ABSTRACT
Prior to characterization of antifungal inhibitors that target CYP51, Trichophyton rubrum CYP51 was expressed in Escherichia coli, purified, and characterized. T. rubrum CYP51 bound lanosterol, obtusifoliol, and eburicol with similar affinities (dissociation constant [Kd] values, 22.7, 20.3, and 20.9 μM, respectively) but displayed substrate specificity, insofar as only eburicol was demethylated in CYP51 reconstitution assays (turnover number, 1.55 min−1; Km value, 2 μM). The investigational agent VT-1161 bound tightly to T. rubrum CYP51 (Kd = 242 nM) with an affinity similar to that of clotrimazole, fluconazole, ketoconazole, and voriconazole (Kd values, 179, 173, 312, and 304 nM, respectively) and with an affinity lower than that of itraconazole (Kd = 53 nM). Determinations of 50% inhibitory concentrations (IC50s) using 0.5 μM CYP51 showed that VT-1161 was a tight-binding inhibitor of T. rubrum CYP51 activity, yielding an IC50 of 0.14 μM, whereas itraconazole, fluconazole, and ketoconazole had IC50s of 0.26, 0.4, and 0.6 μM, respectively. When the activity of VT-1161 was tested against 34 clinical isolates, VT-1161 was a potent inhibitor of T. rubrum growth, with MIC50, MIC90, and geometric mean MIC values of ≤0.03, 0.06, and 0.033 μg ml−1, respectively. With its selectivity versus human CYP51 and drug-metabolizing cytochrome P450s having already been established, VT-1161 should prove to be safe and effective in combating T. rubrum infections in patients.
KEYWORDS: VT-1161, CYP51, Trichophyton rubrum, azole resistance, substrate specificity
INTRODUCTION
Infections with the ascomycete fungi Trichophyton spp. (e.g., onychomycosis or nail fungus, tinea pedis or athlete's foot, tinea corporis or ringworm) are some of the oldest human dermatological afflictions. While not life-threatening, these infections can be of significant annoyance to the sufferer. Trichophyton rubrum is the most common dermatophyte infection in healthy individuals, accounting for up to 70% of skin infections (1) and up to 90% of nail infections (2, 3). Nail infections caused by T. rubrum affect about 10% of the population and are frequently intractable and prone to relapse upon termination of antifungal therapy (4, 5). T. rubrum infections of hair, skin, and nails have increased over the past 70 years, especially in the elderly and in some countries also in children (6–8). Chronic skin infections caused by T. rubrum can become sites for secondary infection by other microorganisms, such as Candida spp., Cryptococcus spp., Aspergillus spp., and Staphylococcus aureus, which can become life-threatening in immunocompromised and immunosuppressed patients if the secondary infection becomes systemic (9–12).
Current therapeutic treatments against T. rubrum infection include azole antifungal agents, allylamines, and thiocarbamates (all of which inhibit ergosterol biosynthesis) administered orally or applied topically in creams and lotions. In chronic invasive and systemic fungal infections, especially among immunocompromised patients, amphotericin B (which disrupts fungal cell membranes) can be utilized intravenously. These antimycotic agents are most effective against the growing organism but are often ineffective against static phases of the organism, such as T. rubrum conidia, leading to reinfection, unless prolonged treatment regimens are adopted. Recently, photodynamic treatments have been developed using photosensitizers in combination with UV-A1 radiation (340 to 400 nm) to kill both the mycelial form and the conidia of T. rubrum (13) in topical dermal infections. The antifungal agents most commonly used against T. rubrum are ketoconazole, fluconazole, terbinafine, and flucytosine (13). The prolonged treatment regimens often required have led to the emergence of azole-resistant T. rubrum strains, especially strains resistant to fluconazole (14–17).
In this study, we characterized the catalytic properties of recombinant T. rubrum CYP51 and compared the novel antifungal VT-1161 (18, 19) with clinical azole antifungal drugs in terms of potency and selectivity of binding to and inhibition of recombinant T. rubrum CYP51 and in terms of inhibition of fungal growth in broth microdilution assays.
RESULTS
Expression and purification of Trub51.
Following heterologous expression in Escherichia coli, the Trub51 protein was extracted by sonication in 2% (wt/vol) sodium cholate, which yielded 240 ± 80 nmol per liter culture, as determined by carbon monoxide difference spectroscopy (20). Purification by Ni2+-nitrilotriacetic acid (NTA) agarose chromatography resulted in an 84% recovery of native Trub51 protein, yielding a stock 48 μM solution after dialysis. SDS-polyacrylamide gel electrophoresis confirmed the purity of the Ni2+-NTA agarose-purified Trub51 to be greater than 90% when assessed by staining intensity.
Spectral properties of Trub51.
The absolute spectrum of the resting oxidized form of Trub51 (Fig. 1A) was typical for a low-spin ferric cytochrome P450 (CYP) enzyme (21, 22), with α, β, Soret (γ), and δ spectral bands being found at 567, 540, 420, and 361 nm, respectively. Reduced carbon monoxide difference spectra for Trub51 (Fig. 1B) gave the red-shifted heme Soret peak at 447 nm, characteristic of P450 enzymes, indicating that the Trub51 protein was isolated in the native form. The formation of the reduced CO-P450 complex with Trub51 was rapid (half-life = 0.18 ± 0.06 min), although it did not proceed to completion (a hump was visible at 422 nm).
FIG 1.
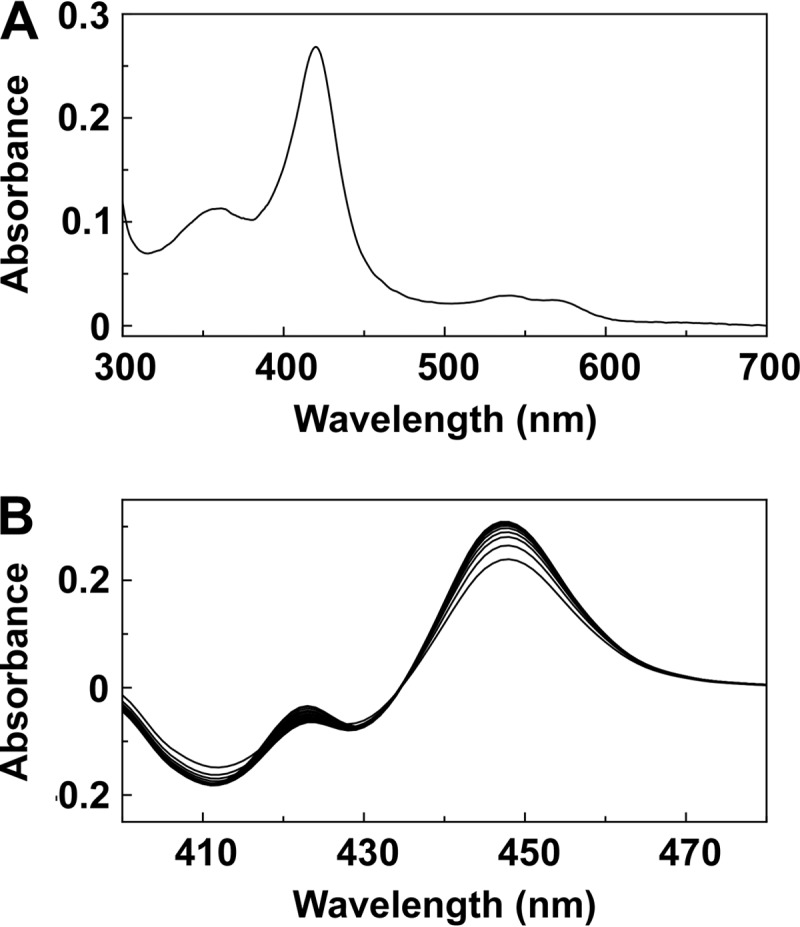
Spectral characteristics of Trub51. (A) Absolute spectra were determined using 3 μM purified Trub51 in the oxidized resting state. (B) Reduced carbon monoxide difference spectra were determined using 3 μM purified Trub51, with sequential measurements being made every 45 s.
Sterol binding properties of Trub51.
Progressive titration of Trub51 with lanosterol, eburicol, and obtusifoliol gave type I difference spectra with a peak at 388 nm and a trough at 421 nm (Fig. 2). Type I binding spectra occur when the substrate or another molecule displaces the water molecule coordinated as the sixth ligand to the low-spin hexa-coordinated heme prosthetic group, causing the heme to adopt the high-spin penta-coordinated conformation (22). The intensity (change in the maximum absorbance [ΔAmax]) of the type I binding spectra obtained with lanosterol was 7-fold lower than that obtained with eburicol and 3-fold lower than that obtained with obtusifoliol, suggesting that eburicol was the preferred substrate. However, dissociation constant (Kd) values of 20.3 ± 1.2 μM, 22.7 ± 3.6 μM, and 20.9 ± 0.3 μM were obtained for eburicol, lanosterol, and obtusifoliol, respectively, indicating that all three sterols bound with a similar affinity.
FIG 2.
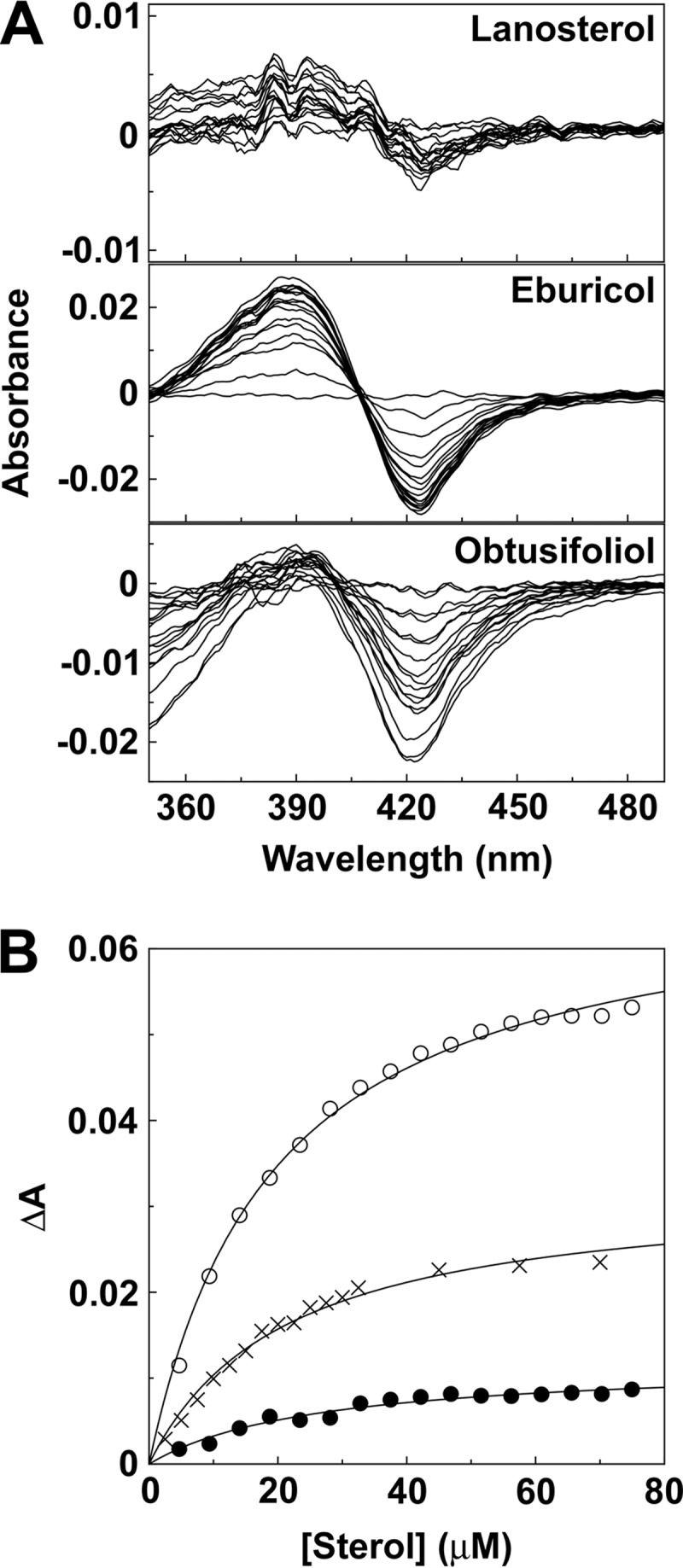
Sterol binding properties of Trub51. (A) Absorbance difference spectra were measured during the progressive titration of 5 μM Trub51 with lanosterol, eburicol, and obtusifoliol. (B) Saturation curves for lanosterol (filled circles), eburicol (hollow circles), and obtusifoliol (crosses) were constructed from the absorbance difference (ΔA388–421) of the type I difference spectra observed. Sterol binding data were fitted using the Michaelis-Menten equation.
CYP51 reconstitution assays.
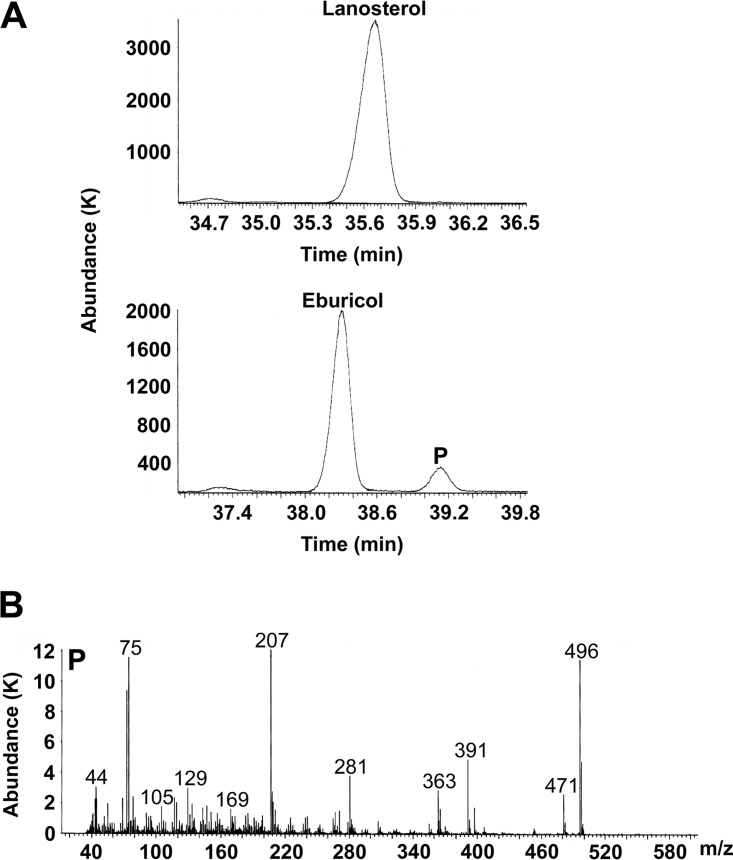
Trub51 did not catalyze the 14α-demethylation of lanosterol under the stated assay conditions. Gas chromatography (GC) traces for tetramethylsilane (TMS)-derivatized CYP51 assay metabolites showed lanosterol and eburicol emerging from the GC column after 35.65 and 38.25 min, respectively (Fig. 3A), whereas the 14α-demethylated product of eburicol emerged after 39.15 min. Confirmation of the identity of product P was obtained by the mass fragmentation pattern (Fig. 3B) as TMS-derivatized C-14-demethylated eburicol (M+ 496). Trial Trub51 assays using 50 μM obtusifoliol yielded no detectable metabolites (data not shown). This is only the second time that such strict substrate specificity has been observed for a fungal CYP51 enzyme, with Mycosphaerella graminicola CYP51 previously being shown to demethylate eburicol but not lanosterol in vitro (23).
FIG 3.
GC/MS analysis of Trub51 reconstitution assay metabolites. (A) GC traces for Trub51 reconstitution assays (37°C, 15 min) using lanosterol and eburicol as the substrates are shown. (B) In addition, the mass fragmentation pattern for the TMS-derivatized C-14-demethylated eburicol product (M+ 496; product P) is shown. Abundance is expressed in thousands (K).
Mild substrate inhibition was evident from the eburicol velocity curve obtained for Trub51 (Fig. 4), with the calculated Km and Ki values for eburicol being 2 μM and 225 μM, respectively. The maximum eburicol turnover number was 1.55 min−1. The observed substrate inhibition suggests the presence of two distinct eburicol binding sites or binding orientations in Trub51, with one binding site/orientation being catalytically productive, while the other leads to the formation of an unproductive dead-end complex. However, no allosterism was observed in the eburicol type I difference binding spectra (Fig. 2B), suggesting that eburicol binds in only one conformation that causes the displacement of the axial ligated heme water molecule responsible for the transition from the low- to the high-spin state.
FIG 4.
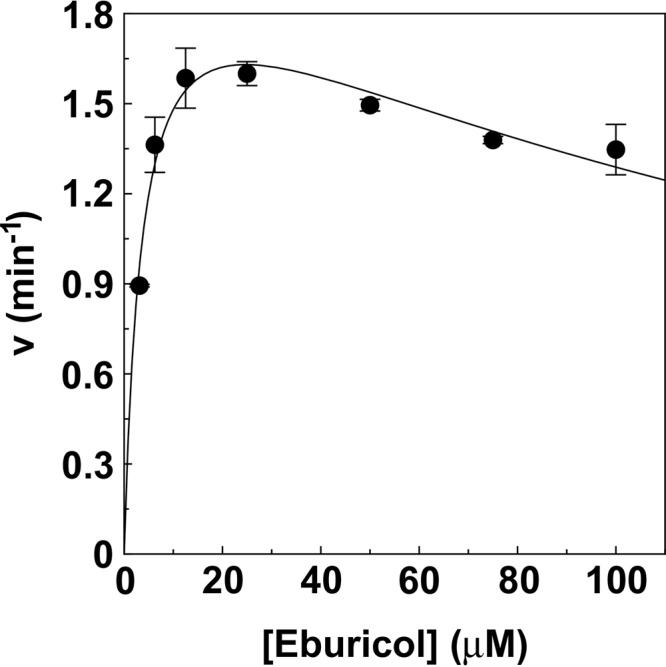
Km determination for eburicol. A velocity curve was constructed for eburicol with Trub51 using the CYP51 reconstitution assay (34, 55). The single substrate inhibition equation v = (Vmax · [S])/{Km + [S] · (1 + [S]/Ki)} (57) was used to fit the velocity curve. Mean values from three replicates and the associated standard deviation bars are shown.
CYP51 inhibitor binding properties of Trub51.
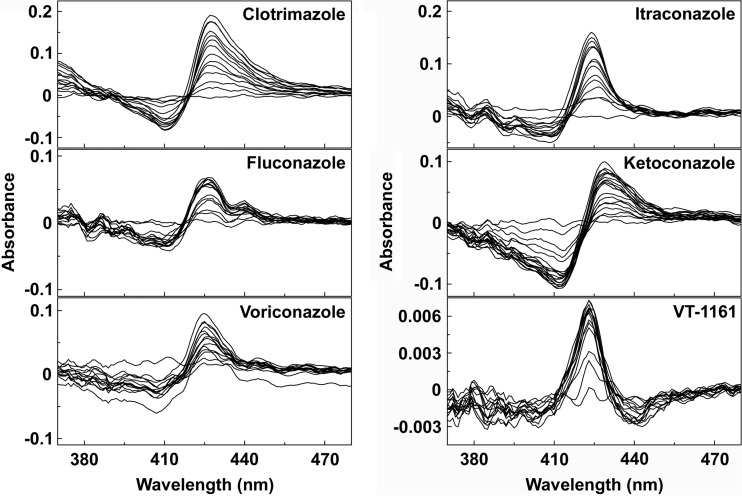
All five marketed imidazole and triazole antifungal agents and the novel tetrazole VT-1161 produced type II binding spectra (24) with Trub51 (Fig. 5). Ligand saturation curves (Fig. 6) confirmed that azole binding was tight, with the rearranged Morrison equation providing the best fit to the data (25, 26). Trub51 bound itraconazole the tightest and had a Kd value of 53 ± 29 nM, while clotrimazole, fluconazole, voriconazole, ketoconazole, and VT-1161 all apparently bound less tightly to Trub51 and had similar Kd values of 179 ± 83, 173 ± 53, 304 ± 64, 312 ± 36, and 242 ± 99, respectively.
FIG 5.
Type II azole binding spectra for Trub51. Clotrimazole, fluconazole, voriconazole, itraconazole, ketoconazole, and VT-1161 were progressively titrated against 2 μM CYP51 protein, with the difference spectra being determined after each addition of azole. The resultant type II difference spectra obtained for each azole are shown. Each experiment was performed in triplicate, although only one replicate is shown.
FIG 6.
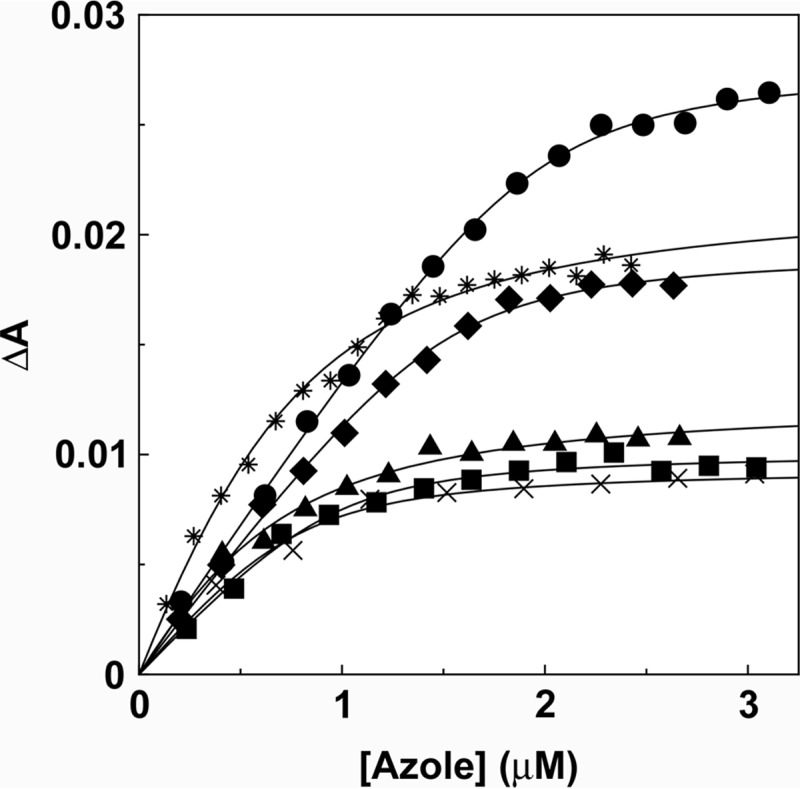
Azole binding saturation curves for Trub51. Saturation curves were constructed from the absorbance difference (ΔApeak-trough) of the type II difference spectra (Fig. 5) for clotrimazole (circles), fluconazole (squares), voriconazole (triangles), itraconazole (diamonds), ketoconazole (asterisks), and VT-1161 (crosses). A rearrangement of the Morrison equation (25) was used to fit the tight ligand binding observed. Each experiment was performed in triplicate, although only one replicate is shown.
CYP51 inhibitor IC50 determinations.
Determinations of 50% inhibitory concentrations (IC50s) (Fig. 7) confirmed that fluconazole, itraconazole, ketoconazole, and VT-1161 all inhibited T. rubrum CYP51 activity in vitro. VT-1161 caused the strongest inhibition (IC50, 0.14 μM), followed by itraconazole (IC50, 0.26 μM) and then fluconazole and ketoconazole (IC50s, 0.4 and 0.6 μM, respectively). Given that the concentration of CYP51 used in this assay was 0.5 μM, the expected IC50 for an extremely tight-binding azole antifungal would be 0.25 μM. Therefore, both VT-1161 and itraconazole bound extremely tightly to Trub51, while fluconazole and ketoconazole bound less tightly.
FIG 7.
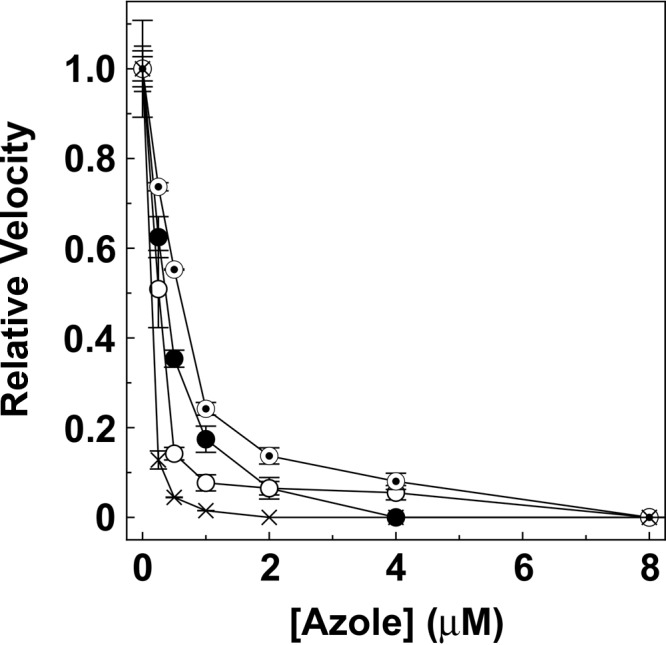
IC50 determinations for antifungal agents. CYP51 reconstitution assays were performed using a CYP51/AfCPR ratio of 1:2 for 0.5 μM Trub51 with 25 μM eburicol as the substrate at various fluconazole (filled circles), itraconazole (hollow circles), ketoconazole (bullets), and VT-1161 (crosses) concentrations ranging from 0 and 8 μM. Mean relative velocity values are shown along with the associated standard deviation values. Relative velocities of 1.00 were equivalent to velocities of 1.55 min−1.
CYP51 inhibitor MIC determinations.
MIC determinations (Table 1) confirmed the potency of VT-1161, as the MICs ranged from less than or equal to the lowest concentration tested (0.03 μg ml−1) to the highest MIC values of 0.06 μg ml−1. VT-1161's MIC50, MIC90, and geometric mean (GM) values of ≤0.03, 0.06, and 0.033 μg ml−1, respectively, were slightly less than those for itraconazole (0.06, 0.06, and 0.052 μg ml−1, respectively), and both of these CYP51 inhibitors were significantly more potent than fluconazole (MIC50, MIC90, and geometric mean values, 2, 16, and 2.3 μg ml−1, respectively). The GM MICs of both VT-1161 and itraconazole were significantly lower than the GM MIC of fluconazole (P = 0.0018 for both comparisons) but were not significantly different from each other.
TABLE 1.
MICs of VT-1161, itraconazole, and fluconazole against 34 clinical isolates of T. rubrum
| Drug | MIC (μg ml−1) |
|||
|---|---|---|---|---|
| 50% | 90% | Geometric mean | Range | |
| VT-1161 | ≤0.03 | 0.06 | 0.033 | ≤0.03–0.06 |
| Itraconazole | 0.06 | 0.06 | 0.052 | ≤0.03–0.12 |
| Fluconazole | 2 | 16 | 2.3 | 0.5–>64 |
Phylogenetic comparison of fungal CYP51 enzymes.
The primary amino acid sequence of T. rubrum CYP51 contained all 23 conserved CYP51 residues previously identified by Lepesheva and Waterman (27), in addition to the conserved heme-binding cysteine residue (see Fig. S1 in the supplemental material). The degree of conservation between the six substrate recognition sites (SRSs) (28) varied (Fig. S1), with SRS-1 being the most conserved and SRS-6 being the least conserved. Both the T. rubrum and M. graminicola CYP51 enzymes can turn over eburicol but not lanosterol (23; this study). Aspergillus fumigatus CYP51 isoenzymes A and B turn over both eburicol and lanosterol, albeit with a 4- to 7-fold preference for eburicol in terms of measured velocity using purified proteins (29) or more than an 18-fold preference for eburicol using membrane fractions. Candida albicans, Cryptococcus neoformans, and Malassezia globosa CYP51 enzymes, on the other hand, all readily turn over both eburicol and lanosterol (29–31). Analysis of the amino acid sequences of the six SRSs between the seven fungal CYP51 enzymes did not identify any residue changes that could be directly linked to the change in substrate specificity observed in the T. rubrum and M. graminicola CYP51 enzymes.
DISCUSSION
In preparation for studying antifungal inhibitors of CYP51, we have fully characterized CYP51 from the most prevalent fungus causing human dermatophytosis, Trichophyton rubrum. The T. rubrum CYP51 (Trub51) Kd values for sterol substrates of 20 to 23 μM were comparable to those of CYP51 enzymes from Candida albicans (11 to 28 μM) (32), Mycosphaerella graminicola (11 to 13 μM) (33), Aspergillus fumigatus CYP51B (9 to 23 μM) (29), Cryptococcus neoformans (12 to 21 μM) (30), and Malassezia globosa (23 to 32 μM) (31). However, the Kd values of Trypanosoma cruzi CYP51 for lanosterol and eburicol were lower at 1.9 and 1.2 μM, respectively (34), and the Kd values of Homo sapiens and Mycobacterium tuberculosis CYP51s for lanosterol were lower than those of Trub51 at 0.5 to 6 μM (28, 35) and 1 μM (21), respectively. However, Trub51 catalyzed the 14α-demethylation only of eburicol and not that of lanosterol and obtusifoliol, and its catalysis mirrors that previously observed for Mycosphaerella graminicola CYP51 (23). This narrow substrate specificity is in contrast to the broad substrate specificity observed previously for CYP51 enzymes from Candida albicans, Mycobacterium tuberculosis, Homo sapiens, Trypanosoma cruzi, Cryptococcus neoformans, and Malassezia globosa, (30, 31, 36). Additional CYP51 enzymes that exhibit narrow substrate specificities include obtusifoliol-specific Trypanosoma brucei CYP51 and plant CYP51 enzymes, such as Sorghum bicolor CYP51 (36), while the Aspergillus fumigatus CYP51A and CYP51B isoenzymes have a strong preference for eburicol (29). The Trub51 Km for eburicol of 2 μM was comparable to the substrate Km values previously obtained for CYP51 enzymes from C. albicans and Saccharomyces cerevisiae (32, 37, 38) but was 5- to 30-fold lower than those determined for CYP51 enzymes from Leishmania infantum, Homo sapiens, Mycosphaerella graminicola, and Malassezia globosa (23, 31, 36, 39). The strict eburicol substrate specificity of Trub51 could not be directly attributable to changes in the primary amino acid sequence of the six substrate recognition sites (28) relative to the primary amino acid sequences of fungal CYP51 enzymes that readily demethylate both eburicol and lanosterol (see Fig. S1 in the supplemental material).
It has been long recognized that fungal CYP51 inhibitors derive much of their binding potency through an azole/heme iron interaction (40) and that this binding can be directly measured spectroscopically (41). Therefore, as expected, Trub51 bound imidazole-based ketoconazole and clotrimazole; triazole-based fluconazole, voriconazole, and itraconazole; and the novel tetrazole-based VT-1161. Each compound displayed a type II binding spectrum caused by the interaction of a heterocyclic ring nitrogen coordinating as the sixth ligand with the heme iron (24) to form the low-spin CYP51-azole complex, resulting in a red shift of the heme Soret peak. Whereas the specific nitrogen is known for the triazole inhibitors (N-4) (42) and imidazole inhibitors (N-3) (28), the interaction of VT-1161 with the heme ferric ion is through either the tetrazole's N-3 or N-4 nitrogen. The N-4 nitrogen was found to be more nucleophilic in heats of formation experiments (data not shown) and would therefore be the atom most likely to interact with the CYP51 heme iron.
The antifungal agents tested in this study bound Trub51 somewhat less tightly than other fungal CYP51 enzymes, with the possible exception of itraconazole. The relative differences observed in the Kd values, however, did not translate into equally large differences in IC50s, with only a 4-fold increase in the IC50 between VT-1161 and ketoconazole being observed and with the IC50 for VT-1161 being numerically but not significantly lower than that for itraconazole. Therefore, for Trub51, the CYP51 reconstitution assay proved to be better at assessing CYP51-inhibitory potency than direct ligand binding to aqueous purified enzyme, and its results were in agreement with the intrinsic antifungal potency measured in broth microdilution assays, which ranked VT-1161 as the most potent T. rubrum inhibitor, closely followed by itraconazole, with fluconazole being the least potent (Table 1).
The performance of the drug candidate VT-1161 against T. rubrum CYP51 and T. rubrum itself was encouraging. We have shown biochemically that VT-1161 bound to the heme iron in the active site of Trub51 and strongly inhibited Trub51 activity through tight ligand binding. The MICs for the cellular potency of VT-1161 against T. rubrum ranged from ≤0.03 to 0.06 μg/ml, making it slightly more potent than itraconazole (MICs, ≤0.03 to 0.12 μg/ml) and significantly more potent than fluconazole (MICs, 0.5 to >64 μg/ml). This MIC potency range for VT-1161 compares favorably to published MIC values for T. rubrum of 0.03 to 256 μg ml−1 for fluconazole, 0.008 to 0.25 μg ml−1 for itraconazole, 0.06 to 2 μg ml−1 for ketoconazole, and 0.06 to 1 μg ml−1 for voriconazole (43–46). In addition, VT-1161 was as effective as itraconazole in treating Trichophyton mentagrophytes-induced dermatophytosis in guinea pig when treatments were orally administered daily and was more effective than itraconazole when administered weekly (47).
Equally important, the use of the tetrazole has allowed for the engineering of an inhibitor more selective for fungal CYP51 enzymes than key human CYP enzymes (range of IC50s against CYPs 3A4, 2C9, 2C19, and 51, 65 to ∼600 μM) (19). This greater selectivity coupled with at least maintained if not improved antifungal potency should translate into a greater clinical therapeutic window, which in turn could allow for higher doses and, possibly, greater efficacy. To this end, VT-1161 has achieved proof-of-concept efficacy (48) in a phase 2a study for the treatment of tinea pedis (ClinicalTrials.gov registration no. NCT01891305), and a phase 2b study of VT-1161 for the treatment of onychomycosis has just been completed (ClinicalTrials.gov registration no. NCT02267356), with interim data demonstrating antifungal and clinical efficacy in conjunction with an excellent safety profile (49). Phase 3 studies are currently being planned to support the registration approval of VT-1161 as a novel agent to treat onychomycosis.
MATERIALS AND METHODS
Construction of the pCWori+:Trub51 expression vector.
The T. rubrum CYP51 gene (Trub51; UniProt accession number F2SHH3) was synthesized by Eurofins MWG Operon (Ebersberg, Germany), incorporating an NdeI restriction site at the 5′ end and a HindIII restriction site at the 3′ end of the gene, which was cloned into plasmid pBSIISK+. In addition, the first 8 amino acids were changed to MALLLAVF (50), and a 4-histidine extension (CATCACCATCAC) was inserted immediately before the stop codon. The Trub51 gene was excised by NdeI/HindIII restriction digestion, followed by cloning into the pCWori+ expression vector. Gene integrity was confirmed by DNA sequencing.
Heterologous expression and purification of recombinant Trub51 protein.
The pCWori+:Trub51 construct was transformed into competent E. coli DH5α cells and expressed as previously described for Candida albicans CYP51 (32). The recombinant Trub51 protein was isolated according to the method of Arase et al. (51), except that 2% (wt/vol) sodium cholate was used as the sole detergent in the sonication buffer with the addition of 0.1 mM phenylmethylsulfonyl fluoride. The solubilized Trub51 protein was purified by affinity chromatography using Ni2+-NTA agarose as previously described (21, 32), followed by dialysis against 20 mM Tris-HCl (pH 8.1) and 10% (wt/vol) glycerol. Protein purity was assessed by SDS-polyacrylamide gel electrophoresis.
Cytochrome P450 protein determinations.
Reduced carbon monoxide difference spectroscopy was performed (20), with carbon monoxide being passed through the cytochrome P450 solution prior to addition of sodium dithionite to the sample cuvette (light path, 10 mm). An extinction coefficient of 91 mM−1 cm−1 (52) was used to calculate cytochrome P450 concentrations from the absorbance difference between 447 and 490 nm. Absolute spectra were determined between 700 and 300 nm (light path, 4.5 mm). All spectral determinations were made using an Hitachi U-3310 UV/visible spectrophotometer (San Jose, CA).
Sterol binding properties of Trub51.
Stock 2-mg ml−1 solutions of lanosterol, obtusifoliol, and eburicol were prepared in 40% (wt/vol) (2-hydroxypropyl)-β-cyclodextrin (HPCD). Sterol was progressively titrated against 5 μM Trub51 in a quartz semimicrocuvette (light path, 4.5 mm), with equivalent amounts of 40% (wt/vol) HPCD being added to the reference cuvette, which also contained 5 μM Trub51. The absorbance difference spectrum between 500 and 350 nm was determined after each incremental addition of sterol (up to 75 μM). The sterol saturation curves were constructed from the change in the absorbance at 388 nm and that at 421 nm (ΔA388–421) derived from the difference spectra. The substrate dissociation constants (Kd values) were determined by nonlinear regression (Levenberg-Marquardt algorithm) using the Michaelis-Menten equation.
Azole binding properties of Trub51.
The binding of clotrimazole, fluconazole, voriconazole, itraconazole, ketoconazole, and the drug candidate VT-1161 to Trub51 was performed as previously described (32, 53) using 4.5-mm-light-path quartz split cuvettes. Stock 0.05-, 0.1-, and 0.2-mg ml−1 solutions of the azoles were prepared in dimethyl sulfoxide (DMSO) and progressively titrated against 2 μM Trub51 in 0.1 M Tris-HCl (pH 8.1) and 25% (wt/vol) glycerol. The difference spectra between 500 and 350 nm were determined after each incremental addition of azole, and binding saturation curves were constructed from the change in the absorbance at the peak and that at the trough (ΔApeak-trough) against the azole concentration. The dissociation constants (Kd values) of the enzyme-azole complex were determined by nonlinear regression (Levenberg-Marquardt algorithm) using a rearrangement of the Morrison equation for tight ligand binding (25, 26). Tight binding is normally observed where the Kd for a ligand is similar to or less than the concentration of the enzyme present (54).
CYP51 reconstitution assays.
The reconstitution assay mixtures (34, 55) contained 0.5 μM Trub51, 1 μM Aspergillus fumigatus cytochrome P450 reductase isoenzyme 1 (AfCPR1; UniProt accession number Q4WM67), 50 μM sterol substrate, 50 μM dilauryl phosphatidylcholine, 4% (wt/vol) HPCD, 0.4 mg ml−1 isocitrate dehydrogenase, 25 mM trisodium isocitrate, 50 mM NaCl, 5 mM MgCl2, and 40 mM MOPS (morpholinepropanesulfonic acid) (pH, ∼7.2). Assay mixtures were incubated at 37°C prior to initiation with 4 mM β-NADPH Na4, followed by shaking at 37°C for 15 min. Sterol metabolites were recovered by extraction with ethyl acetate, followed by derivatization with 0.1 ml N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA)–trimethylchlorosilane (TMCS) (99:1) and 0.3 ml anhydrous pyridine (2 h at 80°C) prior to analysis by gas chromatography-mass spectrometry (GC/MS) (56). The Trub51 Km value for eburicol was determined by varying the eburicol concentration in the CYP51 reconstitution assay mixture between 3 and 100 μM while maintaining a constant HPCD concentration of 4% (wt/vol). The single substrate inhibition equation v = (Vmax · [S])/{Km + [S] · (1 + [S]/Ki)} (57), where v is velocity and [S] is the substrate concentration, was used to fit the data and to determine Km and Ki values.
Azole IC50 determinations.
IC50 determinations were performed using the CYP51 reconstitution assay detailed above, in which various fluconazole, itraconazole, ketoconazole, and VT-1161 concentrations in 2.5 μl dimethyl sulfoxide were added to the assay mixtures prior to incubation at 37°C and addition of β-NADPH Na4. The IC50 assay mixtures contained 25 μM eburicol, 0.5 μM Trub51, 1 μM AfCPR1, and 4 mM β-NADPH Na4.
MIC determinations.
Drug preparations were prepared according to the recommendation outlined in Clinical and Laboratory Standards Institute (CLSI) document M38-A2; this includes testing in RPMI 1640 with l-glutamine, with 0.165 M MOPS as the buffer (pH 7.0), and without bicarbonate; an inoculum size of 1 × 104 to 5 × 104; and incubation at 35°C for 96 h. The MICs were measured visually and were the lowest concentration of each antifungal agent that resulted in an 80% reduction in turbidity compared to that for drug-free, growth control wells. Stock solutions of each agent were prepared in DMSO. Further dilutions were made in RPMI 1640, and the final concentration of DMSO was 1% (vol/vol). The final testing concentrations for VT-1161 and itraconazole ranged from 0.03 to 16 μg ml−1, and those for fluconazole ranged from 0.125 to 64 μg ml−1. Trichophyton mentagrophytes (ATCC MYA-4439) served as the quality control organism, as recommended by CLSI document M38-A2, and was used on each day of testing. Results for this control isolate were within the appropriate range for each agent test. Thirty-four clinical Trichophyton rubrum isolates that were submitted to the Fungus Testing Laboratory (University of Texas Health Science Center at San Antonio, San Antonio, TX) for antifungal susceptibility testing and/or identification were used in this study. All strains were fresh clinical strains that had not been previously frozen. All MICs were measured once.
Phylogenetic analysis of fungal CYP51 proteins.
Selected fungal CYP51 amino acid sequences were obtained from the UniProtKB database (http://www.uniprot.org) and were aligned using ClustalX software (version 2.0.12; http://www.clustal.org/clustal2/). The fungal sequences compared were those of Aspergillus fumigatus CYP51 isoenzyme A (UniProt accession number Q4WNT5), Aspergillus fumigatus CYP51 isoenzyme B (UniProt accession number Q96W81), Candida albicans CYP51 (UniProt accession number P10613), Cryptococcus neoformans CYP51 (UniProt accession number Q5KQ65), Malassezia globosa CYP51 (UniProt accession number A8Q3I7), Mycosphaerella graminicola CYP51 (UniProt accession number Q5XWE5), and Trichophyton rubrum CYP51 (UniProt accession number F2SHH3).
Data analysis.
All ligand binding experiments were performed in triplicate, and curve fitting of the data was performed using the computer program QuantumSoft ProFit (version 6.1.12). Differences in geometric mean (GM) MIC values, calculated following log2 transformation of individual MIC values, between VT-1161, itraconazole, and fluconazole were assessed for significance by analysis of variance with Tukey's posttest for multiple comparisons. A P value of <0.05 was considered statistically significant. For MIC values that were greater than the highest concentration tested, the next higher dilution value was used in the GM MIC calculations (e.g., for a fluconazole MIC of >64 μg ml−1, an MIC of 128 μg ml−1 was used). For MICs that were equal to or lower than the lowest concentration tested, the lowest concentration tested was used (e.g., for a VT-1161 or itraconazole MIC of ≤0.03 μg ml−1, an MIC of 0.03 μg ml−1 was used).
Chemicals.
All chemicals, including clotrimazole, fluconazole, itraconazole, ketoconazole, and voriconazole, were obtained from Sigma Chemical Company (Poole, UK). Growth medium, sodium ampicillin, IPTG (isopropyl-β-d-thiogalactopyranoside), and 5-aminolevulenic acid were obtained from Foremedium Ltd. (Hunstanton, UK). The Ni2+-NTA agarose affinity chromatography matrix was obtained from Qiagen (Crawley, UK). VT-1161 was supplied by Viamet Pharmaceuticals Inc. (Durham, NC, USA).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Engineering and Physical Sciences Research Council National Mass Spectrometry Service Centre at Swansea University and Marcus Hull for assistance in GC/MS analyses and to Annette Fothergill at the University of Texas Health Science Center at San Antonio for assistance with in vitro susceptibility testing.
This work was in part supported by the European Regional Development Fund/Welsh government-funded BEACON research program (Swansea University); National Science Foundation of the United States grant NSF-MCB-09020212, awarded to W. David Nes (Texas Tech University); and Viamet Pharmaceuticals Inc. (Durham, NC, USA).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00333-17.
REFERENCES
- 1.Weitzman I, Summerbell RC. 1995. The dermatophytes. Clin Microbiol Rev 8:240–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summerbell RC, Kane J, Krajden S. 1989. Onychomycosis, tinea pedis and tinea manuum caused by non-dermatophytic filamentous fungi. Mycoses 32:609–619. [DOI] [PubMed] [Google Scholar]
- 3.Tietz HJ, Kunzelmann V, Schonian G. 1995. Changes in the fungal spectrum of dermatomycoses. Mycoses 38(Suppl 1):S33–S39. [DOI] [PubMed] [Google Scholar]
- 4.Santos DA, Hamdan JS. 2006. In vitro antifungal oral drug and drug-combination activity against onychomycosis causative dermatophytes. Med Mycol 44:357–362. [DOI] [PubMed] [Google Scholar]
- 5.Jackson CJ, Barton RC, Kelly SL, Evans EG. 2000. Strain identification of Trichophyton rubrum by specific amplification of subrepeat elements in the ribosomal DNA nontranscribed spacer. J Clin Microbiol 38:4527–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seebacher C, Bouchara J-P, Mignon B. 2008. Updates on the epidemiology of dermatophyte infections. Mycopathologia 166:335–352. doi: 10.1007/s11046-008-9100-9. [DOI] [PubMed] [Google Scholar]
- 7.Adams C, Athanasoula E, Lee W, Mahmudova N, Vlahovic TC. 2015. Environmental and genetic factors on the development of onychomycosis. J Fungi 1:211–216. doi: 10.3390/jof1020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oke OO, Onayemi O, Olasode OA, Omisore AG, Oninla OA. 2014. The prevalence and pattern of superficial fungal infections among school children in Ile-Ife, south-western Nigeria. Dermatol Res Pract 2014:842917. doi: 10.1155/2014/842917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouzaud C, Hay R, Chosidow O, Dupin N, Puel A, Lortholary O, Lanternier F. 2016. Severe dermatophytosis and acquired or innate immunodeficiency: a review. J Fungi 2:4. doi: 10.3390/jof2010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mays SR, Bogle MA, Bodey GP. 2006. Cutaneous fungal infections in the oncology patient. Am J Clin Dermatol 7:31–43. doi: 10.2165/00128071-200607010-00004. [DOI] [PubMed] [Google Scholar]
- 11.Virgili A, Zampino MA, Mantovani L. 2002. Fungal skin infections in organ transplant recipients. Am J Clin Dermatol 3:19–35. doi: 10.2165/00128071-200203010-00003. [DOI] [PubMed] [Google Scholar]
- 12.Ilkit M, Durdu M. 2015. Tinea pedis: the etiology and global epidemiology of a common fungal infection. Crit Rev Microbiol 41:374–388. doi: 10.3109/1040841X.2013.856853. [DOI] [PubMed] [Google Scholar]
- 13.Smijs TG, Pavel S. 2011. The susceptibility of dermatophytes to photodynamic treatment with special focus on Trichophyton rubrum. Photochem Photobiol 87:2–13. doi: 10.1111/j.1751-1097.2010.00848.x. [DOI] [PubMed] [Google Scholar]
- 14.Balkis MM, Leidich SD, Mukherjee PK, Ghannoum MA. 2002. Mechanisms of fungal resistance: an overview. Drugs 62:1025–1040. doi: 10.2165/00003495-200262070-00004. [DOI] [PubMed] [Google Scholar]
- 15.Ishida K, de Mello JC, Cortez DA, Filho BP, Ueda-Nakamura T, Nakamura CV. 2006. Influence of tannins from Stryphnodendron adstringens on growth and virulence factors of Candida albicans. J Antimicrob Chemother 58:942–949. doi: 10.1093/jac/dkl377. [DOI] [PubMed] [Google Scholar]
- 16.Santos DA, Hamdan JS. 2007. In vitro activities of four antifungal drugs against Trichophyton rubrum isolates exhibiting resistance to fluconazole. Mycoses 50:286–289. doi: 10.1111/j.1439-0507.2007.01325.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, Lim YR, Ohk SO, Kim BJ, Chun YJ. 2011. Functional expression and characterization of CYP51 from dandruff-causing Malassezia globosa. FEMS Yeast Res 11:80–87. doi: 10.1111/j.1567-1364.2010.00692.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoekstra WJ, Garvey EP, Moore WR, Rafferty SW, Yates CM, Schotzinger RJ. 2014. Design and optimization of highly-selective fungal CYP51 inhibitors. Bioorg Med Chem Lett 24:3455–3458. doi: 10.1016/j.bmcl.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 19.Warrilow AG, Hull CM, Parker JE, Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Kelly DE, Kelly SL. 2014. The clinical candidate VT-1161 is a highly potent inhibitor of Candida albicans CYP51 but fails to bind the human enzyme. Antimicrob Agents Chemother 58:7121–7127. doi: 10.1128/AAC.03707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estabrook RW, Peterson JA, Baron J, Hildebrandt AG. 1972. The spectrophotometric measurement of turbid suspensions of cytochromes associated with drug metabolism, vol 2 Appleton-Century-Crofts, New York, NY. [Google Scholar]
- 21.Bellamine A, Mangla AT, Nes WD, Waterman MR. 1999. Characterization and catalytic properties of the sterol 14α-demethylase from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 96:8937–8942. doi: 10.1073/pnas.96.16.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jefcoate CR. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol 52:258–279. doi: 10.1016/S0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 23.Price CL, Warrilow AG, Parker JE, Mullins JG, Nes WD, Kelly DE, Kelly SL. 2015. Novel substrate specificity and temperature-sensitive activity of Mycosphaerella graminicola CYP51 supported by the native NADPH cytochrome P450 reductase. Appl Environ Microbiol 81:3379–3386. doi: 10.1128/AEM.03965-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefcoate CR, Gaylor JL, Calabrese RL. 1969. Ligand interactions with cytochrome P-450. I. Binding of primary amines. Biochemistry 8:3455–3463. [DOI] [PubMed] [Google Scholar]
- 25.Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. 2009. Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem Pharmacol 77:258–268. doi: 10.1016/j.bcp.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison JF. 1969. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim Biophys Acta 185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 27.Lepesheva GI, Waterman MR. 2011. Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814:88–93. doi: 10.1016/j.bbapap.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strushkevich N, Usanov SA, Park HW. 2010. Structural basis of human CYP51 inhibition by antifungal azoles. J Mol Biol 397:1067–1078. doi: 10.1016/j.jmb.2010.01.075. [DOI] [PubMed] [Google Scholar]
- 29.Warrilow AG, Parker JE, Price CL, Nes WD, Kelly SL, Kelly DE. 2015. In vitro biochemical study of CYP51-mediated azole resistance in Aspergillus fumigatus. Antimicrob Agents Chemother 59:7771–7778. doi: 10.1128/AAC.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warrilow AG, Parker JE, Price CL, Nes WD, Garvey EP, Hoekstra WJ, Schotzinger RJ, Kelly DE, Kelly SL. 2016. The investigational drug VT-1129 is a highly potent inhibitor of Cryptococcus species CYP51 but only weakly inhibits the human enzyme. Antimicrob Agents Chemother 60:4530–4538. doi: 10.1128/AAC.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warrilow AG, Price CL, Parker JE, Rolley NJ, Smyrniotis CJ, Hughes DD, Thoss V, Nes WD, Kelly DE, Holman TR, Kelly SL. 2016. Azole antifungal sensitivity of sterol 14α-demethylase (CYP51) and CYP5218 from Malassezia globosa. Sci Rep 6:27690. doi: 10.1038/srep27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warrilow AG, Martel CM, Parker JE, Melo N, Lamb DC, Nes WD, Kelly DE, Kelly SL. 2010. Azole binding properties of Candida albicans sterol 14α-demethylase (CaCYP51). Antimicrob Agents Chemother 54:4235–4245. doi: 10.1128/AAC.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker JE, Warrilow AG, Cools HJ, Martel CM, Nes WD, Fraaije BA, Lucas JA, Kelly DE, Kelly SL. 2011. Mechanism of binding of prothioconazole to Mycosphaerella graminicola CYP51 differs from that of other azole antifungals. Appl Environ Microbiol 77:1460–1465. doi: 10.1128/AEM.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, Liu J, Hill GC, Waterman MR. 2006. CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B′ helix defines substrate preferences of sterol 14α-demethylase. J Biol Chem 281:3577–3585. doi: 10.1074/jbc.M510317200. [DOI] [PubMed] [Google Scholar]
- 35.Lepesheva GI, Nes WD, Zhou W, Hill GC, Waterman MR. 2004. CYP51 from Trypanosoma brucei is obtusifoliol-specific. Biochemistry 43:10789–10799. doi: 10.1021/bi048967t. [DOI] [PubMed] [Google Scholar]
- 36.Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, Lepesheva GI. 2011. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (CYP51) from Leishmania infantum. J Biol Chem 286:26838–26848. doi: 10.1074/jbc.M111.237099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trösken ER, Adamska M, Arand M, Zarn JA, Patten C, Völkel W, Lutz WK. 2006. Comparison of lanosterol-14α-demethylase (CYP51) of human and Candida albicans for inhibition by different antifungal. Toxicology 228:24–32. doi: 10.1016/j.tox.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Kitahama Y, Nakamura M, Yoshida Y, Aoyama Y. 2009. The construction and characterization of self-sufficient lanosterol 14-demethylase fusion proteins consisting of yeast CYP51 and its reductase. Biol Pharm Bull 32:558–563. doi: 10.1248/bpb.32.558. [DOI] [PubMed] [Google Scholar]
- 39.Ekins S, Mankowski DC, Hoover DJ, Lawton MP, Treadway JL, Harwood HJ Jr. 2007. Three-dimensional quantitative structure-activity relationship analysis of human CYP51 inhibitors. Drug Metab Dispos 35:493–500. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida Y. 1988. Cytochrome P450 of fungi: primary target for azole antifungal agents. Curr Top Med Mycol 2:388–418. doi: 10.1007/978-1-4612-3730-3_11. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida Y, Aoyama Y. 1987. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol 36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]
- 42.Chen CK, Leung SS, Guilbert C, Jacobson MP, McKerrow JH, Podust LM. 2010. Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and fluconazole. PLoS Negl Trop Dis 4:e651. doi: 10.1371/journal.pntd.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barros MES, Santos DA, Hamdan JS. 2006. In vitro methods for antifungal susceptibility testing of Trichophyton spp. Mycol Res 110:1355–1360. doi: 10.1016/j.mycres.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Barros MES, Santos DA, Hamdan JS. 2007. Antifungal susceptibility testing of Trichophyton rubrum by E-test. Arch Dermatol Res 299:107–109. doi: 10.1007/s00403-006-0731-8. [DOI] [PubMed] [Google Scholar]
- 45.Sarifakioglu E, Seckin D, Demirbilek M, Can F. 2007. In vitro antifungal susceptibility patterns of dermatophyte strains causing tinea unguium. Clin Exp Dermatol 32:675–679. doi: 10.1111/j.1365-2230.2007.02480.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Yang W, Wang K, Zhu J, Shen F, Hu Y. 2012. Synthesis and biological evaluation of vinyl ether-containing azole derivatives as inhibitors of Trichophyton rubrum. Bioorg Med Chem Lett 22:4887–4890. doi: 10.1016/j.bmcl.2012.05.070. [DOI] [PubMed] [Google Scholar]
- 47.Garvey EP, Hoekstra WJ, Moore WR, Schotzinger RJ, Long L, Ghannoum MA. 2015. VT-1161 dosed once daily or once weekly exhibits potent efficacy in treatment of dermatophytosis in a guinea pig model. Antimicrob Agents Chemother 59:1992–1997. doi: 10.1128/AAC.04902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Degenhardt T, Pollak R, Jones T, Jarratt M, Brandt S, Schotzinger R. 2015. Efficacy and safety of oral VT-1161, a novel and selective inhibitor of fungal CYP51, in a randomized, double-blind, placebo-controlled phase 2 study (NCT01891305) in patients with moderate-to-severe interdigital tinea pedis. Abstr Am Podiatr Med Assoc 2015 Annu Meet, Orlando, FL. [Google Scholar]
- 49.Tavakkol A, Pollak R, Reyzelman A, Weisfeld M, Curelop A, Handelsman C, Brand S, Degenhardt T, Schotzinger R. 2016. A randomized, double-blind, placebo-controlled clinical trial of four oral dosing regimens of VT-1161 in the treatment of patients with moderate-severe toenail onychomycosis (RENOVATE): results of a planned week 24 interim analysis, poster 142. Abstr Am Podiatr Med Assoc 2016 Annu Meet, Philadelphia, PA. [Google Scholar]
- 50.Barnes HJ, Arlotto MP, Waterman MR. 1991. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A 88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arase M, Waterman MR, Kagawa N. 2006. Purification and characterization of bovine steroid 21-hydroxylase (P450c21) efficiently expressed in Escherichia coli. Biochem Biophys Res Commun 344:400–405. doi: 10.1016/j.bbrc.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 52.Omura T, Sato R. 1964. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385. [PubMed] [Google Scholar]
- 53.Lamb DC, Kelly DE, Waterman MR, Stromstedt M, Rozman D, Kelly SL. 1999. Characteristics of the heterologously expressed human lanosterol 14α-demethylase (other names: P45014DM, CYP51, P45051) and inhibition of the purified human and Candida albicans CYP51 with azole antifungal agents. Yeast 15:755–763. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Copeland RA. 2005. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists, p 178–213. Wiley-Interscience, New York, NY. [PubMed] [Google Scholar]
- 55.Lepesheva GI, Ott RD, Hargrove TY, Kleshchenko YY, Schuster I, Nes WD, Hill GC, Villalta F, Waterman MR. 2007. Sterol 14α-demethylase as a potential target for antitrypanosomal therapy: enzyme inhibition and parasite cell growth. Chem Biol 14:1283–1293. doi: 10.1016/j.chembiol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker JE, Warrilow AG, Cools HJ, Fraaije BA, Lucas JA, Rigdova K, Griffiths WJ, Kelly DE, Kelly SL. 2013. Prothioconazole and prothioconazole-desthio activities against Candida albicans sterol 14α-demethylase. Appl Environ Microbiol 79:1639–1645. doi: 10.1128/AEM.03246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Copeland RA. 2000. Enzymes: a practical introduction to structure, mechanism and data analysis, 2nd ed, p 137 Wiley-VCH Inc, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.