LETTER
Colistin, representing a family of cationic polypeptide antibiotics, acts as a final resort for treatment of infections by the multidrug-resistant Gram-negative pathogens. The emergence of the mobilized colistin resistance (MCR-1) gene attracted much interest from scientific community. Fast spread of the mcr-1 gene challenged global public health (1). No fewer than 8 types of plasmids (such as those encoding IncI2 [2], IncP [3], and IncX4 [4]) can carry the mcr-1 gene among Enterobacteriaceae, suggesting its complex dissemination (1–13).
Very recently, Quan et al. reported an epidemiological study of mcr-1-positive isolates from patient bloodstreams in China (14), and Wang et al. addressed its clinical impact (15). Given the risk factor of mcr-1 in hospitals, we were interested in examining bacterial isolates (200 Escherichia coli isolates in total) provided by Lishui People's Hospital (Zhejiang Province, China). All the strains were isolated from blood cultures of patients with suspected infectious diseases from January 2014 to December 2015. Further data on the number of patients and possible antimicrobial pretreatments were unfortunately not available. Ten E. coli isolates were mcr-1 positive, but all of them were mcr-2 negative in PCR screens (see Table S1 in the supplemental material). The mcr-1-carrying E. coli isolates were, namely, LS12, LS19, LS26, LS32, LS52, LS96, LS139, LS142, LS156, and LS178 (see Fig. S1 in the supplemental material). Both multilocus sequence typing (MLST) (8) and pulsed-field gel electrophoresis (PFGE) (6, 16) showed that these mcr-1-positive E. coli strains exhibit diversified genetic backgrounds (Table S2 and Fig. S2). In accordance with the genome sequence of the first-identified mcr-1-harboring plasmid, pHNSHP45, we designed a series of primers (corresponding to genes such as tnpA and nikB; Table S1), as we described recently (4, 17), to probe the genetic context that surrounded the mcr-1 gene. Two strains, LS142 and LS178, were positive in all the PCR assays described above (Fig. S1B); all 8 of the remaining E. coli isolates were PCR negative for the target loci that are supposed to occur in the pHNSHP45-like variants (Fig. S1A). PCR profiles suggested the presence of two types of plasmids (pLS142 and pLS178 represented pHNSHP45-like IncI2-type plasmid variants [Fig. 1A; see also Fig. S1]; all eight of the other plasmids, like pLS12, represented non-IncI2-type pHNSHP45 plasmid variants [Fig. 1B; see also Fig. S1]). Following Clinical and Laboratory Standards Institute (CLSI) guidelines, microbroth dilution experiments determined that the colistin MIC of the mcr-1-carrying clinical strains was not less than 4 μg/ml (Table S3).
FIG 1.
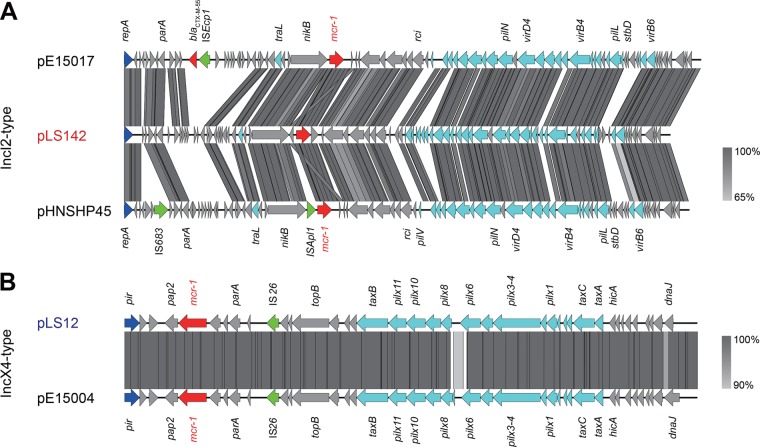
Genomic analyses for the two newly discovered mcr-1-harboring plasmids from inpatient bloodstreams (IncI2-type pLS142 and IncX4-type pLS12). (A) Linear comparison of three mcr-1-positive IncI2-type plasmids (pE15017, pHNSHP45, and pLS142). (B) Linear genome alignment of the IncX4-like pLS12 plasmid with pE15004. Boxed arrows represent the positions and transcriptional directions of ORFs. Gray shading denotes regions of homology shared among different plasmids. Genes associated with the tra, pil, and vir loci are colored light blue; replication-associated genes are colored dark blue; antibiotic resistance genes are colored red; insertion sequences are colored green; and other genes are colored gray.
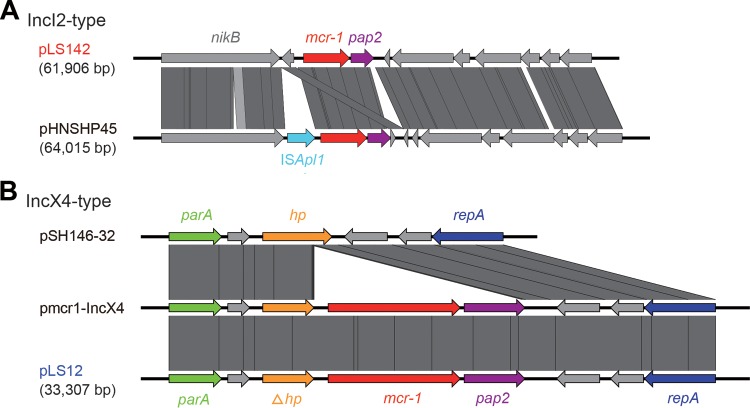
Subsequently, the two representative plasmids (pLS142 and pLS12) were subjected to full-genome sequencing using the strategies of HiSeq 2000 and MiSeq. As expected, a pool of contigs was assembled into two unique plasmids of different sizes (61.9 kb for pLS142 and 33.3 kb for pLS12). Plasmid Finder-based typing (https://cge.cbs.dtu.dk/services/PlasmidFinder/) showed that pLS142 is an IncI2-type plasmid that is closely related to the first-identified mcr-1-positive pHNSHP45 plasmid (GenBank accession no. KP347127), whereas pLS12 is an IncX4-class plasmid (Table 1 and Fig. 1). Using the bacterial mobilome annotation pipeline, it seems likely that the IncI2-type pLS142 plasmid (GenBank accession no. KY075654) contains about 90 open reading frames (ORFs), whereas the IncX4-class pLS12 plasmid (GenBank accession no. KY075653) contains around 50 ORFs. Together with our multiplex PCR typing data (Fig. S1), the results suggested that the IncX4-type mcr-1-harboring small plasmids (such as pLS12) were prevalent in the patient's E. coli isolates. The ISApl1 insertion element was absent in pLS142 compared with pHNSHP45 (Fig. 1A and 2A). The pLS12 plasmid seems identical to the pE15004 plasmid isolated from a diarrhea inpatient's feces that we had analyzed earlier in 2016 (Fig. 1B and 2B) (4, 9). Sequence alignment suggested that the mcr-1-negative pSH146-32 plasmid might be an ancestor for the pLS12 and pmcr-1-IncX4 plasmids (Fig. 2B). With the exception of the mcr-1 gene, ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) failed to detect any resistance gene in either pLS142 or pLS12.
TABLE 1.
Analysis of E. coli plasmids with full-genome sequences, including two decoded in this studya
| Plasmid | Type | Length (bp) | Host | GenBank accession no. | Reference(s) or origin |
|---|---|---|---|---|---|
| pHNSHP45 | IncI2 | 64,015 | Pig | KP347127 | 2 |
| pE15017 | IncI2 | 65,375 | Patient feces | KX772778 | 4, 9 |
| pLS142 | IncI2 | 61,906 | Patient blood | KY075654 | This study |
| pE15004 | IncX4 | 33,309 | Patient feces | KX772777 | 4, 9 |
| pLS12 | IncX4 | 33,307 | Patient blood | KY075653 | This study |
The plasmids we reported here are highlighted in bold.
FIG 2.
Genetic analyses of the mcr-1-containing cassette. (A) Linear comparison of the two IncI2-type plasmids pLS142 and its putative ancestor pHNSHP45. The data suggest that pLS142 lacking the insertion sequence ISApl1 might be a new variant for the prototypical version mcr-1-carrying pHNSHP45 plasmid. (B) Colinear alignments of two mcr-1-postive plasmids (pLS12 and pmcr-1-IncX4) with an mcr-1-negative ancestor plasmid (pSH146-32). Note that all the three plasmids here are IncX4-type plasmids.
In summary, the data from our follow-up study support the general conclusion drawn by Quan et al. (14) and alert us to reconsider the massive use of colistin in veterinary/clinical medicine.
Accession number(s).
Sequence data determined in this study are available at GenBank under accession no. KY075653 and KY075654.
Supplementary Material
ACKNOWLEDGMENTS
Y.F. designed the project; Y.F., J.S., X.-P.L., Q.W., and Y.D. performed experiments; Y.F., J.S., Q.W., and Y.-H.L. analyzed the data and prepared the figures; Y.F. and J.S. drafted the manuscript.
This work was supported by the National Key Basic Research Program of China (2016YFC1200100). Y.F. is a recipient of the “Young 1000 Talents” Award.
We declare that we have no conflict of interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00361-17.
REFERENCES
- 1.Wang X, Zhang H, Sun J, Liu YH, Feng Y. 2017. The MCR-1 colistin resistance: a new challenge to global public health. Chin Sci Bull 62:1018–1029. doi: 10.1360/N972016-01084. [DOI] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Zhao F, Feng Y, Lu X, McNally A, Zong Z. 24 January 2017. IncP plasmid carrying colistin resistance gene mcr-1 in Klebsiella pneumoniae from hospital sewage. Antimicrob Agents Chemother doi: 10.1128/AAC.02229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, Ye H, Liu F, Srinivas S, Li D, Zhu B, Liu YH, Tian GB, Feng Y. 2016. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog 12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao R, Wang Q, Li P, Li Z, Feng Y. 2016. Genome sequence and characteristics of plasmid pWH12, a variant of the mcr-1-harbouring plasmid pHNSHP45, from the multi-drug resistant E. coli. Virulence 7:732–735. doi: 10.1080/21505594.2016.1193279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XP, Fang LX, Song JQ, Xia J, Huo W, Fang JT, Liao XP, Liu YH, Feng Y, Sun J. 2016. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep 6:38511. doi: 10.1038/srep38511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Yang RS, Zhang Q, Feng Y, Fang LX, Xia J, Li L, Lv XY, Duan JH, Liao XP, Liu YH. 2016. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol 1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Li Z, Lin J, Wang X, Deng X, Feng Y. 2016. Complex dissemination of the diversified mcr-1-harbouring plasmids in Escherichia coli of different sequence types. Oncotarget 7:82112–82122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, Gao GF, Hu Q, Feng Y. 2016. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 7:e00177. doi: 10.1128/mBio.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Garch F, Sauget M, Hocquet D, LeChaudee D, Woehrle F, Bertrand X. 2016. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin Microbiol Infect doi: 10.1016/j.cmi.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 11.Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Seward CH, Wu Z, Ye H, Feng Y. 2016. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichiacoli with multi-drug resistance. Sci Bull (Beijing) 61:875–878. doi: 10.1007/s11434-016-1086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xavier BB, Lammens C, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene mcr-1 isolated from Belgian pig farms. J Antimicrob Chemother 71:2342–2344. doi: 10.1093/jac/dkw191. [DOI] [PubMed] [Google Scholar]
- 14.Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, Yu Y. 28 January 2017. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, Fang Y, Ren H, Zhong LL, Shen Z, Zeng KJ, Wang S, Liu JH, Wu C, Walsh TR, Shen J. 28 January 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, Li SM, Liao XP, Feng Y, Liu YH. 2017. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep 7:424. doi: 10.1038/s41598-017-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T; RESET consortium. 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.