Abstract
We analyzed the pathogenicity of various serotypes of Listeria monocytogenes using a Balb/c mouse intravenous injection model. The survival rates of mice inoculated with strains NS1/2b (serotype 1/2b), NS3b (serotype 3b) and NS 4b (serotype 4b) were 60, 63.6 and 63.6%, respectively. Although the survival rates were similar, the bacterial growth in the liver of NS3b-infected mice was 144.5-fold higher than that in the liver of NS4b-infected mice. Histopathological analyses suggest that the NS4b strain replicated more in monocytes/macrophages, whereas the NS3b strain replicated more in hepatocytes. These results raise a possibility that the serotype 4b strains replicated more in monocytes/macrophages compared to the other serotype strains. To assess this, we isolated CD11b-positive cells from mouse livers infected with EGDe (serotype 1/2a), NS1/2b, NS3b, NS4b and the serotype 4b strains 51414 and F17 and counted the number of live bacteria in these cells. CD11b-positive cells from the NS4b-, 51414- and F17-infected mice possessed 24.4- to 42.7-fold higher numbers of live bacteria than those from mice infected with EGDe and NS3b strains. These results suggest that serotype 4b strains replicated more in monocytes/macrophages than the other serotypes, and this may be involved in the pathogenicity of serotype 4b strains, particularly in the dissemination of L. monocytogenes through the host body.
Keywords: Listeria monocytogenes, monocyte/macrophage, pathogenicity, serotype
Listeria monocytogenes (L. monocytogenes) is a gram positive, facultative intracellular bacterium that causes listeriosis in humans and animals. L. monocytogenes infection in healthy adult individuals induces influenza-like symptoms and mild gastroenteritis [3]. Food-borne outbreaks have been reported in many countries since an outbreak due to contaminated coleslaw was reported in Canada in 1981 [22, 24]. The elderly, immunocompromised hosts, pregnant women and neonates exhibit particularly severe symptoms, such as sepsis, meningitis, abortion and neonatal listeriosis [7, 13]. The fatal rate among high-risk patients is approximately 20–30%, which is relatively high among food-borne diseases [10].
L. monocytogenes is divided into 13 serotypes: 1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4ab, 4b, 4c, 4d, 4e and 7 [18]. Most of the clinical isolates in sporadic and outbreak cases in humans belong to serotype 4b, followed by serotypes 1/2a and 1/2b. Nonetheless, several other serotypes, such as serotype 3b, also contribute to the incidence of sporadic listeriosis [14, 19]. Further, an outbreak from contaminated butter in Finland in 1998–1999 was caused by a serotype 3a strain [16]. To date, most of the studies regarding the pathogenicity of L. monocytogenes have been performed using the standard strains EGD, EGDe and 10403S that belong to serotype 1/2a, and some studies used serotypes 4b and 1/2b. A previous study by von Koening et al. analyzed the pathogenicity of serotypes 1/2b, 3a, 4b and 4d in NMRI mice infected with L. monocytogenes serotypes via intraperitoneal and intravenous (i.v.) routes. Their results suggest that the pathogenicity of serotypes 3a and 4d was lower than that of serotypes 4b and 1/2b [27]. However, there are little information about the pathogenicity of serotypes other than 1/2a, 1/2b and 4b. Also, it still remains unclear if there are any differences among serotypes in the pathogenic mechanism.
In the current study, we compared the pathogenicity of 7 serotypes of L. monocytogenes as well as a standard strain EGDe in a Balb/c mouse i.v. injection model, which is an established model for analyzing the virulence of L. monocytogenes [21]. Our data suggest that serotype 3b might be as pathogenic as serotypes 4b and 1/2b. Our data also suggest that serotype 4b strains had a distinct feature in replication in monocytes/macrophages compared to the other serotypes.
MATERIALS AND METHODS
Bacterial strains
L. monocytogenes strains NS1/2a, NS1/2b, NS3a, NS3b, NS3c, NS4a and NS4b (serotypes 1/2a, 1/2b, 3a, 3b, 3c, 4a and 4b, respectively) were provided by the National Institute of Health, which was renamed as the National Institute of Infectious Diseases in 1997. The sources of these strains are unknown. Strains EGDe (serotype 1/2a, a mouse-passaged strain of EGD isolated from guinea pig) and 51414 (serotype 4b, isolated from raw milk associated with listeriosis outbreak in Massachusetts) were purchased from ATCC (Manassas, VA, U.S.A.). Strain F17 (serotype 4b complex) was isolated from the feces of a clinically healthy sheep. Bacteria were grown in Heart Infusion Broth or on Heart Infusion Agar (Nissui, Tokyo, Japan).
Mice
Total 209 of 6- to 7-week-old female Balb/c mice were purchased from CLEA Japan Inc. (Hamamatsu, Japan). All procedures were approved by the Animal Experiment Committee of the Graduate School of Veterinary Medicine, Hokkaido University. Mice were euthanized by overexposure to sevoflurane (Maruishi Phermaceutical Co., Ltd., Osaka, Japan).
Survival rate of mice in a Balb/c intravenous (i.v.) infection model
Seventy two mice were inoculated intravenously with 104 colony forming unit (CFU) of L. monocytogenes strains EGDe (n=8), NS1/2a (n=8), NS1/2b (n=10), NS3a (n=8), NS3b (n=11), NS3c (n=8), NS4a (n=8) or NS4b (n=11) in 50 µl of Hank’s balanced salt solution (HBSS) (Sigma-Aldrich, Tokyo, Japan). Fifty microliters of HBSS was inoculated in 3 mice as a negative control (mock). Mice were observed until 7 days post inoculation (dpi).
Bacterial growth in livers and spleens
Eighty eight mice were inoculated intravenously with L. monocytogenes strains EGDe (n=12), NS1/2a (n=12), NS1/2b (n=10), NS3a (n=11), NS3b (n=11), NS3c (n=11), NS4a (n=10) or NS4b (n=11). HBSS was inoculated in 9 mice for mock. At 1, 3 and 6 dpi, the livers and spleens were collected from 3 to 4 mice and homogenized in phosphate buffered saline (PBS) at 10% w/v. Ten-fold serial dilutions were made with PBS and spread onto the agar plates. The plates were incubated at 37°C for 24 hr, and the colonies were counted.
Histopathological analysis
Thirteen mice were inoculated intravenously with L. monocytogenes strains EGDe (n=4), NS1/2b (n=3), NS3b (n=3) or NS4b (n=3). HBSS was inoculated to 3 mice for mock. At 3 dpi, the livers, spleens, kidneys, hearts, lungs and brains were collected. For analysis of dead mice, we used the tissue from dead mice used for analysis of the survival rate. The number of dead mice available for the examination was 2 for NS1/2b-infected mice, 1 for NS3a-infected mice, 2 for NS3b-infected mice and 2 for NS4b-infected mice. The tissues were fixed with 10% phosphate-buffered formalin and embedded in paraffin. The sections were subjected to hematoxylin and eosin (HE) staining and immunostaining. For immunohistochemistry, anti-L. monocytogenes rabbit polyclonal antibodies (Abcam, Cambridge, U.K., No. 35132) were incubated at a 1:5,000 dilution as primary antibodies at 4°C overnight. EnVision+ System HRP Labeled Polymer Anti-Rabbit (Dako, Tokyo, Japan, no. K4002) were incubated for 30 min at room temperature. The ImmPACT DAB Substrate (Vector Laboratories, Vurlingame, CA, U.S.A.) was reacted to visualize the immunoreactivity. Nuclei were counterstained with hematoxylin. For double immunofluorescence staining, the sections were incubated with anti-L. monocytogenes antibodies as described above, and Alexa Fluor 488-labeled anti-Rabbit IgG (H+L) F (ab’)2 goat polyclonal antibodies (Thermo Fisher Scientific, Yokohama, Japan) were used as secondary antibodies. After being fixed with 4% paraformaldehyde, the sections were incubated with Alexa Fluor 555-labeled anti-Iba1 rabbit IgG (order made by Sigma). Fluorescence labeling was performed using the Alexa Fluor 555 Antibody Labeling Kit (Thermo Fisher Scientific).
Isolation of CD11b-positive cells from livers
Twenty one mice were inoculated intravenously with L. monocytogenes strains EGDe (n=4), NS1/2b (n=4), NS3b (n=4), NS4b (n=3), 51414 (n=3) or F17 (n=3). At 3 dpi, livers were collected, minced and digested with 0.1 U/ml of Dispase II (Roche, Basel, Swetzerland) at 37°C for 10 min. After the cells were dispersed by pipetting, the debris was removed by filtration using 100 µm pore filter. The cell suspension was centrifuged at 300 g for 10 min, and the cells were resuspended in 0.5% bovine serum albumin (Sigma) in HBSS. CD11b-positive cells were isolated using CD11b microbeads, LS column and a MACS separator according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The cells were serially diluted and spread onto the agar plates. After incubation at 37°C for 24 hr, the number of colonies was counted.
RESULTS
Survival rate of Balb/c mice infected intravenously with 7 serotypes of L. monocytogenes
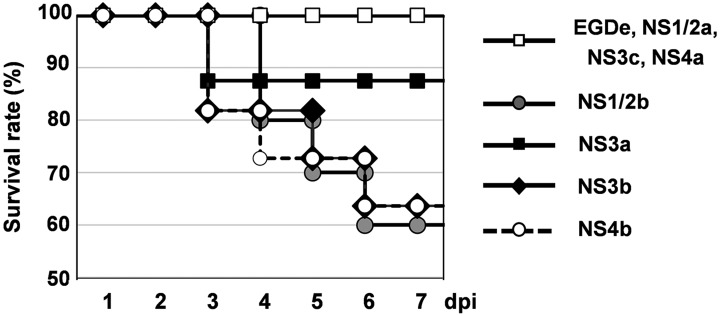
To compare the pathogenicity among serotypes of L. monocytogenes, we inoculated L. monocytogenes to Balb/c mice intravenously (Fig. 1). All mice showed ruffled fur at 2 dpi, but NS3c-, NS4a- and mock-infected mice did not. Mice inoculated with the NS1/2b, NS3b and NS4b strains died from 3 to 6 dpi, and the survival rates were 60% (6/10 mice), 63.6% (7/11 mice) and 63.6% (7/11 mice), respectively. Mice that survived until 7 dpi appeared to recover from the infection as they did not show any clinical signs. One of the eight mice inoculated with the NS3a strain died at 3 dpi, and the survival rate was 87.5%. The survival rates of mice inoculated with the EGDe, NS1/2a, NS3c and NS4a strains were 100% (8/8 mice for each).
Fig. 1.
Survival curve of Balb/c mice infected with 7 serotypes of L. monocytogenes. Eight to 11 mice were inoculated intravenously with each strain. Survival of mice was observed until 7 dpi.
Growth of 7 serotypes of L. monocytogenes in the livers and spleens of Balb/c mice
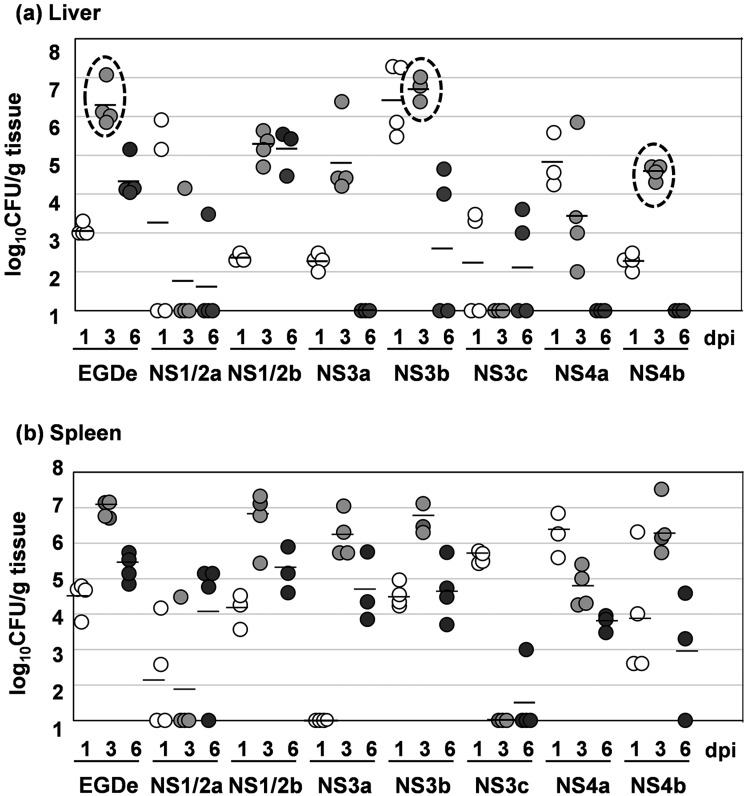
After an i.v. inoculation to mice, L. monocytogenes grew in the liver and spleen [26]. To examine if there were any differences in the bacterial growth among the serotypes, we counted the number of live bacteria in the liver and spleen at 1, 3 and 6 dpi (Fig. 2a and 2b). In the EGDe-, NS1/2b-, NS3a-, NS3b- and NS4b-infected mice, bacterial counts reached a peak at 3 dpi and decreased at 6 dpi both in the livers and spleens. Interestingly, the average bacterial count in the livers of NS3b-infected mice (106.72 CFU/g tissue) was 144.5-fold higher than that of NS4b-infected mice (104.57 CFU/g tissue; Fig. 2a, dashed circles), although their survival rates were the same. Further, the average bacterial count in the livers of EGDe-infected mice (106.25 CFU/g tissue) was 49-fold higher than those in the livers of the 4b-infected mice (Fig. 2a, dashed circles), although no mice died by EGDe infection. The bacterial growth of the NS1/2a, NS3c and NS4a strains was much lower than that of the EGDe, NS1/2b, NS3b and NS4b strains. No bacterial colony was observed in the livers and spleens of mock-infected mice.
Fig. 2.
Bacterial growth of 7 serotypes of L. monocytogenes. (a) Bacterial growth in the liver. (b) Bacterial growth in the spleen. Mice were inoculated intravenously with each strain. At 1, 3 and 6 dpi, the livers and spleens were harvested from 3 or 4 mice for each strain, and live bacteria were counted. The detection limit was 102 CFU/g tissue. The bacterial counts in the livers of EGDe-, NS3b- and NS4b-infected mice at 3 dpi were enclosed by dashed circles for emphasis. White dots, 1 dpi; light gray dots, 3 dpi; dark gray dots, 6 dpi. Bars indicate the means of three or four samples.
Different cell tropism in the liver among the strains EGDe, NS1/2b, NS3b and NS4b
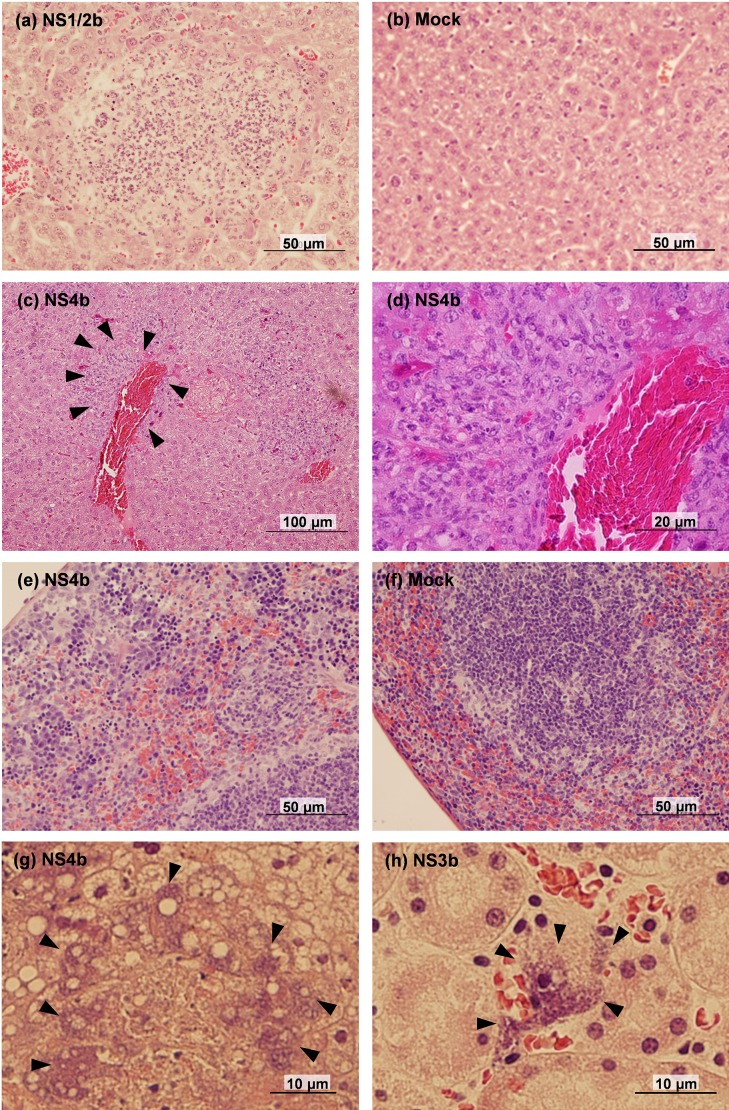
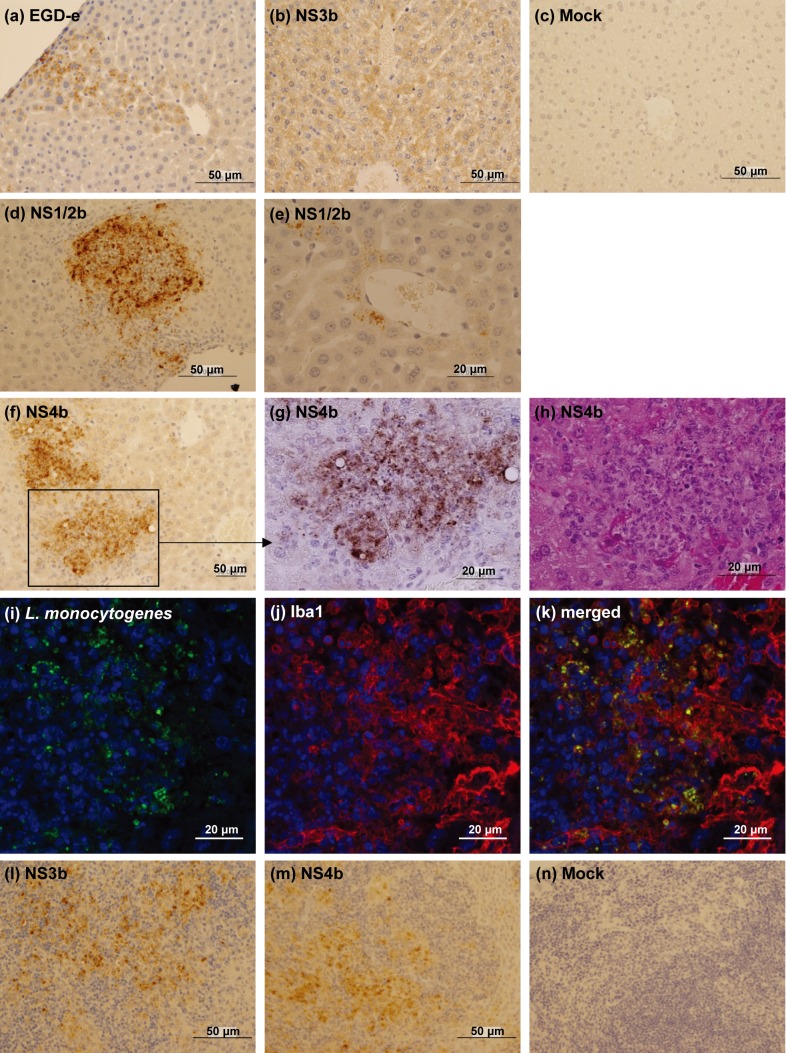
To explore the reason for low bacterial growth in the livers of NS4b-infected mice, we performed a histopathological analysis at 3 dpi (Figs. 3 and 4). In the liver, multiple abscesses were observed in EGDe-, NS1/2b-, NS3b- and NS4b-infected mice (Fig. 3a), although the sizes of the abscesses were smaller in EGDe-infected mice compared to the others. In addition, infiltration of macrophages around blood vessels and bile ducts was prominent in the livers of NS4b-infected mice (Fig. 3c and 3d). In EGDe- and NS3b-infected mice, bacterial antigens were mainly detected in the hepatocytes (Fig. 4a and 4b). On the other hand, in NS4b-infected mice, bacterial antigens were mainly detected in the foci of inflammatory cells (Fig. 4f–4h). The bacterial antigen-positive cells in the livers of NS4b-infected mice were positive for a monocyte/macrophage marker, Iba1 (Fig. 4i–4k). In NS1/2b-infected mice, bacterial antigens were detected in both foci of inflammatory cells (Fig. 4d) and hepatocytes (Fig. 4e). The histological findings were observed in all examined mice. These results suggest that the NS4b strain infected monocytes/macrophages more than the other strains. There was no significant histopathological change and bacterial antigen in the livers of mock-infected mice (Figs. 3b and 4c).
Fig. 3.
Histopathology of L. monocytogenes-infected Balb/c mice. (a) Abscess in the liver of NS1/2b-infected mouse. (b) Liver of mock-infected mouse. (c) Perivascular infiltration of macrophages in the liver of NS4b-infected mouse. Arrowheads surround the area with macrophage infiltration. (d) Magnified image of the area surrounded by arrowheads in (c). (e) Migration of macrophages in the splenic sinus of NS4b-infected mouse. (f) Spleen of mock-infected mouse. (g, h) Bacterial clumps in the dead mice, observed in the liver of NS4b-infected mouse (g) and in the kidney of NS3b-infected mouse (h). Arrowheads show the bacterial clumps.
Fig. 4.
Immunostaining for L. monocytogenes antigens. (a–g) Distribution of the L. monocytogenes antigens in the livers of EGDe-(a), NS3b-(b), mock-(c), NS-1/2b-(d, e) and NS4b-infected (f, g) mice. (g) Magnified image of the enclosed area of (f). (h) HE staining of the serial sections of (g). (i–k) Double immunofluorescence staining of L. monocytogenes antigens and Iba1 in the liver of NS4b-infected mouse. Green, L. monocytogenes; Red, Iba1; Blue, Nuclei. (l–n) Distribution of L. monocytogenes antigens in the spleens of NS3b-(l), NS4b-(m) and mock-infected (n) mice.
Histopathological findings in the tissues other than the livers
We further performed histopathological analysis on the spleens, kidneys, hearts, lungs and brains of EGDe-, NS1/2b-, NS3b- and NS4b-infected mice at 3 dpi (Fig. 3e–3h). In the spleens, migration of monocytes/macrophages was prominent in EGDe-, NS1/2b-, NS3b- and NS4b-infected mice (Fig. 3e), but not in mock-infected mice (Fig. 3f). There were no histopathological lesions in the kidneys, hearts, lungs and brains (data not shown). Bacterial antigens were mainly detected in the dendritic cells of the lymphoid follicles in the spleens of EGDe-, NS1/2b-, NS3b- and NS4b-infected mice (Fig. 4l and 4m). Bacterial antigens were occasionally in monocytes/macrophages of the splenic sinus of EGDe-, NS1/2b-, NS3b- and NS4b-infected mice. No bacterial antigen was detected in mock-infected mice (Fig. 4n). We also examined dead mice infected with NS1/2b, NS3a, NS3b and NS4b strains. Bacterial clumps were observed in the livers, spleens, and kidneys regardless of the L. monocytogenes strain (Fig. 3g and 3h). Multifocal necrosis were observed in the livers and spleens of all dead mice, although there were no histological lesions in the other tissues. Bacterial antigens were also observed in the blood vessels of lungs and meninges of an NS4b-infected mouse, although any histopathological legions were not observed in these tissues (data not shown).
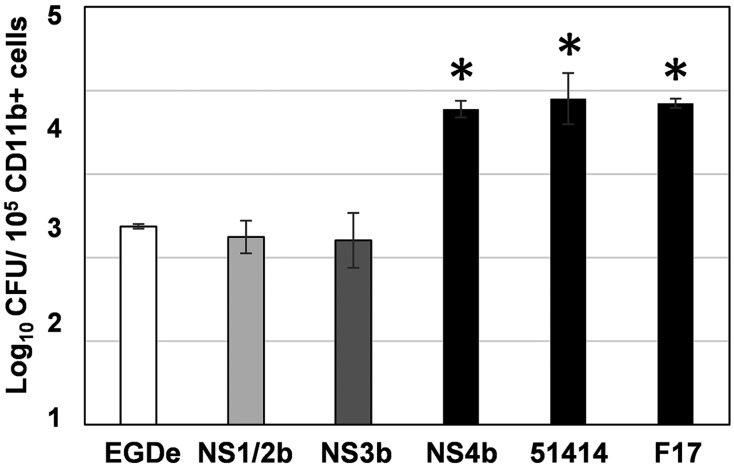
Difference in the replication in monocytes/macrophages between serotype 4b strains and the other serotypes
The histopathological analysis of the livers raised a possibility that the NS4b strain replicated in monocytes/macrophages more than the other strains. We hypothesized that this feature might be common among the serotype 4b strains. To assess this, we counted the number of live bacteria in CD11b-positive cells from the livers of mice infected with strains EGDe, NS1/2b, NS3b, NS4b and the serotype 4b strains 51414 and F17 at 3 dpi (Fig. 5). In 105 of CD11b-positive cells from the livers of EGDe- 1/2b- and NS3b-infected mice, 2.5 × 102, 2.2 × 102 and 2.2 × 102 CFU of live bacteria were detected, respectively. On the other hand, 6.1 × 103 CFU of bacteria were detected in those from NS4b-infected mouse livers, which was significantly higher than those from EGDe- and NS3b-infected mouse livers (P<0.05, Dunnett’s test). Further, as we expected, 9.4 × 103 and 7 × 103 CFU of bacteria were detected in the CD11b-positive cells from mouse livers infected with the serotype 4b strains 51414 and F17, and these bacterial counts were significantly higher than those from EGDe-, NS1/2b- and NS3b-infected mouse livers (P<0.05, Dunnett’s test). These results suggest that L. monocytogenes serotype 4b strains replicated in CD11b-positive monocytes/macrophages more than the other serotypes.
Fig. 5.
Bacterial counts in CD11b-positive cells isolated from L. monocytogenes-infected mouse livers. Three or four mice were inoculated intravenously with each strain. At 3 dpi, CD11b-positive cells were isolated from the liver, and live bacteria were counted. *P<0.05 vs EGDe (Dunnett’s test).
DISCUSSION
In the current study, our data suggest a possibility that the pathogenesis of listeriosis caused by serotype 4b strains may be different from the others, because L. monocytogenes serotype 4b strains replicated more than the other serotypes in CD11b-positive monocytes/macrophages in the liver. This feature may be involved in the pathogenicity of L. monocytogenes serotype 4b strains, particularly in the dissemination of L. monocytogenes through the host body. Drevets et al. [5] suggest that intracellular L. monocytogenes in phagocytes play an important role in the systemic spread of bacteria replicated in the liver and spleen. In the current study, we isolated cells using anti-CD11b antibody-coated microbeads. Although neutrophils as well as monocytes/macrophages are CD11b-positive, the results of Fig. 5 were more likely to be indicative of L. monocytogenes infection to monocytes/macrophages because the antigen-positive cells in the livers of NS4b-infected mice were positive for a monocyte/macrophage marker, Iba1 (Fig. 4i–4k). The importance of monocytes in the dissemination of bacteria during systemic L. monocytogenes infection has been suggested by previous studies. Most of the L. monocytogenes-infected leukocytes in the blood have been morphologically identified as mononuclear phagocytes [4]. Further, Drevets et al. [6] suggested that Ly6Chigh monocytes contributed to the transport of L. monocytes into the brain. In contrast to the livers, there was no difference in the bacterial growth in the spleen between serotype 4b strains and the others. Most of the L. monocytogenes antigen-positive cells appeared to be dendritic cells in the splenic follicles, while some monocytes/macrophages in the sinus were also positive. The identical cell tropism may result in the similar bacterial growth in the spleens of serotype 4b strains and the others.
A serotype 3b strain, NS3b, was as pathogenic as the NS4b strain in Balb/c mice when injected intravenously. Human clinical cases caused by serotype 3b strains are not common, although serotype 3b strains are occasionally isolated from food and the environment [2, 23]. One possible reason for this is that the successful invasion of serotype 3b strain via the oral route may be quite limited compared with that of serotypes 4b, 1/2a and 1/2b strains. Indeed, the serotype 3b strain seems to be less invasive to intestinal epithelial cells than serotypes 4b and 1/2b strains, as Bueno et al. demonstrated using Caco-2 cells [1]. Further, it may be possible that serotype 3b strains are pathogenic only when invasion to the bloodstream has been established. The low infectivity to monocytes/macrophages may result in an insufficient invasion into the bloodstream.
Serotypes 1/2a and 1/2b strains are also often isolated from human clinical cases and foods. The majority of food isolates belong to serotype 1/2a, while food-borne outbreaks of human listeriosis are mainly caused by serotype 4b strains [8, 9, 11, 12, 17, 20, 25]. However, multistate outbreaks by serotypes 1/2a and 1/2b strains occurred in the United States in 2011 [17]. In Canada, serotype 1/2a isolates with very similar pulsed-field gel electrophoresis patterns predominantly caused human listeriosis from 1988 to 2010 [12]. Therefore, the pathogenicity of serotypes 1/2a and 1/2b strains may be variable depending on the strain or host immune condition. In the current study, the NS1/2a strain seemed to be low pathogenic to a Balb/c i.v. injection model, because the mortality late was low, while the NS1/2b strain caused the highest mortality. L. monocytogenes antigens in the livers of NS1/2b-infected mice were detected both in hepatocytes and monocytes/macrophages. However, the NS1/2b strain did not replicate in CD11b-positive macrophages as successfully as serotype 4b strains. Although the reason for unsuccessful replication of the NS1/2b strain in macrophages is unknown for now, it would be of interest to analyze the factors that determine the replication ability of L. monocytogenes in macrophages in the future.
The cause of death for the mice infected with NS1/2b, NS3b and NS4b strains was most likely due to sepsis, because bacterial clumps were often observed in the livers and spleens of dead animals regardless of the strains. Further, L. monocytogenes antigens were detected in the blood vessels in the lungs, kidneys and meninges. These results suggest that replication ability in macrophages did not affect the occurrence of sepsis, once adequate amount of L. monocytogenes invade the blood stream. As we injected L. monocytogenes into the tail vein here, significance of replication ability in macrophages on bacterial dissemination in the host body should be assessed in the future by using an oral exposure of a transgenic mouse model expressing human E-cadherin in the intestinal epithelium, in which L. monocytogenes invade from intestinal epithelium more efficiently than from those of wild type mice [15]. Meningoencephalitis is one of the clinical conditions caused by L. monocytogenes infection in humans and animals. However, meningoencephalitis hardly occurred in the mouse model used in the current study. Although we speculate that the sufficient replication of L. monocytogenes in macrophages might increase the incidence of meningoencephalitis, this should be also examined by using an appropriate model in the future, such as intraperitoneal inoculation of L. monocytogenes-infected macrophages as described by Drevets et al. [5].
Acknowledgments
This work was supported by grants from the Morinaga Foundation for Health and Nutrition, and Hokkaido University Research Center for Zoonosis Control.
REFERENCES
- 1.Bueno V. F., Banerjee P., Banada P. P., José de Mesquita A., Lemes-Marques E. G., Bhunia A. K.2010. Characterization of Listeria monocytogenes isolates of food and human origins from Brazil using molecular typing procedures and in vitro cell culture assays. Int. J. Environ. Health Res. 20: 43–59. doi: 10.1080/09603120903281283 [DOI] [PubMed] [Google Scholar]
- 2.Chambel L., Sol M., Fernandes I., Barbosa M., Zilhão I., Barata B., Jordan S., Perni S., Shama G., Adrião A., Faleiro L., Requena T., Peláez C., Andrew P. W., Tenreiro R.2007. Occurrence and persistence of Listeria spp. in the environment of ewe and cow’s milk cheese dairies in Portugal unveiled by an integrated analysis of identification, typing and spatial-temporal mapping along production cycle. Int. J. Food Microbiol. 116: 52–63. doi: 10.1016/j.ijfoodmicro.2006.12.035 [DOI] [PubMed] [Google Scholar]
- 3.Dalton C. B., Austin C. C., Sobel J., Hayes P. S., Bibb W. F., Graves L. M., Swaminathan B., Proctor M. E., Griffin P. M.1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336: 100–105. doi: 10.1056/NEJM199701093360204 [DOI] [PubMed] [Google Scholar]
- 4.Drevets D. A.1999. Dissemination of Listeria monocytogenes by infected phagocytes. Infect. Immun. 67: 3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drevets D. A., Jelinek T. A., Freitag N. E.2001. Listeria monocytogenes-infected phagocytes can initiate central nervous system infection in mice. Infect. Immun. 69: 1344–1350. doi: 10.1128/IAI.69.3.1344-1350.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drevets D. A., Dillon M. J., Schawang J. S., Van Rooijen N., Ehrchen J., Sunderkötter C., Leenen P. J.2004. The Ly-6Chigh monocyte subpopulation transports Listeria monocytogenes into the brain during systemic infection of mice. J. Immunol. 172: 4418–4424. doi: 10.4049/jimmunol.172.7.4418 [DOI] [PubMed] [Google Scholar]
- 7.Fleming D. W., Cochi S. L., MacDonald K. L., Brondum J., Hayes P. S., Plikaytis B. D., Holmes M. B., Audurier A., Broome C. V., Reingold A. L.1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 312: 404–407. doi: 10.1056/NEJM198502143120704 [DOI] [PubMed] [Google Scholar]
- 8.Gilbreth S. E., Call J. E., Wallace F. M., Scott V. N., Chen Y., Luchansky J. B.2005. Relatedness of Listeria monocytogenes Isolates recovered from selected ready-to-eat foods and listeriosis patients in the United States. Appl. Environ. Microbiol. 71: 8115–8122. doi: 10.1128/AEM.71.12.8115-8122.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilot P., Genicot A., André P.1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34: 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Milian A., Payeras-Cifre A.2014. What is new in listeriosis? Biomed. Res. Int. 2014: 358051. doi: 10.1155/2014/358051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathariou S., Graves L., Buchrieser C., Glaser P., Siletzky R. M., Swaminathan B.2006. Involvement of closely related strains of a new clonal group of Listeria monocytogenes in the 1998-99 and 2002 multistate outbreaks of foodborne listeriosis in the United States. Foodborne Pathog. Dis. 3: 292–302. doi: 10.1089/fpd.2006.3.292 [DOI] [PubMed] [Google Scholar]
- 12.Knabel S. J., Reimer A., Verghese B., Lok M., Ziegler J., Farber J., Pagotto F., Graham M., Nadon C. A. (Canadian Public Health Laboratory, N.), Gilmour M.W. 2012. Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 50: 1748––1751.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linnan M. J., Mascola L., Lou X. D., Goulet V., May S., Salminen C., Hird D. W., Yonekura M. L., Hayes P., Weaver R., Audurier A., Plikaytis B. D., Fannin S. L., Kleks A., Broome C. V.1988. Epidemic listeriosis associated with Mexican-style cheese. N. Engl. J. Med. 319: 823–828. doi: 10.1056/NEJM198809293191303 [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Valladares G., Tham W., Parihar V. S., Helmersson S., Andersson B., Ivarsson S., Johansson C., Ringberg H., Tjernberg I., Henriques-Normark B., Danielsson-Tham M. L.2014. Human isolates of Listeria monocytogenes in Sweden during half a century (1958-2010). Epidemiol. Infect. 142: 2251–2260. doi: 10.1017/S0950268813003385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecuit M., Vandormael-Pournin S., Lefort J., Huerre M., Gounon P., Dupuy C., Babinet C., Cossart P.2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292: 1722–1725. doi: 10.1126/science.1059852 [DOI] [PubMed] [Google Scholar]
- 16.Lyytikäinen O., Autio T., Maijala R., Ruutu P., Honkanen-Buzalski T., Miettinen M., Hatakka M., Mikkola J., Anttila V. J., Johansson T., Rantala L., Aalto T., Korkeala H., Siitonen A.2000. An outbreak of Listeria monocytogenes serotype 3a infections from butter in Finland. J. Infect. Dis. 181: 1838–1841. doi: 10.1086/315453 [DOI] [PubMed] [Google Scholar]
- 17.McCollum J. T., Cronquist A. B., Silk B. J., Jackson K. A., O’Connor K. A., Cosgrove S., Gossack J. P., Parachini S. S., Jain N. S., Ettestad P., Ibraheem M., Cantu V., Joshi M., DuVernoy T., Fogg N. W., Jr, Gorny J. R., Mogen K. M., Spires C., Teitell P., Joseph L. A., Tarr C. L., Imanishi M., Neil K. P., Tauxe R. V., Mahon B. E.2013. Multistate outbreak of listeriosis associated with cantaloupe. N. Engl. J. Med. 369: 944–953. doi: 10.1056/NEJMoa1215837 [DOI] [PubMed] [Google Scholar]
- 18.Mclauchlin J., Rees C. E. D.2009. Phylum XIII. Firmicutes Family III. Lieteriaceae Genus I. Liesteria Pirie 1940a, 383AL pp. 244–255. In: Burgey’s Manual of Systematic Bacteriology, volume 3, 2nd ed. (De Vos, P. G., Garrity, G. M., Jones, D., Krieg, N. R., Ludwig, W., Rainey, F. A., Schleifer, K. H., and Whitman, W. B. eds.), Splinger, New York. [Google Scholar]
- 19.Pontello M., Guaita A., Sala G., Cipolla M., Gattuso A., Sonnessa M., Gianfranceschi M. V.2012. Listeria monocytogenes serotypes in human infections (Italy, 2000-2010). Ann. Ist. Super. Sanita 48: 146–150. doi: 10.4415/ANN_12_02_07 [DOI] [PubMed] [Google Scholar]
- 20.Revazishvili T., Kotetishvili M., Stine O. C., Kreger A. S., Morris J. G., Jr, Sulakvelidze A.2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42: 276–285. doi: 10.1128/JCM.42.1.276-285.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roche S., Velge P., Liu D.2008. Virulence determination. pp. 241–270. In: Handbook of Listeria monocytogenes. (Liu, D. ed.), CRC Press, New York. [Google Scholar]
- 22.Schlech W. F., 3rd, Lavigne P. M., Bortolussi R. A., Allen A. C., Haldane E. V., Wort A. J., Hightower A. W., Johnson S. E., King S. H., Nicholls E. S., Broome C. V.1983. Epidemic listeriosis--evidence for transmission by food. N. Engl. J. Med. 308: 203–206. doi: 10.1056/NEJM198301273080407 [DOI] [PubMed] [Google Scholar]
- 23.Soni D. K., Singh R. K., Singh D. V., Dubey S. K.2013. Characterization of Listeria monocytogenes isolated from Ganges water, human clinical and milk samples at Varanasi, India. Infect. Genet. Evol. 14: 83–91. doi: 10.1016/j.meegid.2012.09.019 [DOI] [PubMed] [Google Scholar]
- 24.Swaminathan B., Gerner-Smidt P.2007. The epidemiology of human listeriosis. Microbes Infect. 9: 1236–1243. doi: 10.1016/j.micinf.2007.05.011 [DOI] [PubMed] [Google Scholar]
- 25.Tresse O., Shannon K., Pinon A., Malle P., Vialette M., Midelet-Bourdin G.2007. Variable adhesion of Listeria monocytogenes isolates from food-processing facilities and clinical cases to inert surfaces. J. Food Prot. 70: 1569–1578. doi: 10.4315/0362-028X-70.7.1569 [DOI] [PubMed] [Google Scholar]
- 26.Vázquez-Boland J. A., Kuhn M., Berche P., Chakraborty T., Domínguez-Bernal G., Goebel W., González-Zorn B., Wehland J., Kreft J.2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14: 584–640. doi: 10.1128/CMR.14.3.584-640.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Koenig C. H., Heymer B., Hof H., Finger H.1983. Course of infection and development of immunity in experimental infection of mice with Listeria serotypes. Infect. Immun. 40: 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]