ABSTRACT
IncX plasmids are receiving much attention as vehicles of carbapenem and colistin resistance genes, such as blaNDM, blaKPC, and mcr-1. Among them, IncX2 subgroup plasmids remain rare. Here, we characterized IncX2 and IncX1-X2 hybrid plasmids coexisting in a FosA6-producing Escherichia coli strain that were possibly generated as a consequence of recombination events between an R6K-like IncX2 plasmid and a pLN126_33-like IncX1 plasmid. Variable multidrug resistance mosaic regions were observed in these plasmids, indicating their potential to serve as flexible carriers of resistance genes. The diversity of IncX group plasmid backbones and accessory genes and the evolution of hybrid IncX plasmids pose a challenge in detecting and classifying them.
KEYWORDS: IncX plasmid, taxC, pir, genotyping, glutathione synthetase
INTRODUCTION
The IncX family plasmids are narrow-host-range plasmids mostly found in Enterobacteriaceae (1–3). IncX plasmids contain a highly syntenic backbone consisting of core genes responsible for plasmid replication, partitioning, maintenance, and conjugal DNA transfer (2). So far, at least eight subgroups of IncX plasmids have been reported, IncX1 to IncX8, carrying various resistance genes, including oqxAB, qnrS1, qnrS2, blaTEM, blaSHV, blaCTX-M, blaKPC, blaNDM, and mcr-1, and mediating resistance to fluoroquinolones, cephalosporins, carbapenems, colistin, or other types of antibiotics (2–9). Plasmid R6K, recovered from an Escherichia coli strain cultured in Greece in the 1960s, is considered to be the prototypical IncX plasmid (1). R6K belongs to a rare IncX subgroup, IncX2, with only two other currently recognized members, pNGX2-QnrS1 (GenBank accession number JQ269335.1) and pEBG1 (GenBank accession number KF738053.1), both discovered in strains from western Nigeria and carrying qnrS1 genes (3, 10). In contrast, the IncX1, IncX3, and IncX4 subgroup plasmids are much more prevalent and are associated with oqxAB, blaNDM, and mcr-1, respectively (2, 3, 7, 8, 11). Here, we describe a new IncX2 plasmid, pYD786-4, that coexisted with a hybrid IncX1-X2 plasmid pYD786-3 in an E. coli ST410 urinary tract strain collected from a patient in the United States (12).
RESULTS AND DISCUSSION
Hybrid IncX1-X2 plasmid pYD786-3.
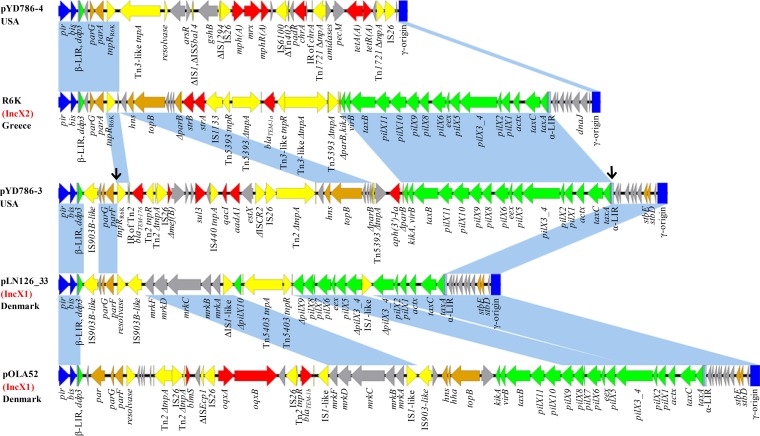
Plasmid pYD786-3 is 44,806 bp in size and shares conserved backbone modules of both IncX1 and IncX2 plasmids. The replication, partitioning, and stability regions are from the IncX1 plasmid, while the maintenance and DNA transfer regions are from the IncX2 plasmid (Fig. 1). pir and bis encode the plasmid initiation replication protein and auxiliary protein that bind to the core region of replication gamma origin (2). parGF and stbED have putative functions of partitioning and stability. Two long inverted repeats (LIRs), α-LIR and β-LIR, contain origins of conjugal transfer (oriT) which act with DNA distortion proteins (DDP) TaxC and TaxA to initiate the relaxation and nicking of oriT (2, 13). There are also genes (hns and topB) encoding putative maintenance functions and genes (kikA, pilX, actX, and taxBCA) encoding conjugative DNA transfer in pYD786-3. An 8,411-bp fragment organized as α-LIR (in part)–stbED–gamma origin–pir–bis–β-LIR–ddp3–parGF is almost identical to the core genes of IncX1 plasmid pLN126_33 (GenBank accession number HE578058.1) (14), except for two point mutations, two indels (insertions and deletions), and insertion of an IS903B-like sequence (Fig. 1). Plasmid pLN126_33 is structurally similar to pOLA52 (GenBank accession number EU370913.1), a representative and well-characterized IncX1 plasmid carrying the type 3 fimbria gene cassette mrkABCDF (2, 14). In contrast, the rest of pYD786-3 (36,395 bp long), including regions of hns-topB and kikA-taxB-pilX-actX-taxCA, differs from the IncX2 plasmid R6K by only three single nucleotides and the presence of two accessory modules composed of mobile genetic elements (MGEs) and antimicrobial resistance genes (Fig. 1). Interestingly, pYD786-4, a small plasmid of 25,678 bp, possesses the 5.3-kb backbone of R6K containing gamma origin–pir–bis–β-LIR–ddp3–parGA, with only one nucleotide difference and one indel. Therefore, pYD786-3 and pYD786-4 were probably generated as a consequence of recombinations between an R6K-like IncX2 plasmid and a pLN126_33-like IncX1 plasmid in E. coli YD786.
FIG 1.
Linear comparison of rearranged IncX1 and IncX2 plasmids including pYD786-3 (GenBank accession number KU254580.1), pYD786-4 (KU254581.1), pLN126_33 (HE578058.1), pOLA52 (EU370913.1), and R6K (NCTC50005 [http://www.sanger.ac.uk/resources/downloads/plasmids/]). Genes are colored based on the predicted function: blue, replication; brown, partition, stability, and maintenance; green, DNA transfer; yellow, mobile genetic elements; red, resistance; light gray, other functions. Black arrows indicate putative recombination junction sites in pYD786-3. Shading between plasmids indicates shared backbone regions with a nucleotide identity of >99.3% to 100%.
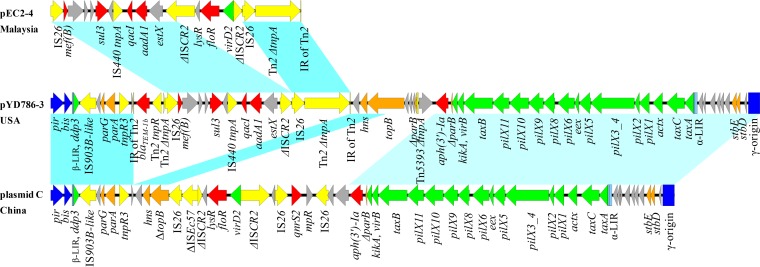
Two putative recombination junction sites are indicated in Fig. 1 (see also Fig. S1 in the supplemental material), one located between parA and the tnpR gene from R6K (tnpRR6K) (Fig. S1A) and the other in α-LIR (Fig. S1B), thus creating two hybrid IncX plasmids. pYD786-3 is characterized by a mosaic IncX1-X2 backbone, a hybrid α-LIR, and a resolvase gene, tnpRR6K, from R6K (Fig. 1). Wider BLASTn comparisons with publicly available plasmids showed that plasmid C (GenBank accession number CP010155.1) in E. coli strain D9 possessed a backbone almost identical to that of pYD786-3, with only five nucleotide differences (Fig. 2). A similar structure was also found in the chromosome of E. coli strain D4 (GenBank accession number CP010143.1); both strains were isolated from Chinese dogs. Interestingly, tnpRR6K, belonging to the serine recombinase family, is specifically located in IncX2 plasmids (R6K, pNGX2-QnrS1, pEBG1, and pYD786-4) or their hybrid derivatives (pYD786-3, plasmid C, and E. coli strain D4), indicating its ancient linkage with this subgroup of plasmids and global dissemination.
FIG 2.
Comparison of backbone and multidrug resistance regions of rearranged plasmid pYD786-3 (GenBank accession number KU254580.1) with those of plasmid C (CP010155.1; rearranged and inversely displayed) and pEC2-4 (CP016184.1; nucleotides 23922 to 39746). Shading between plasmids indicates shared backbone regions with a nucleotide identity of >99.9% to 100%.
Most recently, an IncX4-X3 hybrid plasmid, pCQ02-121 (GenBank accession number KU647721.1), was discovered in an E. coli strain from a cat in China harboring blaNDM-5 and mcr-1 resistance determinants (7). Similar to pYD786-3, pCQ02-121 contained replication and partition regions from a pMCR1_IncX4-like plasmid and DNA transfer and maintenance regions (hns-topB-pilX-taxBCA) from a pNDM5_IncX3-like plasmid (7). Plasmids pNDM5_IncX3 (GenBank accession no. KU761328.1) and pMCR1_IncX4 (KU761327.1) were reported to coexist in the same clinical strain of Klebsiella pneumoniae in China (15), providing another piece of evidence for in vivo rearrangements of IncX plasmids.
The MRR of pYD786-3.
A Tn2 transposon, bearing the β-lactamase gene blaTEM-176, is inserted into the backbone of pYD786-3, followed by recruitment of an IS26-flanked composite transposon harboring an uncommon sul3-type integron and interrupting the tnpA of Tn2 (16). The sul3-type integron carries gene cassettes estX-aadA1-qacI, followed by IS440-sul3-ΔmefB rather than the 3′ conserved segment (CS). The 5′ CS is replaced by a truncated ISCR2, thus generating a multidrug resistance mosaic region (MRR) in pYD786-3. Almost identical MRRs have been identified in two mcr-1-bearing plasmids of E. coli, isolated from Malaysian food animals (pEC2_1-4, GenBank accession number CP016183.1; pEC2-4, GenBank accession number CP016184.1) (Fig. 2), indicating IS26-mediated mobilization of the fragment ΔISCR2-estX-aadA1-qacI-IS440-sul3-ΔmefB among strains though the combined unit has rarely been reported. The other variable region of pYD786-3 is composed of a truncated Tn5393 and an aminoglycoside resistance gene, aph(3′)-Ia, located between hns-topB and the DNA transfer region.
pYD786-4, a new IncX2 plasmid.
Unlike classic IncX2 plasmids, pYD786-4 has lost part of its core DNA transfer and mating pair formation system (pilX and taxCAB) genes, possibly due to the recombination between two IncX plasmids and subsequent recruitment of resistance genes with MGEs. The MRR of pYD786-4 is composed of a Tn3-like transposon, an IS26-flanked composite transposon, and a glutathione synthetase gene (gshB) flanked by truncated IS1294, ISSba14, and IS1 (Fig. 1). Tn1721, carrying tetR(A) and tetA(A), is truncated by IS26 and an inverted repeat carrying the chrA gene (IRchrA)-chrA module, followed by IRt-IS6100 (where IRt is an inverted repeat of class 1 integrons) and a macrolide resistance module bound at one end by another IS26, as described previously (17). Since glutathione is a substrate of the glutathione S-transferase FosA6, which inactivates fosfomycin by forming a glutathione-fosfomycin adduct (12), the glutathione synthetase here may play a role in modulating the function of FosA6. The gshB genes have been found on 13 plasmids and one chromosome, with 9 of them coexisting with the fosfomycin resistance gene fosA3 or fosA6 (Table S1). The gshB genes are carried by E. coli or Salmonella species isolated from humans, animals, forest soil, or sewage, mostly in China, with only two found in urinary tract E. coli strains from Pennsylvania (USA) (Table S1) (12, 18). Eleven of the genes possess an identical combination of IS26-ΔIS1294-gshB-ΔISSba14, suggesting the presence of a common origin of gshB which has disseminated across several plasmid families (Table S1).
Subtyping of IncX group plasmids.
IncX group plasmids are highly diverse between subgroups, thus posing a great challenge in plasmid typing (3). IncX plasmids were initially categorized into IncX1 and IncX2 subgroups, based on restriction analysis and replicon probes (1). In 2005, Carattoli et al. developed a PCR-based replicon typing method (PBRT) for plasmid typing which included the IncX group, with primers targeting the replication region (gamma origin) of IncX2 plasmid R6K (19). As more IncX plasmids were fully sequenced, a revised PBRT was developed to classify subgroups IncX1 to IncX4 on the basis of taxC genes (3), and then the replication genes (pir and bis) were used for the typing of subgroups IncX1 to IncX8 in PlasmidFinder, a Web tool for in silicon detection and typing of plasmids (5). Recently, Chen et al. and Du et al. reported blaKPC-bearing IncX5 and IncX6 plasmids (pBK31567, GenBank accession number JX193302.1; pKPC3_SZ, KU302800.1), classified by taxC genes and conserved backbone sequences, which are now included in PlasmidFinder as IncX5 and IncX6 plasmids on the basis of pir genes (Fig. S2) (4, 6). We therefore performed a phylogenetic analysis to compare the backbone genes taxC, pir, and bis of 33 IncX plasmids, including pYD786-4, pYD786-3, plasmid C, pMCR1_IncX4, pNDM5_IncX3, pCQ02-121, pESTMCR (IncX4; GenBank accession number KU743383.1), pHS696_34 (IncX4; JX258654.1), pJARS35 (IncX7; NC_015054.1), pHI4320 (IncX8; AM942760.1), and 23 IncX plasmids, as described by Du et al. (6). All of them were subtyped based on the criteria of PlasmidFinder, with four reference bis genes for the IncX1 subgroup and nine reference pir genes for the subgroups IncX2 to IncX8. The phylogeny of 32 taxC genes was almost consistent with that of bis, except that the IncX1-X2 hybrid plasmids (pYD786-3 and plasmid C) and the IncX4-X3 hybrid plasmid pCQ02-121 were grouped into IncX2 and IncX3 clusters, respectively, in the taxC tree (Fig. S2). pYD786-4 was not included in the taxC tree due to the loss of taxC. In the pir tree, IncX3 and IncX4 plasmids were grouped into two clusters which were consistent with the criteria of PlasmidFinder as two kinds of IncX3 and IncX4 plasmids were used as references, and IncX1 plasmids were grouped into four clusters based on the diversity of pir genes in this subgroup. Hybridization events between IncX1-X2 and IncX4-X3 plasmids have led to a separation of the DNA transfer gene locus (including taxC) from the replication region of the same subgroup (Fig. S2), resulting in confusion when subtyping is performed with different typing markers. Also, we found an IncX2 plasmid without the pilX-tax module, providing additional evidence that replication genes might be the preferred targets for discrimination of the IncX group plasmids as used in the PlasmidFinder Web tool (5).
MATERIALS AND METHODS
E. coli YD786 carries four plasmids including fosA6-harboring pYD786-2 (GenBank accession no. KU254579.1), blaCTX-M-2-harboring pYD786-1 (KU254578.1), and two IncX plasmids (pYD786-3 and pYD786-4), which were sequenced and assembled as reported before (12, 20). BLAST tools (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search for homologues of IncX plasmid backbone. The reference genome of plasmid R6K is available via the Sanger Institute's website (http://www.sanger.ac.uk/resources/downloads/plasmids/). Easyfig, version 2.2, was used for comparison of related plasmids. Phylogenetic analysis was performed using MEGA, version 7 (21).
Accession number(s).
The nucleotide sequences of pYD786-3 and pYD786-4 are available in the GenBank database under accession numbers KU254580.1 and KU254581.1, respectively.
Supplementary Material
ACKNOWLEDGMENTS
Q.G. was supported by grant numbers 81673479 and 81120108024 from the National Natural Science Foundation of China and grant number 16PJD010 from the Shanghai Pujiang Program. N.S. is funded by a University of Oxford Clinical Lectureship. Part of the analysis was supported by a research grant from the National Institutes of Health (R21AI123747).
The funders had no role in study design, analysis, or preparation of the manuscript.
Y.D. has served on advisory boards for Achaogen, Allergan, Curetis, The Medicines Company, and Roche and received research funding from The Medicines Company for studies unrelated to this work. No other authors have potential conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00536-17.
REFERENCES
- 1.Jones CS, Osborne DJ, Stanley J. 1993. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J Gen Microbiol 139:735–741. doi: 10.1099/00221287-139-4-735. [DOI] [PubMed] [Google Scholar]
- 2.Norman A, Hansen LH, She Q, Sorensen SJ. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. 2013. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother 57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang YW, Kreiswirth BN. 2016. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother 60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Yang RS, Zhang Q, Feng Y, Fang LX, Xia J, Li L, Lv XY, Duan JH, Liao XP, Liu YH. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol 1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]
- 8.Brauer A, Telling K, Laht M, Kalmus P, Lutsar I, Remm M, Kisand V, Tenson T. 2016. Plasmid with colistin resistance gene mcr-1 in extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob Agents Chemother 60:6933–6936. doi: 10.1128/AAC.00443-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, Fang Y, Ren H, Zhong LL, Shen Z, Zeng KJ, Wang S, Liu JH, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 10.Sumrall ET, Gallo EB, Aboderin AO, Lamikanra A, Okeke IN. 2014. Dissemination of the transmissible quinolone-resistance gene qnrS1 by IncX plasmids in Nigeria. PLoS One 9:e110279. doi: 10.1371/journal.pone.0110279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Que TL, Chow KH. 2012. Identification and characterization of a novel incompatibility group X3 plasmid carrying blaNDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:e39. doi: 10.1038/emi.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. 2016. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 71:2460–2465. doi: 10.1093/jac/dkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunez B, Avila P, de la Cruz F. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol Microbiol 24:1157–1168. doi: 10.1046/j.1365-2958.1997.4111778.x. [DOI] [PubMed] [Google Scholar]
- 14.Burmolle M, Norman A, Sorensen SJ, Hansen LH. 2012. Sequencing of IncX-plasmids suggests ubiquity of mobile forms of a biofilm-promoting gene cassette recruited from Klebsiella pneumoniae. PLoS One 7:e41259. doi: 10.1371/journal.pone.0041259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curiao T, Canton R, Garcillan-Barcia MP, de la Cruz F, Baquero F, Coque TM. 2011. Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-β-lactamase-producing Escherichia coli clones from humans. Antimicrob Agents Chemother 55:2451–2457. doi: 10.1128/AAC.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge SR. 2011. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev 35:820–855. doi: 10.1111/j.1574-6976.2011.00277.x. [DOI] [PubMed] [Google Scholar]
- 18.McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE. 2016. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: First report of mcr-1 in the United States. Antimicrob Agents Chemother 60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Guo Q, Ding B, Jove T, Stoesser N, Cooper VS, Wang M, Doi Y. 2016. Characterization of a novel IncHI2 plasmid carrying tandem copies of blaCTX-M-2 in a fosA6-harboring Escherichia coli sequence type 410 strain. Antimicrob Agents Chemother 60:6742–6747. doi: 10.1128/AAC.01173-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.