ABSTRACT
With the rapid spread of antimicrobial resistance in Gram-negative pathogens, biofilm-associated infections are increasingly harder to treat and combination therapy with colistin has become one of the most efficient therapeutic strategies. Our study aimed to evaluate the potential for the synergy of colistin combined with CHIR-090, a potent LpxC inhibitor, against in vitro and in vivo Pseudomonas aeruginosa biofilms. Four P. aeruginosa isolates with various colistin susceptibilities were chosen for evaluation. The tested isolates of P. aeruginosa exhibited MIC values ranging from 1 to 64 and 0.0625 to 0.5 μg/ml for colistin and CHIR-090, respectively. Against 24-h static biofilms, minimum biofilm eradication concentration values ranged from 256 to 512 and 8 to >128 μg/ml for colistin and CHIR-090, respectively. Interestingly, subinhibitory concentrations of CHIR-090 contributed to the eradication of subpopulations of P. aeruginosa with the highest colistin MIC values. The combination of colistin and CHIR-090 at subinhibitory concentrations demonstrated synergistic activity both in vivo and in vitro and prevented the formation of colistin-tolerant subpopulations in in vitro biofilms. In summary, our study highlights the in vivo and in vitro synergistic activity of the colistin and CHIR-090 combination against both colistin-susceptible and -nonsusceptible P. aeruginosa biofilms. Further studies are warranted to investigate the clinical relevance of the combination of these two antimicrobials and outline the underlying mechanism for their synergistic action.
KEYWORDS: LpxC inhibitor, Pseudomonas aeruginosa, biofilms, colistin
INTRODUCTION
Infections caused by Gram-negative bacteria, including Pseudomonas aeruginosa, are increasingly becoming harder to treat with existing antimicrobials (1). As a versatile pathogen, P. aeruginosa plays an important role in a broad range of infections, including those that are biofilm associated. Biofilm-associated infections are particularly problematic because biofilm-embedded cells are more resistant to antibiotic treatments than their planktonic counterparts and represent the main cause of chronic infections (2). When multidrug-resistant Gram-negative bacteria are involved, colistin often remains the only available therapeutic option (3, 4).
Colistin (or polymyxin E) is a natural antibiotic that interacts with lipopolysaccharide (LPS) molecules in the outer membrane of Gram-negative bacteria, causing increased cell membrane permeability and leakage of cell contents, leading to cell death (5). In recent years, increasing numbers of in vitro and in vivo studies have reported on the emergence of colistin resistance (6–11). With practically no new antibiotics being introduced on the market, this underlines the need for effective combinatorial treatment strategies involving compounds with new modes of action.
In the past decade, a lipid membrane enzyme, LpxC, was identified to be a potential and attractive target for antibiotic development (12). LpxC is a zinc-dependent enzyme that catalyzes the deacetylation of UDP-3-O-acyl-GlcNAc, the first step of lipid A biosynthesis (13). Lipid A is the membrane anchor for LPSs and an essential component of the outer membrane of Gram-negative bacteria that protects cells from external agents (13). So far, several LpxC inhibitors, such as L-161,240, TU-514, BB-78485, and CHIR-090, have been identified and extensively characterized (12, 14–16). Among all these inhibitors, CHIR-090 was the first known LpxC inhibitor that was shown to be active against P. aeruginosa and exhibited the most potent inhibitory effect against a wide range of LpxC enzymes with a subnanomolar affinity (13). This compound demonstrated antibacterial activity similar to that of ciprofloxacin against Escherichia coli and P. aeruginosa (16). However, the efficacy of CHIR-090 alone or in combination against biofilms has not yet been examined.
Combination therapy is a powerful but also controversial strategy in infectious diseases used to extend the antimicrobial spectrum of activity, enhance the bactericidal effect, and reduce the risk of the emergence of resistance. We hypothesized that colistin and CHIR-090 may work synergistically in eradicating bacterial biofilms through a self-promoted uptake mechanism, where colistin, as a cationic peptide, causes distortion of the outer membrane, thus facilitating the uptake of CHIR-090 (17). Hence, in our study, we aimed at evaluating the antimicrobial synergy of colistin in combination with CHIR-090 against both colistin-susceptible and -resistant P. aeruginosa biofilms using an in vitro flow cell biofilm model and an in vivo (mouse) biofilm implant model of infection.
RESULTS
Antimicrobial susceptibility under planktonic conditions.
The MIC values of colistin and CHIR-090 are reported in Table 1. Colistin and CHIR-090 exhibited MIC values of 1 and 0.5 μg/ml for strain PAO1, respectively (Table 1). Interestingly, the in vitro-selected colistin-resistant derivative of the PAO1 strain was more susceptible to CHIR-090 (CHIR-090 MIC = 0.25 μg/ml) than its parental strain (Table 1). Two clinical isolates, SCV-1 and SCV-2, exhibited reduced susceptibility to colistin, with MIC values of 16 and 8 μg/ml, respectively, but were highly susceptible to CHIR-090, with MIC values of 0.0625 μg/ml for each strain (i.e., the MICs were 8-fold less than those for PAO1) (Table 1).
TABLE 1.
In vitro susceptibility to colistin and CHIR-090 alone or in combination of the four tested P. aeruginosa strains
| Strain | MIC (μg/ml) |
ΣFICa | |
|---|---|---|---|
| Colistin | CHIR-090 | ||
| PAO1 | 1 | 0.5 | 0.75 (0.5 + 0.25) |
| PAO1-TJH | 64 | 0.25 | 0.13 (1 + 0.0625) |
| SCV-1 | 16 | 0.0625 | 0.5 (4 + 0.015) |
| SCV-2 | 8 | 0.0625 | 0.75 (4 + 0.015) |
The values in parentheses are the concentrations of colistin + CHIR-090 (in micrograms per milliliter) for the reported ΣFIC values.
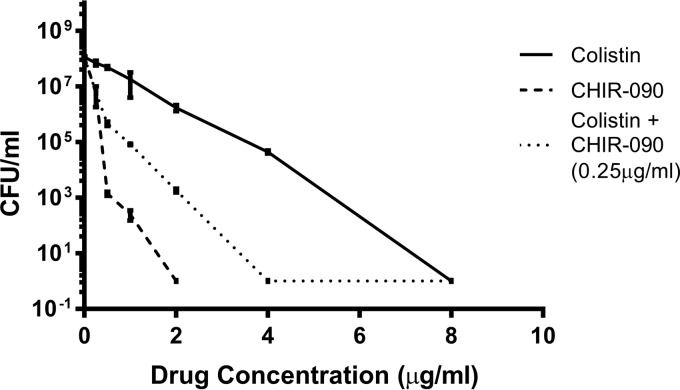
Strain PAO1 demonstrated heterogeneous resistance to both colistin and CHIR-090, with subpopulations growing in the presence of concentrations equivalent to 4× MIC (4 and 2 mg/liter for colistin and CHIR-090, respectively) (Fig. 1). Interestingly, when CHIR-090 (at 0.25 mg/liter, which is equal to 0.5× MIC) was added to colistin, we observed a population shift toward lower colistin MIC values, with a subpopulation growing in the presence of concentrations up to 2× MIC only (Fig. 1), suggesting that the drug combination has a potential for synergy. The potential for the synergy of colistin combined with CHIR-090 was further examined by a checkerboard assay. The synergistic effect of colistin combined with CHIR-090 was confirmed for two colistin-resistant isolates, PAO1-TJH and SCV-1 (index of the fractional inhibitory concentration [ΣFIC] ≤ 0.5), but no interaction against PAO1 and SCV-2 (ΣFIC > 0.5) was found (Table 1).
FIG 1.
Population analysis profiles of PAO1 obtained using colistin monotherapy, CHIR-090 monotherapy, and colistin–CHIR-090 combination therapy. PAO1 (108 CFU/ml) was plated on LB plates containing 0 to 8 μg/ml of colistin alone, 0 to 2 μg/ml CHIR-090 alone, or 0 to 8 μg/ml of colistin with 0.25 μg/ml (1/4 MIC) of CHIR-090.
Antimicrobial susceptibility of P. aeruginosa biofilms.
The colistin minimum biofilm eradication concentration (MBEC) values were significantly higher than the MIC values (256 μg/ml for PAO1, SCV-1, and SCV-2 and 512 μg/ml for PAO1-TJH) (Table 2). In contrast, CHIR-090 exhibited lower MBEC values against all tested strains, with the values ranging from 8 to >128 μg/ml (Table 2). Interestingly, using a checkerboard assay and biofilm conditions, synergy (ΣFIC range, 0.13 to 0.34) was observed against both colistin-susceptible and -resistant P. aeruginosa biofilms. Of note, biofilm eradication could be achieved at lower concentrations for SCV-1 than for the other strains (2 and 1 μg/ml of colistin and CHIR-090, respectively).
TABLE 2.
In vitro MBEC values of colistin and CHIR-090 alone and in combination against four P. aeruginosa strains
| Strain | MBEC (μg/ml) |
ΣFICa | |
|---|---|---|---|
| Colistin | CHIR-090 | ||
| PAO1 | 256 | >128 | 0.34 (64 + 1) |
| PAO1-TJH | 512 | 64 | 0.34 (64 + 16) |
| SCV-1 | 256 | 8 | 0.13 (2 + 1) |
| SCV-2 | 256 | 128 | 0.31 (64 + 8) |
The values in parentheses are the concentrations of colistin + CHIR-090 (in micrograms per milliliter) for the reported ΣFIC values.
Colistin and CHIR-090 efficacy against PAO1 biofilms grown in a biofilm flow cell model.
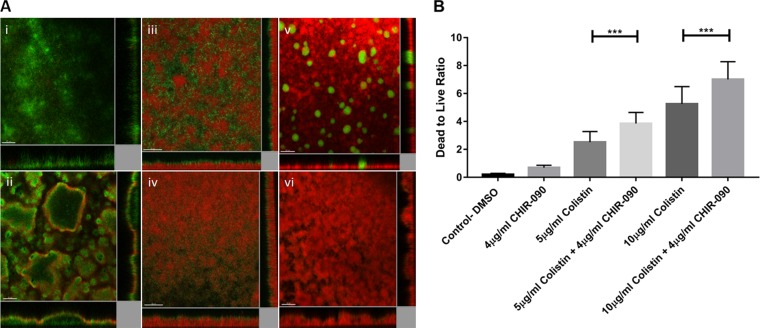
The efficacy of the colistin and CHIR-090 combination against 3-day-old biofilms of PAO1 was evaluated in a flow cell chamber. When dimethyl sulfoxide (DMSO; used as a control) was added, a minimal killing effect was observed (Fig. 2Ai). CHIR-090 monotherapy at 4 μg/ml failed to eradicate the metabolically inactive layer of the biofilm, and only the metabolically active population was affected (Fig. 2Aii). Using colistin at 5 μg/ml, a killing effect was observed only at the bottom layer of the biofilm, as reported by the increase in the propidium iodide (PI) fluorescence (Fig. 2Aiii). Treatment with colistin at 10 μg/ml resulted in a much higher level of bacterial eradication. The results for the remaining populations observed in the flow cells support the population analysis profiles (PAP), suggesting the presence of a colistin-tolerant subpopulation (Fig. 2Av). Combination of colistin at 10 μg/ml with CHIR-090 allowed for the complete eradication of these subpopulations with higher colistin MICs (Fig. 2Avi) Interestingly, by combining colistin at 5 μg/ml with CHIR-090 at 4 μg/ml, an increase in the killing effect was observed on the top layer of the biofilm (Fig. 2Aiv). These microscopic observations were supported by the evaluation of the killing efficacy of the different treatment regimens using the dead/live ratio (Fig. 2B). Lastly, we also used a bead biofilm assay to confirm the synergistic effect of colistin and CHIR-090 by direct counting of the number of CFU, and similar results were obtained (see Fig. S1 in the supplemental material).
FIG 2.
In vitro efficacy of colistin and CHIR-090 alone or in combination against 3-day-old PAO1 biofilms in biofilm flow cells. (A) Confocal imaging of the killing effects of DMSO (control) (i), CHIR-090 at 4 μg/ml (ii), colistin at 5 μg/ml (iii), colistin at 5 μg/ml combined with CHIR-090 at 4 μg/ml (iv), colistin at 10 μg/ml (v), and colistin at 10 μg/ml combined with CHIR-090 at 4 μg/ml (vi). Bars = 50 μm. (B) Dead-to-live ratio of colistin and CHIR-090 alone and in combination. Statistical significance was analyzed by Student's t test (***, P < 0.01).
In vivo evaluation of colistin and CHIR-090 efficacies, alone or in combination, against PAO1 biofilms grown in a mouse implant model of infection.
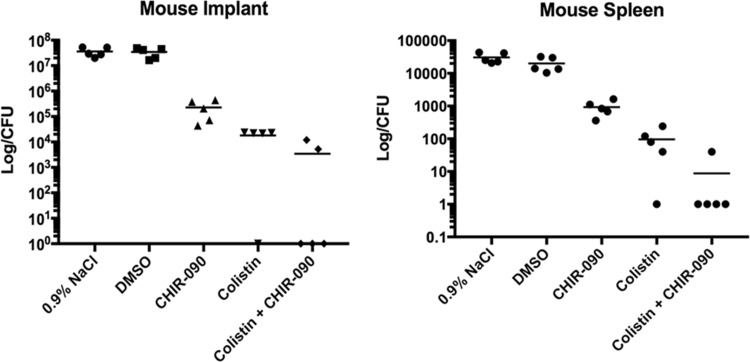
The in vivo efficacy of colistin and CHIR-090, alone and in combination, was evaluated using a mouse implant model of infection. The results are presented in Fig. 3. Both in the implant and in the spleen, similar log counts were observed for the mice from the control group (treated with DMSO) and the untreated group (treated with 0.9% NaCl) (Fig. 3). For both the implant and spleen, CHIR-090 used as monotherapy resulted in an approximately 2-log10 kill, whereas colistin caused a 3-log10 reduction compared to the counts in the untreated or control group (Fig. 3). The combination of colistin and CHIR-090 resulted in a greater reduction in the number of CFU (up to 4 log10) (Fig. 3).
FIG 3.
In vivo evaluation of colistin (10 μg/ml) and CHIR-090 (4 μg/ml) alone and in combination using a mouse implant model of PAO1 biofilm. Each experimental group included five replicates.
DISCUSSION
Colistin, also known as polymyxin E, is a cationic peptide often used in combination therapy as the last-resort option to treat such infections (3). However, due to its extensive use, bacterial strains with reduced susceptibility to colistin have begun to emerge in clinical settings (6–9).
Combination therapy is used empirically to (i) ensure that at least one of the agents is active against the responsible pathogen, (ii) aim for potential synergy and therefore enable faster killing, and (iii) mitigate the development of resistance during treatment. As a result, this strategy is recognized to increase the likelihood of faster clearance of the infection and, therefore, recovery of the patients.
Researchers have been interested in the microbiological and therapeutic effects of the combination of colistin with selected antimicrobials over the last 2 decades, and such combinations have been well studied (11). This combination treatment strategy has shown encouraging results; however, well-designed clinical studies supporting the in vitro and in vivo observations obtained with new antimicrobial compounds in combination with colistin are still warranted. With respect to bacterial biofilms, LpxC inhibitors, such as CHIR-090, have drawn recent attention. These compounds target the LpxC enzyme, involved in lipid A (the anchor membrane for LPS) synthesis. While CHIR-090 has previously been demonstrated to be highly effective against planktonic cells of P. aeruginosa, this class of antibiotic has not been examined for its antibiofilm properties (16). Given that colistin and CHIR-090 target different components of the LPS of Gram-negative bacteria, we hypothesized that there might be a potential for synergy when combining colistin and CHIR-090.
Of interest and as never reported before, we did not observe cross-resistance between colistin and the tested LpxC inhibitor. Indeed, CHIR-090 demonstrated low MIC values (less than 1 μg/ml) against both colistin-susceptible and -nonsusceptible P. aeruginosa strains. The combination of colistin with subinhibitory concentrations of CHIR-090 also resulted in a shift in the colistin MICs for the PAO1 population toward lower values, suggesting that (i) CHIR-090 may have the potential to eradicate PAO1 subpopulations with lower colistin susceptibility and (ii) the combination may demonstrate synergistic effects against P. aeruginosa. This is of particular importance, since conventional dosing of antibiotics most likely selects for the more resistant subpopulations if highly resistant subpopulations of a heteroresistant isolate are not considered. The combination of colistin with CHIR-090 is therefore less likely to select for colistin resistance over time. Using static and flow cell conditions of growth, a synergistic effect against all tested strains grown as planktonic and biofilm cells was observed. As we hypothesized, the effect was greater when cells were grown as a biofilm than when they were grown in the planktonic state. The combination of CHIR-090 and colistin also exhibited a stronger bactericidal effect against colistin-nonsusceptible strains than colistin-susceptible strains. Altogether, these observations support a potential mechanism by which CHIR-090 targets subpopulations with decreased colistin MIC values, with a greater impact on cells growing in biofilms.
To gain a better spatial understanding of the killing dynamics of colistin and CHIR-090, we tested the combinatorial treatment in a biofilm flow cell system. On the basis of our observations, CHIR-090 seems to target cells that are metabolically active. The reasons for this could be either that cells on the top layer of the biofilm have a different phenotype that is favorable to CHIR-090 activity or that the CHIR-090 penetration ability might be reduced due to the presence of the biofilm matrix (18). In contrast and as reported previously, colistin targeted the subpopulation located at the substratum of the biofilm (19). This observation was associated with the migration of the tolerant subpopulation at the top layer of the biofilm exposed to colistin (19). These distinct killing dynamics of colistin and CHIR-090 support our initial hypothesis and findings that the combination of the two antimicrobials has the potential to be synergistic against a P. aeruginosa biofilm by targeting different subpopulations. Interestingly, this effect was observed with subinhibitory concentrations of colistin that are also clinically achievable. In vivo animal experiments supported the in vitro observations, with a combination of colistin and CHIR-090 being more effective than monotherapy. If the effect of the combination did not result in total eradication of the biofilms, it is important to keep in mind that we tested only a single-dose regimen. The administration of repeated dosages over a course of 5 to 7 days, which would be used in practice, would provide us with a better prognosis of the potential therapeutic outcomes of such a combined treatment. The testing of additional regimens, including those using clinically relevant dosages based on the area under the concentration-time curve/MIC parameter, is now warranted to confirm these preliminary findings and further evaluate the therapeutic interest in colistin combined with CHIR-090. Finally, and of great interest, the combined treatment contributed to control of the spread of the bacteria to the spleen.
In conclusion, our study demonstrates for the first time the in vitro and in vivo antimicrobial efficacies of CHIR-090 alone and in combination with colistin against biofilms of both colistin-susceptible and -nonsusceptible P. aeruginosa. While our data suggested that CHIR-090 has a limited ability to eradicate P. aeruginosa biofilms by itself, the compound seems to be capable of suppressing colistin-tolerant subpopulations. A better understanding of the molecular mechanisms responsible for the reduced colistin susceptibility in the subpopulations of each strain with higher colistin MICs is now of great interest to better understand the mechanism of synergy observed with CHIR-090. Inactivation of lipid A biosynthesis has already been reported to be associated with reduced susceptibility to colistin in Acinetobacter baumannii and recently in P. aeruginosa (20, 21) Of interest, a complex and reversible mechanism involving the regulatory network, genetic alterations, and lipid A biosynthesis was suggested in P. aeruginosa (22). These findings highlight the great interest in inhibitors of lipid A biosynthesis and their therapeutic potential in combination with colistin for the treatment of Gram-negative bacterial infections associated or not associated with biofilms. In-depth studies are therefore required to (i) elucidate the mechanisms of synergy and (ii) investigate the therapeutic potential of CHIR-090 in combination with colistin. Nevertheless, our results suggest the strong potential of CHIR-090 and maybe other LpxC inhibitors to be adjunct treatments to prevent the emergence of colistin resistance during therapy (23, 24).
MATERIALS AND METHODS
Bacterial strains and media.
Two colistin-resistant clinical isolates of P. aeruginosa described previously (25) (isolates SCV-1 [CF173] and SCV-2 [CF273]), the lab strain PAO1, the green fluorescent protein-tagged PAO1 strain, and its colistin-resistant derivative (PAO1-TJH) were tested in this study. PAO1-TJH was generated by successive exposure to increased colistin concentrations for 7 days. Muller-Hinton (MH) broth (MNB; Difco, Fisher Scientific, SAS, Illkirch, France) was used for all in vitro experiments except the flow cell experiments, where ABT minimum medium supplemented with 0.5% glucose (ABTG) was employed. LB agar, Miller (Luria-Bertani) (Difco, Sparks, Glencoe, USA), was used for the colistin population analysis profiles (PAP) and colony counting.
Antimicrobial compounds.
Colistin sulfate and CHIR-090 were commercially obtained from Sigma-Aldrich, France, and MedChem Express, NJ, USA, respectively. Each compound was freshly prepared in water for colistin or dimethyl sulfoxide (DMSO) for CHIR-090 according to Clinical and Laboratory Standard Institute (CLSI) guidelines (26).
Population analysis profiles (PAP).
The presence of colistin-resistant subpopulations of strain PAO1 or colistin heteroresistance in PAO1 was examined according to the protocol described elsewhere, with some modifications (10). An initial inoculum of 109 CFU/ml was prepared in sterile normal saline using an overnight culture grown in LB medium. Serial dilutions of this inoculum were then plated onto LB agar plates containing concentrations of colistin and CHIR-090 ranging from 0 to 16 and 0 to 2 μg/ml, respectively. The plates were incubated for 36 h at 37°C. The number of CFU per milliliter was plotted versus time using Prism (version 7.0) software (GraphPad Software, Inc., San Diego, CA, USA). The experiment was performed in duplicate to ensure reproducibility. Heterogeneous resistance to colistin was defined as the presence of detectable subpopulations growing in the presence of concentrations greater than the MIC, as described elsewhere (27).
Antimicrobial susceptibility.
The MIC values of CHIR-090 and colistin for each tested P. aeruginosa strain were determined according to CLSI guidelines using a starting inoculum of ∼106 CFU/ml (26). The potential for the synergy of colistin combined with CHIR-090 against all tested strains was examined using a checkerboard assay and concentrations ranging from 0 to 64 and 0 to 16 μg/ml for colistin and CHIR-090, respectively (28). Bacterial growth was assessed visually after 18 h of incubation at 37°C. Synergistic activity was interpreted using the index of the fractional inhibitory concentration (ΣFIC), as follows: for a ΣFIC of >0.5, synergy; for 0.5 < ΣFIC < 4, no interaction; and for a ΣFIC of >4, antagonism (29). All the experiments were repeated three times.
Antimicrobial susceptibility of biofilm-embedded cells (MBEC).
The minimum biofilm eradication concentration (MBEC) values of colistin and CHIR-090 against P. aeruginosa biofilms were evaluated in triplicate, as described by Naparstek et al. (30). Briefly, biofilms were grown in 96-well microplates for 24 h at 37°C in MH medium and then washed twice with 0.9% NaCl to remove the planktonic cells. Attached biofilms were then subjected to treatment with combinations of different antimicrobial concentrations in a checkerboard assay format. The antimicrobial concentrations tested varied from 0 to 512 and 0 to 128 μg/ml for colistin and CHIR-090, respectively. The plates were incubated for 24 h at 37°C. After antimicrobial challenge, the wells were thoroughly washed with 0.9% NaCl to remove any antimicrobial residue. Two hundred microliters of fresh MH growth medium was added to the washed wells, and then the plates were incubated for 24 h at 37°C. MBEC values were determined to be the lowest antibiotic concentrations that prevented bacterial growth from the treated biofilm. The potential for synergy was also evaluated by calculating the index of the fractional inhibitory concentration (ΣFIC) from the resulting biofilm checkerboard assay. All experiments were performed in triplicate.
Bead biofilm assay.
The effect of the colistin and CHIR-090 combination was evaluated using a bead biofilm assay, as described by Konrat et al. (31). A diluted (optical density at 600 nm, 0.002) overnight PAO1 culture was added to each well of a 24-well microplate containing 3-mm glass beads (EMD Millipore Corporation, Merck, Darmstadt, Germany) immersed in 1 ml of MH medium. The PAO1 biofilm was allowed to grow at 37°C for 72 h, with fresh medium being replenished every 24 h. The beads were then washed with 0.9% NaCl to remove planktonic cells and loose biofilm and immersed in fresh MH II medium. The following treatments were tested: DMSO (0.0005%), colistin (at 5 and 10 μg/ml), and CHIR-90 (at 4 μg/ml) alone or combined with colistin (at 5 and 10 μg/ml). After 24 h of treatment, the beads were removed with sterile forceps at 0, 4, 8, and 24 h and placed into Eppendorf tubes containing 0.9% NaCl. Biofilm cells were recovered following 4 alternating cycles of vortexing (10 s) and sonication (37 Hz). The samples were serially diluted, spotted onto LB agar plates, and incubated at 37°C for 16 h. All experiments were conducted in triplicate.
Biofilms in flow cell chambers.
Biofilms of gfp-tagged P. aeruginosa strains were cultivated in a well-described 40- by 4- by 1-mm three-channel flow cell base (BioCentrum-DTU). The flow chamber was set up according to a previously described protocol (32) and filled with ABTG medium. The flow channels were each inoculated with 500 μl of 1:1,000-diluted overnight culture and then left to incubate for 1 h without any medium flow. Medium flow was initiated using a Cole-Palmer peristaltic pump with a flow velocity of 0.2 mm/s. Three replicate channels were used for each experimental condition. After 72 h of incubation in the flow cells, the flow cells were treated with the following regimens: DMSO (0.0005%), colistin (at 5 and 10 μg/ml), and CHIR-90 (at 4 μg/ml) alone or combined with colistin (at 5 and 10 μg/ml). After 24 h of antimicrobial exposure, 200 μl of 20 μM propidium iodide (PI; Sigma-Aldrich) was injected into each individual flow channel to stain the dead cells of the biofilm. Microscopic images were captured and acquired using a Zeiss LSM 780 confocal laser scanning microscope (Carl Zeiss, Germany) with a 20× objective for monitoring the green fluorescent protein (GFP) and PI fluorescence. Ten images from each sample were used to calculate the dead-to-live ratio using Imaris (version 7.7) software (Bitplane AG, Switzerland), and the mean values of the dead-to-live ratio of the sample are presented.
Mouse biofilm implant model of infection.
Experiments with the mouse biofilm implant model of infection were performed according to the rules of the NACLAR Guidelines and Animal and Birds (Care and Use of Animals for Scientific Purposes) by the Agri-Food & Authority of Singapore (AVA), with authorization and approval being provided by the Institutional Care and Use Committee (IACUC) and Nanyang Technological University under permit number A-0191. Seven-week-old female BALB/c mice (Taconic M&B A/S) were used in this study. The experimental setup and preparation were conducted on the basis of the protocol described by Chua et al. (33). In summary, PAO1 biofilms were grown on cylindrical implants (3 mm by 5 mm in diameter) in 0.9% NaCl at 37°C with shaking at 110 rpm for 20 h. After incubation, biofilm-coated implants were washed three times with 0.9% NaCl and transplanted into the peritoneum of anesthetized mice. Antibiotic regimens were injected as a single dose at the implantation site of groups of five mice. Treatments and the associated groups included the control group, which was treated with no antibiotic (injection of 0.2 ml of 0.04% DMSO); test group 1, which was treated with 10 mg kg−1 of body weight colistin (which is well below the lethal dose of 86 mg kg−1 in mice); test group 2, which was treated with 4 mg kg−1 CHIR-090; and test group 3, which was treated with 10 mg kg−1 colistin and 4 mg kg−1 CHIR-090. After 24 h, mice were euthanized and the implants were retrieved from the peritoneum and washed with 0.9% NaCl. The implants were then sonicated in an ice water bath using an Elmasonic P120H sonicator (Elma, Germany) at 50% power and 37 KHz for 10 min, followed by vortexing of the samples three times for 10 s each time. The spleens were also collected and homogenized using a Bio-Gen PRO200 homogenizer (Pro Scientific, USA) at maximum power on ice. The implants and spleen tissue samples were then serially diluted in 0.9% NaCl, plated onto LB agar, and incubated overnight at 37°C. The number of CFU was calculated and plotted versus treatment using Prism (version 7.0) software (GraphPad Software, Inc., San Diego, CA, USA). Results are presented as means ± standard deviations.
Statistical analysis.
The killing effects of each antimicrobial regimen tested in vitro were analyzed using a t test. All statistical analyses were performed using Prism (version 7.0) software (GraphPad Software, Inc., San Diego, CA, USA). A P value of <0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Research Foundation and the Ministry of Education of Singapore under its Research Centre of Excellence Programme (AcRF Tier 2, MOE2014-T2-2-172).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02223-16.
REFERENCES
- 1.Slama TG. 2008. Gram-negative antibiotic resistance: there is a price to pay. Crit Care 12(Suppl 4):S4. doi: 10.1186/cc6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 4.Falagas ME, Grammatikos AP, Michalopoulos A. 2008. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev Anti Infect Ther 6:593–600. doi: 10.1586/14787210.6.5.593. [DOI] [PubMed] [Google Scholar]
- 5.Gupta S, Govil D, Kakar PN, Prakash O, Arora D, Das S, Govil P, Malhotra A. 2009. Colistin and polymyxin B: a re-emergence. Indian J Crit Care Med 13:49–53. doi: 10.4103/0972-5229.56048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansen HK, Moskowitz SM, Ciofu O, Pressler T, Hoiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J Cyst Fibros 7:391–397. doi: 10.1016/j.jcf.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Denton M, Kerr K, Mooney L, Keer V, Rajgopal A, Brownlee K, Arundel P, Conway S. 2002. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr Pulmonol 34:257–261. doi: 10.1002/ppul.10166. [DOI] [PubMed] [Google Scholar]
- 8.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. 2012. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock RE. 2010. Adaptive resistance to the last hope antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidaillac C, Benichou L, Duval RE. 2012. In vitro synergy of colistin combinations against colistin-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae isolates. Antimicrob Agents Chemother 56:4856–4861. doi: 10.1128/AAC.05996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrosillo N, Ioannidou E, Falagas ME. 2008. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin Microbiol Infect 14:816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 12.Onishi HR, Pelak BA, Gerckens LS, Silver LL, Kahan FM, Chen MH, Patchett AA, Galloway SM, Hyland SA, Anderson MS, Raetz CR. 1996. Antibacterial agents that inhibit lipid A biosynthesis. Science 274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 13.Barb AW, Zhou P. 2008. Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr Pharm Biotechnol 9:9–15. doi: 10.2174/138920108783497668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackman JE, Fierke CA, Tumey LN, Pirrung M, Uchiyama T, Tahir SH, Hindsgaul O, Raetz CR. 2000. Antibacterial agents that target lipid A biosynthesis in gram-negative bacteria. Inhibition of diverse UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylases by substrate analogs containing zinc binding motifs. J Biol Chem 275:11002–11009. [DOI] [PubMed] [Google Scholar]
- 15.Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG. 2002. Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob Agents Chemother 46:1793–1799. doi: 10.1128/AAC.46.6.1793-1799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CR. 2005. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry 44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock RE, Patrzykat A. 2002. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr Drug Targets Infect Disord 2:79–83. doi: 10.2174/1568005024605855. [DOI] [PubMed] [Google Scholar]
- 18.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann G, Yang L, Wu H, Song Z, Wang H, Hoiby N, Ulrich M, Molin S, Riethmuller J, Doring G. 2010. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J Infect Dis 202:1585–1592. doi: 10.1086/656788. [DOI] [PubMed] [Google Scholar]
- 20.Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JY, Park YK, Chung ES, Na IY, Ko KS. 2016. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:25543. doi: 10.1038/srep25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pamp SJ, Gjermansen M, Johansen HK, Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol Microbiol 68:223–240. doi: 10.1111/j.1365-2958.2008.06152.x. [DOI] [PubMed] [Google Scholar]
- 23.Tomaras AP, McPherson CJ, Kuhn M, Carifa A, Mullins L, George D, Desbonnet C, Eidem TM, Montgomery JI, Brown MF, Reilly U, Miller AA, O'Donnell JP. 2014. LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in Gram-negative pathogens. mBio 5:e01551-14. doi: 10.1128/mBio.01551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown MF, Reilly U, Abramite JA, Arcari JT, Oliver R, Barham RA, Che Y, Chen JM, Collantes EM, Chung SW, Desbonnet C, Doty J, Doroski M, Engtrakul JJ, Harris TM, Huband M, Knafels JD, Leach KL, Liu S, Marfat A, Marra A, McElroy E, Melnick M, Menard CA, Montgomery JI, Mullins L, Noe MC, J O'Donnell J, Penzien J, Plummer MS, Price LM, Shanmugasundaram V, Thoma C, Uccello DP, Warmus JS, Wishka DG. 2012. Potent inhibitors of LpxC for the treatment of Gram-negative infections. J Med Chem 55:914–923. doi: 10.1021/jm2014748. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108:7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliopoulos GM, Moellering RC Jr. 1991. Antimicrobial combinations, p 432–492. In Lorian V. (ed), Antibiotics in laboratory medicine, 3rd ed The Williams & Wilkins Co., Baltimore, MD. [Google Scholar]
- 29.Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 30.Naparstek L, Carmeli Y, Navon-Venezia S, Banin E. 2014. Biofilm formation and susceptibility to gentamicin and colistin of extremely drug-resistant KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 69:1027–1034. doi: 10.1093/jac/dkt487. [DOI] [PubMed] [Google Scholar]
- 31.Konrat K, Schwebke I, Laue M, Dittmann C, Levin K, Andrich R, Arvand M, Schaudinn C. 2016. The bead assay for biofilms: a quick, easy and robust method for testing disinfectants. PLoS One 11:e0157663. doi: 10.1371/journal.pone.0157663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sternberg C, Tolker-Nielsen T. 2006. Growing and analyzing biofilms in flow cells. Curr Protoc Microbiol Chapter 1:Unit 1B.2. doi: 10.1002/9780471729259.mc01b02s00. [DOI] [PubMed] [Google Scholar]
- 33.Chua SL, Yam JK, Hao P, Adav SS, Salido MM, Liu Y, Givskov M, Sze SK, Tolker-Nielsen T, Yang L. 2016. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat Commun 7:10750. doi: 10.1038/ncomms10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.