ABSTRACT
Rubicon is part of a Beclin-1-Vps34-containing autophagy complex. Rubicon induces antimicrobial responses upon Toll-like receptor (TLR) stimulation and functions as a feedback inhibitor to prevent unbalanced proinflammatory responses depending on dectin-1 signaling. However, the role played by Rubicon during antiviral immune responses, particularly the type I interferon (IFN) responses, remains largely unknown. Here, we report that Rubicon acts as a negative regulator for virus-triggered IFN signaling. Knockdown of Rubicon promoted type I interferon signaling and inhibited virus replication, while overexpression of Rubicon had the opposite effect. Rubicon specifically interacts with the interferon regulatory factor (IRF) association domain (IAD) of IRF3, and this interaction leads to inhibition of the dimerization of IRF3, which negatively regulates IFN-mediated antiviral response. Thus, our findings suggest the novel additional role of Rubicon as a negative regulator that inhibits the IFN signaling and cellular antiviral responses, providing a novel cellular mechanism of IRF3 inhibition.
IMPORTANCE The type I IFN system is a critical innate immune response that protects organisms against virus infection. However, type I IFN signaling must be tightly regulated to avoid excessive production of IFNs. Hence, negative regulatory mechanisms for type I IFN signaling are important, and to date, several related molecules have been identified. Here, we show that Rubicon is a major negative regulator of type I IFN signaling, and unlike previous reports of cellular molecules that inhibit IRF3 activation via proteasomal degradation or dephosphorylation of IRF3, we show that Rubicon interacts with IRF3 and that ultimately this interaction leads to inhibition of the dimerization of IRF3. Thus, we identified a novel negative regulator of type I IFN signaling pathways and a novel cellular mechanism of IRF3 inhibition. The results of this study will increase our understanding of the role of negative-feedback mechanisms that regulate type I IFN signaling and maintain immune homeostasis.
KEYWORDS: IRF3 dimerization, interferon, Rubicon
INTRODUCTION
Upon virus infection, a series of cellular signaling events are induced, and ultimately, proinflammatory cytokines and type I interferons (IFNs), such as alpha interferon (IFN-α) and IFN-β, are produced.
These cytokines play a crucial role in the innate immune responses, which are involved in the inhibition of virus replication and induction of adaptive immune responses (1–6). In particular, type I IFNs are potent inhibitors of viral replication; they act as such by promoting apoptosis of infected cells and stimulating surrounding cells to induce transcription of IFN-related genes, which work to synergistically inhibit viral replication and to activate innate and adaptive immune cells (7, 8). Type I IFNs are transcriptionally regulated and induced by latent transcription factors that are activated via type I IFN signaling pathways.
Interferon regulatory factor 3 (IRF3) is a member of the transcriptional factor family called interferon regulatory factors (IRF) and a major transcription factor that induces production of type I IFNs (9–12). After viral infection, pattern recognition receptors (PRRs), including retinoic acid-inducible gene 1 (RIG-I), melanoma differentiation-associated protein 5 (MDA5), and Toll-like receptor 3 (TLR3), detect viral molecular features and activate IRF3 via phosphorylation of its C-terminal trans-activation domain by tank-binding kinase 1 (TBK1) and inhibitor-κB kinase ε (IKKε) (13, 14). Phosphorylated IRF3 forms dimers via its IRF association domain (IAD) region; these dimers then translocate from the cytoplasm to the nucleus (15–18), where IRF3 induces transcription of type I IFN genes.
Production of type I IFNs must be tightly regulated and terminated in an effective and timely manner to avoid problems, such as inflammatory diseases, septic shock, and autoimmune disorders (19–22). The activation of IRF3 is also tightly controlled by regulatory cellular molecules. Some negative regulatory molecules restrain IRF3 activity via proteasomal degradation or dephosphorylation. For example, peptidylprolyl cis-trans isomerase NIMA-interacting 1 (Pin 1), E3 ligase Ro52, Cullin-based ubiquitin ligases, and RBCC protein interacting with PKC1 (RBCK1) all trigger proteasomal degradation of IRF3 (23–26). A recent study reported that RTA-associated ubiquitin ligase (RAUL) is the major endogenous ubiquitin E3 ligase responsible for the degradation of IRF3 and IRF7 (27). In addition, protein phosphatase 2A (PP2A) and its adaptor molecule, RACK1, dephosphorylate IRF3 (28). Although dimerization of IRF3 is a critical step for activating the type I IFN signaling pathway, the cellular molecules that inhibit IRF3 dimerization are unknown.
Rubicon (RUN domain Beclin-1 interacting cysteine-rich domain containing) is a RUN domain protein, like Beclin-1-interacting and cysteine-rich-domain-containing autophagy protein, and it regulates autophagy (29, 30). Upon microbial infection or TLR2 activation, Rubicon activates phagocytosis by interacting with the p22phox molecule in the NADPH oxidase complex to generate reactive oxygen species and inflammatory cytokines (31). Rubicon also acts as a negative regulator of responses against fungal infections by interacting with CARD9 (32). However, the role of Rubicon during antiviral immune responses, particularly the type I IFN pathway, remains largely unknown.
In this study, we identified Rubicon as a negative regulator for virus-triggered type I IFN signaling pathways. Rubicon specifically interacts with IRF3, and this interaction leads to inhibition of the dimerization of IRF3, thereby inhibiting excessive cellular antiviral immune responses. Our findings reveal the additional role of Rubicon as a novel cellular molecule for IRF3 inhibition.
RESULTS
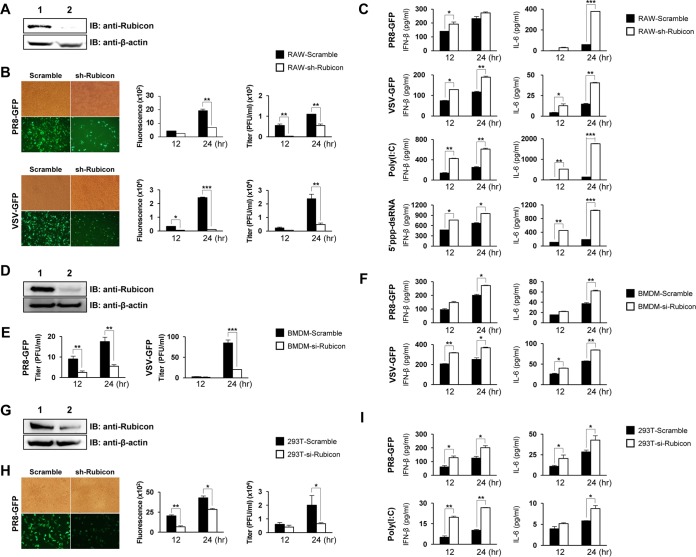
Knockdown of Rubicon increases type I IFN secretion and inhibits viral replication.
To examine whether Rubicon affects antiviral responses, we first generated a cell line exhibiting suppressed expression of endogenous Rubicon. This was achieved by infecting RAW264.7 cells with lentivirus carrying Rubicon-specific short hairpin RNA (shRNA). Immunoblotting revealed that these cells showed markedly lower expression of Rubicon than the parental cells (Fig. 1A). Control and Rubicon knockdown RAW264.7 cells were then infected with green fluorescent protein (GFP)-expressing H1N1 influenza virus (PR8-GFP) or GFP-expressing vesicular stomatitis virus (VSV-GFP), followed by measurement of virus replication. GFP expression was visualized by fluorescence microscopy and measured in a luminometer. The number of GFP-expressing cells among Rubicon knockdown cells was less than that among control cells (Fig. 1B). Additionally, a plaque assay revealed that the virus titer in Rubicon knockdown RAW264.7 cells was lower than that in control cells. To identify a link between inhibited antiviral responses and innate cytokine responses, we collected culture supernatants from the cells at 12 and 24 h postinfection (hpi) with VSV-GFP and PR8-GFP and measured IFN-β and interleukin-6 (IL-6) concentrations by enzyme-linked immunosorbent assay (ELISA). Rubicon knockdown led to increased levels of IFN-β and IL-6 upon infection with PR8-GFP or VSV-GFP (Fig. 1C). In addition, control and Rubicon knockdown RAW 264.7 cells were exposed to poly(I·C) (a TLR3 ligand) and 5′-triphosphate double-stranded RNA (5′ppp-dsRNA) (a RIG-I ligand), and IFN-β and IL-6 secretion was measured by ELISA. As shown in Fig. 1C, Rubicon knockdown cells produced significantly more IFN-β and IL-6 upon exposure to 5′ppp-dsRNA and poly(I·C) than control cells. Moreover, to investigate the role of Rubicon in antiviral responses by primary mouse bone marrow-derived macrophages (BMDMs), we isolated BMDMs from the femurs and tibiae of C57BL/6 mice, transfected them with scrambled small interfering RNA (siRNA) or siRNA which specifically interferes with Rubicon expression, and then confirmed the knockdown of Rubicon expression (Fig. 1D). The cells were then infected with PR8-GFP or VSV-GFP, and the virus titer and IFN-β and IL-6 levels were measured. Knockdown of Rubicon significantly inhibited viral replication and increased IFN-β and IL-6 production (Fig. 1E and F). These results demonstrate that Rubicon negatively regulates virus-induced IFN-β production in RAW264.7 cells, as well as primary immune cells. To examine potential cell-type-specific differences in Rubicon function, we next assessed the role of Rubicon in HEK293T cells after stimulation of type I IFN signaling. Rubicon knockdown HEK293T cells obtained by transfection of Rubicon-specific siRNA showed lower Rubicon expression levels than control cells (Fig. 1G). Rubicon knockdown HEK293T cells were then infected with PR8-GFP. Knockdown of Rubicon led to a significant reduction in viral replication and increased production of IFN-β and IL-6 (Fig. 1H and I). Rubicon knockdown also reduced poly(I·C)-induced production of IFN-β and IL-6 in HEK293T cells (Fig. 1I). Taken together, these results suggest that, irrespective of cell type, knockdown of Rubicon inhibits virus replication by increasing type I IFN secretion.
FIG 1.
Knockdown of Rubicon increases antiviral responses. (A) Rubicon expression in RAW264.7 cells infected with control lenti-shRNA (lane 1; RAW-Scramble) or Rubicon lenti-shRNA (lane 2; RAW-sh-Rubicon) was measured by immunoblotting (IB). β-Actin was used to confirm equal loading of proteins. (B and C) RAW-Scramble and RAW-sh-Rubicon cells were infected with PR8-GFP (MOI = 3) or VSV-GFP (MOI = 1) or treated with poly(I·C) (20 μg/ml) or 5′ppp-dsRNA (1 μg/ml). (B) Virus replication was examined by measuring GFP expression levels under a fluorescence microscope at 24 hpi and quantified using a fluorescence modulator at 12 and 24 hpi. Virus titers were determined by plaque assay. (C) IFN-β and IL-6 concentrations in the supernatants of RAW-Scramble and RAW-sh-Rubicon cells were measured by ELISA at 12 and 24 hpi. (D) Rubicon expression in BMDMs transfected with control siRNA (lane 1; BMDM-Scramble) or Rubicon siRNA (lane 2; BMDM-si-Rubicon) was measured by immunoblotting. β-Actin was used to confirm equal loading of proteins. (E and F) Virus titers in BMDM-Scramble and BMDM-si-Rubicon cells were measured by plaque assay at 12 and 24 hpi with PR8-GFP (MOI = 3) or VSV-GFP (MOI = 1) (E), and IFN-β and IL-6 levels in the supernatants were measured by ELISA (F). (G) Rubicon expression in HEK293T cells transfected with control siRNA (lane 1; 293T-Scramble) or Rubicon siRNA (lane 2; 293T-si-Rubicon) was measured by immunoblotting. β-Actin was used to confirm equal loading of proteins. (H and I) 293T-Scramble and 293T-si-Rubicon cells were infected with PR8-GFP (MOI = 1) or treated with poly(I·C) (20 μg/ml). GFP expression by infected cells was visualized by fluorescence microscopy at 24 hpi and quantified using a fluorescence modulator at 12 and 24 hpi. (H) Virus titers were determined by plaque assay. (I) IFN-β and IL-6 levels in the cell supernatants were measured by ELISA. The data are expressed as means and SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test). The data are representative of the results of at least two independent experiments.
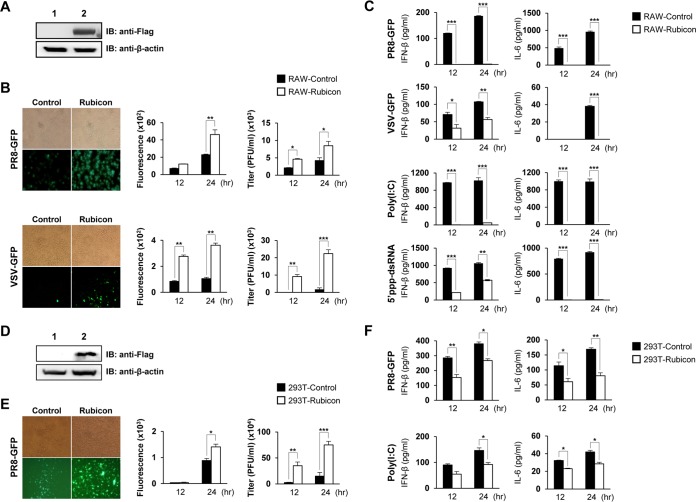
Overexpression of Rubicon inhibits type I IFN secretion and increases viral replication.
To investigate and compare the effects of Rubicon overexpression and knockdown on type I IFN-β signaling, we next constructed a stable Rubicon-overexpressing RAW264.7 cell line by transfecting cells with a Rubicon-Flag expression plasmid, followed by selection with puromycin. Overexpression of Rubicon was then confirmed by immunoblotting with an anti-Flag antibody (Fig. 2A). After infection with PR8-GFP and VSV-GFP, the GFP expression level and the titer were measured. Rubicon-overexpressing RAW264.7 cells expressed markedly higher levels of GFP expression and showed a higher virus titer than control cells (Fig. 2B). As shown in Fig. 2C, overexpression of Rubicon suppressed secretion of IFN-β and IL-6 upon stimulation by PR8-GFP, VSV-GFP, poly(I·C), and 5′ppp-dsRNA. Additionally, we prepared Rubicon-overexpressing HEK293T cells and examined Rubicon expression by immunoblotting (Fig. 2D). Replication of PR8-GFP was much higher in Rubicon-overexpressing HEK293T cells than in control HEK293 cells (Fig. 2E), and IFN-β and IL-6 secretion was suppressed in Rubicon-overexpressing RAW264.7 cells upon PR8-GFP infection or poly(I·C) treatment (Fig. 2F). Taken together, these results suggest that overexpression of Rubicon inhibits type I IFN secretion and increases viral replication, which is in agreement with the results shown in Fig. 1.
FIG 2.
Overexpression of Rubicon inhibits antiviral responses. (A) Overexpression of Rubicon was confirmed by immunoblotting of RAW264.7 cells stably transfected with an IRES vector (lane 1; RAW-Control) or Rubicon-Flag (lane 2; RAW-Rubicon). β-Actin was used to confirm equal loading of proteins. (B and C) RAW-Control and RAW-Rubicon cells were infected with PR8-GFP (MOI = 3) or VSV-GFP (MOI = 1) or treated with poly(I·C) (20 μg/ml) or 5′ppp-dsRNA (1 μg/ml). Virus replication was examined at 24 hpi by measuring GFP expression levels using fluorescence microscopy and quantified at 12 and 24 hpi using a fluorescence modulator. (B) Virus titers were determined by plaque assay. (C) IFN-β and IL-6 in cell culture supernatants were measured by ELISA at 12 and 24 hpi. (D) Overexpression of Rubicon was confirmed by immunoblotting of HEK293T cells transfected with an IRES vector (lane 1; 293T-Control) or Rubicon-Flag (lane 2; 293T-Rubicon). β-Actin was used to confirm equal loading of proteins. (E and F) 293T-Control and 293T-Rubicon cells were infected with PR8-GFP (MOI = 1) or treated with poly(I·C) (20 μg/ml). GFP expression by infected cells was visualized by fluorescence microscopy at 24 hpi and quantified using a fluorescence modulator at 12 and 24 hpi. (E) Virus titers were determined by plaque assay. (F) Levels of IFN-β and IL-6 in the supernatants were measured by ELISA. The data are expressed as means and SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student's t test). The data are representative of the results of at least two independent experiments.
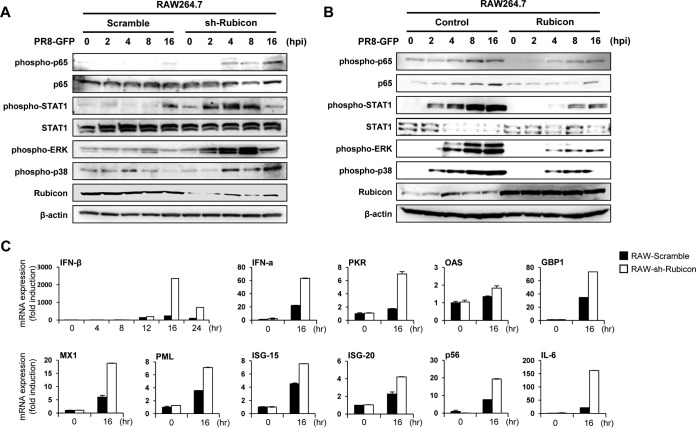
Rubicon inhibits type I IFN signaling and transcription of IFN-related genes.
To examine whether Rubicon affects the type I IFN signaling pathway, we confirmed phosphorylation of molecules (p65, STAT1, ERK, and p38) involved in signaling cascades triggered by virus infection. RAW264.7 cells were infected with PR8-GFP and collected at 0, 2, 4, 8, and 16 hpi for immunoblotting. First, we examined the phosphorylation levels of the indicated proteins in control and Rubicon knockdown RAW264.7 cells after PR8-GFP infection. Phosphorylation in Rubicon knockdown RAW264.7 cells was initiated more quickly and strongly than in control cells, and these differences became more distinct over time (i.e., greater at 8 or 16 hpi) (Fig. 3A). The phosphorylation levels in Rubicon-overexpressing RAW264.7 cells were lower than those in control cells. (Fig. 3B). These results suggest that Rubicon inhibits signal transduction of the type I IFN signaling pathway. We also examined the mRNA expression levels of IFN (IFN-β and IFN-α) and IFN-related (PKR, OAS, GBP1, MX1, PML, ISG-15, ISG-20, p56, and IL-6) genes to determine whether the inhibitory effect of Rubicon on type I IFN signaling influences the transcription levels of these genes (Fig. 3C). We isolated mRNA from control and Rubicon knockdown RAW 264.7 cells at 16 hpi with PR8-GFP. Expression of mRNA-encoding IFN and IFN-related genes was higher in Rubicon knockdown RAW264.7 cells than in control cells. Collectively, these results indicate that suppressed and enhanced antiviral responses were related to overexpression and knockdown of Rubicon, respectively. This in turn means that Rubicon acts as a negative regulator of the type I IFN signaling pathway and inhibits transcription of IFN and IFN-related genes.
FIG 3.
Rubicon inhibits the type I IFN signaling pathway and suppresses expression of IFN-related genes. (A and B) Control RAW264.7 (RAW-Scramble) and Rubicon knockdown RAW264.7 (RAW-sh-Rubicon) cells (A) or control RAW264.7 (RAW-Control) and Rubicon-overexpressing RAW264.7 (RAW-Rubicon) cells (B) were infected with PR8-GFP (MOI = 2). At the indicated times after infection, IRF3, p65, and STAT1 and phosphorylated IRF3, p65, STAT1, p38, and TBK1 in cell extracts were measured by immunoblotting. β-Actin was used to confirm equal loading of proteins. (C) Total RNAs were extracted from RAW-Scramble and RAW-sh-Rubicon cells after PR8-GFP (MOI = 3) infection. Expression of the indicated mRNA-encoding genes was analyzed by real time quantitative reverse transcription-PCR (qRT-PCR) at 0, 4, 8, 12, 16, and 24 hpi (IFN) or at 0 and 16 hpi (IFN-α, PKR, OAS, GBP1, MX1, PML, ISG-15, ISG-20, p56, and IL-6). The data are expressed as means and SEM and are representative of the results of at least two independent experiments.
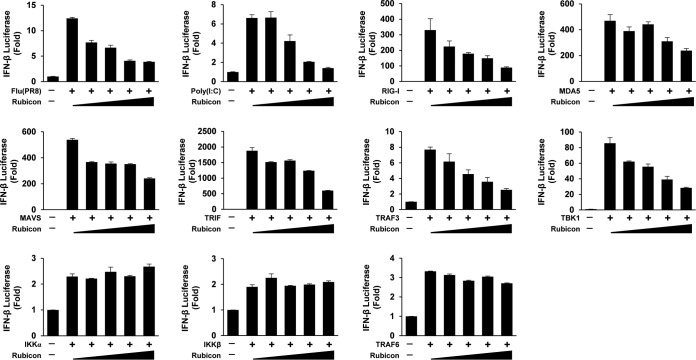
Rubicon inhibits IRF3-mediated IFN-β promoter activity.
To examine how Rubicon affects IFN-β promoter activation, luciferase assays were performed by stimulating type I IFN signaling with several stimulants and proteins (Fig. 4). First, we transfected cells with an IFN-β luciferase reporter plasmid and with increasing amounts of Rubicon plasmid and stimulated them with PR8-GFP or poly(I·C). The luciferase assays revealed that Rubicon inhibited PR8-GFP- and poly(I·C)-mediated IFN-β promoter activity in a dose-dependent manner. Next, to check whether Rubicon affects RIG-I- and TLR3-mediated type I IFN signaling, we induced type I IFN signaling of cells by overexpression of RIG-I, MDA5, mitochondrial antiviral signaling protein (MAVS), and TIR domain-containing adapter-inducing interferon-β (TRIF). The activity of the IFN-β promoter (induced by RIG-I, MDA5, MAVS, or TRIF) was reduced as Rubicon expression levels increased. To identify the stage at which Rubicon inhibits type I IFN signaling, we stimulated type I IFN signaling by overexpression of downstream molecules, tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3), and TBK1. We found that Rubicon decreased TRAF3- and TBK1-induced IFN-β promoter activity in a dose-dependent manner. In addition, to confirm whether Rubicon regulates type I IFN via NF-κB signaling, we measured IKKα-, IKKβ-, and TRAF6-induced IFN-β promoter activities. The results showed that Rubicon did not affect NF-κB-mediated IFN-β promoter activity. Therefore, we suggest that Rubicon influences MAVS-mediated type I IFN signaling and targets molecules downstream of TBK1 to inhibit signal transduction.
FIG 4.
Rubicon inhibits IFN-β promoter activity induced by virus and IRF3-mediated type I IFN signaling molecules. HEK293 cells were transfected with an IFN-β reporter plasmid and increasing amounts of a Rubicon plasmid. The cells were then infected with PR8-GFP (MOI = 3) or stimulated with poly(I·C) (20 μg/ml) at 24 h posttransfection. After another 12 h, luciferase activity was analyzed in a luminometer. To induce luciferase activity with type I IFN signaling molecules, HEK293T cells were transfected with an IFN-β reporter plasmid and increasing amounts of a Rubicon plasmid, along with overexpression plasmids for RIG-I, MDA5, or MAVS, or TRIF, TRAF3, or TBK1 and IKKα, IKKβ, or TRAF6. At 24 h posttransfection, luciferase activity was analyzed in a luminometer. The data are expressed as means and SEM and are representative of the results of at least two independent experiments.
Rubicon targets IRF3 upon virus infection.
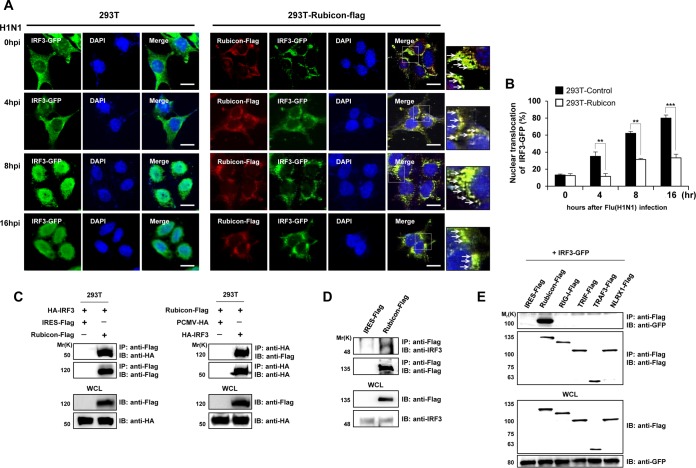
IRF3, a molecule downstream of TBK1, is a key transcription factor for IFN-β during TLR3/4- and RIG-I-mediated signaling. Activation of TLR3/4 and RIG-I phosphorylates IRF3, resulting in IRF3 dimerization. The IRF3 dimer subsequently translocates to the nucleus, where it binds the IFN-β promoter. To examine the effect of Rubicon on IRF3 activation, we first examined IRF3 localization in control and Rubicon-overexpressing HEK293T cells after virus infection. The cells were transfected with GFP-tagged IRF3, which was then detected by confocal microscopy, together with overexpressed Rubicon (Fig. 5A). After H1N1 infection of control HEK293T cells, the percentage of cells containing IRF3 in the nucleus increased to 80% at 16 hpi (Fig. 5B). In contrast, IRF3 in Rubicon-overexpressing HEK293T cells colocalized with Rubicon and was retained in the cytoplasm, even at 16 hpi (Fig. 5A), and only 30% of the cells harbored IRF3 in the nucleus (Fig. 5B).
FIG 5.
Rubicon binds to IRF3. (A) Control and Rubicon-Flag-overexpressing HEK293T cells were transfected with a GFP-IRF3 plasmid, followed by infection with H1N1 virus. Rubicon-Flag was then detected by an anti-Flag antibody and labeled with a Cy3-conjugated antibody. At the indicated times after infection, IRF3 and Rubicon were identified as green and red signals, respectively, by confocal microscopy. Nuclei were stained with DAPI (blue). Yellow represents colocalization and is marked with arrows in the enlarged images on the right. Bars, 10 μm. (B) The percentages of cells showing nuclear translocation of IRF3 were calculated by dividing the number of cells exhibiting nuclear expression of GFP-IRF3 by the total number of GFP-positive cells. At least 100 cells were counted per random field. (C) HEK293 cells were cotransfected with the control vector (IRES-Flag or PCMV-HA) and Rubicon-Flag and HA-IRF3 plasmids. The cell lysates were subjected to immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-HA antibody (left). Alternatively, the cell lysates were immunoprecipitated with an anti-HA antibody, followed by immunoblotting with an anti-Flag antibody (right). WCL were immunoblotted with anti-Flag and -HA antibodies. (D) Lysates of HEK293T cells transfected with an IRES-Flag vector or with a Rubicon-Flag plasmid were subjected to immunoprecipitation (IP) with an anti-Flag antibody, followed by immunoblotting with anti-IRF3 or -Flag antibodies. WCL were then immunoblotted with anti-Flag and -IRF3 antibodies. (E) IRES-Flag, Rubicon-Flag, and Flag-tagged RIG-I, TRIF, TRAF3, and NLRX1 (negative controls) were overexpressed in HEK293 cells, along with GFP-IRF3. The lysates were subjected to immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-GFP antibody. WCL were immunoblotted with anti-Flag and -GFP antibodies. The data are expressed as means and SEM. **, P < 0.01; ***, P < 0.001 (Student's t test). The data are representative of the results of at least two independent experiments.
To further confirm the relationship between Rubicon and IRF3, we examined the physical interaction between the two molecules. HEK293 cells were cotransfected with Rubicon-Flag and hemagglutinin (HA)-IRF3 and subsequently subjected to immunoprecipitation with anti-Flag or anti-HA antibodies, followed by detection of HA-IRF3 or Rubicon-Flag, respectively (Fig. 5C). In addition, Rubicon-Flag was immunoprecipitated and then detected by immunoblotting with an anti-IRF3 antibody; a band corresponding to IRF3 was detected (Fig. 5D). Moreover, to confirm the specificity of Rubicon-IRF3 binding, cells were cotransfected with Flag-tagged RIG-I, TRIF, TRAF3, or NLRX1 and with GFP-IRF3 as a negative control. As seen in Fig. 5E, GFP-IRF3 coprecipitated only with Rubicon-Flag. Therefore, we speculate that Rubicon targets IRF3 to inhibit type I IFN signal transduction.
Rubicon inhibits dimerization of IRF3 upon virus infection.
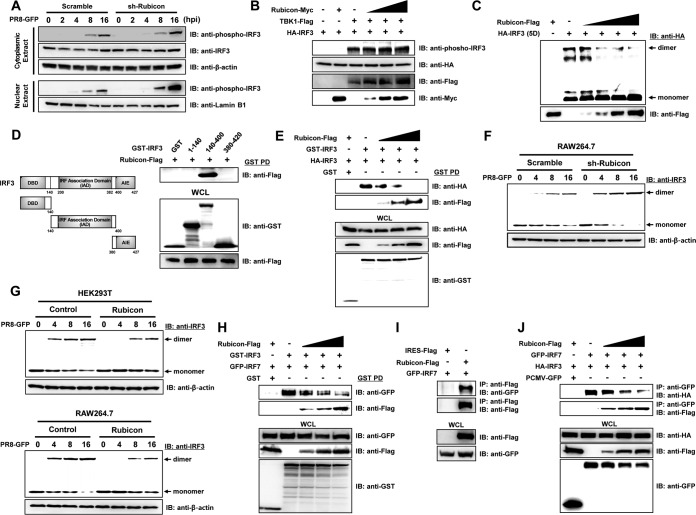
Control and Rubicon knockdown RAW264.7 cells were infected with PR8-GFP, and cytoplasmic and nuclear fractions were obtained; phosphorylated IRF3 was then detected by immunoblotting at the indicated times (Fig. 6A). Phosphorylation of IRF3 was observed in both the cytoplasm and nuclear fraction at 8 hpi, and the level increased up to 16 hpi. Interestingly, the amounts of phosphorylated IRF3 in the cytoplasm of control and Rubicon knockdown RAW264.7 cells were similar, whereas the amount of phosphorylated IRF3 in the nuclear fraction from Rubicon knockdown RAW264.7 cells was significantly higher than in that from control cells. From these data, we assumed that Rubicon disrupts the nuclear translocation of IRF3, probably not by inhibiting phosphorylation, but by inhibiting dimerization. To confirm exactly whether Rubicon affects the phosphorylation of IRF3, we next induced phosphorylation of IRF3 by overexpressing TBK1, and Rubicon was overexpressed in a dose-dependent manner. Under these conditions, we observed that increasing overexpression of Rubicon had no effect on IRF3 phosphorylation. Then, we examined the inhibitory effect of Rubicon on IRF3 dimerization by transfecting cells with IRF3-5D, a constitutively active phosphomimetic mutant of IRF3 that can dimerize in the absence of stimulation. As shown in Fig. 6C, dimerization of IRF3-5D was observed; however, it was reduced as Rubicon expression increased. To further elucidate the mechanism by which Rubicon inhibits IRF3 dimerization, we examined the IRF3 domain responsible for the interaction with Rubicon using glutathione S-transferase (GST)-tagged IRF3 domain constructs (amino acids [aa] 1 to 141 of the DNA-binding domain [DBD], 140 to 400 of the IAD, and 380 to 420 of the autoinhibition element [AIE]) (Fig. 6D). The results showed that Rubicon interacted with GST-IAD, but not with GST-DBD and GST-AIE, suggesting that Rubicon inhibits IRF3 dimerization by interacting with the IAD region. Additionally, we performed a competition assay by coexpressing GST-IRF3 and HA-IRF3 proteins in HEK293T cells, along with increasing amounts of Rubicon-Flag (Fig. 6E). A GST pulldown assay showed that the levels of GST-IRF3 and HA-IRF3 homodimers fell as Rubicon expression increased, whereas the interaction between GST-IRF3 and Rubicon-Flag increased in a dose-dependent manner. Based on this finding, we examined the time course of endogenous IRF3 dimerization after PR8-GFP infection of Rubicon knockdown and -overexpressing cells (Fig. 6F and G). IRF3 dimerization in Rubicon knockdown RAW 264.7 cells was higher than that in control RAW 264.7 cells, especially at 8 and 16 dpi. Also, overexpression of Rubicon in HEK293T and RAW 264.7 cells almost completely blocked IRF3 dimerization. Additionally, to confirm whether Rubicon also inhibits dimerization of IRF3/IRF7 (heterodimer), competition assays were performed by coexpressing GST-IRF3 and GFP-IRF7 proteins, along with increasing amounts of Rubicon-Flag. We observed that the amount of binding of IRF3 and IRF7 was reduced as Rubicon increasingly bound with IRF3 in a dose-dependent manner (Fig. 6H). To further confirm whether Rubicon has the ability to target IRF7, binding between Rubicon and IRF7 was investigated by overexpressing Rubicon-Flag and GFP-IRF7. Immunoprecipitation was conducted with Flag antibody, followed by immunoblotting with GFP antibody, and we confirmed that Rubicon binds to IRF7 (Fig. 6I). Moreover, we observed that Rubicon inhibits IRF3/IRF7 dimerization by binding to IRF7 (Fig. 6J). Collectively, these data suggest that Rubicon inhibits IRF3/IRF3 homodimerization and IRF3/IRF7 heterodimerization by binding to the IAD region of IRF3. Furthermore, Rubicon inhibits IRF3/IRF7 dimerization by binding to IRF7, and there is a possibility that Rubicon can target IRF7 to inhibit IRF7 homodimerization.
FIG 6.
Rubicon inhibits dimerization of IRF3. (A) Control (Scramble) and Rubicon knockdown (sh-Rubicon) RAW264.7 cells were infected with PR8-GFP (MOI = 2) and harvested at the indicated times (0, 2, 4, 8, and 16 hpi). Cytoplasmic and nuclear extracts were then subjected to immunoblotting with anti-phospho-IRF3 and -IRF3 antibodies. β-Actin and lamin B1 were used to confirm equal loading of proteins. (B) TBK1-Flag and HA-IRF3 were coexpressed, along with increasing levels of Rubicon-Myc, in HEK293T cells. The lysates were then subjected to immunoblotting with anti-phospho-IRF3, -HA, -Flag, and -Myc antibodies. (C) An HA-tagged active phosphomimetic mutant of IRF3 (HA-IRF3-5D) and increasing levels of Rubicon-Flag were overexpressed in HEK293T cells. The cell lysates were then resolved in native polyacrylamide gels, and both monomeric and dimeric forms of IRF3-5D were detected using an anti-HA antibody. Rubicon-Flag was detected by immunoblotting with an anti-Flag antibody. (D) Schematic representation of GST-IRF3 constructs comprising amino acids 1 to 140 (containing the DBD), amino acids 140 to 400 (containing the IAD), and amino acids 380 to 427 (containing the AIE). HEK293T cells were transfected with the indicated GST-IRF3 constructs and Rubicon-Flag plasmids. GST pulldown (PD) was then conducted, followed by immunoblotting with an anti-Flag antibody. WCL were immunoblotted with anti-GST and -Flag antibodies. (E) Control vector (GST), HA-IRF3, or GST-IRF3 was coexpressed with increasing levels of Rubicon-Flag in HEK293T cells. Immunoprecipitation of GST-IRF3 from the cell lysates was conducted in a GST pulldown assay, followed by immunoblotting with anti-HA and -Flag antibodies. WCL were immunoblotted with anti-HA, -Flag, and -GST antibodies. (F and G) Together with control cells (Scramble and Control), Rubicon knockdown RAW264.7 cells (sh-Rubicon) or Rubicon-overexpressing HEK293T and RAW264.7 cells (Rubicon) were infected with PR8-GFP. At the indicated times (0, 4, 8, and 16 hpi), cells were harvested and subjected to native polyacrylamide gel electrophoresis to detect monomeric and dimeric forms of IRF3, followed by immunoblotting with an anti-IRF3 antibody. β-Actin was used to confirm equal loading of proteins. (H) Control vector (GST), GFP-IRF7, and GST-IRF3 were coexpressed with increasing levels of Rubicon-Flag in HEK293T cells. Immunoprecipitation of GST-IRF3 from the cell lysates was conducted in a GST pulldown assay, followed by immunoblotting with anti-GFP and -Flag antibodies. WCL were immunoblotted with anti-GFP, -Flag, and -GST antibodies. (I) Cell lysates of HEK293T cells that were overexpressed with control vector (IRES-Flag), Rubicon-Flag, and GFP-IRF7 were subjected to immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with an anti-GFP antibody. WCL were immunoblotted with anti-Flag and -GFP antibodies. (J) HEK293T cells were cotransfected with control vector (PCMV-GFP), GFP-IRF7, HA-IRF3, and increasing doses of Rubicon-Flag plasmids. IRF7-binding proteins were detected by immunoprecipitation with anti-GFP antibody, followed by immunoblotting with anti-HA and anti-Flag. WCL were immunoblotted with anti-HA, anti-Flag, and anti-GFP antibodies.
DISCUSSION
Rubicon engages the Beclin-1/Vps34-PI3K/UVRAG complex and inhibits the fusion of endosomes and autophagosomes with lysosomes (29, 30). It is a multidomain adaptor protein that assembles different protein complexes in macrophages under normal and PRR-activated conditions. These complexes govern diverse host immune response pathways that impact the ability to control different pathogen classes, such as bacteria, viruses, and fungi. Upon microbial infection or TLR activation, Rubicon interacts with p22phox of the NADPH oxidase complex to positively regulate its assembly, thereby generating superoxide and controlling bacterial pathogens (31). Furthermore, Dectin-1-mediated activation of Rubicon upon viral and fungal infection leads to negative regulation of the CARD9/Bcl10–MALT-1 complex and downstream inflammatory cytokine production (32).
Here, we report that Rubicon is a potent negative regulator of the virus-mediated type I IFN signaling pathway. First, knockdown of endogenous Rubicon in immune cells or epithelial cells promoted virus-induced IFN-β and proinflammatory cytokine secretion and inhibited viral replication. Second, overexpression of Rubicon decreased RIG-I-like receptor (RLR)-mediated antiviral responses against virus infection. Third, Rubicon interacts with IRF3, and the IAD of IRF3 is responsible for the interaction between Rubicon and IRF3. Finally, this interaction leads to inhibition of the dimerization of IRF3 and blocks the translocation of IRF3 into the nucleus. Taken together, these findings indicate the novel additional role of Rubicon as a negative regulator that inhibits the IFN signaling and cellular antiviral responses.
Virus infection triggers innate immune responses that lead to the induction of type I IFNs and other cytokines to inhibit viral replication and spreading. Specifically, type I IFNs are responsible for the inhibition of viral replication, clearance of virus-infected cells, and induction of adaptive immune responses (1–6). However, uncontrolled immune responses via excessive production of type I IFNs have a detrimental effect on the host (19, 33). Therefore, the host innate immune system has diverse molecular strategies to control the proper production of type I IFNs.
IRF3 is a critical transcription factor that regulates type I IFN production in response to invasion by pathogens (9–12). IRF3 is located in the cytoplasm of unstimulated cells and is phosphorylated by TBK1 and IKKε (at Ser/Thr residues on amino acids 396 to 405) upon viral infection (13, 14). Phosphorylation of IRF3 induces conformational changes that expose the DBD and IAD regions via disassociation of the C-terminal autoinhibition domain; IRF3 then forms homodimers via the IAD region (15–18). IRF3 dimers translocate to the nucleus and bind to the IFN stimulation response element and PRDI/PRDIII-containing promoters to induce transcription of the sequence of genes, including the IFN-β gene (34).
During the type I IFN signaling process, IRF3 is regulated by cellular molecules, such as Pin 1, Ro52, Cullin-based ubiquitin ligases, RBCK1, and RAUL (which induce proteosomal degradation of IRF3) (23–27) and by PP2A and RACK1 (which dephosphorylate IRF3) (28) to control excessive production of type I IFNs. On the other hand, viruses can also inhibit IRF3 activation to defend themselves against host type I IFN responses. For example, severe acute respiratory syndrome (SARS) coronavirus PLpro interacts with IRF3 to prevent its phosphorylation, while bovine herpesvirus bICP0, rotavirus NSP1, and classical swine fever virus Npro target IRF3 for proteasomal degradation (35–38). Specifically, Theiler's virus L protein and Epstein-Barr virus LF2 inhibit dimerization of IRF3 and IRF7, respectively (39, 40).
In this study, we found that the novel cellular mechanism of IRF3 inhibition is unlike previously reported cellular molecules that inhibit IRF3 activation via proteasomal degradation or dephosphorylation of IRF3.
After evaluation of antiviral phenotypes by Rubicon expression, we hypothesized that Rubicon targets IRF3 to inhibit type I IFN signaling, since Rubicon did not affect IFN-β luciferase activity mediated by molecules upstream of IRF3 (i.e., RIG-I to TBK1) (Fig. 4). Also, binding assays revealed that IRF3 binds to Rubicon (Fig. 5C to E). Importantly, after virus infection, we observed that IRF3 was retained in the cytoplasm of Rubicon-overexpressing cells. Moreover, the amount of phosphorylated IRF3 that translocated to the nucleus in Rubicon knockdown cells was significantly higher than that in control cells (Fig. 5A and B). Therefore, we postulated that two mechanisms, inhibition of IRF3 phosphorylation and dimerization of IRF3, may be responsible for the inhibition of IRF3 by Rubicon. Ultimately, we found that Rubicon did not affect the phosphorylation of IRF3; however, it did inhibit dimerization. Rubicon had no effect on TBK1-mediated phosphorylation of IRF3, as the levels of phosphorylated IRF3 in the cytoplasm of control and Rubicon knockdown cells were similar (Fig. 6A and B). However, Rubicon competitively binds to the IAD region of IRF3, and this binding leads to inhibition of the dimerization of IRF3 (Fig. 6D and E). Furthermore, Rubicon inhibits dimerization of IRF3-5D, which mimics the C-terminal phosphorylated serine of active IRF3 by mutating five C-terminal serine residues to aspartic acid (Fig. 6C). These results were confirmed by observation of higher and lower dimer-IRF3 levels in Rubicon knockdown and -overexpressing cells, respectively, after virus infection (Fig. 6F and G). Moreover, IRF3/IRF7 dimerization was also inhibited by Rubicon via binding to IRF3 (Fig. 6H). IRF7 is highly homologous with IRF3 and is also known as a key regulator of type I IFN gene expression upon virus infection, which undergoes phosphorylation, dimerization, and nuclear translocation, similar to IRF3 (12, 41, 42). Here, we confirmed that Rubicon can bind to IRF7 (Fig. 6I). Furthermore, Rubicon inhibits IRF3/IRF7 dimerization by binding to IRF7 (Fig. 6J). Based on these data and the well-known fact that the IADs of IRF3 and IRF7 are both highly conserved (15, 18, 43), we could anticipate that Rubicon might target the IAD of IRF7 to inhibit dimerization of IRF7, as with IRF3.
Collectively, these results indicate a novel role for Rubicon during antiviral immune responses; Rubicon acts as a negative regulator of type I IFN signaling by inhibiting the dimerization and nuclear translocation of IRF3. Unlike previously reported cellular molecules that regulate proteasomal degradation and dephosphorylation of IRF3, we identified the novel cellular molecule that inhibits dimerization of IRF3 or IRF3/IRF7, which could contribute to our understanding of negative regulatory mechanisms acting on the type I IFN pathway to maintain cellular homeostasis.
MATERIALS AND METHODS
Cell culture.
Mouse leukemic monocyte macrophage (Raw264.7) and human embryonic kidney 293 (HEK293T) cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco-BRL Life Technologies) and 1% antibiotic-antimycotic (Gibco) (10% FBS DMEM) under 37°C and 5% CO2 conditioned incubation. The BMDMs were isolated from femurs and tibias of C57BL/6 mice (4 to 6 weeks of age) aseptically. After removing the muscles, the bone marrow was obtained by flushing the bone with DMEM using a syringe connected to a 26G × 1/2 (0.45-mm-diameter) needle at least 3 times. The red blood cells in the bone marrow were lysed with ammonium-chloride-potassium (ACK) lysis buffer (Gibco), and the cell pellets were collected after centrifugation. The bone marrow was differentiated into BMDMs with 10% FBS DMEM and 10 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) for 5 days. All animal experiments were managed in strict accordance with the Guide for the Care and Use of Laboratory Animals (45) and performed in BSL-2 laboratory facilities with the approval of the Institutional Animal Care and Use Committee of Chungnam National University (reference number CNU-00766).
Plasmids.
Rubicon tagged with Flag- and Myc-expressing plasmids (pIRES-Rubcion-Flag and pIRES-Rubicon-Myc) were cloned into the pIRES-Flag and pIRES-Myc vectors by inserting the complete Rubicon open reading frame (ORF). Full-length IRF3 inserted into the pCMV-HA vector and GFP-tagged IRF3 plasmid was kindly donated by Jae U. Jung (Department of Molecular Microbiology and Immunology, University of Southern California). Full-length GST-tagged IRF3 (427 aa) and 3 fragments (aa 1 to 140, 140 to 400, and 380 to 420) were constructed by cloning into the pEBG vector. Flag-tagged RIG-I, TRIF, TRAF3, NLRX1, and TBK1 were generated by insertion of full-length cDNA into the pIRES-Flag vector. Full-length IRF7 was inserted into the pCMV-GFP vector.
Lentiviral shRNA.
The pGIPZ lentiviral vector containing Rubicon-specific shRNA (pGIPZ-shRubicon) was purchased from Open Biosystems. To produce lentiviruses, pGIPZ-shRubicon and packaging plasmids (psPAX2 and pMD2.VSV-G, purchased from Addgene) were transfected into HEK293T cells using Lipofectamine 2000. After 72 h, virus-containing supernatants were collected and used to infect RAW264.7 cells with 8 μg/ml Polybrene (Sigma) after filtration (0.45-μm filter; Millipore). This lentivirus has the ability to express GFP in infected cells. Every 2 days after the transduction process (12 h), medium was replaced with puromycin-containing medium to select resistant cells that contained Rubicon-specific shRNA. Similarly, lentiviruses containing a control vector (pGIPZ) were infected, and control RAW264.7 cells were selected with puromycin.
siRNA experiments.
To knock down Rubicon gene expression, siRNA oligonucleotides targeting Rubicon (siRubicon-sense, 5′-CUCAGAGUAACAGGACCUU-3′, and siRubicon-antisense, 5′-AAGGUCCUGUUACUCUGAG-3′) were transfected into BMDMs using Lipofectamine RNAiMax (Invitrogen).
Generation of stable cell lines.
To generate stable overexpressing cell lines, Raw264.7 and HEK293T cells were transfected with the pIRES-Flag vector or pIRES-Rubicon-Flag with Lipofectamine 2000 (Invitrogen). The cells were cultured with 10% FBS DMEM containing 2 μg/ml puromycin (Gibco) for 2 weeks to select Flag or Rubicon-Flag stably expressing cells.
Virus infection and stimulant transfection.
To infect cells with VSV-GFP and PR8-GFP, 10% FBS DMEM was replaced with DMEM supplemented with 1% FBS and 1% antibiotic-antimycotic (Gibco) (1% FBS DMEM) and infected virus at different multiplicities of infection (MOIs). After 2 h of incubation, extracellular virus was removed and replaced with 10% FBS DMEM. To stimulate cells with poly(I·C) (Invivogen) or 5′ppp-dsRNA (InvivoGen), poly(I·C) was transfected with Lipofectamine 2000 into HEK293T cells or used to treat Raw264.7 cells without transfection, and 5′ppp-dsRNA was transfected into both cell lines with Lipofectamine RNAiMax.
Virus titer determination.
Supernatants of growing cells or freeze-thawed cells were used for titration of VSV-GFP or PR8-GFP, respectively. The supernatants were serially 10-fold diluted and inoculated into Cercopithecus aethiops epithelial kidney (Vero) cells with medium containing 1% FBS. After 2 h at 37°C, the medium was replaced with DMEM containing 0.1% agarose (Sigma). After 48 h of incubation at 37°C and 5% CO2, the cells were stained with crystal violet to visualize plaques. Virus titers were determined by calculation using the number of plaques and the dilution factor. GFP expression levels were visualized under a fluorescence microscope (×200 magnification) and digitized using a fluorescence modulator (GloMax-Multi detection system; Promega).
ELISA.
After stimulation of the cells, cytokine production levels were analyzed using cell supernatants collected at the indicated times. Commercial kits were used to detect mouse IFN-β (PBL interferon source), mouse IL-6 (BD Biosciences), human IFN-β (PBL interferon source), and human IL-6 (BD Biosciences) in the collected supernatants according to the manufacturer's protocol.
Immunoblot analysis.
For immunoblot analysis, cell lysates or immunoprecipitated beads were mixed with 2× sample buffer (Sigma), and samples were loaded onto SDS-PAGE and separated by molecular weight. Proteins on the SDS-PAGE were transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) using a Trans-Blot semidry transfer cell (Bio-Rad). The membranes were blocked with Tris-buffered saline containing 0.05% Tween 20 (TBST) and 5% bovine serum albumin (BSA) for 1 h and incubated with target antibodies overnight at 4°C. To detect target proteins, the membranes were washed with phosphate-buffered saline containing 0.05% Tween 20 (PBST) or TBST and incubated with a 1:3,000 dilution of horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h, and the HRP on the membranes was developed with Western blotting detection reagents (GE Healthcare; ECL Select Western blotting detection reagent) and visualized using an ImageQuant LAS 4000 (GE Healthcare).
mRNA expression analysis by real time quantitative reverse transcription-PCR (qRT-PCR).
Total RNA was isolated from cells and tissues using an RNeasy minikit (Qiagen), and cDNA was synthesized using a ReverTra Ace kit (Toyobo). The quantification of selected mRNA transcripts in a particular cDNA was performed using gene-specific primer pairs (44) from a QuantiTect SYBR green PCR kit (Qiagen), and reactions were conducted on a Rotor-Gene Q (Qiagen) instrument. Relative mRNA expression levels of those genes were calculated using the ΔΔCT method after normalizing each gene with GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Luciferase reporter assays.
HEK293 cells were transfected with IFN-β and thymidine kinase promoter-renilla (TK-Renilla) luciferase reporter plasmids using Lipofectamine 2000. To stimulate IFN-β promoter luciferase, plasmids carrying the RIG-I, MDA5, MAVS, TRIF, TRAF3, TBK1, IKKα, IKKβ, or TRAF6 gene were transfected together with luciferase reporter plasmids, or PR8-GFP infection and poly(I·C) transfection were conducted at 12 h posttransfection. At 12 h after stimulation, the cells were washed with PBS, lysed with 1× passive lysis buffer (Promega), and assayed for dual-luciferase activity with lysate using a dual-luciferase assay reagent kit (Promega; E1980) according to the manufacturer's instructions. Luciferin buffers and stop solution were added to the lysate, and relative light units were measured on a luminometer (Promega). All firefly luciferase values were normalized to renilla luciferase to control for transfection efficiency.
Immunofluorescence and confocal analysis.
To confirm the nuclear translocation of IRF3, IRF3-GFP plasmids were transfected into normal or Rubicon-Flag-overexpressing HEK293T cells in collagen-coated chamber slides (LabTek; Nunc). At 24 h posttransfection, the cells were infected with H1N1 for the indicated times. The cells were fixed with 4% paraformaldehyde for 20 min and permeabilized by −20°C incubation with 100% methanol for 20 min. The fixed cells were blocked with 2% FBS diluted in PBS for 1 h, and Rubicon-Flag was detected through incubation with the primary Flag antibody (Cell Signaling; 8146; diluted 1:100 in 2% BSA) for 12 h at 4°C. After 3 washes with PBST, the cells were incubated with the secondary Cy3-conjugated donkey anti-mouse IgG antibody (Jackson Laboratory; 715-165-150; diluted 1:100 in 2% BSA) for 1 h at room temperature and washed three times PBST, followed by 10 min of incubation with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) (Sigma) containing 0.01% RNase A. The chamber slides were mounted with mounting solution (Vector), and the locations of IRF3-GFP and Rubicon-Flag were confirmed under fluorescence microscopy. Images were captured using a Nikon C2 Plus confocal microscope (Nikon), consisting of a Nikon Eclipse Ti inverted microscope with a confocal scanning system (Nikon) in conjunction with a C-HGFIE precentered fiber illuminator (Nikon). Fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isocyanate (TRITC) fluorescence was detected using the 488-nm and 561-nm laser lines of a Sapphire driver unit (Coherent), respectively, and DAPI fluorescence was detected using the 405-nm laser line of a Cube laser system (Coherent). The image data were analyzed using NIS Elements microscope imaging software (Nikon).
GST pulldown and immunoprecipitation.
Cells were harvested at 48 h posttransfection of target plasmids, and whole-cell lysates (WCL) were obtained after lysis with protease inhibitor cocktail- and phosphatase inhibitor cocktail (Sigma)-containing radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% sodium deoxycholate, 1% IGEPAL, 1 mM NaF, 1 mM Na3VO4) and sonication with a sonicator (Sonics). The WCL were precleared with Sepharose 6B (GE Healthcare Life Science) at 4°C for 2 h. After preclearing, for GST pulldown, the WCL were incubated with a 50% slurry of glutathione-conjugated Sepharose (GST) beads (Amersham Biosciences) for 12 h or, for immunoprecipitation of HA or Flag protein, incubated with HA or Flag antibody (1.0 μg/ml) for 12 h and then incubated with Protein A/G Plus agarose beads (Santa Cruz) for 4 h. The immunoprecipitated beads collected after centrifugation were washed with lysis buffer under different washing conditions.
IRF3 dimerization assay.
Cells were lysed with RIPA lysis buffer supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail. The lysates were electrophoresed on an 8% Tris-glycine gel (Invitrogen) in Tris-glycine native running buffer (25 mM Tris, pH 8.3, 192 mM glycine), and monomeric and dimerized IRF3 proteins were detected by immunoblot analysis.
Antibodies.
The antibodies used in this study were as follows: anti-GST (Santa Cruz; SC-138), anti-Flag (Cell Signaling; 8146), anti-HA (Covance; 16B12), anti-Rubicon (Abcam; ab92388), anti-IRF3 (Abcam; ab25950), anti-NF-κB p65 (Cell Signaling; 4764), anti-phospho-NF-κB p65 (Ser536) (Cell Signaling; 3031), anti-STAT1 (Cell Signaling; 9175), anti-phospho-STAT1 (Cell Signaling; 9167), anti-phospho-ERK (Cell Signaling; 9101S), anti-phospho-p38 (Cell Signaling; 9216), anti-β-actin (Santa Cruz; SC-7199), and anti-lamin B1 (Santa Cruz; SC-20682).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism version 6 for Windows (GraphPad Software). All the data were from at least two independent experiments, and the data are shown as means and standard errors of the mean (SEM). The mean values of the results of all the experiments were compared by Student's t test. Comparisons between multiple time points were analyzed by one-way analysis of variance (ANOVA). In all experiments, P values of less than 0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We declare that we have no competing interests.
This work was supported by the Ministry for Food, Agriculture, Forestry and Fisheries (315044031, 316043-3, and 112013033), the National Research Foundation (2015020957), the Internal Research Fund of the Animal Quarantine and Inspection Agency (QIA) (2013-1181), the Technology Innovation Program (10046418.TGM0721311), and the Korean Institute of Oriental Medicine (KIOM) (grant no. K16281), Republic of Korea.
REFERENCES
- 1.O'Shea JJ, Murray PJ. 2008. Cytokine signaling modules in inflammatory responses. Immunity 28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Q, Ding JL. 2016. The molecular mechanisms of TLR-signaling cooperation in cytokine regulation. Immunol Cell Biol 94:538–542. doi: 10.1038/icb.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22:240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Mogensen TH, Paludan SR. 2001. Molecular pathways in virus-induced cytokine production. Microbiol Mol Biol Rev 65:131–150. doi: 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkin J, Cohen B. 2001. An overview of the immune system. Lancet 357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 7.Seo YJ, Hahm B. 2010. Type I interferon modulates the battle of host immune system against viruses. Adv Appl Microbiol 73:83–101. doi: 10.1016/S0065-2164(10)73004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fensterl V, Sen GC. 2009. Interferons and viral infections. BioFactors 35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 9.Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17:251–263. doi: 10.1016/S1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 10.Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 11.Hiscott J. 2007. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539–548. doi: 10.1016/S1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4:491–496. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 15.Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, Derynck R, Lin K. 2003. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol 10:913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- 16.Takahasi K, Suzuki NN, Horiuchi M, Mori M, Suhara W, Okabe Y, Fukuhara Y, Terasawa H, Akira S, Fujita T, Inagaki F. 2003. X-ray crystal structure of IRF-3 and its functional implications. Nat Struct Biol 10:922–927. doi: 10.1038/nsb1001. [DOI] [PubMed] [Google Scholar]
- 17.Ikushima H, Negishi H, Taniguchi T. 2013. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harbor Symp Quant Biol 78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 18.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Trinchieri G. 2010. Type I interferon: friend or foe? J Exp Med 207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salloum R, Niewold TB. 2011. Interferon regulatory factors in human lupus pathogenesis. Transl Res 157:326–331. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 22.Decker T, Muller M, Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol 5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 23.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol 181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibeau-Poirier A, Gravel SP, Clement JF, Rolland S, Rodier G, Coulombe P, Hiscott J, Grandvaux N, Meloche S, Servant MJ. 2006. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol 177:5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, Diao FC, Chen DY, Zhai ZH, Shu HB. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 18:1096–1104. doi: 10.1038/cr.2008.277. [DOI] [PubMed] [Google Scholar]
- 26.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol 7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 27.Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long L, Deng Y, Yao F, Guan D, Feng Y, Jiang H, Li X, Hu P, Lu X, Wang H, Li J, Gao X, Xie D. 2014. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40:515–529. doi: 10.1016/j.immuni.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, Zhong Q. 2011. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem 286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. 2010. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A 107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, Kim HJ, Lee KH, Kim CJ, Oh B, Zandi E, Yue Z, Kramnik I, Liang C, Jung JU. 2012. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe 11:264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CS, Rodgers M, Min CK, Lee JS, Kingeter L, Lee JY, Jong A, Kramnik I, Lin X, Jung JU. 2012. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe 11:277–289. doi: 10.1016/j.chom.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs JL, Coyne CB. 2013. Mechanisms of MAVS regulation at the mitochondrial membrane. J Mol Biol 425:5009–5019. doi: 10.1016/j.jmb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem 273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 35.Matthews K, Schafer A, Pham A, Frieman M. 2014. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J 11:209. doi: 10.1186/s12985-014-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saira K, Zhou Y, Jones C. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol 81:3077–3086. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen A, Feng N, Ettayebi K, Hardy ME, Greenberg HB. 2009. IRF3 inhibition by rotavirus NSP1 is host cell and virus strain dependent but independent of NSP1 proteasomal degradation. J Virol 83:10322–10335. doi: 10.1128/JVI.01186-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauhofer O, Summerfield A, Sakoda Y, Tratschin JD, Hofmann MA, Ruggli N. 2007. Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J Virol 81:3087–3096. doi: 10.1128/JVI.02032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ricour C, Delhaye S, Hato SV, Olenyik TD, Michel B, van Kuppeveld FJ, Gustin KE, Michiels T. 2009. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J Gen Virol 90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, Hutt-Fletcher LM, Hass J, Jung JU. 2009. Epstein-Barr virus LF2: an antagonist to type I interferon. J Virol 83:1140–1146. doi: 10.1128/JVI.00602-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, Okabe Y, Namiki H, Fujita T. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- 42.Ning S, Pagano JS, Barber GN. 2011. IRF7: activation, regulation, modification and function. Genes Immun 12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nehyba J, Hrdlickova R, Bose HR. 2009. Dynamic evolution of immune system regulators: the history of the interferon regulatory factor family. Mol Biol Evol 26:2539–2550. doi: 10.1093/molbev/msp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J-H, Park M-E, Nikapitiya C, Kim T-H, Uddin MB, Lee H-C, Kim EH, Ma JY, Jung JU, Kim C-J, Lee J-S. 2017. FAS-associated factor-1 positively regulates type I interferon response to RNA virus infection by targeting NLRX1. PLoS Pathog 13:e1006398. doi: 10.1371/journal.ppat.1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]