ABSTRACT
The pathogenic yeast Candida albicans can develop resistance to the widely used antifungal agent fluconazole, which inhibits ergosterol biosynthesis. Resistance is often caused by gain-of-function mutations in the transcription factors Mrr1 and Tac1, which result in constitutive overexpression of multidrug efflux pumps, and Upc2, which result in constitutive overexpression of ergosterol biosynthesis genes. However, the deregulated gene expression that is caused by hyperactive forms of these transcription factors also reduces the fitness of the cells in the absence of the drug. To investigate whether fluconazole-resistant clinical C. albicans isolates have overcome the fitness costs of drug resistance, we assessed the relative fitness of C. albicans isolates containing resistance mutations in these transcription factors in competition with matched drug-susceptible isolates from the same patients. Most of the fluconazole-resistant isolates were outcompeted by the corresponding drug-susceptible isolates when grown in rich medium without fluconazole. On the other hand, some resistant isolates with gain-of-function mutations in MRR1 did not exhibit reduced fitness under these conditions. In a mouse model of disseminated candidiasis, three out of four tested fluconazole-resistant clinical isolates did not exhibit a significant fitness defect. However, all four fluconazole-resistant isolates were outcompeted by the matched susceptible isolates in a mouse model of gastrointestinal colonization, demonstrating that the effects of drug resistance on in vivo fitness depend on the host niche. Collectively, our results indicate that the fitness costs of drug resistance in C. albicans are not easily remediated, especially when proper control of gene expression is required for successful adaptation to life within a mammalian host.
KEYWORDS: Candida albicans, drug resistance evolution, fitness costs, gain-of-function mutation, transcription factors
INTRODUCTION
The yeast Candida albicans is a member of the microbiota in the oral cavity and the gastrointestinal and urogenital tracts of most healthy individuals. Especially in immunocompromised patients, C. albicans can also become a pathogen and cause superficial as well as systemic infections. Such infections are usually treated with the antimycotic drug fluconazole, which inhibits ergosterol biosynthesis. C. albicans can develop resistance to fluconazole, particularly after long-term therapy of oropharyngeal candidiasis in AIDS patients (1). The most commonly observed resistance mechanisms are mutations in the drug target enzyme Erg11, which reduce drug binding, and gain-of-function (GOF) mutations in the zinc cluster transcription factors (ZnTFs) Mrr1, Tac1, and Upc2, which result in changes in gene expression that promote drug resistance (2–4). Specifically, GOF mutations in Mrr1 cause overexpression of the multidrug efflux pump MDR1 and additional genes that contribute to fluconazole resistance (5–11). Similarly, GOF mutations in Tac1 confer fluconazole resistance by mediating overexpression of the multidrug efflux pumps CDR1 and CDR2 and other Tac1 target genes (12–17). Finally, GOF mutations in Upc2 result in upregulation of ERG11 and other ergosterol biosynthesis genes (18–21). Many fluconazole-resistant clinical C. albicans strains exhibit several of these resistance mechanisms, which results in high levels of drug resistance and therapy failure (22–25).
While the overexpression of genes that confer increased fluconazole resistance is advantageous in the presence of the drug, the deregulated gene expression caused by constitutively active transcription factors (TFs) should reduce the fitness of the cells under nonselective conditions, because it limits their adaptive flexibility and represents an unnecessary waste of energy. Indeed, using isogenic strains with defined GOF mutations in MRR1, TAC1, and UPC2, we previously found that each of the hyperactive TFs decreased the competitive fitness of the strains in coculture experiments with the parental wild-type reference strain SC5314, especially during growth in rich medium (26). The competitive fitness was further reduced in strains containing two or all three hyperactive TFs, and the latter also exhibited a strong fitness defect in a mouse model of gastrointestinal colonization. A separate study similarly reported on the decreased virulence of genetically engineered strains with hyperactive forms of Upc2 and, to a lesser extent, Tac1 in a mouse model of disseminated candidiasis (8). Some fluconazole-resistant clinical C. albicans isolates with GOF mutations in MRR1 or TAC1 were also found to be less virulent than fluconazole-susceptible isolates from the same patients in mouse models of systemic or oral candidiasis (27, 28). On the other hand, many fluconazole-resistant isolates did not exhibit a fitness defect in mixed cultures with matched susceptible isolates during in vitro growth or in animal models (29), suggesting that they overcame the fitness costs of drug resistance during further evolution by compensatory mutations, a phenomenon that is well-known from antibiotic-resistant bacteria (30, 31). Support for this assumption comes from a recent study that investigated the fitness of several series of clinical C. albicans isolates that developed fluconazole resistance over time. The authors found that drug resistance often was accompanied by an initial decrease in fitness, followed by a recovery of fitness in later isolates (32).
In the present study, we have further investigated this aspect of the evolution of fluconazole resistance. We compared the competitive fitness of fluconazole-resistant clinical C. albicans isolates containing known GOF mutations in MRR1, TAC1, and/or UPC2 and matched fluconazole-susceptible isolates from the same patients to find out if the resistant isolates mitigated the fitness costs caused by the hyperactive TFs. Our results show that drug-resistant strains with hyperactive forms of these TFs usually have fitness defects in vitro and/or in a mammalian host but can successfully compete with their drug-susceptible counterparts in some host niches.
RESULTS
Identification of GOF mutations in MRR1, TAC1, and UPC2 of fluconazole-resistant clinical C. albicans isolates.
For the competitive fitness assays, we used 10 matched pairs of fluconazole-susceptible and -resistant isolates from the same patients, which were available in our strain collection in Würzburg (Table 1). The genetic relatedness of these isolate pairs has been demonstrated in previous studies by highly sensitive fingerprinting methods and subsequent sequence analyses of specific genes (see the references cited in Tables 1 and 2). For most of the fluconazole-resistant clinical isolates of this set, GOF mutations in MRR1, TAC1, and/or UPC2 have been described previously (Table 2). Earlier work has shown that the MDR1-overexpressing isolate F5, which contains an MRR1 GOF mutation (9), also overexpresses ERG11 (22). As ERG11 overexpression is frequently caused by UPC2 mutations (19), we sequenced the UPC2 alleles of isolate F5 and the matched susceptible isolate F2. We found that F5 has additionally acquired a G1942A exchange in UPC2 and become homozygous for the mutated allele. The resulting G648S mutation in Upc2 has previously been found in other fluconazole-resistant isolates and shown to cause ERG11 overexpression and increased drug resistance (19). Isolate Gu5 overexpresses CDR1/CDR2 (33); therefore, we sequenced the TAC1 alleles of isolate Gu5 and matched susceptible isolate Gu4. We found that Gu5 had acquired a G2939A substitution in TAC1 and become homozygous for the mutation. The resulting G980E exchange in Tac1 has also been found in other fluconazole-resistant isolates and shown to mediate CDR1/CDR2 overexpression and increased drug resistance (12, 14). Isolate TW17 has long been known to overexpress MDR1 (25). We sequenced the MRR1 alleles of isolate TW17 and matched susceptible isolate TW1 and found that TW17 had become homozygous for a G2839A substitution (the position corresponds to that in the MRR1 reference sequence) that was not present in isolate TW1 and resulted in a G947S mutation in Mrr1. We also found that isolate TW2 from the same series was heterozygous for the mutated allele and a loss of heterozygosity occurred in the next isolate of the series, TW3, in accordance with the increasing MDR1 expression levels reported for these isolates (25).
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Relevant characteristics or genotype | Reference or source |
|---|---|---|---|
| SC5314 | Wild-type reference strain | 46 | |
| Matched clinical isolate pairs | |||
| F2 | FLUs clinical isolate from patient F | 22 | |
| F5 | FLUr clinical isolate from patient F | 22 | |
| G2 | FLUs clinical isolate from patient G | 22 | |
| G5 | FLUr clinical isolate from patient G | 22 | |
| B3 | FLUs clinical isolate from patient B | 33 | |
| B4 | FLUr clinical isolate from patient B | 33 | |
| Gu4 | FLUs clinical isolate from patient Gu | 33 | |
| Gu5 | FLUr clinical isolate from patient Gu | 33 | |
| 1442 | FLUs clinical isolate from patient 9 | 47 | |
| 2271 | FLUr clinical isolate from patient 9 | 47 | |
| 1490 | FLUs clinical isolate from patient 40 | 47 | |
| 1587 | FLUr clinical isolate from patient 40 | 47 | |
| 5833 | FLUs clinical isolate from patient II | 48 | |
| 6692 | FLUr clinical isolate from patient II | 48 | |
| DSY294 (C43)a | FLUs clinical isolate | 49 | |
| DSY296 (C56) | FLUr clinical isolate | 49 | |
| DSY2285 (26) | FLUs clinical isolate | 50 | |
| DSY2286 (91) | FLUr clinical isolate | 50 | |
| TW1 (#1) | FLUs clinical isolate | 51 | |
| TW17 (#17) | FLUr clinical isolate | 51 | |
| RFP-labeled strains | |||
| SCADH1R1A | SC5314 | ADH1/adh1::PADH1-RFP-caSAT1 | 26 |
| F2ADH1R1A and -B | F2 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| F5ADH1R1A and -B | F5 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| G2ADH1R1A and -B | G2 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| G5ADH1R1A and -B | G5 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| B3ADH1R1A and -B | B3 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| B4ADH1R1A and -B | B4 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| Gu4ADH1R1A and -B | Gu4 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| Gu5ADH1R1A and -B | Gu5 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 1442ADH1R1A and -B | 1442 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 2271ADH1R1A and -B | 2271 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 1490ADH1R1A and -B | 1490 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 1587ADH1R1A and -B | 1587 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 5833ADH1R1A and -B | 5833 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| 6692ADH1R1A and -B | 6692 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| DSY294ADH1R1A and -B | DSY294 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| DSY296ADH1R1A and -B | DSY296 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| DSY2285ADH1R1A and -B | DSY2285 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| DSY2286ADH1R1A and -B | DSY2286 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| TW1ADH1R1A and -B | TW1 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| TW17ADH1R1A and -B | TW17 | ADH1/adh1::PADH1-RFP-caSAT1 | This study |
| Genetically engineered strains with MRR1 GOF mutations | |||
| SCMRR1R24A and -B | SC5314 | MRR1-FRT/MRR1-FRT | 26 |
| SCMRR1R34A and -B | SC5314 | MRR1P683S-FRT/MRR1P683S-FRT | 10 |
| SCMRR1R44A and -B | SC5314 | MRR1G997V-FRT/MRR1G997V-FRT | —b |
| SCMRR1R54A and -B | SC5314 | MRR1G878E-FRT/MRR1G878E-FRT | — |
| SCMRR1R64A and -B | SC5314 | MRR1Q350L-FRT/MRR1Q350L-FRT | — |
| SCMRR1R74A and -B | SC5314 | MRR1N803D-FRT/MRR1N803D-FRT | — |
| SCMRR1R84A and -B | SC5314 | MRR1T360I-FRT/MRR1T360I-FRT | — |
| SCMRR1R94A and -B | SC5314 | MRR1K335N-FRT/MRR1K335N-FRT | — |
| SCMRR1R982A and -B | SC5314 | MRR1K335N-FRT/MRR1T360I-FRT | — |
| SCMRR1R104A and -B | SC5314 | MRR1T896I-FRT/MRR1T896I-FRT | — |
| F2MRR1R34A and -B | F2 | MRR1P683S-FRT/MRR1P683S-FRT | This study |
| 5833MRR1R892A and -B | 5833 | MRR1T360I-FRT/MRR1K335N-FRT | This study |
Names in parentheses are previously used strain designations.
—, Hampe and Morschhäuser, unpublished.
TABLE 2.
ZnTF mutations in fluconazole-resistant clinical isolates
| Isolate | MIC (μg/ml) | ZnTF mutationa |
Reference(s) or source | ||
|---|---|---|---|---|---|
| MRR1 | TAC1 | UPC2 | |||
| F2 | 8 | −/− | −/− | ||
| F5 | 128 | P683S/P683S | G648S/G648S | 9, this study | |
| G2 | 1 | −/− | |||
| G5 | 128 | G997V/G997V | 9 | ||
| B3 | 1 | −/− | |||
| B4 | 32 | G878E/G878E | 6 | ||
| 1442 | 0.5 | −/− | |||
| 2271 | 16 | Q350L/Q350L | 6 | ||
| 1490 | 1 | −/− | |||
| 1587 | 4 | N803D/N803D | 6 | ||
| DSY2285 | 1 | −/− | |||
| DSY2286 | 16 | T896I/T896I | 6 | ||
| 5833 | 1 | −/− | |||
| 6692 | 64 | K335N/T360I | 6 | ||
| Gu4 | 4 | −/− | |||
| Gu5 | >256 | G980E/G980E | This study | ||
| DSY294 | 1 | −/− | |||
| DSY296 | 128 | N977D/N977D | 13 | ||
| TW1 | 1 | −/− | −/− | −/− | |
| TW17 | >256 | G947S/G947Sb | A736V/Δ962–969 | −/A643V | 8, 12, 21, this study |
Shown are the amino acid exchanges in the proteins encoded by the two alleles of the genes.
This mutation has recently been reported as G963S (8) but corresponds to G947S in the SC5314 reference sequence, because Mrr1 of TW17 contains 5 tandem repeats of the NPQS sequence (positions 165 to 168 in Mrr1), as confirmed by sequencing in the present study.
Competitive fitness of fluconazole-resistant clinical C. albicans isolates with different resistance mechanisms.
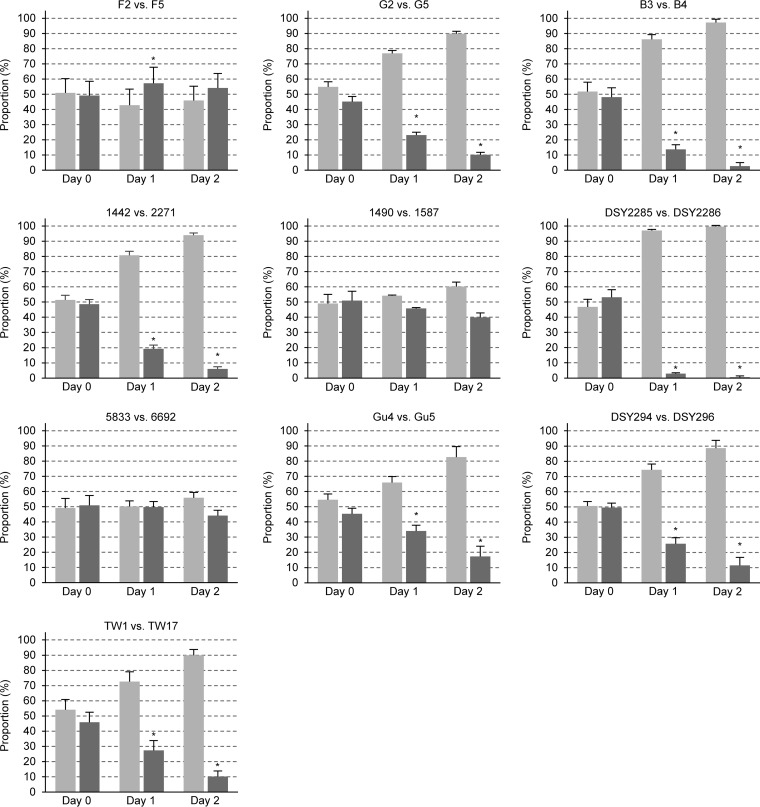
In order to assess the competitive fitness of the fluconazole-resistant clinical isolates containing hyperactive ZnTFs, we generated RFP-labeled derivatives of these isolates and their matched fluconazole-susceptible isolates. Each fluconazole-resistant clinical isolate was tested in competition experiments with two independently constructed RFP-labeled derivatives of the matched fluconazole-susceptible isolate, and, vice versa, each fluconazole-susceptible clinical isolate was tested with two independently constructed RFP-labeled derivatives of the matched fluconazole-resistant isolate. The use of four different mixtures for each isolate pair, combined with marker swapping, ensured that reduced or increased fitness was indeed an intrinsic characteristic of the clinical isolate and not caused by the genetic manipulation. The strains were mixed in roughly equal proportions, and the cultures were grown for 24 h in rich yeast extract-peptone-dextrose (YPD) medium, diluted in fresh medium, and grown for another 24 h in the same medium. The proportions of each strain in the inoculum and after 24 h and 48 h of growth were determined by plating appropriate dilutions and counting the number of white (unlabeled strain) and red (labeled strain) colonies. The results of these competition experiments are summarized in Fig. 1; details of each individual experiment (number of colonies counted, proportions of each strain) can be found in Data Set S1 in the supplemental material. Interestingly, 7 of the 10 fluconazole-resistant isolates had a clear fitness defect under these conditions and were outcompeted by their matched fluconazole-susceptible isolate. These included isolates G5, B4, 2271, and DSY2286 containing different GOF mutations in MRR1; isolates Gu5 and DSY296 with hyperactive TAC1 alleles; and isolate TW17, which contained GOF mutations in MRR1, TAC1, and UPC2. The reduced fitness that is associated with drug resistance in these isolates may be caused by the hyperactive transcription factors, but other genomic alterations may also play a role. In contrast, isolates F5, 1587, and 6692 did not exhibit a significant fitness defect in these experiments, although all of them contained GOF mutations in MRR1, and isolate F5 additionally contained a GOF mutation in UPC2. Apparently, these isolates could somehow compensate for the negative effect caused by Mrr1 hyperactivity.
FIG 1.
Competitive fitness of matched pairs of fluconazole-resistant isolates (dark gray bars) and fluconazole-susceptible isolates (light gray bars) from the same patients. In each case, the susceptible isolate was mixed with two independently generated RFP-labeled derivatives of the matched resistant isolate and the resistant isolate was mixed with two independently generated RFP-labeled derivatives of the matched susceptible isolate. Shown are the relative proportions of the two strains in the inoculum (day 0) and after two rounds of coculture (day 1 and day 2) at 30°C in YPD medium. Results are the means and standard deviations for the four cocultures. For isolate pair F2/F5, each combination was tested four times; i.e., results are from 16 cocultures. Significant differences in the proportions of the resistant isolates at day 1 and day 2 compared to day 0 are indicated by asterisks (P < 0.05). See Data Set S1 for details.
Fitness effects of MRR1 GOF mutations in isogenic strains.
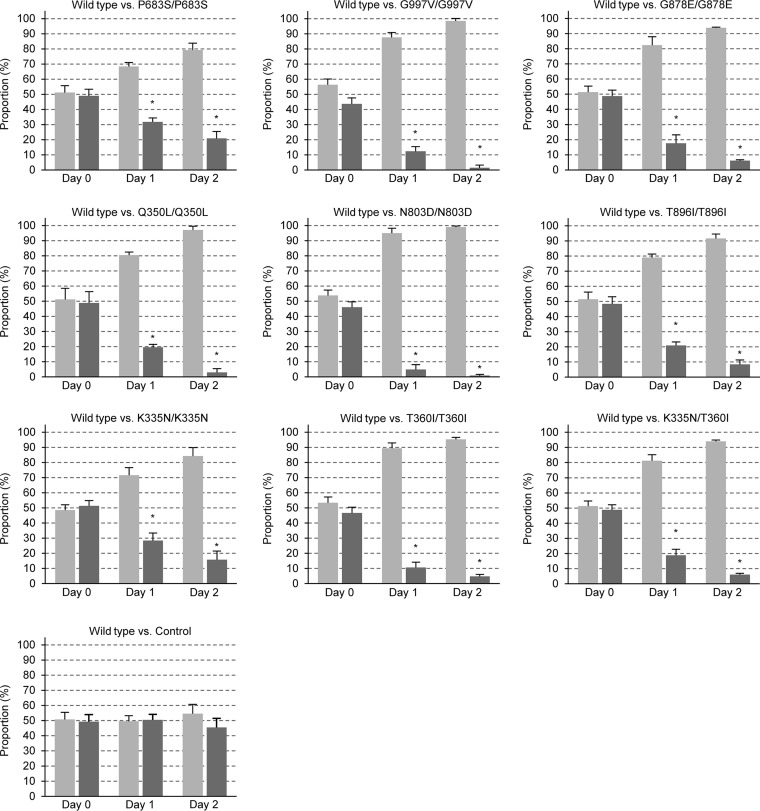
Although the P683S mutation in Mrr1 of isolate F5 had previously been shown to cause a mild fitness defect when introduced into the fluconazole-susceptible C. albicans reference strain SC5314 (26), it remained possible that the N803D mutation contained in Mrr1 of isolate 1587 and the K335N and T360I mutations found in Mrr1 of isolate 6692 (this isolate contained two different hyperactive MRR1 alleles) do not cause such a fitness defect, which would also explain the unaltered fitness of these fluconazole-resistant isolates. We therefore assessed the effects of these and various other MRR1 GOF mutations found in fluconazole-resistant clinical isolates in an isogenic background. For this purpose, we used a set of SC5314 derivatives in which both endogenous wild-type MRR1 alleles had been replaced by different hyperactive alleles to compare their effects on the expression of specific Mrr1 target genes (I. Hampe and J. Morschhäuser, unpublished data). The competitive fitness of these strains, each of which was constructed two times independently, was then tested in coculture experiments with an RFP-labeled derivative of the parental strain SC5314 in the same way as for the clinical isolate pairs. Figure 2 shows that all MRR1 GOF mutations caused a significant fitness defect during growth in rich YPD medium; in fact, the previously tested P683S mutation had the weakest effect, and all other tested mutations reduced the fitness of strain SC5314 even more strongly. Importantly, this was also true for the MRR1 GOF mutations found in isolates 1587 (N803D) and 6692 (K335N and T360I, either alone or in combination, as in the clinical isolate), demonstrating that the impact of a hyperactive Mrr1 on fitness depends on the strain background. Two independently constructed control strains, in which the endogenous MRR1 alleles of strain SC5314 were replaced in an identical fashion by a nonmutated allele, exhibited wild-type fitness, in agreement with our previous findings (26), confirming that fitness defects were caused by the MRR1 mutations.
FIG 2.
Competitive fitness of isogenic strains containing mutated MRR1 alleles. An RFP-labeled derivative of strain SC5314 (SCADH1R1A, light gray bars) was mixed with two independently generated strains in which the endogenous MRR1 alleles in strain SC5314 were replaced by the indicated mutant alleles or a wild-type copy without a mutation (dark gray bars). Shown are the relative proportions of the strains in the inoculum (day 0) and after two rounds of coculture (day 1 and day 2) at 30°C in YPD medium. Results are the means and standard deviations from four coculture experiments (two with strain A and two with strain B). The control strains were each tested six times; i.e., results are from 12 cocultures. Significant differences in the proportions of the test strains at day 1 and day 2 compared to day 0 are indicated by asterisks (P < 0.05). See Data Set S2 for details.
Fitness effects of MRR1 GOF mutations in clinical isolates.
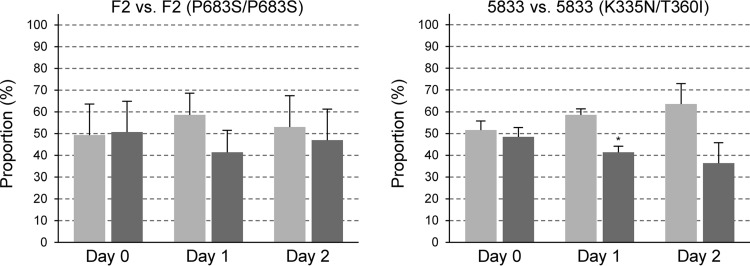
Our finding that the P683S, N803D, and K335N/T360I GOF mutations in Mrr1 caused a fitness defect when introduced into strain SC5314 but not in the fluconazole-resistant isolates F5, 1587, and 6692, in which they were originally found, suggested that the last three strains had overcome the fitness costs imposed by the hyperactive Mrr1 during further evolution. Alternatively, the MRR1 mutations may not have caused a fitness defect when they were acquired by the clinical isolates because of previous adaptations that allowed the strains to tolerate the presence of a hyperactive Mrr1. To distinguish between these possibilities, we introduced the MRR1 mutations found in the fluconazole-resistant isolates F5 and 6692 into the matched susceptible isolates F2 and 5833, respectively. As expected, the introduction of the P683S mutation into both MRR1 alleles of isolate F2 resulted in enhanced fluconazole resistance (the MIC increased from 8 μg/ml to 64 μg/ml). Similarly, the introduction of the K335N and T360I mutations into the MRR1 alleles of isolate 5833 also increased the MIC of fluconazole from 1 μg/ml to 16 μg/ml. To assess the effect of the mutations on the fitness of the strains, two independently constructed strains with the MRR1 GOF mutations were used in competition experiments with each of the two independently generated RFP-labeled derivatives of the respective parental clinical isolate. Surprisingly, the introduction of the MRR1 GOF mutations into the clinical isolates did not affect or only weakly affected the fitness of the strains (Fig. 3), indicating that adaptation mechanisms that were already in place in these strains circumvented the fitness costs that are normally caused by a hyperactive Mrr1.
FIG 3.
Competitive fitness of fluconazole-susceptible clinical isolates F2 and 5833 (light gray bars) and genetically engineered derivatives carrying the MRR1 GOF mutations from matched resistant isolates F5 and 6692, respectively (dark gray bars). Two independently generated RFP-labeled derivatives of isolates F2 and 5833 were each mixed with two independently constructed corresponding mutants; i.e., four coculture experiments were performed for each comparison. Shown are the relative proportions of the strains in the inoculum (day 0) and after two rounds of coculture (day 1 and day 2) at 30°C in YPD medium. Results are the means and standard deviations from the four coculture experiments. Significant differences in the proportions of the strains with MRR1 GOF mutations at day 1 and day 2 compared to day 0 are indicated by asterisks (P < 0.05). See Data Set S3 for details.
In vivo fitness of fluconazole-resistant clinical C. albicans isolates.
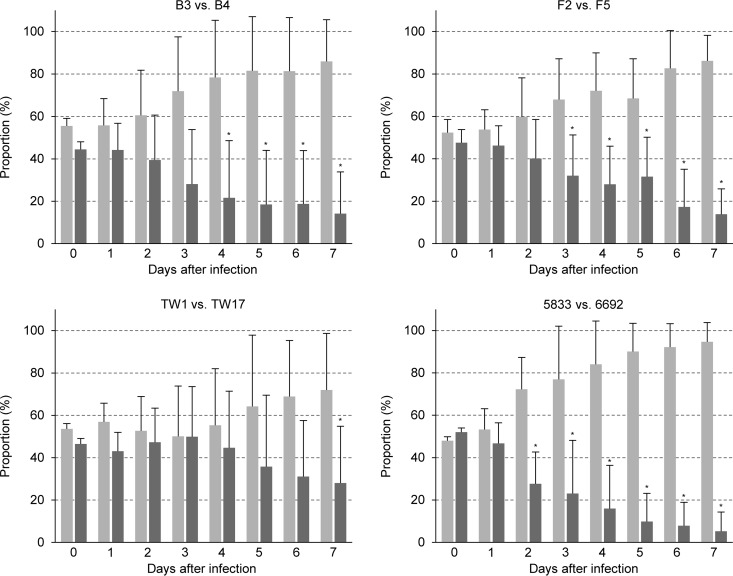
The fitness tests described above were performed in rich YPD medium, which provides optimal conditions that allow fast growth of C. albicans but is highly different from the environments encountered by the fungus in a mammalian host. Indeed, in a mouse model of gastrointestinal colonization, a significantly reduced fitness was previously observed only for genetically engineered strains containing several hyperactive ZnTFs and not for strains containing only one of the three hyperactive TFs, Mrr1, Tac1, or Upc2 (26). We therefore tested the competitive fitness of several fluconazole-resistant clinical C. albicans isolates and their matched susceptible isolates in the same model. We selected two fluconazole-resistant isolates that displayed reduced fitness in vitro (B4, TW17) and two fluconazole-resistant isolates that did not exhibit a fitness defect in YPD medium (F5, 6692) for the in vivo fitness tests. Isolate B4 also exhibited a significantly reduced fitness compared with its matched susceptible isolate B3 in this in vivo model, whereas only a slight fitness defect was observed for isolate TW17 (Fig. 4, left). Notably, both isolates F5 and 6692, which did not exhibit a fitness defect in the in vitro experiments, showed significantly reduced competitive fitness during colonization of the mouse gastrointestinal tract (Fig. 4, right). Therefore, the relative fitness of fluconazole-resistant C. albicans isolates during growth in rich YPD medium in vitro does not predict their fitness in a mammalian host.
FIG 4.
Competitive fitness of fluconazole-susceptible isolates F2, B3, 5833, and TW1 (light gray bars) and matched fluconazole-resistant isolates F5, B4, 6692, and TW17 (dark gray bars), respectively, in a mouse model of gastrointestinal colonization. In each case, the susceptible isolate was mixed with two independently generated RFP-labeled derivatives of the matched resistant isolate and the resistant isolate was mixed with two independently generated RFP-labeled derivatives of the matched susceptible isolate. For all comparisons, each of the four mixtures was used to infect 3 mice (4 mice were infected with the 5833/6692ADH1R1A and TW1/TW17ADH1R1A pairs); i.e., the competitive fitness of each isolate pair was tested in 12 mice (13 mice for 5833/6692 and TW1/TW17). Shown are the relative proportions of the strains in the inoculum (day 0) and in samples recovered at the indicated times from the feces of the animals. Results are the means and standard deviations from the 12 (or 13) coinfection experiments. Significant differences in the proportions of resistant isolates after passage through the gastrointestinal tract compared to the inoculum are indicated by asterisks (P < 0.05). See Data Set S4 for details.
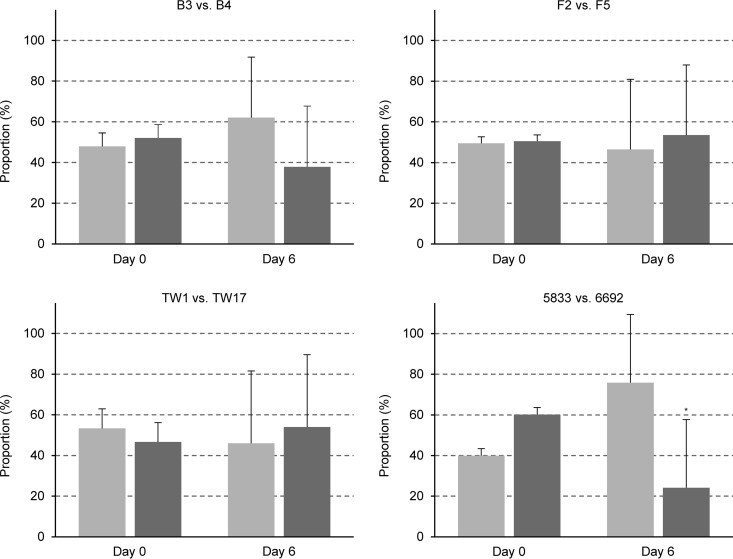
As C. albicans is exposed to highly variable environments during colonization and infection, we also investigated the competitive fitness of the same drug-resistant isolates in a mouse model of disseminated candidiasis. Interestingly, three of the four resistant isolates (B4, F5, TW17) did not exhibit a fitness defect in this model, because their relative proportion in samples recovered after 6 days from infected kidneys was similar to that in the inoculum, and only isolate 6692 displayed a slightly reduced fitness (Fig. 5). These results demonstrate that the fitness costs of drug resistance may become obvious in some but not all host niches colonized or infected by C. albicans.
FIG 5.
Competitive fitness of fluconazole-susceptible isolates F2, B3, 5833, and TW1 (light gray bars) and matched fluconazole-resistant isolates F5, B4, 6692, and TW17 (dark gray bars), respectively, in a mouse model of disseminated candidiasis. In each case, the susceptible isolate was mixed with two independently generated RFP-labeled derivatives of the matched resistant isolate and the resistant isolate was mixed with two independently generated RFP-labeled derivatives of the matched susceptible isolate. For all comparisons, each of the four mixtures was used to infect 3 mice (5 mice were infected with the B3/B4ADH1R1B pair); i.e., the competitive fitness of each isolate pair was tested in 12 mice (14 mice for B3/B4). Shown are the relative proportions of the strains in the inoculum (day 0) and in the kidneys after 6 days of infection. Results are the means and standard deviations of the 12 (or 14) coinfection experiments. Significant differences in the proportions of the resistant isolates in the kidneys compared to the inoculum are indicated by asterisks (P < 0.05). See Data Set S5 for details.
DISCUSSION
In a previous study, using genetically engineered, isogenic strains, we showed that hyperactive forms of the ZnTFs Mrr1, Tac1, and Upc2, which confer increased fluconazole resistance, also cause a mild but significant competitive fitness defect during growth in standard YPD medium in the absence of the drug (26). The reduced fitness of strains with a single hyperactive TF was conditional, because it was not observed during growth in minimal medium or in a mouse model of gastrointestinal colonization. However, the fitness defect was exacerbated in strains containing two or all three hyperactive TFs and then became apparent under all tested in vitro and in vivo conditions. Other studies found that many fluconazole-resistant clinical C. albicans isolates did not exhibit a fitness defect during growth in vitro or in a mammalian host (29, 32), suggesting that drug-resistant strains can overcome the fitness costs imposed by hyperactive TFs. Here, we assessed the competitive fitness of a set of fluconazole-resistant clinical isolates containing different GOF mutations in Mrr1, Tac1, and Upc2 to investigate if the mitigation of fitness costs is a common phenomenon in such strains. Interestingly, we found that most fluconazole-resistant isolates containing hyperactive forms of these TFs also displayed reduced competitive fitness when grown in vitro in rich YPD medium, indicating that the negative effects of the deregulated gene expression are not easily circumvented. It should be pointed out, however, that the clinical strains isolated from patients came from a different environment, the human oral cavity, and reduced fitness during cultivation in rich medium may not be relevant in their natural habitat. Nevertheless, some of the fluconazole-resistant clinical isolates did not exhibit a detectable fitness defect in YPD medium. All these isolates contained GOF mutations in MRR1 that caused a significant fitness defect when they were introduced into the reference strain SC5314. We hypothesized that the clinical isolates had overcome the fitness costs incurred by these mutations through compensatory changes during further evolution. However, we found that the same mutations did not significantly affect fitness when introduced into the matched fluconazole-susceptible isolates. Therefore, these clinical isolates already exhibited a genetic background in which the acquisition of a hyperactive Mrr1 had no negative impact on their ability to grow in rich medium. Previous work has shown that the effect of a particular hyperactive MRR1 allele on gene expression depends on the strain background. Although a core set of genes was upregulated after introduction of the P683S and G997V mutations both into strain SC5314 and into the fluconazole-resistant clinical isolates F5 and G5 containing the same mutations, not all Mrr1 target genes were upregulated in the clinical isolates, which in turn differentially expressed other genes in an Mrr1-dependent manner as well as Mrr1-independent genes (9, 10). It is conceivable that such differences may modulate the impact of a hyperactive Mrr1 on the fitness of the strains.
Some apparent discrepancies between the findings of our previous study (26) and the present studies, on the one hand, and two reports by other researchers, on the other hand, should be discussed. We found that, when introduced into strain SC5314, all tested Mrr1 GOF mutations (eight different mutations) as well as GOF mutations in Tac1 and Upc2 caused a significant fitness defect during growth in YPD medium. In contrast, Lohberger et al. did not observe an effect of GOF mutations in these TFs on in vitro fitness in the same strain background (8). Of note, these authors used a modified yeast extract-peptone-dextrose medium containing only half the standard amount of peptone (1% instead of 2%) and yeast extract (0.5% instead of 1%), so the absence of an observable fitness defect in this medium may reflect the absence of a fitness defect in minimal medium in the study by Sasse et al. (26). In addition, cocultures were started with 3.7 × 106 cells/ml in the study by Lohberger et al. (8), which amounts to only ca. 7 generations until stationary phase is reached, whereas the cocultures were grown for ca. 14 to 15 generations (starting from an optical density of 0.002) in the study by Sasse et al. (26). In the present study, we performed two rounds of coculture (2 × 14 generations), which potentiated the growth differences of the competing strains and made fitness defects even more obvious (Fig. 2). In our present study, we also observed a significant fitness defect of clinical isolate TW17 compared to the fitness of the initial isolate of this series, isolate TW1, during coculture in YPD medium. Considering that TW17 contains hyperactive forms of all three ZnTFs, Mrr1, Tac1, and Upc2, this was not unexpected. In contrast, a recent study by Ford et al. reported a similar in vitro fitness of these two isolates (32). There are several possible explanations for the conflicting results. In the study by Ford et al. (32), isolates TW1 and TW17 were not directly competed with one another but were separately tested in competition experiments with an unrelated, genetically marked strain, whereas we directly compared the fitness of these two isolates and labeled derivatives in coculture experiments. Furthermore, in the study by Ford et al. (32), competition experiments were performed in RPMI medium, a poorer growth medium in which the strains grow much more slowly, and strains were grown for only 5 to 10 generations, which may additionally mask fitness defects.
An important finding of our present study was that the relative fitness of the fluconazole-resistant clinical isolates during growth in YPD medium did not correlate with their competitive fitness in a mammalian host. Even isolates F5 and 6692, which did not exhibit a fitness defect in the in vitro experiments, had significantly reduced fitness in the mouse model of gastrointestinal colonization. Conversely, isolate TW17, which displayed a clear fitness defect in YPD medium, could compete comparatively well with its matched fluconazole-susceptible isolate TW1 in this in vivo model. Furthermore, in the mouse model of disseminated candidiasis, all tested resistant isolates showed no fitness defects or only a minor fitness defect and infected the kidneys as efficiently as the matched susceptible isolates or only slightly less well. It should also be noted that there was a high variability in the observed relative fitness of a particular C. albicans isolate between individual animals in both infection models, something that has been observed in similar studies by other researchers (34–37). Even in animals infected with the same mixed inoculum, the generally less fit isolate sometimes dominated the population (see Data Sets S4 and S5 in the supplemental material). This may have been caused by infection bottlenecks, but it is also possible that the constitutive overexpression of the target genes of a hyperactive TF may confer a selective advantage under particular conditions encountered in individual animals, similar to the advantage conferred in the presence of the drug fluconazole. Another aspect that must be stressed is that the environment and the selective pressures encountered by the fungus in a human host are different from those encountered in mouse colonization and infection models. All the clinical isolates used in our present study were obtained from HIV-infected patients with oropharyngeal candidiasis. Even a mouse model of oral candidiasis lacks certain aspects of a human infection, for example, the presence of histatin 5, an antimicrobial peptide that is highly active against C. albicans and produced only by humans and other primates (38). In addition, the ability of C. albicans to colonize the oral cavity and other host niches depends on the immune status and the composition of the competing bacterial microflora, which differs not only between mice and humans but also between individuals. Therefore, the fitness costs of drug resistance probably also depend on the individual host.
Although the fitness defects of fluconazole-resistant isolates depended on the growth conditions and the colonization/infection model, it is noteworthy that we did not observe an increased fitness of any of the clinical isolates that had acquired drug resistance. This is in contrast to what has been reported for C. glabrata, where GOF mutations in Pdr1 cause not only azole resistance but also increased virulence (39–42). Considering that the fluconazole-resistant C. albicans isolates displayed reduced fitness under at least some conditions but never showed increased fitness compared to that of the matched susceptible isolates under any of the tested conditions, it is likely that, in most cases, fluconazole-resistant mutants will be outcompeted in the long run by the susceptible cells in a population in the absence of the drug.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 17.2% glycerol at −80°C and subcultured on YPD agar plates (10 g yeast extract, 20 g peptone, 20 g glucose, 15 g agar per liter) at 30°C. The strains were routinely grown in YPD liquid medium at 30°C in a shaking incubator. Selection of nourseothricin-resistant transformants and recycling of the SAT1 flipper cassette were performed as described previously (43).
Strain constructions.
C. albicans strains were transformed by electroporation (44) with gel-purified inserts from the following plasmids: pADH1R1 contains the Candida-adapted SAT1 (caSAT1) marker and the RFP gene (which encodes red fluorescent protein [RFP]) under the control of the ADH1 promoter (26). The insert from this plasmid was used to label C. albicans clinical isolates with RFP. Plasmid pMRR1R3 contains a mutated MRR1 allele with the P683S GOF mutation and the recyclable SAT1 flipper cassette and has been previously described (10). Plasmids pMRR1R8 and pMRR1R9 are otherwise identical but contain the T360I and K335N GOF mutations, respectively, instead of the P683S mutation (Hampe and Morschhäuser, unpublished). The insert from pMRR1R3 was used to replace both endogenous MRR1 alleles in clinical isolate F2 by the MRR1 allele with the P683S mutation (the MRR1P683S allele), and the inserts from pMRR1R8 and pMRR1R9 were used to replace one of the endogenous MRR1 alleles in clinical isolate 5833 by the MRR1K335N allele and the other endogenous allele by the MRR1T360I allele. The correct integration of each construct as well as recycling of the SAT1 flipper cassette was confirmed by Southern hybridization. Introduction of the MRR1 GOF mutations was verified by reamplification of the genes from heterozygous and homozygous mutants and sequencing of the PCR products. In each case, two independent series of strains (A and B; see Table 1) were generated and used for further analysis.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from the C. albicans strains was isolated as described previously (43). The DNA was digested with appropriate restriction enzymes, separated on a 1% agarose gel, transferred by vacuum blotting onto a nylon membrane, and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence-labeled probes was performed with an Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, UK) according to the instructions of the manufacturer.
Sequence analysis of MRR1, TAC1, and UPC2 in clinical isolates.
To identify the UPC2 GOF mutation in ERG11-overexpressing isolate F5, the UPC2 coding sequence was amplified from genomic DNA of RFP-labeled F2 and F5 derivatives with primers UPC2-1 and UPC2-2, and the PCR products were sequenced with primers UPC2-1, UPC2-4B, and UPC2-9 (the oligonucleotide primers used in this study are listed in Table 3). To identify the TAC1 GOF mutation in CDR1/CDR2-overexpressing isolate Gu5, a part of the TAC1 coding sequence was amplified from genomic DNA of isolates Gu4 and Gu5 with primers TAC1-7 and TAC1-14, and the PCR products were sequenced with primer TAC1-12A. To identify the MRR1 GOF mutation in MDR1-overexpressing isolates of the TW series, a part of the MRR1 coding sequence was amplified from genomic DNA of an RFP-labeled TW17 derivative and from isolates TW1, TW2, and TW3 with primers ZCF36-3 and ZCF36-6, and the PCR products were sequenced with primer ZCF36seq4.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| TAC1-7 | TTTTGGGCCCTGGTGAAATTCCGAACC |
| TAC1-12A | GTTGAGAGTAGTGATCAAAG |
| TAC1-14 | TTAAGAGCTCGCAGTACATATAATAAAGTGGG |
| UPC2-1 | ATATCTCGAGAATGATGATGACAGTGAAACAAGAATC |
| UPC2-2 | ATATAGATCTATTTCATATTCATAAACCCATTATC |
| UPC2-4B | GCATTCAATACTTGCCTTTAGTGC |
| UPC2-9 | ATATGGCGCCCCAACTAATCCACTTAGTGCTTTG |
| ZCF36-3 | GAATAATTCGGAGCTCAATTTGCGTTTAGCC |
| ZCF36-6 | ATATTGGGCCCGCTACCATAAGCCTCGCTCG |
| ZCF36seq4 | GTTGGAATTGCAGCTGTATCC |
Fluconazole susceptibility assays.
The fluconazole susceptibilities of the strains were determined by a previously described broth microdilution method (45), with slight modifications. A 2-day-old colony from a YPD agar plate was suspended in 2 ml of a 0.9% NaCl solution, and 4 μl of the suspension was mixed with 2 ml 2× SD-CSM medium (13.4 g yeast nitrogen base with ammonium sulfate [YNB; MP Biomedicals, Illkirch, France], 40 g glucose, 1.58 g complete supplement medium [CSM; MP Biomedicals]). A 2-fold dilution series of fluconazole (Sigma GmbH, Deisenhofen, Germany) was prepared in water, starting from an initial concentration of 512 μg/ml. One hundred microliters of each fluconazole solution was then mixed with 100 μl of the cell suspension in a 96-well microtiter plate, and the plates were incubated for 48 h at 37°C. The MIC of fluconazole was defined as the drug concentration that abolished or drastically reduced visible growth compared to that of a drug-free control.
In vitro competition experiments.
Strains were grown overnight in YPD medium at 30°C. The fluconazole-susceptible clinical isolates were mixed in a 1:1 ratio with RFP-labeled derivatives of the matched fluconazole-resistant isolates, and the fluconazole-resistant clinical isolates were mixed in a 1:1 ratio with RFP-labeled derivatives of the matched fluconazole-susceptible isolates. Similarly, genetically engineered derivatives of strain SC5314 or clinical isolates F2 and 5833 containing introduced MRR1 GOF mutations were mixed in a 1:1 ratio with RFP-labeled derivatives of their parental strains. The mixtures were diluted in fresh YPD medium at a starting optical density of 0.002 and grown at 30°C. After 24 h, the cocultures were diluted to an optical density of 0.002 in fresh YPD medium and grown for another 24 h at 30°C. Dilution series were prepared from each coculture at the beginning and after 24 h and 48 h of coincubation, spread on YPD plates, and grown for 2 days at 30°C. The percentage of red colonies (RFP-labeled strains) and white colonies (unlabeled strains) was determined after storage of the plates at 4°C, which enhanced the color of the RFP-expressing colonies.
Fitness test in a mouse model of gastrointestinal colonization.
Female BALB/c mice (6 to 8 weeks old; Janvier Labs, Saint-Berthevin, France) were fed 1 mg/ml tetracycline, 2 mg/ml streptomycin, and 0.1 mg/ml gentamicin in their drinking water, starting from day 4 prior to infection. C. albicans strains were grown overnight in YPD medium at 30°C, washed two times in phosphate-buffered saline (PBS), and adjusted to a density of 109 cells per ml. Competing strains were mixed in equal proportions, and 50 μl containing approximately 5 × 107 cells of the mixed suspensions was intragastrically applied to the mice (usually three mice per strain pair). After 24 h and on the following days, the feces of the mice were collected and homogenized in PBS. A dilution series was prepared and spread on YPD plates containing 50 μg/ml chloramphenicol. The proportions of both strains in the inoculum and in the populations recovered on each following day from the feces were determined by counting the number of red and white colonies, as for the in vitro competition experiments. It has been shown previously that the numbers of CFU in fecal pellets reflect the colonization of different parts of the gastrointestinal tract in this model (34). For each comparison of a matched fluconazole-susceptible and fluconazole-resistant isolate pair, two additional mice were infected with single RFP-labeled and unlabeled strains to verify that they maintained their characteristic colony phenotypes during the infection and exclude the possibility of contaminations.
Fitness test in a mouse model of disseminated candidiasis.
C. albicans strains were grown overnight in YPD medium at 30°C, washed three times in PBS, and adjusted to a density of 2 × 106 cells per ml. Competing strains were mixed in a 1:1 ratio, and 100 μl of the mixed suspensions was used for infection of at least 3 female BALB/c mice via the lateral tail vein. As a control, one mouse was infected with a single RFP-labeled strain for each experiment. Six days after the infection, the kidneys were collected and homogenized in sterile PBS. A dilution series was prepared and spread on YPD plates containing 50 μg/ml chloramphenicol. The proportions of both strains in the inoculum and in the populations recovered from the kidneys were determined by counting the number of red and white colonies.
Statistical analysis.
Since in vitro fitness assays were performed under controlled conditions (in YPD medium at 30°C), we assumed that the collected data fit a standard normal distribution. The paired t test (two-tailed) was used to compare the relative proportions of the strains in the inoculum and after 1 and 2 days of coculture. For the in vivo competition experiments, the Wilcoxon signed-rank test was applied to compare the relative proportions of the strains in the inoculum and in the populations recovered from infected mice. All statistical tests were conducted using GraphPad Prism (version 6.07) software.
Ethics statement.
The animal studies and protocols (permission number AZ 2531.01-07/12) were approved by the local government of Lower Franconia, Germany. All animal studies were performed in strict accordance with the guidelines for animal care and experimentation of the German Animal Protection Law and with EU directive 2010/63/EU.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alix Coste for providing C. albicans isolates TW1, TW2, TW3, and TW17.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grant MO 846/6) and the National Institutes of Health (NIH grant AI058145).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00584-17.
REFERENCES
- 1.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11:382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morschhäuser J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim Biophys Acta 1587:240–248. doi: 10.1016/S0925-4439(02)00087-X. [DOI] [PubMed] [Google Scholar]
- 3.Morschhäuser J. 2010. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet Biol 47:94–106. doi: 10.1016/j.fgb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Sanglard D, Coste A, Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res 9:1029–1050. doi: 10.1111/j.1567-1364.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- 5.Branco J, Silva AP, Silva RM, Silva-Dias A, Pina-Vaz C, Butler G, Rodrigues AG, Miranda IM. 2015. Fluconazole and voriconazole resistance in Candida parapsilosis is conferred by gain-of-function mutations in MRR1 transcription factor gene. Antimicrob Agents Chemother 59:6629–6633. doi: 10.1128/AAC.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunkel N, Blaß J, Rogers PD, Morschhäuser J. 2008. Mutations in the multi-drug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol Microbiol 69:827–840. doi: 10.1111/j.1365-2958.2008.06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddouzi J, Parker JE, Vale-Silva LA, Coste A, Ischer F, Kelly S, Manai M, Sanglard D. 2013. Molecular mechanisms of drug resistance in clinical Candida species isolated from Tunisian hospitals. Antimicrob Agents Chemother 57:3182–3193. doi: 10.1128/AAC.00555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohberger A, Coste AT, Sanglard D. 2014. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot Cell 13:127–142. doi: 10.1128/EC.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morschhäuser J, Barker KS, Liu TT, Blaß-Warmuth J, Homayouni R, Rogers PD. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog 3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, Morschhäuser J. 2011. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 55:2212–2223. doi: 10.1128/AAC.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert S, Rogers PD, Morschhäuser J. 2008. Gain-of-function mutations in the transcription factor MRR1 are responsible for overexpression of the MDR1 efflux pump in fluconazole-resistant Candida dubliniensis strains. Antimicrob Agents Chemother 52:4274–4280. doi: 10.1128/AAC.00740-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coste A, Selmecki A, Forche A, Diogo D, Bougnoux ME, d'Enfert C, Berman J, Sanglard D. 2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot Cell 6:1889–1904. doi: 10.1128/EC.00151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. 2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coste AT, Crittin J, Bauser C, Rohde B, Sanglard D. 2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot Cell 8:1250–1267. doi: 10.1128/EC.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coste AT, Karababa M, Ischer F, Bille J, Sanglard D. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot Cell 3:1639–1652. doi: 10.1128/EC.3.6.1639-1652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhäuser J, Nantel A, Raymond M, Rogers PD. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell 6:2122–2138. doi: 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Znaidi S, De Deken X, Weber S, Rigby T, Nantel A, Raymond M. 2007. The zinc cluster transcription factor Tac1p regulates PDR16 expression in Candida albicans. Mol Microbiol 66:440–452. doi: 10.1111/j.1365-2958.2007.05931.x. [DOI] [PubMed] [Google Scholar]
- 18.Dunkel N, Liu TT, Barker KS, Homayouni R, Morschhäuser J, Rogers PD. 2008. A gain-of-function mutation in the transcription factor Upc2p causes upregulation of ergosterol biosynthesis genes and increased fluconazole resistance in a clinical Candida albicans isolate. Eukaryot Cell 7:1180–1190. doi: 10.1128/EC.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, Gygax SE, Morschhäuser J, Rogers PD. 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11:1289–1299. doi: 10.1128/EC.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilmann CJ, Schneider S, Barker KS, Rogers PD, Morschhäuser J. 2010. An A643T mutation in the transcription factor Upc2p causes constitutive ERG11 upregulation and increased fluconazole resistance in Candida albicans. Antimicrob Agents Chemother 54:353–359. doi: 10.1128/AAC.01102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoot SJ, Smith AR, Brown RP, White TC. 2011. An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob Agents Chemother 55:940–942. doi: 10.1128/AAC.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franz R, Kelly SL, Lamb DC, Kelly DE, Ruhnke M, Morschhäuser J. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother 42:3065–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perea S, Lopez-Ribot JL, Kirkpatrick WR, McAtee RK, Santillan RA, Martinez M, Calabrese D, Sanglard D, Patterson TF. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother 45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard D, Ischer F, Koymans L, Bille J. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother 42:241–253. doi: 10.1093/jac/42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41:1482–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasse C, Dunkel N, Schäfer T, Schneider S, Dierolf F, Ohlsen K, Morschhäuser J. 2012. The stepwise acquisition of fluconazole resistance mutations causes a gradual loss of fitness in Candida albicans. Mol Microbiol 86:539–556. doi: 10.1111/j.1365-2958.2012.08210.x. [DOI] [PubMed] [Google Scholar]
- 27.Hayama K, Ishibashi H, Ishijima SA, Niimi K, Tansho S, Ono Y, Monk BC, Holmes AR, Harding DR, Cannon RD, Abe S. 2012. A d-octapeptide drug efflux pump inhibitor acts synergistically with azoles in a murine oral candidiasis infection model. FEMS Microbiol Lett 328:130–137. doi: 10.1111/j.1574-6968.2011.02490.x. [DOI] [PubMed] [Google Scholar]
- 28.Schulz B, Weber K, Schmidt A, Borg-von Zepelin M, Ruhnke M. 2011. Difference in virulence between fluconazole-susceptible and fluconazole-resistant Candida albicans in a mouse model. Mycoses 54:e522–. doi: 10.1111/j.1439-0507.2010.01970.x. [DOI] [PubMed] [Google Scholar]
- 29.Andes D, Forrest A, Lepak A, Nett J, Marchillo K, Lincoln L. 2006. Impact of antimicrobial dosing regimen on evolution of drug resistance in vivo: fluconazole and Candida albicans. Antimicrob Agents Chemother 50:2374–2383. doi: 10.1128/AAC.01053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/S1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 31.Maisnier-Patin S, Andersson DI. 2004. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol 155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 32.Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao RP, Berman J, Thompson DA, Regev A. 2015. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 4:e00662. doi: 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franz R, Ruhnke M, Morschhäuser J. 1999. Molecular aspects of fluconazole resistance development in Candida albicans. Mycoses 42:453–458. doi: 10.1046/j.1439-0507.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- 34.White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, Whiteway M, Mecsas J, Kumamoto CA. 2007. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog 3:e184. doi: 10.1371/journal.ppat.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez JC, Kumamoto CA, Johnson AD. 2013. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11:e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce JV, Dignard D, Whiteway M, Kumamoto CA. 2013. Normal adaptation of Candida albicans to the murine gastrointestinal tract requires Efg1p-dependent regulation of metabolic and host defense genes. Eukaryot Cell 12:37–49. doi: 10.1128/EC.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce JV, Kumamoto CA. 2012. Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations. mBio 3:e00117-12. doi: 10.1128/mBio.00117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puri S, Edgerton M. 2014. How does it kill?: understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell 13:958–964. doi: 10.1128/EC.00095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrari S, Ischer F, Calabrese D, Posteraro B, Sanguinetti M, Fadda G, Rohde B, Bauser C, Bader O, Sanglard D. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog 5:e1000268. doi: 10.1371/journal.ppat.1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari S, Sanguinetti M, Torelli R, Posteraro B, Sanglard D. 2011. Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS One 6:e17589. doi: 10.1371/journal.pone.0017589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vale-Silva L, Ischer F, Leibundgut-Landmann S, Sanglard D. 2013. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infect Immun 81:1709–1720. doi: 10.1128/IAI.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vale-Silva LA, Moeckli B, Torelli R, Posteraro B, Sanguinetti M, Sanglard D. 2016. Upregulation of the adhesin gene EPA1 mediated by PDR1 in Candida glabrata leads to enhanced host colonization. mSphere 1:e00065-15. doi: 10.1128/mSphere.00065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuß O, Vik Å, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 44.Köhler GA, White TC, Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol 179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhnke M, Eigler A, Tennagen I, Geiseler B, Engelmann E, Trautmann M. 1994. Emergence of fluconazole-resistant strains of Candida albicans in patients with recurrent oropharyngeal candidosis and human immunodeficiency virus infection. J Clin Microbiol 32:2092–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Ribot JL, McAtee RK, Lee LN, Kirkpatrick WR, White TC, Sanglard D, Patterson TF. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother 42:2932–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saidane S, Weber S, De Deken X, St-Germain G, Raymond M. 2006. PDR16-mediated azole resistance in Candida albicans. Mol Microbiol 60:1546–1562. doi: 10.1111/j.1365-2958.2006.05196.x. [DOI] [PubMed] [Google Scholar]
- 49.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother 39:2378–2386. doi: 10.1128/AAC.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calabrese D, Bille J, Sanglard D. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 146:2743–2754. doi: 10.1099/00221287-146-11-2743. [DOI] [PubMed] [Google Scholar]
- 51.White TC, Pfaller MA, Rinaldi MG, Smith J, Redding SW. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis 3(Suppl 1):S102–S109. doi: 10.1111/j.1601-0825.1997.tb00336.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.