Abstract
Proteomic and genomic techniques have reached full maturity and are providing unforeseen details for the comprehensive understanding of disease pathologies at a fraction of previous costs. However, for kidney diseases, many gaps in such information remain to inhibit major advances in the prevention, treatment and diagnostics of these devastating diseases, which have enormous global impact. The discovery of ubiquitous extracellular vesicles (EV) in all bodily fluids is rapidly increasing the fundamental knowledge of disease mechanisms and the ways in which cells communicate with distant locations in processes of cancer spread, immunological regulation, barrier functions and general modulation of cellular activity. In this review, we describe some of the most prominent research streams and findings utilizing urinary extracellular vesicles as highly versatile and dynamic tools with their extraordinary protein and small regulatory RNA species. While being a highly promising approach, the relatively young field of EV research suffers from a lack of adherence to strict standardization and carefully scrutinized methods for obtaining fully reproducible results. With the appropriate guidelines and standardization achieved, urine is foreseen as forming a unique, robust and easy route for determining accurate and personalized disease signatures and as providing highly useful early biomarkers of the disease pathology of the kidney and beyond.
Keywords: Kidney disease, Biomarkers, MicroRNA, Extracellular vesicles, Urine
Introduction
Following extensive genome-wide analyses and the full maturation of integrative technologies such as RNA-sequencing (RNA-Seq) techniques to reveal the crucial regulatory functions of the “non-coding” DNA bulk of human and other species (Lee et al. 2010), recent years have seen strong advances in a variety of organ functions with unprecedented molecular accuracy. Nevertheless, an extensive understanding of, for example, key kidney functions, including mechanisms of glomerular filtration and the formation of urine from primary urine to the final voided urine, remains to be achieved (Saritas et al. 2015; Maas et al. 2016). The combined use of proteomics, lipidomics, metabolomics and comprehensive genomic analyses, together with advanced systems biology algorithms, offers the promise of major new discoveries for the benefit of patients with kidney disease.
Urine formation provides a key route for waste removal from the body. In addition to immediate metabolic waste products, variable amount of electrolytes, small peptides and larger functional proteins (Hildonen et al. 2016) are removed. Based, for example, on hydration status, diet and excercise, the electrolyte concentration may variate considerably. For years, this dynamic nature of urine has presented a challenge for the full exploitation of the potential of this bodily fluid, which is easy to collect.
The structural basis of glomerular filtration has been defined down to the fine molecular level, including the identification of the respective signaling and functional pathways, whereas the functional mechanisms remain to be fully understood (Patrakka and Tryggvason 2010; Pollak et al. 2014; Scott and Quaggin 2015). At the same time, however, the continuous global increase of chronic kidney diseases (CKD) attributable to a variety of causes involves over 10 % of most populations and geographical areas (El Nahas 2005; Remuzzi et al. 2006). These alarming numbers call for practical advances leading to early diagnostics, better therapies and cost-efficient personalized disease management even in less fortunate areas and countries. Many of the technologies required are beginning to reach their full maturity for the achievement of these goals.
The increase of CKD is mostly the result of the rapid increase of diabetes world-wide (El Nahas 2005). However, patient subpopulations susceptible to life-threatening diabetic end-organ damage mostly cannot be identified early enough to prevent the progress of disease. Apart from the medicinal modulation of the renin-angiotensin axis (Remuzzi et al. 2006), only a few other CKD disease mechanisms have been identified and, accordingly, the repertoire of targeted medication available to halt disease progression is not satisfactory. Currently, the exhaustive genetic data produced have provided few clues to the factors and mechanisms for the identification of the vulnerable target subpopulations or even of those who might benefit from the currently available treatment options. In spite of numerous proposed early biomarkers to monitor disease progression and activity, none have been fully validated for wide clinical use.
The discovery of vesicles of various sizes secreted from practically all cell types in the body (Thery et al. 2009; Yanez-Mo et al. 2015) is rapidly revolutionizing our understanding of key biological phenomena including organ growth and development, cancer and metastasis, cellular homeostasis, immunological defense, barrier functions to the exterior and, importantly, intercellular communication (for references, see Thery et al. 2009, van der Pol et al. 2012). Although this is a relatively young field, many new cellular-intercellular mechanisms including the spread of viral infections (Nour and Modis 2014) and the spread of cancer cells (Tomasetti et al. 2017) can be explained by messages from extracellular vesicles (EV).
Extracellular vesicles
EV are a heterogenous group of membrane-coated particles of various sizes (Fig.1) either actively or passively secreted from cells by well-established mechanisms (Thery et al. 2009; van der Pol et al. 2012; Akers et al. 2013; Ciardiello et al. 2016; Morrison et al. 2016; Table 1).
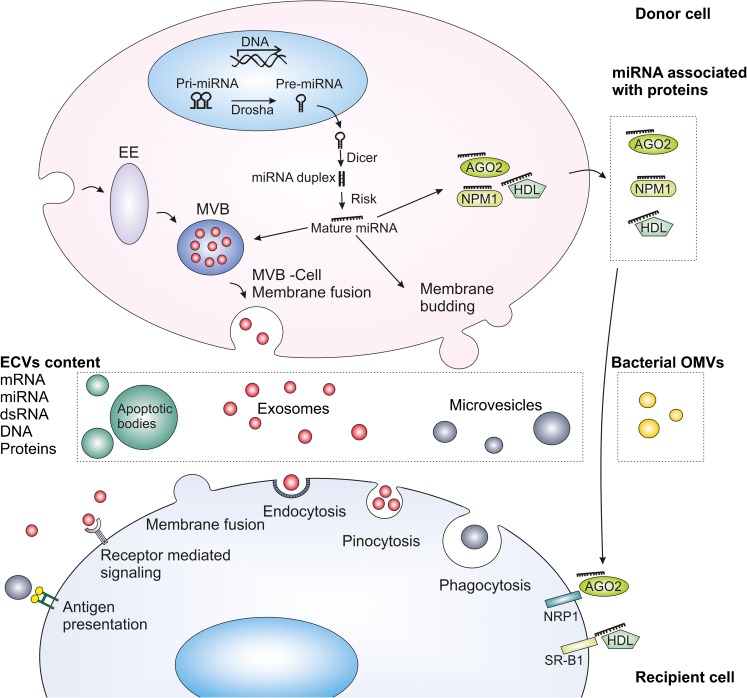
Fig. 1.
Biogenesis of microRNA (miRNA), extracellular vesicles (EV) and uptake mechanisms. miRNAs are transcribed by RNA polymerase II from chromosomal DNA into primary RNA (Pri-miRNA; 1-3 kb). Pri-miRNA is processed by Drosha into precursor miRNA (Pre-miRNA). Pre-miRNA is transported to the cytoplasm and cleeved into miRNA/miRNA duplices (∼22 bp) by Dicer. miRNA duplex strands separate with the incorporation of the protein Argonaute (AGO) and RNA-induced silencing complex (Risk; Xu et al. 2013; Sohel 2016; Tomasetti et al. 2017). miRNA can be packed in the EV or be exported as protein-miRNA complexes (Arroyo et al. 2011; Canfran-Duque et al. 2014). Exosomes are derived from the endocytic pathway and their biogenesis requires multiprotein complexes called Endosomal Sorting Complex Required for Transport. Microvesicles are formed by the outward budding of the plasma membrane, whereas apoptotic bodies are vesicles released from cells that undergo apoptosis (Akers et al. 2013; Morrison et al. 2016). EV are taken up by recipient cells by a variety of mechanisms including endocytosis (mediated by lipid rafts, clathrin or caveolin), pinocytosis, phagocytosis and membrane fusion (Mulcahy et al. 2014). Proteins present on the EV can trigger signaling pathways in the target cells or be involved in antigen presentation (Thery et al. 2009; El Andaloussi et al. 2013). miRNA-protein complexes are also internalized by interaction with specific receptors on the recipient cell. High-density lipoprotein (HDL) associated with miRNA (miRNA-HDL) interacts with scavenger receptor class B type 1 (SR-B1; Canfran-Duque et al. 2014). miRNA-AGO-2 interacts with neuropilin-1 (NRP1; Prud’homme et al. 2016). EE Early endosome, MVB multivesicular body, NPM1 nucleophosmin 1, OMVs outer membrane vesicles, dsRNA double-stranded RNA
Table 1.
Classification and characteristics (Thery et al. 2009; van der Pol et al. 2012; Akers et al. 2013; Ciardiello et al. 2016) of extracellular vesicles (EM electron microscopy, ND not determined, TNFRI tumour necrosis factor receptor I)
| Characteristic | Exosomes | Microvesicles | Ectosomes | Membrane particles | Exosome-like vesicles | Apoptotic bodies |
|---|---|---|---|---|---|---|
| Size | 50-100 nm | 20-1000 nm | 50-200 nm | 50–80 to 600 nm | 20-50 nm | 50-500 nm and 500-4000 nm |
| Density in sucrose | 1.13–1.19 g/ml | ND | ND | 1.04–1.07 g/ml | 1.1 g/ml | 1.16–1.28 g/ml |
| EM morphology | Cup shape | Irregular shape and electron-dense | Bilamellar round structures | Round | Irregular shape | Heterogeneous |
| Sedimentation | 100,000 g | 10,000–20,000 g | 160,000–200,000 g | 100,000–200,000 g | 175,000 g | 1200 g, 10,000 g or 100,000 g |
| Lipid composition | Enriched in cholesterol, sphingomyelin and ceramide; contain lipid rafts; expose phosphatidylserine | Expose phosphatidylserine | Enriched in cholesterol and diacylglycerol; expose phosphatidylserine | ND | Do not contain lipid rafts | Expose phosphatidylserine |
| Main protein markers | Tetraspanins (CD63, CD9), Alix and TSg101 | Integrins, selectins and CD40 ligand, VCAMP3, ARF6 | CR1 and proteolytic enzymes; no CD63 | CD133; no CD63 | TNFRI | Histones, caspase3, C3b |
| Intracellular origin | Internal compartments (endosomes) | Plasma membrane | Plasma membrane | Plasma membrane | Internal compartments | Plasma membrane, endoplasmic reticulum |
EVs have a distinct surface coat with an abundance of membrane-associated proteins, glycoproteins and lipids, whereas the EV interior (“cargo”) consists in structural and functional proteins, enzymes, lipids and peptides of various lengths. Notably, DNA (including mitochondrial DNA) and a variety of RNA species are transported within EVs. These important RNA classes have now been convincingly shown to regulate all cellular functions efficiently and include small RNAs, microRNAs and messager RNAs (Thery et al. 2009; El Andaloussi et al. 2013; Liu et al. 2015; Zaborowski et al. 2015; Morrison et al. 2016), see Fig. 1. Originally mostly ignored in electron micrographs or described as “cell-derived dust” from megakaryocytes (Wolf 1967), their original role was supposed to be passive cellular waste management. However, intriguing findings of active and orderly secretion mechanisms have emerged during the last few years. Interestingly, a wealth of information is now available that confirms that the vesicles are indeed taken up by target cells via a variety of mechanisms (summarized in Mulcahy et al. 2014) revealing their key role in intercellular communication, although targeted uptake mechanisms in any particular organ systems, except for the brain, remain to be studied in detail.
Crude urine or urinary EV (uEV), with their protein, enzyme or RNA content, have been discovered over the last few years (Alter et al. 2012; Alvarez et al. 2012; Cheng et al. 2014). In this review, we summarize some recent discoveries and highlight especially the distinct value of urinary RNA for biomarker purposes (Alter et al. 2012; Alvarez and Distefano 2013; Yang et al. 2013; Argyropoulos et al. 2015). With all the present data available, especially of the surprisingly rich and variable content of uEV, urine now appears as a very promising and completely uninvasive source for new information reflecting accurately the pathophysiology of the kidney and, most likely, also of other organ systems. Despite all the enthusiasm in this rapidly growing field, we aim to pinpoint some of the caveats, misinterpretations and limitations of current approaches and to emphasize the importance of the appropriate standardization needed.
EV classification
Since the first descriptions of EV, a variety of vesicle categories has been described. The classification was based initially on their cellular or subcellular sources, such as prostasomes, exosomes, membrane vesicles and others (see van der Pol et al. 2012, 2016; Thery et al. 2009; Wang and Sun 2014) and basically their physico-chemical properties. These include the vesicle size, density, morphology, lipid composition, protein composition, subcellular origin and light scattering (Thery et al. 2009; van der Pol et al. 2012). As seen in Table 1, most of the physico-chemical EV characteristics overlap significantly. This also makes a reliable categorization and their isolation a challenge yet to be fully overcome. Notably, in addition to the size and density of vesicles, the efficiency of isolating these vesicles depends on the shape and volume fraction of the vesicles, the viscosity of the fluid in which they lie, the temperature and presence of other confounding factors such as proteins, peptides or pigments in the fluid, the centrifugation time and the type of rotor used for centrifugation (fixed angle or swing-out; Cvjetkovic et al. 2014; Livshits et al. 2015). Based on these variables and the current lack of thoroughly standardized isolation protocols, considerable cross-contamination of vesicle types can obviously occur in any sample studied and reported. Indeed, this overlap between vesicle categories and the variety of isolation methods might have led to “published artifacts, over-interpretation and non-comparable results between laboratories” (van der Pol et al. 2016), despite continuous attempts at standardization by international organizations (Witwer et al. 2013). On the other hand, especially for biomarker identification, the current trend is leaning towards the use of methods providing the best total EV yield, which may ignore strict EV categorization. Depending on downstream uses and goals, this should be considered acceptable, in particular as most vesicle classes share, to a significant degree, the same surface and cargo contents (even if in different ratios).
As stated above, urine contains a variety of vesicles of variable sizes (Pisitkun et al. 2004; Miranda et al. 2010; van Balkom et al. 2011; Alvarez et al. 2012; Alvarez et al. 2013). Notably, however, urinary contents, including vesicles can be strongly influenced by factors such as diet (Garcia-Perez et al. 2017), exercise (Alter et al. 2012; Yang et al. 2013; Mansueto et al. 2017) and medication and by the presence of urinary pigments. As discussed below in more detail, the lack of standardization and variables in nomenclature might have led to reported artifacts and challenges of reproducibility. Furthermore, free urine contains free and active proteases and RNases, whereas within the vesicles, both proteins and RNA are protected against ubiquitous proteases and RNases (Cheng et al. 2014). Thus, only results of crude urines are comparable with each other, whereas studies utilizing the much richer contents of EV should be preferred because of their mostly unmodified contents.
Interestingly, the bulk of urinary vesicles is considered to derive from the epithelial lining of nephrons and kidney parenchyme (Turco et al. 2016) but ample evidence has been presented that urinary vesicles also originate from the circulation (Miranda et al. 2010; Ma et al. 2016; Pazourkova et al. 2016). This offers an exciting opportunity to develop easy, non-invasive and easy-to-repeat diagnostics for remote tissue-specific markers (Ma et al. 2016) present in the urine. However, whether uEVs are actively secreted through the glomerular filtration barrier or through the epithelial cell lining of nephrons is not known, although this could involve many active mechanisms (for a variety of the mechanisms proposed, see Mulcahy et al. 2014).
Evidence is increasing for uEV usefulness in the biomarker search for kidney diseases such as minimal change disease and focal segmental glomerulosclerosis (Ramezani et al. 2015), diabetic nephropathy (Barutta et al. 2013; Musante et al. 2015; Delic et al. 2016) and others (for an excellent comprehensive review of uEV findings in kidney diseases, see Erdbrugger and Le 2016).
Although proteins within uEVs have been widely recognized and reported (see, for example, www.exocarta.org, the dedicated database for vesicle contents), the recent technical advances in RNA-sequencing (Wang et al. 2009; Lee et al. 2010) highlight the usefulness especially of urinary miRNA for biomarker purposes (Van Roosbroeck et al. 2013). Interestingly, both uEV proteins and miRNAs are products that not only are secreted by all the epithelial cell types along the nephrons and lower urinary tract but are also filtered or secreted from the circulation by the kidney parenchyme.
In contrast to earlier studies also utilizing urinary sediment cells for miRNA isolation, the protocols presently call for them being discarded as these most likely represent contents from apoptotic cells. The information available concerning specific downstream target effects of uEV miRNAs remains limited (however, see Bellingham et al. 2012; Alvarez-Erviti et al. 2011).
Interestingly, evidence is accumulating of yet another potential source of urinary RNA, namely the resident bacteria (Table 2). Accordingly, inherent but well-constrained bacterial colonization can be found in the urinary bladder (Brubaker and Wolfe 2015). For anatomical reasons, the female urinary bladder and urine are more prone to urinary tract infections (UTI). Whether this represents an escape of normal bacterial flora from the host or the entry of more pathogenic urinary pathogens, such as the uropathogenic fimbriated strains of Escherichia coli (Korhonen et al. 1988), remains unknown. Interestingly, the most common bacterial strains in urinary bladder associated with UTI (both Gram negative and Gram positives) have been well-established, whereas recent studies by using RNA-Seq of the 16S RNA isolated from urine have shown a wide variety of additional bacterial strains in urine (Valadi et al. 2007; Brubaker and Wolfe 2015; Koeppen et al. 2016; Tataruch-Weinert et al. 2016). At the same time, bacteria have been shown to actively use their outer membrane vesicles (OMVs) for packaging and sending their genetic material for interaction with the immediate environment. The OMV contents also include abundant small RNA species (Wang et al. 2012; Koeppen et al. 2016), which mediate interactions with host cells and tissues. Thus, urine unavoidably also contains vesicles from rich bacterial sources, with their respective RNA contents. Although little is still known of the exact roles of OMVs, they are clearly involved in the constant interplay with the host defense system and in maintaining the barrier function to prevent ascending infections.
Table 2.
Characteristics of bacterial extracellular vesicles (ND not determined, EM electron microscopy)
| Characteristics | Bacterial extracellular vesicles | |
|---|---|---|
| Gram-negative | Gram-positive | |
| Size | 20-300 nm | 20-100 nm |
| Density in sucrose | 1.20–1.22 g/ml | ND |
| EM morphology | Round | Round |
| Sedimentation | 150,000 g | 150,000 g |
| Lipid compositiona | Phosphatidylglycerol, phosphatidylethanolamine (E. coli) | Palmitic acid, myristic acid (B. anthracis and S. pneumoniae) |
| Main protein markers | Outer membrane proteins, virulence factors | Bacterial adhesion and invasion proteins, host cell modulation proteins |
| Intracellular origin | Bacterial outer membrane | Cell membrane |
| References | Lee et al. 2007; Bai et al. 2014; Kim et al. 2015; Watanabe 2016 | Gurung et al. 2011; Brown et al. 2015; Kim et al. 2015 |
aVariability between strains, species
For practical purposes in uEV studies, the OMVs and their protein and RNA content are of special importance. Several questions arise. How can excessive bacterial products in the analyses and respective artifacts in the reported proteomes and RNA be avoided (Shmaryahu et al. 2014; Cheung et al. 2016)? Should samples from male and female subjects be differently interpreted as male samples present with less abundant bacteria and their OMV products? Carefulness at all steps in sample collection, storage, analysis and data interpretation is mandatory and, notably, calls for rigorous technical controls to pinpoint excessive bacterial RNA and protein products. This, in turn, may require a much larger volume of urine samples to allow all the necessary orthogonal controls (Tataruch-Weinert et al. 2016).
The capacity of bacteria to exchange functional genetic information between host cells is a widely unexplored area that has profound repercussions in our understanding of clinical UTI and beyond. The presence of bacterial proteins, metabolites and RNA products may generate misleading data and this possibility should always be critically considered in studies with uEV analytics.
Isolation methods of uEV have been extensively reviewed (Musante et al. 2014b; Wang and Sun 2014; Gamez-Valero et al. 2015). The most widely used methods are still the differential (ultra)centrifugation–based methods, often with modifications (see Table 3; Gardiner et al. 2016).
Table 3.
Advantages and disadvantages of isolation methods for urinary extracellular vesicles (DC differential centrifugation, CHAPS 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate, DTT dithiothreitol, SEC size exclusion chromatography, THP Tamm-Horsfall glycoprotein, uEV urinary extracellular vesicles)
| Method | Advantages | Disadvantages | |
|---|---|---|---|
| DC | Vesicle enrichment as a pellet | No standard conditions for: 1. number of centrifugations; 2. relative centrifugation force; 3. time; 4. rotor type; 5. sample volume; 6. temperature during centrifugation; 7. presence/absence of protease inhibitors |
Not applicable for large volume of samples, not suitable for samples from large cohort of patients. Relatively expensive because of devise setup/reagent price |
| DC + CHAPS treatment | Protein activity prevention | ||
| DC + DTT treatment | Removal of, for example, THP excess in sample | Not suitable for protein activity assessment designated samples | |
| DC + SEC | |||
| Nano-membrane filtration | Removal of cell debris and urinary casts | Differences in removal of larger (>0.22 μm or more) vesicles without assessment of their importance for biomarkers screening; major loss of uEVs on the filter | |
| DC + microfiltration | |||
| DC + nanofiltration | |||
| DC + ultrafiltration | |||
| DC + sucrose gradient | Vesicle separation according to density | Highly time consuming | |
| Ultrafiltration + DC | |||
| Exoquick | No need for ultracentrifugation step | Overtly expensive when applied to large volumes | |
| Total exosome isolation reagent | |||
| Hydrostatic filtration dialysis (HFD) | Inexpensive, quick, versatile. Applicable to large sample volumes and large sample numbers; no need for special machinery or highly trained personnel | ||
To isolate urinary EVs, a minimum of two steps are usually used, including low-speed centrifugation to pellet any cellular debris in the urine for discard.
Variations and combinations of other methods as “add-ons” to differential centrifugation have been introduced to overcome the observed limitations, including the aggregation and unnecessary loss of valuable pellets still containing EVs during the process. Notably, a number of practical parameters for the ultracentrifugation-based methods should always be carefully considered and details including relative centrifugal force, centrifugation time, temperature and the rotor type used should be recorded since variation in these parameters will lead to substantial quantitative and qualitative differences in the final EV yield (Thery et al. 2009). A detailed discussion of caveats in many of the currently used protocols can be found in recent critical reviews (Gardiner et al. 2016; van der Pol et al. 2016).
Reducing agents including dithiothreitol (DTT) or CHAPS (3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) are used to prevent excessive protein complexing during EV isolation, especially in order to regulate polymerization of Tamm-Horsfall glycoprotein (THP; Fernandez-Llama et al. 2010; Musante et al. 2012), the most abundant normal urinary protein distorting many downstream analyses. Notably, an intact urinary THP meshwork efficiently entraps vesicles of all sizes at all steps of EV harvesting, causes clogging and a seriously reduced isolation capacity of in-filtration-based methods and may result in a loss of up to 30 % of the final uEV yield (Fernandez-Llama et al. 2010). Notably, however, the use of reducing agents may also release miRNA bound to protective circulating proteins or from the surface of uEVs (Wachalska et al. 2016).
The current golden standard, namely serial (ultra)centrifugation, has been shown to easily miss up to 20–30 % of vesicles in the pre-purification steps (Musante et al. 2014a). Furthermore, recent results demonstrate that the final pellet after the ultracentrifugation steps for vesicle isolation may have missed yet another 20–30 % of vesicles (Musante et al. 2017). These alarming facts should be carefully taken into consideration in order to include EV isolation methods that have been selected and modified and published results that have been critically evaluated.
The most commonly used current methods and their combinations for EV isolation from urine are listed in Table 3. Notably, many of these methods can easily clog because of protein aggregates or the capacity of the method might be limited. For practical isolation purposes and for the amounts necessary especially for uEVs, large sample volumes are a definite benefit as this allows the inclusion of the necessary quality control of the samples. An additional source of artifacts, namely variations in urinary electrolyte concentrations, e.g., resulting from hydration status because of exercise (Maughan 1991), may likewise change the sample conditions and distort EV yield. Furthermore, urine is abundant in pigments, a recognized major source of artifacts in all downstream analyses. An interesting approach might be to use fluorescence-activated cell sorting (FACS) for direct vesicle isolation into various categories, although the resolution of FACS is presently limited mostly to vesicle size above 100 nm and thus misses a major part of of vesicles, especially exosomes.
Musante et al. (2014a) developed an alternative isolation method for the comprehensive catching of urinary vesicles. This method is called hydrostatic filtration dialysis (HFD) and avoids the recognized limitations of most EV isolation methods. It uses simple and quick low-speed centrifugation (2000 g) to remove, for example, bacteria, cellular debris and excessive polymers of THP before filter-dialysing the sample (especially urine).
In HFD, the hydrostatic pressure of the sample pushes it through the dialysis membrane tubing. For recovery of urinary proteins and miRNA, the HFD method is clearly superior to the ultracentrifugation-based methods and provides superior quality of protein and RNA yield (Musante et al. 2014a; Tataruch-Weinert et al. 2016).
HFD is versatile, simple and inexpensive and can be used for a variety of samples including urine, plasma, cell culture medium and saliva (Musante et al. 2014a). It is based on sample dialysis in a tube with a defined-cutoff pore-size membrane (Musante et al. 2012). This membrane allows the passage of small solutes and peptides, effectively standardizing the solute environment, while retaining the EVs because of their larger size. HFD easily provides normalization and collection of the yield and washes away electrolytes while efficiently retaining the vesicles. Notably, the major contaminant in all urinary downstream analyses, namely urinary pigments, are also washed though the membrane and thus are avoided. The capacity of this method is adequate, with up to 1000 ml sample volumes being easily handled. Furthermore, this method requires little or no previous knowledge or training, is extremely cost-efficient and can readily process a number of samples in parallel during a workday (Musante et al. 2014a). The HFD method is not suitable for distinguishing between defined vesicle populations, which, for most diagnostic, prognostic or biomarker searches is, however, irrelevant.
Antibodies and synthetic peptides with affinity for EV membrane proteins have been used for EV isolation (Ghosh et al. 2014; Wang and Sun 2014). Although these methods may preferentially yield exosomes or other EV types with distinct surface antigens as required, additional EV subpopulations sharing the same membrane proteins may be isolated. Moreover, their capacity for EV catching is limited making them more suitable for smaller sample volumes.
Specific kits to precipitate exosomes have been developed, e.g., ExoSpin Exosome Purification Kit (Cell Guidance Systems, USA), Invitrogen Total Exosome Isolation Kit (Life Technologies, USA) and others. These products have been tested with solutions of liposomes and exosomes and have shown not only a robust isolation capacity for EV but also the co-isolation of molecules with similar physical properties. Thus, other additional purification methods may need to be used (Lane et al. 2015).
RNA species in vesicles
Intracellular RNA species are involved in the translation of DNA information to proteins at ribosomes and in the general regulation of RNA translation (see Grosshans and Filipowicz 2008, Xu et al. 2013). MiRNA, with the size of 20-25 nt, has a distinct role in gene regulation (Xu et al. 2013). Its formation is regulated by a specific intracellular pathway distinguishing it from other small RNA species present both in pro- and eukaryotic cells (Xu et al. 2013; see Fig. 1).
Interestingly, all RNA classes can also be found extracellularly, in all bodily fluids, including serum and plasma, saliva, tear fluid and urine (Wang et al. 2012; Patton et al. 2015; Yanez-Mo et al. 2015). The way in which the cellular RNA secretion itself is regulated and the relative proportions of the various small RNA species secreted are not exactly known. The intracellular RNA profile may differ from that of the secreted profile, suggesting the possibility of the overflow being secreted. Packaged into EV but also free, RNA appears as a unique and archaic method of cell-to-cell communication found throughout living cells (Brown et al. 2015; Yanez-Mo et al. 2015). Interestingly, the RNA secretome appears to reflect accurately the physiological state of the cell of origin and is highly valuable and accurate for the mapping of any personalized functions.
Although free extracellular RNA was identified years ago, the extracellular milieu consists in complex proteases and ubiquitous RNases actively splicing any target secreted from the cells (Sorrentino 1998; DeClerck et al. 2004; Ricard-Blum and Vallet 2016). The splicing process produces the extracellular fluid proteome (peptidome) and RNome. What is even more exciting is that cells ranging from archaic bacteria to the most highly sophisticated cell types have developed methods to protect these valuable entities from degradation by packaging them into vesicles and, indeed, extracellular RNAs appear to be universally associated instead with carrier vehicles (Patton et al. 2015), probably because of the rapid degradation of unprotected RNAs in biofluids.
Exosomes appear as a dominant pathway and vehicle for RNA secretion from cells (Crescitelli et al. 2013; Lunavat et al. 2015; Willms et al. 2016), although other vesicle types also contain RNA. This fact reduces the value of any method designed strictly for the purification of a distinct EV class. Moreover, these methods always end up in losing up in 40 % of EV because of losses in the process up until the last separation steps (Musante et al. 2017). Furthermore, the membrane layer of all vesicle types has been shown to contain a distinct surface coat that may be quite different from the vesicle interior, i.e., the “vesicle cargo” (Liu et al. 2015). This is interesting, as many lines of evidence now suggest different roles for these regions: the surface proteins and even EV surface RNA may serve as an “address code” for targeting (for references, see Mulcahy et al. 2014). As the vesicle attaches and fuses with the target cell membrane, its contents are released for intracellular interaction. This appears as a finely tuned and highly sophisticated system for target cell regulation in processes such as immunological defense, cancer spread and the modulation of the metabolic state of cells in a specific manner (El Andaloussi et al. 2013; Yanez-Mo et al. 2015; Zaborowski et al. 2015).
The gut, skin, urine and mucous membrane microbiomes appear to have developed methods to efficiently interfere with the host defense systems by releasing outer membrane vesicles (OMVs), with distinct RNA contents and constant interaction with the host defense system. As a result, a strong, although less-well understood barrier function is achieved.
Future aspects
The number of studies on EV, especially exosomes, has been sky-rocketing during the last few years. This is because their extraordinary potential is not limited to the dissection of disease mechanisms to identify new druggable targets but can also provide early biomarkers of unforeseen accuracy. EV, which are also abundantly present in the urine, are an unparalleled resource for individualized molecular fingerprints reflecting cellular pathophysiology at their site of origin upstream in the kidney and beyond (Miranda et al. 2010; Alvarez and Distefano 2013). Thus, EV are derived not only from all cell types along the nephron and kidney parenchyme (Salih et al. 2014) but also from the circulation and normal bacterial colonization in the urinary bladder.
The potential of EV, especially for early diagnostics and as novel biomarkers for kidney diseases, is exciting. With full systems biology integration, combining their proteome, RNome and metabolomics information, EV now show great promise in dissecting these thus far less-well understood disease mechanisms in an individualized way (Erdbrugger and Le 2016; van der Pol et al. 2016). The field still lacks comprehensive standardization, starting from the nomenclature and methods used for efficient EV harvesting and extending up to their critical downstream analysis. Misleading or false results in the field may lead to wrong conclusions (van der Pol et al. 2016). However, the challenges have been recognized and respective international organizations are committed to continuous standardization (Witwer et al. 2013). Novel, inexpensive and critically scrutinized methods are being developed in order to avoid the gaps of the earlier ones and to match expectations, for example, for personalized markers.
Whereas the physical properties and protein and RNA content of the various EV types significantly overlap, a careful selection of methods available should provide a rational basis for an appropriate selection of each type of application (Musante et al. 2014b; Rupert et al. 2017).
Notably, methods for single vesicle detection are developing rapidly, including those utilizing microfluidics and nano-sized beads. These will be highly useful, especially for linking the different vesicle types to specific contents and, thus, for revealing details of their functions. The respective compact systems and techniques including micro-nuclear magnetic resonanace (Shao et al. 2012), magneto-electrochemical sensor systems (Jeong et al. 2016) and nanoplasmonic chips (Im et al. 2014) are expected to develop into practical and rapid point-of-care detection for diagnostics, especially for uEV-based protein miRNA biomarkers (Shao et al. 2015). Flow cytometry is currently developing as an exciting extension for EV isolation directly in suspension and provides a means for the efficient identification of EV based on their type-specific epitopes and sequestration into subpopulations. However, currently, the size-range of flow cytometric detection starts at 200 nm going upwards, although a detection limit down to 100 nm has been achieved (Pospichalova et al. 2015; Rupert et al. 2017). Thus, FACS detection, especially of exosomes, is currently beyond the detection limit (Shao et al. 2015).
The massively parallel Next-Generation Sequencing as applied also to uEV analytics is a welcome extension to the repertoire of the novel robust and non-biased methods for urinary analytics (Renkema et al. 2014; Khurana et al. 2017). Carefully planned studies, meticulous controls and critical evaluation of results will provide an unforeseen accuracy to mechanisms of health and disease beyond the kidney parenchyme and urinary tract. The obvious caveat of genetic material as a “contaminant” from bacterial OMVs should be noted. Moreover, novel mechanisms to distinguish bacterial from “non-bacterial” cystitis, an important clinical problem, are expected to be discovered with emerging new therapeutic options.
An exciting extension to utilizing EV biology relates to bioengineered nanoparticles. These can serve as EV mimics and can be released into the circulation with a defined target cell “address code”. Upon attachment, the engineered protein, regulatory RNA or selected medication contents can be released as previously exemplified by the delivery of targeted chemotherapeutics (Jang et al. 2013). These treatments will have a huge influence, including on kidney diseases, with next-generation targeted therapies fully utilizing the obtained knowledge of EV biology and their bioengineering capacities.
Acknowledgements
This study was supported by the EU-Innovative Medicines Initiative (IMI) Program “Biomarker Enterprise to Attack Diabetic Kidney Disease” (BEAt-DKD, Grant number 115974) and the FRIAS program of the Albert-Ludwigs University, Freiburg.
Contributor Information
Karina Barreiro, Email: karina.barreiro@helsinki.fi.
Harry Holthofer, Email: harry.holthofer@helsinki.fi.
References
- Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neuro-Oncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter ML, Kretschmer A, Von Websky K, Tsuprykov O, Reichetzeder C, Simon A, Stasch JP, Hocher B. Early urinary and plasma biomarkers for experimental diabetic nephropathy. Clin Lab. 2012;58:659–671. [PubMed] [Google Scholar]
- Alvarez ML, Distefano JK. The role of non-coding RNAs in diabetic nephropathy: potential applications as biomarkers for disease development and progression. Diabetes Res Clin Pract. 2013;99:1–11. doi: 10.1016/j.diabres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- Alvarez S, Suazo C, Boltansky A, Ursu M, Carvajal D, Innocenti G, Vukusich A, Hurtado M, Villanueva S, Carreno JE, Rogelio A, Irarrazabal CE. Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplant Proc. 2013;45:3719–3723. doi: 10.1016/j.transproceed.2013.08.079. [DOI] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Argyropoulos C, Wang K, Bernardo J, Ellis D, Orchard T, Galas D, Johnson JP. Urinary microRNA profiling predicts the development of microalbuminuria in patients with type 1 diabetes. J Clin Med. 2015;4:1498–1517. doi: 10.3390/jcm4071498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Kim SI, Ryu S, Yoon H. Identification and characterization of outer membrane vesicle-associated proteins in Salmonella enterica serovar typhimurium. Infect Immun. 2014;82:4001–4010. doi: 10.1128/IAI.01416-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutta F, Tricarico M, Corbelli A, Annaratone L, Pinach S, Grimaldi S, Bruno G, Cimino D, Taverna D, Deregibus MC, Rastaldi MP, Perin PC, Gruden G. Urinary exosomal microRNAs in incipient diabetic nephropathy. PLoS One. 2013;8:e73798. doi: 10.1371/journal.pone.0073798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol. 2012;3:124. doi: 10.3389/fphys.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol. 2015;13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker L, Wolfe AJ. The new world of the urinary microbiota in women. Am J Obstet Gynecol. 2015;213:644–649. doi: 10.1016/j.ajog.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfran-Duque A, Ramirez CM, Goedeke L, Lin CS, Fernandez-Hernando C. microRNAs and HDL life cycle. Cardiovasc Res. 2014;103:414–422. doi: 10.1093/cvr/cvu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86:433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- Cheung KH, Keerthikumar S, Roncaglia P, Subramanian SL, Roth ME, Samuel M, Anand S, Gangoda L, Gould S, Alexander R, Galas D, Gerstein MB, Hill AF, Kitchen RR, Lotvall J, Patel T, Procaccini DC, Quesenberry P, Rozowsky J, Raffai RL, Shypitsyna A, Su AI, Thery C, Vickers K, Wauben MH, Mathivanan S, Milosavljevic A, Laurent LC. Extending gene ontology in the context of extracellular RNA and vesicle communication. J Biomed Semantics. 2016;7:19. doi: 10.1186/s13326-016-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardiello C, Cavallini L, Spinelli C, Yang J, Reis-Sobreiro M, de Candia P, Minciacchi VR, Di Vizio D. Focus on extracellular vesicles: new frontiers of cell-to-cell communication in cancer. Int J Mol Sci. 2016;17:175. doi: 10.3390/ijms17020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvjetkovic A, Lotvall J, Lasser C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles. 2014;3:23111. doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeClerck YA, Mercurio AM, Stack MS, Chapman HA, Zutter MM, Muschel RJ, Raz A, Matrisian LM, Sloane BF, Noel A, Hendrix MJ, Coussens L, Padarathsingh M. Proteases, extracellular matrix, and cancer: a workshop of the path B study section. Am J Pathol. 2004;164:1131–1139. doi: 10.1016/S0002-9440(10)63200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic D, Eisele C, Schmid R, Baum P, Wiech F, Gerl M, Zimdahl H, Pullen SS, Urquhart R. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- El Nahas M. The global challenge of chronic kidney disease. Kidney Int. 2005;68:2918–2929. doi: 10.1111/j.1523-1755.2005.00774.x. [DOI] [PubMed] [Google Scholar]
- Erdbrugger U, Le TH. Extracellular vesicles in renal diseases: more than novel biomarkers? J Am Soc Nephrol. 2016;27:12–26. doi: 10.1681/ASN.2015010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T, Knepper MA. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77:736–742. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamez-Valero A, Lozano-Ramos SI, Bancu I, Lauzurica-Valdemoros R, Borras FE. Urinary extracellular vesicles as source of biomarkers in kidney diseases. Front Immunol. 2015;6:6. doi: 10.3389/fimmu.2015.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH, Vestergaard H, Hansen T, Beckmann M, Pedersen O, Elliott P, Stamler J, Nicholson JK, Draper J, Mathers JC, Holmes E, Frost G. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol. 2017;5:184–195. doi: 10.1016/S2213-8587(16)30419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, Barnett D, Crapoulet N, Fournier S, Joy A, Caissie MC, Ferguson AD, Daigle M, Meli MV, Lewis SM, Ouellette RJ. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS One. 2014;9:e110443. doi: 10.1371/journal.pone.0110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One. 2011;6:e27958. doi: 10.1371/journal.pone.0027958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildonen S, Skarpen E, Halvorsen TG, Reubsaet L. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci Rep. 2016;6:36331. doi: 10.1038/srep36331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lotvall J, Kim YK, Gho YS. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano. 2013;7:7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano. 2016;10:1802–1809. doi: 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana R, Ranches G, Schafferer S, Lukasser M, Rudnicki M, Mayer G, Huttenhofer A. Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease. RNA (New York) 2017;23:142–152. doi: 10.1261/rna.058834.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee J, Park J, Gho YS. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol. 2015;40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, Stanton BA. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 2016;12:e1005672. doi: 10.1371/journal.ppat.1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen TK, Virkola R, Westerlund B, Tarkkanen AM, Lahteenmaki K, Sareneva T, Parkkinen J, Kuusela P, Holthofer H. Tissue interactions of Escherichia coli adhesins. Antonie Van Leeuwenhoek. 1988;54:411–420. doi: 10.1007/BF00461859. [DOI] [PubMed] [Google Scholar]
- Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, Park KS, Lee JO, Kim YK, Kwon KH, Kim KP, Gho YS. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA (New York) 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chinello C, Musante L, Cazzaniga M, Tataruch D, Calzaferri G, James Smith A, De Sio G, Magni F, Zou H, Holthofer H. Intraluminal proteome and peptidome of human urinary extracellular vesicles. Proteomics Clin Appl. 2015;9:568–573. doi: 10.1002/prca.201400085. [DOI] [PubMed] [Google Scholar]
- Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, Generozov EV, Govorun VM. Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci Rep. 2015;5:17319. doi: 10.1038/srep17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunavat TR, Cheng L, Kim DK, Bhadury J, Jang SC, Lasser C, Sharples RA, Lopez MD, Nilsson J, Gho YS, Hill AF, Lotvall J. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells—evidence of unique microRNA cargos. RNA Biol. 2015;12:810–823. doi: 10.1080/15476286.2015.1056975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhang XQ, Zhou DX, Cui Y, Deng LL, Yang T, Shao Y, Ding M. Feasibility of urinary microRNA profiling detection in intrahepatic cholestasis of pregnancy and its potential as a non-invasive biomarker. Sci Rep. 2016;6:31535. doi: 10.1038/srep31535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas RJ, Deegens JK, Smeets B, Moeller MJ, Wetzels JF. Minimal change disease and idiopathic FSGS: manifestations of the same disease. Nat Rev Nephrol. 2016;12:768–776. doi: 10.1038/nrneph.2016.147. [DOI] [PubMed] [Google Scholar]
- Mansueto G, Armani A, Viscomi C, D’Orsi L, De Cegli R, Polishchuk EV, Lamperti C, Di Meo I, Romanello V, Marchet S, Saha PK, Zong H, Blaauw B, Solagna F, Tezze C, Grumati P, Bonaldo P, Pessin JE, Zeviani M, Sandri M, Ballabio A. Transcription factor EB controls metabolic flexibility during exercise. Cell Metab. 2017;25:182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan RJ. Fluid and electrolyte loss and replacement in exercise. J Sports Sci. 1991;9:117–142. doi: 10.1080/02640419108729870. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N, Brown D, Russo LM. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Bailey MA, Dear JW. Renal extracellular vesicles: from physiology to clinical application. J Physiol (Lond) 2016;594:5735–5748. doi: 10.1113/JP272182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Saraswat M, Duriez E, Byrne B, Ravida A, Domon B, Holthofer H. Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS One. 2012;7:e37279. doi: 10.1371/journal.pone.0037279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Tataruch D, Gu D, Benito-Martin A, Calzaferri G, Aherne S, Holthofer H. A simplified method to recover urinary vesicles for clinical applications, and sample banking. Sci Rep. 2014;4:7532. doi: 10.1038/srep07532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Tataruch DE, Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol (Lausanne) 2014;5:149. doi: 10.3389/fendo.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Tataruch D, Gu D, Liu X, Forsblom C, Groop PH, Holthofer H. Proteases and protease inhibitors of urinary extracellular vesicles in diabetic nephropathy. J Diabetes Res. 2015;2015:289734. doi: 10.1155/2015/289734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Tataruch-Weinert D, Kerjaschki D, Henry M, Meleady P, Holthofer H. Residual urinary extracellular vesicles in ultracentrifugation supernatants after hydrostatic filtration dialysis enrichment. J Extracell Vesicles. 2017;6:1267896. doi: 10.1080/20013078.2016.1267896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour AM, Modis Y. Endosomal vesicles as vehicles for viral genomes. Trends Cell Biol. 2014;24:449–454. doi: 10.1016/j.tcb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrakka J, Tryggvason K. Molecular make-up of the glomerular filtration barrier. Biochem Biophys Res Commun. 2010;396:164–169. doi: 10.1016/j.bbrc.2010.04.069. [DOI] [PubMed] [Google Scholar]
- Patton JG, Franklin JL, Weaver AM, Vickers K, Zhang B, Coffey RJ, Ansel KM, Blelloch R, Goga A, Huang B, L’Etoille N, Raffai RL, Lai CP, Krichevsky AM, Mateescu B, Greiner VJ, Hunter C, Voinnet O, McManus MT. Biogenesis, delivery, and function of extracellular RNA. J Extracell Vesicles. 2015;4:27494. doi: 10.3402/jev.v4.27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazourkova E, Pospisilova S, Svobodova I, Horinek A, Brisuda A, Soukup V, Hrbacek J, Capoun O, Mares J, Hanus T, Babjuk M, Korabecna M. Comparison of microRNA content in plasma and urine indicates the existence of a transrenal passage of selected microRNAs. Adv Exp Med Biol. 2016;924:97–100. doi: 10.1007/978-3-319-42044-8_18. [DOI] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak MR, Quaggin SE, Hoenig MP, Dworkin LD. The glomerulus: the sphere of influence. Clin J Am Soc Nephrol. 2014;9:1461–1469. doi: 10.2215/CJN.09400913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme GJ, Glinka Y, Lichner Z, Yousef GM. Neuropilin-1 is a receptor for extracellular miRNA and AGO2/miRNA complexes and mediates the internalization of miRNAs that modulate cell function. Oncotarget. 2016;7:68057–68071. doi: 10.18632/oncotarget.10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani A, Devaney JM, Cohen S, Wing MR, Scott R, Knoblach S, Singhal R, Howard L, Kopp JB, Raj DS. Circulating and urinary microRNA profile in focal segmental glomerulosclerosis: a pilot study. Eur J Clin Invest. 2015;45:394–404. doi: 10.1111/eci.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi G, Benigni A, Remuzzi A. Mechanisms of progression and regression of renal lesions of chronic nephropathies and diabetes. J Clin Invest. 2006;116:288–296. doi: 10.1172/JCI27699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema KY, Stokman MF, Giles RH, Knoers NV. Next-generation sequencing for research and diagnostics in kidney disease. Nat Rev Nephrol. 2014;10:433–444. doi: 10.1038/nrneph.2014.95. [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S, Vallet SD. Proteases decode the extracellular matrix cryptome. Biochimie. 2016;122:300–313. doi: 10.1016/j.biochi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- Rupert DL, Claudio V, Lasser C, Bally M. Methods for the physical characterization and quantification of extracellular vesicles in biological samples. Biochim Biophys Acta. 2017;1861:3164–3179. doi: 10.1016/j.bbagen.2016.07.028. [DOI] [PubMed] [Google Scholar]
- Salih M, Zietse R, Hoorn EJ. Urinary extracellular vesicles and the kidney: biomarkers and beyond. Am J Physiol Renal Physiol. 2014;306:F1251–F1259. doi: 10.1152/ajprenal.00128.2014. [DOI] [PubMed] [Google Scholar]
- Saritas T, Kuppe C, Moeller MJ. Progress and controversies in unraveling the glomerular filtration mechanism. Curr Opin Nephrol Hypertens. 2015;24:208–216. doi: 10.1097/MNH.0000000000000116. [DOI] [PubMed] [Google Scholar]
- Scott RP, Quaggin SE. Review series: the cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmaryahu A, Carrasco M, Valenzuela PD. Prediction of bacterial microRNAs and possible targets in human cell transcriptome. J Microbiol (Seoul, Korea) 2014;52:482–489. doi: 10.1007/s12275-014-3658-3. [DOI] [PubMed] [Google Scholar]
- Sohel MH. Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achievem Life Sci. 2016;10:175–186. doi: 10.1016/j.als.2016.11.007. [DOI] [Google Scholar]
- Sorrentino S. Human extracellular ribonucleases: multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell Mol Life Sci. 1998;54:785–794. doi: 10.1007/s000180050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataruch-Weinert D, Musante L, Kretz O, Holthofer H. Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination. J Extracell Vesicles. 2016;5:30281. doi: 10.3402/jev.v5.30281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp Mol Med. 2017;49:e285. doi: 10.1038/emm.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco AE, Lam W, Rule AD, Denic A, Lieske JC, Miller VM, Larson JJ, Kremers WK, Jayachandran M. Specific renal parenchymal-derived urinary extracellular vesicles identify age-associated structural changes in living donor kidneys. J Extracell Vesicles. 2016;5:29642. doi: 10.3402/jev.v5.29642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Balkom BW, Pisitkun T, Verhaar MC, Knepper MA. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 2011;80:1138–1145. doi: 10.1038/ki.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Boing AN, Gool EL, Nieuwland R. Recent developments in the nomenclature, presence, isolation, detection and clinical impact of extracellular vesicles. J Thromb Haemost. 2016;14:48–56. doi: 10.1111/jth.13190. [DOI] [PubMed] [Google Scholar]
- Van Roosbroeck K, Pollet J, Calin GA. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev Mol Diagn. 2013;13:183–204. doi: 10.1586/erm.12.134. [DOI] [PubMed] [Google Scholar]
- Wachalska M, Koppers-Lalic D, van Eijndhoven M, Pegtel M, Geldof AA, Lipinska AD, van Moorselaar RJ, Bijnsdorp IV. Protein complexes in urine interfere with extracellular vesicle biomarker studies. J Circ Biomark. 2016;5:4. doi: 10.5772/62579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sun W. Urinary extracellular microvesicles: isolation methods and prospects for urinary proteome. Proteomics. 2014;14:1922–1932. doi: 10.1002/pmic.201300371. [DOI] [PubMed] [Google Scholar]
- Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One. 2012;7:e51009. doi: 10.1371/journal.pone.0051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. Bacterial membrane vesicles (MVs): novel tools as nature- and nano-carriers for immunogenic antigen, enzyme support, and drug delivery. Appl Microbiol Biotechnol. 2016;100:9837–9843. doi: 10.1007/s00253-016-7916-7. [DOI] [PubMed] [Google Scholar]
- Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, El Andaloussi S, Wood MJ, Vader P. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–1719. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xiao L, Li J, Kanwar YS, Liu F, Sun L. Urine miRNAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Med Hypotheses. 2013;81:274–278. doi: 10.1016/j.mehy.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]