ABSTRACT
In a Mycobacterium smegmatis mutant library screen, transposon mutants with insertions in fhaA, dprE2, rpsT, and parA displayed hypersusceptibility to antibiotics, including the β-lactams meropenem, ampicillin, amoxicillin, and cefotaxime. Sub-MIC levels of octoclothepin, a psychotic drug inhibiting ParA, phenocopied the parA insertion and enhanced the bactericidal activity of meropenem against Mycobacterium tuberculosis in combination with clavulanate. Our study identifies novel factors associated with antibiotic resistance, with implications in repurposing β-lactams for tuberculosis treatment.
KEYWORDS: Mycobacterium tuberculosis, Mycobacterium smegmatis, β-lactams, meropenem, octoclothepin, ParA
TEXT
The global effort to eradicate tuberculosis (TB) has been hampered by the emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB (1). The current treatment for MDR TB includes administration of second-line drugs of the aminoglycoside and fluoroquinolone classes for 20 to 28 months, and XDR TB requires a substantially longer period of treatment involving third-line drugs, such as clofazimine and clarithromycin (2). However, the success rates for MDR and XDR TB treatments are 50% and 26%, respectively, indicating an urgent need for novel and more effective treatment regimens (1). Repurposing preexisting drugs with known safety and toxicity profiles for MDR and XDR TB treatment may be an effective solution to this problem, as it saves money and time involved in the identification and validation of novel anti-TB compounds (3). Based on this strategy, existing antibiotics, such as gatifloxacin, rifapentine, linezolid, and a few β-lactams, are currently being evaluated for MDR and XDR TB treatment (2). Among these, the β-lactam antibiotics deserve special attention because of their well-established clinical safety and mechanism of action, which involves inhibition of bacterial peptidoglycan biosynthesis by the inactivation of l,d-transpeptidases and/or d,d-transpeptidases (4). Mycobacterium tuberculosis, the causative organism of human tuberculosis, is intrinsically resistant to β-lactams, primarily because of the presence of β-lactamases and the l,d-transpeptidases (4). The latter class of enzymes, which are resistant to penicillins and cephalosporins, is used by M. tuberculosis to generate 3→3 transpeptide cross-links in its peptidoglycan (4–7). In addition, its hydrophobic cell envelope, which acts as a permeability barrier for several antibiotics, also confers resistance to β-lactams (4). Supporting this view, a synthetic lethality screen with an M. tuberculosis CDC1551 transposon mutant library for hypersusceptibility to imipenem, a β-lactam antibiotic of the carbapenem class, revealed that the majority of hypersusceptible mutants had mutations in cell envelope-associated genes (8). Despite the possibility that targeting such mycobacterial cell envelope-associated factors with inhibitors may sensitize M. tuberculosis to β-lactams, to our knowledge, no studies have been carried out thus far to validate this hypothesis. Our objective was therefore to identify novel mycobacterial cell envelope-associated factors linked with resistance to antibiotics, including penicillin and nonpenicillin β-lactams, and to assess the effect of chemical inhibition of such factors in enhancing the effectiveness of β-lactams against M. tuberculosis.
To identify novel cell envelope-related mycobacterial genes associated with β-lactam resistance, we assessed the ampicillin sensitivity of 69 colony morphotype mutants of Mycobacterium smegmatis mc26 that were originally isolated in a colony morphology screen from a library of 5,000 transposon mutants (9). The mutants and wild-type M. smegmatis were grown to log phase as described previously (9), and 1 μl of each of these cultures was spotted on Middlebrook 7H10 agar plates containing 50 mg/liter of ampicillin, a concentration lower than the reported MIC of 200 to 400 mg/liter for M. smegmatis (10). As a control for growth, all cultures were spotted onto agar plates lacking ampicillin. Since changes in colony morphologies were likely to result from variations in cell envelope composition, we initially hypothesized that these mutants might show differential susceptibilities to ampicillin by virtue of their altered permeability. Earlier observations that defects in the mycobacterial cell envelope are frequently associated with altered colony morphology and hypersusceptibility to antibiotics supported our hypothesis (11, 12). We obtained four mutants sensitive to ampicillin, and mapping of their transposon insertion sites with a modified genome walking protocol revealed the identity of the disrupted genes to be homologs of the M. tuberculosis genes dprE2, fhaA, rpsT, and parA (13) (Table 1). M. tuberculosis dprE2 encodes decaprenylphosphoryl-d-2-keto erythro pentose reductase, which with DprE1 catalyzes the epimerization of decaprenylphosphoryl ribose (DPR) to decaprenylphosphoryl arabinose (DPA), a precursor in cell wall arabinan synthesis (14). M. tuberculosis fhaA codes for a conserved protein with a C-terminal forkhead-associated domain functionally linked to cell wall peptidoglycan biosynthesis (15, 16). M. tuberculosis rpsT codes for a 30S ribosomal protein S20 (http://tuberculist.epfl.ch/), and we suspect that the change in the colony morphology and the β-lactam sensitivity observed in the corresponding M. smegmatis mutant might be due to an indirect effect caused by diminished translational efficiency, leading to a reduction in the levels of factors associated with cell envelope homeostasis and proteins being targeted by β-lactams. M. tuberculosis parA codes for a chromosome partitioning protein ParA, which with ParB is involved in the segregation of genomic DNA during bacterial cell division (17). Its interaction with Wag31, a protein involved in polar peptidoglycan biosynthesis, indicates that ParA coordinates segregation with cell wall biosynthesis (18). Thus, the annotated/reported functions of these M. tuberculosis homologs revealed their association with cell envelope homeostasis, providing validation for our hypothesis.
TABLE 1.
Transposon insertion sites of M. smegmatis mc26 mutants hypersusceptible to ampicillin, their gene lengths, M. tuberculosis homologs, and predicted functions
| Mutant | Disrupted gene | Gene length (bp) | POIa | M. tuberculosis homolog | Predicted function |
|---|---|---|---|---|---|
| TR37 | MSMEG_6385 | 765 | 359 | dprE2 | Decaprenylphosphoryl-d-2-keto erythro pentose reductase |
| TR49 | MSMEG_0035 | 1,464 | 194 | fhaA | FHA domain protein |
| TR58 | MSMEG_4571 | 261 | 88 | rpsT | 30S ribosomal protein S20 |
| TR62 | MSMEG_6939 | 972 | 650 | parA | Chromosome partitioning protein |
POI, point of transposon insertion relative to the 5′ end of a specific open reading frame used to examine the disruptive nature of the transposon insertion on gene function. Insertions occurring in the proximal 80% region of the gene are assumed to eliminate activity in the gene product (13).
Since meropenem, a carbapenem, is one of the foremost β-lactams being evaluated for TB treatment (2), we determined the meropenem MIC values for these four mutants through a resazurin-based microplate assay as described previously (19, 20). Consistent with their susceptibility to ampicillin, we observed a 2-fold reduction in MIC values for all of these mutants in comparison with those of the wild-type strain (Table 2). This result was further confirmed by CFU enumeration, where we observed a log10 reduction in the range of 1.87 to 2.6 for meropenem-treated mutants versus the wild-type control (Fig. 1a; see also Fig. S1 in the supplemental material). In addition, three of these mutants (TR37 [dprE2Ms::Tn], TR49 [fhaAMs::Tn], and TR62 [parAMs::Tn]) were observed to be hypersusceptible to amoxicillin (a moderate-spectrum β-lactam), cefotaxime (belonging to the cephalosporin class of β-lactams), and the non-β-lactam antibiotics isoniazid and rifampin (Table 2). The observations indicate that the mutants exhibit a generalized susceptibility to these antibiotics possibly due to their altered cell envelope permeability as discussed above. On the other hand, the rpsT mutation did not confer a generalized susceptibility phenotype but caused a significant reduction in the MIC level (2- to >64-fold) to diverse classes of β-lactams (Table 2). This suggests that TR58 (rpsTMs::Tn) is primarily susceptible to β-lactams, an observation that warrants further investigation.
TABLE 2.
Antibiotic susceptibility profiles of wild-type M. smegmatis mc26, its transposon mutants, the corresponding M. smegmatis complement of TR62 (parAMs::Tn), and M. tuberculosis H37Ra
| Strain | Susceptibility profile (MIC in mg/liter)a: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Meropenem | Octoclothepin | Meropenem in presence of octoclothepin (28 mg/liter [60.7 μM]) | Meropenem in presence of clavulanate (5 mg/liter [21.1 μM]) | Meropenem in presence of octoclothepin (7 mg/liter [15.2 μM]) | Amoxicillin | Cefotaxime | Isoniazid | Rifampin | |
| M. smegmatis | |||||||||
| Wild type | 4 | 112 | 2 (ΣFIC 0.75) | – | – | 32 | 64 | 8 | 8 |
| TR37 (dprE2Ms::Tn) | 2 | – | – | – | – | 8 | 2 | <1 | <1 |
| TR49 (fhaAMs::Tn) | 2 | – | – | – | – | 8 | 2 | 4b | <1c |
| TR58 (rpsTMs::Tn) | 2 | – | – | – | – | 16 | <1 | 4 | 8 |
| TR62 (parAMs::Tn) | 2 | – | 1 | – | – | 16 | 4 | 2 | <1 |
| TR62:parAMs | 4 | – | – | – | – | – | – | – | – |
| M. tuberculosis H37Ra | 4 | 28 | – | 2 | 2 (ΣFIC 0.75) | – | – | – | – |
Results shown are representative of two biological and four technical replicates. –, Data not applicable; ΣFIC, fractional inhibitory concentration value; ΣFIC value of >0.5 and ≤4 is considered nonantagonistic (23).
The M. smegmatis fhaA deletion mutant (ΔfhaAMs) exhibited an MIC of 4 mg/liter (16).
ΔfhaAMs exhibited an MIC of <1 mg/liter (16).
FIG 1.
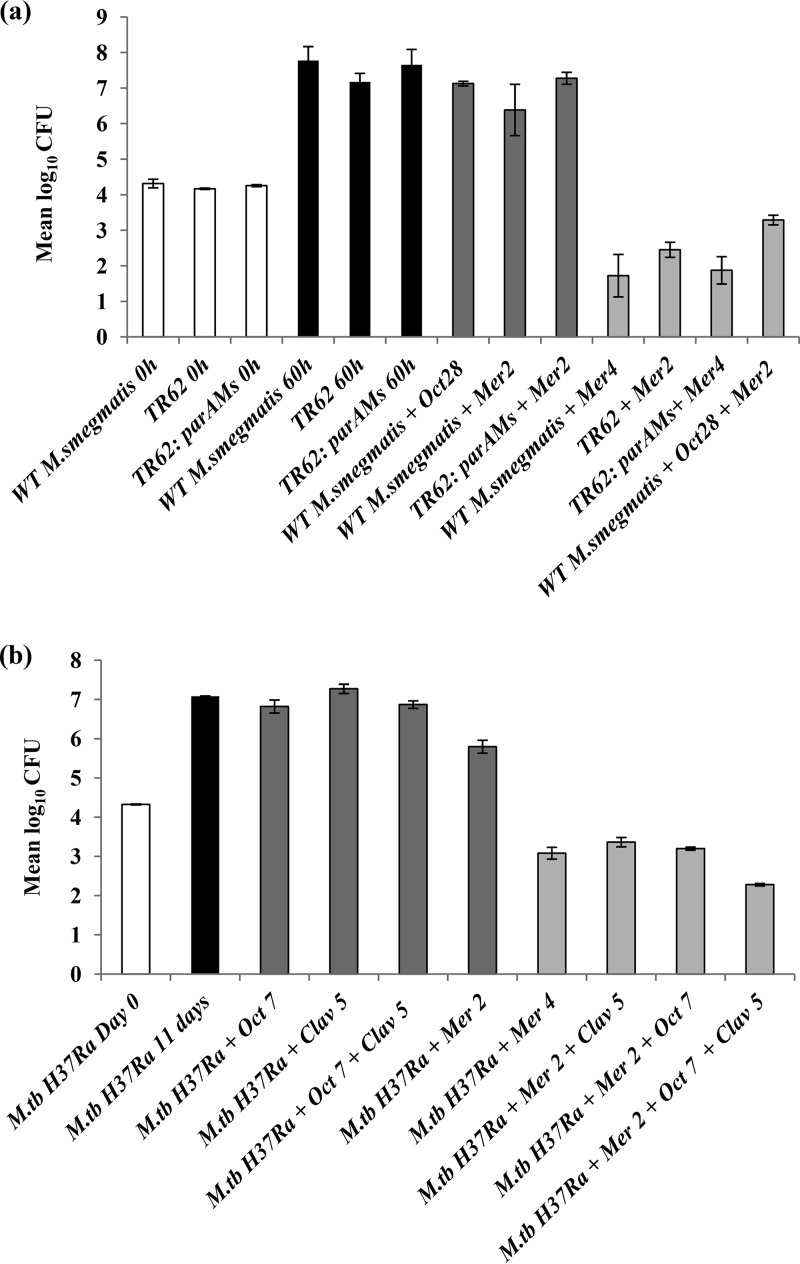
Potentiating effect of octoclothepin (Oct) on meropenem (Mer) or meropenem-clavulanate (Clav) combinations against mycobacteria as determined by mean log10CFU counts. (a) Evaluation of potentiating effect of octoclothepin on meropenem against M. smegmatis (light-gray bars). TR62 (parAMs::Tn) was included in the experiment to determine the extent to which octoclothepin phenocopies the parAMs mutation. TR62:parAMs was included as a complementation control. Strains were incubated with the above-mentioned antibiotic/inhibitors for 60 h. Untreated strains were plated at 0 h (open bars) and 60 h (black bars) as controls. As an additional control, the designated strains were treated with sublethal concentrations of the above inhibitors (dark-gray bars). WT, wild type. (b) Evaluation of potentiating effect of octoclothepin on meropenem and meropenem-clavulanate combinations against M. tuberculosis H37Ra (light-gray bars). M. tuberculosis H37Ra was incubated with the above-mentioned antibiotic/inhibitor for 11 days. Untreated M. tuberculosis H37Ra was plated on day 0 (open bar) and day 11 (black bar) as controls. As an additional control, M. tuberculosis H37Ra was treated with sublethal concentrations of the above inhibitors (dark-gray bars). Compound concentrations are represented in mg/liter. The error bars represent standard deviations. Results shown are representative of two biological replicates.
We chose to further characterize TR62 (parAMs::Tn), since robust functional data are available for ParA (17, 21). For complementation analysis, the open reading frame of M. smegmatis parA was amplified by PCR using its corresponding forward (5′-AGGGATCCATGGGTTCGGGTCAGAACAAA-3′) and reverse (5′-AGTAAGCTTTGCTGCACTACTGCTGGC-3′) primers and cloned between the BamHI and HindIII sites of the mycobacterium-E. coli shuttle vector pMV261h (22), an episomal plasmid carrying an hsp60 promoter and hygromycin resistance marker. Transformation of TR62 with the recombinant plasmid led to restoration of the meropenem MIC to wild-type levels (Table 2), confirming the association of parA with the observed phenotype and ruling out the possibility of a polar effect due to the transposon insertion.
This result led us to hypothesize that inhibition of ParA may further potentiate the action of meropenem against wild-type M. smegmatis. Based on a previous study describing octoclothepin, a potential neuroleptic drug, as an inhibitor of the ATPase activity of mycobacterial ParA (21), we assessed the effect of the octoclothepin-meropenem combination on M. smegmatis survival. Octoclothepin at a sub-MIC value of 28 mg/liter was observed to cause a 2-fold reduction in the meropenem MIC, a phenocopy of the meropenem sensitivity of TR62 (Table 2), thereby validating our hypothesis. In addition, the meropenem-octoclothepin combination exhibited a potentiating and nonantagonistic effect against M. smegmatis (Table 2), as deduced from fractional inhibitory concentration (ΣFIC) calculations (23). These results were validated by enumerating CFU after subjecting M. smegmatis to the above-mentioned treatments (Fig. 1a). Because bacterial cell division involves the coordinated action of multiple proteins associated with events, including DNA replication, divisome assembly, chromosome segregation, and cell wall synthesis, the loss of function of proteins involved in any of these processes is likely to affect cell division as a whole (24). Because ParA coordinates chromosome segregation with cell wall biosynthesis (18), we propose that by inhibiting the function of ParA, octoclothepin might affect cell wall and cell envelope-associated processes. This may potentiate the activity of meropenem via an enhancement of outer membrane permeability, leading to an increased accumulation of this drug in the periplasmic space, where the targets for meropenem (d,d-transpeptidases and l,d-transpeptidases) are localized (25). Furthermore, octoclothepin at the sub-MIC of 28 mg/liter also caused a 2-fold reduction in meropenem MIC for TR62 (Table 2), suggesting that in the absence of parA, octoclothepin may act on unknown secondary targets, leading to a further reduction in the meropenem MIC value.
To test whether our observation in M. smegmatis held true for M. tuberculosis, we assessed the effect of the octoclothepin-meropenem pair on the survival of M. tuberculosis by using a modification of the resazurin-based microplate assay described previously (19). For this, M. tuberculosis H37Ra was cultured as described previously (26) and incubated along with the corresponding antibiotic/inhibitor without replenishment for 11 days before the addition of resazurin, followed by a visual assessment of color change. We found that octoclothepin at a sub-MIC of 7 mg/liter caused a 2-fold reduction in the meropenem MIC value, consistent with our observation in M. smegmatis (Table 2). Importantly, octoclothepin effectively potentiated the activity of meropenem against M. tuberculosis H37Ra at a lower concentration than with clavulanate (Table 2). These results were further verified by CFU assays (Fig. 1b), through which we also tested the effect of the octoclothepin-meropenem pair in combination with the β-lactamase inhibitor clavulanate on M. tuberculosis survival (27). We included clavulanate at 5 mg/liter in these experiments because this compound is currently being evaluated for its activity against MDR TB isolates in combination with β-lactams (2, 28) and has been found to be optimally active at this concentration in enhancing the activity of carbapenems against M. tuberculosis (27, 29). Moreover, the meropenem-clavulanate combination is clinically relevant because its efficacy and safety were established in previous studies (30–32). We found that octoclothepin at the above-mentioned sub-MIC showed a nonantagonistic effect in combination with clavulanate and meropenem, leading to a 1.08 log10 reduction in CFU compared with the value in meropenem-clavulanate-treated M. tuberculosis (Fig. 1b).
In conclusion, our study identifies novel factors that can be targeted to enhance the activity of β-lactams and possibly other antibiotics against M. tuberculosis. The corresponding M. tuberculosis homologs of three (dprE2, rpsT, parA) of the four disrupted M. smegmatis genes were reported to be indispensable for M. tuberculosis growth (http://tuberculist.epfl.ch/), implying that in addition to potentiating β-lactam activity that occurs by cytolysis of M. tuberculosis (33), targeting their corresponding proteins may in principle lead to the killing of M. tuberculosis. In addition to ParA, DprE2 possesses enzymatic activity and hence is a potential target for inhibition by small molecules. Our demonstration that the ParA inhibitor octoclothepin showed a potentiating effect on meropenem activity, as well as the meropenem-clavulanate combination, against M. tuberculosis provides a valid proof of principle for the hypothesis on which this study is based. Chemical modifications of octoclothepin, such as those described in reference 34, may lead to an improvement in its specificity against M. tuberculosis and its in vivo effectiveness. By identifying a novel β-lactam-potentiating agent and new factors associated with antibiotic resistance, our study provides new modalities to enhance the activity of β-lactams and probably other antibiotics against M. tuberculosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Starting Research Grant from Institut Merieux (to T.R.R.) and the Council of Scientific and Industrial Research (CSIR), Government of India.
G.V. was supported by a Senior Research Fellowship from CSIR. S.Y. was supported by a fellowship from Institut Merieux.
G.V. and T.R.R. designed the study; G.V. and S.Y. performed the experiments; G.V., S.Y., and T.R.R. analyzed the data; and G.V. and T.R.R. wrote the paper.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00425-17.
REFERENCES
- 1.World Health Organisation. 2015. Global tuberculosis report. World Health Organisation, Geneva, Switzerland. [Google Scholar]
- 2.Zumla A, Nahid P, Cole ST. 2013. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 12:388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 3.Maitra A, Bates S, Kolvekar T, Devarajan PV, Guzman JD, Bhakta S. 2015. Repurposing—a ray of hope in tackling extensively drug resistance in tuberculosis. Int J Infect Dis 32:50–55. doi: 10.1016/j.ijid.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Wivagg CN, Bhattacharyya RP, Hung DT. 2014. Mechanisms of β-lactam killing and resistance in the context of Mycobacterium tuberculosis. J Antibiot (Tokyo) 67:645–654. doi: 10.1038/ja.2014.94. [DOI] [PubMed] [Google Scholar]
- 5.Dubee V, Triboulet S, Mainardi JL, Etheve-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet JE, Arthur M. 2012. Inactivation of Mycobacterium tuberculosis l,d-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob Agents Chemother 56:4189–4195. doi: 10.1128/AAC.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta R, Lavollay M, Mainardi J-L, Arthur M, Bishai WR, Lamichhane G. 2010. The Mycobacterium tuberculosis gene, ldtMt2, encodes a non-classical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16:466–469. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. 2008. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J Bacteriol 190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lun S, Miranda D, Kubler A, Guo H, Maiga MC, Winglee K, Pelly S, Bishai WR. 2014. Synthetic lethality reveals mechanisms of Mycobacterium tuberculosis resistance to β-lactams. mBio 5:e01767-14. doi: 10.1128/mBio.01767-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viswanathan G, Joshi SV, Sridhar A, Dutta S, Raghunand TR. 2015. Identifying novel mycobacterial stress associated genes using a random mutagenesis screen in Mycobacterium smegmatis. Gene 574:20–27. doi: 10.1016/j.gene.2015.07.063. [DOI] [PubMed] [Google Scholar]
- 10.Billman-Jacobe H, Haites RE, Coppel RL. 1999. Characterization of a Mycobacterium smegmatis mutant lacking penicillin binding protein 1. Antimicrob Agents Chemother 43:3011–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen L, Chinnapapagari S, Thompson CJ. 2005. FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J Bacteriol 187:6603–6611. doi: 10.1128/JB.187.19.6603-6611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philalay JS, Palermo CO, Hauge KA, Rustad TR, Cangelosi GA. 2004. Genes required for intrinsic multidrug resistance in Mycobacterium avium. Antimicrob Agents Chemother 48:3412–3418. doi: 10.1128/AAC.48.9.3412-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manina G, Pasca MR, Buroni S, De Rossi E, Riccardi G. 2010. Decaprenylphosphoryl-beta-d-ribose 2′-epimerase from Mycobacterium tuberculosis is a magic drug target. Curr Med Chem 17:3099–3108. doi: 10.2174/092986710791959693. [DOI] [PubMed] [Google Scholar]
- 15.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. 2012. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal 5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan G, Yadav S, Joshi SV, Raghunand TR. 2017. Insights into the function of FhaA, a cell division-associated protein in mycobacteria. FEMS Microbiol Lett 364:fnw294. doi: 10.1093/femsle/fnw294. [DOI] [PubMed] [Google Scholar]
- 17.Maloney E, Madiraju M, Rajagopalan M. 2009. Overproduction and localization of Mycobacterium tuberculosis ParA and ParB proteins. Tuberculosis (Edinb) 89(Suppl 1):S65–S69. doi: 10.1016/S1472-9792(09)70015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginda K, Bezulska M, Ziolkiewicz M, Dziadek J, Zakrzewska-Czerwinska J, Jakimowicz D. 2013. ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol Microbiol 87:998–1012. doi: 10.1111/mmi.12146. [DOI] [PubMed] [Google Scholar]
- 19.Taneja NK, Tyagi JS. 2007. Resazurin reduction assays for screening of anti-tubercular compounds against dormant and actively growing Mycobacterium tuberculosis, Mycobacterium bovis BCG and Mycobacterium smegmatis. J Antimicrob Chemother 60:288–293. doi: 10.1093/jac/dkm207. [DOI] [PubMed] [Google Scholar]
- 20.Viswanathan G, Yadav S, Raghunand TR. 2016. Identification of novel loci associated with mycobacterial isoniazid resistance. Tuberculosis 96:21–26. doi: 10.1016/j.tube.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Nisa S, Blokpoel MC, Robertson BD, Tyndall JD, Lun S, Bishai WR, O'Toole R. 2010. Targeting the chromosome partitioning protein ParA in tuberculosis drug discovery. J Antimicrob Chemother 65:2347–2358. doi: 10.1093/jac/dkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 23.Heifets L. 1988. Qualitative and quantitative drug-susceptibility tests in mycobacteriology. Am Rev Respir Dis 137:1217–1222. doi: 10.1164/ajrccm/137.5.1217. [DOI] [PubMed] [Google Scholar]
- 24.Ryan KR, Shapiro L. 2003. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu Rev Biochem 72:367–394. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- 25.Kumar P, Kaushik A, Lloyd EP, Li SG, Mattoo R, Ammerman NC, Bell DT, Perryman AL, Zandi TA, Ekins S, Ginell SL, Townsend CA, Freundlich JS, Lamichhane G. 2017. Non-classical transpeptidases yield insight into new antibacterials. Nat Chem Biol 13:54–61. doi: 10.1038/nchembio.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiwari BM, Kannan N, Vemu L, Raghunand TR. 2012. The Mycobacterium tuberculosis PE proteins Rv0285 and Rv1386 modulate innate immunity and mediate bacillary survival in macrophages. PLoS One 7:e51686. doi: 10.1371/journal.pone.0051686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solapure S, Dinesh N, Shandil R, Ramachandran V, Sharma S, Bhattacharjee D, Ganguly S, Reddy J, Ahuja V, Panduga V, Parab M, Vishwas KG, Kumar N, Balganesh M, Balasubramanian V. 2013. In vitro and in vivo efficacy of β-lactams against replicating and slowly growing/nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:2506–2510. doi: 10.1128/AAC.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang D, Wang Y, Lu J, Pang Y. 2015. In vitro activity of beta-lactams in combination with beta-lactamase inhibitors against multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 60:393–399. doi: 10.1128/AAC.01035-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaushik A, Makkar N, Pandey P, Parrish N, Singh U, Lamichhane G. 2015. Carbapenems and rifampin exhibit synergy against Mycobacterium tuberculosis and Mycobacterium abscessus. Antimicrob Agents Chemother 59:6561–6567. doi: 10.1128/AAC.01158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dauby N, Muylle I, Mouchet F, Sergysels R, Payen MC. 2011. Meropenem/clavulanate and linezolid treatment for extensively drug-resistant tuberculosis. Pediatr Infect Dis J 30:812–813. doi: 10.1097/INF.0b013e3182154b05. [DOI] [PubMed] [Google Scholar]
- 31.De Lorenzo S, Alffenaar JW, Sotgiu G, Centis R, D'Ambrosio L, Tiberi S, Bolhuis MS, van Altena R, Viggiani P, Piana A, Spanevello A, Migliori GB. 2013. Efficacy and safety of meropenem-clavulanate added to linezolid-containing regimens in the treatment of MDR-/XDR-TB. Eur Respir J 41:1386–1392. doi: 10.1183/09031936.00124312. [DOI] [PubMed] [Google Scholar]
- 32.Sotgiu G, D'Ambrosio L, Centis R, Tiberi S, Esposito S, Dore S, Spanevello A, Migliori GB. 2016. Carbapenems to treat multidrug and extensively drug-resistant tuberculosis: a systematic review. Int J Mol Sci 17:373. doi: 10.3390/ijms17030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhar N, Dubee V, Ballell L, Cuinet G, Hugonnet JE, Signorino-Gelo F, Barros D, Arthur M, McKinney JD. 2015. Rapid cytolysis of Mycobacterium tuberculosis by faropenem, an orally bioavailable β-lactam antibiotic. Antimicrob Agents Chemother 59:1308–1319. doi: 10.1128/AAC.03461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen JL, Puschl A, Jensen M, Risgaard R, Christoffersen CT, Bang-Andersen B, Balle T. 2010. Exploring the neuroleptic substituent in octoclothepin: potential ligands for positron emission tomography with subnanomolar affinity for α(1)-adrenoceptors. J Med Chem 53:7021–7034. doi: 10.1021/jm100652h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.