ABSTRACT
Sequence analysis of 79 ciprofloxacin-resistant Campylobacter jejuni isolates collected in China showed resistance-related sequence variations in gyrA and CmeR-Box. All the isolates contain an identical Thr-86-Ile substitution in GyrA. Several novel CmeR-Box variations, including point substitutions, deletion, and insertion, were identified. The point insertion or deletion led to dramatically reduced binding of CmeR to the cmeABC promoter, which significantly increases the expression of cmeABC and contributes to the high fluoroquinolone resistance.
KEYWORDS: Campylobacter jejuni, DNA gyrase, cmeABC, fluoroquinolone resistance
TEXT
Campylobacter jejuni is the major foodborne pathogen that causes human bacterial gastroenteritis (1, 2). Chicken is a major reservoir of C. jejuni (3), and contaminated chicken products are recognized as the main source of C. jejuni-related human infections (4). Moreover, the extensive use of antibiotics in the poultry industry has led to the increasing prevalence of antibiotic-resistant C. jejuni strains. Some isolates are especially resistant to clinically used antibiotics, such as fluoroquinolones (FQ), which have been commonly used to treat acute bacterial diarrhea (3, 5, 6). Therefore, surveillance of the FQ susceptibility of C. jejuni is important for not only the purpose of animal breeding but also public health (7, 8). In this study, in order to uncover the underlying genetic mechanisms of FQ resistance in C. jejuni, we undertook an investigation of the genetic polymorphism of gyrA and the promoter region of the cmeABC operon and examined the correlation between the promoter polymorphism and the expression of cmeABC, as well as the fluoroquinolone resistance.
From 2014 to 2016, 79 ciprofloxacin-resistant C. jejuni strains in China were isolated from poultry-related samples, including intestinal tracts, anal swabs, feces, and poultry meat. To investigate the molecular basis of high FQ resistance in the C. jejuni isolates, we analyzed the full gene sequences encoding the DNA gyrase subunits (gyrA and gyrB) (9, 10) and the topoisomerase IV subunits (parC and parE) (11, 12), which are known targets of FQ, the regulatory protein CmeR, and the sequence of the promoter region of cmeABC, which encodes an efflux pump (13, 14), by DNA sequencing as previously described (15, 16). The MICs of ciprofloxacin against the 39 isolates which represent different sequence variants were tested according to CLSI guidelines (17, 28). C. jejuni strain ATCC 33560 was used as the quality control. Statistical significance was analyzed using Student's t test.
As shown in Table 1, a series of point substitutions were identified in the gyrA gene from the C. jejuni isolates, and among these substitutions, two nonsynonymous sequence substitutions were found. Each of the isolates had a Thr-86-Ile substitution, which was reported as the most common cause of FQ resistance in C. jejuni strains (10, 18). Val-149-Ile substitution was found in 6 isolates, and the MICs of ciprofloxacin against these isolates ranged from 4 to 128 μg/ml. As codon 149 is not in the FQ resistance-determining region of gyrA (codons 69 to 120) (10), we take it as a circumstantial sequence variation. Another two reported substitutions, Asp-90-Asn and Ala-70-Thr, which might cause medium-level FQ resistance (19), were not found in the gyrA gene of our isolates. All of the point substitutions in gyrB were synonymous substitutions which did not cause any changes in amino acid sequence. The Arg-139-Gln variation of parC was reported in C. jejuni by Gibreel et al. (20). However, we failed to amplify the parC and parE genes by PCR in our isolates, and this result was consistent with those of other studies (21).
TABLE 1.
Sequence polymorphisms of gyrA in 79 ciprofloxacin-resistant C. jejuni isolates
| Type of variation | No. of strains | Percentage | Substitutions in different pointsa |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gly-78 (GGT) | His-81 (CAC) | Thr-86 (ACA) | Gly-110 (GGC) | Ser-119 (AGT) | Ala-120 (GCC) | Val-149 (GTT) | Ser-157 (AGC) | Val-161 (GTT) | |||
| 1 | 62 | 78.5 | — | (−T) | Ile(-T-) | — | (−C) | (−T) | — | (−T) | (−C) |
| 2 | 3 | 3.8 | — | (−T) | Ile(-T-) | (−T) | (−C) | (−T) | — | (−T) | (−C) |
| 3 | 6 | 7.6 | — | (−T) | Ile(-T-) | — | (−C) | (−T) | Ile(A−) | (−T) | (−C) |
| 4 | 1 | 1.3 | (−C) | — | Ile(-T-) | — | (−C) | (−T) | — | (−T) | (−C) |
| 5 | 1 | 1.3 | (−C) | — | Ile(-T-) | — | (−C) | — | — | (−T) | — |
| 6 | 1 | 1.3 | — | (−T) | Ile(-T-) | — | (−C) | — | — | (−T) | — |
| 7 | 5 | 6.3 | — | — | Ile(-T-) | — | — | — | — | — | — |
—, no change.
The cmeABC operon encodes an important drug efflux pump in C. jejuni (22), and it is negatively regulated by CmeR, which binds to a 16-base inverted repeat (IR) sequence (TGTAATA[or T]TTTATTACA; the repeat sequences are underlined) named CmeR-Box located in the promoter region of the cmeABC operon (16, 23). In contrast with previous studies (16, 24), we did not find mutations inactivating CmeR (16). Instead, several nucleotide variations were found in the CmeR-Box of our isolates, making 12 sequence variants (listed in Table 2). Compared with the conserved CmeR-Box (TGTAATA[or T]TTTATTACA), up to 48.1% of our isolates contained one point substitution, and 2.5% contained two point substitutions in the IR sequence. The most frequent substitution site was the second nucleotide of the IR sequence (26.6% G to A, 13.9% C to T in the reverse repeat). In addition, 15.2% of the isolates contained a point deletion, and one isolate contained a point insertion between the inverted sequences.
TABLE 2.
Sequence variations in CmeR-Box of ciprofloxacin-resistant C. jejuni isolates
| Type of variation | Sequence variant | Sequence of CmeR-Boxa | No. of strainsb | Percentage | MICs (μg/ml) (related expression levels of cmeA)c |
|
|---|---|---|---|---|---|---|
| Of tested strains | Average | |||||
| Conserved sequences | 1 | TGTAATTTTTATTACA | 6 (5) | 7.6 | 4 (0.82 ± 0.07), 4 (0.86 ± 0.11), 4 (0.79 ± 0.08), 8 (1.11 ± 0.14), 8 (0.93 ± 0.11) | 5.6 (0.90 ± 0.09) |
| 2 | TGTAATATTTATTACA | 20 (6) | 25.3 | 4 (1.04 ± 0.09), 4 (1.07 ± 0.04), 4 (1.21 ± 0.07), 8 (1.12 ± 0.11), 8 (1.37 ± 0.19), 8 (1.17 ± 0.21) | 6 (1.16 ± 0.09) | |
| Point substitution | 3 | TATAATTTTTATTACA | 17 (5) | 21.5 | 4 (1.32 ± 0.26), 16 (1.28 ± 0.17), 32 (1.37 ± 0.22), 64 (2.38 ± 0.19), 64 (2.11 ± 0.22) | 36 (1.69 ± 0.44) |
| 4 | TGTAGTTTTTATTACA | 6 (3) | 7.6% | 32 (2.43 ± 0.31), 32 (2.55 ± 0.18), 64 (3.43 ± 0.25) | 42.7 (2.80 ± 0.42) | |
| 5 | TGTAATTTTTATTATA | 5 (3) | 6.3 | 16 (3.21 ± 0.33), 64 (2.11 ± 0.31), 64 (2.73 ± 0.33) | 48 (2.68 ± 0.38) | |
| 6 | TATAATATTTATTACA | 4 (3) | 5.1 | 64 (3.14 ± 0.48), 64 (2.81 ± 0.32), 128 (4.22 ± 0.29) | 85.3 (3.39 ± 0.56) | |
| 7 | TATAATATTTATTACA | 1 (1) | 1.3 | 64 (2.12 ± 0.11) | 64 (2.12) | |
| 8 | TGTAATATTTATTGCA | 1 (1) | 1.3 | 32 (1.70 ± 0.24) | 32 (1.70) | |
| 9 | TGTAATATTTATTATA | 4 (3) | 5.1 | 8 (1.81 ± 0.22), 8 (2.28 ± 0.35), 16 (2.11 ± 0.31) | 10.7 (2.07 ± 0.17) | |
| 10 | TGTAATATCTATTATA | 2 (2) | 2.5 | 32 (3.49 ± 0.51), 32 (5.08 ± 0.83) | 32 (4.29 ± 0.79) | |
| Point deletion | 11 | TGTAAT-TTTATTACA | 12 (6) | 15.2 | 128 (13.32 ± 0.41), 128 (16.39 ± 0.93), 128 (15.67 ± 1.11), 128 (16.80 ± 0.68), 128 (9.71 ± 0.77), 128 (10.58 ± 0.38) | 128 (13.75 ± 2.54) |
| Point insertion | 12 | TGTAATATTTTATTACA | 1 (1) | 1.3 | 128 (8.08 ± 1.63) | 128 (8.08) |
The positions underlined indicate sequence variations in CmeR-Box.
The number of isolates randomly chosen for MIC and cemA expression level test are in parentheses.
Relative expression based on C. jejuni NCTC 11168 was calculated using the 2–-ΔΔCT method. 16S rDNA was used as the internal reference.
To analyze the correlation between the sequence variations in the CmeR-Box and ciprofloxacin resistance levels, we tested the MICs of the isolates containing each different sequence variation. In instances where there were more than 3 isolates per sequence variation, 3 to 6 isolates were selected randomly for testing. As shown in Table 2, the MICs of isolates with point substitutions, insertion, or deletion were significantly higher than those of the isolates with a conserved CmeR-Box (P < 0.05). In particular, the MICs of isolates with the point insertion or deletion between the two halves of the inverted sequences showed the highest MIC values (P < 0.05). We further examined the expression level of cmeA in these isolates by real-time reverse transcriptase PCR. The mRNA was extracted from 2 ml of bacteria in exponential phase (optical density at 600 nm [OD600] = 0.5), and the relative expression based on C. jejuni NCTC 11168 was calculated using the comparative threshold (2–ΔΔCT) method (25). As shown in Table 2, the expression of cmeA in the isolates with point substitution, insertion, or deletion was higher than that in the isolates with the conserved CmeR-Box, and the cmeA in the isolates with a point insertion or deletion showed the highest expression levels (P < 0.05). These results are similar to those for ciprofloxacin resistance in these isolates.
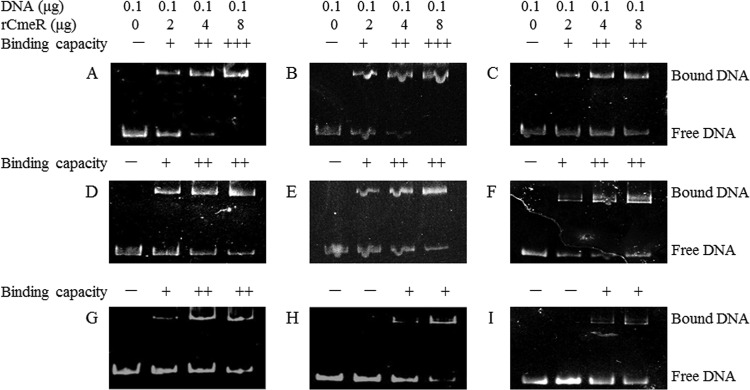
The electrophoretic mobility shift assay (EMSA) was then carried out to further assess the binding of CmeR to the CmeR-Box sequence variants, as previously described (22). As shown in Fig. 1, the CmeR-Box with the point insertion or deletion showed significantly decreased binding by CmeR. This result suggests that the cmeABC operon with the point insertion or deletion in its CmeR-Box was highly derepressed. The functional CmeR is a dimeric two-domain molecule, and the dimer could bind the CmeR-Box to repress expression of the cmeABC operon. The N-terminal domain of each monomer binds to one half of the IR in the CmeR-Box (26, 27). We suppose that the point insertion or deletion identified here may change the distance between two consecutive binding sequences, which leads to a dramatically decreased binding affinity of the CmeR dimer to the CmeR-Box.
FIG 1.
Binding of CmeR to the variant cmeABC promoter DNA. The serial numbers of CmeR-Box sequence variants are listed in Table 2. The tested promoters included conserved CmeR-Box (variation types 1 [A] and 2 [B]), point substitutions (variation types 3 [C], 4 [D], 7 [E], 8 [F], and 10 [G]), point deletion (variation type 11 [H]), and point insertion (variation type 12 [I]). Considering that the CmeR-Box is an inverted repeat, we chose variation type 3 to represent point substitution at the second position of the forward or inverted repeat in our test. +++, all of the DNA was bound; ++, some of the DNA was bound; +, little DNA was bound; −, none of the DNA was bound.
In summary, all of our ciprofloxacin-resistant C. jejuni isolates possessed a Thr-86-Ile substitution in GyrA. Most of the resistant strains contained extra variations in the CmeR-Box, including point substitutions, point insertion, or point deletion, among which the strains with the point insertion or deletion had high ciprofloxacin resistance levels. In this work, we identified several new sequence variations of the CmeR-Box. The point insertion or deletion in the CmeR-Box led to reduced binding by CmeR, which might significantly increase the expression of cmeABC and enhance the ciprofloxacin resistance level in the C. jejuni isolates. This study helps us to further understand the roles of the CmeR-CmeABC efflux pump system in fluoroquinolone resistance in C. jejuni.
ACKNOWLEDGMENTS
This work was supported by the Chinese Key Research and Development Plan (grant 2016YFD0501305), the Chinese Special Fund for Agro-scientific Research in the Public Interest (grant 201303044), and the China Agriculture Research System (grant CARS-42-G11).
REFERENCES
- 1.Huang JL, Xu HY, Bao GY, Zhou XH, Ji DJ, Zhang G, Liu PH, Jiang F, Pan ZM, Liu XF, Jiao XA. 2009. Epidemiological surveillance of Campylobacter jejuni in chicken, dairy cattle and diarrhoea patients. Epidemiol Infect 137:1111–1120. doi: 10.1017/S0950268809002039. [DOI] [PubMed] [Google Scholar]
- 2.Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B, Ma L, Li Y, Jia H, Wei J, Shao D, Liu K, Shi Y, Qiu Y, Ma Z. 2017. Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in Shanghai, China. Foodborne Pathog Dis 14:96–102. doi: 10.1089/fpd.2016.2186. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Luo Q, Chen Y, Li T, Wen G, Zhang R, Luo L, Lu Q, Ai D, Wang H, Shao H. 2016. Molecular epidemiology, virulence determinants and antimicrobial resistance of Campylobacter spreading in retail chicken meat in central China. Gut Pathog 8:48. doi: 10.1186/s13099-016-0132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan H, Ge Y, Xu H, Zhang J, Kuang D, Yang X, Su X, Huang Z, Shi X, Xu X, Meng J. 2016. Molecular characterization, antimicrobial resistance and Caco-2 cell invasion potential of Campylobacter jejuni/coli from young children with diarrhea. Pediatr Infect Dis J 35:330–334. doi: 10.1097/INF.0000000000001016. [DOI] [PubMed] [Google Scholar]
- 6.Yao H, Shen Z, Wang Y, Deng F, Liu D, Naren G, Dai L, Su CC, Wang B, Wang S, Wu C, Yu EW, Zhang Q, Shen J. 2016. Emergence of a potent multidrug efflux pump variant that enhances Campylobacter resistance to multiple antibiotics. mBio 7:pii:e01543-16. doi: 10.1128/mBio.01543-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham NT, Ushijima H, Trinh QD, Khamrin P, Komine-Aizawa S, Okitsu S, Maneekarn N, Hayakawa S. 2015. Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from adult hospitalized patients with diarrhea in Thailand. Clin Lab 61:1809–1812. doi: 10.7754/Clin.Lab.2015.150415. [DOI] [PubMed] [Google Scholar]
- 8.Hou FQ, Sun XT, Wang GQ. 2012. Clinical manifestations of Campylobacter jejuni infection in adolescents and adults, and change in antibiotic resistance of the pathogen over the past 16 years. Scand J Infect Dis 44:439–443. doi: 10.3109/00365548.2011.652163. [DOI] [PubMed] [Google Scholar]
- 9.Power EG, Munoz Bellido JL, Phillips I. 1992. Detection of ciprofloxacin resistance in Gram-negative bacteria due to alterations in gyrA. J Antimicrob Chemother 29:9–17. doi: 10.1093/jac/29.1.9. [DOI] [PubMed] [Google Scholar]
- 10.Hakanen A, Jalava J, Kotilainen P, Jousimies-Somer H, Siitonen A, Huovinen P. 2002. gyrA polymorphism in Campylobacter jejuni: detection of gyrA mutations in 162 C. jejuni isolates by single-strand conformation polymorphism and DNA sequencing. Antimicrob Agents Chemother 46:2644–2647. doi: 10.1128/AAC.46.8.2644-2647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SY, Lee SK, Park MS, Na HT. 2016. Analysis of the fluoroquinolone antibiotic resistance mechanism of Salmonella enterica isolates. J Microbiol Biotechnol 26:1605–1612. doi: 10.4014/jmb.1602.02063. [DOI] [PubMed] [Google Scholar]
- 12.Domenech A, Tirado-Velez JM, Fenoll A, Ardanuy C, Yuste J, Linares J, de la Campa AG. 2014. Fluoroquinolone-resistant pneumococci: dynamics of serotypes and clones in Spain in 2012 compared with those from 2002 and 2006. Antimicrob Agents Chemother 58:2393–2399. doi: 10.1128/AAC.02669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iovine NM. 2013. Resistance mechanisms in Campylobacter jejuni. Virulence 4:230–240. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo B, Wang Y, Shi F, Barton YW, Plummer P, Reynolds DL, Nettleton D, Grinnage-Pulley T, Lin J, Zhang Q. 2008. CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J Bacteriol 190:1879–1890. doi: 10.1128/JB.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corcoran D, Quinn T, Cotter L, O'Halloran F, Fanning S. 2005. Characterization of a cmeABC operon in a quinolone-resistant Campylobacter coli isolate of Irish origin. Microb Drug Resist 11:303–308. doi: 10.1089/mdr.2005.11.303. [DOI] [PubMed] [Google Scholar]
- 16.Grinnage-Pulley T, Zhang Q. 2015. Genetic basis and functional consequences of differential expression of the CmeABC efflux pump in Campylobacter jejuni isolates. PLoS One 10:e0131534. doi: 10.1371/journal.pone.0131534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge B, McDermott PF, White DG, Meng J. 2005. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother 49:3347–3354. doi: 10.1128/AAC.49.8.3347-3354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Changkwanyeun R, Yamaguchi T, Kongsoi S, Changkaew K, Yokoyama K, Kim H, Suthienkul O, Usui M, Tamura Y, Nakajima C, Suzuki Y. 2016. Impact of mutations in DNA gyrase genes on quinolone resistance in Campylobacter jejuni. Drug Test Anal 8:1071–1076. doi: 10.1002/dta.1937. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Huang WM, Taylor DE. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother 37:457–463. doi: 10.1128/AAC.37.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibreel A, Sjogren E, Kaijser B, Wretlind B, Skold O. 1998. Rapid emergence of high-level resistance to quinolones in Campylobacter jejuni associated with mutational changes in gyrA and parC. Antimicrob Agents Chemother 42:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piddock LJ, Ricci V, Pumbwe L, Everett MJ, Griggs DJ. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J Antimicrob Chemother 51:19–26. doi: 10.1093/jac/dkg033. [DOI] [PubMed] [Google Scholar]
- 22.Lin J, Akiba M, Sahin O, Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan M, Sahin O, Lin J, Zhang Q. 2006. Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J Antimicrob Chemother 58:1154–1159. doi: 10.1093/jac/dkl412. [DOI] [PubMed] [Google Scholar]
- 24.Hanninen ML, Hannula M. 2007. Spontaneous mutation frequency and emergence of ciprofloxacin resistance in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother 60:1251–1257. doi: 10.1093/jac/dkm345. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Ding Y, Li T, Wan Y, Li W, Chen H, Zhou R. 2012. A Fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis. BMC Microbiol 12:85. doi: 10.1186/1471-2180-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu R, Su CC, Shi F, Li M, McDermott G, Zhang Q, Yu EW. 2007. Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni. J Mol Biol 372:583–593. doi: 10.1016/j.jmb.2007.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei HT, Shen Z, Surana P, Routh MD, Su CC, Zhang Q, Yu EW. 2011. Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni. Protein Sci 20:712–723. doi: 10.1002/pro.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI document M100-S26 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]