ABSTRACT
Eastern equine encephalitis virus (EEEV) is a representative member of the New World alphaviruses. It is pathogenic for a variety of vertebrate hosts, in which EEEV induces a highly debilitating disease, and the outcomes are frequently lethal. Despite a significant public health threat, the molecular mechanism of EEEV replication and interaction with hosts is poorly understood. Our previously published data and those of other teams have demonstrated that hypervariable domains (HVDs) of the alphavirus nsP3 protein interact with virus-specific host factors and play critical roles in assembly of viral replication complexes (vRCs). The most abundantly represented HVD-binding proteins are the FXR and G3BP family members. FXR proteins drive the assembly of vRCs of Venezuelan equine encephalitis virus (VEEV), and G3BPs were shown to function in vRC assembly in the replication of chikungunya and Sindbis viruses. Our new study demonstrates that EEEV exhibits a unique level of redundancy in the use of host factors in RNA replication. EEEV efficiently utilizes both the VEEV-specific FXR protein family and the Old World alphavirus-specific G3BP protein family. A lack of interaction with either FXRs or G3BPs does not affect vRC formation; however, removal of EEEV's ability to interact with both protein families has a deleterious effect on virus growth. Other identified EEEV nsP3 HVD-interacting host proteins are also capable of supporting EEEV replication, albeit with a dramatically lower efficiency. The ability to use a wide range of host factors with redundant functions in vRC assembly and function provides a plausible explanation for the efficient replication of EEEV and may contribute to its highly pathogenic phenotype.
IMPORTANCE Eastern equine encephalitis virus (EEEV) is one of the most pathogenic New World alphaviruses. Despite the continuous public health threat, to date, the molecular mechanisms of its very efficient replication and high virulence are not sufficiently understood. The results of this new study demonstrate that North American EEEV exhibits a high level of redundancy in using host factors in replication complex assembly and virus replication. The hypervariable domain of the EEEV nsP3 protein interacts with all of the members of the FXR and G3BP protein families, and only a lack of interaction with both protein families strongly affects virus replication rates. Other identified HVD-binding factors are also involved in EEEV replication, but their roles are not as critical as those of FXRs and G3BPs. The new data present a plausible explanation for the exceptionally high replication rates of EEEV and suggest a new means of its attenuation and new targets for screening of antiviral drugs.
KEYWORDS: eastern equine encephalitis virus, FMR1, FXR1, FXR2, G3BP1, G3BP2, chikungunya virus, nsP3, replication complex, virus-host interactions
INTRODUCTION
The Alphavirus genus of the Togaviridae family contains more than 30 currently known members (1). On the basis of their geographical distribution, they are divided into the New World (NW) and the Old World (OW) alphaviruses. Most of the NW alphaviruses are known for their encephalitogenic phenotype, while the diseases associated with the OW alphaviruses are less severe and are characterized by rash, fever, and arthritis (1). The NW encephalitogenic representatives include Venezuelan equine encephalitis virus (VEEV), eastern equine encephalitis virus (EEEV), and western equine encephalitis virus (WEEV) (2–6). The overall mortality rates from infections caused by VEEV, EEEV, and WEEV are ∼1% (7), 30 to 80% (8), and 1 to 5% (9), respectively. However, these numbers can be higher following aerosol infection (10). The North American (NA) strains of EEEV are in continuous circulation in the United States (5), and NA EEEV represents the most pathogenic NW alphavirus (11). EEEV can be easily propagated to titers above 1010 PFU/ml in many commonly used cell lines and is listed as a select agent which can potentially be applied by bioterrorists. Despite the continuous public health threat, to date, the molecular mechanisms underpinning EEEV's high replication rates and virulence are not sufficiently understood. No safe and efficient vaccines or therapeutic means against EEEV infection have been developed.
The EEEV genome is a single-stranded RNA of positive polarity. It mimics the structure of cellular mRNAs, in that it has a cap at the 5′ terminus and a poly(A) tail at the end of the 3′ untranslated region (3′ UTR) (1). The genome encodes only a few proteins. As in the case of other alphaviruses, the nonstructural proteins are translated directly from the genomic RNA (G RNA) as polyprotein precursors P123 and P1234, which are later processed by the nsP2-associated protease activity into individual nsP1, nsP2, nsP3, and nsP4 proteins. Previous studies performed on Sindbis virus (SINV) demonstrated that differential processing of the nonstructural polyproteins regulates the synthesis of the negative-strand genome intermediate, new viral G RNA, and subgenomic RNA (SG RNA) at different times postinfection (12–14). The SG RNA serves as a template for translation of the structural proteins, the capsid, E2, and E1 proteins, which ultimately form the G RNA-containing infectious virions.
The wide geographical distribution of the alphaviruses requires adaptation to replication in different mosquito vectors and vertebrate hosts. This host-specific adaptation is strongly determined by the evolution of alphavirus glycoproteins E2 and E1, which mediate the interaction with cellular receptors and virus entry (1). Ultimately, it resulted in separation of the genus into six major serocomplexes (15). The nonstructural proteins nsP1, nsP2, and nsP4 demonstrate lower rates of evolution, which is likely restricted by the defined enzymatic functions of the nsPs in G RNA replication and transcription of the SG RNA. However, nsP3 is an exception. Similar to other nsPs, the two N-terminal domains of nsP3 are conserved among all of the alphavirus species. Moreover, one of the domains, the macrodomain, demonstrates a significant level of sequence identity with domains identified in many other positive-sense RNA viruses, such as rubella virus, astrovirus, calicivirus, and numerous plant viruses (16). The structure of the macrodomain is also similar to the structures of the macrodomains found in histones, macroH2A1 and macroH2A2, of vertebrates (17). Recently, it has been demonstrated that the nsP3 macrodomain has mono-ADP-ribosylhydrolase activity (18–20). The following second conserved N-terminal domain was shown to bind Zn2+, but its function(s) in virus replication remains to be determined (21). In contrast to the high degree of sequence conservation observed in the N-terminal domains, the C-terminal domain of nsP3 is intrinsically disordered, is highly phosphorylated, and demonstrates very low levels of amino acid sequence identity between the known alphavirus representatives. This approximately 200-amino-acid (aa)-long domain is usually referred to as the hypervariable domain (HVD) (1, 22).
Our recent studies demonstrated that the nsP3-specific HVD serves as a critical determinant of alphavirus replication (23–25). It functions as an assembly hub for recruiting different host factors which are required for viral replication complex (vRC) assembly and initiation of RNA replication (23). Accumulating data suggest that the nsP3 HVDs of different alphaviruses interact with distinct sets of cellular proteins which are specific for particular virus species. Replication of the OW alphaviruses, such as Semliki Forest virus (SFV), Sindbis virus (SINV), and chikungunya virus (CHIKV), is critically dependent on interaction of their HVDs with both members of the cellular G3BP protein family, G3BP1 and G3BP2 (23, 26). The HVD-G3BP interaction is determined by short repeating amino acid sequences, located at the carboxy terminus of the HVD (27). The knockout (KO) of both G3bp genes but not the individual family members in murine cells almost completely abrogates CHIKV replication and reduces the rates of SINV replication by a few orders of magnitude (23). However, this double G3bp1 and G3bp2 KO has no negative effect on the replication of VEEV (23). The nsP3 HVD of the latter representative member of the NW alphaviruses does not interact with G3BPs. However, its HVD also contains a repeating peptide which demonstrates no similarity to the repeating amino acid sequence of the OW alphaviruses and binds all of the members of the fragile X syndrome (FXR) protein family (23, 25). Each of these proteins can independently support VEEV replication but not replication of the OW alphaviruses. However, KO of all of the FXR family members has a dramatic negative effect on VEEV infectivity, its ability to initiate RNA replication, and RNA replication rates (23). Despite belonging to different families and demonstrating no identity on the amino acid level, the FXR and G3BP proteins exhibit a common ability to interact with RNAs and to self-assemble into higher-order complexes, such as cellular stress granules, which are composed of a wide variety of cellular proteins (28).
In this study, we continued investigation of the role of HVDs and host proteins in alphavirus replication. Our data demonstrate that, in addition to using all of the members of a particular family, the most pathogenic alphavirus, NA EEEV, exhibits another level of redundancy in recruiting host factors. In contrast to the previously characterized VEEV HVD (HVDveev) and CHIKV HVD (HVDchikv), the HVD of EEEV (HVDeeev) has evolved to interact with both FXR and G3BP family members. Moreover, in the absence of an interaction with both FXRs and G3BPs, the HVD interaction with other host factors supports virus replication, albeit with a dramatically lower efficiency and in a cell-specific mode. The use of diverse sets of cellular proteins represents a significant evolutionary advantage that likely promotes efficient EEEV replication in a wide variety of cells and tissues, in which the presence of each host factor may vary.
RESULTS
The EEEV nsP3 HVD interacts with a specific combination of cellular factors.
To identify combinations of cellular proteins which interact with the EEEV HVD, we fused the nsP3 HVD-coding sequence of North American EEEV strain Florida93 (FL93) with a Flag-green fluorescent protein (GFP) tag and cloned the resulting cassette into an SINV replicon under the control of an SG RNA promoter. These SINrep/Flag-GFP-HVDeeev and control SINrep/Flag-GFP replicons (Fig. 1) were packaged into SINV particles by coelectroporation of the in vitro-synthesized replicon genomes and helper RNAs into BHK-21 cells. Next, NIH 3T3 cells were infected with the harvested infectious viral particles at a multiplicity of infection (MOI) of 20 infectious units per cell (inf.u/cell). After incubation for 2 h at 37°C, cells were lysed and protein complexes were isolated using Flag-specific monoclonal antibody (MAb)-loaded magnetic beads, electrophoretically separated, and then analyzed by mass spectrometry. The results of the analysis are presented in Table 1. They were compared to the spectra of proteins binding to Flag-GFP-HVDchikv, Flag-GFP-HVDsinv, and Flag-GFP-HVDveev upon their expression from similar constructs, which were designed and used in previous studies (Fig. 1 and Table 1). To avoid overproduction of the proteins and formation of nonspecific complexes, all of the affinity purifications were performed within 2 to 3 h postinfection (p.i.) with replicons. At this early stage, the concentration of recombinant proteins and replicon-encoded nsPs is not high enough to promote the formation of nonspecific protein aggregates, which have been observed in previous studies, when protein synthesis is allowed to continue for periods exceeding 3 h (data not shown). Therefore, it is possible to confidently ascribe a functional relevance to the protein interactions detected in these experiments.
FIG 1.
Schematic representation of replicons encoding Flag-GFP fusions with the nsP3 HVDs derived from the indicated alphaviruses. Infection of NIH 3T3 cells with the indicated packaged replicons and immunoprecipitations were performed as described in Materials and Methods. SG pr, SG RNA promoter; p(A), poly(A) tail.
TABLE 1.
Host proteins interacting with nsP3 HVDs of different alphaviruses
| Protein type and protein | Protein definition | Total no. of spectra identified in: |
|||
|---|---|---|---|---|---|
| EEEV | VEEV | CHIKV | SINV | ||
| RNA binding proteins | |||||
| FXR1 | FMR1 autosomal homolog 1 | 267 | 374 | 0 | 0 |
| FMR1 | Fragile X mental retardation 1 | 160 | 230 | 0 | 0 |
| FXR2 | FMR1 autosomal homolog 2 | 95 | 137 | 0 | 0 |
| G3BP1 | Ras GTPase-activating protein-binding protein 1 | 192 | 2 | 217 | 69 |
| G3BP2 | Ras GTPase-activating protein-binding protein 2 | 35 | 0 | 118 | 54 |
| IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 | 21 | 4 | 6 | 14 |
| IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 10 | 2 | 0 | 14 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA-binding protein 2 | 9 | 1 | 0 | 11 |
| YBX1 | Y-box-binding protein 1 | 11 | 2 | 0 | 0 |
| YBX3 | Y-box-binding protein 3 | 0 | 0 | 7 | 12 |
| DDX17 | DEAD box polypeptide 17 | 0 | 0 | 0 | 19 |
| DDX5 | DEAD box polypeptide 5 | 0 | 0 | 0 | 25 |
| DDX6 | DEAD box polypeptide 6 | 0 | 0 | 0 | 11 |
| Actin-interacting proteins | |||||
| CD2AP | CD2-associated protein | 134 | 158 | 13 | 0 |
| SH3KBP1 | SH3 domain-containing kinase-binding protein 1 | 82 | 158 | 8 | 0 |
| CAPZB | Capping actin protein of muscle Z-line beta subunit | 12 | 32 | 2 | 0 |
| CAPZA1 | Capping actin protein of muscle Z-line alpha subunit 1 | 6 | 12 | 0 | 0 |
| CAPZA2 | Capping actin protein of muscle Z-line alpha subunit 2 | 5 | 6 | 0 | 0 |
| RDX | Radixin | 0 | 0 | 11 | 0 |
| BAR domain-containing proteins | |||||
| SNX9 | Sorting nexin-9 | 52 | 2 | 0 | 0 |
| SNX33 | Sorting nexin-33 | 40 | 0 | 0 | 0 |
| BIN1 | Bridging integrator 1 | 0 | 0 | 49 | 43 |
| Others | |||||
| HSPA1B | Heat shock protein 1B | 33 | 25 | 22 | 34 |
| S100A4 | S100 calcium binding protein A4 | 27 | 5 | 0 | 0 |
| SLC25A13 | Solute carrier family 25 member 13 | 19 | 6 | 0 | 0 |
| CLTC | Clathrin heavy chain | 19 | 0 | 0 | 0 |
| SLC25A5 | Solute carrier family 25 member 5 | 10 | 3 | 0 | 0 |
| PGAM5 | PGAM family member 5, serine/threonine phosphatase | 5 | 4 | 14 | 0 |
| DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 | 0 | 0 | 12 | 4 |
| FHL2 | Four and a half LIM domains protein 2 | 0 | 0 | 17 | 0 |
| HIST1H1C | Histone cluster 1 H1 family member C | 0 | 0 | 27 | 0 |
| JUN | Jun proto-oncogene | 0 | 0 | 11 | 0 |
| MYBBP1A | MYB binding protein 1A | 0 | 0 | 34 | 10 |
| NAP1L1 | Nucleosome assembly protein 1-like 1 | 0 | 0 | 99 | 0 |
| NAP1L4 | Nucleosome assembly protein 1-like 4 | 0 | 0 | 44 | 0 |
| WDR48 | WD repeat domain 48 | 0 | 0 | 0 | 13 |
In agreement with the previously published data (23), the HVDs of the representative NW alphavirus VEEV and the OW alphaviruses CHIKV and SINV were found to interact with FXR and G3BP family members, respectively. No G3BPs were detected in samples from Flag-GFP-HVDveev-expressing cells, and no FXRs were present in the samples isolated from Flag-GFP-HVDchikv- and Flag-GFP-HVDsinv-expressing cells. Interestingly, Flag-GFP-HVDeeev coprecipitated with all of the members of both the G3BP and FXR families, and they were the most abundant proteins among other coisolated host factors (Table 1). Other identified HVDeeev-interacting, RNA-binding host proteins included the members of the IGF2BP protein family and YBX1. However, they were present at lower levels.
The nsP3 proteins of the OW alphaviruses were previously shown to bind the BAR domain-containing protein BIN1 or amphiphysin II (23, 29). The latter protein was proposed to play a role in the formation of membrane curvature during assembly of viral double-stranded RNA (dsRNA)-containing spherules (29). In agreement with these data, BIN1 was reproducibly coimmunoprecipitated with the CHIKV HVD and the SINV HVD from both NIH 3T3 and BHK-21 cells (Table 1) (23). No BAR domain-containing proteins were identified by immunoprecipitation with the VEEV HVD either in our previous study or in these new experiments (Table 1) (23). However, two other BAR domain-containing host factors, SNX9 and SNX33, were abundantly present in the Flag-GFP-HVDeeev complexes (Table 1). As was proposed for BIN1 (29), SNX9 and SNX33 likely interact with the polyproline motif in the EEEV HVD through their SH3 domains. Detection of SNX9 and SNX33 in the HVDeeev complexes is an additional indication of the importance of BAR domain-containing host factors in alphavirus replication. However, understanding of their function requires more experimental data.
In our previous study, we found a group of proteins implicated in actin remodeling which specifically interacted with the VEEV HVD but not with the HVD derived from SINV or CHIKV (23). These protein factors included CD2AP, CAPZB, and CAPZA1. In addition to them, we also identified CAPZA2 and SH3KBP1 to be VEEV HVD-interacting host factors. The same proteins were also immunoprecipitated with the EEEV HVD in the pulldown experiments of this study. Surprisingly, they were also isolated with the CHIKV HVD, although at a lower concentration. This suggests that the indicated host factors may be specific not only to the NW alphaviruses. CD2AP, SH3KBP1, and BAR domain-containing proteins BIN1, SNX9, and SNX33 contain the SH3 domain and, thus, may compete for binding with the same polyproline motifs in HVDs.
Other proteins unambiguously identified in the Flag-GFP-HVDeeev samples included heat shock protein 1B (HSPA1B), S100 calcium binding protein A4 (S100A4), and solute carrier family 25 members 5 (SLC25A5) and 13 (SLC25A13) (Table 1).
Thus, the EEEV-specific nsP3 HVD demonstrated an interaction with a diverse combination of cellular protein factors. The unique characteristic of this set was that it combined proteins which have previously been identified to be specific for the NW alphavirus VEEV or for the OW alphaviruses CHIKV and SINV. Other host factors, which included the BAR domain-containing proteins SNX9 and SNX33, were found to specifically interact only with the EEEV nsP3 HVD and not with the HVDs of the NW and OW alphaviruses used in these experiments as controls.
EEEV nsP3-specific complexes in virus-infected cells contain both FXRs and G3BPs.
The detection of high levels of FXRs and G3BPs in protein complexes coisolated with the EEEV HVD in the coimmunoprecipitation (co-IP) experiments described above was unexpected. This finding reveals a unique characteristic of EEEV which differentiates it from the other better-studied NW and OW alphaviruses which utilize a single protein family for nsP3 complex formation, either FXRs or G3BPs. This was suggestive that FXRs and G3BPs may have redundant functions in EEEV replication. To experimentally interrogate this possibility, we designed a chimeric virus, EEE/nsP3-Cherry/SINV (Fig. 2C). The genome of this recombinant virus combines EEEV-specific nonstructural genes and cis-acting RNA elements with the structural genes of the laboratory strain of SINV Toto1101. Thus, the genome of EEE/nsP3-Cherry/SINV encodes neither the OW alphavirus nsP2 nor the NW alphavirus-specific capsid protein, which could interfere with cellular transcription and activation of the type I interferon (IFN) response. A lack of transcription-inhibitory functions makes this chimeric virus very poorly cytopathic and incapable of developing a spreading infection in cells proficient in type I IFN production and signaling (30, 31). However, it remains an adequate model to study the structure and mechanism of formation of EEEV vRCs. The EEE/nsP3-Cherry/SINV genome also contains a fluorescent Cherry protein gene, which was cloned in frame into the beginning of the nsP3 HVD. Expression of this fluorescent tag allowed us to monitor the formation of the nsP3-specific complexes in the infected cells.
FIG 2.
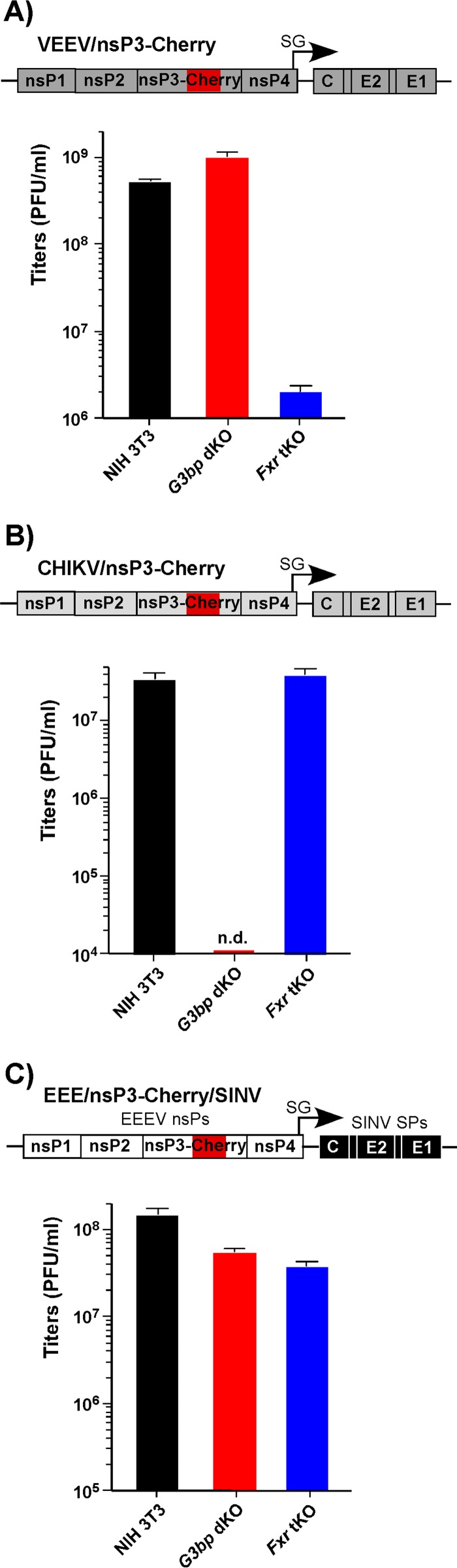
A recombinant alphavirus with EEEV replication machinery efficiently replicates in the absence of expression of either G3BPs or FXRs. Parental NIH 3T3 cells and their G3bp dKO and Fxr tKO derivatives were infected with VEEV/nsP3-Cherry (A), CHIKV/nsP3-Cherry (B), and EEE/nsP3-Cherry/SINV (C) at an MOI of 0.1 PFU/cell. Media were harvested at 7 h p.i., and virus titers were determined by plaque assay on BHK-21 cells. The experiment was repeated three times with reproducible results. SPs, structural proteins; n.d., the titer was below the limit of detection, which was 50 PFU/ml.
Parental NIH 3T3 cells and their derivatives with triple KO (tKO) of the FXR gene (Fxr tKO cells) and double KO (dKO) of the G3BP gene (G3bp dKO cells) were infected with VEEV/nsP3-Cherry, CHIKV/nsP3-Cherry, and EEE/nsP3-Cherry/SINV. In accordance with our previous data (23), Fxr tKO cells supported the replication of VEEV/nsP3-Cherry, albeit with an efficiency reduced by a few orders of magnitude compared to the level of replication in parental NIH 3T3 cells (Fig. 2A). CHIKV/nsP3-Cherry was incapable of replication in G3bp dKO cells (Fig. 2B). However, EEE/nsP3-Cherry/SINV replicated almost as efficiently in the G3bp dKO and Fxr tKO cells as in the parental NIH 3T3 cells (Fig. 2C).
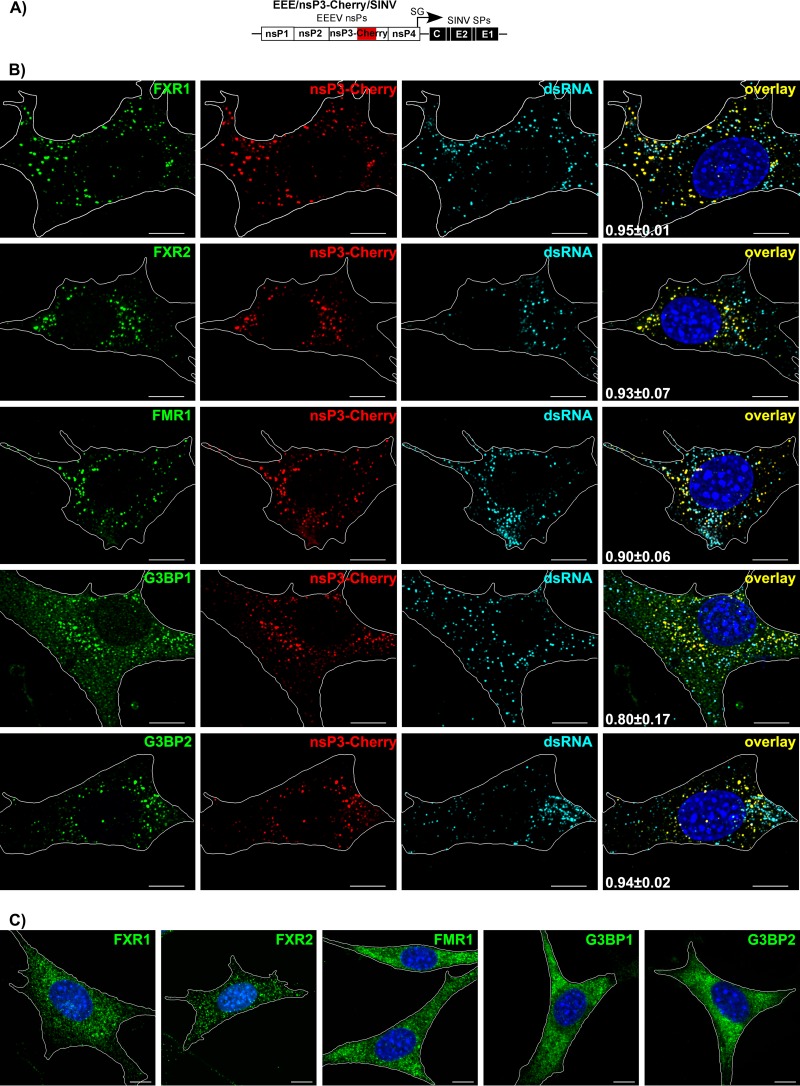
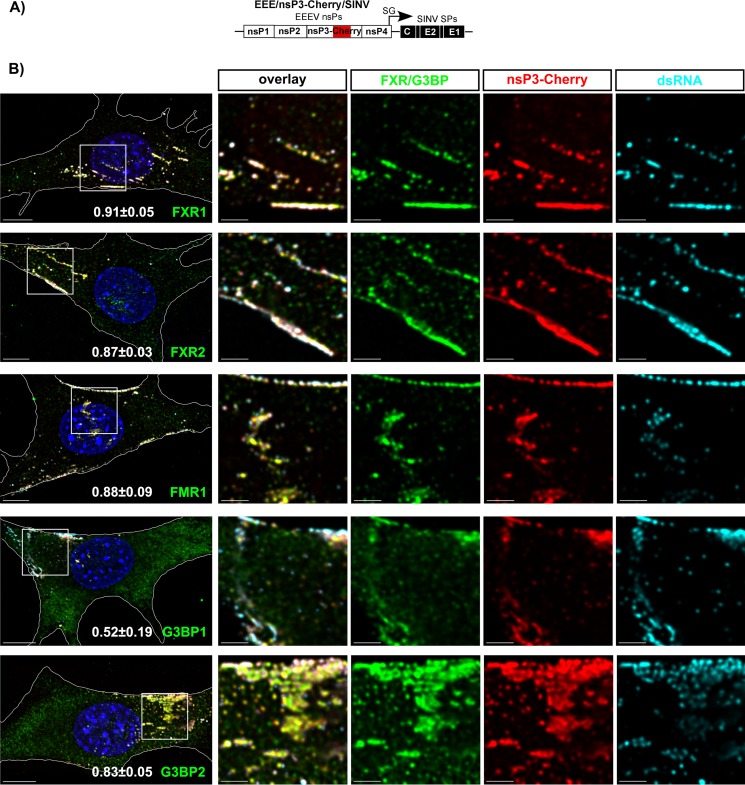
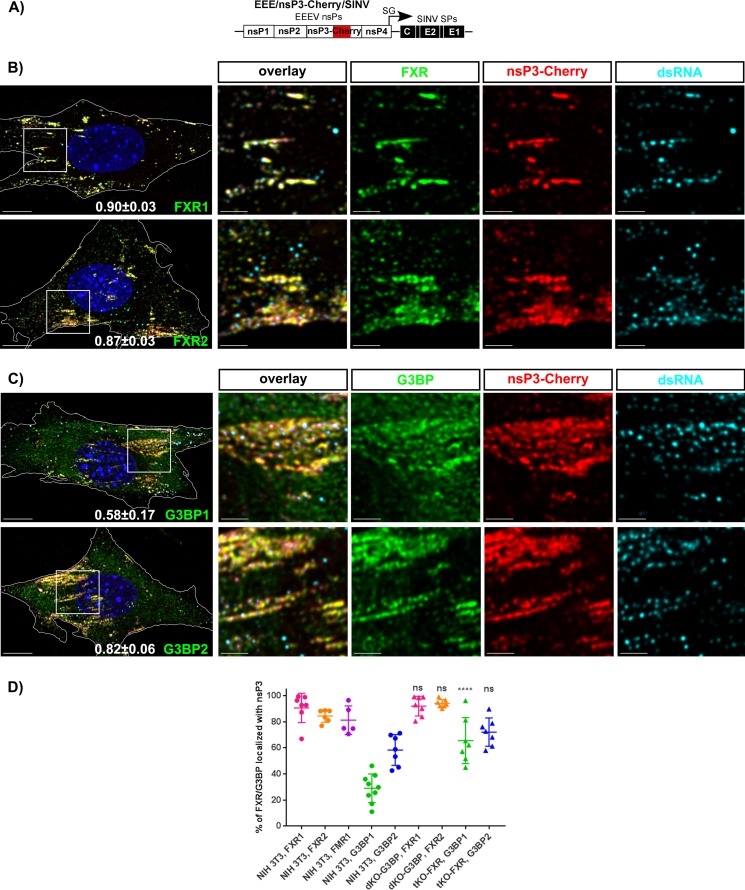
Next, we examined the distribution of FXRs and G3BPs in NIH 3T3 cells infected with EEE/nsP3-Cherry/SINV (Fig. 3A). Similar to our results with other alphaviruses (23, 24, 32), by 7 h p.i., EEEV nsP3-Cherry formed large cytoplasmic complexes. These complexes accumulated almost entire pools of FXR proteins (more than 90% of total detectable protein) (Fig. 3B and Table 2). G3BP1 and G3BP2 clearly colocalized with EEEV nsP3 in the same complexes. As we previously described for other alphaviruses (23, 24, 33), at this late time p.i., most of the virus-specific dsRNAs did not colocalize with these large nsP3 complexes (Fig. 3B and Table 2), additionally indicating that these large structures are not vRCs.
FIG 3.
At late times p.i., FXR and G3BP proteins efficiently accumulate in large cytoplasmic EEEV nsP3-containing complexes. (A) Schematic representation of the EEE/nsP3-Cherry/SINV genome. (B) NIH 3T3 cells were infected with chimeric EEE/nsP3-Cherry/SINV. At 7 h p.i., they were fixed with PFA and stained with antibodies specific to FXR1, FXR2, FMR1, G3BP1, G3BP2, and dsRNA. Nuclei were stained with Hoechst dye. Images are presented as multiple-image projections of the entire cells. Pearson's colocalization coefficients are shown in the final overlay panels (mean ± SD, n > 7 cells). (C) Distribution of FXR1, FXR2, FMR1, G3BP1, and G3BP2 in mock-infected cells. Images are presented as multiple-image projections of 6 optical sections. Bars, 10 μm.
TABLE 2.
| Cell (time p.i.) and protein | Pearson's coefficient for F/Ga-nsP3 | % of F/G colocalized with nsP3 | % of nsP3 colocalized with F/G | Pearson's coefficient for F/G-dsRNA | % of F/G colocalized with dsRNA | % of dsRNA colocalized with F/G | Pearson's coefficient for nsP3-dsRNA | % of nsP3 colocalized with dsRNA | % of dsRNA colocalized with nsP3 |
|---|---|---|---|---|---|---|---|---|---|
| NIH 3T3 cells (3 h) | |||||||||
| FXR1 | 0.91 ± 0.5 | 95 ± 5 | 80 ± 10 | 0.27 ± 0.14 | 48 ± 7 | 75 ± 11 | 0.46 ± 0.18 | 50 ± 9 | 94 ± 3 |
| FXR2 | 0.87 ± 0.03 | 78 ± 19 | 86 ± 8 | 0.34 ± 0.19 | 45 ± 14 | 79 ± 11 | 0.55 ± 0.17 | 63 ± 13 | 91 ± 10 |
| FMR1 | 0.88 ± 0.09 | 73 ± 22 | 87 ± 4 | 0.29 ± 0.16 | 36 ± 14 | 84 ± 10 | 0.56 ± 0.14 | 54 ± 11 | 95 ± 6 |
| G3BP1 | 0.52 ± 0.19 | 29 ± 11 | 97 ± 2 | 0.40 ± 0.18 | 9 ± 4 | 91 ± 7 | 0.62 ± 0.23 | 56 ± 12 | 93 ± 6 |
| G3BP2 | 0.83 ± 0.05 | 59 ± 11 | 81 ± 9 | 0.43 ± 0.13 | 22 ± 7 | 85 ± 11 | 0.56 ± 0.24 | 43 ± 11 | 98 ± 2 |
| Fxr tKO cells (3 h) | |||||||||
| G3BP1 | 0.58 ± 0.17 | 65 ± 18 | 91 ± 4 | 0.28 ± 0.14 | 21 ± 7 | 86 ± 8 | 0.49 ± 0.15 | 48 ± 12 | 95 ± 4 |
| G3BP2 | 0.82 ± 0.06 | 72 ± 11 | 85 ± 4 | 0.31 ± 0.14 | 34 ± 9 | 85 ± 6 | 0.44 ± 0.12 | 53 ± 13 | 94 ± 6 |
| G3bp dKO cells (3 h) | |||||||||
| FXR1 | 0.90 ± 0.03 | 92 ± 8 | 87 ± 7 | 0.23 ± 0.18 | 37 ± 16 | 70 ± 14 | 0.32 ± 0.2 | 42 ± 17 | 80 ± 17 |
| FXR2 | 0.87 ± 0.03 | 94 ± 3 | 83 ± 8 | 0.19 ± 0.12 | 35 ± 13 | 64 ± 13 | 0.30 ± 0.11 | 40 ± 13 | 81 ± 10 |
| NIH 3T3 cells (7 h) | |||||||||
| FXR1 | 0.95 ± 0.01 | 95 ± 2 | 95 ± 3 | −0.09 ± 0.06 | 15 ± 8 | 30 ± 8 | 0.11 ± 0.13 | 21 ± 10 | 52 ± 10 |
| FXR2 | 0.93 ± 0.07 | 92 ± 9 | 92 ± 4 | −0.06 ± 0.06 | 12 ± 6 | 30 ± 12 | 0.15 ± 0.10 | 18 ± 14 | 55 ± 10 |
| FMR1 | 0.90 ± 0.06 | 98 ± 1 | 79 ± 4 | 0.01 ± 0.14 | 17 ± 5 | 35 ± 15 | 0.36 ± 0.20 | 34 ± 9 | 72 ± 11 |
| G3BP1 | 0.80 ± 0.17 | 37 ± 24 | 98 ± 3 | 0.04 ± 0.05 | 9 ± 3 | 70 ± 17 | 0.14 ± 0.09 | 21 ± 7 | 56 ± 13 |
| G3BP2 | 0.94 ± 0.02 | 76 ± 16 | 95 ± 3 | 0.08 ± 0.15 | 15 ± 9 | 35 ± 11 | 0.14 ± 0.21 | 21 ± 11 | 45 ± 9 |
| Fxr tKO cells (7 h) | |||||||||
| G3BP1 | 0.73 ± 0.15 | 58 ± 29 | 86 ± 10 | 0.06 ± 0.06 | 14 ± 4 | 77 ± 9 | 0.47 ± 0.15 | 37 ± 9 | 84 ± 13 |
| G3BP2 | 0.87 ± 0.12 | 91 ± 8 | 76 ± 9 | −0.06 ± 0.09 | 16 ± 7 | 25 ± 23 | 0.28 ± 0.27 | 34 ± 13 | 70 ± 21 |
| G3bp dKO cells (7 h) | |||||||||
| FXR1 | 0.92 ± 0.03 | 90 ± 8 | 89 ± 11 | −0.01 ± 0.06 | 25 ± 11 | 35 ± 16 | 0.03 ± 0.09 | 29 ± 8 | 45 ± 11 |
| FXR2 | 0.94 ± 0.02 | 95 ± 3 | 94 ± 3 | 0.01 ± 0.08 | 32 ± 14 | 36 ± 11 | 0.07 ± 0.13 | 36 ± 11 | 48 ± 11 |
F/G, FXR or G3BP.
In our previous study, we demonstrated that assembly of alphavirus vRCs proceeds within the first 4 h p.i. through formation of G RNA- and nsP3-containing pre-vRCs at the plasma membrane (23, 33, 34). We also showed that the FXR and G3BP proteins play critical roles in the formation of VEEV- and CHIKV-specific vRCs, respectively. Thus, in the next experiments, we examined the colocalization of FXRs and G3BPs with EEEV nsP3-Cherry at the plasma membrane of the cells at 3 h p.i. with EEE/nsP3-Cherry/SINV (Fig. 4). All of the G3BPs and FXRs strongly colocalized with nsP3-Cherry (see the Pearson's coefficients in Fig. 4B and Table 2). Even at this early time p.i., nsP3 complexes had already accumulated large fractions of FXRs (95% ± 5% FXR1, 78% ± 19% FXR2, and 73% ± 22% FMR1) and smaller but clearly detectable fractions of the G3BP pools (29% ± 11% G3BP1 and 59% ± 11% G3BP2) (Table 2). As we previously described for VEEV and CHIKV (23), at this early time p.i., the majority of the plasma membrane-specific EEEV nsP3/FXR/G3BP complexes frequently overlapped with dsRNA, which indicates active vRCs (Fig. 4B).
FIG 4.
At early times p.i., all FXR and G3BP proteins colocalize with membrane-bound EEEV nsP3-containing pre-vRCs. (A) Schematic representation of the EEE/nsP3-Cherry/SINV genome. (B) NIH 3T3 cells were infected with EEE/nsP3-Cherry/SINV. At 3 h p.i., they were fixed and stained with antibodies specific to FXR1, FXR2 FMR1, G3BP1, G3BP2, and dsRNA. Nuclei were stained with Hoechst dye. Images are presented as multiple-image projections of 1-μm x-y sections at the plasma membrane. Pearson's colocalization coefficients are shown in the panels in the left column (mean ± SD, n > 7 cells). The four right columns present enlargements of the boxed regions in the panels in the left column. Bars, 10 μm (left column) and 3 μm (four right columns).
We next assessed the colocalization of FXRs and G3BPs with nsP3 in G3bp dKO (Fig. 5B) and Fxr tKO (Fig. 5C) cells, respectively, infected with EEE/nsP3-Cherry/SINV (Fig. 5A). In the absence of G3BP1 and G3BP2 expression, by 3 h p.i., the FXR pools still efficiently relocalized into nsP3 complexes (Fig. 5B and Table 2). The difference in FXR colocalization between the parental NIH 3T3 and G3bp dKO cells was not statistically significant (Fig. 5D and Table 2). Similarly, in the absence of FXRs, in Fxr tKO cells, G3BPs efficiently accumulated in the nsP3 complexes (Fig. 5C and D and Table 2). Importantly, compared to NIH 3T3 cells, in the Fxr tKO cells, assembly of G3BPs into nsP3 complexes proceeded more efficiently (Fig. 5D). This was particularly evident for G3BP1. An almost 2-fold larger fraction of this protein was associated with nsP3 in Fxr tKO cells than parental NIH 3T3 cells. Similarly, at 7 h p.i., larger fractions of cellular G3BP1 and G3BP2 were found in the large cytoplasmic complexes (Table 2). The increase in the level of accumulation of FXRs or G3BPs in nsP3 complexes in KO cell lines which do not express the second protein family suggests that many membrane-bound nsP3 complexes may not necessarily contain the same levels of FXRs and G3BPs.
FIG 5.
FXR and G3BP proteins independently accumulate in the membrane-bound EEEV nsP3-containing pre-vRCs. (A) Schematic representation of the EEE/nsP3-Cherry/SINV genome. (B and C) G3bp dKO (B) or Fxr tKO (C) cells were infected with chimeric EEE/nsP3-Cherry/SINV. At 3 h p.i., they were fixed and stained with antibodies specific to FXR1, FXR2, G3BP1, or G3BP2 and with antibodies against dsRNA. Nuclei were stained with Hoechst dye. Images are presented as multiple-image projections of x-y sections (1 μm) at the plasma membrane. Pearson's colocalization coefficients are shown in the left overlay panels (mean ± SD, n > 7 cells). The four right columns present enlargements of the boxed regions in the panels in the left column. Bars, 10 μm (left column) and 3 μm (four right columns). (D) Percentage of FXR and G3BP proteins colocalized with nsP3 at 3 h p.i. in the indicated cell lines. The statistical significance of the difference in the colocalization of nsP3 and each protein (G3BP or FXR) in NIH 3T3 cells and cells of the indicated KO cell lines was evaluated using a Kruskal-Wallis test by ranks followed by Dunn's test. ****, <0.0001; ns, not significant.
Taken together, these results demonstrate that, in contrast to VEEV and CHIKV, which utilize the FXR and G3BP protein families, respectively, for vRC assembly, EEEV has evolved to use all of the members of both families. All of the FXRs and G3BPs efficiently accumulated in both the membrane-bound and large cytoplasmic nsP3 complexes. Importantly, KO of either entire family had no deleterious effect on replication mediated by EEEV nsPs, indicating that FXRs and G3BPs have redundant functions in EEEV infection. Moreover, the data also suggest that the FXR and G3BP families function independently of each other. However, the possibilities of some synergistic or additive effects in their function cannot be completely ruled out.
FXR- and G3BP-binding sites are located in different fragments of the EEEV nsP3 HVD.
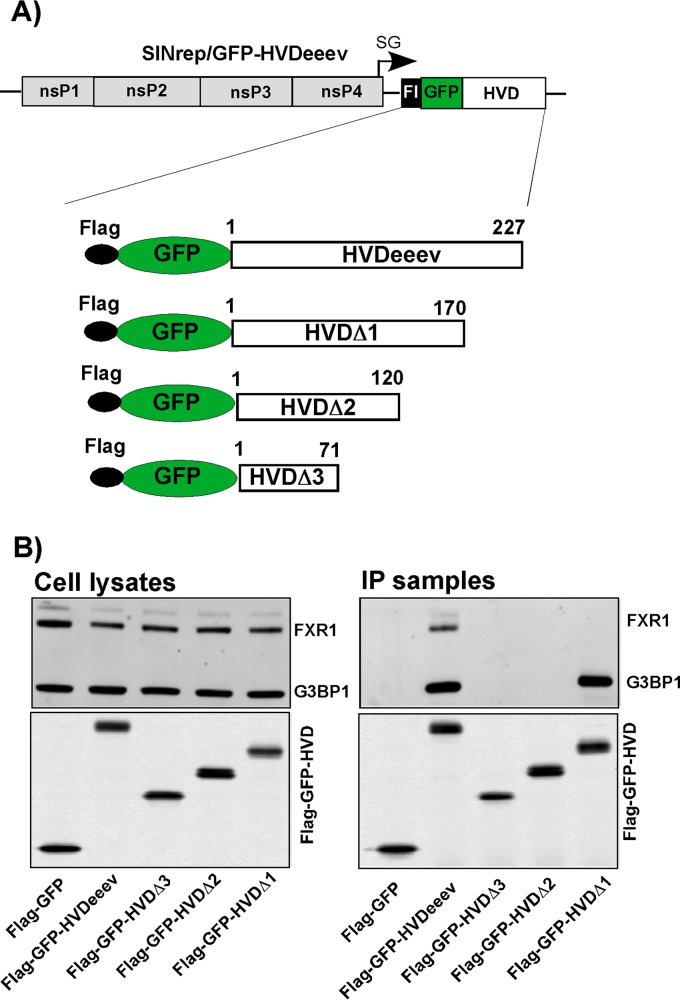
Recent studies have demonstrated that G3BP and FXR proteins interact with short repeating amino acid sequences located in the carboxy termini of the CHIKV and VEEV HVDs, respectively (23). However, a detailed analysis of the amino acid sequence of EEEV's HVD revealed that no such repeats are present in its carboxy terminus. We also failed to identify peptides with clear levels of identity with G3BP- and FXR-binding sequences which had been previously identified in CHIKV or SINV and VEEV, respectively (23, 25, 27). Therefore, in the next experiments, we performed a preliminary mapping of the FXR- and G3BP-binding fragments in the EEEV HVD. The HVD sequence was divided into 4 fragments, which were sequentially deleted in the Flag-GFP-HVDeeev cassette (Fig. 6A). These fusion proteins were expressed from SINV replicons, and protein complexes were immunoprecipitated as described above and analyzed by Western blotting using FXR1- and G3BP1-specific antibodies (Fig. 6B). The FXR-binding sequence was found to be located close to the carboxy terminus, between aa 170 and 227, as the HVDΔ1 fragment failed to bind FXR1. The G3BP1-binding site was identified in the fragment located between aa 120 and 170. The finding of FXR- and G3BP1-binding sites in spatially distinct regions of the HVD was in agreement with the data derived from the experiments with the G3bp dKO and Fxr tKO cell lines. It additionally demonstrates that the FXR and G3BP protein families bind to the EEEV HVD and support RNA replication independently of each other.
FIG 6.
G3BP- and FXR-binding sites are located in different fragments of the EEEV HVD. (A) Schematic representation of the SINV-based replicon and Flag-GFP-HVDeeev cassettes applied for mapping of G3BP- and FXR-binding sites. (B) NIH 3T3 cells were infected with the indicated replicons at an MOI of 20 inf.u/cell. Cells were harvested at 2.5 h p.i., and their lysates were used for immunoprecipitation of protein complexes on magnetic beads with anti-Flag MAbs as described in Materials and Methods. Cell lysates and IP samples were analyzed by Western blotting using anti G3BP1- and FXR1-specific antibodies.
FXR and G3BP binding is mediated by short, nonrepeating peptides in the EEEV nsP3 HVD.
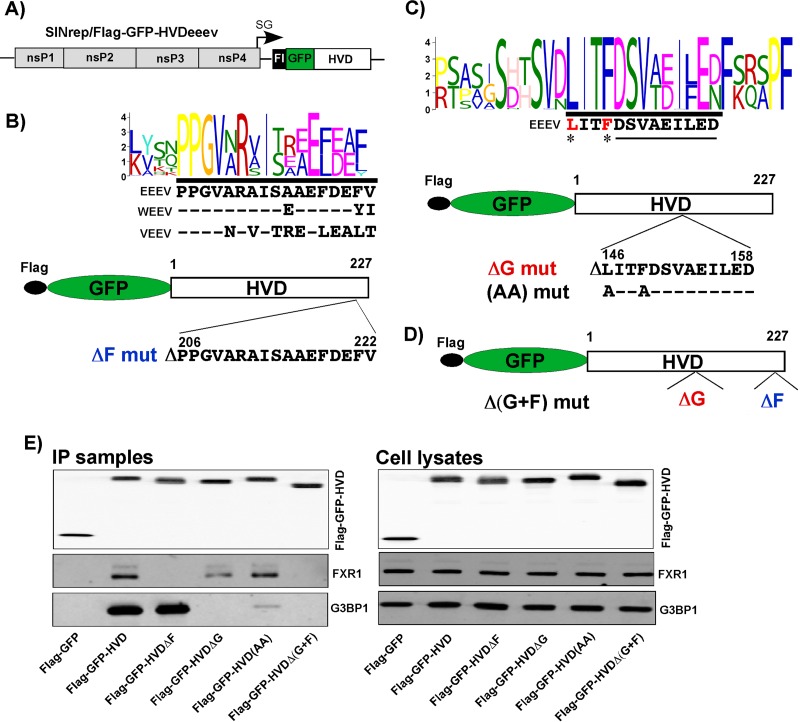
To identify potential binding sites for FXRs and G3BPs in the EEEV HVD, we performed a motif search using MEME software (http://meme-suite.org/tools/meme). Analysis of 242 HVD sequences of VEEV, EEEV, and WEEV suggested a short amino acid sequence in the carboxy terminus of the EEEV HVD which has some degree of similarity to the FXR-binding repeating peptide of VEEV's HVD (Fig. 7A and B). Screening of 143 HVD sequences of EEEV and WEEV defined a peptide located between aa 120 and 170 of the EEEV HVD to be a potential G3BP-binding site (Fig. 7C). This peptide was absent in the EEEV HVDΔ2 mutant, which exhibited no interaction with G3BP1. Although this short motif is very different from the G3BP-binding sites of the OW alphaviruses, such as CHIKV and SFV, it shares common characteristic features, such as hydrophobic amino acids at the amino terminus and negatively charged amino acids at the carboxy terminus (Fig. 7C). To experimentally test the possibility of interaction of putative binding sites with FXRs and G3BPs, we introduced both small deletions and point mutations into the HVD of the Flag-GFP-HVDeeev fusion (Fig. 7B to D) expressed from the SINV replicon (Fig. 7A). The subsequent co-IP experiments confirmed that these small deletions abrogated binding of the corresponding FXR or G3BP protein in NIH 3T3 cells (Fig. 7E). Moreover, just two point mutations in the putative G3BP-binding sequence of the Flag-GFP-HVD(AA) mutant strongly reduced the amount of precipitated G3BP1. Deletion of both predicted G3BP- and FXR-binding sequences rendered the Flag-GFP-HVDΔ(G+F) mutant incapable of interacting with the members of either protein family.
FIG 7.
FXR- and G3BP-binding sites in the EEEV nsP3 HVD. (A) Schematic representation of the SINV replicon encoding the Flag-GFP-HVDeeev fusion protein. (B to D) Composite motif logos of multiple-sequence alignments of the peptides containing putative G3BP- and FXR-binding sites, the schemes of the designed Flag-GFP-HVDeeev cassettes, and the amino acid sequences of the deleted or mutated peptides. (E) NIH 3T3 cells were infected with the indicated packaged replicons at an MOI of 20 inf.u/cell. Cells were harvested at 2.5 h postinfection, and their lysates were used for immunoprecipitation of protein complexes. The original cell lysates and IP samples were analyzed by Western blotting using Flag-, G3BP1-, and FXR1-specific antibodies. F, FXR binding site; G, G3BP binding site.
The amino-terminal NTF2-like and Agenet-like domains mediate the interaction of G3BP1 and FXR1 with the EEEV nsP3 HVD.
In another set of experiments, we took advantage of previously generated stable cell lines which express deletion mutants of either G3BP1 (Fig. 8A) or FXR1 (Fig. 8B) fused in frame with a GFP-coding sequence. These cell lines represent an experimental system for identification of FXR- and G3BP-specific domains which interact with the EEEV HVD. To avoid the effect of endogenous proteins on binding of the deletion protein variants under examination, the G3BP1 mutant- and FXR1 mutant-expressing cell lines were established in G3bp dKO and Fxr tKO NIH 3T3 cells, respectively. Previously published data from this group showed that the replication of SINV replicons is G3BP dependent and G3bp dKO cells do not support the replication of SINrep/Flag-GFP-HVD constructs (23). Therefore, in G3bp dKO cells and in the derived stable cell lines expressing G3BP1 fragments, Flag-GFP-HVDeeev was expressed from a VEEV replicon. In Fxr tKO cells and their derivatives, Flag-GFP-HVDeeev was produced by the SINrep/Flag-GFP-HVDeeev construct (Fig. 1). Protein complexes were isolated at 2.5 h p.i. and analyzed by Western blotting using GFP-specific antibodies. The results presented in Fig. 8C demonstrate that the FXR1Δ1-GFP variant, which lacks the Agenet-like domain, and the G3BP1Δ1-GFP variant, which lacks the NTF2-like domain, were the only proteins absent from the co-IP samples. Thus, these domains appear to be the main contributors to FXR and G3BP binding to the EEEV nsP3 HVD. In addition, despite demonstrating very low similarity in the G3BP-binding peptide, EEEV- and CHIKV-specific HVDs interact with the same NTF2-like domain of G3BP (Fig. 8C) (26).
FIG 8.
Binding of G3BP1 and FXR1 to the EEEV nsP3 HVD is mediated by their NTF2-like and Agenet-like domains, respectively. (A and B) Schematic representations of VEEV (A) and SINV (B) replicons encoding the Flag-GFP-HVDeeev fusion protein and schematic representation of full-length and deletion mutant G3BP1-GFP (A) and FXR1-GFP (B) fusions stably expressed in G3bp dKO- and Fxr tKO-based cell lines. (C) The indicated cell lines were infected with packaged VEErep/Flag-GFP-HVDeeev or SINrep/Flag-GFP-HVDeeev replicons at an MOI of 20 inf.u/cell. At 2.5 h p.i., cells were harvested, and after lysis, protein complexes were isolated on magnetic beads loaded with Flag-specific MAbs. The presence of G3BP1 and FXR1 variants in isolated complexes and cell lysates was analyzed by Western blotting using GFP-specific MAbs.
HVD-FXR and HVD-G3BP interactions play critical roles in EEE/SINV and wild-type (wt) EEEV replication.
The above-described experiments provided new critical data about EEEV HVD-G3BP and EEEV HVD-FXR interactions but did not present direct evidence that these interactions play an essential role(s) in RNA and virus replication. Therefore, we sought to evaluate the effects of deletions of FXR- and G3BP-binding sites on the replication of EEE/nsP3-GFP/SINV. The ΔG, ΔF, and Δ(G+F) mutants contained deletions of the G3BP-, FXR-, and both G3BP- and FXR-binding sequences, respectively (Fig. 9A). The in vitro-synthesized RNAs were electroporated into BHK-21 cells. In the infectious center assay, the in vitro-synthesized RNAs, which contained deletions of single sites in the nsP3-coding sequence, displayed infectivities similar to the infectivity of the original EEE/nsP3-GFP/SINV (Fig. 9A). This was an indication that the new constructs did not need to acquire additional adaptive mutations for their viability. At 24 h postelectroporation, the titers of these mutants were also similar to the titer of parental EEE/nsP3-GFP/SINV (Fig. 9A). The electroporation-derived EEE/nsP3-GFPΔG/SINV was able to replicate with essentially the same efficiency in both NIH 3T3 and G3bp dKO cells, but its replication rates in Fxr tKO cells were dramatically lower (Fig. 9B). EEE/nsP3-GFPΔF/SINV, in turn, replicated at similar rates in NIH 3T3 and Fxr tKO cells, but its replication in G3bp dKO cells was a few orders of magnitude less efficient (Fig. 9B). Thus, the lack of an EEEV nsP3 interaction with both the G3BP and FXR protein families strongly affects the rates of virus replication.
FIG 9.
Lack of interaction with both the G3BP and FXR families of proteins profoundly reduces the efficiency of replication of chimeric viruses encoding EEEV nsPs. (A) Schematic representations of the EEE/nsP3-GFP/SINV chimeric virus genome and mutated nsP3-GFP fusion proteins. The indicated deletions were the same as those presented in Fig. 7. The table presents the infectivities of the in vitro-synthesized RNAs in the infectious center assay and the titers of infectious viruses harvested at 24 h postelectroporation (PEP) of 1 μg of the in vitro-synthesized RNAs into BHK-21 cells. (B) Fxr tKO, G3bp dKO, and parental NIH 3T3 cells were infected with the indicated chimeric viruses at an MOI of 0.5 PFU/cell. Titers were determined at 8 and 24 h postinfection. The results represent averages from three experiments.
In agreement with these data, the in vitro-synthesized RNA of the double deletion mutant EEE/nsP3-GFPΔ(G+F)/SINV performed with a 2-order-of-magnitude lower efficiency in the infectious center assay. The rescued virus also formed pinpoint-size plaques, and the BHK-21 cell-derived stocks had titers below 106 PFU/ml (Fig. 9A). Thus, the double deletion did not make EEE/nsP3-GFPΔ(G+F)/SINV nonviable but did have a deleterious effect on replication rates. This suggested that other cellular factors, for example, those presented in Table 1, also appear to be capable of supporting virus replication. However, in the absence of an HVD interaction with both FXRs and G3BPs, the replication rates were insufficient to produce titers high enough for further analysis. The attempts to select more efficiently replicating variants by serial passage of EEE/nsP3-GFPΔ(G+F)/SINV in BHK-21 or NIH 3T3 cells were unsuccessful. Titers remained very low, and no further experiments with this double mutant were performed.
In parallel experiments, the effect of HVD interactions with different host factors was evaluated in the context of wt EEEV FL93. For this study, we designed an EEEV genome in which the natural nsP3 HVD was replaced with an artificial HVD variant (EEEV/artHVD) containing the amino acids of the wt, but in a different order (Fig. 10A). Thus, this artificial HVD had the same amino acid content as wt HVD and a hydrophobicity profile similar to that of wt HVD, but all of the protein-binding sites identified above and any as yet unidentified protein-binding sites were mutated. Another variant, EEEV/artHVD+F+G, had the same disordered HVD, but the G3BP- and FXR-binding sites were cloned back into positions similar to those in the wt sequence. EEEV/ΔG, EEEV/ΔF, and EEEV/Δ(G+F) encoded wt HVDs in which short sequences of the G3BP-, the FXR-, and both the G3BP- and FXR-binding sites, respectively, were deleted (see Fig. 7 for details). The in vitro-synthesized RNAs were transfected into BHK-21 cells. All of the designed variants were viable, but the titer of the EEEV/artHVD stock harvested at 24 h posttransfection was 2 orders of magnitude lower than that of the wt virus. Nevertheless, it was sufficient for further analysis of virus replication in different cell types.
FIG 10.
EEEV nsP3 HVD interactions with G3BPs and FXRs and other host proteins determine the rates of virus replication. (A) Schematic representation of the EEEV genome and modifications introduced into the nsP3 HVD. (B) The indicated cell lines were infected with the designed viruses at an MOI of 1 PFU/cell. Then, the cells were washed 5 times with complete medium and incubated for 7 h (NIH 3T3 and Vero cells) or 8 h (BHK-21 cells). The titers of harvested samples were determined by plaque assay on BHK-21 cells. n.d., the titer was below the limit of detection, which was 50 PFU/ml.
Vero, NIH 3T3, and BHK-21 cells were infected at an MOI of 0.5 PFU/cell, and the titers of the released viruses were determined at 7 h p.i. The data presented in Fig. 10B demonstrate that replacement of the wt HVD with the designed, randomized sequence made EEEV/artHVD essentially nonviable in Vero and NIH 3T3 cells, despite its ability to replicate in BHK-21 cells. Addition of FXR- and G3BP-binding sequences to artHVD allowed EEEV/artHVD+F+G to replicate in Vero and NIH 3T3 cells; however, the titers remained lower than those of wt EEEV. As in the case of the EEE/nsP3-GFP/SINV chimeric virus, deletion of either the G3BP- or FXR-binding site produced only small negative effects, if any, on EEEV replication in the indicated cell types. However, the deletion of both sites affected virus release from Vero and NIH 3T3 cells at 7 h p.i. by more than 3 and 2 orders of magnitude, respectively.
Taken together, the results demonstrate that the deletion of both binding sites while having other HVD sequences intact has a strong negative effect on EEEV replication. However, HVD interactions with proteins other than the G3BP and FXR host factors were able to support virus viability in the cell lines used in these experiments. Similarly, in the absence of other amino acid sequences which could putatively interact with as yet unknown host factors, FXR-HVD and G3BP-HVD interactions were able to drive EEEV/artHVD+F+G replication but not as efficiently as in the context of the wt EEEV HVD.
DISCUSSION
The results of our previous study demonstrated that the intrinsically disordered, hypervariable domains of alphavirus nsP3 proteins are critical players in the formation of viral vRCs and viral replication (23–25). HVDs interact with sets of host factors which are specific for each alphavirus representative. To date, among the varied host proteins pulled down in co-IP experiments with alphavirus nsP3 HVDs, G3BPs and FXRs were the most abundant (23). The CHIKV and SINV nsP3 HVDs were found to interact with both G3BP1 and G3BP2 but not with FXRs, while the VEEV nsP3 HVD binds all three FXRs but not G3BPs. Accordingly, G3BPs were found to function in the replication of the OW arthritogenic alphaviruses through binding to the carboxy-terminal repeating amino acid sequences of the nsP3 HVD, resulting in assembly of viral pre-vRCs at the plasma membrane (23, 27, 35). Interference with HVD's ability to interact with G3BPs, caused either by deletion of the identified carboxy-terminal repeating peptides or by KO of both G3bp genes in NIH 3T3 cells, strongly affected the rates of SINV replication and made CHIKV essentially nonviable (23). Similarly, the VEEV nsP3 HVD interaction with FXRs is also determined by a unique carboxy-terminal amino acid repeat (23, 25). This interaction is a critical determinant of VEEV RC assembly, and its alteration by deletion of FXR-binding sites in the VEEV HVD or by KO of all three FXR genes has a strong negative effect on both the infectivity of VEEV and its replication rates.
The use of entire families of G3BP and FXR proteins by SINV/SFV/CHIKV and VEEV, respectively, appears to be an excellent evolutionary strategy to employ multiple proteins with redundant functions for recruitment of viral G RNA to pre-vRCs and efficient vRC formation. It is also tempting to speculate that the encephalitogenic phenotype of the NW alphaviruses and arthritogenic phenotype of the OW alphaviruses are at least partially determined by the abilities of these viruses to utilize different families of cellular factors. However, the results of this study demonstrate that this is not entirely true. In contrast to previously studied members of the alphavirus genus, the EEEV nsP3 HVD interacts with both FXRs and G3BPs and a number of other cellular proteins (Table 1). Moreover, the data also show that efficient EEEV replication can be supported by one of these cellular protein families in the absence of the other. However, disruption of both G3BP-HVD and FXR-HVD interactions has profound negative effects on virus replication. Thus, NA EEEV, which is the most pathogenic representative of the NW encephalitogenic alphaviruses, exhibits an additional level of redundancy in utilizing host factors and is capable of exploiting two protein families, FXRs and G3BPs, for efficient replication.
In our previous studies, it was found that replacement of the VEEV (a NW alphavirus) HVD with that of an OW alphavirus, e.g., SINV, allowed efficient replication and led to the production of virus at titers similar to those of wt VEEV (24). However, the opposite approach yielded different results. In the case of SINV, replacement of G3BP-binding sequences with FXR-binding sequences had a deleterious effect on SINV replication. In a similar experiment, replacement of G3BP-binding sites in the CHIKV HVD by the FXR-binding amino acid sequence made the chimeric CHIKV nonviable (23, 24). Thus, it is reasonable to speculate that the FXR-HVD interactions developed later in alphavirus evolution than binding of G3BPs to the HVD. This assumption provides a plausible explanation for why G3BP-dependent CHIKV and SINV cannot utilize FXRs even if the binding sequences are inserted into their HVDs. VEEV has apparently already lost the G3BP-binding sites in its HVD but retains an ability to use these proteins in vRC formation, if the corresponding binding sites are inserted to replace those interacting with FXRs (24). EEEV, however, appears to represent an evolutionary stepping stone in nsP3 HVD development. It has retained the G3BP-binding site and has attained the FXR-binding peptide in its HVD. The use of two cellular protein families known for their ability for self-assembly into higher-order complexes may be a factor contributing to efficient replication and the highly cytopathic phenotype of NA EEEV. Moreover, utilization of two protein families may be beneficial for virus replication in different cell types which have various levels of expression of each of these different family members.
The FXR and G3BP proteins demonstrate similarities in their domain structures (36, 37). Their very amino-terminal domains (an NTF2-like domain in G3BP and an Agenet-like domain in FXR) are responsible for interaction with the EEEV nsP3 HVD (Fig. 8). On the basis of our previous results (23) and those from this study, the downstream carboxy-terminal domains, with the exception of the acidic domain of G3BP, are essential for these proteins' function in vRC formation and RNA replication but are dispensable for binding to HVD. In contrast to HVD interactions with G3BPs and FXRs, described for CHIKV and VEEV, the EEEV HVD contains single binding sites for both FXRs and G3BPs, and interaction with the proteins belonging to one family does not depend on binding to those belonging to another family. This independent binding is another factor differentiating EEEV and CHIKV, in which two G3BP molecules bind to the peptide repeats with different affinities (26).
Importantly, G3BPs and FXRs are the most abundant, but they were not the only cellular proteins identified in the co-IP experiments as interacting with the EEEV nsP3 HVD (Table 1). Defined combinations of the RNA-binding, BAR domain-containing, and actin-interacting proteins were readily detectable in the co-IP samples, suggesting that they are also involved in virus replication. Indeed, removal of EEEV's ability to interact with both the FXR and G3BP proteins did not prevent EEE/SINV or EEEV replication in mouse, primate, or hamster cells, though its efficiency was reduced by a few orders of magnitude (Fig. 10). In agreement with these results, EEEV/artHVD+F+G, which encodes FXR- and G3BP-binding sites in an otherwise disordered HVD, replicated with dramatically less efficiency than the wt virus. This is an additional indication that other HVD interactions are important in facilitating optimal EEEV replication.
Previous studies on BHK-21 cells have generated somewhat surprising data (25). These cells are far more amenable to replication of VEEV nsP3 HVD mutants than other cell types. VEEV TC-83 retains the ability to replicate in BHK-21 cells, albeit with a lower efficiency, but not in any other tested cell lines, even in the complete absence of its nsP3 HVD. To achieve an RNA replication level which was sufficient for virus spread, it had to acquire only a single adaptive mutation in the nsP2 or nsP3 protein (25). In the case of EEEV, randomization of the entire nsP3 HVD also made the corresponding mutant virus nonviable in Vero and NIH 3T3 cells but not in BHK-21 cells. In the latter cell line, it is able to achieve infectious titers close to 108 PFU/ml. This suggests the existence of additional proviral interactions of nsPs with host factors, at least in BHK-21 cells. The extraordinary permissiveness of BHK-21 cells in supporting the replication of alphaviruses and other viruses and their mutants certainly deserves further investigation. It also raises a concern about the applicability of BHK-21 cell-derived data to other cell lines.
In summary, the results of this study demonstrate that (i) EEEV occupies an intermediate position between the previously studied OW and NW alphavirus representatives. Its nsP3-specific HVD contains binding sites for both FXR and G3BP proteins, and these sites are presented in single copies. (ii) The FXR and G3BP protein families independently interact with the EEEV HVD, and each of them can support efficient EEEV replication. The proviral effect is likely determined by the ability of G3BP and FXR proteins to bind RNA and to self-assemble into higher-order complexes. (iii) Abrogation of the EEEV interaction with both protein families simultaneously has a dramatic negative effect on EEEV replication. (iv) There are other EEEV nsP3 HVD-interacting host factors identified in this study that appear to support EEEV replication, but with a lower efficiency than FXRs and G3BPs. These results suggest a new means for attenuation of EEEV by manipulating combinations of HVD-binding host factors, resulting in an alteration of this virus's ability to replicate in different cell types. FXR and G3BP binding to their corresponding sites in the EEEV HVD can also be exploited in the screening of drugs which specifically alter the assembly of EEEV replication complexes. Moreover, further in vitro studies of EEEV HVD interactions with host factors will provide an opportunity to identify new drugs without working under biosafety level 3 (BSL3) conditions.
MATERIALS AND METHODS
Cell cultures.
NIH 3T3 and Vero cells were obtained from the American Type Culture Collection (Manassas, VA). BHK-21 cells were kindly provided by Paul Olivo (Washington University, St. Louis, MO). These cell lines were maintained at 37°C in alpha minimum essential medium (αMEM) supplemented with 10% fetal bovine serum (FBS) and vitamins. The G3bp dKO and Fxr tKO cell lines have been described elsewhere (23). They were generated by making sequential knockouts (KO) of all of the members of the G3BP and FXR families in NIH 3T3 cells. The cell lines expressing deletion mutants of G3BP1 and FXR1 in the corresponding dKO and tKO cells were designed in the previous study (23).
Plasmid constructs.
The original plasmids containing the infectious cDNAs of the NA EEEV Florida93 (FL93) strain and the CHIKV 181/25 strain were kindly provided by Scott Weaver (University of Texas Medical Branch, Galveston, TX). Plasmids containing infectious cDNA of VEEV TC-83 and EEE/SINV have been described elsewhere (30). The cDNAs of the genomes of the EEE/nsP3-Cherry/SINV and EEE/nsP3-GFP/SINV chimeric viruses encoded EEEV-specific nonstructural proteins of the Florida91 (FL91) strain, while the structural genes were derived from the laboratory strain SINV Toto1101 (38). They had the same design, in that the fluorescent Cherry- and GFP-coding sequences were inserted in frame into the amino-terminal fragment of the HVDs. Other plasmids containing infectious cDNAs of VEEV/nsP3-Cherry and CHIKV/nsP3-Cherry had essentially the same design, and the Cherry protein-coding sequence was inserted in frame into the amino termini of the corresponding HVDs. Plasmids carrying cDNAs of SINV and VEEV replicons containing Flag-GFP or Flag-GFP fused with HVDs were described elsewhere (23). Plasmids carrying SINV and VEEV replicons contained Flag-GFP or Flag-GFP, which was fused in frame with either full-length EEEV HVD or its modified versions, under the control of the subgenomic promoter to replace the structural genes. They were designed using standard PCR-based techniques. Plasmids carrying the genomes of helper constructs, which were used for packaging of replicon RNAs into infectious viral particles, were described elsewhere (39, 40). Plasmids containing cDNAs of the EEEV and EEE/nsP3-GFP/SINV genomes with modified HVDs were also designed using standard PCR-based techniques. Schematic representations of all of the modified genomes are shown in the corresponding figures. The sequences of the plasmids and details of the cloning procedures can be provided upon request.
In vitro RNA transcription and transfection.
Plasmids were purified by ultracentrifugation in CsCl gradients. Then they were linearized using unique restriction sites located downstream of the poly(A) sequence. RNAs were synthesized by the use of SP6 RNA polymerase in the presence of a cap analog (New England BioLabs) according to the manufacturer's recommendations (Invitrogen). Aliquots of the transcription reaction mixtures were used for electroporation without additional purification. Electroporation of BHK-21 cells by in vitro-synthesized viral genomes was performed under previously described conditions (41, 42). Replicon genomes were packaged by coelectroporation of their in vitro-synthesized RNA and homologous helper RNAs. Viruses and replicons were harvested at 20 to 24 h postelectroporation. Virus titers were determined by a plaque assay on BHK-21 cells (43). The infectious titers of packaged replicons were determined by infecting BHK-21 cells (5 × 105 cells/well) in 6-well Costar plates with 10-fold dilutions of the samples and counting the number of GFP-positive cells at 6 h postinfection (p.i.). The titers of all of the packaged replicons were between 4 × 109 and 10 × 109 infectious units per ml, and these stocks were directly used in further experiments. Wild-type EEEV FL93 and its HVD mutants were rescued in BHK-21 cells using the TransIT-mRNA reagent according to the manufacturer's instructions (Mirus). In vitro RNA synthesis, virus rescue, and analysis of EEEV replication were performed in the BSL3 SEBLAB of the University of Alabama at Birmingham.
Infectious center assay.
To compare the infectivities of the viral RNAs, BHK-21 cells were electroporated with 1 μg of the in vitro-synthesized genomic RNAs. Tenfold dilutions of electroporated cells were seeded in 6-well Costar plates containing subconfluent monolayers of naive BHK-21 cells. After 2 h of incubation at 37°C, cells were overlaid with agarose supplemented with minimal essential medium and 3% FBS. Plaques were stained after 2 days of incubation at 37°C, and RNA infectivity was determined as the number of PFU per microgram of transfected RNA.
Isolation and identification of cellular proteins that interact with alphavirus HVDs.
NIH 3T3 cells (1 × 107) were infected with viral particles containing replicon genomes encoding Flag-GFP, Flag-GFP-HVDeeev, Flag-GFP-HVDveev, Flag-GFP-HVDchikv, and Flag-GFP-HVDsinv at a multiplicity of the infection (MOI) of 20 infectious units per cell. Cells were harvested at 2 to 3 h p.i., when GFP expression remained barely detectable and, thus, the concentrations of the expressed fusion proteins were low. Protein complexes were isolated from the post-nuclear fraction using magnetic beads loaded with a Flag-specific monoclonal antibody (MAb) as described elsewhere (23). Proteins were separated on 4 to 12% NuPAGE gels (Invitrogen) and, after staining with Coomassie blue, identified by mass spectrometry as previously described (23). An identified protein was selected as being specifically bound to particular HVDs if it was not present in control immunoprecipitations (IPs) with Flag-GFP and the total number of spectra for a protein was more than 6 in at least one sample.
Analysis of virus replication.
Cells were seeded into 35-mm dishes and infected at the MOIs indicated in the figure legends. At the indicated times, media were harvested and the virus titers in the samples were determined by plaque assay on BHK-21 cells.
Confocal microscopy.
Cells were seeded in 8-well Ibidi chambers (5 × 103/well) and incubated overnight at 37°C. They were then infected with the viruses indicated in the figures. At the times postinfection indicated in the figure legends, cells were fixed with 4% paraformaldehyde (PFA) for 15 min, permeabilized, and stained with the antibodies indicated above. The following primary antibodies were used: an anti-dsRNA mouse monoclonal antibodies (MAb J2 or MAb K1; Scicons, Hungary), anti-G3BP1 rabbit polyclonal antibodies (a gift from Richard Lloyd), anti-G3BP2 rabbit polyclonal antibodies (catalog number A302-040; Epitomics), anti-FXR1 rabbit monoclonal antibodies (catalog number 12295; Cell Signaling), anti-FXR2 rabbit monoclonal antibodies (catalog number 7098; Cell Signaling), and anti-FMR1 rabbit monoclonal antibodies (catalog number 7104; Cell Signaling). Cell nuclei were stained with Hoechst dye. Three-dimensional images were acquired on a Zeiss LSM700 confocal microscope with a 63× PlanApochromat oil objective (numerical aperture, 1.4). The image stacks were deconvolved using the point spread function value measured in Huygens software (Scientific Volume Imaging), and images were assembled using Imaris software (Bitplane AG). Colocalization parameters were determined using Huygens and Imaris software. The images of mock-infected cells were acquired as stacks of 6 optical sections and not subjected to deconvolution.
ACKNOWLEDGMENTS
We thank Niall J. Foy for helpful discussions and critical reading and editing of the manuscript.
This work was supported by Public Health Service grants AI118867 to E.I.F. and AI095449 to I.F.
REFERENCES
- 1.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zacks MA, Paessler S. 2010. Encephalitic alphaviruses. Vet Microbiol 140:281–286. doi: 10.1016/j.vetmic.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. 2004. Venezuelan equine encephalitis. Annu Rev Entomol 49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- 4.Gibbs EP. 1976. Equine viral encephalitis. Equine Vet J 8:66–71. doi: 10.1111/j.2042-3306.1976.tb03293.x. [DOI] [PubMed] [Google Scholar]
- 5.Gibney KB, Robinson S, Mutebi JP, Hoenig DE, Bernier BJ, Webber L, Lubelczyk C, Nett RJ, Fischer M. 2011. Eastern equine encephalitis: an emerging arboviral disease threat, Maine, 2009. Vector Borne Zoonotic Dis 11:637–639. doi: 10.1089/vbz.2010.0189. [DOI] [PubMed] [Google Scholar]
- 6.Go YY, Balasuriya UB, Lee CK. 2014. Zoonotic encephalitides caused by arboviruses: transmission and epidemiology of alphaviruses and flaviviruses. Clin Exp Vaccine Res 3:58–77. doi: 10.7774/cevr.2014.3.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver SC. 2001. Venezuelan equine encephalitis, p 539–548. In Service MW. (ed), The encyclopedia of arthropod-transmitted infections. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 8.Weaver SC. 2001. Eastern equine encephalitis, p 151–159. In Service MW. (ed), The encyclopedia of arthropod-transmitted infections. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 9.Reisen WK. 2001. Western equine encephalitis, p 558–563. In Service MW. (ed), The encyclopedia of arthropod-transmitted infections. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 10.Reed DS, Larsen T, Sullivan LJ, Lind CM, Lackemeyer MG, Pratt WD, Parker MD. 2005. Aerosol exposure to western equine encephalitis virus causes fever and encephalitis in cynomolgus macaques. J Infect Dis 192:1173–1182. doi: 10.1086/444397. [DOI] [PubMed] [Google Scholar]
- 11.Vogel P, Kell WM, Fritz DL, Parker MD, Schoepp RJ. 2005. Early events in the pathogenesis of eastern equine encephalitis virus in mice. Am J Pathol 166:159–171. doi: 10.1016/S0002-9440(10)62241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemm JA, Rice CM. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol 67:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus strand RNA synthesis. J Virol 185:1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol 82:6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver SC, Dalgarno L, Frey TK, Huang HV, Kinney RM, Rice CM, Roehrig JT, Shope RE, Strauss EG. 2000. Family Togaviridae, p 879–889. In van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeogh DJ, Pringle CR, Wickner RB (ed), Virus taxonomy. Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 16.Neuvonen M, Ahola T. 2009. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol 385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rack JG, Perina D, Ahel I. 2016. Macrodomains: structure, function, evolution, and catalytic activities. Annu Rev Biochem 85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- 18.McPherson RL, Abraham R, Sreekumar E, Ong SE, Cheng SJ, Baxter VK, Kistemaker HA, Filippov DV, Griffin DE, Leung AK. 2017. ADP-ribosylhydrolase activity of chikungunya virus macrodomain is critical for virus replication and virulence. Proc Natl Acad Sci U S A 114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckei L, Krieg S, Butepage M, Lehmann A, Gross A, Lippok B, Grimm AR, Kummerer BM, Rossetti G, Luscher B, Verheugd P. 2017. The conserved macrodomains of the non-structural proteins of chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci Rep 7:41746. doi: 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. 2016. Viral macro domains reverse protein ADP-ribosylation. J Virol 90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. 2012. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A 109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, Frolova EI. 2016. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog 12:e1005810. doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, Frolova EI. 2013. Hypervariable domains of nsP3 proteins of New World and Old World alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol 87:1997–2010. doi: 10.1128/JVI.02853-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foy NJ, Akhrymuk M, Shustov AV, Frolova EI, Frolov I. 2013. Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J Virol 87:7569–7584. doi: 10.1128/JVI.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte T, Liu L, Panas MD, Thaa B, Dickson N, Gotte B, Achour A, McInerney GM. 2016. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol 6:160078. doi: 10.1098/rsob.160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panas MD, Ahola T, McInerney GM. 2014. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 88:5888–5893. doi: 10.1128/JVI.00439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panas MD, Varjak M, Lulla A, Eng KE, Merits A, Karlsson Hedestam GB, McInerney GM. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuvonen M, Kazlauskas A, Martikainen M, Hinkkanen A, Ahola T, Saksela K. 2011. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog 7:e1002383. doi: 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DY, Atasheva S, Foy NJ, Wang E, Frolova EI, Weaver S, Frolov I. 2011. Design of chimeric alphaviruses with a programmed, attenuated, cell type-restricted phenotype. J Virol 85:4363–4376. doi: 10.1128/JVI.00065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frolova E, Gorchakov R, Garmashova N, Atasheva S, Vergara LA, Frolov I. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol 80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholte FE, Tas A, Albulescu IC, Zusinaite E, Merits A, Snijder EJ, van Hemert MJ. 2015. Stress granule components G3BP1 and G3BP2 play a proviral role early in chikungunya virus replication. J Virol 89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamanini F, Van Unen L, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. 1999. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. Biochem J 343(Pt 3):517–523. [PMC free article] [PubMed] [Google Scholar]
- 37.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Rice CM, Levis R, Strauss JH, Huang HV. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol 61:3809–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol 67:6439–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkova E, Gorchakov R, Frolov I. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315–327. doi: 10.1016/j.virol.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J Virol 65:4107–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorchakov R, Hardy R, Rice CM, Frolov I. 2004. Selection of functional 5′ cis-acting elements promoting efficient Sindbis virus genome replication. J Virol 78:61–75. doi: 10.1128/JVI.78.1.61-75.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemm JA, Durbin RK, Stollar V, Rice CM. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol 64:3001–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]