Abstract
Due to its sensitivity and specificity, real-time quantitative PCR (qRT-PCR) is a popular technique for investigating gene expression levels in plants. Based on the Minimum Information for Publication of Real-Time Quantitative PCR Experiments (MIQE) guidelines, it is necessary to select and validate putative appropriate reference genes for qRT-PCR normalization. In the current study, three algorithms, geNorm, NormFinder, and BestKeeper, were applied to assess the expression stability of 10 candidate reference genes across five different tissues and three different abiotic stresses in Isatis indigotica Fort. Additionally, the IiYUC6 gene associated with IAA biosynthesis was applied to validate the candidate reference genes. The analysis results of the geNorm, NormFinder, and BestKeeper algorithms indicated certain differences for the different sample sets and different experiment conditions. Considering all of the algorithms, PP2A-4 and TUB4 were recommended as the most stable reference genes for total and different tissue samples, respectively. Moreover, RPL15 and PP2A-4 were considered to be the most suitable reference genes for abiotic stress treatments. The obtained experimental results might contribute to improved accuracy and credibility for the expression levels of target genes by qRT-PCR normalization in I. indigotica.
Keywords: qRT-PCR, reference gene, Isatis indigotica Fort., gene expression, normalization
Introduction
Gene expression analysis is an effective strategy that offers certain important information related to gene functions in the plant life cycle and the signal pathways that regulate effective component accumulations in response to environmental changes (Miao et al., 2015; Shu et al., 2015; Yue et al., 2015; Vojta et al., 2016). Selected methods can be applied to detect gene expression levels, including microarray/gene chips, RNA sequencing, semi-quantitative PCR (semi-qPCR), Northern blot, RNase protection analysis (RPA), and quantitative real-time PCR (qRT-PCR) (VanGuilder et al., 2008). Among these methods, qRT-PCR is considered one of the most popular techniques for analyzing gene expression levels due to its high sensitivity, specificity, reproducibility, accuracy, and high efficiency (Hu et al., 2014). However, qRT-PCR results are affected by the amount and source of the original sample, RNA integrity, cDNA quality and quantity, primer specificity, stability of the reference gene, efficiency of reverse transcription, and experimental conditions, among other factors (Kumar and Singh, 2015). To obtain reliable qRT-PCR analysis results, it is crucial to normalize the raw data for gene expression and minimize variations for the different samples and experimental conditions according to the Minimum Information for Publication of Real-Time Quantitative PCR Experiments (MIQE) guidelines (Han et al., 2012; Martins et al., 2016). These guidelines are most commonly used to select and normalize the reference gene for qRT-PCR analysis (Chao et al., 2012).
If qRT-PCR is used to analyze the target gene expression levels, the reference genes must be stably expressed for different samples and different experimental conditions (Wong and Medrano, 2005). In practice, the transcription levels of the traditional reference genes, even certain housekeeping genes, could not be expressed stably for different samples and experimental conditions (Kozera and Rapacz, 2013), i.e., no universal reference genes have been confirmed up to now (Radonić et al., 2004), a situation well documented in the published literature. For Setaria viridis, Protein Kinase, RNA Binding Protein, and SDH could be used as the most stable reference genes across different developmental stages and under drought or aluminum stress (Martins et al., 2016). For the tung tree, ACT7, UBQ, GAPDH, and EF1α were the most stable reference genes for all samples and developing seeds, and ACT7, EF1β, GAPDH, and TEF1 were the top four candidates for different tissues, whereas LCR69 and ALB did not favor qRT-PCR normalization under the same conditions (Han et al., 2012). For Pinus massoniana L., ACT1 was confirmed as the optimal reference gene for all samples, UBI4 was stably expressed in various tissues and under zinc stress, CYP was the best gene for insect-resistant leaves and Pb stress, and Fbox and UBI11 could be used as the most stable reference genes for salt stress. Additionally, certain best reference groups have been identified, namely, Fbox + RRP8, ARF + TUBA, and EF1B + IDH for drought and low temperature stress and flowers in different developmental stages, respectively (Chen et al., 2016). Therefore, different reference genes should be used in different samples and experimental conditions. Furthermore, a reference gene with lower stability might lead to a biased understanding of qRT-PCR (Gutierrez et al., 2008) and mask the true nature of gene expression (Bustin, 2002; Bustin and Nolan, 2004).
Several prevalent mathematical algorithms are available, such as geNorm, NormFinder, and BestKeeper (Pfaffl, 2001; Vandesompele et al., 2002; Andersen et al., 2004), and can be used to explore novel reference genes for qRT-PCR. For geNorm and NormFinder analysis, all raw Ct values must be transferred to relative expression values according to the Delta Ct method, whereas no transformation is needed for BestKeeper. The geNorm algorithm considers both the average expression stability value (M-value) and pairwise variation (Vn/Vn+1) to assess the expression stability and determine the optimal number of the candidate reference genes (Vandesompele et al., 2002). NormFinder focuses attention on both intra- and inter-group variations and estimates the expression levels of the candidate reference genes using the stability value, with lower values indicating higher stability (Andersen et al., 2004). BestKeeper obtains the expression variability of reference genes through the standard deviation (SD), coefficient of variance (CV), or coefficient of correlation (Zhang W.X. et al., 2016). In general, different algorithms are usually combined together for qRT-PCR evaluation and normalization (Demidenko et al., 2011; Qi et al., 2016; Reddy et al., 2016).
The biennial herb Isatis indigotica Fort. (Cruciferae) is a traditional medical plant widely cultivated in China (Angelini et al., 2007) and is popular in pharmaceuticals, food additives, flavors, and other industrial materials due to abundant bio-active components, including alkaloids (Wu et al., 1997, 2011), flavonoids (Deng et al., 2008), organic acids, and glycosides (Liau et al., 2007). Until now, only a few references mentioned gene expression and functions in the developmental process and accumulation of metabolites responding to abiotic or biotic stress treatments in which the reference genes were nothing more than UBQ, ACT and 18s r RNA (Han et al., 2013; Li Q. et al., 2014; Chen et al., 2015; Dong et al., 2015; Han, 2015; Hu et al., 2015). The literature does not mention the selection and validation of accurate reference genes for gene expression in different samples and experimental conditions by qRT-PCR in this plant, nevertheless, it is a vital step for further gene function research.
In this study, 10 candidate reference genes (UBC22, UBC29, Act7, PP2A-4, eIF2, APT3, AP-2, RPL15, TUB4, and UBC19) from the transcriptome sequencing data in our lab were selected for validation of their expression stabilities for different tissues and abiotic stresses using qRT-PCR. Three mathematical algorithms (geNorm, NormFinder, and BestKeeper) were applied to analyze the raw Ct value and assess the reference gene expression stability. In addition, the candidate reference genes were tested using a target gene IiYUC6, which is involved in the pathway of indole-3-acetic acid (IAA) biosynthesis. According to the references (Yamamoto et al., 2007; Expósito-Rodríguez et al., 2011; Liu et al., 2014; Li et al., 2015), the expression patterns of CsYUCs in cucumber, FvYUCs in strawberry, ToFZY (YUCCA-like flavin monooxygenases, FMOs) in tomato, and OsYUCCA in rice were analyzed and the results revealed that the expression patterns of YUCCA varied in different species. Moreover, functional studies on YUCCA6 were performed in strawberry (Liu et al., 2014), Arabidopsis (Kim et al., 2007; Im Kim et al., 2011; Cha et al., 2015), poplar (Ke et al., 2015), and potato (Im Kim et al., 2013). All of these research studies produced valuable references. Until recently, no research was available on YUCCA in I. indigotica Fort. or whether it played important roles in the synthesis of IAA, a topic that deserves further study. However, discussion of the expression patterns was the first step. In this work, we offer a full length CDS of IiYUC6 in our transcriptome database that can be used to validate the candidate reference genes. The results are expected to act as a general guideline for reference gene selection in normalization and quantification of transcription levels for gene expression and function research in I. indigotica.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of I. indigotica purchased from Shaanxi Geo-Authentic Medicinal Plant Co. Ltd. (Xi’an, China) were germinated on a seedling bed. When the first pair of true leaves appeared, the seedlings were transferred to pots filled with soil (4–5 seedlings per pot). The entire germination and growth process was conducted in the greenhouse in our lab under the conditions of a 16-18 h photoperiod, 2000 Lx irradiance level, 25 ± 2°C temperature, and 60–80% humidity. Furthermore, irrigation, regular weeding, and other management measures were performed every 2 days. Three-month-old seedlings were used as the abiotic treatment materials. At the same time, different tissues (including roots, stems, leaves, flowers, and fruits) were collected in the experimental field, frozen immediately in liquid nitrogen and stored at –80°C. All samples were gathered in three biological replications.
Abiotic Stress Treatments
Three-month-old seedlings were randomly assigned to the control and treatment groups. Three types of abiotic stress treatments were applied: ABA, wounding, and cold stress treatments. The first treatment was performed using a foliar ABA spray solution (0.2 μM), and the control seedlings received water only. In addition, mechanical wounding treatment was applied by scratching approximately 40% of the total leaf surfaces with a blade (Ge et al., 2015). Finally, the cold treatments consisted of seedlings exposed to 4°C for different durations. For the wounding and cold treatment samples, the control groups received non-treatments. All of the above samples were harvested 6 h after the corresponding treatments, frozen into liquid nitrogen and stored at –80°C for further use.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from all samples using the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Dalian, China) with genomic DNA removed by the gDNA Eraser Spin Column and DNase I treatment according to the manufacturer’s instructions. The RNA integrity was examined using 1 % agarose gel electrophoresis. The concentration and purity of the total RNA were detected by a NanoDrop 2000c Spectrophotometer (Thermo Scientific, United States). The A260/A280 ratio of extracted total RNA ranged from 1.90 to 2.10, indicating high purity and no protein contamination. The first-strand cDNA was synthesized using the PrimeScriptTM II 1st Strand cDNA Synthesis Kit (TaKaRa) with 1.0 μg total RNA in a 20 μL volume according to the manufacturer’s protocols. Amplification using oligo (dT) 18 as primer was performed with the following thermal cycling: 42°C for 50 min and 70°C for 10 min. The RT-PCR products were diluted (1:20) for qRT-PCR assays.
Selection of Candidate Reference Genes and Primer Design
Based on previous research studies and the transcriptome sequencing data of our lab (unpublished), 10 candidate genes were selected to normalize and validate the qRT-PCR, including 3 homologous genes (UBC19, UBC22, and UBC29), 2 traditional housekeeping genes (ACT7 and TUB4) and five novel genes (PP2A-4, eIF2, APT3, AP-2, RPL15). To ensure the reliability and accuracy of the putative reference genes, the nucleotide sequences of 10 candidate genes were BLAST-searched against the Arabidopsis genome database to identify their homologs in I. indigotica. Forward and reverse primers of all candidate genes were designed using Primer Premier v5.0 software using Tm values ranging from 58°C to 62°C, GC % (45–60), primer length (19–23 bp), and product length (85–225 bp). All primers were synthesized by Beijing Genomics Institute. The ortholog sequences of Arabidopsis, primer information, and gene characteristics are presented in Table 1. Moreover, the primer specificity was validated using melting curve analysis.
Table 1.
Details of the primers used for qRT-PCR normalization.
| Gene abbreviations | Gene name | Accession No. | Length (bp) | Primer sequence U/L [5′–3′] | Arabidopsis ortholog No | Identity (%) | Tm°C | E (%) | R2 |
|---|---|---|---|---|---|---|---|---|---|
| eIF2 | Eukaryotic transation initiation factor | c40298.graph_c0 | 117 | For:TACCAGTGGCTCGCTTGAC | At5g20920.3 | 87 | 83 | 1.20 | 0.9583 |
| Rev:CAACCAAAGCAAATGACGTACTC | |||||||||
| ACT7 | Actin7 | AY870652.1 | 181 | For: AGGAATCGCTGACCGTATG | At5g09810.1 | 93 | 85.5 | 1.15 | 0.9829 |
| Rev : TGGACCCGACTCATCGTATT | |||||||||
| PP2A-4 | Protein phosphatase 2A-4 | c41201.graph_c0 | 151 | For: GAATGCCTGCGAAAGTATGG | At3g58500.1 | 93 | 83 | 1.12 | 0.9859 |
| Rev: TCCTAATGTTGTCAAGGGTCTC | |||||||||
| AP-2 | AP-2 complex | c47959.graph_c0 | 137 | For: AAGACCTTCAGCCTTACGCC | At5g22780.1 | 92 | 83 | 1.35 | 0.9849 |
| Rev: ACTGCATCGAGGTTGTCTGG | |||||||||
| APT3 | Adenine phosphoribosyl transferase | c37104.graph_c0 | 178 | For:CCGTGTCGTTCCAGATTTTC | At4g22570.1 | 93 | 82 | 1.04 | 0.9983 |
| Rev:GATTGGTGGACCGAATAGGA | |||||||||
| UBC29 | Ubiquitin-conjugating enzyme 29 | c38774.graph_c0 | 148 | For:TTCTTAAAGACCAGTGGAGC | At2g16740.1 | 90 | 83.5 | 0.97 | 0.9996 |
| Rev:ATGGCTTCGTATTTGACCTT | |||||||||
| RPL15 | Ribosomal protein L | c42633.graph_c1 | 133 | For:GGGCGAAGAGGAAAGGTAG | At3g25920.1 | 87 | 84.6 | 1.05 | 0.9920 |
| Rev: CGGAAGACGGCGATAAAGAG | |||||||||
| UBC22 | Ubiquitin-conjugating enzyme 22 | c38263.graph_c0 | 225 | For:GTATGAAGTTGGCATTGTCGC | At5g05080.2 | 89 | 83 | 0.89 | 0.9990 |
| Rev:TCTTTCCAGCCTGTTCGTTT | |||||||||
| UBC19 | Ubiquitin-conjugating enzyme 19 | c39354.graph_c0 | 188 | For: TGAGACATGCTGCTTCCATC | At3g20060.1 | 92 | 84 | 0.94 | 0.9845 |
| Rev: CTTCTTGGTTGCTCCAAAGC | |||||||||
| TUB4 | Tubulin | c43494.graph_c0 | 85 | For:GCCCCGATAACTTCGTCTT | At5g44340.1 | 93 | 85.8 | 1.12 | 0.9790 |
| Rev: TCAATCAACTCCGCTCCCT | |||||||||
qRT-PCR Conditions and Amplification Efficiency
The qRT-PCR was performed on a 7500 ABI Real-time PCR system (Applied Biosystems, United States) using SYBR Premix Ex Taq (Takara Bio, Kusatsu, Shiga, Japan). The total 20 μL reaction mixture, including 10 μL SYBR Premix Ex Taq, 2 μL cDNA, 0.8 μL forward primer, 0.8 μL reverse primer, and 6.4 μL dd H2O in a 96-well optical plate, was amplified according to the following thermal cycling conditions: 95°C for 10 s, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. To draw the melting curve, the PCR product was heated from 65 to 95°C (0.5°C/5 s), and the raw Ct values were obtained. Each qRT-PCR reaction was conducted in technical triplicate. The slope of the standard curve generated through serial 10-fold dilutions of the cDNA samples was used to calculate the amplification efficiency for each candidate gene.
Assessment of the Expression Stability of the Reference Gene
Three software packages, namely, geNorm, NormFinder, and BestKeeper, were used to analyze the gene expression stability based on their own algorithms. geNorm assesses the expression stability of the candidate reference gene using the M-value, which refers to the average pairwise variation between each reference gene and the other reference genes. A gene with M < 1.5 is usually treated as the stable reference gene (Guo et al., 2010; Han et al., 2012). Furthermore, the normalization factor obtained by calculating the pairwise variation (Vn/Vn+1) of the two sequential normalization factors (NFn and NFn+1) can be used to determine the most suitable numbers of the reference genes. If the value of Vn/Vn+1 is lower than 0.15, then the value of “n” is the most appropriate number of reference genes. The stability value is the important factor for NormFinder in estimating the top internal control genes, and the intra- and inter-group variations are also necessary factors that must be considered. The lower the stability value and inter- and intra-group variation, the more stable the reference gene will be. The SD value set at 1.0 by BestKeeper represents the Ct variation. If the value is less than 1.0, the candidate gene is considered a stable inter-control gene and vice versa. Furthermore, the coefficient of variation (CV value) is used to rank the candidate reference genes.
Validation of the Candidate Reference Genes
The selected reference genes, including the most and least stable genes, were validated for different tissue samples (root, stem, leaf, flower and fruit) using the relative expression level analysis of IiYUC6, a key gene associated with IAA biosynthesis in I. indigotia. The 2-ΔΔct method was used to calculated the relative expression level of the target gene.
Statistical Analysis
Statistical analyses were performed using SPSS 22.0, and all data for the relative expression level of the target gene were subjected to one-way analysis of variance (ANOVA) and presented as the means ± standard error (SE). The statistical level was assessed using ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Results
Primer Specificity and Expression Level Analysis of 10 Candidate Reference Genes
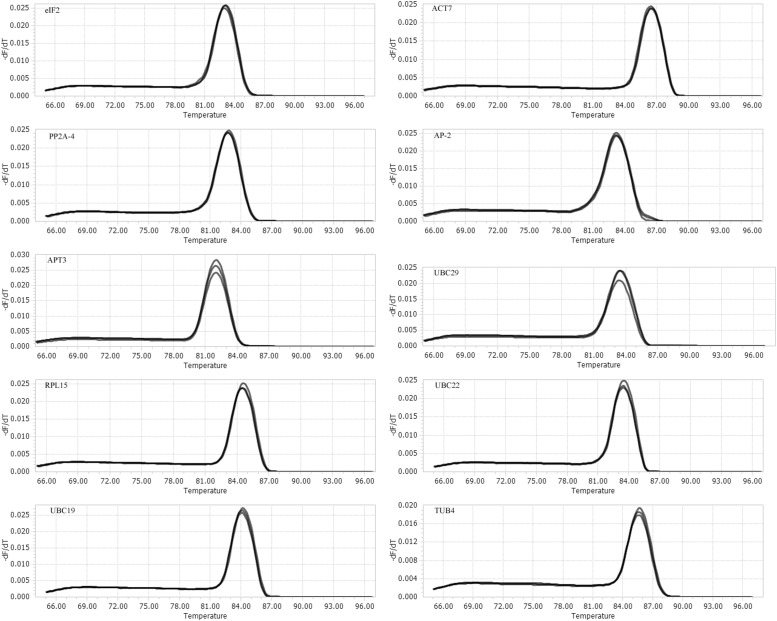
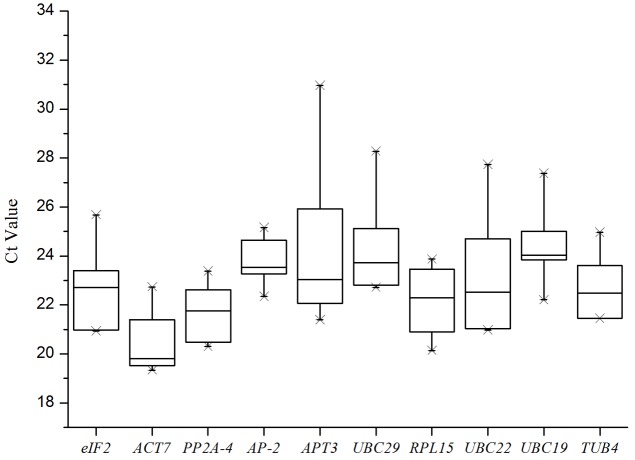
A total of 10 putative reference genes (UBC19, UBC22, UBC29, ACT7, PP2A-4, eIF2, APT3, AP-2, RPL15, and TUB4) from the transcriptome sequencing data of I. indigotica were selected as candidates for qRT-PCR normalization. The gene names and abbreviation, accession number, primer sequences, Arabidopsis orthologous, identity (compared to Arabidopsis), amplification efficiency (E) and length, Tm value and correlation coefficient (R2) are listed in Table 1. The Tm values ranged from 83°C to 85.8°C, the amplification efficiency varied from 89% for UBC22 to 135% for AP-2, and the correlation coefficients (R2) ranged from 0.9583 for eIF2 to 0.9996 for UBC29. Moreover, the amplifications were validated using the single peak of the melting curve for each primer (Figure 1). The gene expression levels were determined by the cycle threshold (Ct) values, and the Ct value distribution of the 10 candidate reference genes varied from 19.34 to 30.97 (Figure 2), which represents the different expression levels of the candidates. Among them, APT3 (for wounding stress treatment) showed the lowest expression level with the highest Ct value (30.97), and ACT7 (in root) presented the highest expression level with the lowest Ct value (19.34).
FIGURE 1.
Melting curves for the candidate reference genes. Melting temperatures were visualized by plotting the negative first derivative of fluorescence relative to the temperature in Celsius (–(d/dT)). Melting curves of 10 reference genes show a simple peak.
FIGURE 2.
Ct values for 10 candidate reference genes. Expression data are displayed as Ct values for each reference gene in all combined experimental conditions. A line across the box depicts the median. The vertical sides of the box indicate the 25th and 75th percentiles, and the whiskers show the caps for maximum and minimum values.
Expression Stability Analysis of 10 Candidate Reference Genes
The performance of 10 candidate reference genes in I. indigotica was assessed via qRT-PCR in different samples, which were divided into three experimental sets (total, tissues and stresses), five different tissues (roots, stems, leaves, flowers, and fruits), and three stress treatments (ABA, wounding, and cold).
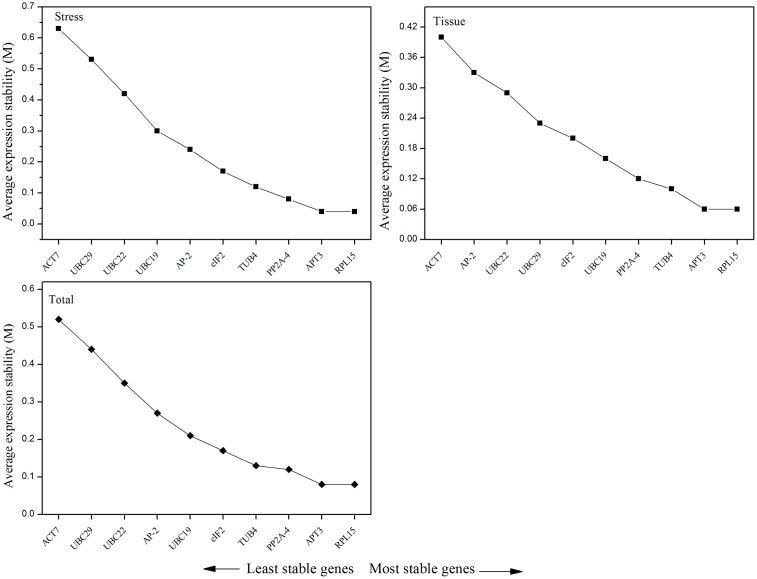
According to the M-value, geNorm depicted the ranking of 10 candidate reference genes for the different tissues and experimental conditions (Table 2 and Figure 3). For the five tissue samples, the expression stability ranking of the 10 reference genes was arranged as follows: RPL15 > APT3 > TUB4 > PP2A-4 > UBC19 > eIF2 > UBC29 > UBC22 > AP-2 > ACT7. For the abiotic stress samples, the most stable and relatively unstable genes were RPL15 (M = 0.04) and ACT7 (M = 0.63), respectively. Based on all data sets, RPL15 (M = 0.08) was still the most stable reference gene, followed by APT3 (M = 0.08) and PP2A-4 (M = 0.12). Additionally, ACT7 (M = 0.52) was the most variable reference gene for the same samples and experimental conditions (Table 2).
Table 2.
Ranking of the candidate reference genes and their M-values calculated by geNorm.
| Total | Tissue | Abiotic stress | ||||
|---|---|---|---|---|---|---|
| Rank | Gene | M-value | Gene | M-value | Gene | M-value |
| 1 | RPL15 | 0.08 | RPL15 | 0.06 | RPL15 | 0.04 |
| 2 | APT3 | 0.08 | APT3 | 0.06 | APT3 | 0.04 |
| 3 | PP2A-4 | 0.12 | TUB4 | 0.10 | PP2A-4 | 0.08 |
| 4 | TUB4 | 0.13 | PP2A-4 | 0.12 | TUB4 | 0.12 |
| 5 | eIF2 | 0.17 | UBC19 | 0.16 | eIF2 | 0.17 |
| 6 | UBC19 | 0.21 | eIF2 | 0.20 | AP-2 | 0.24 |
| 7 | AP-2 | 0.27 | UBC29 | 0.23 | UBC19 | 0.30 |
| 8 | UBC22 | 0.35 | UBC22 | 0.29 | UBC22 | 0.42 |
| 9 | UBC29 | 0.44 | AP-2 | 0.33 | UBC29 | 0.53 |
| 10 | ACT7 | 0.52 | ACT7 | 0.40 | ACT7 | 0.63 |
FIGURE 3.
Average expression stability (M-value) of 10 candidate reference genes calculated by geNorm and ranking of the candidate reference genes in different tissues, abiotic treatments, and total group, respectively. A lower value of average expression stability (M) indicates a more stable expression level.
Table 3 shows the ranking of the 10 candidate reference genes and their expression stability values as calculated by NormFinder. RPL15 and APT3 were the top two most suitable genes for normalization if both different tissue (stability value = 0.019) and abiotic stress (stability value = 0.011) samples were considered. Moreover, according to all data sets, RPL15 (stability value = 0.038) and PP2A-4 (stability value = 0.073) were the top two best reference genes for normalization of qRT-PCR, but the most variable reference gene was UBC29 (stability value = 0.571).
Table 3.
Ranking of the candidate reference genes and their expression stability values calculated by NormFinder.
| Total | Tissue | Abiotic stress | ||||
|---|---|---|---|---|---|---|
| Rank | Gene | Stability | Gene | Stability | Gene | Stability |
| 1 | RPL15 | 0.038 | RPL15 | 0.019 | RPL15 | 0.011 |
| 2 | PP2A-4 | 0.073 | APT3 | 0.019 | APT3 | 0.011 |
| 3 | APT3 | 0.076 | TUB4 | 0.023 | PP2A-4 | 0.023 |
| 4 | eIF2 | 0.097 | PP2A-4 | 0.042 | TUB4 | 0.030 |
| 5 | TUB4 | 0.105 | eIF2 | 0.125 | eIF2 | 0.046 |
| 6 | AP-2 | 0.149 | UBC29 | 0.142 | UBC19 | 0.189 |
| 7 | UBC19 | 0.154 | UBC19 | 0.158 | AP-2 | 0.224 |
| 8 | ACT7 | 0.250 | AP-2 | 0.189 | ACT7 | 0.389 |
| 9 | UBC22 | 0.337 | ACT7 | 0.269 | UBC22 | 0.761 |
| 10 | UBC29 | 0.571 | UBC22 | 0.392 | UBC29 | 1.151 |
In addition, the ranking results examined by BestKeeper are presented in Table 4 based on calculation of the CV and SD values. The results revealed that AP-2 (CV = 3.08, SD = 0.75) and PP2A-4 (CV = 4.30, SD = 0.94) were the first and second most stable reference genes for the abiotic stress samples. UBC29 (CV = 2.02, SD = 0.47) was the most stable reference gene, followed by AP-2 (CV = 3.06, SD = 0.72) if different tissue samples were considered. According to all data sets, the top two most stable reference genes were AP-2 (CV = 3.49, SD = 0.84) and PP2A-4 (CV = 4.47, SD = 0.97), and the least stable reference gene was APT3 (CV = 10.41, SD = 2.59).
Table 4.
Ranking of the candidate reference genes and their expression stability values calculated using BestKeeper.
| Gene | UBC29 | AP-2 | TUB4 | ACT7 | UBC19 | APT3 | eIF2 | PP2A-4 | RPL15 | UBC22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | SD[±CP] | 1.46 | 0.84 | 1.10 | 1.07 | 1.10 | 2.59 | 1.14 | 0.97 | 1.08 | 1.94 |
| CV | 5.96 | 3.49 | 4.80 | 5.18 | 4.47 | 10.41 | 5.01 | 4.47 | 4.82 | 8.32 | |
| Tissue | SD[±CP] | 0.47 | 0.72 | 0.76 | 0.81 | 0.86 | 0.86 | 0.99 | 0.99 | 1.09 | 1.63 |
| CV | 2.02 | 3.06 | 3.37 | 3.99 | 3.59 | 3.77 | 4.47 | 4.57 | 4.85 | 7.11 | |
| Abiotic stress | SD[±CP] | 1.20 | 0.75 | 1.38 | 1.10 | 1.31 | 1.76 | 1.16 | 0.94 | 1.01 | 2.54 |
| CV | 4.54 | 3.08 | 5.86 | 5.14 | 5.15 | 6.22 | 4.86 | 4.30 | 4.51 | 10.62 | |
The value of SD[±CP] below 1.0, marked in bold, indicate that the reference gene is qualified, and the value of CV is used to rank the reference genes.
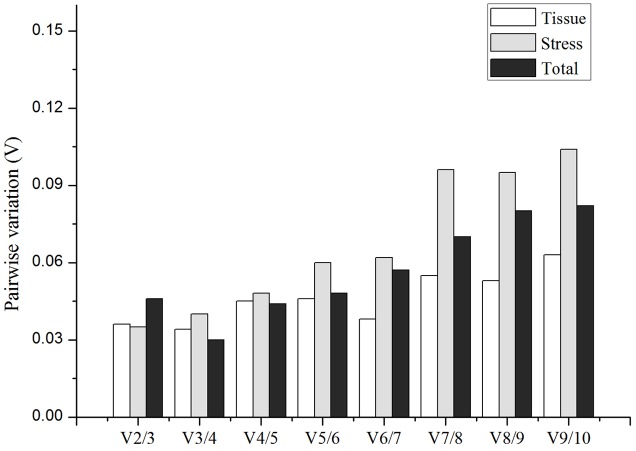
Furthermore, after identification of the best candidate gene for each experimental set, the pairwise variations Vn/Vn+1 were investigated by geNorm to normalize qRT-PCR for the different tissues and abiotic stresses (Figure 4). The results revealed that the V2/3 value between the two sequential normalization factors (NF2 and NF3) was the lowest and was less than the cutoff threshold of 0.15 in all of the experimental sets. Therefore, the optimal number of the candidate reference genes should be 2, showing that the best combination of the reference genes for normalization of qRT-PCR were RPL15 and TUB4 for different tissues and RPL15 and PP2A-4 for abiotic stress samples in I. indigotica.
FIGURE 4.
Pairwise variation (V-value) calculated by geNorm showing the optimal number of reference genes required for qRT-PCR normalization in different tissues, abiotic treatments and total group, respectively. The average pairwise variations Vn/Vn+1 were analyzed between the normalization factors NFn and NFn+1.
Validation of the Selected Reference Genes for Different Tissues
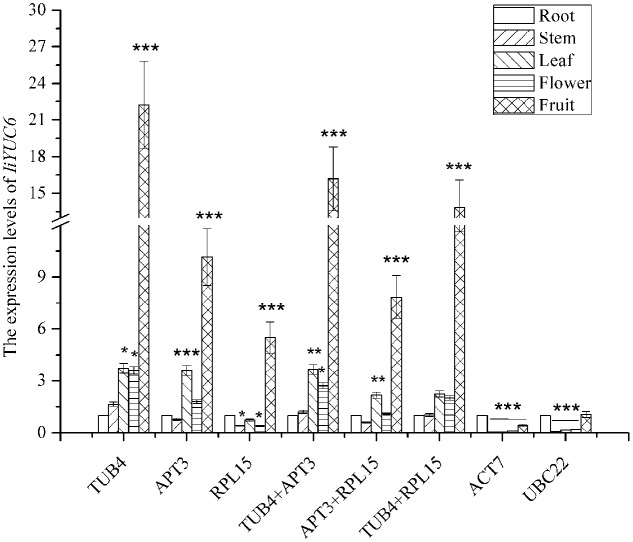
To evaluate the effects of suitable candidate reference genes on assessment of target gene expression, we examined the expression patterns of I. indigotica YUCCA (IiYUC6), a key gene involved in IAA synthesis (Kim et al., 2007) in different tissues. The relative expression levels of the three top ranking reference genes (RRL15, APT3, and TUB4) and two least stable genes (ACT7 and UBC22) were validated according to geNorm and NormFinder.
As shown in Figure 5, the transcription level of IiYUC6 was upregulated compared with the results in root when normalized with TUB4. Similar expression patterns were observed when normalized by TUB4 combined with APT3. When normalized with APT3, the expression level of IiYUC6 was still upregulated in most of the tissues except for stem, but the difference was not apparent. However, downregulation of the transcription level of IiYUC6 was noted in stem, leaf and root when normalized with RPL15, and similar expression patterns were also observed when normalized with APT3 and RPL15. In addition, the highest expression level occurred for IiYUC6 in fruit when normalized with the most stable reference genes, including TUB4, APT3, and RPL15 or their pairwise combinations. In contrast, when normalized with the least stable reference genes of ACT7 or UBC22, the expression profiles were more variable, with obvious downregulated expression levels in different tissues compared with the root sample. In conclusion, similar expression levels of the target gene were obtained when normalized with RPL15, APT3, and TUB4. However, when unstable reference genes including ACT7 and UBC22 were used, the relative expression level was underestimated, which might lead to a biased result. Hence, the analysis results of expression patterns on IiYUC6 revealed that the candidate reference gene selection was feasible and credible.
FIGURE 5.
Relative expression levels of IiYUC6 in different tissues (root, stem, leaf, flower, and fruit) were normalized using different reference genes or a combination including the three most stable (TUB4, APT3, and RPL15) and two least stable genes (ACT7 and UBC22). Error bars show the mean standard error calculated from three biological replicates. The statistical level was assessed with ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
Discussion
Currently, qRT-PCR plays an important role in quantifying gene expression level because of its sensitivity, specificity, reproducibility and accuracy (Bustin, 2002; Bustin et al., 2005; Nolan et al., 2006). To obtain reliable results, the appropriate reference gene must be selected, which is a key factor in assessing the expression level of the target gene by qRT-PCR. In previous studies, certain traditional reference genes were selected and validated for different samples and various experimental conditions in certain species (Kumar et al., 2011), such as in Arabidopsis thaliana (Czechowski et al., 2005; Dekkers et al., 2012), Salvia miltiorrhiza (Yang et al., 2010), Glycine max (Jian et al., 2008; Libault et al., 2008), Brachiaria grass (Silveira et al., 2009), Cucumis sativus (Wan et al., 2010), etc. Until now, no systematic study was available on the reliable reference gene selection and validation for qRT-PCR normalization in I. indigotica. Previously, 18S rRNA (Hu et al., 2015), IiActin (Han et al., 2013; Li Q. et al., 2014; Dong et al., 2015; Han, 2015) and Ubiquitin (Chen et al., 2015) had been used as internal control genes to normalize the target gene (IiFeSOD, IiHCT, IiGGPPS1, and selected relative genes involved in biosynthesis of lignans) expression levels in I. indigotica. However, it is well known that a universal reference gene does not exist. Therefore, it is important and necessary to select and validate the best suitable reference genes for target gene function research in I. indigotica.
RNA-seq is one of the most effective high-throughput sequencing methods, and the sequencing data can be used to explore the specific functional genes or candidate reference genes. For instance, suitable reference genes were selected and validated from the transcriptome data of Gentiana macrophylla (He et al., 2016), Fagopyrum esculentum (Demidenko et al., 2011), and Pinus massoniana (Chen et al., 2016). Therefore, the reported literature on the transcriptome information of I. indigotica (Chen et al., 2013; Tang et al., 2014; Chen et al., 2015; Zhou et al., 2015; Zhang L. et al., 2016), combined with our lab’s transcriptome data on this plant (unpublished), had well served as the effective resources for finding the candidate reference genes. In this work, 10 candidate reference genes were chosen, and their expression stabilities were validated for five different tissues and three abiotic stress treatments for qRT-PCR normalization.
In the current paper, the candidate reference genes primarily included 3 homologous genes (UBC19, UBC22, and UBC29), 2 traditional housekeeping genes (ACT7 and TUB4) and 5 other novel genes (PP2A-4, eIF2, APT3, AP-2, RPL15). Among these, three homologous genes, i.e., UBC19, UBC22, and UBC29, played different roles in normalization of qRT-PCR, and UBC22 and UBC29 were used as the least stable reference genes, but UBC19 was the moderate performer. Furthermore, as novel reference genes, RPL15 and PP2A-4 were the most stable candidate reference genes for different tissues and experimental conditions. As traditional housekeeping genes, ACT7 and TUB4 were average performers and the same as the other novel genes, such as eIF2 and AP-2. APT3, which was an ambivalent gene, ranked in the second and third places for geNorm and NormFinder, respectively, whereas it showed the least performance for BestKeeper. Therefore, the different candidate reference genes showed different expression characteristics for the different tissues and experimental conditions according to the corresponding algorithms in I. indigotica.
Homologous genes have been commonly used as reference genes for gene expression analysis. Taking the Actin gene in Fortunella crassifolia as an example, ACT6, ACT8, and ACT7 were selected as internal control genes for salt, drought and heavy metal stress treatments, respectively (Hu et al., 2014). For Glycine max, the expressions of EF1A 2a, EF1A 2b, and EF1A1a1 were the most stable under all tested conditions and therefore, these genes were always included in all gene combinations (Costa et al., 2016). However, our experimental results revealed that UBC19, UBC22, and UBC29, as homologous genes, exhibited different expression levels and relatively least stabilities, especially UBC22 and UBC29. Hence, although the homologous genes have similar sequences, they play various roles in gene expression quantification.
PP2A-4 heterotrimeric protein phosphatase is a ubiquitous and conserved serine/threonine phosphatase with broad substrate specificity and diverse cellular functions. As another suitable reference gene in the current paper, PP2A-4 was a good performer for different tissues and abiotic stress treatments in I. indigotica. Moreover, LsPP2A-1 was the most stable in diurnal and developmental timecourse experiments in Lactuca sativa (Sgamma et al., 2016). Similar results were observed in Malus domestica, PP2A-4 was recommended as the most suitable reference gene for all tissues and different biotic stresses (Kumar and Singh, 2015) and for abiotic stress and all sample sets in Sorghum bicolor (Reddy et al., 2016). Additionally, PP2Acs was the best performer in heat and waterlogging stress in Chrysanthemum (Gu et al., 2011). In general, PP2A-4 could be a potential reference gene for optimization of qRT-PCR normalization.
In conjunction with rRNA, ribosomal protein makes up the ribosomal subunits involved in the cellular process of translation, and its gene is ubiquitously expressed and often used as a good reference gene. RPL2 was found to be the best reference gene for all tissue types and different biotic treatments in Malus domestica (Kumar and Singh, 2015). In the same way, RPL15 exhibited the best stable expression levels for different tissues and abiotic stress treatments in I. indigotica. However, RPL was considered the most unstable gene across the developmental stages of P. tomentosa stems (Wang et al., 2016). In summary, the ribosomal protein showed expression diversity for different species and different experimental conditions, and our findings enrich the overall knowledge of ribosomal protein expression patterns.
ACT7, a member of the Actin gene families (Ge et al., 2015; Tian et al., 2015), is commonly used as a common reference gene but showed the least stability for different tissues and abiotic stresses according to geNorm in this study. TUB4, a member of the Tubulin gene families, is also used as common reference gene for qRT-PCR analysis, but for I. indigotica, it presented different expression characteristics for different tissues and abiotic stresses. The results revealed that not all of the common reference genes can be used as suitable internal control genes without selection and validation, and the same reference gene might have different expression levels for different samples and experiment conditions.
The three algorithms of geNorm, NormFinder and BestKeeper were applied to identify the suitable candidate reference genes in this study. Previous research reported the ranking results of suitable reference genes according to different software analyses (Wang et al., 2016), and the candidates of reference genes selected by different algorithms were varied (Li et al., 2016). Our results revealed that the top three rankings for the candidate reference gene were RPL15, APT3, and PP2A-4 for all samples according to geNorm and were RPL15, PP2A-4, and APT3 according to NormFinder. Nevertheless, PP2A-4 remained in second place, and first place went to AP-2 in BestKeeper analysis. The ranking order calculated by BestKeeper was slightly different from the results generated by geNorm and NormFinder, and a similar result was found by Qi’s research (Qi et al., 2016). PP2A-4 was considered the acceptable reference gene for all samples according to the three algorithms. However, it is worth mentioning that the stability ranking of PP2A-4 was slightly inconsistent if considering different tissues and different abiotic stress samples.
YUCCA (YUC) encodes FMOs that could convert indole-3-pyruvate (IPA) to indole-3-acetic acid (IAA) and was identified in Arabidopsis (Kim et al., 2007), Zea mays (Li et al., 2015), Cucumis sativus (Yan et al., 2016), Glycine max (Costa et al., 2016), Triticum aestivum (Li N. et al., 2014), Fragaria × ananassa Duch (Liu et al., 2012), and Oryza sativa (Abu-Zaitoon, 2014). Based on YUCCA protein, a similar sequence from the transcriptome data of I. indigotia was found in the Full-Length CDS DataBase, and the expression patterns of IiYUC6 were discussed. In this work, IiYUC6 presented the highest expression level in fruit compared with other tissues. Similar results were observed from strawberry in which FaYUC1-2 might be involved in flower and fruit development processes (Liu et al., 2012). Therefore, our results suggested that IiYUC6 could play an important role in fruit development processes in I. indigotica.
In brief, this work examined the selection and validation of candidate reference genes for qRT-PCR normalization for different tissues and abiotic stress treatments in I. indigotica. Identification of a suitable reference gene depends on the types of tissues, experimental conditions and analysis methods, and therefore, it is vital and essential to highlight the appropriate reference genes for corresponding samples in different species. The obtained results offer effective information related to the excavation and functional analysis of important gene resources in I. indigotica.
Author Contributions
TL managed and organized the experimental studies. JW performed the qRT-PCR experiments and led the overall data analysis. ML designed the primers and performed the qRT-PCR. TZ and XQ directed the abiotic stress treatments. TL and JW drafted the manuscript, and ZW revised it. All authors discussed and commented on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (31200221), the Fundamental Research Funds for the Central Universities (GK201703075), and the Natural Science Foundation of Shaanxi (2016JQ3007).
References
- Abu-Zaitoon Y. M. (2014). Phylogenetic analysis of putative genes involved in the tryptophan-dependent pathway of auxin biosynthesis in rice. Appl. Biochem. Biotechnol. 172 2480–2495. 10.1007/s12010-013-0710-4 [DOI] [PubMed] [Google Scholar]
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Angelini L. G., Tozzi S., o Di Nasso N. N. (2007). Differences in leaf yield and indigo precursors production in woad (Isatis tinctoria L.) and Chinese woad (Isatis indigotica Fort.) genotypes. Field Crops Res. 101 285–295. 10.1016/j.fcr.2006.12.004 [DOI] [Google Scholar]
- Bustin S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29 23–39. 10.1677/jme.0.0290023 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Nolan T., Pfaffl M. W. (2005). Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 34 597–601. 10.1677/jme.1.01755 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Nolan T. (2004). Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 15 155–166. [PMC free article] [PubMed] [Google Scholar]
- Cha J. Y., Kim W. Y., Kang S. B., Im Kim J., Baek D., Jung I. J., et al. (2015). A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 6:8041 10.1038/ncomms9041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W. S., Doǧramaci M., Foley M. E., Horvath D. P., Anderson J. V. (2012). Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS ONE 7:e42839 10.1371/journal.pone.0042839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yang Z., Hu Y., Tan J., Jia J., Xu H., et al. (2016). Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees 30 685–696. 10.1007/s00468-015-1311-3 [DOI] [Google Scholar]
- Chen J., Dong X., Li Q., Zhou X., Gao S., Chen R., et al. (2013). Biosynthesis of the active compounds of Isatis indigotica based on transcriptome sequencing and metabolites profiling. BMC Genomics 14:857 10.1186/1471-2164-14-857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Li Q., Tan H., Chen J., Xiao Y., Ma R., et al. (2015). Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front. Plant Sci. 6:952 10.3389/fpls.2015.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J. H., Saraiva K. D., Morais V. D., Oliveira J. T., Sousa D. O., de Melo D. F., et al. (2016). Reference gene identification for real-time PCR analyses in soybean leaves under fungus (Cercospora kikuchii) infection and treatments with salicylic and jasmonic acids. Australas. Plant Pathol. 45 191–199. 10.1007/s13313-016-0403-x [DOI] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B. J., Willems L., Bassel G. W., van Bolderen-Veldkamp R. M., Ligterink W., Hilhorst H. W., et al. (2012). Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 53 28–37. 10.1093/pcp/pcr113 [DOI] [PubMed] [Google Scholar]
- Demidenko N. V., Logacheva M. D., Penin A. A. (2011). Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE 6:e19434 10.1371/journal.pone.0019434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gao G., Zheng S., Li F. (2008). Qualitative and quantitative analysis of flavonoids in the leaves of Isatis indigatica Fort. by ultra-performance liquid chromatography with PDA and electrospray ionization tandem mass spectrometry detection. J. Pharm. Biomed. Anal. 48 562–567. 10.1016/j.jpba.2008.05.020 [DOI] [PubMed] [Google Scholar]
- Dong H., Yang J., Huang L., Jia J., Tang J. (2015). Cloning and expression analysis of a hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferasegene (IiHCT) from Isatis indigotica. China J. Chin. Mater. Med. 40 4149–4154. [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A. A., Borges-Pérez A., Pérez J. A. (2011). Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol. Biochem. 49 782–791. 10.1016/j.plaphy.2011.02.022 [DOI] [PubMed] [Google Scholar]
- Ge Q., Zhang Y., Hua W. P., Wu Y. C., Jin X. X., Song S. H., et al. (2015). Combination of transcriptomic and metabolomic analyses reveals a JAZ repressor in the jasmonate signaling pathway of Salvia miltiorrhiza. Sci. Rep. 5:14048 10.1038/srep14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Chen S., Liu Z., Shan H., Luo H., Guan Z., et al. (2011). Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Mol. Biotechnol. 49:192 10.1007/s12033-011-9394-6 [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen J. X., Yang S., Fu X. P., Zhang Z., Chen K. H., et al. (2010). Selection of reliable reference genes for gene expression study in nasopharyngeal carcinoma. Acta Pharmacol. Sin. 31 1487–1494. 10.1038/aps.2010.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., Mauriat M., Guénin S., Pelloux J., Lefebvre J. F., Louvet R., et al. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6 609–618. 10.1111/j.1467-7652.2008.00346.x [DOI] [PubMed] [Google Scholar]
- Han L. (2015). Clone and expression characterization of a new gene encoding geranylgeranyl pyrophosphate synthase from Isatis indigotica fortune. Genomics Appl. Biol. 34 1172–1178. 10.13417/j.gab.034.001172 [DOI] [Google Scholar]
- Han L.-M., Hua W.-P., Wang Z.-Z. (2013). Clone and stress expression analysis of iron superoxide dismutase from Isatis indigotica. Guihaia 33 663–668. 10.3969/j.issn.1000-3142 [DOI] [Google Scholar]
- Han X., Lu M., Chen Y., Zhan Z., Cui Q., Wang Y. (2012). Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS ONE 7:e43084 10.1371/journal.pone.0043084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Yan H., Hua W., Huang Y., Wang Z. (2016). Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Front. Plant Sci. 7:945 10.3389/fpls.2016.00945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen H., Luo C., Dong L., Zhang S., He X., et al. (2014). Selection of reference genes for real-time quantitative PCR studies of kumquat in various tissues and under abiotic stress. Sci. Hortic. 174 207–216. 10.1016/j.scienta.2013.12.003 [DOI] [Google Scholar]
- Hu Y., Zhang L., Chen W. (2015). Molecular cloning and expression analysis of cinnamic acid 4-hydroxylase gene from Isatis indigotica. Chin. Tradit. Herb. Drugs 46 101–106. 10.7501/j.issn.0253-2670.2015.01.020 [DOI] [Google Scholar]
- Im Kim J., Baek D., Park H. C., Chun H. J., Oh D. H., Lee M. K., et al. (2013). Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 6 337–349. 10.1093/mp/sss100 [DOI] [PubMed] [Google Scholar]
- Im Kim J., Murphy A. S., Baek D., Lee S. W., Yun D. J., Bressan R. A., et al. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 62 3981–3992. 10.1093/jxb/err094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian B., Liu B., Bi Y., Hou W., Wu C., Han T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9:59 10.1186/1471-2199-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Q., Wang Z., Ji C. Y., Jeong J. C., Lee H. S., Li H., et al. (2015). Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 94 19–27. 10.1016/j.plaphy.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Kim J. I., Sharkhuu A., Jin J. B., Li P., Jeong J. C., Baek D., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145 722–735. 10.1104/pp.107.104935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozera B., Rapacz M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54 391–406. 10.1007/s13353-013-0173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Singh A. K. (2015). Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci. Hortic. 197 597–606. 10.1016/j.scienta.2015.10.025 [DOI] [Google Scholar]
- Kumar V., Sharma R., Trivedi P. C., Vyas G. K., Khandelwal V. (2011). Traditional and novel references towards systematic normalization of qRT-PCR data in plants. Aust. J. Crop Sci. 5 1455–1468. [Google Scholar]
- Li J., Han J., Hu Y., Yang J. (2016). Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 7:516 10.3389/fpls.2016.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Yin N., Niu Z., Hui W., Song J., Huang C., et al. (2014). Isolation and characterization of three TaYUC10genes from wheat. Gene 546 187–194. 10.1016/j.gene.2014.06.020 [DOI] [PubMed] [Google Scholar]
- Li Q., Chen J., Xiao Y., Di P., Zhang L., Chen W. (2014). The dirigent multigene family in Isatis indigotica: gene discovery and differential transcript abundance. BMC Genomics 15:388 10.1186/1471-2164-15-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhao X., Zhang X. (2015). Genome-wide analysis and expression patterns of the YUCCA genes in maize. J. Genet. Genomics 42 707–710. 10.1016/j.jgg.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Liau B. C., Jong T. T., Lee M. R., Chen S. S. (2007). LC-APCI-MS method for detection and analysis of tryptanthrin, indigo, and indirubin in Daqingye and Banlangen. J. Pharm. Biomed. 43 346–351. 10.1016/j.jpba.2006.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Thibivilliers S., Bilgin D. D., Radwan O., Benitez M., Clough S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome 1 44–54. 10.3835/plantgenome2008.02.0091 [DOI] [Google Scholar]
- Liu H., Xie W. F., Zhang L., Valpuesta V., Ye Z. W., Gao Q. H., et al. (2014). Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 56 350–363. 10.1111/jipb.12150 [DOI] [PubMed] [Google Scholar]
- Liu H., Ying Y. Y., Zhang L., Gao Q. H., Li J., Zhang Z., et al. (2012). Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 31 1425–1435. 10.1007/s00299-012-1258-4 [DOI] [PubMed] [Google Scholar]
- Martins P. K., Mafra V., De Souza W. R., Ribeiro A. P., Vinecky F., Basso M. F., et al. (2016). Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 6:28348 10.1038/srep28348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Xu W., Li D., Hu X., Liu J., Zhang R., et al. (2015). De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress-responsive pathway. BMC Genomics 16:818 10.1186/s12864-015-2019-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T., Hands R. E., Bustin S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1 1559–1582. 10.1038/nprot.2006.236 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Yang L., Wen X., Hong Y., Song X., Zhang M., et al. (2016). Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 7:287 10.3389/fpls.2016.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonić A., Thulke S., Mackay I. M., Landt O., Siegert W., Nitsche A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys. Res. Commun. 313 856–862. 10.1016/j.bbrc.2003.11.177 [DOI] [PubMed] [Google Scholar]
- Reddy P. S., Reddy D. S., Sivasakthi K., Bhatnagar-Mathur P., Vadez V., Sharma K. K. (2016). Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 7:529 10.3389/fpls.2016.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgamma T., Pape J., Massiah A., Jackson S. (2016). Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods 12 21 10.1186/s13007-016-0121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Liu Y., Zhang J., Song L., Guo C. (2015). Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 6:1247 10.3389/fpls.2015.01247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira É. D., Alves-Ferreira M., Guimar ães L. A., da Silva F. R., Carneiro V. T. (2009). Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol. 9:84 10.1186/1471-2229-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Xiao Y., Lv T., Wang F., Zhu Q., Zheng T., et al. (2014). High-throughput sequencing and de novo assembly of the Isatis indigotica transcriptome. PLoS ONE 9:e102963 10.1371/journal.pone.0102963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X. J., Long Y., Wang J., Zhang J. W., Wang Y. Y., Li W. M., et al. (2015). De novo transcriptome assembly of common wild rice (Oryza rufipogon Griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. PLoS ONE 10:e0131455 10.1371/journal.pone.0131455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGuilder H. D., Vrana K. E., Freeman W. M. (2008). Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44 619–626. 10.2144/000112776 [DOI] [PubMed] [Google Scholar]
- Vojta P., Kokáš F., Husičková A., Grúz J., Bergougnoux V., Marchetti C. F., et al. (2016). Whole transcriptome analysis of transgenic barley with altered cytokinin homeostasis and increased tolerance to drought stress. New Biotechnol. 33 676–691. 10.1016/j.nbt.2016.01.010 [DOI] [PubMed] [Google Scholar]
- Wan H., Zhao Z., Qian C., Sui Y., Malik A. A., Chen J. (2010). Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399 257–261. 10.1016/j.ab.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen Y., Ding L., Zhang J., Wei J., Wang H. (2016). Validation of reference genes for gene expression by quantitative real-time RT-PCR in stem segments spanning primary to secondary growth in Populus tomentosa. PLoS ONE 11:e0157370 10.1371/journal.pone.0157370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. L., Medrano J. F. (2005). Real-time PCR for mRNA quantitation. Biotechniques 39 75–85. 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- Wu X., Qin G., Cheung K. K., Cheng K. F. (1997). New alkaloids from Isatis indigotica. Tetrahedron 53 13323–13328. 10.1016/S0040-4020(97)00846-6 [DOI] [Google Scholar]
- Wu Y., Zhang Z. X., Hu H., Li D., Qiu G., Hu X., et al. (2011). Novel indole C-glycosides from Isatis indigotica and their potential cytotoxic activity. Fitoterapia 82 288–292. 10.1016/j.fitote.2010.10.016 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143 1362–1371. 10.1104/pp.106.091561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Che G., Ding L., Chen Z., Liu X., Wang H., et al. (2016). Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 6:20760 10.1038/srep20760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Hou S., Cui G., Chen S., Wei J., Huang L. (2010). Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 37 507–513. 10.1007/s11033-009-9703-3 [DOI] [PubMed] [Google Scholar]
- Yue R., Tie S., Sun T., Zhang L., Yang Y., Qi J., et al. (2015). Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS ONE 10:e0118751 10.1371/journal.pone.0118751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen J., Li Q., Chen W. (2016). Transcriptome-wide analysis of basic helix-loop-helix transcription factors in Isatis indigotica and their methyl jasmonate responsive expression profiling. Gene 576 150–159. 10.1016/j.gene.2015.09.083 [DOI] [PubMed] [Google Scholar]
- Zhang W. X., Fan J., Ma J., Rao Y. S., Zhang L., Yan Y. E. (2016). Selection of suitable reference genes for quantitative real-time PCR normalization in three types of rat adipose tissue. Int. J. Mol. Sci. 17:968 10.3390/ijms17060968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Kang L., Liao S., Pan Q., Ge X., Li Z. (2015). Transcriptomic analysis reveals differential gene expressions for cell growth and functional secondary metabolites in induced autotetraploid of Chinese woad (Isatis indigotica Fort.). PLoS ONE 10:e0116392 10.1371/journal.pone.0116392 [DOI] [PMC free article] [PubMed] [Google Scholar]