ABSTRACT
The echinocandin susceptibilities of 122 Candida glabrata complex strains (including 5 Candida nivariensis and 3 Candida bracarensis strains) were evaluated by microdilution and compared with the results from a molecular tool able to detect FKS mutations. No echinocandin resistance was detected. The PCR results coincide with the MIC data in 99.25% of the cases (1 C. glabrata strain was misidentified as resistant) but were 20 h faster. C. nivariensis FKS genes were sequenced and showed differences with C. glabrata FKS genes.
KEYWORDS: Candida glabrata species complex, Candida nivariensis, Candida bracarensis, echinocandin resistance, hot spot
TEXT
The epidemiology of Candida infections has undergone recent changes due to the description of cryptic species. Candida glabrata species complex includes three human-pathogenic species: C. glabrata sensu stricto, C. nivariensis, and C. bracarensis (1, 2). Candida glabrata sensu stricto accounts for 15 to 20% of all cases of Candida infections worldwide, and it is the second most common cause of candidemia in the United States (3, 4). In Latin American countries, such as Argentina, C. glabrata ranked fourth, representing 4% of the candidemia cases (5).
C. glabrata infection treatment is often difficult due to the increasing prevalence of azole resistance. Thus, the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and the Infectious Diseases Society of America (IDSA) have proposed echinocandins as the treatment of choice for these infections (6–8). In recent years, the prevalence of echinocandin resistance in C. glabrata sensu stricto has increased (9, 10). On the other hand, there are little data about echinocandin susceptibilities for the other two species of the complex, and no treatment recommendations have been released. However, the few available data support them being susceptible to echinocandins (11–14). Clinical resistance to echinocandins has been associated with hot spot FKS mutations (15, 16). Recently, our group published a set of classical PCRs able to detect these mutations (17). The aims of this study were to evaluate the in vitro activities of echinocandins against Argentinian C. glabrata sensu lato strains and to compare the obtained results with those obtained with the described molecular tool.
We analyzed a collection of 122 C. glabrata complex clinical strains, including: (i) 114 C. glabrata sensu stricto strains (40 isolated from blood, 17 from other normally sterile sites, 20 from the vagina, 14 from urine, 7 from the oral cavity, 2 from a catheter, 1 from stool, and 13 with no origin data), (ii) five C. nivariensis strains (throat, renal catheter, urine, continuous peritoneal dialysis bag, and 1 isolate with no isolation data available), and (iii) three C. bracarensis strains (urine, throat, and 1 with no data). All strains were randomly referred from several medical centers from 1984 to 2014 to the Argentinian National Culture Collection of Instituto Nacional de Microbiología Dr. Carlos G. Malbrán. The isolates were identified by classical, molecular (internal transcribed spacer [ITS] sequencing and multiplex PCR), and proteomic (matrix-assisted laser desorption ionization–time of flight mass spectrometry [MALDI-TOF MS]) methods (18–20). Also, 17 characterized echinocandin-resistant C. glabrata sensu stricto strains harboring different FKS mutations were used as PCR control (17), together with Candida parapsilosis sensu stricto ATCC 22019 and Candida krusei ATCC 6258, which were used as MIC controls (21, 22). Anidulafungin (ANF) and caspofungin (CSF) MICs were determined by broth microdilution in accordance with the CLSI M27-A3 and M27-S4 documents (21, 22). Three of the isolates (C. nivariensis DMic 144820 and C. bracarensis DMic 144819 and DMic 144835) did not grow in RPMI 1640 broth, and susceptibility testing was performed by agar diffusion using Etest strips in RPMI 1640 agar. Echinocandin resistance molecular mechanisms were evaluated by using a recently published set of classical PCRs able to identify hot spot mutations at the FKS1 and FKS2 genes. FKS gene sequencing was used to confirm the obtained results (17).
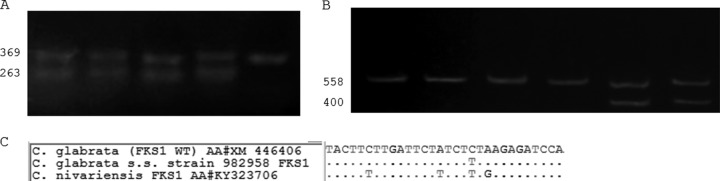
Echinocandins showed good in vitro activity against all the studied isolates (for C. nivariensis, ANF, 0.015 to 0.03 mg/liter, and CSF, 0.06 to 0.13 mg/liter; for C. bracarensis, ANF, 0.03 to 0.06 mg/liter, and CSF, 0.015 to 0.06 mg/liter; and for C. glabrata sensu stricto, ANF, 0.015 to 0.06 mg/liter, and CSF, 0.015 to 0.25 mg/liter). Turning to the molecular detection of FKS mutants, all but one C. glabrata sensu stricto strain showed a wild-type band pattern for the FKS1 and FKS2 hot spot regions (17). Strain DMic 982958 showed a molecular profile consistent with a substitution at Fks1p (S629). The FKS genes of this strain were sequenced. FKS1 showed a silent mutation (T1888C and no amino acid substitution) that was detected but represents a false-positive result (minor error) (Fig. 1A).
FIG 1.
(A) Electrophoresis gel (1.5% agarose) using 1-1670F, 1-S629R, and 1-2225R primers from Dudiuk et al. (16). Lanes 1 to 4, four different C. glabrata sensu stricto clinical strains showing wild-type FKS1 hot spot 1 (TTC TTG ATT CTA TCT CTA AGA GAT CCA); lane 5, C. glabrata sensu stricto strain 982958 showing false-resistant genotype due to a silent mutation (underlined) (TTC TTG ATT CTA TCT TTA AGA GAT CCA). (B) Electrophoresis gel (1.5% agarose) using 2-1619F, 2-S663R, and 2-2177R primers from Dudiuk et al. (16). Lanes 1 to 4, C. nivariensis strains; lanes 5 and 6, C. glabrata sensu stricto. C. nivariensis strains harbor silent FKS2 hot spot 1 mutations compared with C. glabrata sensu stricto (TTT TTG ATT CTT TCT TTG AGA GAT CCA versus TTC TTG ATT TTG TCT CTA AGA GAC CCT, respectively). Underlined sequences are the silent mutations. (C) Sequence alignments of the FKS1 hot spot 1 regions of C. glabrata sensu stricto (wild-type and silent mutant strains) and C. nivariensis. WT, wild type; s.s. sensu stricto. The numbers on the left of each panel are the size of the PCR bands (in bp).
All C. nivariensis strains showed a molecular profile consistent with a C. glabrata sensu stricto FKS mutant (a single band, using the Dudiuk et al. method [17]) (Fig. 1B). These C. nivariensis FKS genes were sequenced by using FKS universal primers (23) and showed several nucleotide differences compared with C. glabrata sensu stricto FKS genes. However, all the described naturally occurring polymorphisms yielded no amino acid changes. For C. bracarensis, it was not possible to obtain PCR bands by using the same primers.
C. glabrata strains with FKS mutations and reduced echinocandin susceptibilities have been described worldwide (10, 11, 15). The prevalences of such mutations ranged from 2.9% to 18% in different reports from different U.S. centers (9, 24, 25). Neither elevated MIC values nor FKS hot spot mutations were detected in our strains. These results are in accordance with other reports from Latin America (26). These geographical differences might be due to the higher use of echinocandin drugs in the United States. Beyda et al. suggested that the unique predictor for echinocandin resistance related to FKS mutations is the use of echinocandin drugs in clinical practice (24). In Argentina, the use of these drugs is very scarce and could greatly contribute to the inexistence of such resistance in our collection.
All C. bracarensis and C. nivariensis isolates showed low echinocandin MIC values and were consistent with previous reports (11, 12, 14).
Shields et al. considered that FKS mutation detection is the most significant risk factor for therapy failure for C. glabrata infections (16). The PCR method used in this work showed 99.25% accordance with susceptibility testing for C. glabrata sensu stricto, and it is at least 20 h faster (17) (95.1% accordance if cryptic species were included). The unique false results (minor error) obtained for the described C. glabrata sensu stricto strain were due to a silent mutation at the hot spot 1 region of FKS1. In addition, this PCR method would be a suitable tool to circumvent the technical problems reported for caspofungin susceptibility testing (27).
Turning to the cryptic species of the C. glabrata complex, the PCR set was unable to correctly classify them regarding echinocandin susceptibility. All C. nivariensis strains were incorrectly considered echinocandin resistant, owing to naturally occurring silent substitutions, while no PCR bands were obtained for C. bracarensis. Thus, we suggest identifying the cryptic species using PCR (18) or MALDI-TOF MS in order to improve the specificity and sensitivity of the molecular detection of echinocandin resistance.
Accession number(s).
The nucleotide sequences for C. nivariensis FKS1 and FKS2 genes were deposited in GenBank under the accession numbers KY323706 and KY494841, respectively (Fig. 1C).
ACKNOWLEDGMENTS
This study was supported in part by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) grant PIP2011/331 to G.G.-E. and by a Universidad Nacional del Litoral (UNL) grant (CAID-PIRCA) to G.G.-E. C.D. has a doctoral fellowship from CONICET.
REFERENCES
- 1.Alcoba-Flórez J, Mendez-Alvarez S, Cano J, Guarro J, Perez-Roth E, del Pilar AM. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol 43:4107–4111. doi: 10.1128/JCM.43.8.4107-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Correia A, Sampaio P, James S, Pais C. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol 56:313–317. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin-Fukuhara M, Fairhead C. 2014. Candida glabrata: a deadly companion? Yeast 31:279–288. doi: 10.1002/yea.3019. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland AA, Harrison LH, Farley MM, Hollick R, Stein B, Chiller TM, Lockhart SR, Park BJ. 2015. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008–2013: results from population-based surveillance. PLoS One 10:e0120452. doi: 10.1371/journal.pone.0120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Córdoba S, Vivot W, Bosco-Borgeat ME, Taverna C, Szusz W, Murisengo O, Isla G, Davel G, Red Nacional De Laboratorios De Micologia. 2011. Species distribution and susceptibility profile of yeasts isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev Argent Microbiol 43:176–185. [DOI] [PubMed] [Google Scholar]
- 6.Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Florl C, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Ullmann AJ, ESCMID Fungal Infection Study Group. 2012. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 18(Suppl 7):19–37. doi: 10.1111/1469-0691.12039. [DOI] [PubMed] [Google Scholar]
- 7.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Executive summary: clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:409–417. doi: 10.1093/cid/civ1194. [DOI] [PubMed] [Google Scholar]
- 8.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham CD, Iqbal N, Bolden CB, Kuykendall RJ, Harrison LH, Farley MM, Schaffner W, Beldavs ZG, Chiller TM, Park BJ, Cleveland AA, Lockhart SR. 2014. Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob Agents Chemother 58:4690–4696. doi: 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop JA, Chase N, Magill SS, Kurtzman CP, Fiandaca MJ, Merz WG. 2008. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J Clin Microbiol 46:443–446. doi: 10.1128/JCM.01986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuenca-Estrella M, Gomez-Lopez A, Isla G, Rodriguez D, Almirante B, Pahissa A, Rodriguez-Tudela JL, Barcelona Candidemia Project Study Group. 2011. Prevalence of Candida bracarensis and Candida nivariensis in a Spanish collection of yeasts: comparison of results from a reference centre and from a population-based surveillance study of candidemia. Med Mycol 49:525–529. doi: 10.3109/13693786.2010.546373. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo-Carvalho MH, Ramos LS, Barbedo LS, Chaves AL, Muramoto IA, Santos AL, Almeida-Paes R, Zancope-Oliveira RM. 2016. First description of Candida nivariensis in Brazil: antifungal susceptibility profile and potential virulence attributes. Mem Inst Oswaldo Cruz 111:51–58. doi: 10.1590/0074-02760150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren TA, McTaggart L, Richardson SE, Zhang SX. 2010. Candida bracarensis bloodstream infection in an immunocompromised patient. J Clin Microbiol 48:4677–4679. doi: 10.1128/JCM.01447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlin DS. 2015. Echinocandin resistance in Candida. Clin Infect Dis 61(Suppl 6):S612–S617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields RK, Nguyen MH, Press EG, Kwa AL, Cheng S, Du C, Clancy CJ. 2012. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob Agents Chemother 56:4862–4869. doi: 10.1128/AAC.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudiuk C, Gamarra S, Leonardeli F, Jimenez-Ortigosa C, Vitale RG, Afeltra J, Perlin DS, Garcia-Effron G. 2014. Set of classical PCRs for detection of mutations in Candida glabrata FKS genes linked with echinocandin resistance. J Clin Microbiol 52:2609–2614. doi: 10.1128/JCM.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudiuk C, Morales-Lopez SE, Podesta V, Macedo D, Leonardelli F, Vitale RG, Tosello ME, Cabeza MS, Biasoli M, Gamarra S, Garcia-Effron G. 2016. Multiplex PCR designed to differentiate species within the Candida glabrata complex. Rev Iberoam Micol doi: 10.1016/j.riam.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. 2013. The yeasts: a taxonomic study. Elsevier, London, United Kingdom. [Google Scholar]
- 20.Morales-López SE, Taverna CG, Bosco-Borgeat ME, Maldonado I, Vivot W, Szusz W, Garcia-Effron G, Cordoba SB. 2016. Candida glabrata species complex prevalence and antifungal susceptibility testing in a culture collection: first description of Candida nivariensis in Argentina. Mycopathologia 181:871–878. doi: 10.1007/s11046-016-0052-1. [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th informational supplement CLSI document M27-S4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Garcia-Effron G, Katiyar SK, Park S, Edlind TD, Perlin DS. 2008. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob Agents Chemother 52:2305–2312. doi: 10.1128/AAC.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyda ND, John J, Kilic A, Alam MJ, Lasco TM, Garey KW. 2014. FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin Infect Dis 59:819–825. doi: 10.1093/cid/ciu407. [DOI] [PubMed] [Google Scholar]
- 25.Farmakiotis D, Tarrand JJ, Kontoyiannis DP. 2014. Drug-resistant Candida glabrata infection in cancer patients. Emerg Infect Dis 20:1833–1840. doi: 10.3201/eid2011.140685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi AM, Pignatari AC, Edmond MB, Marra AR, Camargo LF, Siqueira RA, da Mota VP, Colombo AL. 2016. Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian national surveillance program. PLoS One 11:e0146909. doi: 10.1371/journal.pone.0146909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. Using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]