ABSTRACT
The extended-spectrum-β-lactamase (ESBL)- and Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae represent serious and urgent threats to public health. In a retrospective study of multidrug-resistant K. pneumoniae, we identified three clinical isolates, CN1, CR14, and NY9, carrying both blaCTX-M and blaKPC genes. The complete genomes of these three K. pneumoniae isolates were de novo assembled by using both short- and long-read whole-genome sequencing. In CR14 and NY9, blaCTX-M and blaKPC were carried on two different plasmids. In contrast, CN1 had one copy of blaKPC-2 and three copies of blaCTX-M-15 integrated in the chromosome, for which the blaCTX-M-15 genes were linked to an insertion sequence, ISEcp1, whereas the blaKPC-2 gene was in the context of a Tn4401a transposition unit conjugated with a PsP3-like prophage. Intriguingly, downstream of the Tn4401a-blaKPC-2-prophage genomic island, CN1 also carried a clustered regularly interspaced short palindromic repeat (CRISPR)-cas array with four spacers targeting a variety of K. pneumoniae plasmids harboring antimicrobial resistance genes. Comparative genomic analysis revealed that there were two subtypes of type I-E CRISPR-cas in K. pneumoniae strains and suggested that the evolving CRISPR-cas, with its acquired novel spacer, induced the mobilization of antimicrobial resistance genes from plasmids into the chromosome. The integration and dissemination of multiple copies of blaCTX-M and blaKPC from plasmids to chromosome depicts the complex pandemic scenario of multidrug-resistant K. pneumoniae. Additionally, the implications from this study also raise concerns for the application of a CRISPR-cas strategy against antimicrobial resistance.
KEYWORDS: CRISPR-Cas, CTX-M, carbapenem-resistance, chromosomal beta-lactamases, Klebsiella pneumoniae, blaKPC
INTRODUCTION
The acquisition and spread of β-lactamases among bacterial species has led to increased resistance to β-lactam antibiotics. Such resistance constitutes an urgent threat to patient management and public health (1). In the mid-1980s, extended-spectrum β-lactamases (ESBLs) were first detected. To date, the Centers for Disease Control and Prevention (CDC) estimate that ESBL-producing Enterobacteriaceae cause at least 26,000 health care-associated infections and 1,700 deaths per year in the United States (1). Among the many types of ESBLs reported (2, 3), CTX-M enzymes are the most prevalent (3). Although the dominant variants of CTX-M are geographically different, CTX-M-15 and CTX-M-14 are the most common ones identified worldwide (3). Moreover, the genes encoding CTX-M enzymes (blaCTX-M) can be horizontally mobilized by various genetic elements. An insertion sequence (IS), ISEcp1, is often associated with the region upstream of blaCTX-M genes and provides promoters to regulate blaCTX-M expression (3).
Carbapenem-resistant Enterobacteriaceae (CRE) have also emerged and rapidly spread worldwide. Each year, approximately 600 deaths result from CRE infections in the United States (1). Carbapenemases are a diverse group of β-lactamases that are active against oxyimino-cephalosporins and cephamycins as well as carbapenems. Among these, Klebsiella pneumoniae carbapenemase (KPC) is currently the most prevalent (4), with KPC-2 and KPC-3 being the most common variants in the United States (5). Genetically, the gene encoding KPC (blaKPC) has been identified within a ∼10-kb Tn3-family transposon, Tn4401 (6). The mobile genetic element Tn4401 possesses a transposase gene (tnpA) and a resolvase gene (tnpR), in addition to two unrelated ISs, ISKpn6 and ISKpn7.
Klebsiella pneumoniae is an opportunistic pathogen causing severe infections in patients, especially neonates, the elderly, and immunocompromised individuals (7). Emergence and facilitated spread of multidrug-resistant (MDR) K. pneumoniae, such as the ESBL (8–10)- and KPC-producing strains (4, 5, 11), are often responsible for the failure of antibiotic treatment (1). K. pneumoniae isolates carrying both blaCTX-M and blaKPC were first reported in New York City in 2013 (12). However, the genetic structure of these coexisting β-lactamases and mechanism of transmission are unclear. Limited data suggest that blaKPC and blaCTX-M are most commonly carried by promiscuous plasmids that readily transfer among Enterobacteriaceae species (13–16). K. pneumoniae strains containing multiple copies of β-lactamase genes in the chromosome, especially the same β-lactamase gene, are rare.
We previously examined K. pneumoniae clinical isolates collected between 2005 and 2012 at the Westchester Medical Center (WMC), New York, for the presence of ESBL and KPC β-lactamase genes, including blaTEM, blaSHV, blaCTX-M, and blaKPC, by PCR and DNA sequencing (17). In a subsequent study using next-generation sequencing, we analyzed additional K. pneumoniae isolates collected at the WMC between 2012 and 2014 and at two New York City hospitals collected in 2013. We obtained the complete genome sequences of three K. pneumoniae clinical isolates harboring both blaCTX-M and blaKPC. In this report, we present the comparative genomic analysis data that reveal the emergence and potential evolution of K. pneumoniae strains carrying multiple chromosomal β-lactamase genes. We also investigated the molecular mechanism potentially associated with such chromosomal integration of antibiotic resistance genes.
(Part of this study was presented in a poster at the Association of Molecular Pathology 2016 Annual Meeting, Charlotte, NC, 10 to 12 November 2016.)
RESULTS
Whole-genome sequences of K. pneumoniae CN1, NY9, and CR14.
Three K. pneumoniae isolates, CN1, NY9, and CR14, were confirmed to carry both blaCTX-M and blaKPC genes. CN1 and NY9 were recovered from two patients in New York City in 2013, whereas CR14 was cultured from a WMC patient in 2012. To obtain complete genome sequence for structural analysis, we employed both short- and long-read sequencing and conducted de novo assembly. The genomic characteristics based on final complete genome sequences of these three isolates are summarized in Table 1. Briefly, K. pneumoniae isolates CN1, NY9, and CR14 belong to ST392, ST340, and ST258, with corresponding genome sizes of 5.44, 5.88, and 6.06 Mb, respectively. Each strain contains a circular chromosome, with a GC content of about 57.5%, similar to those of other K. pneumoniae strains publicly available. A comparison of their chromosomal architectures is shown in Fig. 1A. Compared to CN1 and NY9, CR14 had a relatively larger genome, with a large insertion (∼171-kb) and a large inversion (∼245-kb) in the chromosome. While CR14 and NY9 contained five and six circular plasmids, respectively, CN1 had only two circular plasmids.
TABLE 1.
Genomic characteristics of K. pneumoniae clinical isolates CR14, NY9, and CN1 with both blaCTX-M and blaKPC genes
| Genomic characteristica | Finding for strain: |
||
|---|---|---|---|
| CR14 | NY9 | CN1 | |
| MLST | ST258 | ST340 | ST392 |
| Chromosome size (Mb) | 5.47 | 5.35 | 5.24 |
| GC (%) | 57.3 | 57.5 | 57.5 |
| No. of coding genes | 5,884 | 5,776 | 5,224 |
| RNAs (no.) | |||
| rRNA | 25 | 25 | 25 |
| tRNA | 85 | 88 | 87 |
| Plasmids | |||
| No. of plasmids | 5 | 6 | 2 |
| Plasmid (size) | pCR14_1 (203 kb) | pNY9_1 (199 kb) | pCN1_1 (183 kb) |
| pCR14_2 (154 kb) | pNY9_2 (140 kb) | pCN1_2 (15 kb) | |
| pCR14_3 (116 kb) | pNY9_3 (89 kb) | ||
| pCR14_4 (110 kb) | pNY9_4 (53 kb) | ||
| pCR14_5 (9 kb) | pNY9_5 (39 kb) | ||
| pNY9_6 (11 kb) | |||
| ESBL and KPC β-lactamases | |||
| blaCTX-M | |||
| Allele (no. of copies) | blaCTX-M-2 (1) | blaCTX-M-15 (1) | blaCTX-M-15 (3) |
| Genetic location | pCR14_2 | pNY9_3 | Chromosome |
| blaKPC | |||
| Allele (no. of copies) | blaKPC-2 (1) | blaKPC-3 (1) | blaKPC-2 (1) |
| Genetic location | pCR14_3 | pNY9_2 | Chromosome |
| CRISPR on chromosome | No | No | Yes |
Refer to supplemental materials for a complete list of antimicrobial resistance genes, including other non-ESBL and KPC β-lactamases (Table S1) and antimicrobial susceptibility profiles (Table S2) of these three clinical isolates. MLST, multilocus sequence typing; CRISPR: clustered regularly interspaced short palindromic repeats.
FIG 1.
Overview of whole-chromosome sequence comparisons using progressive Mauve. (A) Chromosome comparison of K. pneumoniae isolates CR14, NY9, and CN1. Indicated are locations of β-lactamase genes (blaCTX-M-15 and blaKPC-2), prophages (▼), and the CRISPR site, as well as a large inversion in the CR14 chromosome. The boxed areas and CRISPR site are shown in more detail in later figures. (B) Chromosome comparison of K. pneumoniae isolates MS6671, CN1, and KPNIH31. Indicated are multiple β-lactamase genes, blaCTX-M-15, blaKPC-2, and blaOXA-181, and a large inversion in the KPNIH31 chromosome. A homologous PsP3-like prophage (▼) and a similar CRISPR-cas array (labeled in red font) are shared by these chromosomes. For a better view of sequence alignment and chromosomal architecture in comparison, genome sequences of KPNIH31 (2013, United States) and MS6671 (2014, United Arab Emirates) are adjusted to dnaA as the first gene.
From the whole-genome sequences, we identified multiple antimicrobial resistance genes, in either chromosomes or plasmids (see Table S1 in the supplemental material). Among these, multiple β-lactamase genes were identified in each of the three K. pneumoniae isolates: CR14 carried blaCTX-M-2, blaOXA-2, and blaTEM-1B in plasmid pCR14_2 (154 kb) with an A/C2 replicon, as well as blaKPC-2, blaOXA-9, and blaTEM-1A in another plasmid, pCR14_3 (116 kb), with two replicons, FIB and FIIK. Isolate NY9 carried blaSHV-11 in the chromosome; blaKPC-3, blaOXA-9, and blaTEM-1A in plasmid pNY9_2 (140 kb) with two replicons, FIA and FII; and blaCTX-M-15, blaOXA-1, and blaTEM-1B in another plasmid, pNY9_3 (89 kb), with an FII replicon. Interestingly, both blaKPC-carrying plasmids contained two replicons, whereas both blaCTX-M-carrying plasmids only had one. More remarkably, CN1 carried three copies of blaCTX-M-15, in addition to one copy of blaSHV-11 and blaKPC-2, in the chromosome (Fig. 1A), with only blaOXA-1 in plasmid pCN1_1 (183 kb).
Chromosomal blaCTX-M-15 and blaKPC-2 in K. pneumoniae.
The coexistence of chromosomal blaCTX-M-15 and blaKPC-2 is rare and raised our interest. In search of homologous genome sequence similar to that of the CN1 chromosome, we identified K. pneumoniae isolate KPNIH31, which was recovered from a patient in the United States in 2013 (15). Both CN1 and KPNIH31 belonged to ST392, with the same GC content of 57.5%. These two isolates shared a small, 15.1-kb plasmid (pCN1_2 or pAAC154-a9e). Their largest plasmid (pCN1_1, 183 kb; and pKPN-c22, 179 kb) and chromosome (CN1, 5.24 Mb; and KPNIH31, 5.23 Mb) also had similar sizes (Table S3) and showed high nucleotide sequence identity of more than 99% over the coverage of more than 98% of chromosomal and plasmid sequences. However, KPNIH31 carried an extra plasmid, pKPN-852, in which no antimicrobial resistance gene was found. Genome sequence alignment demonstrated that compared to CN1, KPNIH31 had a 60-kb inversion in the chromosome (Fig. 1B). Interestingly, KPNIH31 also harbored both blaCTX-M-15 and blaKPC-2 in the chromosome.
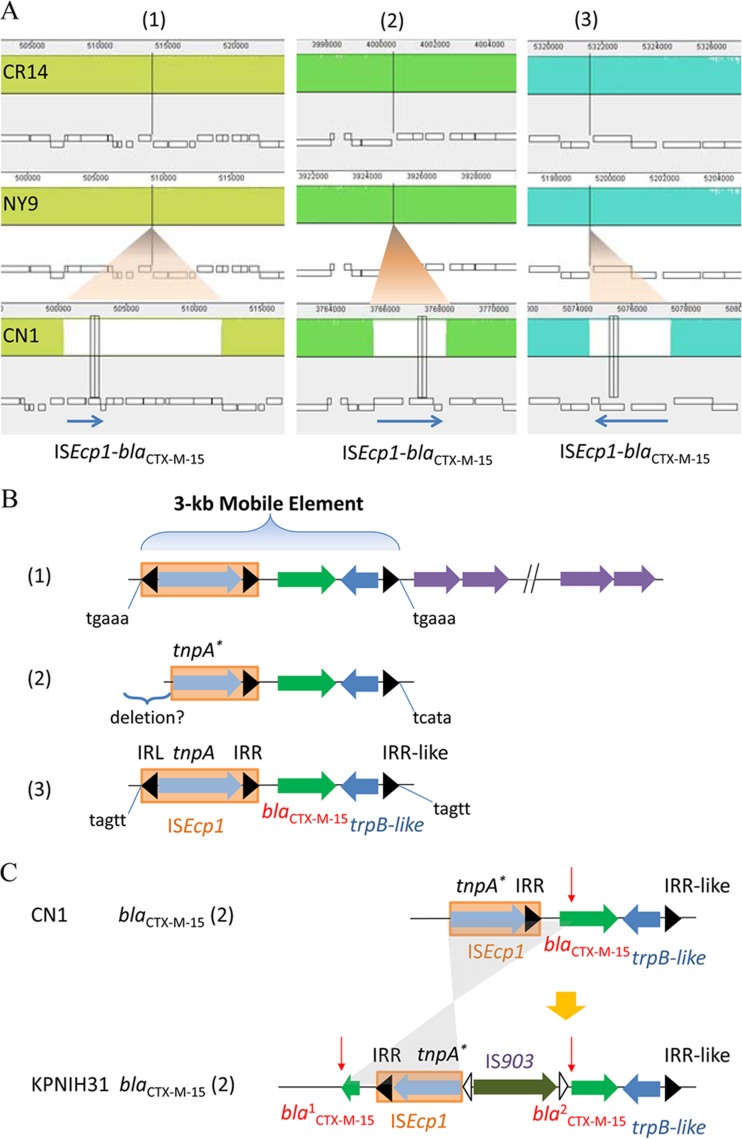
Insertions of three copies of blaCTX-M-15 in the CN1 chromosome were found to be associated with the same mobile element, ISEcp1, despite their different insertion sizes (Fig. 2A and B). This feature was consistent with transposition events of blaCTX-M-15 in plasmids, as seen in plasmid pNY9_3 of isolate NY9. In the context of these blaCTX-M-15 genes, we identified a 5-bp target site duplication (TSD) sequence for each ISEcp1 copy (Fig. 2B). Interestingly, the second ISEcp1-blaCTX-M-15 copy in isolate CN1 was not complete, with the left TSD eliminated and the ISEcp1 transposase truncated at the 5′ end (Fig. 2B), suggesting a sequence deletion occurred after ISEcp1-blaCTX-M-15 insertion. Unlike isolate CN1, KPNIH31 had only one complete copy of the ISEcp1-blaCTX-M-15 unit. While the third ISEcp1-blaCTX-M-15 unit was totally eliminated in KPNIH31, the second ISEcp1-blaCTX-M-15 unit was also truncated and further interrupted by an insertion of IS903 in the middle of blaCTX-M-15, followed by an inversion (Fig. 2C).
FIG 2.
Integration of blaCTX-M-15 in the chromosome by mobile element ISEcp1. (A) A zoomed-in view of three blaCTX-M-15 genes inserted in the CN1 chromosome, compared with chromosomes of CR14 and NY9. The blue arrow indicates a mobile element of ISEcp1-blaCTX-M-15. (B) A schematic view of genetic elements in 3-kb mobile elements of ISEcp1-blaCTX-M-15 in the CN1 chromosome. IRL, left inverted repeat; IRR, right inverted repeat; tnpA, gene encoding transposase A type; trpB-like, gene encoding tryptophan synthase subunit beta-like protein. The target site duplication (TSD) sequences are shown at the ends. The suggested deletion was marked on the second copy of ISEcp1-blaCTX-M-15. (C) A schematic view of the second copies of ISEcp1-blaCTX-M-15 in CN1 and KPNIH31 chromosomes. The second copy of ISEcp1-blaCTX-M-15 in KPNIH31 was disrupted by IS903 insertion. Indicated with a red arrow is the interruption site. Insertion of IS903 interrupts the blaCTX-M-15 gene in the middle and further inverts a truncated part of ISEcp1 (shaded gray). IRR, right inverted repeat.
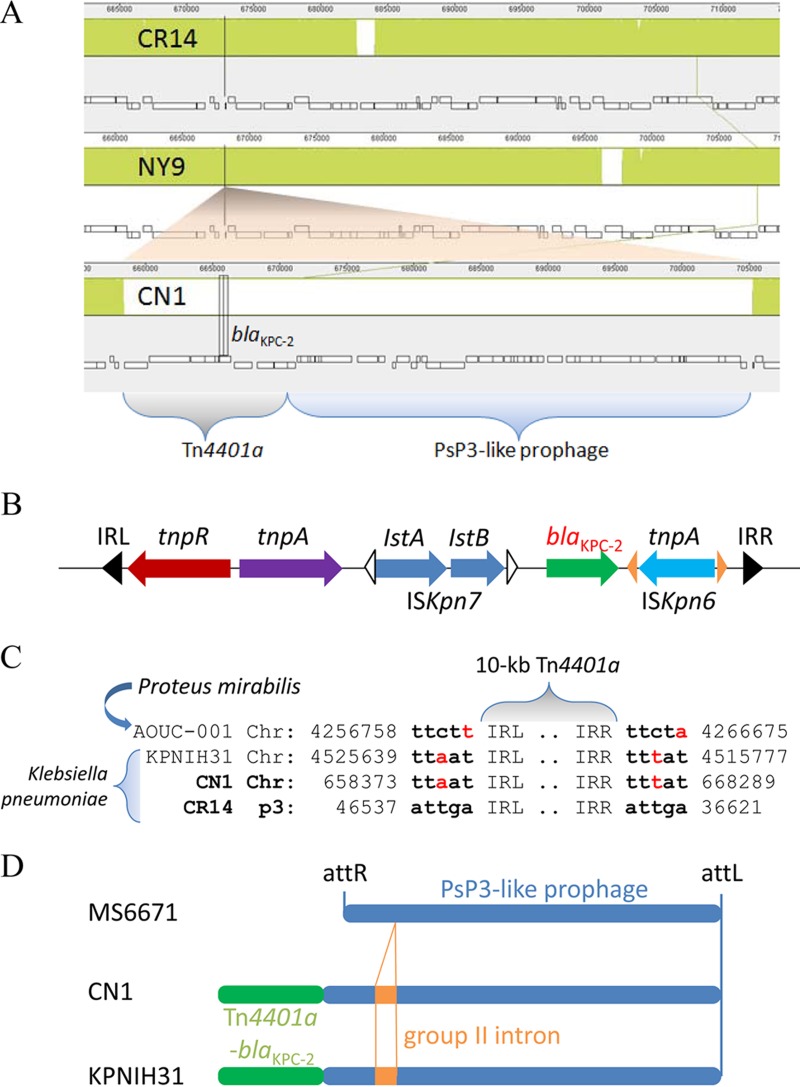
In addition to the three insertions of ISEcp1-blaCTX-M-15, the CN1 chromosome retained an insertion of Tn4401a-blaKPC-2 in a genomic island (Fig. 3A). There were only two additional Enterobacteriaceae strains, the aforementioned K. pneumoniae KPNIH31 (15) and Proteus mirabilis AOUC-001, isolated from Italy in 2013 (18), which carry the chromosomal blaKPC-2 gene. The blaKPC-2 gene in these chromosomes shared the same mobile genetic element as the plasmids, a 10-kb Tn4401a flanked by two 38-bp imperfect but conserved inverted repeats (IRL and IRR) (Fig. 3B). However, we observed that the paired 5-bp TSD sequences of transposon Tn4401a-blaKPC-2 in chromosomes did not match perfectly those in plasmids, such as in pCR14_3 of isolate CR14 (Fig. 3C). This feature of chromosomal Tn4401a-blaKPC-2 differed from that of chromosomal ISEcp1-blaCTX-M-15.
FIG 3.
Integration of blaKPC-2 in the chromosome by a mobile element, Tn4401a. (A) A zoomed-in view of blaKPC-2 insertion in the CN1 chromosome by a 10-kb Tn4401a conjugated with a 43-kb PsP3-like prophage and compared with chromosomes of CR14 and NY9. (B) A schematic view of genetic elements in a 10-kb mobile element of Tn4401a-blaKPC-2, including transposase tnpA, resolvase tnpR, and insertion sequences ISKpn6 and ISKpn7. IRL, left inverted repeat; IRR, right inverted repeat. (C) Comparison of target site duplication (TSD) sequences of Tn4401a-blaKPC-2 transposon in plasmid pCR14_3 (p3) with those in chromosomes of K. pneumoniae CN1 and KPNIH31 (2013, United States) and Proteus mirabilis AOUC-001 (2013, Italy). The numbers shown indicate nucleotide positions at either chromosome or plasmid. (D) A schematic view of PsP3-like prophage region in K. pneumoniae MS6671, CN1, and KPNIH31. MS6671 contained both 32-bp phage attachment sites (attL and attR) at the ends, whereas CN1 and KPNIH31 lost attR at the site of Tn4401a-blaKPC-2 (green bar) conjugating with the PsP3-like prophage (blue bar). Additionally, CN1 and KPNIH31 had identical sequences in the region, and both contained an insertion of group II intron (orange bar) in the prophage.
Both CN1 and KPNIH31 had an identical genomic island, with an aforementioned 10-kb Tn4401a-blaKPC-2 transposon conjugated to a 43-kb Enterobacteriaceae phage PsP3-like prophage. The whole prophage region contained about 50 genes coding for one tRNA and 49 proteins. Among these, 44 proteins (90%) had high similarity to phage proteins, and 30 proteins (61%) were highly homologous to the proteins in bacteriophage PsP3, isolated from Salmonella. Sequence homology search further revealed that this prophage region could also be found in the chromosomes of K. pneumoniae MS6671 (2014, United Arab Emirates), SKGH01 (2015, United Arab Emirates), and AATZP (2014, United States) but without a 2-kb insertion of a retroelement containing a group II intron reverse transcriptase (Fig. 3D). Notably, these chromosomes contained both 32-bp phage attachment sites (attL and attR) with a consensus sequence of TGGGTTTGAACCAACGACCAAGCGATTATGAG. In contrast, with the Tn4401a-blaKPC-2 transposon integrated into the chromosomes of CN1 and KPNIH31, one phage attachment site (attR) was eliminated (Fig. 3D).
Association of CRISPR with chromosomal integration of multiple β-lactamase genes.
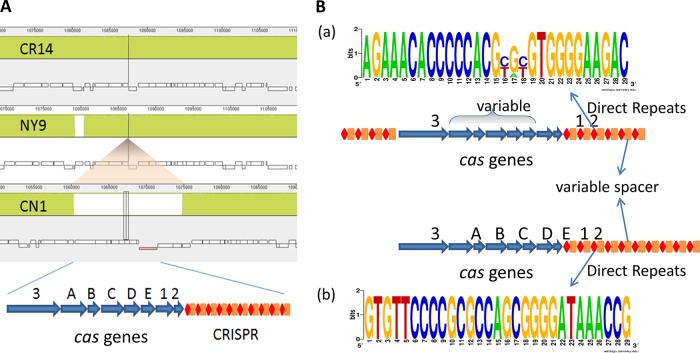
About 360 kb downstream of the prophage site in CN1 chromosome (1069045..1071450), we identified a CRISPR locus with a consensus direct repeat (DR), CGGTTTATCCCCGCTGGCGCGGGGAACAC (29 bp in length), and 39 spacers. Proximate to and upstream of the CRISPR sequences, there was a cluster of eight CRISPR-associated cas genes (cas operon) (Fig. 4A). We then conducted a survey of the CRISPR-cas system on 64 complete genomes of K. pneumoniae publicly available at the NCBI genome database (as of May 2016, including our own three genomes). We identified 22 (34%) strains harboring the CRISPR-cas array in the chromosome. All of the CRISPR-cas arrays in K. pneumoniae strains belonged to type I-E in the classification of CRISPR-cas systems (19, 20). On the basis of their distinguishable features, we further classified them into two distinctive subtypes, a and b (Table S4 and Fig. 4B). In subtype a, two CRISPRs were separated by the cas operon; the DR sequences were either 28 or 29 bp in length and had consensus with 2 to 3 variables in the middle; also, the cas operons were variable in cas genes and their order, occasionally with a transposase gene integrated in the middle (Fig. 4B). In contrast, subtype b was quite consistent and stable, containing the same DR (29 bp in length) and cas operon with only one CRISPR downstream of the cas operon (Fig. 4B). In both subtypes a and b, the number and sequence of spacers varied in each strain, which might represent its own established immune system.
FIG 4.
CRISPR-cas system in K. pneumoniae. (A) A zoomed-in view of CRISPR-cas array insertion in the CN1 chromosome, showing the cas operon of eight cas genes followed by sequence repeats with spacers. (B) A schematic view of two subtypes (a and b) of the CRISPR-cas system in K. pneumoniae. The consensus sequences were generated in WebLogo in analysis of direct repeats collected from 22 CRISPR arrays and grouped into two subtypes. Numbers and sizes of direct repeats, spacers, and cas genes are not to scale.
Eight of 22 (36.4%) K. pneumoniae isolates harboring the CRISPR-cas array carried multiple β-lactamase genes in their chromosomes. In contrast, multiple chromosomal β-lactamase genes were detected in only 3 of 42 (7.1%) K. pneumoniae strains without CRISPR-cas in their chromosomes (P = 0.0164) (Table S5).
There were five K. pneumoniae strains containing both the PsP3-like prophage site and subtype b CRISPR-Cas array in the chromosome, i.e., CN1, KPNIH31, AATZP, MS6671, and SKGH01. Intriguingly, four of them carried multiple β-lactamase genes in the chromosome. Instead of multiple copies of blaCTX-M-15 observed in CN1 and KPNIH31, three copies of blaOXA-181 (a carbapenem-hydrolyzing oxacillinase) were carried by MS6671 and SKHG01, in addition to one blaCTX-M-15 copy and one blaSHV-11 copy, in the chromosome (Table S4). Strain AATZP was exceptional, carrying only one chromosomal blaSHV-11 copy but blaOXA-9 and blaTEM-1A in one plasmid (pKPN-041) and blaCTX-M-15, blaOXA-1, and blaNDM-1 in another (pNDM-1fa) (21). Upon investigating spacer sequences in these five K. pneumoniae strains, we found that they shared identical spacers, except that four “old” spacers in the 3′ extremities of CRISPR arrays were eliminated from CN1 and KPNIH31 (Table S6). Sequence homology search further demonstrated that four spacers (1, 28, 29, and 38 in CN1) targeted a variety of K. pneumoniae plasmids, of which the “newest” one (1; GAGCAGGCACCCGCCGCAACGACGAAGAGCGC) at the 5′ extremities of CRISPR arrays targeted more than 200 plasmids. Some of these plasmids were hit more than one time, and most of them carried antimicrobial resistance genes, including blaKPC-2 (e.g., p628-KPC) and blaCTX-M-15 (e.g., pKpN01-CTX). Notably, of 43 spacers in AATZP, none were homologous to any sequence of its plasmid pKPN-1fa carrying blaCTX-M-15.
DISCUSSION
To date, the ESBL and KPC β-lactamase genes blaCTX-M and blaKPC have been reported to be carried on separate plasmids in most Enterobacteriaceae strains (13–16), just as we observed in clinical K. pneumoniae isolates CR14 and NY9 in this study. Since plasmids can be horizontally transferred across strains, species, and even genera, plasmid-borne antimicrobial resistance is of concern. Moreover, we report here a clinical K. pneumoniae isolate, CN1, with multiple copies of blaCTX-M-15 and blaKPC-2 integrated in the chromosome. Our whole-genome sequence analysis suggests that these chromosomal blaCTX-M-15 and blaKPC-2 elements mobilized from plasmids. Such integration and dissemination of antimicrobial resistance genes illustrates the currently complex pandemic scenario of MDR K. pneumoniae isolates.
It is uncommon for K. pneumoniae to harbor multiple β-lactamase genes in its chromosome, especially multiple copies of the same β-lactamase gene. Most recently, it has been reported that strain KPNIH31 carries two chromosomal blaCTX-M-15 copies with ISEcp1 (although one is disrupted by IS903, as shown in Fig. 2C) (15), KPNIH33 carries two chromosomal blaKPC-3 copies with Tn4401b (14, 15), and MS6671 carries three chromosomal blaOXA-181 copies with ISEcp1 (Fig. 1B) (22). We describe that the CN1 chromosome carries three blaCTX-M-15 copies with ISEcp1 and one blaKPC-2 copy with Tn4401a. Most interestingly, our comparative genomic analysis demonstrates that strains KPNIH31, MS6671, and CN1 share the same chromosomal architecture feature: multiple copies of a β-lactamase gene linked with ISEcp1, a large PsP3-like prophage sequence, and a CRISPR-cas array with the same DR sequence. The CRISPR-cas system is a prokaryotic immune system that confers resistance to foreign genetic elements, such as those present within plasmids and phages, and provides a form of acquired immunity in its spacers (19, 20). Coexistence of a CRISPR-cas array and multiple copies of the same β-lactamase genes in the chromosome lead us to hypothesize a mechanism for antimicrobial resistance genes jumping from plasmids to the chromosome, along with their associated mobile genetic elements. We postulate that the novel acquired spacer in the CRISPR-cas array could induce degradation of its targeting plasmids in the host or in an environmental niche and promote antimicrobial resistance gene mobilization from plasmids into the chromosome when the bacteria are under the selective force of antimicrobial agents. Notably, isolate CN1 has relatively fewer plasmids than isolates CR14 and NY9.
Recently, the CRISPR-cas system has been proposed and tested for application against antimicrobial resistance using bacteriophage as a vector (23–25). However, our whole-genome analysis reveals that both CRISPR-cas array and bacteriophage could be integrated into the chromosome, as well as the antimicrobial resistance genes along with their mobile genetic elements. This suggests that CRISPR targeting on the plasmids carrying antimicrobial resistance leads to resistance genes being mobilized into the chromosome. Efficient CRISPR targeting should be on sequences specific to the antimicrobial resistance gene itself. It also suggests that vector sequences, such as bacteriophages or plasmids, invade the chromosome. Thus, the genetic components of the vector should be carefully considered to avoid long-term genetic contamination when designing the CRISPR-cas strategies against antimicrobial resistance. Additionally, the existing CRISPR-cas array in some bacteria should be taken into account and utilized wisely.
In conclusion, this study describes three complete genomes of MDR K. pneumoniae carrying both blaCTX-M and blaKPC. Consistent with previous observations, transpositions of blaCTX-M-15 and blaKPC-2 in chromosomes or plasmids are linked to the mobile genetic elements ISEcp1 and Tn4401a, respectively. Such mobile elements provide opportunities for horizontal gene transfer of antibiotic resistance genes among plasmids and between plasmid and chromosome, posing complications for the efficient prevention and control of MDR bacterial infections in health care. Our comparative genomic analysis further reveals that genomic characteristics of CRISPR-cas arrays, ISEcp1-blaCTX-M-15 and Tn4401a-blaKPC-2 mobile elements, and prophage sites in K. pneumoniae strains allow differentiation and tracking of the prevalent MDR K. pneumoniae strains and give us insights into the molecular mechanisms of bacterial evolution and adaptation.
MATERIALS AND METHODS
Clinical isolates.
Three K. pneumoniae isolates carrying both blaCTX-M and blaKPC (CR14, NY9, and CN1) were included in this study. These three isolates were identified from 394 nonduplicate K. pneumoniae clinical isolates from the WMC (2005 to 2014; n = 374) and two New York City hospitals (2013; n = 20) that were analyzed by next-generation sequencing. The clinical isolates from two academic hospitals in metropolitan New York City, Bellevue Hospital (n = 11) and New York-Presbyterian Hospital (n = 9), were included for purposes of comparison. All K. pneumoniae isolates were recovered from patient specimens and identified by standard microbiology procedures at the clinical microbiology laboratories of the three hospitals. The WMC is a 643-bed academic tertiary-care medical center in Westchester County, New York. The Institutional Review Board of New York Medical College approved this study.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed for all antimicrobial agents, other than those listed below, with the MicroScan Walk-Away 96 system (Beckman Coulter) (17). Etests (bioMérieux) were employed for susceptibility assessment of cefotaxime, ceftazidime, imipenem, meropenem, tigecycline, and colistin, and results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (26).
Whole-genome sequencing.
Genomic DNA was extracted from each isolate using a QIAamp DNA kit (Qiagen) and subjected to both short- and long-read massively parallel sequencing. Short-read sequencing was performed in the MiSeq system using the Nextera XT sample preparation kit (Illumina), reaching an averaged depth of more than 250× in coverage for each isolate. Long-read sequencing was performed using the Pacific Biosciences (PacBio) RSII single-molecule real-time (SMRT) sequencing system after processing the SMRTbell library using g-TUBE fragmentation (Covaris), the BluePippin size selection system for DNA fragments of 7 to 50 kb (Sage Science), and the SMRTbell template preparation kit (PacBio), reaching an averaged depth of more than 50× in coverage.
Whole-genome de novo assembly.
Sequence assembly was conducted de novo on Illumina short reads using Velvet v.1.2.10 (27) and on the PacBio long reads using the Hierarchical Genome Assembly Process 3, version 2.3.0, pipeline (HGAP3) and Quiver tools in SMRT Analysis v.2.3.0 (PacBio) (28). Sequence assembly was also conducted with the SPAdes Genome Assembler (v3.5.0) for the combination of both Illumina short reads and PacBio long reads (29). Both BWA (30) and Bowtie2 (31) algorithms were employed for short reads aligned to the draft of the whole-genome assembly, whereas BLASR (32) was employed for long-read alignment. The final whole-genome sequences were reviewed under the Integrative Genomics Viewer (33, 34), manually modified, and trimmed into circular chromosome and plasmids.
Whole-genome analysis.
The whole-genome sequences were annotated by the NCBI Prokaryotic Genome Automatic Annotation Pipeline (35). Multilocus sequence typing of each isolate was analyzed using SRST2 (36). Chromosome architecture comparisons between strains were carried out using Mauve 2.3.1 (37). Insertion sequences, CRISPR, and prophage sites were characterized by IslandViewer 3 (38), CRISPRFinder (39), and PHASTER (40), respectively. Antibiotic resistance genes were scanned by ResFinder 2.1 (41). Plasmid incompatibility (Inc.) groups were assessed by BLASTn and by using the PlasmidFinder database from the Center for Genomic Epidemiology (42). BLASTn was also used for other sequence homolog searches.
Nucleotide sequence accession numbers.
The complete genome sequences have been deposited in GenBank under BioProject identifier PRJNA319426 and accession numbers CP015382 to CP015397.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by research funds from the Department of Pathology at New York Medical College and Philips Research North America.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00076-17.
REFERENCES
- 1.CDC. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.Paterson DL, Bonomo RA. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev 18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao WH, Hu ZQ. 2013. Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Cuzon G, Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 5.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structures at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canton R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, Coque TM. 2008. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect 14(Suppl 1):S144–S153. [DOI] [PubMed] [Google Scholar]
- 9.Bush K. 2008. Extended-spectrum beta-lactamases in North America, 1987-2006. Clin Microbiol Infect 14(Suppl 1):S134–S143. [DOI] [PubMed] [Google Scholar]
- 10.Hawkey PM. 2008. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect 14(Suppl 1):S159–S165. [DOI] [PubMed] [Google Scholar]
- 11.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopacz J, Mariano N, Colon-Urban R, Sychangco P, Wehbeh W, Segal-Maurer S, Urban C. 2013. Identification of extended-spectrum-beta-lactamase-positive Klebsiella pneumoniae urinary tract isolates harboring KPC and CTX-M beta-lactamases in nonhospitalized patients. Antimicrob Agents Chemother 57:5166–5169. doi: 10.1128/AAC.00043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coelho A, Gonzalez-Lopez JJ, Miro E, Alonso-Tarres C, Mirelis B, Larrosa MN, Bartolome RM, Andreu A, Navarro F, Johnson JR, Prats G. 2010. Characterisation of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int J Antimicrob Agents 36:73–78. doi: 10.1016/j.ijantimicag.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Conlan S, Deming C, Tsai YC, Lau AF, Dekker JP, Korlach J, Segre JA. 2014. Complete genome sequence of a Klebsiella pneumoniae isolate with chromosomally encoded carbapenem resistance and colibactin synthesis loci. Genome Announc 2:e01332-14. doi: 10.1128/genomeA.01332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Program NCS, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mshana SE, Fritzenwanker M, Falgenhauer L, Domann E, Hain T, Chakraborty T, Imirzalioglu C. 2015. Molecular epidemiology and characterization of an outbreak causing Klebsiella pneumoniae clone carrying chromosomally located bla(CTX-M-15) at a German university-hospital. BMC Microbiol 15:122. doi: 10.1186/s12866-015-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, Huang T, Surendraiah PK, Wang K, Komal R, Zhuge J, Chern CR, Kryszuk AA, King C, Wormser GP. 2013. CTX-M beta-lactamase-producing Klebsiella pneumoniae in suburban New York City, New York, U S A. Emerg Infect Dis 19:1803–1810. doi: 10.3201/eid1911.121470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Pilato V, Chiarelli A, Boinett CJ, Riccobono E, Harris SR, D'Andrea MM, Thomson NR, Rossolini GM, Giani T. 2016. Complete genome sequence of the first KPC-type carbapenemase-positive Proteus mirabilis strain from a bloodstream infection. Genome Announc 4:e00607-16. doi: 10.1128/genomeA.00607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlan S, Lau AF, Program NCS, Palmore TN, Frank KM, Segre JA. 2016. Complete genome sequence of a Klebsiella pneumoniae strain carrying blaNDM-1 on a multidrug resistance plasmid. Genome Announc 4:e00664-16 doi: 10.1128/genomeA.00664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, Yin WF, Chan KG, Li J, Schembri MA, Beatson SA, Paterson DL. 2015. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 5:15082. doi: 10.1038/srep15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol 32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci U S A 112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clinical Laboratory and Standard Institute. 2015. Performance standards for antimicrobial susceptibility testing: 25th supplemental information, M100-S25. Clinical Laboratory and Standards Institute, Wayne, PA. [Google Scholar]
- 27.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 29.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity G, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O. 2008. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS 12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MG, Brinkman FS. 2015. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 43:W104–W108. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.