ABSTRACT
The emergence of pan-resistant Klebsiella pneumoniae strains is an increasing concern. In the present study, we describe a cluster of 9 pan-resistant K. pneumoniae sequence type 147 (ST147) isolates encountered in 4 patients over nearly 1 year in 3 hospitals of the United Arab Emirates (UAE). The isolates exhibited highly similar genotypes. All produced chromosomally encoded OXA-181, and the majority also produced the NDM-5 carbapenemase. As with the previously described single isolate from the UAE, MS6671, the mgrB was disrupted by a functional, ISEcp1-driven blaOXA-181 insertion causing resistance to carbapenems. The mutation was successfully complemented with an intact mgrB gene, indicating that it was responsible for colistin resistance. blaNDM-5 was located within a resistance island of an approximately 100-kb IncFII plasmid carrying ermB, mph(A), blaTEM-1B, rmtB, blaNDM-5, sul1, aadA2, and dfrA12 resistance genes. Sequencing this plasmid (pABC143-NDM) revealed that its backbone was nearly identical to that of plasmid pMS6671E from which several resistance genes, including blaNDM-5, had been deleted. More extensive similarities of the backbone and the resistance island were found between pABC143C-NDM and the blaNDM-5-carrying IncFII plasmids of two K. pneumoniae ST147 isolates from South Korea, one of which was colistin resistant, and both also produced OXA-181. Notably, one of these strains was isolated from a patient transferred from the UAE. Our data show that this pan-resistant clone has an alarming capacity to maintain itself over an extended period of time and is even likely to be transmitted internationally.
KEYWORDS: pan-resistance, Klebsiella pneumoniae, carbapenems, colistin
INTRODUCTION
Klebsiella pneumoniae has long been recognized to cause severe community or hospital-acquired infections (1, 2) and also to gain resistance relatively easily to multiple antibiotics (2, 3). Certain clones (e.g., clonal complex [CC] 258 or sequence type [ST] 147) are particularly important in disseminating multidrug resistance globally, including to carbapenems. Resistance to these later drugs is commonly due to the production of various carbapenemases, such as KPC, NDM, VIM, and various alleles of OXA-type enzymes (3–6).
Management of infections caused by carbapenem-resistant K. pneumoniae often includes polymyxins, typically colistin (7). Over the last decades, strains with decreased susceptibility to polymyxins have started to emerge (8). Colistin resistance is commonly due to the altered physicochemical features of the bacterial cell wall that limit the capacity of the cationic polymyxin molecules to bind to it. This can be the result of mutations in several chromosomal genes, of which the most common is mgrB (9). Recently, genes responsible for colistin resistance located on mobile genetic elements have also been described, further increasing the chance to spread resistance to polymyxins (10).
Although the lack of susceptibility to polymyxins in carbapenem-resistant strains does not mean resistance to all antibiotics, per se, it carries an increased risk of the emergence of pan-drug-resistant (PDR) clones. Indeed, several PDR K. pneumoniae strains, some even causing outbreaks, have already been described (11, 12).
In the present study, we report on a cluster of NDM-5- and OXA-181-producing, pan-resistant K. pneumoniae ST147 strains encountered in two emirates of the United Arab Emirates (UAE) over a period of 1 year, with possible connections to strains subsequently isolated in South Korea. Furthermore, we also provide evidence that the molecular basis of colistin resistance in the cluster was due to the insertion of a functional blaOXA-181 gene into mgrB.
(Part of this work was presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, 2016, Amsterdam, the Netherlands.)
RESULTS
Collection of strains.
In May 2013, four K. pneumoniae isolates (ABC143A to -D) were received for molecular typing. They were recovered from various samples from the same patient in an Abu Dhabi hospital (AD hospital) and were resistant to all antibiotics tested by the hospital laboratory. The patient, who had peritonitis from a leaking anastomosis after an operation for obstruction and perforation of the small intestine, had been transferred from an Umm Al Quwain hospital (UAQ hospital A). A week later another pan-resistant isolate (ABC146) was obtained from the same Abu Dhabi hospital, this one recovered from the sputum of a 75-year-old male patient treated during the same period of time, in the same ward. In late May, two more isolates were recovered from the first patient's blood and peritoneal aspirate (ABC143E and ABC143F).
In August, a PDR strain from UAQ hospital A, i.e., the same hospital from which the first patient had been transferred to Abu Dhabi, was sent to our laboratory. Strain ABC164 was isolated from a sample taken from the mastectomy wound of a 75-year-old, septic, ventilated female patient with advanced breast cancer. According to the hospital records, all three patients succumbed due to sepsis.
We compared these isolates to another PDR K. pneumoniae strain (MS6671) described earlier (12) from a different hospital at Um al Quwain, i.e., UAQ hospital B; this strain was isolated 7 months after the first isolate in UAQ hospital A had been encountered. The characteristics of the isolates are shown in Table 1.
TABLE 1.
Characteristics of the pan-drug-resistant K. pneumoniae isolates
| Isolate | Hospital of isolation | Specimen | Date of isolation | Virulence factor genes | Resistance genesa |
|---|---|---|---|---|---|
| ABC143A | Abu Dhabi hospital | Body fluid | 12 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| ABC143B | Abu Dhabi hospital | Sputum | 11 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| ABC143C | Abu Dhabi hospital | Blood | 12 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| ABC143D | Abu Dhabi hospital | Perianal swab | 11 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| ABC143E | Abu Dhabi hospital | Blood | 29 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, blaSHV-36, blaCTX-M-15, blaOXA-181, rmtF, aac6′-1b-cr, qacΔE, cepA |
| ABC143F | Abu Dhabi hospital | Body fluid | 29 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaTEM-1B, blaSHV-36, blaCTX-M-15, blaOXA-181, rmtF, aac6′-1b-cr, cepA |
| ABC146 | Abu Dhabi hospital | Sputum | 21 May 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| ABC164 | Um al Quwain hospital A | Pus | 18 August 2013 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaNDM-5, blaTEM-1B, rmtB, rmtF, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, qacΔE, cepA |
| MS6671b | Um al Quwain hospital B | Urine | 26 March 2014 | fimH, traT, ureA, uge, wabG, mrkD, fyuA | blaSHV-36, blaCTX-M-15, blaOXA-181, aac6′-1b-cr, cepA, rmtF |
As detected by PCR amplification and sequencing. The genes in bold were also detected in the pABC143C-NDM complete sequence by next-generation sequencing.
See reference 12.
Antibiotic susceptibility.
All strains exhibited nonsusceptibility to all antibiotics tested, including colistin, fosfomycin, and tigecycline. The MICs of benzalkonium and irgasan for the strains were similar to the MICs for the Escherichia coli K-12 laboratory strain; however, the chlorhexidine MICs for the clinical isolates were considerably higher (Table 2).
TABLE 2.
Antibiotic and disinfectant susceptibility of pan-drug-resistant K. pneumoniae isolates and the transconjugant
| Strain | MIC (mg/liter)a |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMI | MEM | ERT | CAZ | CTX | AZT | CIP | GM | AM | SXT | TIG | FOS | COL | BAC | IRG | CHX | |
| ABC143A | >128 | >128 | >64 | >128 | >128 | 128 | >64 | >256 | >256 | >256/4864 | 8 | 128 | 16 | 128 | 1 | 32 |
| ABC143B | >128 | >128 | >64 | >128 | >128 | 128 | 64 | >256 | >256 | >256/4864 | 2 | 128 | 8 | 64 | <0.5 | 32 |
| ABC143C | >128 | >128 | >64 | >128 | >128 | 128 | 64 | >256 | >256 | >256/4864 | 2 | 128 | 8 | 64 | <0.5 | 32 |
| ABC143D | >128 | >128 | >64 | >128 | >128 | 128 | >64 | >256 | >256 | >256/4864 | 4 | 128 | 8 | 64 | 1 | 32 |
| ABC143E | >128 | >128 | >64 | >128 | >128 | >128 | 64 | >256 | >256 | >256/4864 | 4 | 64 | >256 | 64 | <0.5 | 32 |
| ABC143F | 16 | 32 | >64 | 32 | >128 | 128 | >64 | >256 | >256 | 4/76 | 8 | 256 | 4 | 64 | <0.5 | 16 |
| ABC146 | 16 | 128 | >64 | >128 | >128 | >128 | 64 | >256 | >256 | >256/4864 | 2 | 64 | 8 | 64 | <0.5 | 32 |
| ABC164 | 128 | >128 | >64 | >128 | >128 | >128 | >64 | >256 | >256 | >256/4864 | 2 | 64 | 32 | 64 | <0.5 | 32 |
| MS6671 | 16 | >128 | >64 | 64 | >128 | >128 | >64 | >256 | >256 | 8/152 | 2 | 64 | 32 | 64 | 1 | 32 |
| J53RAZ(pABC143C/1) | 4 | 16 | 8 | >128 | 64 | <0.25 | <0.125 | 256 | 256 | <0.5/9.5 | <0.125 | 1 | <0.5 | 128 | <0.5 | 2 |
| J53RAZ | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.25 | <0.125 | <0.5 | <0.5 | <0.5/9.5 | <0.125 | 1 | <0.5 | 128 | <0.5 | 2 |
IMI, imipenem; MEM, meropenem; ETP, ertapenem; CAZ, ceftazidime; CTX, cefotaxime; AZT, aztreonam; CIP, ciprofloxacin; GM, gentamicin; AM, amikacin; SXT, trimethoprim-sulfamethoxazole; TIG, tigecycline; COL, colistin; FOS, fosfomycin; BAC, benzalkonium; IRG, irgasan; CHX, chlorhexidine.
Molecular characterization.
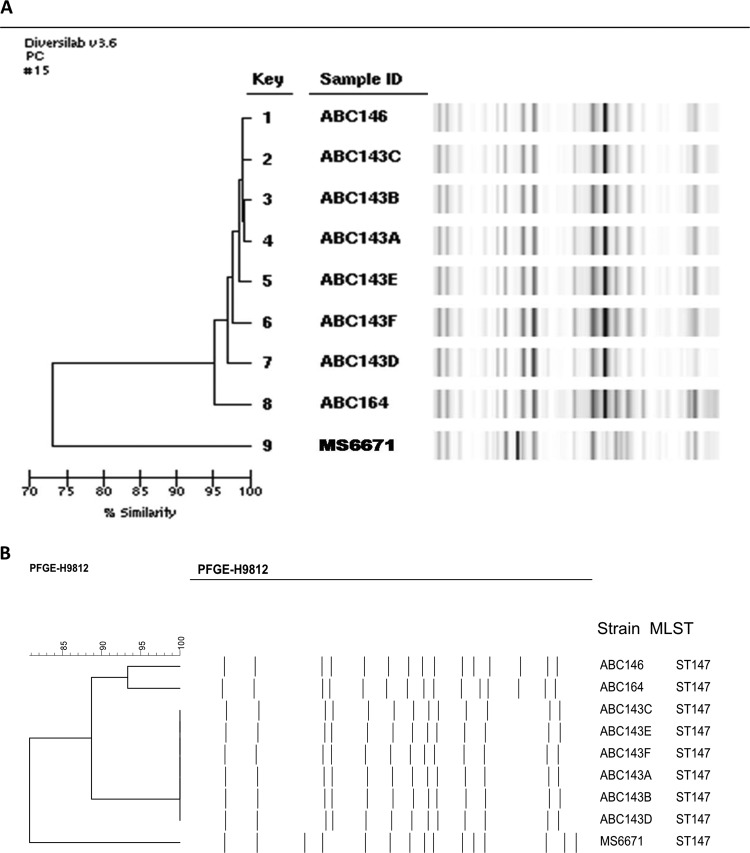
The isolates from the AD hospital and from UAQ hospital A clustered by both repetitive elements PCR (rep-PCR; DiversiLab, bioMérieux) and pulsed-field gel electrophoresis (PFGE). Although K. pneumoniae MS6671 exhibited a considerably different pattern by rep-PCR, its PFGE pattern was still >80% similar to the PFGE patterns of the K. pneumoniae isolated from the other 3 patients (Fig. 1). All isolates tested belonged to ST147. The genotypes of the strains were also highly similar (Table 1).
FIG 1.
Comparative typing of members of the cluster. (A) rep-PCR (DiversiLab); (B) PFGE.
All strains carried both blaOXA-181 and blaNDM-5 carbapenemase genes, with the exception of MS6671 and one isolate (ABC143F) from the first patient, which were positive for blaOXA-181 only. By hybridization, the blaNDM-5 gene could be located on an approximately 100-kb plasmid, but the OXA-181 probe did not hybridize with any of the plasmids (data not shown). On the other hand, hybridization of the XbaI-digested and PFGE-separated full genome of the strains with an OXA-181 probe revealed 3 bands of the same sizes as those seen in MS6671, reported to carry three chromosomal copies of blaOXA-181 (12). One isolate from the first patient (ABC143F) also exhibited an additional, fourth, smaller fragment hybridizing with the OXA probe (Fig. 2).
FIG 2.
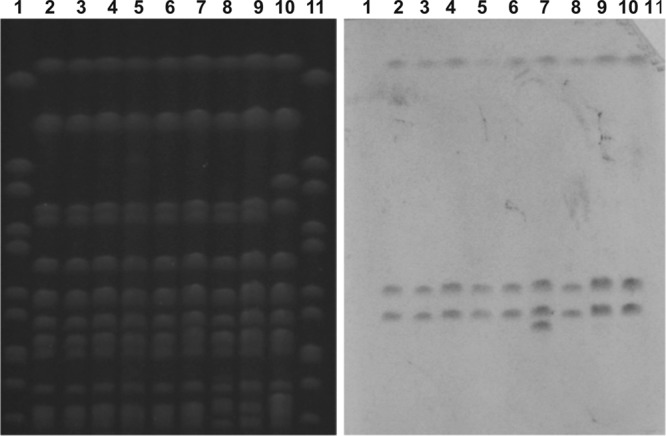
Hybridization of the genomic DNA fragments of strains with an OXA-181 probe. Samples: 1 and 11, Salmonella enterica serovar Braenderup strain H9812, molecular mass standard; 2, ABC143A; 3, ABC143B; 4, ABC143C; 5, ABC143D; 6, ABC143E; 7, ABC143F; 8, ABC146; 9, ABC164; 10, MS6671.
PCR of the mgrB gene of all isolates resulted in an approximately 3,000-bp amplicon, suggesting a large insertion into the gene. Sequencing of the amplicons from all nine isolates revealed an ISEcp1-associated blaOXA-181 gene inserted into the mgrB (Fig. 3). In order to prove the functionality of the inserted carbapenemase gene, the entire mgrB::blaOXA-181 amplicons from ABC143C and MS6671 were cloned into pUC19, resulting in plasmids pCoxABC143C and pCoxMS6671. These plasmids were transformed into E. coli DH5α. The transformants exhibited ertapenem and meropenem resistance while maintaining susceptibility to ceftazidime, cefotaxime, and aztreonam, respectively, suggestive of a functional OXA-181 carbapenemase (Table 3).
FIG 3.
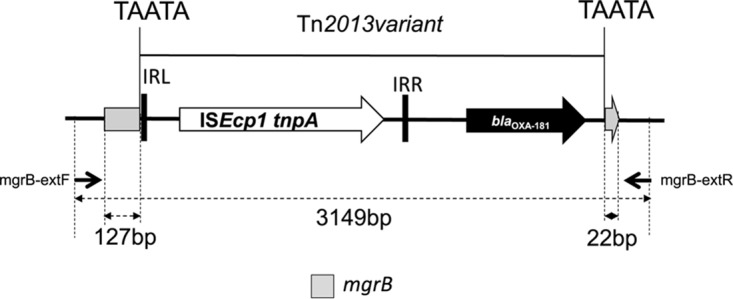
Structure of the mgrB::blaOXA-181 insert.
TABLE 3.
Carbapenemase activity of the mgrB::blaOXA-181 structure cloned from ABC143C and MS6671
| Strain | MIC (mg/liter)a |
|||||
|---|---|---|---|---|---|---|
| IMI | MEM | ERT | CAZ | CTX | AZT | |
| DH5α(pUC19) | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.25 |
| ABC143C | >128 | >128 | >64 | >128 | >128 | 128 |
| DH5α(pCoxABC143C#13) | 1 | 2 | 1 | <0.25 | <0.25 | <0.25 |
| MS6671 | 16 | >128 | >64 | 64 | >128 | >128 |
| DH5α(pCoxMS6671#6) | 1 | 2 | 2 | <0.25 | <0.25 | <0.25 |
IMI, imipenem; MEM, meropenem; ETP, ertapenem; CAZ, ceftazidime; CTX, cefotaxime; AZT, aztreonam.
The effect of the mgrB::blaOXA-181 insertion on the strains' susceptibility to colistin was successfully reversed by introducing pComgrB, i.e., the pDG106 vector carrying an intact mgrB gene. Both ABC143C(pComgrB) and MS6671(pComgrB) regained their susceptibility to colistin, while the transformants carrying the vector only did not (Table 4).
TABLE 4.
Complementation of the mgrB::blaOXA-181 mutation
| Strain | MIC of colistin (mg/liter)a |
|---|---|
| ABC143C(pDG106) | 16 |
| ABC143C(pComgrB) | 1 |
| MS6671(pDG106) | 16 |
| MS6671(pComgrB) | 1 |
Colistin susceptibility tests were carried out in the presence of 100 μM HgCl2.
Plasmid pABC143C-NDM carrying the blaNDM-5 gene.
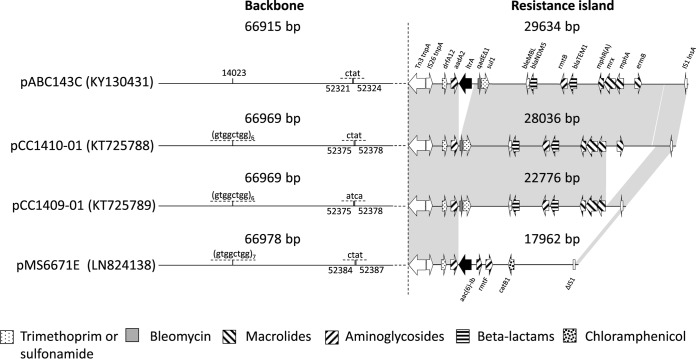
We obtained the complete sequence of the plasmid present in most strains of the cluster carrying the blaNDM-5 gene from K. pneumoniae ABC143C. pABC143C-NDM (GenBank accession no. KY130431) is an IncFII incompatibility type plasmid of 96,549 bp with a GC content of 52.8%. It has 119 coding sequences, of which eight encode antibiotic resistance: ermB, mph(A), blaTEM-1B, rmtB, blaNDM-5, sul1, aadA2, and dfrA12 (Fig. 4). These genes are located in a resistance island inserted into the 66,915-bp-long plasmid backbone, which is highly similar to that of plasmid pMS6671E of MS6671 (GenBank accession no. LN824138), i.e., the blaNDM-negative strain recovered in UAQ hospital B (12). Even more similarities were revealed with two other IncFII-type plasmids, both carrying blaNDM-5, i.e., pCC1410-1 and pCC1409-1 (GenBank accession no. KT725788 and KT725789, respectively) from strains isolated in South Korea (5, 13). The pMS6671E plasmid carried a resistance island different from those of the other three plasmids, as it did not contain the ermB, mph(A), blaTEM-1B, rmtB, blaNDM-5, or sul1 genes, although it also harbored the ltrA gene, which is present in the other Emirati plasmids but notably absent from the Korean ones. On the other hand, the resistance islands of the three other plasmids exhibited high levels of similarity, especially in pCC1410-1 and pABC143C-NDM, which were, with the exception of the presence of ltrA, identical (Fig. 4).
FIG 4.
Structure of pABC143C. Note that the structures are represented disproportionately to their real size. The GenBank numbers of the plasmids are shown in parentheses.
DISCUSSION
Pan-drug-resistant K. pneumoniae infections have been reported since 2005 (14), and most recently, even clusters or outbreaks caused by the KPC-2 producer K. pneumoniae ST258 have been recorded (11, 15). In Greece, a single pan-drug-resistant K. pneumoniae ST147 isolate was also encountered among K. pneumoniae ST258 strains (11) representing the majority of local pan-drug-resistant isolates. Here, we report on a cluster of highly similar, pan-drug-resistant K. pneumoniae ST147 strains isolated from three cases in two emirates of the UAE. The short time between the isolation of strains from the first two patients in the same AD hospital and the association of the first patient with UAQ hospital A, where the third case was identified, make the connection likely. Nevertheless, the paucity of epidemiological data prevents us from substantiating this assumption. We assume that these isolates are also closely related to a pan-drug-resistant K. pneumoniae ST147 isolate from UAQ hospital B, MS6671, carrying three chromosomal copies of blaOXA-181 isolated a few months later (12). In addition to belonging to the same ST and having highly similar genotypes (Table 1) and similarities in their pulsed-field gel electrophoresis (PFGE) patterns (Fig. 1), our isolates also carried three copies of the OXA gene in fragments of their XbaI-restricted genomes of molecular masses identical to that of MS6671 (Fig. 2). In one strain, ABC143F, a fourth fragment of the PFGE pattern was also labeled, suggestive of the presence of another copy of blaOXA-181. As with isolate MS6671, colistin resistance of the current strains was also due to an mgrB::blaOXA-181 insertion. Furthermore, the backbone of the blaNDM-5-containing plasmid pABC143C-NDM exhibited extensive similarities to that of pMS6671E. A possible explanation for the differences between these two plasmids might be that in MS6671, a recombination event between the chromosomally located class I integron and the integron located on the IncFII plasmid took place, resulting in the loss of the ermB, mph(A), blaTEM-1B, rmtB, blaNDM-5, and sul1 resistance genes. It was noteworthy, however, that this event, i.e., the loss of blaNDM-5, did not change the susceptibility profile of K. pneumoniae MS6671, since its remarkable array of resistance genes (12) still made it pan-resistant. A similar scenario might have occurred in ABC143F, one of the isolates from the first patient, which lost the rmtB and blaNDM-5 genes. However, investigating this was beyond the scope of the present study.
Plasmid structures highly similar to pABC143C (pCC1410-1 and pCC1409-1 [GenBank accession no. KT725788 and KT725789]) were previously revealed in two strains isolated in South Korea (5). Like members of the UAE cluster, they belonged to K. pneumoniae ST147 and both carried the blaOXA-181 and blaNDM-5 carbapenemases. Notably, the first strain (CC1409-1) was recovered from a patient who had been transferred in 2014 from an undisclosed hospital in Abu Dhabi to Seoul, South Korea. While CC1409-1 was still susceptible to colistin, the strain isolated 4 months later in the same hospital (CC1410-1) was resistant to polymyxins, although the basis of its colistin resistance has not been disclosed (13). Although these strains were not available to us for a more detailed comparison, on the basis of all the similarities between their genotypes and the nearly identical structures of their blaNDM-5-carrying plasmids (Fig. 3), we surmise that these South Korean strains also closely related to this specific cluster of K. pneumoniae ST147 isolates.
Enterobacteriaceae possessing two carbapenemases have previously been observed worldwide (16) and in the region as well (6, 17, 18). It is of particular concern that representatives of the K. pneumoniae ST147 clone, well known for its epidemic potential (4, 6), not only are present in the UAE as dual-carbapenemase-producing isolates but also develop resistance to the last-resort antibiotic, colistin, by insertion of a functional carbapenemase gene into the mgrB gene. Apart from the earlier report on MS6671 (12), a similar ISEcp1-driven insertional mutation of mgrB in K. pneumoniae has been observed in a single isolate in France, but in that case, an extended-spectrum beta-lactamase gene (blaCTX-M-15) was coded on the insertion, and the strain remained susceptible to an array of antibiotics (19).
It is not known to what extent the reduced susceptibility to chlorhexidine contributed to the spread or reoccurrence of this pan-drug-resistant K. pneumoniae ST147 clone in multiple hospitals. Carrying the cepA and qacΔE genes has been associated previously with such a phenotype (20). On the other hand, the insertional mutation leading to colistin resistance in our study was different from those recently described in colistin- and chlorhexidine-cross-resistant K. pneumoniae isolates, and the level of chlorhexidine nonsusceptibility did not reach the levels noted in that study (21).
Important limitations of the study are the lack of available more-detailed epidemiological data revealing the actual links and exploring the possible modes of transmission between the cases. Furthermore, as currently there are no nationwide antibiotic surveillance data and the laboratories are under no obligation to submit multidrug-resistant isolates for molecular typing, it is not known whether the cluster described represents its real size or additional such isolates are still present in the region.
Nevertheless, our data show that this pan-resistant K. pneumoniae ST147 clone, with a unique, ISEcp1-directed blaOXA-181 insertion in the mgrB gene, i.e., a single genetic event causing resistance to two agents used to treat severe Gram-negative infections, carries a considerable outbreak potential, as evidenced by its repeated occurrence in multiple hospitals, including possible international spread, over an extended period of time. Beyond enforced infection control measures, only extensive, national, and region-wide antimicrobial resistance surveillance with molecular typing investigations may carry the promise to contain the spread of such strains.
MATERIALS AND METHODS
Bacterial isolates.
Strains were submitted to our laboratory for molecular typing according to the nonmandatory recommendation of the Health Authority Abu Dhabi regarding carbapenem-resistant Enterobacteriaceae (CRE) isolates. Strains were received with basic demographic and clinical information only, i.e., strictly without any patient identifiers. The isolates were preserved in duplicate at −80°C.
Antimicrobial susceptibility testing.
Antibiotic susceptibility was routinely tested by microdilution according to the CLSI standards (22), while in the case of colistin, tigecycline, and fosfomycin, the EUCAST guidelines (23) were applied. Agar dilution was used to establish susceptibilities to chlorhexidine, benzalkonium, and irgasan.
Genotyping.
The presence of virulence-related genes magA, allS, mrkD, rmpA, kfuBC, fimH, ureA, uge, wabG, fyuA, iutA, ireA, iroN, cnf1, traT, and clpK (24, 25), of those determining the capsular types (26), and of antibiotic and disinfectant resistance genes blaTEM, blaCTX-M, blaSHV, blaPER, blaAmpC, blaNDM, blaOXA-48-like, blaKPC, blaVIM, blaIMP, armA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, qnrS, qepA, aac6′-1b-cr, emrE, qacDE, qacF, qacE, mdfA, ydgE, ydgF, sugE(c), sugE(p), and cepA was tested by PCR as previously described (27). The allele of the OXA-48-like enzyme was determined by direct sequencing of the amplicon containing the mgrB::blaOXA structure, while that of blaNDM was revealed by PCR and direct sequencing with primers AS-ndm1 and AS-ndm2 (28).
Molecular typing.
Isolates were compared by using rep-PCR (DiversiLab; bioMérieux) and applying a Klebsiella kit, according to the manufacturer's recommendation, and by XbaI-restricted genome digestion followed by pulsed-field gel electrophoresis (PFGE), as described previously (29). For hybridization of the PFGE gel, the amount of bacterial cells for plug preparation was doubled. PFGE patterns were analyzed with GelCompar II software (Applied Maths, Belgium) and by applying the unweighted pair group method with arithmetic mean (UPGMA) with the Dice similarity coefficient (SD) with a 1.5% position tolerance. The sequence type of the isolates was determined by multilocus sequence typing (MLST) (30) using the MLST webpage http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html.
Plasmid work.
Extraction and electrophoresis of the plasmids were carried out by the method of Kado and Liu (31), using plasmids in E. coli 39R861 and E. coli V517 as molecular mass markers.
From K. pneumoniae ABC143C, the pABC143C-NDM-5 plasmid was transferred by conjugation into E. coli J53RAZ as described previously (32). From the transconjugant, the plasmid was purified by using a plasmid maxiprep kit (Qiagen, Germany), and its complete sequence was determined commercially at the CCIB DNA Core Facility at Massachusetts General Hospital (Cambridge, MA). PCR mapping and direct sequencing were used to close the gaps between the contigs and to unambiguously determine the sequence of the plasmid. Antibiotic resistance genes on the assembled plasmid were identified by ResFinder 2.1 (33). The sequences were annotated using Sequin (http://www.ncbi.nlm.nih.gov/Sequin) and submitted to GenBank. The Inc type of the plasmid was deduced from its complete sequence (34).
Hybridization.
Hybridization of the plasmid and PFGE gel transferred to Hybond N+ (Roche) membranes was carried out as described previously (32).
Cloning and complementing the mgrB gene with a blaOXA-181 insertion.
To PCR clone the entire mgrB gene, with or without insertion, external primers (8) were used with additional BamHI plus CGCCGC overhangs. The mgrB::blaOXA-181 structure amplified was cloned into the BamHI-digested pUC19 and transformed into E. coli DH5α (28), and transformants were selected on plates containing 2 mg/liter ertapenem.
To complement the mgrB::blaOXA-181 insertion, an intact mgrB gene was amplified from ABC193, a colistin-susceptible K. pneumoniae strain, and cloned into the BamHI site of pDG106, i.e., a vector containing the mercury resistance merR gene, obtained from the National Institute of Genetics, Japan (35). This construct was transformed (28) into the colistin-resistant wild-type strains ABC143C and MS6671, with selection of transformants on plates containing 100 μM HgCl2. The colistin susceptibilities of the transformants were compared to those of similarly prepared derivatives containing the pDG106 vector without the mgrB insert by using serial dilution of colistin sulfate with a constant concentration of 100 μM HgCl2 in order to maintain the plasmid in the derivatives.
Accession number(s).
The sequences of the mgrB PCR product of ABC143C and pABC143C-NDM were deposited in GenBank under accession numbers KY426739 and KY130431, respectively.
ACKNOWLEDGMENTS
This work was supported by United Arab Emirates University grants UPAR-25143 and CMHS NP/14/07 and Center-Based grant 31R061 to T.P. and United Arab Emirates University grants UPAR-31M235 and CMHS-31M251 to A.S.
We acknowledge the contribution of Brian Forde and Scott Beatson in the elucidation of the mechanism of colistin resistance in the previously reported isolate (12). The support and critical comments during the project by Jens Thomsen (HAAD, Abu Dhabi) is highly appreciated.
D.P. has received honoraria for attendance at advisory boards of Merck, AstraZeneca, Shionogi, Achaogen, and GlaxoSmithKline. The other authors declare no conflicts of interest.
REFERENCES
- 1.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennequin C, Robin F. 2016. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 3.Pitout JD, Nordmann P, Poirel L. 2015. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messaoudi A, Haenni M, Mansour W, Saras E, Bel Haj Khalifa A, Chaouch C, Naija W, Boujaafar N, Bouallegue O, Madec JY. 2017. ST147 NDM-1-producing Klebsiella pneumoniae spread in two Tunisian hospitals. J Antimicrob Chemother 72:315–316. doi: 10.1093/jac/dkw401. [DOI] [PubMed] [Google Scholar]
- 5.Shin J, Baek JY, Cho SY, Huh HJ, Lee NY, Song JH, Chung DR, Ko KS. 2016. blaNDM-5-bearing IncFII-type plasmids of Klebsiella pneumoniae sequence type 147 transmitted by cross-border transfer of a patient. Antimicrob Agents Chemother 60:1932–1934. doi: 10.1128/AAC.02722-15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Sonnevend A, Ghazawi AA, Hashmey R, Jamal W, Rotimi VO, Shibl AM, Al-Jardani A, Al-Abri SS, Tariq WU, Weber S, Pal T. 2015. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS One 10:e0131372. doi: 10.1371/journal.pone.0131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baron S, Hadjadj L, Rolain JM, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 11.Bathoorn E, Tsioutis C, da Silva Voorham JM, Scoulica EV, Ioannidou E, Zhou K, Rossen JW, Gikas A, Friedrich AW, Grundmann H. 2016. Emergence of pan-resistance in KPC-2 carbapenemase-producing Klebsiella pneumoniae in Crete, Greece: a close call. J Antimicrob Chemother 71:1207–1212. doi: 10.1093/jac/dkv467. [DOI] [PubMed] [Google Scholar]
- 12.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, Yin WF, Chan KG, Li J, Schembri MA, Beatson SA, Paterson DL. 2015. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 5:15082. doi: 10.1038/srep15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki CS, Chung DR, Lee NY, Song JH. 2015. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg Infect Dis 21:1088–1089. doi: 10.3201/eid2106.150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falagas ME, Bliziotis IA, Kasiakou SK, Samonis G, Athanassopoulou P, Michalopoulos A. 2005. Outcome of infections due to pandrug-resistant (PDR) Gram-negative bacteria. BMC Infect Dis 5:24. doi: 10.1186/1471-2334-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, Veenemans J. 2015. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis 34:1647–1655. doi: 10.1007/s10096-015-2401-2. [DOI] [PubMed] [Google Scholar]
- 16.Meletis G, Chatzidimitriou D, Malisiovas N. 2015. Double- and multi-carbapenemase-producers: the excessively armored bacilli of the current decade. Eur J Clin Microbiol Infect Dis 34:1487–1493. doi: 10.1007/s10096-015-2379-9. [DOI] [PubMed] [Google Scholar]
- 17.Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin Microbiol Infect 18:E144-8. doi: 10.1111/j.1469-0691.2012.03796.x. [DOI] [PubMed] [Google Scholar]
- 18.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al-Abri S, Al Salman J, Dashti AA, Kutbi AH, Schlebusch S, Sidjabat HE, Paterson DL. 2014. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf Cooperation Council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother 58:3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayol A, Nordmann P, Desroches M, Decousser JW, Poirel L. 2016. Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3199–3201. doi: 10.1128/AAC.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abuzaid A, Hamouda A, Amyes SG. 2012. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J Hosp Infect 81:87–91. doi: 10.1016/j.jhin.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Wand ME, Bock LJ, Bonney LC, Sutton JM. 2017. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother 61:e01162-16. doi: 10.1128/AAC.01162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. 2014. Performance standard for antimicrobial susceptibility testing; M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.EUCAST. 2016. Breakpoint tables for interpretation of MICs and zone diameters. http://www.eucast.org Accessed 15 January 2016.
- 24.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. 2009. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 4:e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 26.Turton JF, Perry C, Elgohari S, Hampton CV. 2010. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol 59:541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 27.Sonnevend A, Ghazawi A, Alqahtani M, Shibl A, Jamal W, Hashmey R, Pal T. 2016. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis 50:85–90. doi: 10.1016/j.ijid.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Ghazawi A, Sonnevend A, Bonnin RA, Poirel L, Nordmann P, Hashmey R, Rizvi TA, M BH, Pal T. 2012. NDM-2 carbapenemase-producing Acinetobacter baumannii in the United Arab Emirates. Clin Microbiol Infect 18:E34–6. doi: 10.1111/j.1469-0691.2011.03726.x. [DOI] [PubMed] [Google Scholar]
- 29.Gautom RK. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol 35:2977–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, Al Haj M, Pal T. 2013. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol 62:1044–1050. doi: 10.1099/jmm.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 33.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gambill BD, Summers AO. 1985. Versatile mercury-resistant cloning and expression vectors. Gene 39:293–297. doi: 10.1016/0378-1119(85)90326-9. [DOI] [PubMed] [Google Scholar]