macleod JM, Loureiro ME, Gomez GA, Lopez N. 2009. The RING domain and the L79 residue of Z protein are involved in both the rescue of nucleocapsids and the incorporation of glycoproteins into infectious chimeric arenavirus-like particles. J Virol 83:7029–7039. doi: 10.1128/JVI.00329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Antuono A, Loureiro ME, Foscaldi S, Marino-Buslje C, Lopez N. 2014. Differential contributions of tacaribe arenavirus nucleoprotein N-terminal and C-terminal residues to nucleocapsid functional activity. J Virol 88:6492–6505. doi: 10.1128/JVI.00321-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anson DS, Choo KH, Rees DJ, Giannelli F, Gould K, Huddleston JA, Brownlee GG. 1984. The gene structure of human anti-haemophilic factor IX. EMBO J 3:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moss B, Elroy-Stein O, Mizukami T, Alexander WA, Fuerst TR. 1990. Product review. New mammalian expression vectors. Nature 348:91–92. [DOI] [PubMed] [Google Scholar]
- 42.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castelló A, Alvarez E, Carrasco L. 2006. Differential cleavage of eIF4GI and eIF4GII in mammalian cells. Effects on translation. J Biol Chem 281:33206–33216. [DOI] [PubMed] [Google Scholar]
- 44.Rossi C, Rey O, Jenik P, Franze-Fernandez MT. 1996. Immunological identification of Tacaribe virus proteins. Res Virol 147:203–211. [DOI] [PubMed] [Google Scholar]
- 45.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 46.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]