Abstract
Haematopoietic stem cells (HSC) are situated at the apex of the haematopoietic differentiation hierarchy, ensuring the life-long supply of mature haematopoietic cells and forming a reservoir to replenish the haematopoietic system in case of emergency such as acute blood loss. To maintain a balanced production of all mature lineages and at the same time secure a stem cell reservoir, intricate regulatory programs have evolved to control multi-lineage differentiation and self-renewal in haematopoietic stem and progenitor cells (HSPCs). Leukaemogenic mutations commonly disrupt these regulatory programs causing a block in differentiation with simultaneous enhancement of proliferation. Here, we briefly summarize key aspects of HSPC regulatory programs, and then focus on their disruption by leukaemogenic fusion genes containing the mixed lineage leukaemia (MLL) gene. Using MLL as an example, we explore important questions of wider significance that are still under debate, including the importance of cell of origin, to what extent leukaemia oncogenes impose specific regulatory programs and the relevance of leukaemia stem cells for disease development and prognosis. Finally, we suggest that disruption of stem cell regulatory programs is likely to play an important role in many other pathologies including ageing-associated regenerative failure.
Keywords: HSPC, MLL gene, AML, ALL
Regulatory programs in normal haematopoietic stem and progenitor cells (HSPCs)
HSCs reside in the bone marrow, where they represent in the mouse approximately 1 in 20,000 nucleated haematopoietic cells. Though mostly quiescent [1], HSCs actively contribute to steady state haematopoiesis [2], which in turn is largely driven by long-lived multipotent progenitor cells [3, 4]. To maintain a balanced production of the more than ten distinct mature haematopoietic cell types throughout adult life, both HSC self-renewal and multi-lineage differentiation need to be tightly controlled. Despite their rarity, powerful protocols for the prospective isolation of HSCs have been developed, with the latest protocols providing over 60% purity when assayed by single cell transplantation [5]. Nevertheless, their low abundance has been a major obstacle towards the generation of detailed insights into the regulatory programs defining HSC function, because many classical biochemical assays require 1000 or 100,000 of cells that simply are not available for highly purified HSCs. There is renewed hope that new single cell profiling technologies will provide a step-change in our understanding of molecular processes controlling HSC function. This is based on a number of recent publications that (i) defined the transcriptional landscape at single cell resolution [6], (ii) used single cell profiling to train literature-curated network models [7], as well as validating an HSPC regulatory network model built on detailed characterization of gene regulatory sequences [8] and (iii) characterized the transcriptional status of multipotent myeloid precursor cells at single cell resolution [9].
Despite the clear challenges in deciphering regulatory programs operating in HSPCs, some key concepts have nevertheless been identified over the past two decades. Firstly, HSC self-renewal most likely requires extracellular external signals (for example from the bone marrow niche), since HSCs cannot be propagated efficiently in vitro, where a drive to differentiate outstrips any inherent self-renewal capability [10]. Several HSC niches have been proposed with most research focussed on the endosteal [11] and vascular niches [12]. In vivo live imaging [13] partially unified these two concepts by showing that HSCs expansion after BM impairment is taking place in a highly vascularized area where osteoblasts are surrounded by blood vessels. Of note, it has been reported that the majority of HSCs need, for their maintenance, factors secreted by perivascular stromal and endothelial cells [14–16]. Secondly, core circuits of transcription factors define the cellular identity of haematopoietic lineages, such as erythroid, megakaryocyte, or granulocytic cells [17–19]. Multipotent cells, on the other hand, are characterized by low-level co-expression of genes affiliated with distinct lineage programs, such as the co-expression of erythroid and myeloid genes within the same single cells [20–24]. Thirdly, the classical haematopoietic tree with a sequence of binary fate choices is most likely not an accurate reflection of in vivo haematopoietic production, since for example HSCs with restricted or even uni-lineage long-term reconstitution ability have been identified [25–27], and there seem to be multiple differentiation routes that can converge on some of the mature lineages [28]. As with any other biological system, much can be learnt about the properties of HSPCs by studying systemic perturbations. In this context, particular attention has been paid to studying leukaemia models as system-wide perturbations since direct translational relevance will come from a better understanding of the molecular processes that operate in normal HSPCs and are disrupted in leukaemia.
Perturbation of HSPC regulatory programs in acute myeloid leukaemia
Research during the past 15 years has established that the cell of origin receiving the initial leukaemogenic mutation in acute myeloid leukaemia (AML) is situated within the HSPC compartment [29–31]. Of interest, while full-blown frank leukaemia may be characterized by the expression of surface markers associated with more mature progenitors such as the granulocyte-macrophage progenitor (GMP) [32, 33], it is now thought that the initial mutation more likely occurs within cells residing in the most immature stem and progenitor compartment [34, 35]. Moreover, it has been shown that the ability of an oncogene to transform cells at increasingly mature HSPC stages is oncogene specific [36]. It has been hypothesised that these oncogene specific effects are at least in part related to the degree to which the transforming gene can induce self-renewal in maturing HSPCs that are increasingly distant from the most immature stem cells that naturally possess such activity.
Despite the apparent phenotypic variety of different types of AML driven by both oncogene and cell-of-origin specific effects, common themes have emerged of how HSPC regulatory programs need to be perturbed to cause the leukaemic AML phenotype. This has lead to a classification into types 1 and 2 AML oncogenes: drivers of proliferation or of the block in differentiation, respectively [37, 38]. A combination of types 1 and 2 oncogenes would then create a perfect storm for leukaemia development, as illustrated for example by the combination of activating growth factor mutations (Flt3 or c-Kit) in combination with translocations involving the Runx1 transcription factor. Here, the growth factor mutations cause enhanced proliferation and survival of progenitors, thought to be simultaneously blocked from differentiation by the dominant-negative action of Runx1 fusion proteins [39–42]. However, it needs to be noted that with more detailed molecular characterization of additional AML leukaemogenic mutations, a more complex picture is emerging where individual mutations cannot be classified into strict categories, as they may affect both proliferation and differentiation, and may do so differently depending on which other mutations are present. This suggests that the original model is not only simplistic but in fact may not be useful any longer.
Of particular interest are leukaemogenic mutations commonly associated with childhood acute leukaemias, because it has become apparent that these tumours often have a comparatively low genetic complexity, where two (or even one) genetic hits are thought to be sufficient to cause malignant transformation [43, 44]. Consequently, these diseases offer a more readily interpretable model for the analysis of perturbed HSPC regulatory programs. In the subsequent sections, we will explore in more detail the acute leukaemia cell of origin, and then focus on leukaemogenic perturbations involving the expression of fusion proteins that contain the N-terminal portion of the MLL gene.
Leukaemia cell of origin
Disease development is characterized by dynamic processes that often begin years before clinical onset. If genetic alterations give survival advantages, the consequence is the growth of a prevalent population, as introduced by Peter Nowell [45] in 1976. Accumulation of sequential somatic mutations in a single clone and subclonal selection lead to the growth of a predominant population with survival advantages. This model has been revisited over the years [46, 47]. For many cancers, the target cell of the transformation events is still unknown. The first evidence for a stem cell origin of cancer was demonstrated by Philip Fialkow in Chronic Myelogenous Leukaemia (CML) [48]. Ten years later, L. J. Smith and colleagues, showed that blast cells from patients with acute leukaemia can co-express markers of both myelopoiesis and lymphopoiesis [32]. The hypothesis for the existence of leukaemia stem cells (LSCs) able to extensively proliferate and sustain leukaemia, came from the observation that proliferation of the majority of leukaemic blasts in AML is finite [49], and only some of the leukaemic cells could form spleen colonies when transplanted in vivo [30]. In the late ‘90s, the use of mouse xenograft models demonstrated that only CD34+CD38− human AML purified cells (which can be as low as 0.2% of the total leukaemic cells) were able to repopulate non-obese diabetic mice with severe combined immunodeficiency disease (NOD/SCID mice) and transfer AML. This subset of LSC cells was defined as SCID leukaemia-initiating cells (SL-IC). A significant number of those (2%) retained the CD34+CD38− phenotype in vivo confirming their self-renewal property [34]. These observations suggest that for most AML subtypes, immature normal HSPCs rather than committed progenitors, are the target for leukaemic transformation.
However, several studies support an alternative model whereby AML LSCs derive from a downstream progenitor phenotypically identified as CD34+CD38−CD90− [50]. Of note, fusion transcripts for the AML1-ETO leukaemia oncogene were detected in both blast cells (CD90−) and normal HSCs (CD90+). However, the latter could differentiate into normal lineage committed cells in vitro without expanding the pool of leukaemic CD90− blasts, indicating that chromosomal translocation may occur in the HSC compartment but transformation into frank AML may require additional mutations that take place in a more downstream population being CD90−. Additionally, primary human CD34+ AML samples were shown to cluster into two CD90− immunophenotypic groups: GMP-like and more immature LMPP-like cells [51]. Interestingly, gene expression profiles of these LSC populations showed higher similarity with their respective normal counterparts rather than the normal HSC profile.
Further evidence that there is not just a single HSPC regulatory program that is susceptible to leukaemic transformation comes from Acute Promyelocytic Leukaemia (APML) patients where the PML-RARα fusion gene is present in a more differentiated population being CD34−CD38+ [52]. Jamieson et al., showed that non self-renewing cells were transformed into LSC in human blast crisis chronic myeloid leukaemia (CML), where activation of the Wnt/β-catenin pathway enhanced self-renewal ability of BCR-ABL expressing granulocyte-macrophage progenitors [53]. This study further shows that LSCs could also derive from a differentiated mature cell which can re-acquire self-renewal properties to generate a tumorigenic cell. Furthermore, in vivo transplantation mouse models using primary human MLL-AF4 and -ENL leukaemias have demonstrated that committed progenitors with a CD34+CD38+CD19+ surface marker phenotype were able to give rise to infant ALL [54]. Finally, exciting studies have also provided a definition of the leukaemia cell of origin based on the chromatin landscape, which represents a powerful platform for the identification of new epigenetic biomarkers [55, 56].
However, identification of LSCs cannot be achieved solely by means of cell surface markers, but critically requires functional assays. Thus, understanding the pathways controlling LSCs properties is crucial for the development of new therapies. Oxidative stress variations have been observed to differently correlate with the self-renewal potential of LSC and HSC [57]; inhibitors of the aryl- hydrocarbon receptor (AhR) pathway have been described to block AML cell differentiation [58]; epigenetic inhibitors have been developed to selectively cause apoptosis of AML LSC [59]. Mutations in genes involved in epigenetic processes are indeed involved in the early event of AML evolution, also defined as pre-leukaemic state [60]. These genes are involved in DNA methylation such as DNA methyltransferase 3 alpha (DNMT3A), histones modifications like sex comb like 1 (ASXL1) and chromatin looping like IKAROS family zinc finger 1(IKZF1). Finally, global chromatin changes result also in a complex deregulation of transcriptional programs. It has been recently described that even in the absence of MLL rearrangements, MEIS1 transcriptional program can be activated, as a consequence of its promoter hypomethylation, by DNMT3A mutations [61].
Cell of origin for pre-leukaemia development
The emergence of subclones after treatment [62, 63] causing relapse has been described in several leukaemias including paediatric leukaemia [64] and CML [65]. Hope and colleagues observed in 2004 proliferation rate heterogeneity in LSCs of AML patients [66]. Quiescence of a subset of LSCs may not only explain the recent discovery that mutations in genes regulating proliferation may be a late event during leukaemogenesis [60], but is also consistent with a model whereby the quiescent normal HSCs are the cell of origin for at least some initiating pre-leukaemic mutations. The earliest clonality studies suggesting the existence of pre-leukaemic HSCs were published by Philip J. Fialkow and colleagues, where X chromosome-linked glucose-6-phosphate dehydrogenase (G6PD) was used as a marker to study clonal remission [67]. At remission, blasts no longer carried the cytogenetic aberration but expressed the “leukaemic” G6PD allele suggesting a leukaemic clonal remission. Long-term remission in AML patients with AML1-ETO translocation similarly showed fusion gene levels in normal single cell derived myeloid and erythroid colonies obtained via isolation of purified HSCs [68]. Further insights into the pre-leukaemic clonal evolution model come from the comparison of DNA copy number abnormalities between diagnostic and relapse samples of paediatric patients with ALL. Fifty-two percent of clones at relapse were antecedent to clones at diagnosis, and the relationship between them was further confirmed via Ig and T cell antigen receptor (TCR) deletions analysis. This unequivocally confirms that the majority of relapse clones derive from a common ancestral one present in small number at diagnosis that acquires additional genetic alterations before emerging as relapse clone [64].
The first study that isolated pre-leukaemic cells in AML patients was published in 2012 [69]. Target exome sequencing was applied on human residual HSC Lin−CD34+CD38− lacking expression of the AML markers TIM3 and CD99. Even though these cells appeared functionally normal, generating long-term engraftment into NSG mice with both myeloid and lymphoid lineage, they were also demonstrated to be the cellular reservoir causing relapse. In fact, “silent” mutations in critical epigenetic regulators like TET2 were identified in five out of six cases analysed both in purified leukaemic cells and in a fraction of residual HSCs. However, in all cases FLT3-ITD and IDH1 genes were found mutated in AML cells only. Finally, through single cells analysis, sequential acquisition of those mutations in pre-leukaemic cells was demonstrated. Taken together therefore, current evidence suggests that pre-leukaemic cells are closely related to normal HSC and form a cellular reservoir where primary mutations (“silent” mutations) accumulate until secondary events (as FLT3-ITD mutation) confer proliferative advantage that causes frank leukaemia. Perturbation of regulatory programs during leukaemogenesis therefore is likely to be a multi-step process, suggesting that detailed knowledge of the stepwise subversion from normal to pre-leukaemic to leukaemic will be required to obtain a detailed molecular understanding of the underlying processes. As outlined in the previous section, knowing the fusion oncogenes is only part of the story, because the cellular context within which they are activated is equally important.
MLL-rearranged leukaemia
Leukaemias characterized by chromosomal translocations affecting the MLL gene, encoding a histone H3 lysine 4 (H3K4) methyltransferase, on chromosome segment 11q23 [70], have poor prognosis [71]. MLL rearrangements are responsible for more than 70% of infant (<1 year) leukaemias with either myeloid (AML), or lymphoid (ALL) immonophenotype [43]. MLL translocations occur also in 10% of adult AML [72] and in therapy related acute leukaemias (t-AL), often characterized as tAML, following treatment with topoisomerase II inhibitors [73]. The MLL gene has been found rearranged with multiple partners (more than 50 translocation partners have been identified). Among the most common, MLL-AF9 t(9;11) is mainly associated with AML in both paediatric and adult patients; MLL-AF4 t(4;11) is associated with a lymphoid/mixed-lineage phenotype (mainly ALL), and MLL-ENL t(11;19) drives paediatric ALL and adult AML (only a small fraction of adult patients develop MLL-ENL ALL) [70]. The dismal prognosis of MLL-rearranged (MLL-r) leukaemia is associated with disease relapse [74].
Retroviral mouse models of MLL-r leukaemia
Several mouse models bearing MLL fusion proteins have been developed in order to understand MLL fusion mediated leukaemogenesis. However, discrepancies between mouse leukaemic models and human leukaemias in terms of ability of MLL fusions to generate the same lineage leukaemia (AML or ALL), as observed in patients, and latency in leukaemia development, make the deconstruction of MLL-r leukaemia development a challenge. One of the first mouse leukaemic models with MLL fusion, was described by Lavau et al. in 1997 [75]. Retroviral transduction of MLL-ENL into lineage-depleted or c-kit sorted mouse bone marrow (BM) HSPCs was followed by culture in methylcellulose. Infected haematopoietic progenitors maintained self-renewal potential in vitro. MLL-ENL expressing progenitors were phenotypically immature myelomonocytic cells being c-kit+, Mac1+ and Sca-1−, and when cultured with granulocyte colony-stimulating factor (G-CSF), terminally differentiated in mature granulocytes. Moreover, their injection into SCID mice, caused death due to AML development. However, while the MLL-ENL translocation gives rise also to human ALL [70], no lymphoid markers were expressed on MLL-ENL expressing cells in the retroviral mouse model. More recent studies demonstrated transforming ability of MLL fusions (both in vitro and in vivo) also in more differentiated haematopoietic progenitor cells [76] such as common myeloid progenitors (CMPs) and granulocytic/monocytic-restricted progenitors (GMPs). By contrast, megacaryocytic/erythroid-restricted progenitors (MEPs) injected mice did not develop AML. Overall, this suggests that MLL-ENL is able to confer a self-renewal regulatory program to some but not all committed progenitors [77]. Subsequent genome wide studies demonstrated that MLL-ENL transcriptional reprogramming happens fast during transformation, and the immediate phase of leukaemogenesis is similar to the progression phase characterized not only by HoxA cluster up-regulation but also by general transcriptional downregulation involving key haematopoietic transcription factors Gata2, Gfi1b and Zfpm1 [78].
Of note, murine models [76, 79, 80] often fail to reproduce the biphenotypic feature observed in MLL-r leukaemia patients with co-expression of some myeloid and lymphoid genes. Zeisig et al. [81] first reported a MLL-ENL transformation model based on a biphenotypic lymphoid/myeloid phenotype. Similarly, when infected murine BM cells were cultured in methylcellulose with Flt3-ligand, stem cell factor (SCF) and interleukin-7 (IL-7) to sustain lymphopoiesis [82], B220+CD19+ and B220+CD19− B cells appeared after 4 weeks. The latter generated leukaemia in vivo characterized by splenomegaly, lymph node enlargement and an overgrown thymus, where cells showed a myeloid morphology, yet expressed the B220 lymphoid marker.
Non-retroviral models of MLL-r leukaemia
All the studies described so far used retroviral models, which may not generate expression levels representative of the endogenous gene loci involved in the translocation events. Since expression levels are critical determinants of cellular programming, it is not surprising that constitutive and conditional knock-in mouse models have provided another powerful approach to analyse MLL-r leukaemias. In 1996, Corral et al. [83] engineered expression of the MLL-AF9 oncogene via homologous recombination [84]. Engineered mice, bearing the MLL-AF9 fusion, developed leukaemia restricted to the myeloid lineage despite of the widespread expression of the fusion gene. Moreover, AML development was characterized by long latency suggesting the need of genetic alterations for complete leukaemic transformation. A Cre-Lox recombination approach generated MLL-AF9 [85] and MLL-ENL [86] mouse models able to rapidly develop AML. Furthermore, de novo MLL-ENL translocations caused myeloproliferative-like myeloid leukaemia development in all mice in which Cre recombinase was expressed from Lmo2, Lck and Rag1 genes (expressed in non-differentiated cells, T-cell linage and early staged of lymphoid lineage, respectively); while no haematological malignancies were observed in MLL-ENL Cd19-Cre (gene expressed in B cell lineage) [87]. Overall, these data demonstrate that the MLL-ENL fusion is leukaemic when expressed in stem cells and progenitors excluding the B-cell compartment [87, 88].
Importantly, an endogenous knock-in mouse model using the MLL-AF9 oncogene [89], demonstrated that GMPs were refractory to leukaemic transformation in complete contrast to previous retroviral studies [76, 79]. While only 100 HSCs and 2500 CMPs from knock-in mice were able to produce AML in the majority of the recipients, all mice transplanted with higher doses of GMPs did not develop any disease. The ability of GMPs to be transformed in retroviral studies seems to be related to the different levels of oncogene expressed (170-fold higher then knock-in GMPs). By contrast, a doxycycline (Dox) inducible mouse model, targeting MLL-ENL to the 3′ UTR of the Col1a1 gene [90], showed that both HSCs and MPPs failed to induce leukaemia in vivo [33]. AML development was observed only when Dox was administrated to mice 4 weeks after HSC transplantation suggesting that MLL-ENL expression could interfere with in vivo homing. The authors therefore suggested that granulocyte-monocyte-lymphoid progenitors (GMLPs), as a subpopulation of the wider Lineage−Sca-1+c-kit+ (LSK) population [91], and GMP precursors (pGMs) represent the most permissive cellular environment for regulatory program perturbations that can cause leukaemia development. Nevertheless, a recent study published in 2016 [92] using another MLL-AF9 Dox inducible mouse model (67), showed that both long-term HSCs (LT-HSCs) and GMP were transformed by MLL-AF9 induction, where transformation in LT-HSCs resulted in a more aggressive AML phenotype. Taken together, rather than providing conclusive answers to the molecular processes underlying AML development in patients, these studies further highlight the intricacies of perturbing regulatory programs and the complex interplay of parameters such as cellular context and oncogene expression level. Given that no mouse model seems perfect, it may be argued that research efforts need to be refocused onto molecular studies with human patient samples, especially since genome engineering has become so much easier with the new clustered regulatory interspaced short palindromic repeats (CRISR) system [93].
Concluding remarks
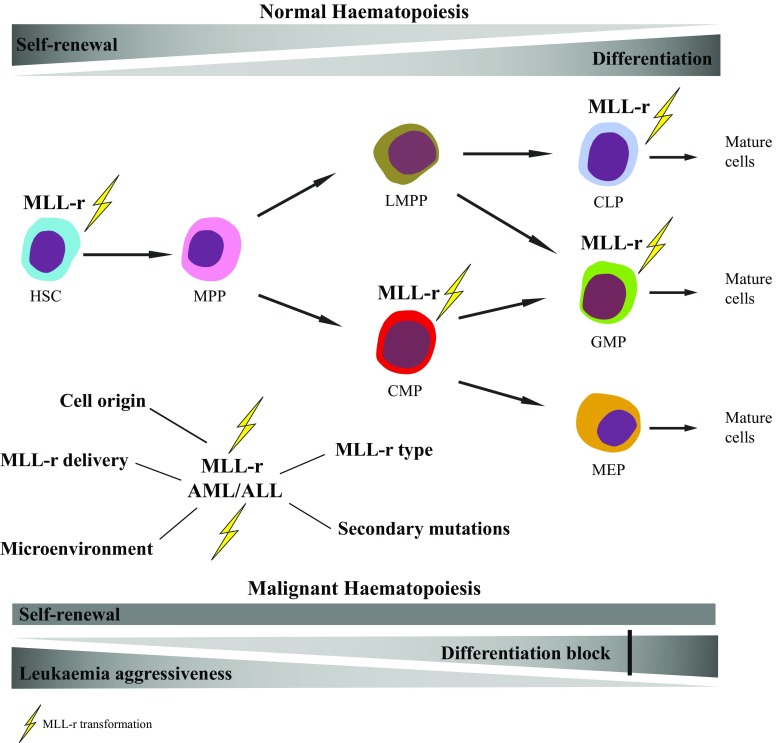
Regulatory programs in HSPCs need to be finely balanced to maintain normal haematopoiesis and are vulnerable to genetic perturbations that result in the development of malignant disease. Despite the remaining disagreements between the various retroviral and transgenic models, it is clear that the cell of origin influences leukaemia biology and prognosis (see Fig. 1). Secondly, even within a group of related oncogenes such as all the MLL fusions, the type of MLL translocation together with the nature of the cellular environment strongly influences leukaemia onset and phenotype [54, 88].
Fig. 1.
Perturbation of haematopoietic development by MLL rearrangements (MLL-r). MLL-r impair self-renewal and differentiation properties of HSCs and HSPCs. MLL driven leukaemic transformation has been mainly described in HSC, CMP, GMP and CLP [31, 33, 75, 76, 81, 86, 87, 92]. MPP and LMPP progenitors are also targets of MLL transformation as cellular permissiveness might be influenced by the specific strategy used to purify HSPCs [5, 91]. Biology and prognosis of AML and ALL bearing MLL translocations depend on multiple factors: cell of origin of leukaemic transformation, type of MLL-r, oncogene delivery methods, microenvironment and secondary mutations.
Further difficulties in understanding this interplay come from inconsistencies between human disease and the murine models. MLL-ENL expressing cells give rise to AML in mouse models [75, 76, 94] while in human patients, this translocation is mainly involved in paediatric ALL [70]. Moreover, mouse models reconstructing MLL-AF4 ALL have been difficult to develop, which may at least partly be caused by limitations of retroviral technology [95]. While MLLAF4 is associated with paediatric and human ALL [70] and can cause transformation of early B -cells, constitutive knock-in mouse models and mice conditionally expressing MLL-AF4 fusion develop only mature B-cell lymphomas [96, 97].
Overall, these discrepancies are likely to not only be a consequence of intrinsic differences between the human and the mouse system but also because of the many variables associated with both in vitro and in vivo studies. Cellular permissiveness might be influenced by the specific strategy used to purify HSPCs [33] as well as oncogene delivery methods [91, 92]. Moreover, a preferential association of some MLL fusions with specific leukaemia subtypes could be under microenvironmental influence or dependent on cytokine signalling sensitivity [98]. The involvement of secondary mutations associated with MLL translocations will have major impacts on the way regulatory programs are perturbed, and therefore will be linked with the aggressiveness of the disease. Although MLL-r AML had been considered to have a low mutation frequency [44], some MLL leukaemia models show long latency preceded by a pre-leukaemic phase [75, 99]. Accordingly, a recent cancer genome sequencing study on MLL-r AML patients has identified mutations in SPI1gene [100], a powerful HSPC transcription factor whose role in murine AML had already been defined at genomic scale [101].
Much remains to be learnt about the complex regulatory programs that are responsible for stem cell function during both tissue maintenance and repair. The haematopoietic system offers exciting opportunities to not only define these processes in normal cells, but also to learn how system perturbations can lead to disease development. While current research efforts are largely aimed at improving our understanding of perturbations that corrupt normal HSPCs towards creating a malignant state, many of the underlying principles will be widely applicable to other instances of stem cell state subversion. In non-haematopoietic tissues, adult stem cell deficiencies, particularly in old age, are associated with both imbalances in tissue maintenance as well as regenerative failure. A better understanding of the mechanisms that underlie the corruption of stem cell regulatory programs is therefore of broad therapeutic relevance [102].
Acknowledgement
SB is supported by the Cambridge Cancer Centre PhD fellowship (grant code: A19405).
References
- 1.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 2.Sawai CM, Babovic S, Upadhaya S, Knapp DJ, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity. 2016;45:597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Hofer T, Rodewald HR. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sanchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 2015;16:712–724. doi: 10.1016/j.stem.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, Wilson NK, Kent DG, Gottgens B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–e31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim CY, Wang H, Woodhouse S, Piterman N, Wernisch L, Fisher J, Gottgens B. BTR: training asynchronous Boolean models using single-cell expression data. BMC Bioinformatics. 2016;17:355. doi: 10.1186/s12859-016-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schutte J, Wang H, Antoniou S, Jarratt A, Wilson NK, Riepsaame J, Calero-Nieto FJ, Moignard V, Basilico S, Kinston SJ, et al. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. elife. 2016;5:e11469. doi: 10.7554/eLife.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 12.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Cote D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, Morrison SJ. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dore LC, Crispino JD. Transcription factor networks in erythroid cell and megakaryocyte development. Blood. 2011;118:231–239. doi: 10.1182/blood-2011-04-285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Gottgens B. Regulatory network control of blood stem cells. Blood. 2015;125:2614–2620. doi: 10.1182/blood-2014-08-570226. [DOI] [PubMed] [Google Scholar]
- 20.Moignard V, Macaulay IC, Swiers G, Buettner F, Schutte J, Calero-Nieto FJ, Kinston S, Joshi A, Hannah R, Theis FJ, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol. 2013;15:363–372. doi: 10.1038/ncb2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautreau L, Boudil A, Pasqualetto V, Skhiri L, Grandin L, Monteiro M, Jais JP, Ezine S. Gene coexpression analysis in single cells indicates lymphomyeloid copriming in short-term hematopoietic stem cells and multipotent progenitors. J Immunol. 2010;184:4907–4917. doi: 10.4049/jimmunol.0902184. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/S1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 23.Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 25.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 26.Takano H, Ema H, Sudo K, Nakauchi H. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J Exp Med. 2004;199:295–302. doi: 10.1084/jem.20030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruce WR, Van Der Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 1963;199:79–80. doi: 10.1038/199079a0. [DOI] [PubMed] [Google Scholar]
- 31.Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJ, Delwel R, Dohner K, Bullinger L, Kung AL, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27:852–860. doi: 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith LJ, Curtis JE, Messner HA, Senn JS, Furthmayr H, McCulloch EA. Lineage infidelity in acute leukemia. Blood. 1983;61:1138–1145. [PubMed] [Google Scholar]
- 33.Ugale A, Norddahl GL, Wahlestedt M, Sawen P, Jaako P, Pronk CJ, Soneji S, Cammenga J, Bryder D. Hematopoietic stem cells are intrinsically protected against MLL-ENL-mediated transformation. Cell Rep. 2014;9:1246–1255. doi: 10.1016/j.celrep.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 34.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 35.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–596. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 38.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program. 2004 doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, Marin-Padilla M, Tenen DG, Speck NA, Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 41.Dash A, Gilliland DG. Molecular genetics of acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:49–64. doi: 10.1053/beha.2000.0115. [DOI] [PubMed] [Google Scholar]
- 42.Care RS, Valk PJ, Goodeve AC, Abu-Duhier FM, Geertsma-Kleinekoort WM, Wilson GA, Gari MA, Peake IR, Lowenberg B, Reilly JT. Incidence and prognosis of c-KIT and FLT3 mutations in core binding factor (CBF) acute myeloid leukaemias. Br J Haematol. 2003;121:775–777. doi: 10.1046/j.1365-2141.2003.04362.x. [DOI] [PubMed] [Google Scholar]
- 43.Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research N Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 46.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 47.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fialkow PJ. The origin and development of human tumors studied with cell markers. N Engl J Med. 1974;291:26–35. doi: 10.1056/NEJM197407042910109. [DOI] [PubMed] [Google Scholar]
- 49.Griffin JD, Lowenberg B. Clonogenic cells in acute myeloblastic leukemia. Blood. 1986;68:1185–1195. [PubMed] [Google Scholar]
- 50.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci U S A. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 52.Turhan AG, Lemoine FM, Debert C, Bonnet ML, Baillou C, Picard F, Macintyre EA, Varet B. Highly purified primitive hematopoietic stem cells are PML-RARA negative and generate nonclonal progenitors in acute promyelocytic leukemia. Blood. 1995;85:2154–2161. [PubMed] [Google Scholar]
- 53.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 54.Aoki Y, Watanabe T, Saito Y, Kuroki Y, Hijikata A, Takagi M, Tomizawa D, Eguchi M, Eguchi-Ishimae M, Kaneko A, et al. Identification of CD34+ and CD34- leukemia-initiating cells in MLL-rearranged human acute lymphoblastic leukemia. Blood. 2015;125:967–980. doi: 10.1182/blood-2014-03-563304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.George J, Uyar A, Young K, Kuffler L, Waldron-Francis K, Marquez E, Ucar D, Trowbridge JJ. Leukaemia cell of origin identified by chromatin landscape of bulk tumour cells. Nat Commun. 2016;7:12166. doi: 10.1038/ncomms12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polak P, Karlic R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahovicek K, Stamatoyannopoulos JA, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herault O, Hope KJ, Deneault E, Mayotte N, Chagraoui J, Wilhelm BT, Cellot S, Sauvageau M, Andrade-Navarro MA, Hebert J, et al. A role for GPx3 in activity of normal and leukemia stem cells. J Exp Med. 2012;209:895–901. doi: 10.1084/jem.20102386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pabst C, Krosl J, Fares I, Boucher G, Ruel R, Marinier A, Lemieux S, Hebert J, Sauvageau G. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11:436–442. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 59.Guzman ML, Yang N, Sharma KK, Balys M, Corbett CA, Jordan CT, Becker MW, Steidl U, Abdel-Wahab O, Levine RL, et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol Cancer Ther. 2014;13:1979–1990. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreira HJ, Heyn H, Vizoso M, Moutinho C, Vidal E, Gomez A, Martinez-Cardus A, Simo-Riudalbas L, Moran S, Jost E, et al. DNMT3A mutations mediate the epigenetic reactivation of the leukemogenic factor MEIS1 in acute myeloid leukemia. Oncogene. 2016;35:3079–3082. doi: 10.1038/onc.2015.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greaves M. Leukaemia ‘firsts’ in cancer research and treatment. Nat Rev Cancer. 2016;16:163–172. doi: 10.1038/nrc.2016.3. [DOI] [PubMed] [Google Scholar]
- 63.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mullighan CG, Phillips LA, Su X, Ma J, Miller CB, Shurtleff SA, Downing JR. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- 67.Fialkow PJ, Janssen JW, Bartram CR. Clonal remissions in acute nonlymphocytic leukemia: evidence for a multistep pathogenesis of the malignancy. Blood. 1991;77:1415–1417. [PubMed] [Google Scholar]
- 68.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A. 2000;97:7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R (2012) Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 4: 149ra118 [DOI] [PMC free article] [PubMed]
- 70.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 71.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 72.Pui CH, Campana D. Age-related differences in leukemia biology and prognosis: the paradigm of MLL-AF4-positive acute lymphoblastic leukemia. Leukemia. 2007;21:593–594. doi: 10.1038/sj.leu.2404598. [DOI] [PubMed] [Google Scholar]
- 73.Felix CA. Secondary leukemias induced by topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:233–255. doi: 10.1016/S0167-4781(98)00139-0. [DOI] [PubMed] [Google Scholar]
- 74.Pui CH, Sandlund JT, Pei D, Campana D, Rivera GK, Ribeiro RC, Rubnitz JE, Razzouk BI, Howard SC, Hudson MM, et al. Improved outcome for children with acute lymphoblastic leukemia: results of total therapy study XIIIB at St Jude Children’s Research Hospital. Blood. 2004;104:2690–2696. doi: 10.1182/blood-2004-04-1616. [DOI] [PubMed] [Google Scholar]
- 75.Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17:3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Na Nakorn T, Traver D, Weissman IL, Akashi K. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J Clin Invest. 2002;109:1579–1585. doi: 10.1172/JCI0215272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bonadies N, Foster SD, Chan WI, Kvinlaug BT, Spensberger D, Dawson MA, Spooncer E, Whetton AD, Bannister AJ, Huntly BJ, et al. Genome-wide analysis of transcriptional reprogramming in mouse models of acute myeloid leukaemia. PLoS One. 2011;6:e16330. doi: 10.1371/journal.pone.0016330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 80.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 81.Zeisig BB, Garcia-Cuellar MP, Winkler TH, Slany RK. The oncoprotein MLL-ENL disturbs hematopoietic lineage determination and transforms a biphenotypic lymphoid/myeloid cell. Oncogene. 2003;22:1629–1637. doi: 10.1038/sj.onc.1206104. [DOI] [PubMed] [Google Scholar]
- 82.So CW, Karsunky H, Passegue E, Cozzio A, Weissman IL, Cleary ML. MLL-GAS7 transforms multipotent hematopoietic progenitors and induces mixed lineage leukemias in mice. Cancer Cell. 2003;3:161–171. doi: 10.1016/S1535-6108(03)00019-9. [DOI] [PubMed] [Google Scholar]
- 83.Corral J, Lavenir I, Impey H, Warren AJ, Forster A, Larson TA, Bell S, McKenzie AN, King G, Rabbitts TH. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell. 1996;85:853–861. doi: 10.1016/S0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 84.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 85.Collins EC, Pannell R, Simpson EM, Forster A, Rabbitts TH. Inter-chromosomal recombination of Mll and Af9 genes mediated by cre-loxP in mouse development. EMBO Rep. 2000;1:127–132. doi: 10.1093/embo-reports/kvd021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Forster A, Pannell R, Drynan LF, McCormack M, Collins EC, Daser A, Rabbitts TH. Engineering de novo reciprocal chromosomal translocations associated with Mll to replicate primary events of human cancer. Cancer Cell. 2003;3:449–458. doi: 10.1016/S1535-6108(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 87.Cano F, Drynan LF, Pannell R, Rabbitts TH. Leukaemia lineage specification caused by cell-specific Mll-Enl translocations. Oncogene. 2008;27:1945–1950. doi: 10.1038/sj.onc.1210818. [DOI] [PubMed] [Google Scholar]
- 88.Drynan LF, Pannell R, Forster A, Chan NM, Cano F, Daser A, Rabbitts TH. Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis. EMBO J. 2005;24:3136–3146. doi: 10.1038/sj.emboj.7600760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 91.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stavropoulou V, Kaspar S, Brault L, Sanders MA, Juge S, Morettini S, Tzankov A, Iacovino M, Lau IJ, Milne TA, et al. MLL-AF9 expression in hematopoietic stem cells drives a highly invasive AML expressing EMT-related genes linked to poor outcome. Cancer Cell. 2016;30:43–58. doi: 10.1016/j.ccell.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 93.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 95.Lin S, Luo RT, Ptasinska A, Kerry J, Assi SA, Wunderlich M, Imamura T, Kaberlein JJ, Rayes A, Althoff MJ, et al. Instructive role of MLL-fusion proteins revealed by a model of t(4;11) pro-B acute lymphoblastic leukemia. Cancer Cell. 2016;30:737–749. doi: 10.1016/j.ccell.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 96.Chen W, Li Q, Hudson WA, Kumar A, Kirchhof N, Kersey JH. A murine Mll-AF4 knock-in model results in lymphoid and myeloid deregulation and hematologic malignancy. Blood. 2006;108:669–677. doi: 10.1182/blood-2005-08-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Metzler M, Forster A, Pannell R, Arends MJ, Daser A, Lobato MN, Rabbitts TH. A conditional model of MLL-AF4 B-cell tumourigenesis using invertor technology. Oncogene. 2006;25:3093–3103. doi: 10.1038/sj.onc.1209636. [DOI] [PubMed] [Google Scholar]
- 98.Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, Zheng Y, Cancelas JA, Gu Y, Jansen M, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer Cell. 2008;13:483–495. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson JJ, Chen W, Hudson W, Yao Q, Taylor M, Rabbitts TH, Kersey JH. Prenatal and postnatal myeloid cells demonstrate stepwise progression in the pathogenesis of MLL fusion gene leukemia. Blood. 2003;101:3229–3235. doi: 10.1182/blood-2002-05-1515. [DOI] [PubMed] [Google Scholar]
- 100.Lavallee VP, Baccelli I, Krosl J, Wilhelm B, Barabe F, Gendron P, Boucher G, Lemieux S, Marinier A, Meloche S, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–1037. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 101.Sive JI, Basilico S, Hannah R, Kinston SJ, Calero-Nieto FJ, Gottgens B. Genome-scale definition of the transcriptional programme associated with compromised PU.1 activity in acute myeloid leukaemia. Leukemia. 2016;30:14–23. doi: 10.1038/leu.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegue E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543:205–210. doi: 10.1038/nature21388. [DOI] [PMC free article] [PubMed] [Google Scholar]