ABSTRACT
We previously isolated a porcine epidemic diarrhea virus (PEDV) strain, PC177, by blind serial passaging of the intestinal contents of a diarrheic piglet in Vero cell culture. Compared with the highly virulent U.S. PEDV strain PC21A, the tissue culture-adapted PC177 (TC-PC177) contains a 197-amino-acid (aa) deletion in the N-terminal domain of the spike (S) protein. We orally inoculated neonatal, conventional suckling piglets with TC-PC177 or PC21A to compare their pathogenicities. Within 7 days postinoculation, TC-PC177 caused mild diarrhea and lower fecal viral RNA shedding, with no mortality, whereas PC21A caused severe clinical signs and 55% mortality. To investigate whether infection with TC-PC177 can induce cross-protection against challenge with a highly virulent PEDV strain, all the surviving piglets were challenged with PC21A at 3 weeks postinoculation. Compared with 100% protection in piglets initially inoculated with PC21A, 88% and 100% TC-PC177- and mock-inoculated piglets had diarrhea following challenge, respectively, indicating incomplete cross-protection. To investigate whether this 197-aa deletion was the determinant for the attenuation of TC-PC177, we generated a mutant (icPC22A-S1Δ197) bearing the 197-aa deletion from an infectious cDNA clone of the highly virulent PEDV PC22A strain (infectious clone PC22A, icPC22A). In neonatal gnotobiotic pigs, the icPC22A-S1Δ197 virus caused mild to moderate diarrhea, lower titers of viral shedding, and no mortality, whereas the icPC22A virus caused severe diarrhea and 100% mortality. Our data indicate that deletion of this 197-aa fragment in the spike protein can attenuate a highly virulent PEDV, but the virus may lose important epitopes for inducing robust protective immunity.
IMPORTANCE The emerging, highly virulent PEDV strains have caused substantial economic losses worldwide. However, the virulence determinants are not established. In this study, we found that a 197-aa deletion in the N-terminal region of the S protein did not alter virus (TC-PC177) tissue tropism but reduced the virulence of the highly virulent PEDV strain PC22A in neonatal piglets. We also demonstrated that the primary infection with TC-PC177 failed to induce complete cross-protection against challenge by the highly virulent PEDV PC21A, suggesting that the 197-aa region may contain important epitopes for inducing protective immunity. Our results provide an insight into the role of this large deletion in virus propagation and pathogenicity. In addition, the reverse genetics platform of the PC22A strain was further optimized for the rescue of recombinant PEDV viruses in vitro. This breakthrough allows us to investigate other virulence determinants of PEDV strains and will provide knowledge leading to better control PEDV infections.
KEYWORDS: porcine epidemic diarrhea virus, coronavirus, spike protein, pathogenesis, pathogenicity, virulence, reverse genetics, 3D structural modeling, enteric pathogens
INTRODUCTION
Porcine epidemic diarrhea virus (PEDV) is a highly virulent and contagious porcine enteric pathogen. It causes up to 100% mortality in neonatal piglets under 1 week of age and milder enteritis in older pigs (1, 2). Since 2010, the emerging, highly virulent PEDV strains have decimated swine farms in China and spread to other Asian countries, North and South America, and Europe, causing substantial economic losses (3, 4). During the first year of PEDV outbreaks in the United States, about 7 million piglets died, resulting in a $9 million to $1.8 billion annual loss for the U.S. economy (4, 5). These large-scale PEDV outbreaks ended in 2014. Subsequently, PEDV outbreaks occurred sporadically in the United States (Swine Health Monitoring Surveillance, USDA [https://www.aphis.usda.gov/]). However, PEDV outbreaks still cause major economic problems in many Asian countries. For example, it has been estimated that more than 490,000 pigs died due to the outbreak of PEDV during October 2013 to August 2015 in Japan (6). PEDV belongs to the genus Alphacoronavirus within the family Coronaviridae. Its 28-kb positive-sense, single-stranded RNA genome contains six open reading frames (ORFs), encoding polyproteins pp1a and pp1ab that are processed to generate 16 nonstructural proteins (nsp1 to nsp16) and the spike (S), ORF3, envelope (E), membrane (M), and nucleocapsid (N) proteins.
The glycosylated S protein is a receptor binding ligand that can be divided into S1 and S2 subunits based on functional domains (7). The S1 subunit contains the receptor binding domains that bind to the putative receptors aminopeptidase N (APN) and sialic acid (N-acetylneuraminic acid [Neu5Ac], whereas a fusion peptide within the S2 subunit mediates virus-cell membrane fusion during virus entry (8). Although the cellular receptor for PEDV is still under study and controversial (9–11), it is hypothesized that a known neutralizing epitope, the CO-26K equivalent epitope (COE; residues 499 to 638), within the C-terminal domain (CTD) of S1 contains the APN-binding domain (12). The N-terminal domain (NTD; residues 19 to 233) of S1 is responsible for sialic acid binding (8, 13, 14). Also, this NTD is dispensable for PEDV replication in cell culture and pigs since PEDV variants bearing large deletions (ranging from 194 to 216 amino acids [aa]) within the S1 NTD have been reported (6), including the tissue culture-adapted PC177 (TC-PC177) strain (197-aa deletion; residues 34 to 230) (15) and strain TTR-2 (194-aa deletion; residues 23 to 216) (16). The 197-aa deletion of the PC177 strain occurred during the isolation of this virus in Vero cell culture, whereas the TTR-2 strain was detected in a pig farm in Japan. These viruses are designated the S1 NTD deletion (NTD-del) type. Similar large deletions in the S1 NTD (about 200 to 230 aa) were found in porcine respiratory coronavirus ([CoV] PRCV), a naturally occurring mutant of the enteric alphacoronavirus transmissible gastroenteritis virus (TGEV) (17). PRCV is an attenuated variant with reduced enteric tropism and increased tropism for the respiratory tract that causes subclinical to mild respiratory disease (18). Additionally, due to its close antigenic relatedness, infection of PRCV provides partial protective immunity against TGEV infection (19–21).
To date, the pathogenicity of the S1 NTD-del type of PEDV variants has not been well established in vivo. Experimental infection of 6-day-old colostrum-deprived piglets infected with the TTR-2 strain revealed that the virulence of this mutant was reduced compared with that of the highly virulent strain OKN-1 (22). However, in addition to the 194-aa deletion in the S1 NTD, TTR-2 also contains an additional 28-amino-acid substitution compared with the sequence of the virulent control OKN-1 strain. These 28 amino acids are located in the ORF1a/b (21 aa), S (6 aa), and N (1 aa) genes and might also influence the reduced pathogenicity of TTR-2. Whether the large deletion in the NTD of S1 alone is responsible for the reduced virulence of PEDV was not confirmed using standard reverse genetics approaches.
Previously, we constructed a full-length cDNA clone of the highly virulent PEDV strain PC22A (icPEDV, infectious clone PEDV) (23). To increase the stability of plasmids and improve recovery of recombinant viruses, we generated an optimized reverse genetics platform for PEDV, icPC22A. Using this second generation of the reverse genetics platform, we deleted the 197-aa region in the NTD of S1 from a highly virulent PEDV strain to confirm the relationship between the S1 NTD and the pathogenicity of a PEDV strain.
We hypothesized that the 197-aa deletion in S1 NTD can attenuate a PEDV strain in neonatal piglets but still induce protective immunity against highly virulent PEDV strains. To test the hypothesis, we inoculated conventional suckling piglets with the tissue culture-adapted PEDV PC177 (TC-PC177) strain and challenged the surviving pigs at 3 weeks of age with the highly virulent PEDV PC21A strain. Furthermore, we generated a deletion mutant virus by introducing the same 197-aa deletion into the S1 subunit of icPC22A (icPC22A-S1Δ197). We then demonstrated that the pathogenicity of this mutant was reduced in neonatal gnotobiotic (Gn) piglets.
RESULTS
The virulence of TC-PC177 was reduced in conventional suckling piglets.
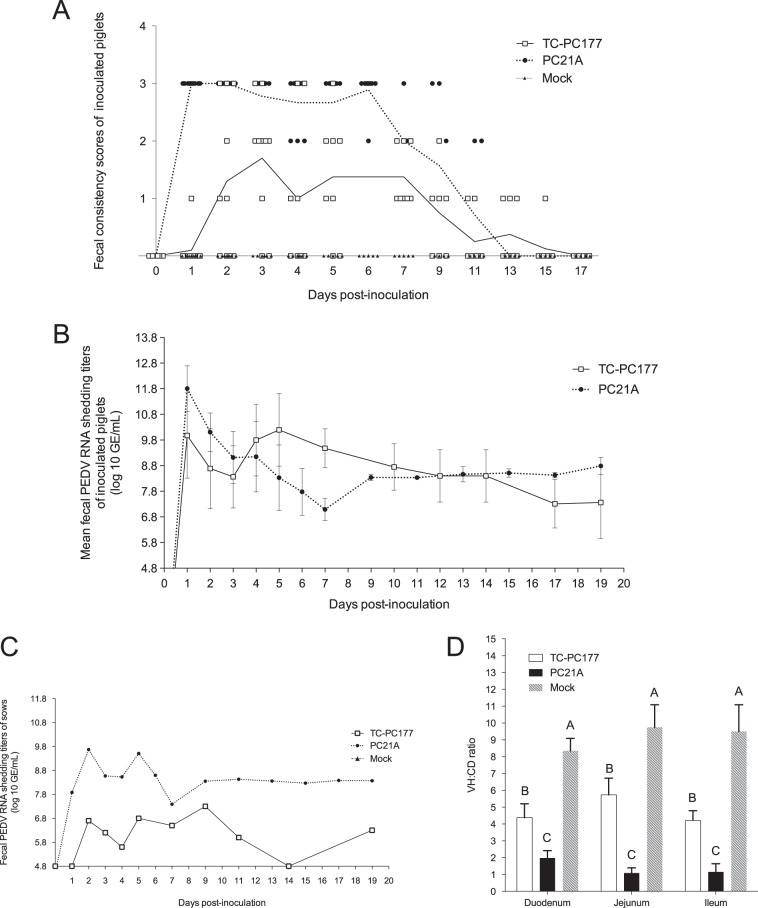
To evaluate the pathogenicity of TC-PC177, 4-day-old conventional suckling piglets were orally inoculated with TC-PC177 P10C8, highly virulent PC21A, or phosphate-buffered saline ([PBS] mock). Clinical signs observed in the acute-infection phase (1 to 11 days postinoculation [dpi]) are summarized in Table 1. In contrast to the PC21A-inoculated group, in which all piglets (11/11) developed diarrhea within 1 dpi, 80% (8/10) of TC-PC177-inoculated piglets had diarrhea with a significantly delayed onset at 2.50 ± 0.53 dpi. No pigs had diarrhea, and no virus shedding was detected in the mock inoculation group. No mortality was observed in the TC-PC177 and mock groups, whereas 55% (6/11) of piglets in the PC21A group died. Evaluation of the fecal consistency (FC) scores (Fig. 1A; Table 1) displayed lower mean scores in the TC-PC177 group than those in the PC21A group in the first 11 dpi. The highest fecal PEDV RNA titer appeared later (5 dpi) than that in the PC21A group, and the titer was significantly lower (P < 0.05) in the TC-PC177 group (Fig. 1B and Table 1). The duration of diarrhea in the TC-PC177-inoculated group was 2.22 ± 2.17 days, which was significantly shorter than that (7.33 ± 1.18 days) of the PC21A-inoculated group (P < 0.05).
TABLE 1.
General litter information and clinical signs of conventional piglets and sows after PEDV inoculation
| Inoculum groupa | Piglet datab |
Sow data |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of pigs | Diarrhea rate (%)c | Mortality rate (%) | Onset of diarrhea (dpi) | Mean cumulative FC score of each pigd | Duration of diarrhea (days) | Onset of fecal RNA shedding (dpi) | Mean highest fecal RNA shedding titer (log10 GE/ml [dpi]e) | Duration of diarrhea (days) | Highest fecal PEDV RNA shedding titer (log10 GE/ml)e | |
| TC-PC177 | 10 | 80 (8/10) A | 0 (0/10) B | 2.50 ± 0.53 A | 7.88 ± 5.51 B | 2.22 ± 2.17 B | 2.00 ± 0.00 | 10.19 ± 0.50 (5) B | ND | 6.80 |
| PC21A | 11 | 100 (11/11) A | 55 (6/11) A | 1.00 ± 0.00 B | 17.00 ± 2.00 A | 7.33 ± 1.18 A | <1 | 11.81 ± 0.27 (1) A | 6.00 | 9.67 |
| Mock | 7 | 0 (0/7) B | 0 (0/7) B | ND | 0.00 ± 0.00 C | 0.00 ± 0.00 C | ND | ND | ND | ND |
Piglets in the TC-PC177 group were inoculated with 5 log10 PFU/pig of TC-PC177 P10C8; piglets in the PC21A group were inoculated with 10 log10 genome equivalents (GE)/pig of wild-type PC21A; piglets in the mock group were inoculated with PBS.
Different uppercase letters indicate a mean significant difference between groups (P < 0.05). Values in parentheses are the number of positive animals/number of animals tested. Data were determined at 1 to 11 dpi. ND, not detected.
Fecal consistency (FC) was scored as follows: 0, solid, 1, pasty, 2, semiliquid; and 3, liquid. An FC score of ≥2 was considered diarrhea.
Mean value = (sum of FC scores for each piglet)/N, where N is the number of surviving piglets in the period. Scores were calculated for piglets at 1 to 5, 7, 9, and 11 days postinoculation (dpi).
The detection limit of PEDV real-time RT-PCR corresponds to 4.8 log10 GE/ml.
FIG 1.
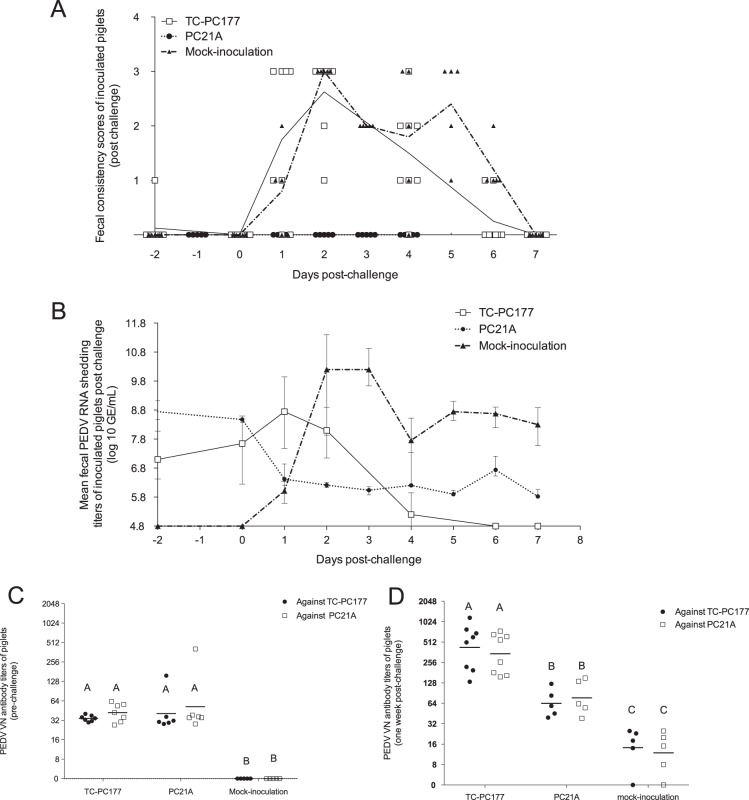
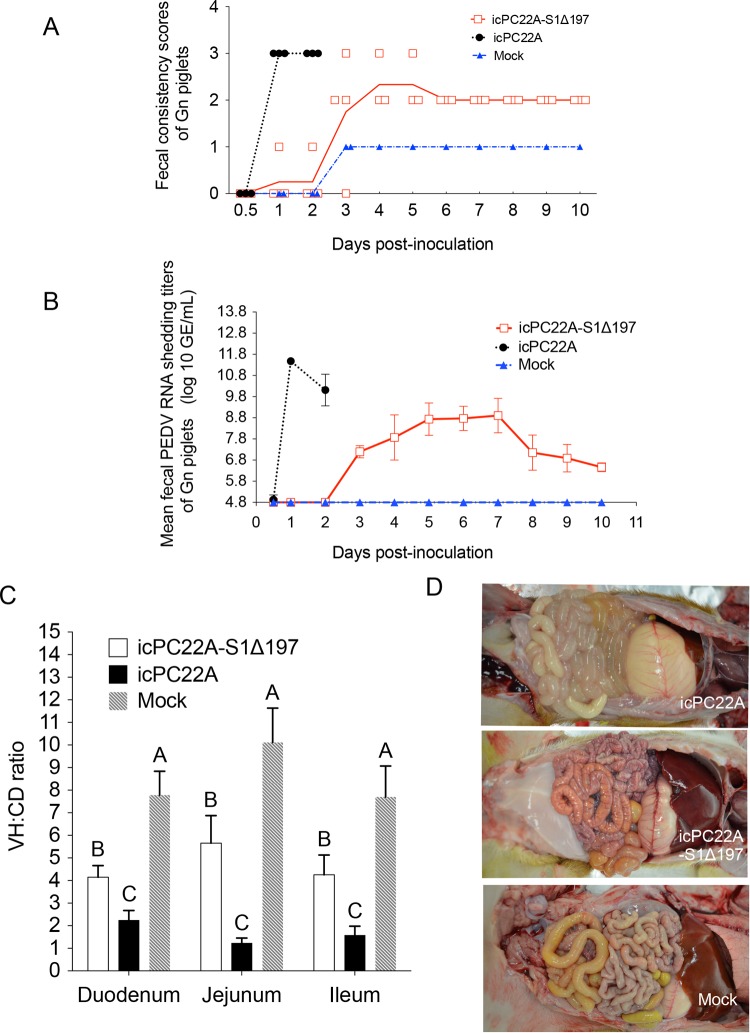
Evaluation of the virulence of the PEDV TC-PC177 (P10C8) strain in neonatal, conventional suckling pigs. PC21A- and mock-inoculated groups were the virulent and negative controls, respectively. (A) Fecal consistency scores postinoculation. Each dot indicates the score of an individual piglet at different dates; each line represents the mean value of scores in each group. (B) Fecal PEDV RNA shedding profile. Data are displayed as mean values ± standard deviations of each group. Values lower than the detection limit (4.8 log10 GE/ml), such as the mock inoculation data, were considered negative samples and were not displayed in this figure. (C) Fecal shedding profile of two PEDV-exposed sows. (D) VH/CD ratios of the small intestine of two (mock) or three piglets (TC-PC177 and PC21A), that died or were euthanized at 3 dpi. Fifteen villi and crypts were measured in the sections of small intestine among different piglets.
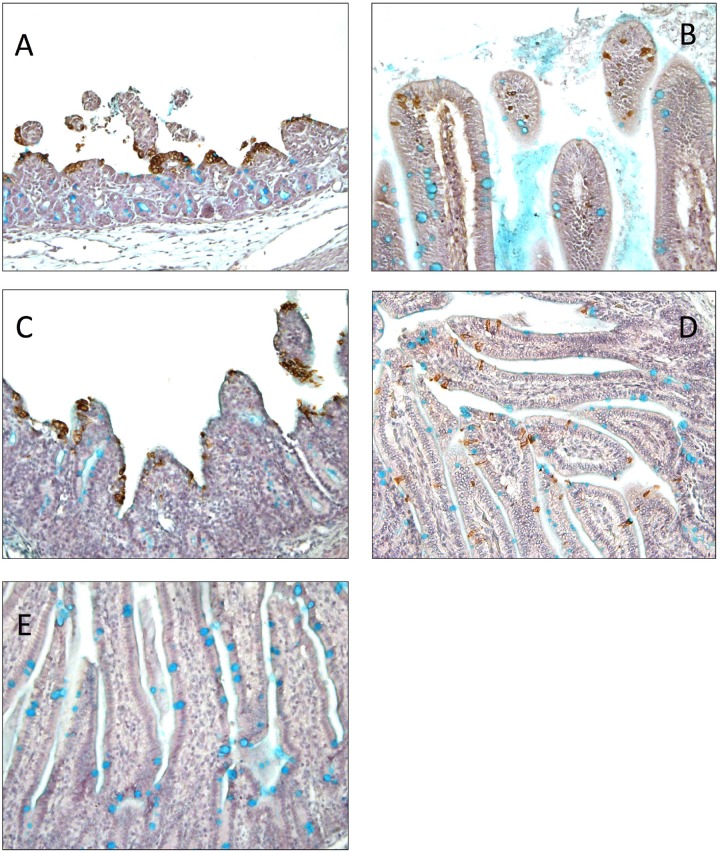
Mild and severe villous atrophy were observed in the small intestines of the TC-PC177- and PC21A-infected piglets, respectively. The villous height-to-crypt depth (VH/CD) ratios of the duodenum, jejunum, and ileum differed significantly among groups: the highest ratio was in the mock infection group, midrange ratios were in the TC-PC177 infection group, and the lowest ratio was in the PC21A infection group (Fig. 1D). Using immunohistochemistry (IHC) staining, we confirmed that the PEDV N antigens were sporadically distributed in the intestine of the TC-PC177 piglets, unlike the extensive antigen signals in the intestine of the PC21A piglets (see Fig. 7A and B). Notably, among all PEDV-infected piglets, no PEDV antigen-positive cells were detected in any other organs (lungs, spleen, kidneys, liver, and heart) other than the intestines and lymphocytes.
FIG 7.
Immunohistochemistry staining of PEDV N proteins in the jejunum sections of piglets that died or were euthanized at 2 to 3 dpi (magnification, ×200): PC21A (A), TC-PC177 (B), icPC22A (C), icPC22A-S1Δ197 (D), mock inoculation (E). The brown signals represent the PEDV antigens in enterocytes. Acidic polysaccharides, including sialylated mucin, were stained with 1% alcian blue and appear as cyan signals, mainly indicating goblet cells in the intestine. Severe villous atrophy can be observed in different sections of intestine of PC21A- and icPC22A-exposed piglets.
The two sows were infected with PC21A or TC-PC177 by being exposed to their virus-shedding piglets. Interestingly, in contrast to 6 days of diarrhea in the PC21A-exposed sow, the sow infected with TC-PC177 did not show any clinical signs (Table 1). The overall fecal PEDV RNA shedding titers of the TC-PC177-infected sow were lower than those of the PC21A-infected sow (Fig. 1C). However, after the PC21A challenge, the sow in the PC21A group did not develop diarrhea and had low fecal RNA shedding titers (Table 2), while sows in both the TC-PC177 and mock infection groups had diarrhea.
TABLE 2.
Clinical signs of pigs postchallenge with the highly virulent strain PC21A
| Inoculum group | Piglet dataa |
Sow data |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of surviving pigsb | Challenge age (days [dpi]) | Diarrhea rate (%)c,d | Mortality rate (%)d | Onset of diarrhea (dpc) | Mean cumulative FC score of each pige | Highest fecal PEDV RNA shedding titer (log10 GE/ml)f | Duration of diarrhea (days) | Duration of diarrhea (days) | Highest fecal PEDV RNA shedding titer (log10 GE/ml)f | |
| TC-PC177 | 8 | 23 (19) | 88 (7/8) A | 0 (0/8) | 1.43 ± 0.53 | 6.00 ± 2.69 A | 8.81 ± 1.65 A | 1.88 ± 1.13 B | 2 | 8.00 |
| PC21A | 4 | 24 (20) | 0 (0/4) B | 0 (0/4) | ND | 0.00 ± 0.00 B | 6.83 ± 0.37 B | ND | ND | 6.61 |
| Mock | 5 | 24 (20) | 100 (5/5) A | 0 (0/5) | 1.80 ± 0.45 | 6.80 ± 1.03 A | 10.23 ± 0.66 A | 3.80 ± 0.84 A | 1 | 10.95 |
Different uppercase letters indicate a mean significant difference between groups (P < 0.05). ND, not detected.
Two PC177-infected piglets, two mock-infected piglets, and one surviving PC21A-infected piglet were euthanized at the acute phase for histopathological examination.
Fecal consistency (FC) was scored as follows: 0, solid, 1, pasty, 2, semiliquid; and 3, liquid. An FC score of ≥2 was considered diarrhea.
Values in parentheses are the number of positive results/number of animals tested.
Mean value = (sum of FC scores for each piglet)/N, where N is the number of surviving piglets in the period. Scores were calculated for piglets at 1, 2 4, and 6 days postchallenge (dpc).
The detection limit of PEDV real-time RT-PCR corresponds to 4.8 log10 GE/ml.
TC-PC177 induced incomplete cross-protection against challenge with the highly virulent PEDV PC21A strain.
All surviving piglets were challenged with 12 log10 genome equivalents (GE)/piglet of PC21A (Table 2) at 3 weeks postinoculation. All pigs (100%) in the mock group, 88% of pigs in the TC-PC177 group, and no pigs in the PC21A group had diarrhea postchallenge. However, the mock-inoculated group exhibited a significantly longer duration of diarrhea (3.80 ± 0.84 days) than the TC-PC177-inoculated group (1.88 ± 1.13 days) (P < 0.05). No mortality was observed in the three litters, similar to previous observations that PEDV infection does not cause mortality in older pigs (2, 24).
Before challenge, pigs in both the TC-PC177- and PC21A-inoculated groups shed PEDV RNA at low titers constantly (ranging from 7 to 8 log10 GE/ml). From 0 to 1 days postchallenge (dpc), the shedding titers in the TC-PC177 group increased from 7.36 ± 1.41 to 8.81 ± 1.64 log10 GE/ml, in contrast to the decreased shedding titers (8.46 ± 0.09 to 6.55 ± 0.51 log10 GE/ml) in the PC21A group (Fig. 2B). The mock-inoculated piglets had increased shedding titers from no shedding at 0 dpc to the peak titer of 10.23 ± 0.66 log10 GE/ml within 2 dpc. However, the PEDV RNA shedding titers were undetectable in the TC-PC177-inoculated group at 7 dpc, they were low in the PC21A-inoculated group (5.89 ± 0.17 log10 GE/ml), and they were moderate in the mock-inoculated group (8.24 ± 0.67 log10 GE/ml). To identify which PEDV strains were shed from the pigs in the TC-PC177 group after challenge, we amplified the S gene fragment covering the 197-aa deletion region from rectal swab (RS) samples using a conventional reverse transcription-PCR (RT-PCR) assay developed in our laboratory (25). We amplified PC21A but not PC177 PEDV from the fecal samples of pigs in the TC-PC177 group after challenge with the PC21A strain, indicating the dominant shedding of PC21A virus after challenge (data not shown).
FIG 2.
Evaluation of the clinical signs and serum virus neutralization (VN) antibody titers of conventional suckling piglets after challenge with the highly virulent PEDV PC21A strain. (A) Fecal consistency scores of piglet postchallenge. Dots and lines represent individual values and mean values, respectively, in each group. (B) Fecal PEDV RNA shedding profile of piglets. Data are shown as mean values ± standard deviations of each group. VN antibody titers of sera collected at 1 day before challenge (C) and at 7 days postchallenge (D) were titrated by CCIF assay. The titers are expressed as the reciprocal of the highest dilution at which the number of PEDV FFU is blocked 80%.
The PEDV virus neutralization (VN) antibody titers in sera collected from piglets at 1 day before challenge (−1 dpc) and at 7 dpc are shown in Fig. 2C and D. Similar to results from our previous report, each serum sample had similar titers against the homologous and heterologous strains (26). The mean PEDV VN antibody titers in TC-PC177- and PC21A-inoculated groups were similar at −1 dpc (Fig. 2C). After PC21A challenge, the titers rose significantly (P < 0.05) by 7 dpc in the TC-PC177 and mock inoculation groups but not in the PC21A group (Fig. 2D).
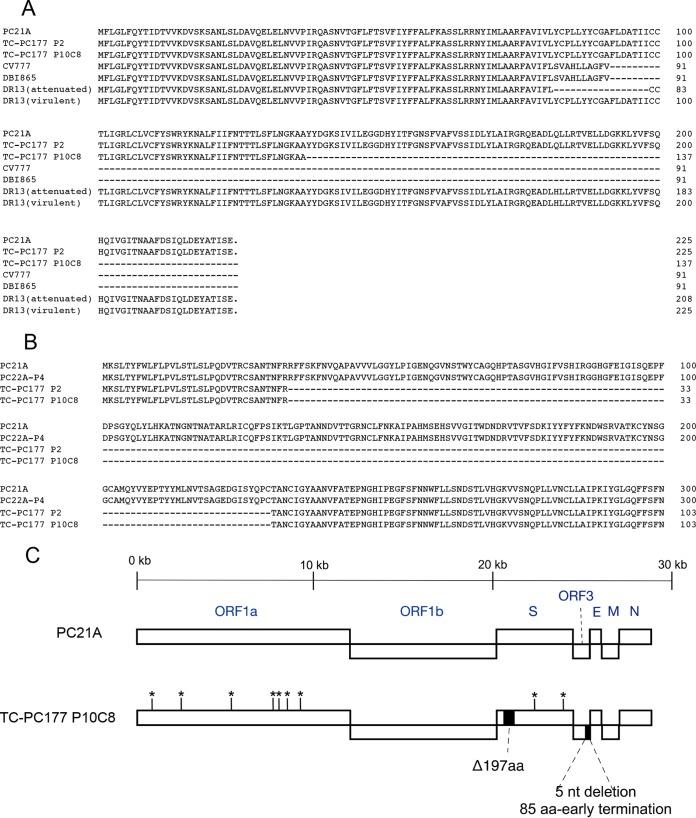
Genomic sequence analysis of TC-PC177 P10C8 revealed additional mutations compared with TC-PC177.
We found an additional 5-nucleotide (nt) deletion in the ORF3 gene of TC-PC177 P10C8 compared with that of TC-PC177 P2 (GenBank accession number KM392229) (15). This deletion resulted in an 85-aa C-terminal truncation in the ORF3 protein (Fig. 3). We also found that this clone has 9 amino acids different from the genome of PC21A, located in the ORF1a (7 aa) and S (2 aa) genes. Therefore, to determine whether the 197-aa deletion in S1 NTD alone can attenuate a PEDV strain, we generated a recombinant virus, icPC22A-S1Δ197, by deleting the 197-aa region from the S gene of the highly virulent icPC22A virus and performed a pathogenesis study in Gn pigs (see below).
FIG 3.
(A) Alignment of amino acids of ORF3 proteins of TC-PC177 P10C8 and other PEDV strains. (B) Alignments of amino acids in the N-terminal sequences (residues 1 to 300) of spike proteins of PEDV strains TC-PC177 P2, TC-PC177 P10C8, wild-type PC21A, and PC22A P4. (C) Schematic amino acid differences between TC-PC177 P10C8 and PC21A. A deletion is indicated by a dash (-).
The optimized full-length infectious cDNA clone of PC22A was more stable and more efficient in rescue of recombinant viruses.
The original six-fragment infectious clone described by Beall et al. (23) was compromised by the low stability of subclones B and E and low virus yields after transfection associated with poor virus replication in cell culture, complicating our ability to isolate attenuated mutants. We determined the boundaries of these new subclone junctions by trial and error to avoid the toxicity effects of PEDV sequences in Escherichia coli. Consequently, the second-generation infectious clone, designated icPC22A, was now captured within five plasmids that replicated more stably in E. coli (data not shown), allowing for more efficient recovery of recombinant viruses. Compared with the genomic sequence of the parental PC22A virus of the previous six-fragment version, our icPC22A contains a GGC insertion (nucleotide positions 22029 to 22031) in the S1 C-terminal domain, resulting in the replacement of an aspartic acid by a glycine and a histidine (D466GH). This insertion first appeared in one batch of the passage 4 (P4) TC-PC22A virus. It replicated more efficiently than previous passages of TC-PC22A. Moreover, this insertion was genetically stable through the 160th passage (27). Therefore, we retained this insertion in plasmid E to increase the efficacy of rescue of the recombinant viruses from Vero cell cultures.
Recombinant viruses were rescued and characterized in Vero cell culture.
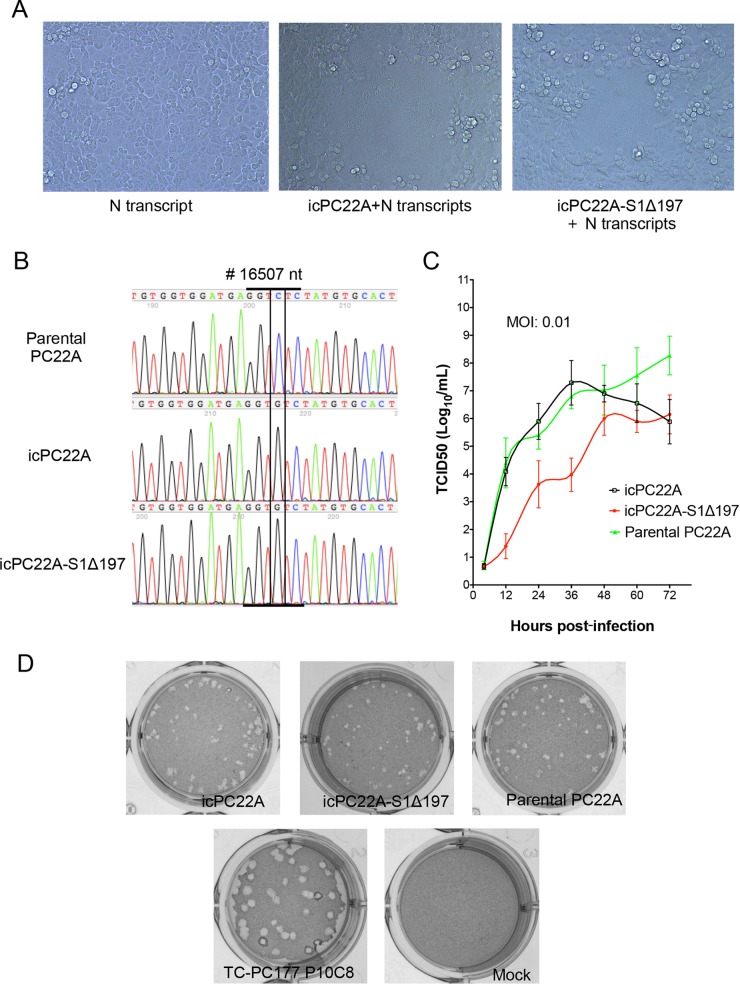
After electroporation of the mixture of full-length genomic and N gene transcripts into Vero cells, we observed the formation of syncytia, which are the typical cytopathic effects (CPE) of PEDV infection, for both recombinant viruses (icPC22A and icPC22A-S1Δ197) at 2 to 3 days posttransfection (Fig. 4A). The 197-aa deletion is the only difference between the viral genomes of icPC22A-S1Δ197 and the virulent control icPC22A. There were no other differences between the genomes of icPC22A-S1Δ197 and icPC22A. CPE developed more slowly with icPC22A-S1Δ197 virus than with the icPC22A virus. We harvested both supernatants at 4 to 5 days posttransfection and passaged them in Vero cells one more time to generate the P1 virus. Titers of these P1 viruses were 7.63 log10 PFU/ml (icPC22A) and 5.51 log10 PFU/ml (icPC22A-S1Δ197). Sanger sequencing data revealed that the appropriate genetic marker, the C-to-G substitution at nucleotide site 16507, existed in the genomes of both icPC22A and icPC22A-S1Δ197 but not in the genome of the parental PC22A P4 virus (Fig. 4B). The 197-aa deletion was confirmed for icPC22A-S1Δ197 virus by RT-PCR, followed by sequence analysis.
FIG 4.
Characterization of recombinant viruses. (A) Cytopathic effects (CPE) of recombinant viruses occurred at 2 days posttransfection of the mixture of full-length genomic RNA and N gene transcripts into Vero cells. (B) Sanger sequencing chromatogram of a silent mutation (nucleotide position 16507; C to G) was identified exclusively in the recombinant viruses icPC22A (P1) and icPC22A-S1Δ197 (P1). (C) Multistep growth kinetics of two recombinant viruses and parental PC22A (P4). Vero cells were inoculated with each virus at an MOI of 0.01. Supernatants were sampled at different time points and titrated for TCID50 values in 96-well plates, and virus titers were calculated using the Reed-Muench method (58). (D) Plaque assay of two recombinant viruses, parental PC22A (P4), TC-PC177 P10C8, and mock inoculation. Images were taken at 3 days postinoculation.
The replication of recombinant viruses in Vero cells was assessed by multistep growth kinetics at a multiplicity of infection (MOI) of 0.01 (Fig. 4C). The replication of icPC22A virus was comparable to that of PC22A P4; however, titers of icPC22A-S1Δ197 virus were about 3 to 4 log10 lower than those of the other two viruses at 36 h postinoculation. The morphology of the plaques of icPC22A was comparable to that of the parental virus (Fig. 4D).
To confirm the loss of the sialic acid binding domain in icPC22A-S1Δ197 virus, we performed neuraminidase (NA) treatment of Vero cells, followed by virus inoculation. Cell surface sialic acids were removed by neuraminidase digestion before virus inoculation, as described previously (14). After PEDV inoculation, Dulbecco's modified Eagle's medium (DMEM) supplemented with 2.5% fetal bovine serum (FBS) was used as the maintenance medium to inhibit proteinase activity to block virus spreading. Compared with the corresponding mock-infected control cells without NA treatment before PEDV inoculation, the numbers of fluorescent focus units (FFU) of both icPC22A and icPC22A-S1Δ197 viruses were decreased in the NA-treated Vero cells (Fig. 5). However, we observed a 92.7% reduction in icPC22A virus, which was significantly higher than the 25.8% reduction in the icPC22A-S1Δ197 virus (P < 0.05).
FIG 5.
Neuraminidase treatment infectivity assay. (A) Indirect immunofluorescent assay of Vero cells pretreated with neuraminidase (NA+) or PBS (NA−) and inoculated with recombinant virus icPC22A or icPC22A-S1Δ197. After inoculation, cells were maintained in DMEM containing 2.5% FBS. At 16 h postinoculation, cells were fixed, and the PEDV antigens were stained using mouse anti-PEDV N antibody as the primary antibody. (B) Percentage of fluorescent focus units (FFU) of sialic acid-dependent infection assay. Neuraminidase- or PBS-treated Vero cell monolayers were inoculated with equal amounts of virus (350 FFU/well).
These results indicate that the recombinant viruses from our optimized infectious clones were effectively rescued in Vero cell cultures. The icPC22A virus propagated similarly to its parental virus, PC22A P4, yet the icPC22A-S1Δ197 virus replicated less efficiently than the other two. We also confirmed the loss of most sialic acid binding activity of icPC22A-S1Δ197 virus in Vero cell culture.
Compared with the virulence of icPC22A virus, the virulence of icPC22A-S1Δ197 was reduced in neonatal Gn piglets.
To determine whether the recombinant icPC22A virus is also highly virulent and whether the 197-aa deletion attenuated the icPC22A-S1Δ197 virus in vivo, 5-day-old Gn piglets were divided into three groups and orally inoculated with icPC22A (n = 3; 100 PFU/pig) or icPC22A-S1Δ197 (n = 4; 100 PFU/pig) virus or mock inoculated (n = 2; PBS).
The clinical signs of the Gn piglets in the acute phase of infection are summarized in Table 3. All icPC22A-infected piglets developed diarrhea within 1 dpi (Table 1 and Fig. 6A), with a peak shedding titer of 11.48 ± 0.17 log10 GE/ml at 1 dpi (Fig. 6B). These piglets developed severe clinical signs, became moribund, and were euthanized at 2 dpi. At necropsy, we observed thin and transparent intestinal walls of different sections of the intestines, which are characteristic for neonatal piglets infected with the highly virulent PEDV (Fig. 6D). Histopathological examination revealed severe villous atrophy in the small intestine. The VH/CD ratios of icPC22A-inoculated piglets (jejunum, 1.23 ± 0.23) were significantly lower than those of the age-matched mock-inoculated piglets (jejunum, 10.11 ± 1.52) (P < 0.05). IHC staining showed icPEDV N proteins extensively in the small intestine (Fig. 7). Comparing these data with results for PC21A-infected conventional piglets euthanized at the acute phase, as well as PC22A-inoculated Gn piglets in our previous study (23), indicates that the icPC22A virus is highly pathogenic in newborn Gn piglets, similar to wild-type PC21A and the PC22A challenge pool (28).
TABLE 3.
General group information and clinical signs of Gn piglets after inoculation with recombinant virusesa
| Inoculum groupb | No. of piglets | Diarrhea rate (%)c,d | Mortality rate (%)d | Onset of diarrhea (dpi) | Highest FC score of each piglet | Duration of diarrhea (days) | Onset of fecal RNA shedding (dpi)f |
|---|---|---|---|---|---|---|---|
| icPC22A-S1Δ197 | 4 | 100 (4/4) A | 0 (0/4) B | 3.25 ± 0.50 A | 2.50 ± 0.58 | 7.67 ± 0.58 | 3.25 ± 0.50 |
| icPC22A | 3 | 100 (3/3) A | 100 (3/3) A | 1.00 ± 0.00 B | 3.00 ± 0.00 | NA (>3)e | <1 |
| Mock | 2 | 0 (0/2) B | 0 (0/2) B | ND | 1.00 | ND | ND |
Different uppercase letters indicate a mean significant difference between groups (P < 0.05). ND, not detected.
Piglets in the icPC22A-S1Δ197 group were inoculated with 100 PFU/pig of virus; piglets in the icPC22A group were inoculated with 100 PFU/pig of virus; piglets in the mock inoculation group were inoculated with PBS.
Fecal consistency (FC) was scored as follows: 0, solid, 1, pasty, 2, semiliquid; and 3, liquid. An FC score of ≥2 was considered diarrhea.
Values in parentheses are the number of positive results/number of animals tested.
NA, not available. Since all icPC22A-inoculated piglets became moribund and were euthanized by 2dpi, this value could not be determined.
The detection limit of PEDV real-time RT-PCR corresponds to 4.8 log10 GE/ml.
FIG 6.
Evaluation of the virulence of recombinant viruses icPC22A and icPC22A-S1Δ197 in 5-day-old Gn piglets. Numbers of piglets in each group were as follows: icPC22A, n = 3; icPC22A-S1Δ197, n = 4; mock, n = 2. (A) Fecal consistency scores of individual Gn piglets. Each line represents the mean value in each group. Three icPC22A piglets became moribund and were euthanized at 2 dpi; one icPC22A-S1Δ197-infected diarrheic piglet and one mock-inoculated piglet were euthanized at 3 dpi for histopathological examination. (B) Fecal PEDV RNA shedding profile of each group. Data are shown as mean values ± standard deviations of groups. (C) VH/CD ratios of Gn piglets euthanized at 2 to 3 dpi. (D) Necropsy images show that transparent intestine was observed in the Gn piglet inoculated with icPC22A but not in icPC22A-S1Δ197-infected or mock-inoculated piglets.
On the other hand, all the icPC22A-S1Δ197-infected piglets showed less severe diarrhea than the icPC22A-infected piglets (Table 3 and Fig. 6B) and significantly delayed onset of fecal virus RNA shedding (3.25 ± 0.50 dpi) compared with the icPC22A-infected piglets (1.00 ± 0.00 dpi) (P < 0.05). The highest viral RNA shedding titer within 10 dpi was 8.88 ± 0.17 log10 GE/ml in the icPC22A-S1Δ197 group, a value which was significantly lower than the peak titer (11.48 ± 0.17 log10 GE/ml) of the icPC22A group (P < 0.05). The intestinal VH/CD ratio of the icPC22A-S1Δ197 group was significantly higher than that of icPC22A group (Fig. 6C), corresponding to less transparency of the small intestine of the icPC22A-S1Δ197-infected piglet than of the icPC22A-infected piglets (Fig. 6D). We detected scattered PEDV N protein antigens in the small intestine (Fig. 7) and colon in this piglet. Similar to the result for TC-PC177, no alteration of tissue tropism was observed in the icPC22A-S1Δ197-infected piglets. These findings suggest that icPC22A-S1Δ197 was less virulent than icPC22A. Collectively, these data support the hypothesis that the 197-aa deletion is responsible for the attenuation of icPC22A-S1Δ197.
3-D structural analyses of the S proteins of icPC22A and icPC22A-S1Δ197.
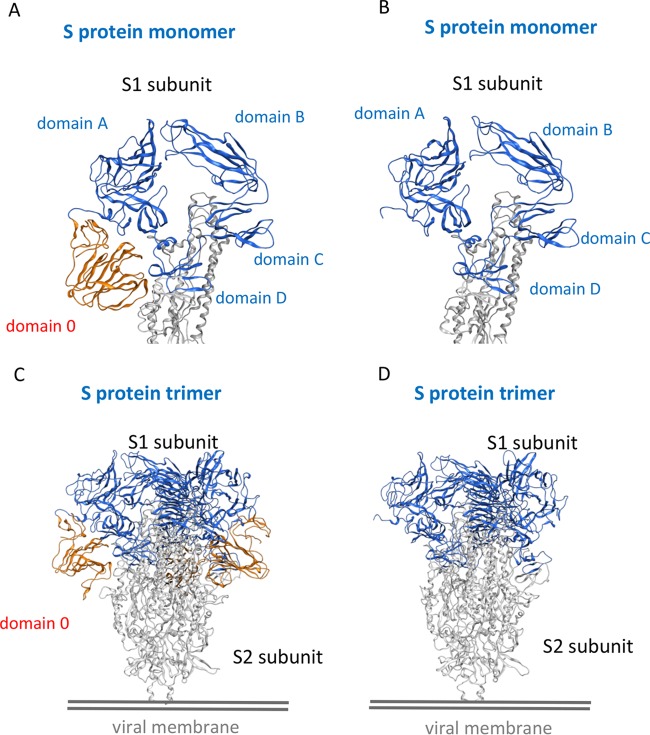
To investigate the structural differences of the S proteins between the two recombinant viruses icPC22A and icPC22A-S1Δ197, we constructed three-dimensional (3-D) models of the S protein trimers by the homology modeling technique. As shown in Fig. 8A, the S1 subunit of icPC22A can be divided into domain 0, domain A, domain B, domain C, and domain D. Domain 0 (residues 34 to 230) is lacking in the S1 subunit of icPC22A-S1Δ197 (Fig. 8B), indicating that the 197-aa deletion corresponds to domain 0. In the S protein trimer of icPC22A, domain 0 is located at the greatest distance from the trimeric axis that does not contribute to the trimer formation (Fig. 8C). Therefore, the deletion of domain 0 in the S protein trimer of icPC22A-S1Δ197 would not interfere with the trimer formation.
FIG 8.
Three-dimensional structural analyses of the S proteins of icPC22A and icPC22A-S1Δ197. The S1 subunit monomers of icPC22A (A) and icPC22A-S1Δ197 (B) and S protein trimers of icPC22A (C) and icPC22A-S1Δ197 (D) were modeled based on the cryo-EM structure of human coronavirus NL63 S protein from the Protein Data Bank (PDB accession number 5SZS) (60). The 197-amino-acid region, designated domain 0, is highlighted in orange, whereas other domains in the S1 subunit are highlighted in blue; the S2 subunit is in gray.
Domain B of the S1 subunit, which includes the known neutralizing epitope COE (residue 499 to 638), is located at the most distal end from the viral membrane. Domain A is located between domain 0 and domain B in the S protein trimer of icPC22A. Thus, the deletion of domain 0 in the S protein trimer of icPC22A-S1Δ197 would not interfere with the neutralizing epitope COE. However, domain 0 in the S protein trimer of icPC22A is located near the nonneutralizing epitopes SS2 (residues 748 to 755) and SS6 (residues 764 to 771), which are the subsequent residues after domain D that are closest to the S2 subunit.
DISCUSSION
In this study, we demonstrated that the pathogenicity of the S1 NTD-del type of PEDV mutants, TC-PC177 and icPC22A-S1Δ197, in neonatal piglets was attenuated, as characterized by no mortality, extended incubation time, milder intestinal lesions, and lower fecal PEDV RNA shedding titers than with the highly virulent PEDV strain. Importantly, no changes in tissue tropism were observed for these viruses. For TC-PC177 P10C8, the 85-aa-truncated ORF3 and other 9-aa alterations in ORF1a and S may also contribute to the attenuated phenotype, as a similar C-terminally truncated ORF3 protein also exists in some attenuated PEDV strains, such as CV777 (GenBank accession number KT323979) and DBI865 (GenBank accession number HQ537432). Therefore, the application of reverse genetics was essential for the final confirmation of the role of the 197-aa deletion in PEDV attenuation.
The milder clinical signs, virus shedding profiles, and histopathological lesions of TC-PC177-inoculated piglets were similar to those of TTR-2-infected neonatal, colostrum-deprived piglets (22). Suzuki et al. did not detect PEDV antigens in the duodenum of the four TTR-2-inoculated pigs. In our study, although PEDV antigens were not detected in the duodenum of the icPC22A-S1Δ197-infected piglet that was euthanized during the acute infection phase, we detected sporadic PEDV-positive enterocytes in the duodenum of TC-PC177-infected cesarean-derived, colostrum-deprived (29), and conventional suckling piglets. Also, we found that the PEDV antigen distributions in different sections of intestines in icPC22A-S1Δ197- and icPC22A-infected piglets were similar (Fig. 7). The discrepant PEDV antigen distributions in different sections of the intestines (from duodenum to colon) may be due to the use of different strains in the two studies, the limited number of animals being evaluated, and the different timing of euthanasia. Overall, icPC22A-S1Δ197 replicated less efficiently than icPC22A in vitro (Fig. 4C and D). This suggests that the NTD of S1 is not essential for PEDV replication in vivo and in vitro; however, this domain increases virus replication levels in vitro and enhances virulence in vivo.
As one of the most variable regions in the PEDV genome, the S1 NTD correlated with different sialic acid binding activities and pathogenicities of PEDV strains in pigs. The S indel (insertion and deletion) type of PEDV strains were characterized by an amino acid insertion (residues 161 to 162) and two deletions (residues 59 to 62 and 140) in the S1 NTD although they shared a similar genomic backbone with the highly virulent strains (30). It has been demonstrated that the virulence of S indel strains is reduced compared with that of the highly virulent strains (31, 32). Interestingly, a recent study revealed that a highly virulent non-S indel strain, GDU, had sialic acid binding activity while an S indel strain, UU, did not (14). This finding was similar to our neuraminidase treatment-dependent infectivity results with icPC22A and icPC22A-S1Δ197 (Fig. 5). For the enteric coronavirus TGEV, virions of the wild-type strain but not an attenuated mutant, m10, of the Purdue-115 strain, bound to mucin-like glycoprotein purified from porcine enteric epithelial cells in vitro (33). The m10 variant contained point mutations in the sialic acid binding domain in S1 NTD and was attenuated in pigs. Therefore, the binding to sialic acid-rich glycoproteins, such as mucus in the intestine, may be important for stabilizing enteric coronaviruses, including the highly virulent PEDV strain, and may facilitate initial virus attachment to cellular receptors in vivo. However, it is difficult to monitor and quantify the initial infection step between the highly virulent and S1 NTD-del strain in vivo. In a previous report, inoculation of a highly virulent U.S. PEDV strain led to the infection of 100% of epithelial cells in the jejunum of a piglet as early as 12 h postinoculation although virus shedding and villous atrophy were not observed at this early time point (1). However, with the delayed virus replication (Fig. 1 and 6), only limited PEDV antigens could be detected in the enterocytes of S1 NTD-del variant TC-PC177- or icPC22A-S1Δ197-infected piglets via IHC (Fig. 7). In IHC of alcian blue-stained tissue, the antigen signals of TC-PC177 or icPC22A-S1Δ197 (Fig. 7) were not necessarily colocalized with the sialylated mucin-rich region, especially goblet cells. The mechanism of sialic acid binding involved in initial PEDV infection in vivo is clearly an important step in pathogenesis and requires more detailed future studies.
Mutants of various species of coronaviruses containing a large deletion in their spike glycoproteins have been identified, including TGEV (34), feline infectious peritonitis virus (FIPV) (35, 36), mouse hepatitis coronavirus (MHV) (37), and bat coronavirus HKU2 (38). In many cases, such mutants display attenuation and/or changes in tissue tropism in the host. For PEDV, similar to the observation in the previous TTR-2 study (22), the loss of S1 NTD also did not change the enteric tropism of TC-PC177 and icPC22A-S1Δ197 in piglets. This phenomenon differs from what occurred with another porcine enteric alphacoronavirus, TGEV: the loss of the S1 NTD of TGEV changed the enteric virus into a respiratory variant virus, PRCV (20). One explanation is that PEDV replicates exclusively in the intestine while TGEV replicates in both the intestine and respiratory tract (21, 39). Such diverse tissue tropisms may be due to different receptor usage and/or proteolytic processing of the S proteins between PEDV and TGEV. For many years, the TGEV receptor, porcine aminopeptidase N (pAPN), was also considered to be the putative receptor of PEDV (40–43). However, recent studies revealed that PEDV could infect pAPN knockout cell lines (11), but the protease activity of pAPN enhanced PEDV infection (10), suggesting that pAPN is not the cellular receptor for PEDV. Moreover, it is well known that extracellular proteases, such as trypsin, enhance the entry and egress of PEDV in cell culture (44, 45), suggesting that PEDV propagation may depend on the protease-rich environment in the intestine. For TGEV, a furin-like protease cleavage motif (KRKYR) is located upstream of the fusion peptide of the S protein (46). Along with recognizing different cellular receptors, TGEV can also bind to two types of sialic acids (47), Neu5Ac and N-glycolylneuraminic acid (Neu5Gc); however, the PEDV S1 NTD attaches only to Neu5Ac (8). Such a difference in carbohydrate binding may also restrict the enteric tropism of S1 NTD-del PEDV variants.
Based on the lower growth kinetics in vitro and reduced pathogenicity of icPC22A-S1Δ197 in piglets, we conclude that the S1 NTD-del type of PEDVs replicate less effectively than the highly virulent strains. Such defective replication of TC-PC177 might induce less robust immune responses than PC21A in conventional suckling piglets. Our results demonstrated that the primary infection with TC-PC177 did not provide significant cross-protection against challenge with the highly virulent strain PC21A in vivo. After each piglet was challenged with a high dose (12 log10 GE) of PC21A virus, none of the piglets in the PC21A group showed clinical signs, but 88% of the TC-PC177-inoculated and 100% of the mock-inoculated piglets developed diarrhea (Table 2) although similar levels of VN antibody titers were detected in the TC-PC177-inoculated and the PC21A-inoculated piglets at 1 day prechallenge in vitro (Fig. 2C). At 7 dpc, the VN antibody titers of piglets in the TC-PC177 group were significantly higher than those in the PC21A group. After challenge, the higher level of PC21A replication in the PC177-inoculated piglets than in the PC21A-inoculated piglets could have been responsible for the induction of the higher VN antibody levels in the TC-PC177 group than in the PC21A group (Fig. 2B and C). It is likely that the existing homologous immunity, induced by PC21A infection, protected more efficiently against PC21A infection after challenge than the heterologous immunity induced by the primary TC-PC177 infection. The robust immune responses likely protected the PC21A-inoculated piglets from homologous challenge. In the three-dimensional structural analysis (Fig. 8), we discovered that domain 0, containing the deleted 197-aa region in the S protein of TC-PC177, potentially does not interfere with trimer formation and the neutralizing epitopes COE (residue 499 to 638) and 2C10 (residues 1368 to 1374) (29). However, the deletion of domain 0 may influence two nonneutralizing B cell epitopes, SS2 and SS6 (48), which may contribute to the conformational change of the S2 subunit during the virus-cell membrane fusion process. Recently, a publication reported that domain 0 of PEDV contains neutralizing epitopes (49). However, we detected that TC-PC177 infection in pigs induced similar VN antibody titers against both homologous and PC21A viruses (Fig. 2C and D), suggesting that the deletion of domain 0 did not significantly affect the induction of serum neutralizing antibodies in pigs and/or that these VN epitopes within domain 0 do not play a predominant role in the induction of serum VN antibodies. Based on the partial cross-protection in the TC-PC177 group piglets, we assume that the deletion of the 197-aa domain may affect the induction of protective mucosal and cellular immune responses, which will be studied in the future.
To date, only a few PEDV reverse genetics platforms have been reported. Li et al. (50) developed a platform for a cell culture-adapted PEDV vaccine strain, DR13, using targeted RNA recombination technology. Jengarn and colleagues constructed a bacterial artificial chromosome (BAC) bearing the genomic cDNA of PEDV strain AVCT12 and recovered the recombinant viruses from a Vero cell line transfected with pAPN (51). However, the infectious cDNA clone contained a 52-nt deletion at the 3′ end of the S gene, resulting in a 7-aa early termination of the S protein and the loss of the start codon of the ORF3 gene. These mutations probably facilitated the rescue of recombinant viruses in Vero cells but may impair the pathogenicity of recombinant AVCT12 (rAVCT12) in vivo. A review article summarizing different reverse genetics platforms for PEDV was recently published (52).
Our second-generation PEDV infectious clone was designed to circumvent problems noted in our previous icPEDV (23), such as low recovery yields of recombinant viruses and a need to increase the stability of subclones B and E. This second-version infectious clone should facilitate increased adaptation in Vero cells as well as maintain the virulence of the recombinant virus in vivo. We noted that a conserved GGC insertion (nucleotide site 22029; D to GH), located upstream of the putative receptor binding domain in the S1 CTD, occurred in the genome of a batch of P4 PC22A virus, corresponding to increased virus titers in Vero cell cultures, but not in the earlier passages or the template of the six-fragment icPEDV generated by Beall et al. (23). After electroporation, the initial infection with the recombinant viruses that contained the GGC insertion caused CPE within 2 to 4 days posttransfection, implying that the rescue of recombination viruses in Vero cells was significantly enhanced compared with that of the previous icPEDV. Particularly, icPC22A was still highly virulent in neonatal piglets, suggesting that the insertion of GGC did not impair the pathogenicity of this recombinant virus in vivo. The biological function of this insertion will be investigated in the future. More recently, an infectious cDNA clone of the highly virulent Chinese strain AH2012/12 was reported (53) using a similar strategy as that used with icPC22A. However, the CPE of rAH2012/12 occurred only after the supernatants of transfected Vero cells were passaged three times, implying less efficiency in rescuing recombinant viruses than that of the optimized icPC22A platform described in this study.
In this study, we confirmed that the virulence of TC-PC177, an S1 NTD-del type of PEDV, was reduced in neonatal piglets without changes in tissue tropism but with induction of cross-neutralizing antibody. Primary infection with TC-PC177 in conventional suckling piglets failed to induce cross-protection against the challenge with the highly virulent strain PC21A at 3 weeks postinfection. Furthermore, we optimized the infectious clone of the highly virulent PEDV strain PC22A and verified that the recombinant virus icPC22A is highly pathogenic in neonatal piglets. Using this reverse genetics platform, we confirmed that the NTD of the S1 subunit of S protein is a virulence determinant of a highly virulent PEDV.
MATERIALS AND METHODS
Cell lines and viruses.
Vero cells (ATCC number CCL81) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 5% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B. After PEDV inoculation, the Vero cells were maintained in DMEM supplemented with 0.3% tryptose phosphate broth, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 μg/ml trypsin (Gibco, Carlsbad, CA, USA) (15, 54). The isolated TC-PC177 was passaged in Vero cells 10 times. A plaque-purified clone 8, designated TC-PC177 P10C8, was used as the inoculum for conventional suckling piglets. Two highly virulent PEDV strains of U.S. origin, PC21A and PC22A, were detected and isolated from the same PED outbreak and have only three single nucleotide polymorphisms (15, 55). PC21A was used in the conventional pig study as the virulent control, whereas PC22A was used to generate the infectious cDNA clone.
Construction of the full-length cDNA clone of PC22A.
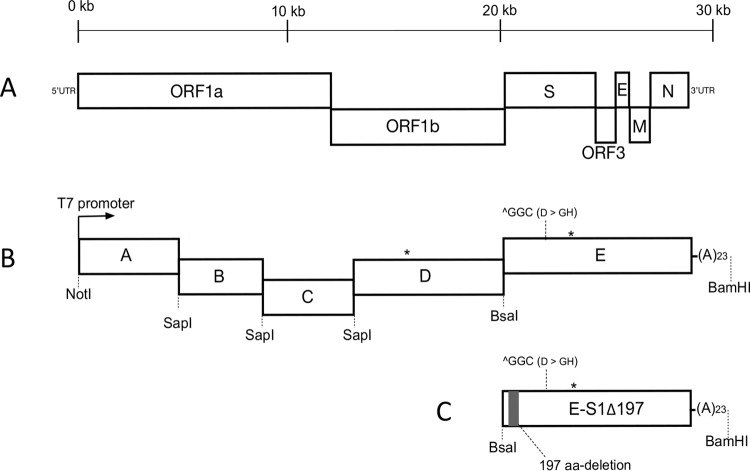
The strategy employed to construct the infectious cDNA clone of PC22A was described previously (23). To increase the stability of plasmids replicating in E. coli and the number of recombinant viruses recovered in cell culture, we divided the genomic cDNA of PC22A at the fourth passage into five fragments. The viral RNA was extracted using an RNeasy minikit (Qiagen, Germany) and reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The five truncated genomic cDNAs, fragments A to E, were amplified by PCR with PrimeSTAR GXL polymerase (TaKaRa, Japan) and were cloned into high-copy-number plasmid pUC19 (NEB, Ipswich, MA) (Fig. 9). Junctions between A/B, B/C, and C/D were joined by a unique nonpalindromic SapI site located at nucleotide positions 5752, 9654, and 14047. Fragments D and E were divided by a BsaI site located at nucleotide position 20418. Two naturally occurring BsaI sites, located within fragment D (nucleotide position 16507) and fragment E (nucleotide position 24335), were removed by introducing synonymous mutations. Fragment A contained a T7 promoter located upstream of the first nucleotide, whereas fragment E harbored a 23-nt poly(A) tail at the 3′ end. We constructed a mutant E plasmid by directly deleting the 591 nt encoding the 197 aa from the S1 region using NEBuilder HiFi DNA assembly master mix (NEB, Ipswich, MA). Together with fragments A to D, this mutant E fragment was used for the generation of recombinant virus icPC22A-S1Δ197.
FIG 9.
Schematic organization of a PEDV genome and the constructions of the full-length infectious cDNA clone. (A) Organization of a PEDV genome, including the ORF1a/b, spike (S), ORF3, envelope (E), membrane (M), and nucleocapsid (N) genes. (B) Five fragments comprising a full-length cDNA clone of PC22A. Two asterisks indicate BsaI sites in the PEDV genome that were removed by introducing silent mutations (nucleotide position 16507, C to G; nucleotide position 24332, A to G). In comparison with the genomic sequence of icPEDV (23), a GGC insertion is located at nucleotide position 22029, as indicated. (C) Mutant fragment E containing the 197-aa deletion in the S1 NTD. UTR, untranslated region; (A)23, 23-nt poly(A) tail.
Recovery of recombinant viruses.
The full-length cDNA was assembled as previously described (56, 57). Briefly, five cDNA fragments (A to E) were digested individually with respective restriction enzymes and gel purified using a QIAquick gel extraction kit (Qiagen, Germany). The five fragments in equal molar ratios were ligated using T4 ligase (NEB, Ipswich, MA) at 4°C overnight. The ligated cDNA was used as the template for in vitro transcription using a mMessage mMachine T7 transcription kit (Ambion, Austin, CA, USA). To increase the efficiency of recovery of recombinant virus as previously described (23), the PEDV nucleocapsid cDNA containing a T7 promoter and poly(A) tail was amplified from plasmid E using primers PEDV-N-F (5′-ATATGGATCCATCGTAATACGACTCACTATAGATGGCTTCTGTCAGTTTTCAGG-3′) and PEDV-N-R (5′-TTTTTTTTAATTTCCTGTGTCGAAGATCTCGTTGATAATTTCAACGG-3′), and then the purified cDNA was transcribed, followed by poly(A) tailing using a HiScribe T7 ARCA mRNA kit (NEB, Ipswich, MA, USA). The mixture of N gene transcripts and the full-length genomic RNA were electroporated into 8 × 106 Vero cells with four pulses at 450 V and 50 μF using a Gene Pulser II electroporator (Bio-Rad, Hercules, CA, USA). After recovery in growth medium for about 24 h, the cells were washed with PBS twice and cultured in the maintenance medium supplemented with 10 μg/ml trypsin for 4 to 6 days.
Identification of the marker mutation.
The supernatant of electroporated Vero cells was harvested at 4 to 6 days postinoculation (dpi) and passaged one more time in Vero cells to generate the first-passage (P1) virus stock. Viral RNA was extracted from the supernatant of P1 virus using an RNeasy minikit and reverse transcribed into cDNA using SuperScript III reverse transcriptase as described above. To identify the presence of the silent mutation of a natural BasI site in fragment D (nucleotide position 16507; nsp13), a fragment was amplified by PCR using primers PEDV-15792-F (5′-CAAGGAGGAGAGCGTTAAGTCATCCTATG-3′) and PEDV-16634-R (5′-TGCGTTGAGTGACAACGTTGTAGTCCT-3′). The fragment was gel-purified and sequenced using primer PEDV-16322-F (5′-GCGGTCGACTCACTTTGTGTGAAAGC-3′).
Growth kinetics.
Vero cell monolayers in six-well plates were infected with each virus at a multiplicity of infection (MOI) of 0.01. After a 1-h absorption, the cell monolayers were washed twice with PBS and then maintained in the maintenance medium as described above. The supernatants were collected at multiple time points, and the virus titers were determined in 96-well plates as the number of 50% tissue culture infective doses (TCID50) by the Reed-Muench method (58).
Study design of experimental infection in pigs.
Three PEDV-naive sows were assigned to three groups (TC-PC177, PC21A, and mock inoculation), and each was housed in individual biosafety level 2 rooms for natural farrowing. At 4 days of age, the piglets were inoculated orally with TC-PC177 P10C8 (n = 10) at a dose of 5 log10 PFU/piglet. Wild-type PC21A infection (n = 11; 10 log10 GE/piglet) and mock infection (n = 9, PBS) were used as the positive and negative controls, respectively (31). We used 10 log10 GE/piglet for PC21A inoculation because 7.7 log10 GE of this strain caused a 10-day-old piglet severe diarrhea within 1 dpi (55). One pig from the mock group, two diarrheic pigs from the TC-PC177 group, and two diarrheic pigs from the PC21A group were randomly selected and euthanized at 3 dpi for histopathological examinations. All other piglets were kept for the evaluation of morbidity, mortality, and fecal PEDV RNA shedding unless piglets became moribund and were euthanized. At 19 dpi (TC-PC177 group) and 20 dpi (mock and PC21A groups), the piglets were challenged orally with the PC21A strain with 12 log10 GE/pig. This challenge dose for 3-week-old PEDV-exposed pigs was optimized in our previously study (31). We observed clinical signs for 7 days postchallenge (dpc). Blood samples of piglets were collected at 1 day prior to the challenge and at 7 dpc for serological assays.
Gnotobiotic (Gn) pigs were used to evaluate the pathogenesis of two infectious clone-derived recombinant viruses, icPC22A and icPC22A-S1Δ197. The Gn pigs were derived and raised as described previously (59). Nine Gn piglets were assigned to three experimental groups, and piglets in each group were housed in the same isolator. At 5 days of age, piglets were orally inoculated with the plaque-purified, first passage of icPC22A (n = 3; 100 PFU/pig) or icPC22A-S1Δ197 (n = 4; 100 PFU/pig) or mock inoculated (n = 2; PBS). This dose corresponded to 1,000 median pig diarrhea doses (PDD50) of PC22A in 4-day-old cesarean-derived, colostrum-deprived piglets, as we determined previously (28). At the acute phase of infection, one diarrheic piglet of each PEDV-inoculated group was euthanized for histopathological examination.
After inoculation, all piglets in these experimental groups were evaluated daily for clinical signs, such as diarrhea, vomiting, anorexia, and depression. Fecal consistency was examined by collection of rectal swabs, and the fecal consistency was scored as follows: 0, solid; 1, pasty; 2, semiliquid (mild diarrhea); and 3, liquid (severe diarrhea). Total fecal RNA was extracted using a MagMax RNA isolation kit (Austin, CA, USA), and the PEDV N gene was detected by TaqMan real-time reverse-transcription quantitative PCR (RT-qPCR) as described previously (55). All animal experiments were performed according to the protocols approved by Institutional Animal Care and Use Committee (IACUC) of the Ohio State University (OSU).
Genomic sequencing of PEDV strains.
The original inoculum of TC-PC177 P10C8 was sent to the University of Minnesota Genomic Center (St. Paul, MN) for next-generation sequencing as described previously (60). We determined the genome sequence of PC22A P4, the template for icPC22A in this study, by Sanger sequencing (Plant-Microbe Genomics Facility, OSU).
Histopathological examination and IHC staining.
At necropsy, duodenum, three sections of the jejunum (proximal, middle, and distal), ileum, colon, and other major organs (heart, liver, lungs, kidneys, spleen, and mesenteric lymph nodes) were collected and fixed in 10% formalin. All tissues were trimmed, processed, embedded in paraffin, and sectioned with routine procedures (55). The immunohistochemical (IHC) staining procedure was performed as described previously, using a nonbiotin polymerized horseradish peroxidase system (BioGenex Laboratories, San Ramon, CA, USA) and monoclonal antibody SD6-29 (gift of Eric Nelson and Steven Lawson, South Dakota State University) targeting PEDV nucleocapsid (N) proteins as the primary antibody. Tissues were counterstained with hematoxylin. To stain the sialylated mucus, the intestine tissues were treated with 1% alcian blue at room temperature for 5 min.
Ratios of villous height to crypt depth (VH/CD) of the duodenum, middle jejunum, and ileum of individual piglets in the acute infection phase were measured using a computerized imaging system (MetaMorph, Olympus, Japan) as described previously (55). For each intestinal section, 15 villi and crypts were measured.
VN assay.
The virus neutralizing (VN) antibody titers against PEDV were evaluated by fluorescent focus reduction VN assay as described previously (31). Briefly, 50 μl of 4-fold-diluted serum was incubated with an equal volume of PC22A or TC-PC177 containing 200 fluorescent focus units (FFU) at 37°C for 1 h. Vero cells were inoculated with the serum-virus mixtures at 37°C for 4 h. After a washing step, the cell monolayers were cultured in DMEM containing 5% FBS to inhibit trypsin activity. At 16 h postinoculation, a PEDV-specific cell culture immunofluorescence (CCIF) assay was conducted by staining the cells with the PEDV N monoclonal antibody SD6-29 followed by fluorescein isothiocyanate (FITC)-labeled goat anti-mouse antibody (Invitrogen, Carlsbad, CA). The VN titer of a serum sample was expressed as the reciprocal of the highest dilution of a sample causing an 80% reduction in the number of FFU.
Neuraminidase treatment infectivity assay.
Monolayers of Vero cells in six-well plates were pretreated with 200 μl of 250 mM neuraminidase (NA) from Clostridium welchii (Sigma, St. Luis, MO) or of PBS at 37°C for 2 h. After treatment, the cells were washed twice, followed by inoculation with 350 FFU of recombinant virus icPC22A or icPC22A-S1Δ197 with trypsin (10 μg/ml) at 37°C for 1 h. Next, the inocula were removed from the cells, DMEM containing 2.5% FBS was added, and cells were incubated at 37°C for 16 h. The number of PEDV-infected Vero cell foci was determined by CCIF assay as described above (26).
Three-dimensional structural analysis of the S proteins of icPC22A and icPC22A-S1Δ197.
The Protein Data Bank cryo-electron microscopy (cryo-EM) structure of the S protein trimer of human coronavirus NL63 at a resolution of 3.4 Å was used (PDB accession number 5SZS) (60). The sequence identities between the S proteins of the two recombinant PEDVs, icPC22A and icPC22A-S1Δ197, and the human coronavirus NL63 S protein were 43.5% and 47.5%, respectively. Sequence alignments were generated using MOE-Align in the Molecular Operating Environment (MOE), version 2015.10 (Chemical Computing Group, Inc., Quebec, Canada). Three-dimensional (3-D) models of the S protein trimers of PEDV icPC22A and icPC22A-S1Δ197 were constructed by the homology modeling technique using MOE-Homology in MOE as previously described (61–63). For each homology modeling in MOE, we obtained 10 intermediate models and selected the intermediate 3-D models with the best scores according to the generalized Born/volume integral methodology (64). The 3-D structures were thermodynamically optimized by energy minimization using the MOE and an Amber 10:EHT force field (65, 66) combined with the generalized Born model of aqueous solvation implemented in MOE.
Statistical analysis.
The statistical analyses were performed using GraphPad Prism, version 6.0. Comparisons of VH/CD ratios, diarrhea rates, mortality rates, mean cumulative FC scores, duration of diarrhea, peak fecal PEDV RNA shedding titers before challenge between different PEDV strains, and percentage of FFU were analyzed by one-way analysis of variance (ANOVA). Values of onset of diarrhea and peak fecal PEDV RNA shedding titers postchallenge were analyzed by Student's t test. A P value of less than 0.05 was considered significantly different.
Accession number(s).
Newly determined sequences were deposited in the GenBank under the following accession numbers: PC22A P4 for icPEDV (23), KX683006; TC-PC177 P10C8 KY499261; and PC22A P4 for icPC22A (this study), KY499262.
ACKNOWLEDGMENTS
We thank Juliette Hanson, Megan Strother, Dennis Hartzler, Ronna Wood, Sara Tallmadge, and Jeff Ogg for animal care assistance and Susan Sommer-Wagner, Xiaohong Wang, Xiang Gao, Zhongyan Lu, and Lanlan Zheng for technical assistance. We thank Anastasia Vlasova, Chang-Won Lee, and Patricia Boley for their critical reviews of the manuscript. The monoclonal antibody SD6-29 was kindly provided by Eric Nelson and Steven Lawson at South Dakota State University.
This study was supported in part by the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award number 2015-67015-23067 (Q.W., principal investigator [PI]; L.J.S., co-PI), the National Institutes of Health, contract number U19 AI109761 (R.S.B., PI), and National Institutes of Health grant AI089728 (R.S.B., co-PI). Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. Y.Q. received the Guangxi Scientist Development Scholarship from the Guangxi Veterinary Institute, China.
REFERENCES
- 1.Madson DM, Arruda PHE, Magstadt DR, Burrough ER, Hoang H, Sun D, Bower LP, Bhandari M, Gauger PC, Stevenson GW, Wilberts BL, Wang C, Zhang J, Yoon KJ. 2016. Characterization of porcine epidemic diarrhea virus isolate US/Iowa/18984/2013 infection in 1-day-old Cesarean-derived colostrum-deprived piglets. Vet Pathol 53:44–52. doi: 10.1177/0300985815591080. [DOI] [PubMed] [Google Scholar]
- 2.Jung K, Annamalai T, Lu Z, Saif LJ. 2015. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet Microbiol 178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Fang L, Xiao S. 2016. Porcine epidemic diarrhea in China. Virus Res 226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhury B, Dastjerdi A, Doyle N, Frossard J-P, Steinbach F. 2016. From the field to the lab—an European view on the global spread of PEDV. Virus Res 226:40–49. doi: 10.1016/j.virusres.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paarlberg PL. 2014. Updated estimated economic welfare impacts of porcine epidemic diarrhea virus (PEDV). Purdue University, W. Lafayette, IN: https://pdfs.semanticscholar.org/919d/0d660cad088e31ff4bbd678442fbfabd6ac6.pdf. [Google Scholar]
- 6.Diep NV, Norimine J, Sueyoshi M, Lan NT, Yamaguchi R. 2017. Novel porcine epidemic diarrhea virus (PEDV) variants with large deletions in the spike (S) gene coexist with PEDV strains possessing an intact S gene in domestic pigs in Japan: a new disease situation. PLoS One 12:e0170126. doi: 10.1371/journal.pone.0170126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan CAM, Haijema BJ, Schellen P, Wichgers Schreur P, te Lintelo E, Vennema H, Rottier PJM. 2008. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J Virol 82:6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L, Li F. 2015. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol 89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Ma Y, Yang Y, Zheng Y, Shang J, Zhou Y, Jiang S, Du L, Li J, Li F. 2016. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J Biol Chem 291:24779–24786. doi: 10.1074/jbc.M116.740746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shirato K, Maejima M, Islam MT, Miyazaki A, Kawase M, Matsuyama S, Taguchi F. 2016. Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J Gen Virol 97:2528–2539. doi: 10.1099/jgv.0.000563. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Luo R, He Q, van Kuppeveled FJM, Rottier PJ, Bosch BJ. 2017. Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res 235:6–13. doi: 10.1016/j.virusres.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang S-H, Bae J-L, Kang T-J, Kim J, Chung G-H, Lim C-W, Laude H, Yang M-S, Jang Y-S. 2002. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol Cells 14:295–299. [PubMed] [Google Scholar]
- 13.Deng F, Ye G, Liu Q, Navid MT, Zhong X, Li Y, Wan C, Xiao S, He Q, Fu ZF, Peng G. 2016. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses 8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch B-J. 2016. Cellular entry of the porcine epidemic diarrhea virus. Virus Res 226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oka T, Saif LJ, Marthaler D, Esseili MA, Meulia T, Lin C-M, Vlasova AN, Jung K, Zhang Y, Wang Q. 2014. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains, including a novel strain with a large deletion in the spike gene. Vet Microbiol 173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda T, Murakami S, Takahashi O, Miyazaki A, Ohashi S, Yamasato H, Suzuki T. 2015. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch Virol 160:2565–2568. doi: 10.1007/s00705-015-2522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hulswit RJG, de Haan CAM, Bosch B-J. 2016. Coronavirus spike protein and tropism changes. Adv Virus Res 96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox E, Pensaert MB, Callebaut P. 1993. Intestinal protection against challenge with transmissible gastroenteritis virus of pigs immune after infection with the porcine respiratory coronavirus. Vaccine 11:267–272. doi: 10.1016/0264-410X(93)90028-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godet M, Grosclaude J, Delmas B, Laude H. 1994. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 68:8008–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brim TA, VanCott JL, Lunney JK, Saif LJ. 1995. Cellular immune responses of pigs after primary inoculation with porcine respiratory coronavirus or transmissible gastroenteritis virus and challenge with transmissible gastroenteritis virus. Vet Immunol Immunopathol 48:35–54. doi: 10.1016/0165-2427(94)05416-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.VanCott JL, Brim TA, Lunney JK, Saif LJ. 1994. Contribution of antibody-secreting cells induced in mucosal lymphoid tissues of pigs inoculated with respiratory or enteric strains of coronavirus to immunity against enteric coronavirus challenge. J Immunol 152:3980–3990. [PubMed] [Google Scholar]
- 22.Suzuki T, Shibahara T, Yamaguchi R, Nakade K, Yamamoto T, Miyazaki A, Ohashi S. 2016. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J Gen Virol 97:1823–1828. doi: 10.1099/jgv.0.000513. [DOI] [PubMed] [Google Scholar]
- 23.Beall A, Yount B, Lin C-M, Hou Y, Wang Q, Saif L, Baric R. 2016. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio 7:e01451-15. doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas JT, Chen Q, Gauger PC, Giménez-Lirola LG, Sinha A, Harmon KM, Madson DM, Burrough ER, Magstadt DR, Salzbrenner HM, Welch MW, Yoon K-J, Zimmerman JJ, Zhang J. 2015. Effect of porcine epidemic diarrhea virus infectious doses on infection outcomes in naïve conventional neonatal and weaned pigs. PLoS One 10:e0139266. doi: 10.1371/journal.pone.0139266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Wang Q. 2016. Reverse transcription-PCR assays for the differentiation of various US porcine epidemic diarrhea virus strains. J Virol Methods 234:137–141. doi: 10.1016/j.jviromet.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C-M, Gao X, Oka T, Vlasova AN, Esseili MA, Wang Q, Saif LJ. 2015. Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J Virol 89:3332–3342. doi: 10.1128/JVI.03196-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CM, Hou Y, Marthaler DG, Gao X, Liu X, Zheng L, Saif LJ, Wang Q. 2017. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet Microbiol 201:62–71. doi: 10.1016/j.vetmic.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Lin C-M, Annamalai T, Gao X, Lu Z, Esseili MA, Jung K, El-Tholoth M, Saif LJ, Wang Q. 2015. Determination of the infectious titer and virulence of an original US porcine epidemic diarrhea virus PC22A strain. Vet Res 46:109. doi: 10.1186/s13567-015-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin C-M, Saif LJ, Marthaler D, Wang Q. 2016. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res 226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. 2014. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg Infect Dis 20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin C-M, Annamalai T, Liu X, Gao X, Lu Z, El-Tholoth M, Hu H, Saif LJ, Wang Q. 2015. Experimental infection of a US spike-insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross-protection to the original US PEDV infection. Vet Res 46:134. doi: 10.1186/s13567-015-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, Gauger PC, Stafne MR, Thomas JT, Madson DM, Huang H, Zheng Y, Li G, Zhang J. 2016. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-indel-variant strains in conventional neonatal piglets. J Gen Virol 97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- 33.Schwegmann-Wessels C, Zimmer G, Schroder B, Breves G, Herrler G. 2003. Binding of transmissible gastroenteritis coronavirus to brush border membrane sialoglycoproteins. J Virol 77:11846–11848. doi: 10.1128/JVI.77.21.11846-11848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pensaert M, Callebaut P, Vergote J. 1986. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q 8:257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- 35.Chang H-W, Egberink HF, Halpin R, Spiro DJ, Rottier PJ. 2012. Spike protein fusion peptide and feline coronavirus virulence. Emerg Infect Dis 18:1089–1095. doi: 10.3201/eid1807.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terada Y, Shiozaki Y, Shimoda H, Mahmoud HY, Noguchi K, Nagao Y, Shimojima M, Iwata H, Mizuno T, Okuda M, Morimoto M, Hayashi T, Tanaka Y, Mochizuki M, Maeda K. 2012. Feline infectious peritonitis virus with a large deletion in the 5′-terminal region of the spike gene retains its virulence for cats. J Gen Virol 93:1930–1934. doi: 10.1099/vir.0.043992-0. [DOI] [PubMed] [Google Scholar]
- 37.Fazakerley JK, Parker SE, Bloom F, Buchmeier MJ. 1992. The V5A13. 1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology 187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau SKP, Woo PCY, Li KSM, Huang Y, Wang M, Lam CSF, Xu H, Guo R, Chan K-H, Zheng B-J, Yuen K-Y. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballesteros ML, Sanchez CM, Enjuanes L. 1997. Two amino acid changes at the N terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology 227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng F, Suo S, Zarlenga DS, Cong Y, Ma X, Zhao Q, Ren X. 2014. A phage-displayed peptide recognizing porcine aminopeptidase N is a potent small molecule inhibitor of PEDV entry. Virology 456–457:20–27. doi: 10.1016/j.virol.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J-E, Park E-S, Yu J-E, Rho J, Paudel S, Hyun B-H, Yang D-K, Shin H-J. 2015. Development of transgenic mouse model expressing porcine aminopeptidase N and its susceptibility to porcine epidemic diarrhea virus. Virus Res 197:108–115. doi: 10.1016/j.virusres.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan Z, Yin J, Wang Z, Chen P, Li Y, Tang L. 2015. Identification of the functional domain of the porcine epidemic diarrhoea virus receptor. J Gen Virol 96:2656–2660. doi: 10.1099/vir.0.000211. [DOI] [PubMed] [Google Scholar]
- 43.Li BX, Ge JW, Li YJ. 2007. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 365:166–172. doi: 10.1016/j.virol.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wicht O, Li W, Willems L, Meuleman TJ, Wubbolts RW, van Kuppeveld FJM, Rottier PJM, Bosch BJ. 2014. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J Virol 88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirato K, Matsuyama S, Ujike M, Taguchi F. 2011. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J Virol 85:7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Licitra BN, Duhamel GE, Whittaker GR. 2014. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses 6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultze B, Krempl C, Ballesteros ML, Shaw L, Schauer R, Enjuanes L, Herrler G. 1996. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J Virol 70:5634–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun D, Feng L, Shi H, Chen J, Cui X, Chen H, Liu S, Tong Y, Wang Y, Tong G. 2008. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet Microbiol 131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Li W, Lucio de Esesarte E, Guo H, van den Elzen P, Aarts E, van den Born E, Rottier PJM, Bosch B-J. 5 April 2017. Cell attachment domains of the PEDV spike protein are key targets of neutralizing antibodies. J Virol doi: 10.1128/JVI.00273-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, Li Z, Zou Y, Wicht O, van Kuppeveld FJM, Rottier PJM, Bosch BJ. 2013. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One 8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jengarn J, Wongthida P, Wanasen N, Frantz PN, Wanitchang A, Jongkaewwattana A. 2015. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J Gen Virol 96:2206–2218. doi: 10.1099/vir.0.000184. [DOI] [PubMed] [Google Scholar]
- 52.Teeravechyan S, Frantz PN, Wongthida P, Chailangkarn T, Jaru-Ampornpan P, Koonpaew S, Jongkaewwattana A. 2016. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res 226:152–171. doi: 10.1016/j.virusres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan B, Yu Z, Pang F, Xu X, Zhang B, Guo R, He K, Li B. 2017. Characterization of a pathogenic full-length cDNA clone of a virulent porcine epidemic diarrhea virus strain AH2012/12 in China. Virology 500:50–61. doi: 10.1016/j.virol.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofmann M, Wyler R. 1988. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol 26:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung K, Wang Q, Scheuer KA, Lu Z, Zhang Y, Saif LJ. 2014. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg Infect Dis 20:662. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang R, Li Y, Cowley TJ, Steinbrenner AD, Phillips JM, Yount BL, Baric RS, Weiss SR. 2015. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J Virol 89:3598–3609. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 59.Saif LJ, Ward LA, Yuan L, Rosen BI, To TL. 1996. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch Virol Suppl 12:153–161. [DOI] [PubMed] [Google Scholar]
- 60.Walls AC, Tortorici MA, Frenz B, Snijder J, Li W, Rey FA, DiMaio F, Bosch B-J, Veesler D. 2016. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takanashi S, Wang Q, Chen N, Shen Q, Jung K, Zhang Z, Yokoyama M, Lindesmith LC, Baric RS, Saif LJ. 2011. Characterization of emerging GII.g/GII.12 noroviruses from a gastroenteritis outbreak in the United States in 2010. J Clin Microbiol 49:3234–3244. doi: 10.1128/JCM.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motomura K, Oka T, Yokoyama M, Nakamura H, Mori H, Ode H, Hansman GS, Katayama K, Kanda T, Tanaka T, Takeda N, Sato H, Norovirus Surveillance Group of Japan. 2008. Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history. J Virol 82:11247–11262. doi: 10.1128/JVI.00897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Motomura K, Yokoyama M, Ode H, Nakamura H, Mori H, Kanda T, Oka T, Katayama K, Noda M, Tanaka T, Takeda N, Sato H, Norovirus Surveillance Group of Japan. 2010. Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J Virol 84:8085–8097. doi: 10.1128/JVI.02125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Labute P. 2008. The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem 29:1693–1698. doi: 10.1002/jcc.20933. [DOI] [PubMed] [Google Scholar]
- 65.Gerber PR, Müller K. 1995. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. J Comp Aided Mol Des 9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 66.Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, We X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. 2008. Amber 10 users' manual. University of California, San Francisco, CA. [Google Scholar]