Abstract
Genetic investigations have provided unique insight into the mechanism of chronic pancreatitis in humans and firmly established that uncontrolled trypsin activity is a central pathogenic factor. Mutations in the PRSS1, SPINK1, and CTRC genes promote increased activation of trypsinogen to trypsin by stimulation of autoactivation or by impairing protective trypsinogen degradation and/or trypsin inhibition. Here we review key genetic and biochemical features of the trypsin-dependent pathological pathway in chronic pancreatitis.
Keywords: Pancreas, Pancreatitis, Digestive enzymes, Trypsinogen activation, Trypsin inhibitor, Chymotrypsin
The 1996 discovery that mutation p.R122H in the serine protease 1 (PRSS1) gene that encodes human cationic trypsinogen causes hereditary chronic pancreatitis irrefutably established the critical role of trypsin in the pathogenesis of chronic pancreatitis [1]. In the 20 years following this breakthrough, a large number of genetic and functional studies confirmed the central role of trypsin in disease development and the surprisingly consistent body of evidence has been amalgamated in what we termed the trypsin-dependent pathological pathway of chronic pancreatitis. This relatively simple model states that elevated levels of active trypsin in the pancreas elicit disease onset and drive progression and the combined effects of mutations in various susceptibility genes determine intra-pancreatic trypsin activity. Trypsin is synthesized as an inactive precursor called trypsinogen which becomes activated by limited proteolysis of its 8 amino acid-long N-terminal activation peptide. The serine protease enteropeptidase is responsible for intestinal activation of trypsinogen, whereas ectopic activation inside the pancreas occurs through trypsin-mediated trypsinogen activation commonly referred to as autoactivation [2]. Mutations in cationic trypsinogen that increase autoactivation are strong risk factors for chronic pancreatitis, typically associated with hereditary pancreatitis. Protective mechanisms in the pancreas that curtail trypsinogen activation and therefore reduce trypsin activity involve either trypsin inhibition or trypsinogen degradation (Fig. 1). The serine protease inhibitor Kazal type 1 (SPINK1, also known as pancreatic secretory trypsin inhibitor) is a 6.2-kDa protein secreted by the pancreatic acinar cells that can potently inhibit trypsin. Levels in the pancreatic juice amount to about 0.1–0.8% of total protein [3] which, assuming that about 25% of the juice proteins is trypsinogen [4, 5] and after correction for the molecular mass difference, should translate to SPINK1 concentrations that can inhibit 2–13% of the potential trypsin content. Rinderknecht et al. [6] reported average SPINK1 concentrations that can inhibit 13% of the potential trypsin content in the pancreatic juice of healthy volunteers and 5% in chronic alcoholics. During autoactivation of trypsinogen, the newly generated trypsin reacts with SPINK1 and becomes unavailable to catalyze further trypsinogen activation. In time, however, SPINK1 reserves become depleted and autoactivation can freely proceed. Thus, the protective role of SPINK1 is to delay trypsinogen autoactivation (Fig. 2). This delay is important as it allows for the digestive proteases to transit from the pancreas to the duodenum in an inactive form. Consequently, a decrease in SPINK1 levels or reduced ductal fluid flow can impair this protective mechanism and increase the risk of premature, intra-pancreatic trypsinogen autoactivation. This concept might also explain how inborn or acquired anatomical changes in duct morphology contribute to pancreatitis risk. Similarly, mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride-bicarbonate channel expressed on the ductal epithelium, can increase pancreatitis risk due to impaired ductal flushing [7 and references therein]. Besides trypsin inhibition, an independent line of defense against trypsinogen autoactivation in the pancreas is degradation of trypsinogen by the concerted actions of chymotrypsin C (CTRC) and trypsin (Fig. 3). CTRC-mediated degradation requires the presence of some active trypsin, first to activate chymotrypsinogen C to active CTRC and then to facilitate degradation through autolytic cleavage of the Arg122-Val123 peptide bond in trypsinogen. Therefore, it stands to reason that protective trypsinogen degradation occurs once the SPINK1-dependent defense mechanism has been exhausted.
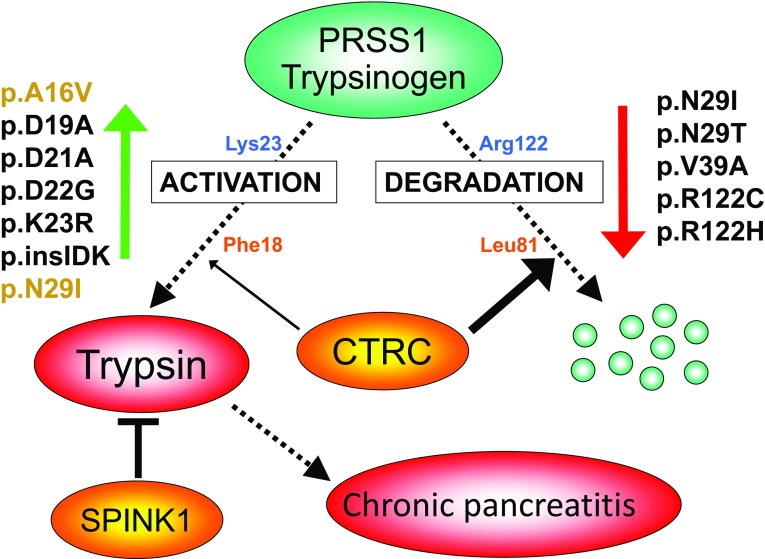
Fig. 1.
Trypsin-dependent pathological pathway in chronic pancreatitis. Activation of PRSS1 trypsinogen to active trypsin in the pancreas is responsible for disease onset and progression. Protective mechanisms to control trypsinogen activation include trypsin inhibition by SPINK1 and trypsinogen degradation by chymotrypsin C (CTRC) and trypsin. CTRC cleaves the Leu81-Glu82 peptide bond, and trypsin cleaves the Arg122-Val123 peptide bond; the combination of these two cleavages results in irreversible trypsinogen degradation. CTRC also stimulates autoactivation of cationic trypsinogen by cleaving the Phe18-Asp19 peptide bond in the activation peptide. The shortened activation peptide is more susceptible to trypsin-mediated activation at the Lys23-Ile24 peptide bond. The indicated hereditary pancreatitis-associated PRSS1 mutations increase trypsinogen autoactivation by inhibition of CTRC-dependent trypsinogen degradation (red arrow) or by increasing CTRC-dependent stimulation of autoactivation (green arrow, mutations in orange type). Activation peptide mutations directly stimulate autoactivation independently of CTRC function (green arrow, mutations in black type). Loss-of-function mutations in SPINK1 reduce inhibitor expression and thus compromise trypsin inhibition. Loss-of function mutations in CTRC reduce secretion, block zymogen activation, diminish catalytic activity or promote degradation by trypsin, and therefore impair protective trypsinogen degradation. p.insIDK indicates mutation p.K23_I24insIDK. Figure modified from Ref. [8]
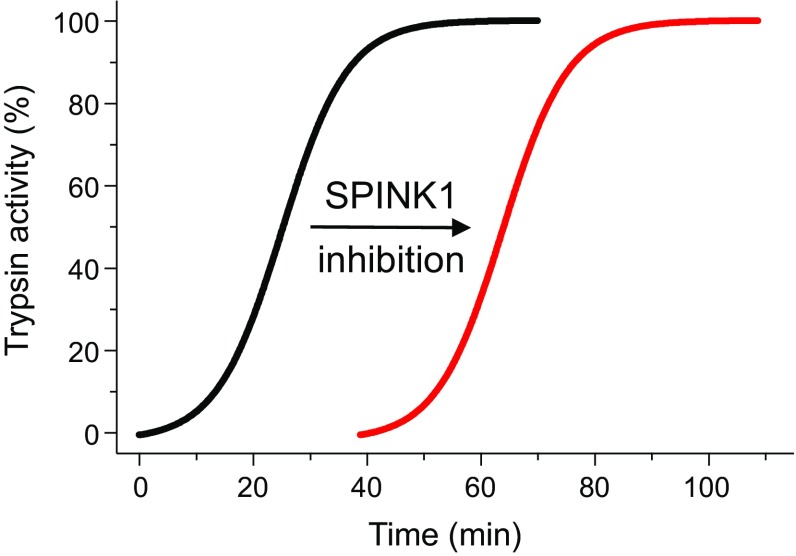
Fig. 2.
Schematic representation of the effect of the SPINK1 trypsin inhibitor on the autoactivation of PRSS1 trypsinogen. Note that SPINK1 delays autoactivation but has no effect on final trypsin levels
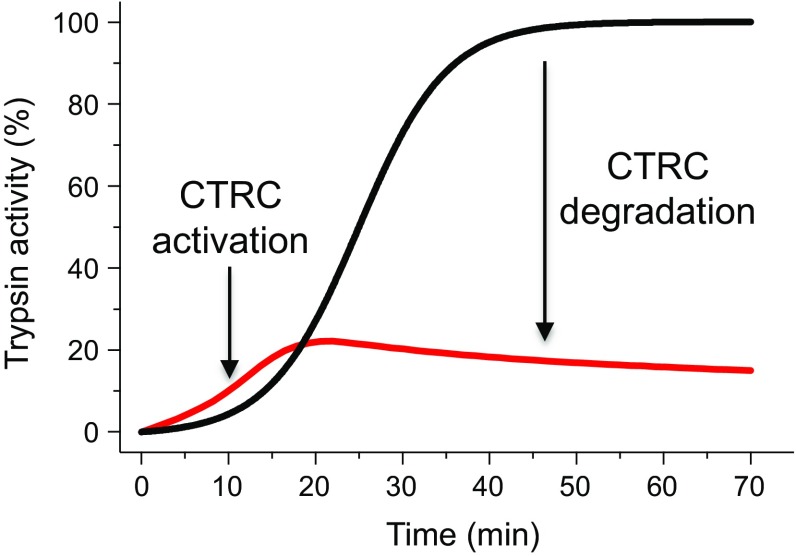
Fig. 3.
Schematic representation of the effect of CTRC on the autoactivation of PRSS1 trypsinogen. CTRC slightly accelerates the onset of autoactivation but markedly reduces final trypsin levels through trypsinogen degradation
Trypsin-Dependent Pathological Pathway in Chronic Pancreatitis
The model posits that genetic risk in chronic pancreatitis is mediated by mutation-associated mechanisms that result in increased activation of trypsinogen in the pancreas (Fig. 1). PRSS1 mutations are gain-of-function mutations that directly or indirectly (e.g., through altering trypsinogen degradation) stimulate autoactivation to trypsin. Heterozygous PRSS1 mutations are often associated with autosomal dominant hereditary disease, indicating that these are strong, essentially causative genetic changes [1, 8 and references therein]. Loss-of-function mutations in SPINK1 that result in reduced inhibitor expression compromise protective trypsin inhibition and increase pancreatitis risk by about tenfold to 40-fold in the heterozygous state [9–12]. Disease risk associated with homozygous SPINK1 variants is markedly higher, and these genetic alterations should be considered causative [9–13]. Loss-of-function mutations in CTRC can lead to diminished expression, resistance to activation by trypsin, defective catalytic activity, or degradation by trypsin, thereby impairing protective CTRC-mediated trypsinogen degradation [14–16]. The majority of heterozygous CTRC variants increase disease risk by about fivefold to tenfold [12, 14–16]. Compound heterozygosity for SPINK1 and CTRC mutations results in highly significant risk. The trypsin-dependent pathogenic model is supported by two additional observations: (1) copy number mutations in the PRSS1-PRSS2 locus which presumably result in elevated trypsinogen expression cause hereditary and sporadic disease [17, 18] and (2) the p.G191R variant in PRSS2 induces autodegradation of human anionic trypsinogen and protects against chronic pancreatitis, decreasing risk by about 2.7-fold [19].
PRSS1 Mutations and the CTRC-PRSS1 Axis in Hereditary Pancreatitis
The clinically most significant, high-penetrance PRSS1 mutations (in the order of frequency: p.R122H > p.N29I > p.A16V ~ p.R122C > p.N29T > p.V39A) are typically found in hereditary pancreatitis families although in the everyday clinical setting the obtainable family history may be limited and patients may present with what appears to be sporadic disease [8 and references therein]. The p.A16V variant exhibits variable penetrance and may be detected in sporadic disease as well as in hereditary pancreatitis families [20, 21]. Mechanistically, PRSS1 mutations stimulate autoactivation of cationic trypsinogen by altering cleavage of CTRC-sensitive and trypsin-sensitive regulatory nick sites (Fig. 1) [22]. The main effect of CTRC is promotion of trypsinogen degradation, which is achieved by cleaving the Leu81-Glu82 peptide bond in the calcium binding loop of trypsinogen [23, 24]. Concurrent autolytic cleavage by trypsin at the Arg122-Val123 peptide bond results in complete inactivation of trypsinogen. Trypsin is also degraded by the same mechanism, however, at a much slower rate because the regulatory nick sites become thermodynamically stable when trypsinogen is activated to trypsin [25]. PRSS1 mutations block or reduce cleavage after Leu81 and/or Arg122 and thereby allow for trypsinogen autoactivation to proceed unfettered [24].
A seemingly paradoxical effect of CTRC is the stimulation of autoactivation through processing of the trypsinogen activation peptide [26]. Cleavage of the Phe18-Asp19 peptide bond shortens the 8 amino acid-long activation peptide to 5 amino acids which is then cleaved by trypsin at an enhanced rate, resulting in an approximately fourfold increase in autoactivation [26, 27]. Mechanistically, this effect is likely due to the mitigation of a repulsive electrostatic interaction between the tetra-Asp motif in the activation peptide and Asp218 in cationic trypsin, which normally curtails autoactivation [26, 28]. As shown in Fig. 3, during autoactivation of wild-type cationic trypsinogen in the presence of CTRC, the effect of the activation peptide processing is seen only as a small initial increase in trypsin levels, which become rapidly quenched by the predominant trypsinogen degradation. The reason for this relatively minor effect is that N-terminal processing by CTRC, activation of trypsinogen to trypsin, and degradation of trypsinogen are all competing reactions and only a small fraction of trypsinogen becomes actually processed at Phe18. However, the situation is drastically different when the activation peptide or the N-terminal region of trypsinogen is mutated and N-terminal processing is stimulated. This is the case with the p.A16V mutation which increases N-terminal processing of cationic trypsinogen by CTRC about sixfold and therefore allows for the development of higher trypsin levels during autoactivation [24, 26]. Similarly, mutation p.N29I increases N-terminal processing by 2.6-fold [24]. Interestingly, mutation p.D19A also stimulates processing of the activation peptide by fourfold but this modification induces only a small further increase in the already higher autoactivation of this mutant. This observation indicates that Asp19 is required to mediate the functional effect of N-terminal processing by CTRC [29]. The physiological rationale for CTRC-mediated N-terminal processing of cationic trypsinogen is unclear but it may be helpful in the generation of an initial “trypsin burst” which can then aid in the full activation of chymotrypsinogen C to active CTRC and to facilitate trypsinogen degradation by cleavage after Arg122.
It is important to note that PRSS1 mutations all cause the same biochemical phenotype of increased autoactivation resulting in high trypsin concentrations. At the molecular level, however, individual mutations may exert their effect differently, either increasing CTRC-mediated N-terminal processing or reducing trypsinogen degradation or both. For a detailed analysis of the mutational effects on cationic trypsinogen, see Table 1 in [24]. The compartment where increased trypsinogen autoactivation takes place is unknown as genetic and biochemical studies provide no information on subcellular compartments and appropriate animal models are lacking. Transfection experiments indicate that the most frequent PRSS1 mutants are secreted normally and do not autoactivate in the secretory pathway [30]. This observation suggests that autoactivation might take place in the pancreatic juice and cause acinar cell damage possibly by ductal obstruction due to cleavage of pancreatic stone protein and pancreatitis-associated protein which precipitate upon tryptic digestion [31, 32 and references therein].
An intriguing subset of high-penetrance PRSS1 mutations (p.D19A, p.D21A, p.D22G, p.K23R, p.K23_I24insIDK) affect the activation peptide of cationic trypsinogen and robustly stimulate autoactivation independently of CTRC [29]. In fact, autoactivation is increased to such an extent that some trypsinogen autoactivates intracellularly and becomes degraded, likely in the endoplasmic reticulum (ER) [33]. As a result, these mutants are secreted poorly from transfected cells and, presumably, from human acinar cells as well. Intracellular autoactivation represents a form of cellular stress, which can lead to acinar cell damage and death [33]. Thus, the mechanism of action of the rare activation peptide mutations may be different from that of the clinically more common PRSS1 mutations.
A common variant in the promoter region of PRSS1 (c.-204C>A) which is part of a larger haplotype in the PRSS1-PRSS2 locus confers a small (1.4-fold to 1.6-fold) protective effect against chronic pancreatitis [34–37]. Interestingly, in the European replication study this effect was best detected in subjects with alcoholic chronic pancreatitis, suggesting that this promoter variant modifies trypsinogen expression in an alcohol-dependent manner [35].
Finally, another subset of PRSS1 mutations (p.K92N, p.D100H, p.L104P, p.R116C, p.S124F, p.C139F, p.C139S, p.G208A) exert their pathogenic effect through a so-called misfolding-dependent pathological pathway [30, 38, 39]. In this case, mutations cause misfolding of cationic trypsinogen and therefore elicit ER stress that may be responsible for pancreas injury and pancreatitis. Misfolding-associated PRSS1 variants are less frequent than the trypsin pathway-associated PRSS1 variants and are more often found in sporadic cases [38], although association with dominant hereditary disease was also described [30, 40].
For a more detailed discussion of PRSS1 variants in chronic pancreatitis, please consult our 2014 review article [8]. For a complete list of published PRSS1 variants, see the www.pancreasgenetics.org Web site.
CTRC Mutations
The critical role of the CTRC-PRSS1 axis in human chronic pancreatitis is reinforced by the observation that carriers of loss-of-function CTRC mutations are at increased risk of chronic pancreatitis even in the absence of trypsinogen mutations [14, 15]. Our initial Nature Genetics report in 2008 suggested that CTRC mutations are relatively rare and altogether found in about 4% of patients with chronic pancreatitis resulting in about fivefold to tenfold increased disease risk in heterozygous carriers [14]. In a German cohort, variants p.R254W (2.1%) and p.K247_R254del (1.2%) were found more frequently, while in an Indian cohort variants p.V235I (4.9%) and p.A73T (3.1%) were more prevalent [41]. The notion that CTRC variants play a relatively minor role in chronic pancreatitis has been refuted by subsequent studies, demonstrating that variant c.180C>T (p.G60=) is one of the most common risk factors in chronic pancreatitis found in more than 30% of patients [15, 41, 42]. In a large Indian cohort, this variant increased risk about 2.5-fold and tenfold in the heterozygous and homozygous states, respectively [41]. According to the ExAC database, variant c.180C>T is associated with reduced mRNA expression, suggesting it might alter pre-mRNA splicing. Other pathogenic variants can impair CTRC function by distinct but mutually non-exclusive mechanisms that result in secretion defect, resistance to activation by trypsin, catalytic deficiency or degradation by trypsin. In two large studies, we characterized properties of 40 CTRC variants and found that 17 exhibited one or more forms of functional impairment [16, 43]. Considering the clinically frequent variants, p.A73T exhibited a marked secretion defect (<10% of wild type), p.V235I showed a 50% decrease in trypsinogen degradation activity, p.K247_R254del was completely inactive and was readily degraded by trypsin, and p.R254W was secreted at slightly reduced levels (60–70% of wild type) and was degraded by trypsin. CTRC mutants with diminished secretion (e.g., p.A73T) also induced ER stress, indicating that reduced secretion was due to intracellular misfolding and degradation [16, 44]. Whether or not ER stress caused by these CTRC variants contributes to pancreatitis risk remains uncertain because CTRC is expressed at relatively low levels when compared to PRSS1 or carboxypeptidase A1 (CPA1), whose mutation-induced misfolding was demonstrated to cause chronic pancreatitis [30, 39, 45]. For a complete list of published CTRC variants, see the www.pancreasgenetics.org Web site.
SPINK1 Mutations
The identification of PRSS1 mutations in hereditary pancreatitis was shortly followed by the candidate-gene analysis of SPINK1. It is of historic interest that the first study which described the c.101A>G (p.N34S) mutation erroneously concluded that it was a harmless variant [46]. Subsequently, Witt et al. [9] demonstrated the association between p.N34S and chronic pancreatitis. A large number of replication studies confirmed that the p.N34S variant and its associated haplotype which includes 4 intronic alterations (c.56-37T>C in intron 1, c.87+268A>G in intron 2, and c.195-606G>A and c.195-66_65insTTTT in intron 3) is the clinically most significant risk factor for chronic pancreatitis. According to a 2008 meta-analysis, the average carrier frequency is 9.7% in patients and 1% in controls and the average odds ratio (OR) is 11. When disease subtypes were considered, the strongest effects were observed in idiopathic chronic pancreatitis (OR 15) and in tropical chronic pancreatitis (OR 19), whereas the effect size in alcoholic chronic pancreatitis was less pronounced (OR 5) [11 and references therein]. Despite efforts to uncover the disease-relevant functional effect of the p.N34S variant-associated haplotype, the underlying molecular mechanism is yet to be identified. Several studies demonstrated that the p.N34S variant has no effect on the trypsin inhibitory activity or the cellular folding and secretion of SPINK1 [47–49]. A functional role for the co-segregating four intronic alterations has been also excluded [50–52]. Thus, experimental evidence to date suggests that the actual pathogenic variant within the p.N34S haplotype is located within the yet unexplored flanking regions of SPINK1. In this regard, a recent study found that in pancreatic cancer cell lines heterozygous for p.N34S, expression of the variant allele was diminished relative to the wild-type allele [53]. Furthermore, the authors identified variant c.-4141G>T in the 5′ upstream region of SPINK1 as part of the p.N34S haplotype and as a potential candidate for the pathogenic effect.
The second most common SPINK1 haplotype associated with chronic pancreatitis contains the c.-215G>A promoter variant and the c.194+2T>C variant in intron 3 [9, 10, 54]. This haplotype is more frequent in East Asia with an average carrier frequency of 6.4% in Japanese subjects with idiopathic (12%), familial (21%), and alcoholic (3%) chronic pancreatitis [55 and references therein]. High carrier frequencies (~10–30%) among patients with idiopathic disease were reported from Korea, with the highest number seen in the pediatric population [56–58]. Reports from a center in Shanghai, China, indicated carrier frequencies of 45 and 57% [59, 60] whereas another study from Taiwan found an average frequency of 8.5% with higher occurrence (29%) among early onset patients [61]. The variant was found in 6.7% of chronic pancreatitis patients in India [62], whereas in Europe and the USA it has been found in about 3% of the cases [10, 12, 63]. In contrast to p.N34S, the c.194+2T>C variant is almost never observed in healthy controls and the effect size (OR) is estimated around 40 in the heterozygous state [12] while homozygosity or transheterozygosity with p.N34S is essentially causative. The c.194+2T>C variant affects the donor splice site in intron 3 and causes skipping of exon 3 [64, 65], resulting in markedly reduced SPINK1 mRNA expression [51, 65]. Thus, in case of the c.194+2T>C variant the clear genetic association with chronic pancreatitis is complemented with convincing functional data demonstrating loss of SPINK1 function. Consequently, this variant offers the strongest evidence for the proposed role of SPINK1 in the trypsin-dependent pathway of chronic pancreatitis pathogenesis. Interestingly, functional studies showed that the c.-215G>A variant increases promoter activity, suggesting that this variant might mitigate the negative effect of the c.194+2T>C variant to some degree [63, 66, 67].
Additional evidence for the pathogenic role of loss-of-function SPINK1 variants came from the identification of a number of rare and private variants in patients with chronic pancreatitis. Although genetic association with the disease phenotype could not be demonstrated due to the low frequency of the individual variants, functional impairment of SPINK1 was either self-evident or demonstrated experimentally. Variants in this category include those affecting splice sites [58, 68, 69], nucleotide insertion or deletions leading to frame shift with premature termination [12, 68, 70, 71], variants causing loss of the initiator methionine codon [9, 12, 13, 72, 73], signal peptide alterations leading to diminished secretion [9, 12, 74–76], and variants that cause loss of SPINK1 secretion due to misfolding, intracellular retention and degradation [48, 49, 75]. Large heterozygous deletions in the SPINK1 gene were also described [77, 78]. Finally, promoter variants that reduce transcriptional activity and presumably result in decreased SPINK1 mRNA expression have been characterized [63, 66, 67].
It is interesting to note that loss-of-function SPINK1 mutations mostly result in diminished SPINK1 levels rather than a decrease in the trypsin inhibitory activity of the protein. So far only two rare mutations, p.K41N and p.I42M, have been reported that directly affect the Lys41-Ile42 reactive-site peptide-bond in SPINK1 [79, 80]. Although the functional effects of these variants have not been characterized yet, mutation p.K41N is expected to cause loss of trypsin inhibition, whereas mutation p.I42M is likely to have a small effect, if any. For a complete list of published SPINK1 variants, see the www.pancreasgenetics.org Web site.
PRSS2 Mutations
The human pancreas secretes two major trypsinogen isoforms: cationic trypsinogen (PRSS1) and anionic trypsinogen (PRSS2) [4, 5]. Even though PRSS2 is 90% identical to PRSS1 in its primary structure and it exhibits an equally strong propensity for autoactivation, mutations in PRSS2 have never been described in association with hereditary pancreatitis. We recently found an explanation for this puzzling observation by demonstrating that PRSS2 is more tightly controlled by CTRC than PRSS1 [27]. Thus, CTRC degrades PRSS2 more effectively and N-terminal processing of the PRSS2 activation peptide slightly inhibits autoactivation. The increased sensitivity of PRSS2 to CTRC-mediated degradation is due to an additional cleavage site at Leu148 in the autolysis loop and the lack of the conserved Cys139-Cys206 disulfide bond which exposes multiple other cleavage sites. As a result, individual natural mutations are unlikely to cause significant stabilization of PRSS2 against CTRC and cannot promote autoactivation to an extent that would cause hereditary pancreatitis.
Although not causative in hereditary pancreatitis, PRSS2 still contributes to pancreatitis risk, as demonstrated by the small protective effect of the self-degrading p.G191R variant [19, 81, 82]. In the largest cohort published, this variant was enriched in the healthy population (frequency 3.4%) relative to subjects with chronic pancreatitis (frequency 1.3%), yielding a protective OR of 2.7 [19]. The mutation introduces a new cleavage site for trypsin which results in rapid degradation of anionic trypsinogen during autoactivation. Since most carriers are heterozygous, this would represent only a 50% loss of their anionic trypsinogen content and about 20–25% reduction in their total trypsinogen. Despite its minimal clinical relevance, the p.G191R PRSS2 variant has been conceptually very important as it supports the significance of trypsinogen degradation as a protective mechanism against chronic pancreatitis.
PRSS1-PRSS2 Copy Number Mutations
The Férec group in 2006 reported that triplication in the PRSS1-PRSS2 locus was associated with hereditary pancreatitis [17]. Subsequently, the same investigators identified duplications in the PRSS1-PRSS2 locus in subjects with idiopathic chronic pancreatitis with no family history [18]. Increased trypsinogen expression due to a gene dosage effect was proposed as pathogenic mechanism. Higher trypsinogen concentrations in the pancreas would increase autoactivation rates and result in elevated intra-pancreatic trypsin activity. Unfortunately, screening for copy number mutations has not become routine in other laboratories and corroborating reports are lacking.
Trypsin-Dependent Pathological Pathway in Alcoholic Chronic Pancreatitis
The absence of high-penetrance PRSS1 mutations in subjects with alcoholic chronic pancreatitis has spawned the erroneous yet persistent notion that trypsin does not play a central role in the pathogenesis of alcoholic chronic pancreatitis. Nothing can be further from the truth. A large number of studies documented that mutations in susceptibility genes that contribute to the trypsin-dependent pathway are associated with alcoholic chronic pancreatitis. Thus, pathogenic CTRC and SPINK1 variants increase disease risk, whereas the p.G191R PRSS2 variant and the PRSS1 promoter variant c.-204C>A are protective against alcoholic chronic pancreatitis [14, 19, 55, 83, 84]. The frequency (~6%) and consequently the effect size (OR ~5) of the SPINK1 p.N34S variant are lower in alcoholic versus idiopathic chronic pancreatitis, suggesting gene-environment interactions [83, 84].
Mouse Models
Attempts to create mouse models for the trypsin-dependent pathological pathway of chronic pancreatitis have failed so far. With respect to PRSS1 mutations, Archer et al. [85] introduced the p.R122H mutation into the native T8 mouse trypsinogen placed under the control of a short elastase promoter and generated a transgenic mouse line with this construct. The mice exhibited signs of focal inflammatory cell infiltration, fibrosis, and enhanced responses to cerulein. However, there was no control strain with a wild-type T8 transgene generated and it remains impossible to interpret whether the observed effects were due to the mutation or the expression of an extra trypsinogen gene. Unfortunately, this strain was lost for further studies and replication. Selig et al. [86] found slightly increased sensitivity to cerulein-induced pancreatitis in transgenic mice carrying the human PRSS1 coding DNA with the p.R122H mutation under the control of the short elastase promoter. Athwal et al. [87] created three transgenic lines using the short elastase promoter to drive expression of wild-type PRSS1 and mutants p.N29I and p.R122H. All three lines developed some spontaneous pancreatic damage (vacuolization, fibrosis, inflammatory cell infiltrates) with low penetrance (10%) and delayed onset (>9 months), but there were no mutation-specific differences. Increased responses to low dose cerulein were also described. These studies suggest that expression of human PRSS1 is somewhat toxic to the mouse pancreas, presumably due to folding problems, which limits the usefulness of the transgenic approach to study PRSS1 mutations. Future studies should focus on the genetic modification of endogenous mouse trypsinogens to mimic the effects of human PRSS1 mutations. To this end, we characterized the expression of mouse trypsinogens and their interactions with mouse Ctrc [88]. We found that the mouse pancreas expresses four of the 20 trypsinogen genes to high levels: T7, T8, T9, and T20. Mouse Ctrc poorly cleaved the calcium binding loop in all mouse trypsinogens and did not promote significant degradation. However, mouse Ctrc rapidly cleaved the Phe150-Gly151 peptide bond in the autolysis loop of T8 and T9 and this regulatory cleavage inhibited autoactivation of these isoforms. The observations indicate that mechanistic details of the CTRC-PRSS1 axis are different in the mouse, and modeling the CTRC-dependent effects of hereditary pancreatitis-associated PRSS1 mutations may not be feasible. Instead, PRSS1 mutations that alter the activation peptide and promote rapid autoactivation independently of CTRC should be utilized to generate mouse models of hereditary pancreatitis. Finally, homozygous deletion of mouse Spink3, the ortholog of human SPINK1, caused rapid autophagic degeneration of pancreatic acinar cells after birth and led to early mortality [89]. Pancreatic acini isolated after birth exhibited elevated trypsin activity relative to acini from wild-type animals [90]. Heterozygous knockout animals showed no overt phenotype and no trypsin activity in isolated acini. These experiments highlight a potential developmental role for Spink3 in the mouse and, however, offer limited insight into the mechanism of human chronic pancreatitis associated with SPINK1 mutations.
Summary
Genetic susceptibility to chronic pancreatitis is mainly caused by variants in the PRSS1, SPINK1, and CTRC genes, which raise intra-pancreatic trypsin activity by increasing trypsinogen activation, reducing trypsinogen degradation or impeding trypsin inhibition. The trypsin-dependent pathological pathway, therefore, is a therapeutic target in chronic pancreatitis. Genetically modified animal models that recapitulate human chronic pancreatitis phenotypically and mechanistically are needed to understand disease onset and progression and to test preventive and therapeutic measures.
Acknowledgments
Work in the senior author’s laboratory has been supported by NIH Grants R01DK058088, R01DK082412, R01DK095753, and DoD grant PR130667. EH was also supported by a grant from the National Pancreas Foundation.
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interest to declare.
References
- 1.Whitcomb DC, Gorry MC, Preston RA, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 2.Rinderknecht H. Activation of pancreatic zymogens. Normal activation, premature intrapancreatic activation, protective mechanisms against inappropriate activation. Dig Dis Sci. 1986;31:314–321. doi: 10.1007/BF01318124. [DOI] [PubMed] [Google Scholar]
- 3.Pubols MH, Bartelt DC, Greene LJ. Trypsin inhibitor from human pancreas and pancreatic juice. J Biol Chem. 1974;249:2235–2242. [PubMed] [Google Scholar]
- 4.Guy O, Lombardo D, Bartelt DC, Amic J, Figarella C. Two human trypsinogens. Purification, molecular properties, and N-terminal sequences. Biochemistry. 1978;17:1669–1675. doi: 10.1021/bi00602a014. [DOI] [PubMed] [Google Scholar]
- 5.Scheele G, Bartelt D, Bieger W. Characterization of human exocrine pancreatic proteins by two-dimensional isoelectric focusing/sodium dodecyl sulfate gel electrophoresis. Gastroenterology. 1981;80:461–473. [PubMed] [Google Scholar]
- 6.Rinderknecht H, Stace NH, Renner IG. Effects of chronic alcohol abuse on exocrine pancreatic secretion in man. Dig Dis Sci. 1985;30:65–71. doi: 10.1007/BF01318373. [DOI] [PubMed] [Google Scholar]
- 7.Hegyi P, Wilschanski M, Muallem S, et al. CFTR: A new horizon in the pathomechanism and treatment of pancreatitis. Rev Physiol Biochem Pharmacol. 2016;170:37–66. doi: 10.1007/112_2015_5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Németh BC, Sahin-Tóth M. Human cationic trypsinogen (PRSS1) variants and chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G466–G473. doi: 10.1152/ajpgi.00419.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 10.Pfützer RH, Barmada MM, Brunskill AP, et al. SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000;119:615–623. doi: 10.1053/gast.2000.18017. [DOI] [PubMed] [Google Scholar]
- 11.Aoun E, Chang CC, Greer JB, Papachristou GI, Barmada MM, Whitcomb DC. Pathways to injury in chronic pancreatitis: Decoding the role of the high-risk SPINK1 N34S haplotype using meta-analysis. PLoS One. 2008;3:e2003. doi: 10.1371/journal.pone.0002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosendahl J, Landt O, Bernadova J, et al. CFTR, SPINK1, CTRC and PRSS1 variants in chronic pancreatitis: is the role of mutated CFTR overestimated? Gut. 2013;62:582–592. doi: 10.1136/gutjnl-2011-300645. [DOI] [PubMed] [Google Scholar]
- 13.Masson E, Chen JM, Audrézet MP, Cooper DN, Férec C. A conservative assessment of the major genetic causes of idiopathic chronic pancreatitis: data from a comprehensive analysis of PRSS1, SPINK1, CTRC and CFTR genes in 253 young French patients. PLoS One. 2013;8:e73522. doi: 10.1371/journal.pone.0073522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosendahl J, Witt H, Szmola R, et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet. 2008;40:78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masson E, Chen JM, Scotet V, Le Maréchal C, Férec C. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum Genet. 2008;123:83–91. doi: 10.1007/s00439-007-0459-3. [DOI] [PubMed] [Google Scholar]
- 16.Beer S, Zhou J, Szabó A, et al. Comprehensive functional analysis of chymotrypsin C (CTRC) variants reveals distinct loss-of-function mechanisms associated with pancreatitis risk. Gut. 2013;62:1616–1624. doi: 10.1136/gutjnl-2012-303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Maréchal C, Masson E, Chen JM, et al. Hereditary pancreatitis caused by triplication of the trypsinogen locus. Nat Genet. 2006;38:1372–1374. doi: 10.1038/ng1904. [DOI] [PubMed] [Google Scholar]
- 18.Masson E, Le Maréchal C, Chandak GR, et al. Trypsinogen copy number mutations in patients with idiopathic chronic pancreatitis. Clin Gastroenterol Hepatol. 2008;6:82–88. doi: 10.1016/j.cgh.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Witt H, Sahin-Tóth M, Landt O, et al. A degradation-sensitive anionic trypsinogen (PRSS2) variant protects against chronic pancreatitis. Nat Genet. 2006;38:668–673. doi: 10.1038/ng1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witt H, Luck W, Becker M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology. 1999;117:7–10. doi: 10.1016/S0016-5085(99)70543-3. [DOI] [PubMed] [Google Scholar]
- 21.Grocock CJ, Rebours V, Delhaye MN, et al. The variable phenotype of the p.A16V mutation of cationic trypsinogen (PRSS1) in pancreatitis families. Gut. 2010;59:357–363. doi: 10.1136/gut.2009.186817. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Sahin-Tóth M. Chymotrypsin C mutations in chronic pancreatitis. J Gastroenterol Hepatol. 2011;26:1238–1246. doi: 10.1111/j.1440-1746.2011.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szmola R, Sahin-Tóth M. Chymotrypsin C (caldecrin) promotes degradation of human cationic trypsin: identity with Rinderknecht’s enzyme Y. Proc Natl Acad Sci USA. 2007;104:11227–11232. doi: 10.1073/pnas.0703714104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabó A, Sahin-Tóth M. Increased activation of hereditary pancreatitis-associated human cationic trypsinogen mutants in presence of chymotrypsin C. J Biol Chem. 2012;287:20701–20710. doi: 10.1074/jbc.M112.360065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabó A, Radisky ES, Sahin-Tóth M. Zymogen activation confers thermodynamic stability on a key peptide bond and protects human cationic trypsin from degradation. J Biol Chem. 2014;289:4753–4761. doi: 10.1074/jbc.M113.538884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoda Z, Sahin-Tóth M. Chymotrypsin C (caldecrin) stimulates autoactivation of human cationic trypsinogen. J Biol Chem. 2006;281:11879–11886. doi: 10.1074/jbc.M600124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jancsó Z, Sahin-Tóth M. Tighter control by chymotrypsin C (CTRC) explains lack of association between human anionic trypsinogen and hereditary pancreatitis. J Biol Chem. 2016;291:12897–12905. doi: 10.1074/jbc.M116.725374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemoda Z, Sahin-Tóth M. The tetra-aspartate motif in the activation peptide of human cationic trypsinogen is essential for autoactivation control but not for enteropeptidase recognition. J Biol Chem. 2005;280:29645–29652. doi: 10.1074/jbc.M505661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geisz A, Hegyi P, Sahin-Tóth M. Robust autoactivation, chymotrypsin C independence and diminished secretion define a subset of hereditary pancreatitis-associated cationic trypsinogen mutants. FEBS J. 2013;280:2888–2899. doi: 10.1111/febs.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kereszturi E, Szmola R, Kukor Z, et al. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: a novel disease mechanism. Hum Mutat. 2009;30:575–582. doi: 10.1002/humu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarles H. Hereditary chronic pancreatitis with radiolucent protein calculi. Pancreas. 1998;17:321–322. doi: 10.1097/00006676-199810000-00020. [DOI] [PubMed] [Google Scholar]
- 32.Medveczky P, Szmola R, Sahin-Tóth M. Proteolytic activation of human pancreatitis-associated protein is required for peptidoglycan binding and bacterial aggregation. Biochem J. 2009;420:335–343. doi: 10.1042/BJ20090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kereszturi E, Sahin-Tóth M. Intracellular autoactivation of human cationic trypsinogen mutants causes reduced trypsinogen secretion and acinar cell death. J Biol Chem. 2009;284:33392–33399. doi: 10.1074/jbc.M109.056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitcomb DC, LaRusch J, Krasinskas AM, et al. Common genetic variants in the CLDN2 and PRSS1-PRSS2 loci alter risk for alcohol-related and sporadic pancreatitis. Nat Genet. 2012;44:1349–1354. doi: 10.1038/ng.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derikx MH, Kovacs P, Scholz M, et al. Polymorphisms at PRSS1-PRSS2 and CLDN2-MORC4 loci associate with alcoholic and nonalcoholic chronic pancreatitis in a European replication study. Gut. 2015;64:1426–1433. doi: 10.1136/gutjnl-2014-307453. [DOI] [PubMed] [Google Scholar]
- 36.Masamune A, Nakano E, Hamada S, Kakuta Y, Kume K, Shimosegawa T. Common variants at PRSS1-PRSS2 and CLDN2-MORC4 loci associate with chronic pancreatitis in Japan. Gut. 2015;64:1345–1346. doi: 10.1136/gutjnl-2015-309802. [DOI] [PubMed] [Google Scholar]
- 37.Boulling A, Sato M, Masson E, Génin E, Chen JM, Férec C. Identification of a functional PRSS1 promoter variant in linkage disequilibrium with the chronic pancreatitis-protecting rs10273639. Gut. 2015;64:1837–1838. doi: 10.1136/gutjnl-2015-310254. [DOI] [PubMed] [Google Scholar]
- 38.Schnúr A, Beer S, Witt H, Hegyi P, Sahin-Tóth M. Functional effects of 13 rare PRSS1 variants presumed to cause chronic pancreatitis. Gut. 2014;63:337–343. doi: 10.1136/gutjnl-2012-304331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balázs A, Hegyi P, Sahin-Tóth M. Pathogenic cellular role of the p.L104P human cationic trypsinogen variant in chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G477–G486. doi: 10.1152/ajpgi.00444.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Németh BC, Patai ÁV, Sahin-Tóth M, Hegyi P. (2016) Misfolding cationic trypsinogen variant p.L104P causes hereditary pancreatitis. Gut. Epub. 12/23/2016. [DOI] [PMC free article] [PubMed]
- 41.Paliwal S, Bhaskar S, Mani KR, et al. Comprehensive screening of chymotrypsin C (CTRC) gene in tropical calcific pancreatitis identifies novel variants. Gut. 2013;62:1602–1606. doi: 10.1136/gutjnl-2012-302448. [DOI] [PubMed] [Google Scholar]
- 42.La Rusch J, Lozano-Leon A, Stello K, et al. The common chymotrypsinogen C (CTRC) variant G60G (C.180T) increases risk of chronic pancreatitis but not recurrent acute pancreatitis in a North American population. Clin Transl Gastroenterol. 2015;6:68. doi: 10.1038/ctg.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szabó A, Ludwig M, Hegyi E, Szépeová R, Witt H, Sahin-Tóth M. Mesotrypsin signature mutation in a chymotrypsin C (CTRC) variant associated with chronic pancreatitis. J Biol Chem. 2015;290:17282–17292. doi: 10.1074/jbc.M114.618439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szmola R, Sahin-Tóth M. Pancreatitis-associated chymotrypsinogen C (CTRC) mutant elicits endoplasmic reticulum stress in pancreatic acinar cells. Gut. 2010;59:365–732. doi: 10.1136/gut.2009.198903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witt H, Beer S, Rosendahl J, et al. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat Genet. 2013;45:1216–2120. doi: 10.1038/ng.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JM, Mercier B, Audrezet MP, Ferec C. Mutational analysis of the human pancreatic secretory trypsin inhibitor (PSTI) gene in hereditary and sporadic chronic pancreatitis. J Med Genet. 2000;37:67–69. doi: 10.1136/jmg.37.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuwata K, Hirota M, Shimizu H, et al. Functional analysis of recombinant pancreatic secretory trypsin inhibitor protein with amino-acid substitution. J Gastroenterol. 2002;37:928–934. doi: 10.1007/s005350200156. [DOI] [PubMed] [Google Scholar]
- 48.Király O, Wartmann T, Sahin-Tóth M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut. 2007;56:1433–1438. doi: 10.1136/gut.2006.115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boulling A, Le Maréchal C, Trouvé P, Raguénès O, Chen JM, Férec C. Functional analysis of pancreatitis-associated missense mutations in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur J Hum Genet. 2007;15:936–942. doi: 10.1038/sj.ejhg.5201873. [DOI] [PubMed] [Google Scholar]
- 50.Masamune A, Kume K, Takagi Y, et al. N34S mutation in the SPINK1 gene is not associated with alternative splicing. Pancreas. 2007;34:423–428. doi: 10.1097/mpa.0b013e3180335fd0. [DOI] [PubMed] [Google Scholar]
- 51.Kereszturi E, Király O, Sahin-Tóth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut. 2009;58:545–549. doi: 10.1136/gut.2008.164947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boulling A, Chen JM, Callebaut I, Férec C. Is the SPINK1 p.Asn34Ser missense mutation per se the true culprit within its associated haplotype? WebmedCentral. Genetics. 2012;3:WMC003084. [Google Scholar]
- 53.Kereszturi E, Sahin-Tóth M. Pancreatic cancer cell lines heterozygous for the SPINK1 p.N34S haplotype exhibit diminished expression of the variant allele. Pancreas. 2017 (accepted). [DOI] [PMC free article] [PubMed]
- 54.Kume K, Masamune A, Mizutamari H, et al. Mutations in the serine protease inhibitor Kazal Type 1 (SPINK1) gene in Japanese patients with pancreatitis. Pancreatology. 2005;5:354–360. doi: 10.1159/000086535. [DOI] [PubMed] [Google Scholar]
- 55.Masamune A. Genetics of pancreatitis: the 2014 update. Tohoku J Exp Med. 2014;232:69–77. doi: 10.1620/tjem.232.69. [DOI] [PubMed] [Google Scholar]
- 56.Oh HC, Kim MH, Choi KS, et al. Analysis of PRSS1 and SPINK1 mutations in Korean patients with idiopathic and familial pancreatitis. Pancreas. 2009;38:180–183. doi: 10.1097/MPA.0b013e31818d1b90. [DOI] [PubMed] [Google Scholar]
- 57.Lee YJ, Kim KM, Choi JH, Lee BH, Kim GH, Yoo HW. High incidence of PRSS1 and SPINK1 mutations in Korean children with acute recurrent and chronic pancreatitis. J Pediatr Gastroenterol Nutr. 2011;52:478–481. doi: 10.1097/MPG.0b013e31820e2126. [DOI] [PubMed] [Google Scholar]
- 58.Cho SM, Shin S, Lee KA. PRSS1, SPINK1, CFTR, and CTRC pathogenic variants in Korean patients with idiopathic pancreatitis. Ann Lab Med. 2016;36:555–560. doi: 10.3343/alm.2016.36.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Sun XT, Weng XL, et al. Comprehensive screening for PRSS1, SPINK1, CFTR, CTRC and CLDN2 gene mutations in Chinese paediatric patients with idiopathic chronic pancreatitis: a cohort study. BMJ Open. 2013;3:e003150. doi: 10.1136/bmjopen-2013-003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun C, Liao Z, Jiang L, et al. The contribution of the SPINK1 c.194+2T>C mutation to the clinical course of idiopathic chronic pancreatitis in Chinese patients. Dig Liver Dis. 2013;45:38–42. doi: 10.1016/j.dld.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Chang YT, Wei SC, PC L, et al. Association and differential role of PRSS1 and SPINK1 mutation in early-onset and late-onset idiopathic chronic pancreatitis in Chinese subjects. Gut. 2009;58:885. doi: 10.1136/gut.2007.129916. [DOI] [PubMed] [Google Scholar]
- 62.Singh S, Choudhuri G, Agarwal S. Frequency of CFTR, SPINK1, and cathepsin B gene mutation in North Indian population: connections between genetics and clinical data. Sci World J. 2014;2014:763195. doi: 10.1155/2014/763195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hegyi E, Geisz A, Sahin-Tóth M, et al. SPINK1 promoter variants in chronic pancreatitis. Pancreas. 2016;45:148–153. doi: 10.1097/MPA.0000000000000412. [DOI] [PubMed] [Google Scholar]
- 64.Kume K, Masamune A, Kikuta K, Shimosegawa T. [-215G>A; IVS3+2T>C] mutation in the SPINK1 gene causes exon 3 skipping and loss of the trypsin binding site. Gut. 2006;55:1214. doi: 10.1136/gut.2006.095752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zou WB, Boulling A, Masson E, et al. Clarifying the clinical relevance of SPINK1 intronic variants in chronic pancreatitis. Gut. 2016;65:884–886. doi: 10.1136/gutjnl-2015-311168. [DOI] [PubMed] [Google Scholar]
- 66.Boulling A, Witt H, Chandak GR, et al. Assessing the pathological relevance of SPINK1 promoter variants. Eur J Hum Genet. 2011;19:1066–1073. doi: 10.1038/ejhg.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Derikx MH, Geisz A, Kereszturi É, Sahin-Tóth M. Functional significance of SPINK1 promoter variants in chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2015;308:G779–G784. doi: 10.1152/ajpgi.00022.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le Maréchal C, Chen JM, Le Gall C, et al. Two novel severe mutations in the pancreatic secretory trypsin inhibitor gene (SPINK1) cause familial and/or hereditary pancreatitis. Hum Mutat. 2004;23:205. doi: 10.1002/humu.9212. [DOI] [PubMed] [Google Scholar]
- 69.Palermo JJ, Lin TK, Hornung L, et al. Genophenotypic analysis of pediatric patients with acute recurrent and chronic pancreatitis. Pancreas. 2016;45:1347–1352. doi: 10.1097/MPA.0000000000000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.LaRusch J, Barmada MM, Solomon S, Whitcomb DC. Whole exome sequencing identifies multiple, complex etiologies in an idiopathic hereditary pancreatitis kindred. JOP. 2012;13:258–262. [PMC free article] [PubMed] [Google Scholar]
- 71.Gaia E, Salacone P, Gallo M, et al. Germline mutations in CFTR and PSTI genes in chronic pancreatitis patients. Dig Dis Sci. 2002;47:2416–2421. doi: 10.1023/A:1020579119691. [DOI] [PubMed] [Google Scholar]
- 72.Keiles S, Kammesheidt A. Identification of CFTR, PRSS1, and SPINK1 mutations in 381 patients with pancreatitis. Pancreas. 2006;33:221–227. doi: 10.1097/01.mpa.0000232014.94974.75. [DOI] [PubMed] [Google Scholar]
- 73.Perri F, Piepoli A, Stanziale P, Merla A, Zelante L, Andriulli A. Mutation analysis of the cystic fibrosis transmembrane conductance regulator (CFTR) gene, the cationic trypsinogen (PRSS1) gene, and the serine protease inhibitor, Kazal type 1 (SPINK1) gene in patients with alcoholic chronic pancreatitis. Eur J Hum Genet. 2003;11:687–692. doi: 10.1038/sj.ejhg.5201035. [DOI] [PubMed] [Google Scholar]
- 74.Király O, Boulling A, Witt H, et al. Signal peptide variants that impair secretion of pancreatic secretory trypsin inhibitor (SPINK1) cause autosomal dominant hereditary pancreatitis. Hum Mutat. 2007;28:447–469. doi: 10.1002/humu.20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boulling A, Keiles S, Masson E, Chen JM, Férec C. Functional analysis of eight missense mutations in the SPINK1 gene. Pancreas. 2012;41:329–330. doi: 10.1097/MPA.0b013e3182277b83. [DOI] [PubMed] [Google Scholar]
- 76.Sultan M, Werlin S, Venkatasubramani N. Genetic prevalence and characteristics in children with recurrent pancreatitis. J Pediatr Gastroenterol Nutr. 2012;54:645–650. doi: 10.1097/MPG.0b013e31823f0269. [DOI] [PubMed] [Google Scholar]
- 77.Masson E, Le Maréchal C, Chen JM, Frebourg T, Lerebours E, Férec C. Detection of a large genomic deletion in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur J Hum Genet. 2006;14:1204–1208. doi: 10.1038/sj.ejhg.5201684. [DOI] [PubMed] [Google Scholar]
- 78.Masson E, Le Maréchal C, Levy P, et al. Co-inheritance of a novel deletion of the entire SPINK1 gene with a CFTR missense mutation (L997F) in a family with chronic pancreatitis. Mol Genet Metab. 2007;92:168–175. doi: 10.1016/j.ymgme.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 79.Terlizzi V, De Gregorio F, Sepe A, et al. Brand new SPINK1 and CFTR mutations in a child with acute recurrent pancreatitis: a case report. Minerva Pediatr. 2013;65:669–672. [PubMed] [Google Scholar]
- 80.Werlin S, Konikoff FM, Halpern Z, et al. Genetic and electrophysiological characteristics of recurrent acute pancreatitis. J Pediatr Gastroenterol. 2015;60:675–679. doi: 10.1097/MPG.0000000000000623. [DOI] [PubMed] [Google Scholar]
- 81.Santhosh S, Witt H, te Morsche RH, et al. A loss of function polymorphism (G191R) of anionic trypsinogen (PRSS2) confers protection against chronic pancreatitis. Pancreas. 2008;36:317–320. doi: 10.1097/MPA.0b013e31815db4b3. [DOI] [PubMed] [Google Scholar]
- 82.Kume K, Masamune A, Takagi Y, et al. A loss-of-function p.G191R variant in the anionic trypsinogen (PRSS2) gene in Japanese patients with pancreatic disorders. Gut. 2009;58:820–824. doi: 10.1136/gut.2008.151688. [DOI] [PubMed] [Google Scholar]
- 83.Witt H, Luck W, Becker M, et al. Mutation in the SPINK1 trypsin inhibitor gene, alcohol use, and chronic pancreatitis. JAMA. 2001;285:2716–2717. doi: 10.1001/jama.285.21.2716-a. [DOI] [PubMed] [Google Scholar]
- 84.Schneider A, Pfützer RH, Barmada MM, Slivka A, Martin J, Whitcomb DC. Limited contribution of the SPINK1 N34S mutation to the risk and severity of alcoholic chronic pancreatitis: a report from the United States. Dig Dis Sci. 2003;48:1110–1115. doi: 10.1023/A:1023768829772. [DOI] [PubMed] [Google Scholar]
- 85.Archer H, Jura N, Keller J, Jacobson M, Bar-Sagi D. A mouse model of hereditary pancreatitis generated by transgenic expression of R122H trypsinogen. Gastroenterology. 2006;131:1844–1855. doi: 10.1053/j.gastro.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 86.Selig L, Sack U, Gaiser S, et al. Characterisation of a transgenic mouse expressing R122H human cationic trypsinogen. BMC Gastroenterol. 2006;6:30. doi: 10.1186/1471-230X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Athwal T, Huang W, Mukherjee R, et al. Expression of human cationic trypsinogen (PRSS1) in murine acinar cells promotes pancreatitis and apoptotic cell death. Cell Death Dis. 2014;5:e1165. doi: 10.1038/cddis.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Németh BC, Wartmann T, Halangk W, Sahin-Tóth M. Autoactivation of mouse trypsinogens is regulated by chymotrypsin C via cleavage of the autolysis loop. J Biol Chem. 2013;288:24049–24062. doi: 10.1074/jbc.M113.478800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ohmuraya M, Hirota M, Araki M, et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology. 2005;129:696–705. doi: 10.1016/j.gastro.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 90.Ohmuraya M, Hirota M, Araki K, Baba H, Yamamura K. Enhanced trypsin activity in pancreatic acinar cells deficient for serine protease inhibitor kazal type 3. Pancreas. 2006;33:104–106. doi: 10.1097/01.mpa.0000226889.86322.9b. [DOI] [PubMed] [Google Scholar]