Abstract
The hydrophobic core, when subjected to analysis based on the fuzzy oil drop model, appears to be a universal structural component of proteins irrespective of their secondary, supersecondary, and tertiary conformations. A study has been performed on a set of nonhomologous proteins representing a variety of CATH categories. The presence of a well-ordered hydrophobic core has been confirmed in each case, regardless of the protein’s biological function, chain length or source organism. In light of fuzzy oil drop (FOD) analysis, various supersecondary forms seem to share a common structural factor in the form of a hydrophobic core, emerging either as part of the whole protein or a specific domain. The variable status of individual folds with respect to the FOD model reflects their propensity for conformational changes, frequently associated with biological function. Such flexibility is expressed as variable stability of the hydrophobic core, along with specific encoding of potential conformational changes which depend on the properties of helices and β-folds.
Keywords: Protein folding, Hydrophobicity, Hydrophobic core
Introduction
The traditional classification of secondary and supersecondary protein structures covers helical and β-fragments along with their mutual arrangement within the protein body. This α/β classification varies with respect to the participation of both folds in proteins, as well as their orientation. Purely helical (hemoglobin, cytochrome) or purely β-shaped (immunoglobulins) structures, as well as proteins characterized by near-equal participation of both types (lactate dehydrogenase or carboxypeptidase), can be identified in protein databases (e.g., PDB). The supersecondary structure, which determines the mutual arrangement of secondary folds can be expressed as follows (the following list is derived from a commonly used biochemistry textbook [1]):
βαβ – helical fragment linking two parallel β-strands
β hairpin – two anti-parallel β-strands linked by a tight U-turn loop
αα motif – two successive helices linked by a tight U-turn loop
Greek key motif – β-strands following one another in an arrangement reminiscent of classic Greek ornaments
β barrels – coaxial β-strands forming a cyclical system reminiscent to the arrangement of planks forming a wooden barrel.
Attempts to introduce a detailed structural classification of domain units in large proteins have resulted in the creation of the CATH/Gene3D (http://www.cathdb.info/ 27 Apr 2016) database, which currently (as of 27 April 2016) contains 26 million domains arranged into 2738 families. Structural classification is performed automatically upon inclusion of a new protein in PDB, via an exhaustive search for proteins homologous to the new entrant [2]. It is immediately apparent that the number of distinct families is quite high, notwithstanding the overall size of the database. This suggests that protein structures are diverse and difficult to classify.
The concept of biological activity likewise remains a mystery [3], despite successful attempts to program it into synthetic proteins. An in-depth analysis of the stability of the backbone and side chains facilitating stabilization of one of two possible rotametric forms, is presented in [4]. The great diversity of geometric forms makes it difficult to propose a single, common classification based on a unified criterion. This, however, does not imply that attempts to identify such a criterion are futile.
Our work focuses on proteins characterized by major supersecondary variability and proposes a coherent classification, covering all cases. The starting point is to acknowledge the common environment in which all proteins undergo folding and gain biological function — water. Water is the immanent condition ensuring proper folding and biological activity. This is why the water participation and its influencing on protein structure is the basis for the fuzzy oil drop model applied to interpret the different structures of proteins. We assume that the proposed model visualizes the effects of the influence of water environment despite the lack of knowledge of its own structuralization.
Our analysis of diverse structures is based on the fuzzy oil drop model and shows that both secondary and supersecondary structural motifs participate in the formation of a domain- or protein-wide hydrophobic core. In addition to enzymes (which are the most widely represented group), the work also discusses proteins capable of binding specific ligands (cytochrome) as well as antigen-binding proteins (immunoglobulin).
We establish that each of the presented molecules (or domains) contains a “seed” (of variable size) representing its hydrophobic core, which, according to textbook knowledge, is responsible for tertiary structural stabilization. As shown in our other papers, local deviations from the “idealized” hydrophobic core structure (i.e., high hydrophobicity density in the central part of the protein body and low hydrophobicity density on the surface) are often linked to the protein’s biological activity. The presence of cavities or exposure of hydrophobic residues on the surface creates suitable conditions for ligand binding and protein complexation respectively. Eliminating such discrepancies from analysis of the hydrophobic core structure invariably leads to identification of parts of the molecule for which the idealized distribution of hydrophobicity closely corresponds to observed values. Such parts are thought to mediate structural stabilization of the protein (or domain) as a whole [5–9].
In this work we show that, regardless of secondary and supersecondary conformational properties, the presence of a more or less prominent hydrophobic core is a common phenomenon. We identify proteins where the core is highly consistent with theoretical values (3D Gaussian) as well as those which exhibit local deviations from the theoretical model, typically associated with the capability to bind ligands or attract complexation partners. The study set is very diverse, yet exhibits common characteristics which point to the necessary presence of water in their environment — this applies to many different molecules encountered in the cell, but particularly to proteins.
Restricting protein structure analysis to topological aspects would disregard the most important factor — the influence of the water environment, which plays an active and often decisive role in the folding process. Hence, our study of the hydrophobic core — a product of the environment, which determines the biological activity of proteins.
Water environment is treated as the external force field which together with the internal force field (non-bonding interaction) participates in structuralization process and ensures the biological activity. The presented paper is an attempt to show the possible interpretation of structural effects which may express the role of water.
Materials and methods
Protein database
Our analysis is based on a set of proteins derived from a popular biochemistry textbook [1]. Table 1 reveals the structural variability observed at the supersecondary level.
Table 1.
Summary of proteins subjected to analysis
| Protein | Name | Source | Class | Ref. |
|---|---|---|---|---|
| α/β-domain fold twisted β-sheet | ||||
| 1A5Z | Lactate dehydrogenase | Bacteria | αβ complex | (Auerbach et al. [10]) |
| 1FW8 | Phosphoglycerate kinase | Yeast | α/β 3-layer (aba) | (Tougard et al. [11]) |
| α,β-domain fold | ||||
| 1AMK | Triose phosphate isomerase | Bacteria | α β barrel | (Williams et al. [12]) |
| 4DRS | Pyruvate kinase | Bacteria | α/β 3-layer (aba) α/β barrel mainly β barrel |
(Cook et al. [13]) |
| All β-domain super-fold | ||||
| 1B4L | Superoxide dismutase | Yeast | Mainly β sandwich | (Hart et al. [14]) |
| 1CON | Concanavalin | Plant | Mainly β sandwich | (Naismith et al. [15]) |
| β-barrel | ||||
| 1RBP | Retinol binding | Human | Mainly β barrel | (Cowan et al. [16]) |
| 1PNG | Aspargine amidase | Bacteria | Mainly β sandwich | (Kuhn et al. [17]) |
| 1TIM | Triose phosphatase isomerase | Chicken | α β barrel | (Banner et al. [18]) |
| Miscellaneous | ||||
| 256B | Cytochrome | E-coli | α Up-down bundle | (Lederer et al. [19]) |
| 7FAB | Immunoglobulin | Human | Mainly β sandwich | (Saul and Poljak [20]) |
| 6LDH | Lactate dehydrogenase | Bacteria | α β complex | (Abad-Zapatero et al. [21]) |
| Cytochrome fold | ||||
| 155C | Cytochrome | Bacteria | α orthogonal bundle | (Timkovich and Dickerson [22]) |
| 1JDL | Cytochrome | Bacteria | α orthogonal bundle | (Camara-Artigas et al. [23]) |
| 2C2C | Cytochrome | Bacteria | α orthogonal bundle | (Bhatia –PDB [24]) |
| 5CYT | Cytochrome | Fish | α orthogonal bundle | (Takano [25]) |
| 4 J20 | Cytochrome | Bacteria | α orthogonal bundle | (Yu et al. [26]) |
The selection is based on supersecondary structural order. The study set is consistent with the one proposed in (Devlin 2011).
The fuzzy oil drop model
An in-depth presentation of the fuzzy oil drop model can be found in an Open Access publication [27, 28]. In the interest of saving space, we will limit ourselves to recapitulating its basic tenets.
The fuzzy oil drop model is a modification of the oil drop model originally introduced by Kauzmann [29]. The original model was discrete, distinguishing a central hydrophobic part and an outer hydrophilic shell. In contrast, the fuzzy oil drop model introduces a 3D Gaussian which peaks at the center of the encapsulating ellipsoid, with values decreasing along with distance from the center, reaching near 0 at a distance of 3σ (where σ is a parameter of the Gaussian expressing the size of the ellipsoid). Distribution along each axis can be characterized by an appropriate sigma parameter. The values of such a function are assumed to represent theoretical (idealized) distribution of hydrophobicity in a protein molecule.
The actual (observed) hydrophobicity distribution depends on the location of each residue in the protein body (in our calculations we apply the function proposed by Levitt [30]). Each residue (represented by its effective atom — averaged-out positions of all atoms belonging to a particular residue) aggregates hydrophobic interactions with other residues separated from it by not more than 9 Å. Clearly, the observed distribution may differ from the idealized (Gaussian) profile. The scope of such interactions is dependent on the relative proximity between both residues and on their intrinsic hydrophobicity, which can be measured experimentally or predicted on theoretical grounds. Different hydrophobicity scales can be applied (a comparison of results obtained for two different scales is presented in [31]). Normalization of both distributions (theoretical and observed) facilitates meaningful comparisons: in particular, we can identify residues for which theoretical (T) values diverge significantly from their observed (O) counterparts.
Both hydrophobicity distribution profiles — expected (T) and observed (O) — can be compared quantitatively. Quantitative expression of the differences between the expected (T) and observed (O) distribution is possible by using the Kullback-Leibler divergence entropy formula [32]:
The value of D KL expresses the distance between the observed (p) and target (p 0) distributions, the latter of which is given by the 3D Gaussian (T). The observed distribution (p) is referred to as O.
For the sake of simplicity, we introduce the following notation:
Since D KL is a measure of entropy it must be compared to a reference value. In order to facilitate meaningful comparisons, we have introduced another opposite boundary distribution (referred to as “uniform” or R) which corresponds to a situation where each effective atom possesses the same hydrophobicity density (1/N, where N is the number of residues in the chain). This distribution is deprived of any form of hydrophobicity concentration at any point in the protein body:
Comparing O|T and O|R tells us whether the given protein (O) more closely approximates the theoretical (T) or uniform (R) distribution. Proteins for which O|T > O|R are regarded as lacking a prominent hydrophobic core. To further simplify matters we introduce the following relative distance (RD) criterion:
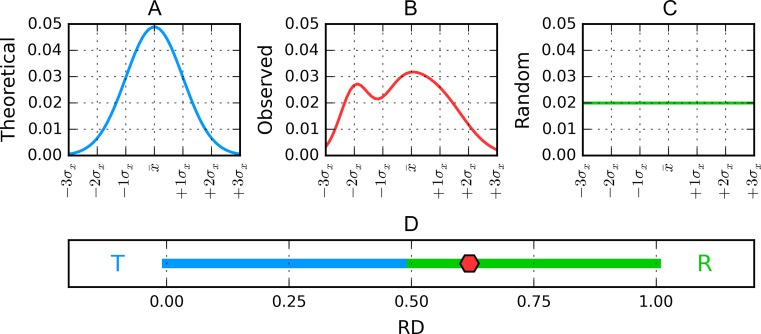
RD < 0.5 is understood to indicate the presence of a hydrophobic core. Figure 1 presents a graphical representation of RD values, restricted (for simplicity) to a single dimension.
Fig. 1.
Graphical representation of fuzzy oil drop model hydrophobicity distributions obtained for a hypothetical protein reduced to a single dimension for simplicity. A is the theorized Gaussian distribution (blue) while chart C corresponds to the uniform distribution (green). Actually observed (red) hydrophobicity density distribution in the target protein B, while its corresponding value of RD (relative distance) in D is marked on the horizontal axis with a red diamond. According to the fuzzy oil drop model this protein does not contain a well-defined hydrophobic core, because its RD value, equal to 0.619, is above the 0.5 threshold (or, generally, closer to R than to T)
D KL (as well as O|T, O|R and RD) may be calculated for specific structural units (protein complex, single molecule, single chain, selected domain etc.) In such cases the bounding ellipsoid is restricted to the selected fragment of the protein. It is also possible to determine the status of polypeptide chain fragments within the context of a given ellipsoid. This procedure requires prior normalization of O and R distributions describing the analyzed fragment, whose length is denoted as N. Note that any selected fragment must be of a reasonable length — calculations cannot be performed for an individual residue.
Generally the idealized distribution (T) is treated as the target for O|T calculation, while the unified distribution (R) is used when computing O|R.
The above procedure will be applied in the analysis of proteins described in this paper. By restricting our analysis to individual fragments, we can determine whether a given fragment participates in the formation of a hydrophobic core. In particular, fragments representing well-defined secondary folds which satisfy RD < 0.5 are thought to contribute to structural stabilization, while fragments for which RD > = 0.5 are less stable. Such fragments, if present on the surface of the protein, may potentially form complexation sites. Specific fragments are derived by analyzing the protein’s secondary conformation. Identification of secondary folds and the composition of protein domains follows the CATH [33] and PDBsum [34] classifications. Likewise, inter-domain/inter-chain contacts have been identified on the basis of the PDBsum distance criteria [34].
A graphical presentation of RD interpretation is shown in Fig. 1.
Results
α/β-domain fold twisted β-sheet
This category is represented by the nonhomologous domain 1 of lactate dehydrogenase (1A5Z – domain 1) and phosphoglycerate kinase (1FW8 – domain 2). The α,β-superfold can be described as a central twisted β-sheet surrounded by a ring of helices. RD values for both domains and their individual secondary folds are listed in Table 2.
Table 2.
Structural properties of the hydrophobic core in two proteins representing the α/β-domain fold twisted β-sheet category
| Lactate dehydrogenase (1A5Z-D1) 3.40.50.720 α-β 3-layer (aba) sandwich | Phosphoglycerate kinase (1FW8-D2) 3.40.50.1260 α-β 3-layer (aba) sandwich | ||||
|---|---|---|---|---|---|
| Structure | Fragment | RD | Structure | Fragment | RD |
| Domain 1 | 0.416 | Domain 2 | 0.523 | ||
| β-sheet | Parallel | 0.298 | β-sheet I and II | 0.516 | |
| Helices | 0.530 | Helices | 0.502 | ||
| Helices No 68–84 |
No ligand binding res. | 0.449 | β-sheet I | Parallel | 0.492 |
| β-conform. | 22–28 | 0.198 | β-conform. | 133–140 | 0.393 |
| 47–52 | 0.259 | 158–163 | 0.127 | ||
| 77–83 | 0.331 | 205–207 | 0.145 | ||
| 92–97 | 0.180 | 210–216 | 0.706 | ||
| 134–138 | 0.243 | 223–229 | 0.455 | ||
| 159–163 | 0.307 | 238–242 | 0.763 | ||
| 258–263 | 0.426 | ||||
| Helices | 30–44 | 0.498 | 294–298 | 0.144 | |
| 55–70 | 0.589 | ||||
| 71–73 | 0.980 | Helices | 146–155 | 0.353 | |
| 84–89 | 0.366 | 166–175 | 0.244 | ||
| 108–131 | 0.444 | 185–202 | 0.347 | ||
| 141–154 | 0.365 | 243–257 | 0.358 | ||
| 271–275 | 0.363 | ||||
| 276–291 | 0.721 | ||||
| 300–309 | 0.378 | ||||
| 311–315 | 0.228 | ||||
| Domain 2 | No β-II | 0.492 | |||
| β-sheet I No 294–298 | No ligand binding | 0.320 | |||
| β-sheet II | Anti-parallel | 0.625 | |||
| Helices No 287–301 |
No ligand binding | 0.455 | |||
Values listed in boldface correspond to RD > 0.5. Items labeled “No (…)” represent shortened fragments where elimination of the indicated residues changes the relation to RD < 0.5. “Ligand binding” stands for fragments directly involved in ligand binding. Our experience with the fuzzy oil drop model indicates that such fragments often exhibit significant deviations from the theoretical hydrophobicity distribution.
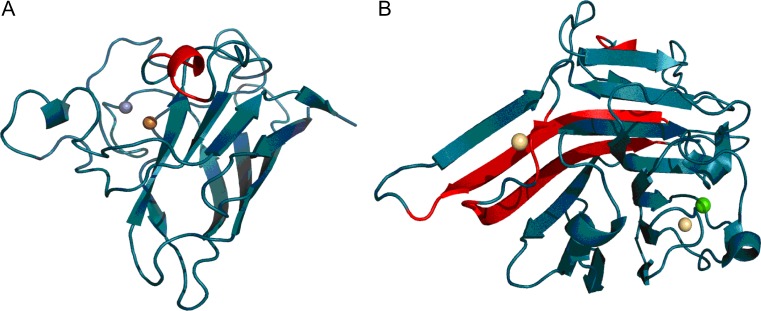
From the structural point of view both proteins provide examples of the so-called flavodoxin fold [35] with a centrally located β-propeller. Domain D1 (1A5Z) appears to include a well-ordered hydrophobic core while domain D2 (1FW8) lacks such a core, as indicated by its RD value, which is in excess of 0.5 (Table 2). It moreover turns out that the extra β-sheet (comprising three separate folds), which is not present in 1A5Z, diverges from the idealized distribution (Fig. 2). Eliminating this fragment from computations (an independent “droplike” capsule constructed for the remainder of the domain) produces a structure which is a good match for the theoretical model. Both domains exhibit similar properties in the scope of their central β-sheet, suggesting that the sheet contributes to structural stabilization (this is based on the assumption that the presence of a prominent hydrophobic core promotes tertiary stabilization). Both domains also contain a single helix (located at a similar distance from the propeller) which diverges from the model (Fig. 3 and Table 2).
Fig. 2.
3D representation of 1A5Z-D1 (A) and 1FW8-D2 (B). Fragments marked in red diverge from the theoretical model (RD > 0.5)
Fig. 3.
Hydrophobicity density distribution profiles (T – theoretical – green; O – observed – red) in 1A5Z-D1 and 1FW8-D2 (charts A and B respectively). Gray areas mark fragments where RD > 0.5
Lactate dehydrogenase (1A5Z) is derived from the hyperthermophilic bacterium Thermotoga maritima and folds near the boiling point of water. Thermostable proteins are the focus of a separate study based on the fuzzy oil drop model (publication currently in preparation).
The catalytic residues in 1A5Z, as well as its disulfide bonds, are all located in domain 2 — thus, we will not consider them in the presented analysis.
Domain 2 of phospoglycerate kinase (1FW8-D2) exhibits RD > 0.5, which means that its hydrophobic core is deformed. The domain contains two β-sheets — a parallel sheet (labeled “I” in Table 2–133-140, 158–163, 205–207, 258–263, 294–298) and an antiparallel sheet (labeled “II” – 210, 223–229, 238–242). Such antiparallel conformation is not present in lactate dehydrogenase (1A5Z-D1). The β-structure which is analogous to 1A5Z is characterized by RD = 0.492, which implies local consistency with the theoretical hydrophobic core model.
As posited by the fuzzy oil drop model, ligand binding residues exhibit deviations from the theoretical status; this is because such residues interact with the ligand, which also participates in the formation of a shared core. In the case of strongly polar ligands, deviations may also be associated with the presence of additional structural elements not directly involved in forming the hydrophobic core, but instead distorting its structure.
In both presented proteins eliminating residues which participate in ligand binding results in a lower RD value. This means that the remainder of the molecule fulfills a stabilizing role by presenting a well-ordered hydrophobic core.
The exposed β-hairpin in D2 of 1FW8 may mediate interaction with other molecules present in the protein’s environment, or it may constitute a dynamic and unstable element (no prominent hydrophobic core) which also potentially affects interaction.
It furthermore appears that the α,β-superfold — particularly the twisted β-sheet — contains a hydrophobic core and therefore improves the stability of its parent domain.
α,β-domain fold
The all-β-sheet superfold is represented by Cu, Zn superoxidase dismutase and concanavalin A. This type of all-β domain has the shape of a β-barrel. Both proteins appear to contain hydrophobic cores (RD < 0.5 in each case).
In 1B4L enzymatic and heavy metal ion binding residues are all located in loops. Table 3 lists positions adjacent to each fragment. The helical fold at 57–61 is locally discordant and contains a catalytic residue which binds Cu2+. Similarly, in 1CON the cadmium ion (Cd2+) is bound by a locally discordant β-fragment.
Table 3.
RD values obtained for 1B4L and 1CON, for their respective secondary folds, for the entire β-sheet and for the fragment bounded by CYS residues which form a disulfide bond
| Cu, Zn superoxide dismutase (1B4L) 2.60.40.200 – mainly β-sandwich | Concanavalin A (1CON) 2.60.120.200 – mainly β-sandwich | ||||
|---|---|---|---|---|---|
| Characteristics | Fragment | RD | Characteristics | Fragment | RD |
| Protein | 153aa | 0.433 | Protein | 237aa | 0.416 |
| 1–7 BI | 0.065 | I | 3–11 B | 0.407 | |
| 14–21 BI | 0.403 | I | 14–18 H | 0.421 | |
| 28–35 BI | 0.468 | 23–30 B | 0.355 | ||
| I | 41–48 BII | 0.246 | 36–40 B | 0.295 | |
| E 63H, I | 57–61 H | 0.724 | 46–56 B | 0.333 | |
| I | 82–89 BII | 0.433 | 59–66 B | 0.338 | |
| 95–101 BI | 0.343 | 73–79 B | 0.335 | ||
| I | 115–120 BII | 0.257 | 80–84 H | 0.218 | |
| 131–135 H | 0.349 | I | 87–97 B | 0.330 | |
| E 143R | 145–148 BII | 0.317 | 103–118 B | 0.601 | |
| 122–130 B | 0.135 | ||||
| 139–145 B | 0.438 | ||||
| 147–149 B | 0.851 | ||||
| 150–152 H | 0.365 | ||||
| 153–156 B | 0.384 | ||||
| 169–176 B | 0.336 | ||||
| 179–181 B | 0.643 | ||||
| I | 185–200 B | 0.762 | |||
| 208–217 B | 0.337 | ||||
| 226–230 H | 0.324 | ||||
| β-sheet | 0.409 | β-sheet | 0.468 | ||
| β -sheet I | 0.368 | Helices | 0.446 | ||
| β -sheet II | 0.432 | ||||
| SS-bond | 57–146 | 0.431 | |||
Values listed in boldface satisfy RD > 0.5. “B” and “H” stand for β-structural and helical forms respectively; “I” indicates an ion-binding fragment while “E” denotes that the given fragment contains an enzymatically active residue (listing its number and type).
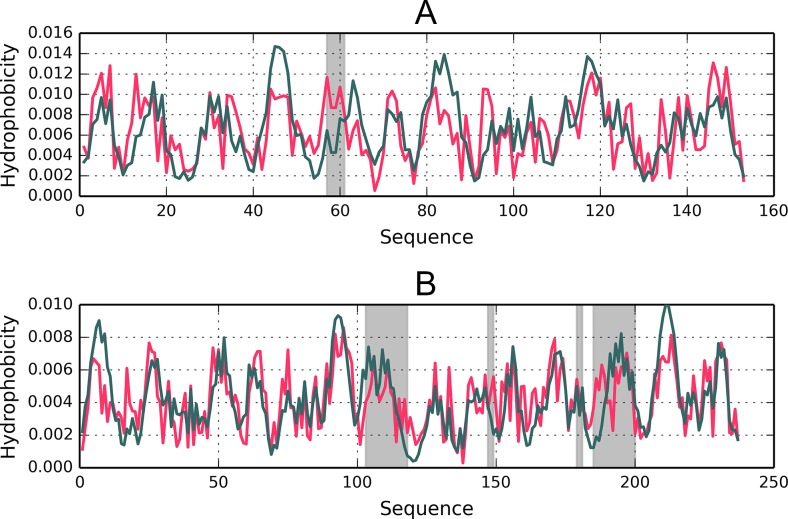
Cu, Zn superoxide dismutase adheres to the theoretical hydrophobicity distribution model as a whole, although some secondary folds diverge from the model. The helical fragment at 57–61 is an example (RD > 0.5). Similarly, in 1CON, despite the overall adherence of the molecule and its β-sheet, four individual β-folds are found to be discordant.
Both proteins (Fig. 4 and Fig. 5) exhibit highly ordered hydrophobic cores in spite of their biological diversity. Note that under the fuzzy oil drop model the concept of a “hydrophobic core” refers to a concentration of hydrophobicity density at the center of the molecule along with the presence of an encapsulating hydrophilic shell.
Fig. 4.
3D structures of 1B4L and 1CON, with fragments exhibiting high discordance (RD > 0.5) marked in red (Table 3). Spheres correspond to ion binding residues
Fig. 5.
Hydrophobicity distribution charts for 1B4L (A) and 1CON (B). Gray bands mark divergent fragments
All β-domain superfold
This category is represented by triose phosphate isomerase and pyruvate kinase domain 1. The core structure comprises a centrally located β-barrel where individual β-folds are interconnected by α-helices located outside of the fold. The sample proteins differ with respect to their composition and biological activity.
Elimination of catalytic residues (Table 4, Fig. 6, and Fig. 7) lowers the RD value to 0.479. This indicates local disorder in the area of the enzymatic active site.
Table 4.
RD values obtained for 1AMK and 4DRS and for their respective secondary folds
| Triose phosphate isomerase (1AMK) 1AMK – E.C.5.3.1.1 | Pyruvate kinase – domain 2 (4DRS) E.C.2.7.1.40 – absent in domain 2 | ||||
|---|---|---|---|---|---|
| Fragment | Characteristics | RD | Fragment | Characteristics | RD |
| Protein | 250 | 0.506 | Protein | 248 | 0.559 |
| 6–12 B | E 11 N, 13 K, | 0.401 | 47–52 B | 0.330 | |
| 17–30 H | 0.402 | 53–57 H | 0.278 | ||
| 38–42 B | 0.452 | 58–69 H | 0.475 | ||
| 44–47 H | 0.747 | 71–77 B | 0.497 | ||
| 48–55 H | 0.336 | 81–98 H | 0.432 | ||
| 59–63 B | 0.263 | 105–110 B | 0.580 | ||
| 79–86 H | 0.330 | 214–224 H | 0.404 | ||
| 89–93 B | 0.148 | 231–235 B | 0.378 | ||
| 95–102 H | E 95H | 0.634 | 240–253 H | 0.387 | |
| 105–119 H | 0.366 | 266–271 B | 0.477 | ||
| 122–127 B | 0.321 | 273–279 H | 0.551 | ||
| 130–136 H | 0.541 | 280–287 H | 0.329 | ||
| 138–152 H | 0.364 | 289–294 B | 0.390 | ||
| 153–154 H | 0.319 | 296–301 H | 0.647 | ||
| 155–160 H | 0.340 | 302–305 H | 0.534 | ||
| 161–167 B | E 167E | 0.317 | 306–321 H | 0.569 | |
| 168–172 H | 0.571 | 323–327 B | 0.395 | ||
| 179–198 H | E 173G | 0.423 | 331–336 H | 0.440 | |
| 199–206 H | 0.654 | 341–355 H | 0.418 | ||
| 207–212 B | 0.542 | 357–361 B | 0.306 | ||
| 218–224 H | 0.382 | 362–367 H | 0.603 | ||
| 229–233 B | 0.524 | 370–387 H | 0.553 | ||
| 234–239 H | 0.222 | ||||
| 240–241 H | 0.600 | ||||
| 242–248 H | 0.160 | ||||
| Β-sheet | β-sheet | 0.429 | |||
| Helices | Helices | 0.482 | |||
| Eliminated | 95–100,198–203 | 0.479 | No ligand | 0.546 | |
| No ligand | 0.498 | No residues | 74–80 L, 229–236 L, 259–262, 264–272 L, 293–296, 319–321 L | 0.486 | |
| NoE | 11,13,95,167,173 | 0.500 | |||
Values listed in boldface satisfy RD > 0.5. “B” and “H” stand for β- and helical forms respectively; “L” indicates a ligand-binding fragment while “E” denotes that the given fragment contains an enzymatically active residue (listing its number and type). The row labeled “NoE” represents the status of each domain following elimination of catalytic residues — the observed reduction in RD values indicates that catalytic residues diverge from the theoretical model.
Fig. 6.
Hydrophobicity distribution profiles (theoretical – blue and observed – red) for 1AMK (A) with indication of eliminated fragments, along with 4DRS (B) with indication of eliminated fragments (blue) and the placement of ligand binding residues (green)
Fig. 7.
3D presentation of 1AMK (A – catalytic residues visualized with CPK; fragments exhibiting RD > 0.5 marked in red) and 4DRS (B – ligand-binding residues colored with CPK; eliminated residues colored pink; fragments exhibiting RD > 0.5 marked in red)
Local deviations from the theoretical model caused by a ligand are commonplace. The ligand — by virtue of its presence — enforces a conformation which represents a balance between the protein’s own folding tendencies and the altered conditions introduced by the ligand’s polarized atoms. The end result of this process depends strongly on the ligand’s hydrophobicity [36].
Residues which diverge from the model, despite not taking part in ligand binding, may represent structurally encoded “readiness” for structural changes in the α/β domain.
Despite the observed differences, both domains share certain similarities. The helical fold at 95–100 in 1AMK exhibits a similar deviation from theoretical values when compared with the folds at 319–321 and 362–364 in 4DRS D2. It seems that local instability in all these areas may promote structural rearrangement.
β-barrel
Three proteins representative of the β-barrel domain are characterized in Table 5 and Fig. 8.
Table 5.
RD values representing individual secondary folds in 1RBP, 1PNG, and 1TIM
| 1RBP Up-and-down β barrel 2.40.128.20 – Mainly ββ-barrel Plasma retinol-binding protein precursor | 1PNG Jelly roll EC 3.5.1.52 2.60.120.230 – Mainly ββ-sandwich Hydrolase | 1TIM α/β barrel EC 5.3.1.1. 3.20.20.70 Αβ-Barrel isomerase | |||
|---|---|---|---|---|---|
| Fragment | RD | Fragment | RD | Fragment | RD |
| Protein 175aa | 0.553 | Domain 2 135aa | 0.694 | Domain 247 aa | 0.493 |
| 142–151 BI | 0.771 | 6–12 B E 11 N | 0.435 | ||
| 154–158 H | 0.472 | 17–31 H | 0.293 | ||
| 159–161 BI | 0.474 | 38–42 B | 0.434 | ||
| 5–9 H | 0.332 | 162–168 L | 0.605 | 46–55 H | 0.223 |
| 22–30 B | 0.567 | 169–175 BII | 0.610 | 58–65 B | 0.510 |
| 39–48 B | 0.431 | 176–180 L | 0.427 | 79–87 H | 0.418 |
| 52–63 B | 0.406 | 181–198 BI | 0.688 | 90–93 B | 0.590 |
| 67–80 B | 0.478 | 199–201 L | 0.560 | 95–103 HE 95H | 0.617 |
| 84–93 B | 0.589 | 202–206 BI | 0.187 | 105–119 H | 0.361 |
| 100–110 B | 0.506 | 207–209 L | 0.415 | 123–129 B | 0.141 |
| 113–124 B | 0.801 | 210–217 BII | 0.436 | 130–137 H | 0.725 |
| 128–139 B | 0.787 | 219–226 BII | 0.649 | 138–154 H | 0.527 |
| 145–159 H | 0.501 | 227–229 L | 0.511 | 159-167B E 165E | 0.431 |
| 166–168 B | 0.404 | 230–234 H | 0.209 | 168–170 H | 0.670 |
| 235–259 L | 0.654 | 177–197 H | 0.551 | ||
| 260–264 BI | 0.832 | 198–204 H | 0.590 | ||
| 265–269 H | 0.381 | 205–209 B | 0.387 | ||
| 271–278 BII | 0.742 | 215–222 H | 0.337 | ||
| 279–289 L | 0.800 | 227–231 B | 0.476 | ||
| 290–303 BI | 0.739 | 232–237 H | 0.221 | ||
| 304–310 L | 0.578 | 238–245 H | 0.291 | ||
| 311–313 BII | 0.508 | ||||
| β-sheet | 0.615 | β - I | 0.688 | β-sheet | 0.438 |
| Helices | 0.522 | β - II | 0.643 | Helices | 0.510 |
| SS 4–160 | 0.566 | SS 51–56 | 0.195 | ||
| SS 70–174 | 0.582 | SS 203–208 | 0.680 | ||
| SS 231–252 | 0.483 | ||||
| Removed: 29–31, 69–71 SS, 91–97 Loop, 117, 127–130 SS, 133–134 L |
0.467 | NoE | 0.478 | ||
Values listed in boldface satisfy RD > 0.5. H – helix; B – β-fold; E # – placement of catalytic residue (with # corresponding to its code); L – ligand present as part of the described fragment
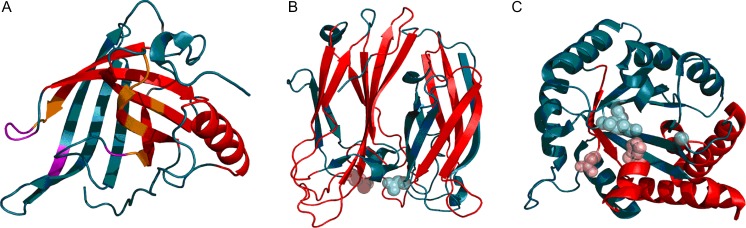
Fig. 8.
3D presentation of 1RBP (A), 1PNG (B), and 1TIM (C) with discordant fragments marked in red. Residues plotted with CPK represent catalytic residues
Each of the above listed proteins represents a different type of β-barrel. The assessment of hydrophobic core structure is, likewise, different in each case. It should be noted that the protein exists to fulfill a specific biological role and that its structure is a means to this end.
1RBP is a retinol binding protein. The presented domain contains three disulfide bonds which promote tertiary structural stability. Two of these bonds stabilize the domain in a “discordant” state, which is very likely their principal role.
In order to facilitate comparisons with other types of β-dominated supersecondary structures, we have included triole phosphate isomerase (1TIM) in our study. The corresponding form is an α/β barrel consisting of eight pairs of alternating β-strands and α-helices rolled up to form an inner barrel of eight parallel β-strands surrounded by an outer barrel of eight parallel helices. Analysis based on the fuzzy oil drop model indicates the presence of a well ordered hydrophobic core. Stabilization is provided by the arrangement of β-folds (RD = 0.493), while catalytic residues deform the domain (their elimination results in RD = 0.478).
Discordant helixes include the fragments at 95–103 (RD = 0.617), 130–137 (RD = 0.725), 138–154 (RD = 0.527), 168–170 (RD = 0.671), 177–197 (RD = 0.551), 198–204 (RD = 0.590). Catalytic residues are located in direct proximity to the active site. Experience with the fuzzy oil drop model indicates that enzymatic active sites are, in most cases, deficient in terms of hydrophobicity and diverge significantly from theoretical values. In the presented cases catalytic residues belong to the helices at 95–103, 168–170, 177–197, and 198–204, with RD values in excess of 0.5 in all cases.
Miscellaneous
The arrangement of helical and β-folds in higher-order structures varies greatly both with respect to the quantity of each and their conformation. An example of a purely α protein is provided by the b562 cytochrome (1APT) from E. coli. Topologically, this protein is a sequence of antiparallel helical fragments described as an up-down-up-down 4-helix bundle.
In contrast, the Fab IgG immunoglobulin domain (7FAB) is comprised almost entirely of β-folds, with its structure characterized as a sandwich of 3- and 4-stranded antiparallel β-sheets.
A combination of helical and β-motifs is found in the 163-residual domain of dogfish lactate dehydrogenase (6LDH), which contains a 6-stranded parallel β-sheet with crossovers between β-strands and α-helices forming a right-handed helical turn with flanking β-strands.
The cytochrome is characterized by RD = 0.426, suggesting the presence of a well-ordered hydrophobic core. All its helical fragments also satisfy RD < 0.5 (3–20: RD = 0.355; 23–43: RD = 0.481; 45–49: RD = 0.342; 57–81: RD = 0.402; 82–94: RD = 0.408). It seems that this protein depends strongly on the stabilizing influence of hydrophobic interactions. Given that the protein’s biological role is to bind heme, its structure provides suitable conditions for firm anchoring of the ligand at its center, with the polypeptide chain forming an “envelope” around the heme molecule.
7FAB
The Fab IgG domain fragment consists of two chains: the light chain (L) and the heavy chain (H), each of which is further composed of two domains (labeled V and C). All these structures exhibit a similar conformation, often referred to as an immunoglobulin-like domain: a sandwich comprising two β-sheets which we will refer to as the upper core and lower core respectively.
Under the CATH classification the Fab fragment is characterized as 2.60.40.10 mainly β-sandwich. Both cores are linked by a centrally placed disulfide bond. The biological role of the Fab fragment is to recognize antigens via so-called CDRs — short variable loops which interact with the antigen, triggering a process called immunological signal transduction. This produces structural changes in the Fc fragment of immunoglobulin, priming it for complexation of the C1q complement, which is a first step in a cascade of reactions resulting in destruction of the cell which originally presented the antigen.
The role of the Fab fragment is therefore to bind the antigen and trigger a signaling pathway. Both processes require a flexible structure, capable of accommodating the antigen and producing structural changes elsewhere in the molecule. The flexibility must, however, remain selective so as to ensure that the correct ligand is recognized and the correct signal sent.
This situation is evidenced by the RD status of individual β-folds in all four domains: VH, CH1, VL, and CL (see Table 6 and Fig. 9).
Table 6.
RD values calculated for ragmentdomains of the Fab fragment of immunoglobulin G, revealing variable stability of individual secondary folds
| Chain L | Chain H | ||||||
|---|---|---|---|---|---|---|---|
| Domain VL | Domain CL | Domain VH | Domain CH1 | ||||
| Fragment | RD | Fragment | RD | Fragment | RD | Fragment | RD |
| Domain | 0.584 | Domain | 0.320 | Domain | 0.601 | DOMAIN | 0.538 |
| 8–12 BI | 0.584 | 110–114 BI | 0.073 | 3–7 BI | 0.583 | 124–128 BI | 0.293 |
| 18–24 BII | 0.649 | 117–123 H | 0.388 | 10–12 BII | 0.673 | 140–149 BI | 0.424 |
| 28–32 H | 0.629 | 126–135 BI | 0.254 | 18–25 BI | 0.682 | 155–159 BII | 0.294 |
| 36–40 BI | 0.437 | 141–147 BII | 0.554 | 33–39 BII | 0.528 | 160–162 H | 0.670 |
| 47–49 BI | 0.281 | 149–151 BII | 0.545 | 46–52 BII | 0.551 | 166–170 BI | 0.808 |
| 56–62 BII | 0.369 | 154–158 BI | 0.525 | 56–60 BII | 0.702 | 173–175 BI | 0.766 |
| 64–71 BII | 0.703 | 161–163 BI | 0.083 | 62–64 H | 0.002 | 179–188 BI | 0.878 |
| 74–78 H | 0.675 | 167–176 BI | 0.555 | 66–72 BI | 0.330 | 189–193 H | 0.556 |
| 79–87 BI | 0.468 | 177–183 H | 0.353 | 76–83 BI | 0.358 | 198–204 BII | 0.283 |
| 90–94 BI | 0.358 | 186–193 BII | 0.394 | 86–90 H | 0.874 | 205–208 H | 0.492 |
| 97–102 BI | 0.710 | 196–202 BII | 0.543 | 91–99 BII | 0.665 | 209–215 BII | 0.367 |
| 103–108 BII | 0.576 | ||||||
| 111–116 BII | 0.689 | ||||||
| SS 22–86 | 0.600 | SS 130–189 | 0.332 | SS 22–95 | 0.568 | SS 144–200 | 0.638 |
| B - I | 0.613 | B - I | 0.331 | B - I | 0.481 | B - I | 0.648 |
| B - II | 0.477 | B - II | 0.482 | B - II | 0.703 | B - II | 0.242 |
Values listed in boldface satisfy RD > 0.5.
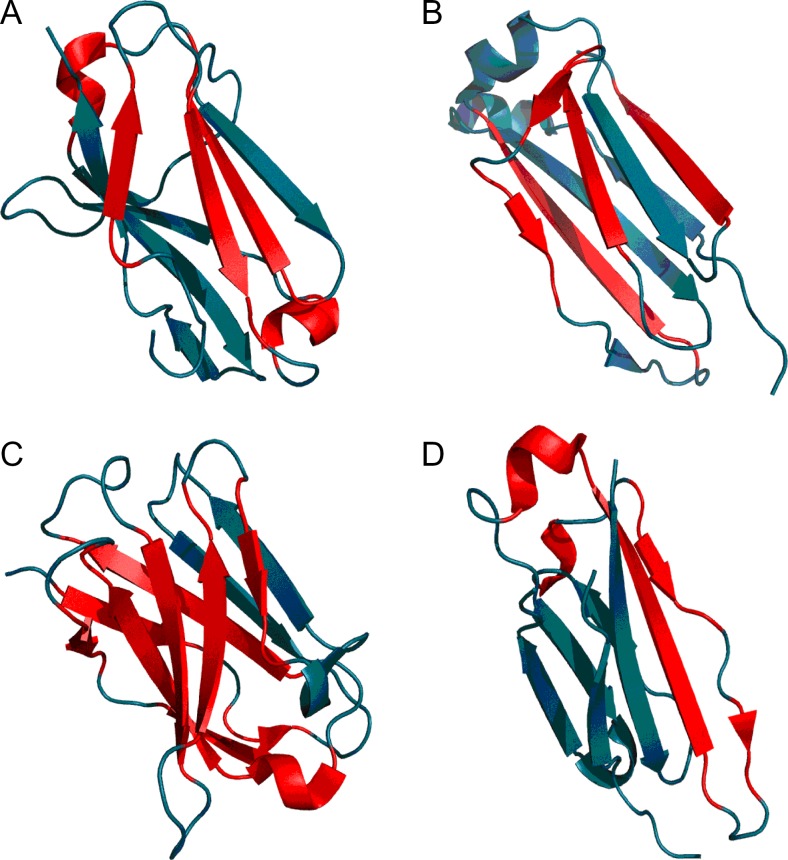
Fig. 9.
3D representation of 7FAB immunoglobulin domains: VL (A), CL (B), VH (C), and CH (D). Fragments marked in red satisfy RD > 0.5
The variable status of individual secondary folds comprising Fab domains suggests local instabilities required for complexation of antigens. One of the domains (CL) is stable as a whole and includes a stable β-I sheet, while all other domains are characterized as relatively unstable and therefore flexible. The stabilizing influence of SS bonds likewise varies: only in the CL domain does the affected fragment remain consistent with theoretical predictions.
Immunoglobulins work in an unpredictable environment — the antigen is never known a priori. Thus, a system characterized by variable local stability creates suitable conditions for targeted signaling while preserving the immunoglobulins ability to interact with a variety of antigens.
A review of immunoglobulin-like domains (which includes domains not associated with immunoglobulin activity, including enzymes and structural proteins) can be found in [8]. The authors reveal a specific arrangement of accordant and discordant fragments comprising the domain, which predispose it toward specific structural changes. Overall, the domain is characterized by poor stability, evidenced by locally high RD values.
The dogfish lactate dehydrogenase N-terminal 163-aa fragment is an example of a 6-strand parallel β-structure linked by a right-handed helical turn. The domain as a whole is characterized by RD = 0.489, while elimination of its N-terminal fragment (1–19 aa), which significantly disrupts the globular form, further reduces RD to 0.428. Evidently, the domain contains a prominent hydrophobic core. Table 7 lists the RD status of its individual folds (compare also Fig. 10 and Fig. 11).
Table 7.
RD values calculated for the N-terminal domain of dogfish lactate dehydrogenase (6LDH)
| 6LDH | |
|---|---|
| Fragment | RD |
| Domain 20–162 | 0.428 |
| 21–27 B | 0.146 |
| 29–41 H | 0.241 |
| 47–52 B | 0.293 |
| 54–68 H | 0.525 |
| 76–80 B | 0.337 |
| 82–87 H | 0.447* |
| 90–95 B | 0.214 |
| 105–128 H | 0.447 |
| 132–136 B | 0.559 |
| 139–152 H | 0.349 |
| 157–161 B | 0.332 |
| β-sheet | 0.366 |
| HELICES | 0.396 |
Values listed in boldface satisfy RD > 0.5. The asterisk marks the right-handed helical fold which causes β-strand flanking
Fig. 10.
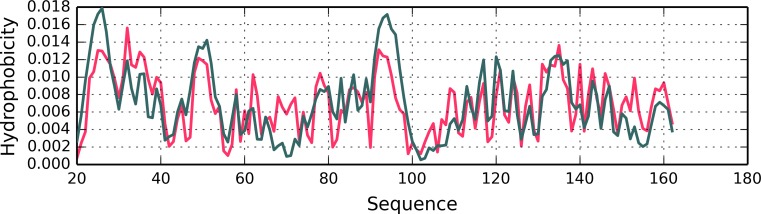
N-terminal domain of dogfish lactate dehydrogenase (6LDH) – theoretical (T – blue) and observed (O – red) distributions indicating the presence of a prominent hydrophobic core
Fig. 11.
3D representation of the N-terminal domain (20–162) of 6LDH. Fragments marked in red exhibit RD > 0.5 while the fragment marked in green corresponds to the right-handed helical fold causing β-strand flanking
Cytochrome fold
The author of [1] also addresses the concept of protein families, with cytochromes discussed as a representative example. This family comprises both eukaryotic cytochromes and c-type cytochromes present in prokaryotes. Both fulfill a similar biological role, acting as electron carriers. Cytochromes derived from various organisms are sequentially dissimilar despite retaining similar 3D forms. The conclusion presented in [1] is that evolution preserves the structural and functional properties of proteins rather than their sequential arrangement. We further propose that the subject of evolutionary conservation is the structure of the proteins hydrophobic core — as we attempt to substantiate in this part.
Table 8 and Table 9 present the properties of cytochromes under consideration. The FOD status of the entire molecule appears preserved despite differences in chain length (88–135 aa) along with the placement and conformation of individual secondary folds. “No ligand” indicates parts of the molecule which do not contact the ligand. Evidently, such fragments are also characterized by consistent RD values.
Table 8.
RD values calculated for cytochromes c550 and c555
| 4 J20 Cytochrome c555 Chlorobium tepidum 1.10.760.10 mainly α orthogonal bundle | 155C Cytochrome c550 Paracoccus 1.10.760.10 mainly α orthogonal bundle | ||
|---|---|---|---|
| Fragment | RD | Fragment | RD |
| Protein | 0.356 | Protein | 0.399 |
| 4–16 H* | 0.434 | 5–13 H | 0.278 |
| 17–21 H* | 0.336 | 14–17 L* | 0.368 |
| 23–27 H* | 0.409 | 18–20 B* | 0.835 |
| 28–31 L* | 0.585 | 21–34 L* | 0.370 |
| 32–41 H* | 0.463 | 35–38 B | 0.661 |
| 43–54 H* | 0.377 | 39–54 L* | 0.306 |
| 55–57 B* | 0.338 | 55–65 H* | 0.198 |
| 60–62 B* | 0.452 | 66–71 L* | 0.385 |
| 63–68 H* | 0.595 | 72–81 H* | 0.385 |
| 72–86 H* | 0.119 | 82–105* | 0.439 |
| 106–118 H | 0.277 | ||
| β-strand 55–62 | 0.290 | β-strand 18–38 | 0.554 |
| β-strand (55–57) + (60–62) | 0.298 | β-strand (18–20) + (35–38) | 0.706 |
| Ligand | 0.338 | Ligand | 0.549 |
| No ligand | 0.356 | No ligand | 0.457 |
Values listed in boldface satisfy RD > 0.5. Asterisks mark fragments involved in interaction with the ligand. The row labeled “Ligand” corresponds to ligand-binding residues, while the row labeled “Noligand” presents residues not involved in ligand interaction.
Table 9.
RD values calculated for cytochromes b562, c2 and c
| 256B Ecoli 1.20.120.10 mainly α up-down bundle Cytochrome b562 | 1JDL Rhodospirillum rubrum 1.10.760.10 α orthogonal bundle Cytochrome c2 | 2C2C Rhodopsin rubrum 1.10.760.10 mainly α orthogonal bundle Cytochrome c2 | 5CYT Thunnus alalunga 1.10.760.10 mainly α orthogonal bundle Cytochrome c | ||||
|---|---|---|---|---|---|---|---|
| Fragment | RD | Fragment | RD | Fragment | RD | Fragment | RD |
| Protein | 0.411 | Protein | 0.476 | Protein | 0.448 | Protein | 0.392 |
| 2–20 H | 0.341 | 4–12 H | 0.295 | 3–11 H | 0.503 | 2–14 H* | 0.312 |
| 22–41 H* | 0.543 | 13–15 H* | 0.229 | 12–15 H* | 0.359 | 15–48 L* | 0.368 |
| 45–49 H* | 0.371 | 16–49 L* | 0.402 | 16–48 L* | 0.488 | 49–54 H* | 0.442 |
| 55–81 H* | 0.424 | 50–59 H* | 0.506 | 49–58 H* | 0.202 | 55–59 L* | 0.643 |
| 83–92 H | 0.300 | 60–63 L* | 0.777 | 59–62 L* | 0.710 | 60–70 H* | 0.254 |
| 93–106* | 0.377 | 64–72 H* | 0.211 | 63–73 H* | 0.345 | 71–75 H* | 0.296 |
| 74–86 H | 0.661 | 74–83 H | 0.493 | 76–86 L* | 0.442 | ||
| 88–93 H | 0.446 | 84–96 L* | 0.561 | 87–102 H* | 0.233 | ||
| 94–103 L* | 0.487 | 97–109 H | 0.192 | ||||
| 104–116 H | 0.236 | ||||||
| 117–119 | 0.146 | ||||||
| Ligand | 0.225 | Ligand | 0.549 | Ligand | 0.593 | Ligand | 0.490 |
| No ligand | 0.408 | No ligand | 0.457 | No ligand | 0.424 | No ligand | 0.376 |
Values listed in boldface satisfy RD > 0.5. Asterisks mark fragments involved in interaction with the ligand. The row labeled “Ligand” corresponds to ligand-binding residues, while the row labeled “No ligand” presents residues not involved in ligand interaction.
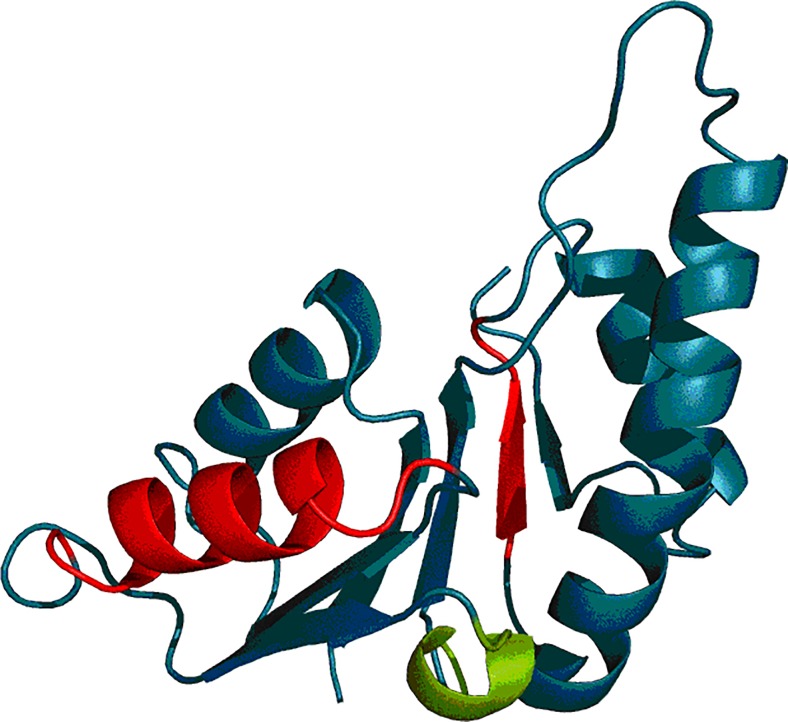
Cytochromes are proteins which act as “envelopes” for their ligand (heme – see Fig. 12). This ligand is a large, flat molecule dominated by hydrophobic interactions. Comparative analysis of cytochromes boils down to comparing various forms of “envelopes” which create suitable conditions for maintaining the intended level of oxidation of iron ions and expose them for participation in biological processes. These forms are briefly outlined in Tables 8 and 9. The status of the fragment which does not participate in binding heme appears not to change. This is due to its primary role — structural stabilization. In contrast, heme-binding residues exhibit variable FOD status, which may be related to the process of exposing the ligand for various types of interactions.
Fig. 12.
3D representation of cytochromes with fragments exhibiting RD > 0.5 marked in red: 4 J20 (A), 155C (B), 256B (C), 1JD2 (D), 2C2C (E), and 5CYT (F). Pink fragments are involved in interaction with the ligand (heme)
The conclusions of [1] also highlight the high structural similarity of the NAD+ binding fragment in enzymes which participate in many different metabolic pathways. These proteins are more widely discussed in [8].
Discussion and conclusions
Applying the fuzzy oil drop model to structures classified using the supersecondary fold criterion reveals similar FOD characteristics, with hydrophobic cores observed either on the domain or on the protein level. A global analysis of a broad nonredundant set of proteins derived from the PDB database indicates that most domains generate prominent hydrophobic cores (RD < 0.5) [37]. It seems that, at least on the level of domains, the water environment greatly influences the folding process.
It is up to each researcher to select fragments of interest, depending on the problem being studied. In our experience, fragments which diverge from theoretical predictions produced by the FOD model are typically associated with biological function. The status of particular secondary fragments in supersecondary structures may quantitatively reveal their contribution to overall supersecondary stabilization.
In summary, we can state that the targeted stability required in many biological processes can be supplied by a combination of nonbinding interactions, structural stabilization mediated by hydrophobic forces, and the presence of disulfide bonds. As shown in [38], SS-bonds may either reinforce or disrupt hydrophobic core stability. The influence of the water environment on protein folding also explains the mechanism behind hydrophobic collapse, which reduces the protein’s conformational space sufficiently to enable a random search for the native fold [39]; this shows that hydrophobic collapse is sufficient to solve Levinthal’s paradox [40].
Discussions concerning the fundamental role of the hydrophobic core have a long history [41] with both experimental [42] and theoretical approaches [43].
In the context of these observations and the analysis presented in this paper we can conclude that the answer to the question posed in the title is “yes”. We can furthermore state that the fuzzy oil drop model is able to describe the hydrophobic core structure in both qualitative and quantitative terms including the substantial and specific difference of hydrophobicity distribution observed in amyloids [31].
Acknowledgements
The authors would like to express their thanks to Piotr Nowakowski and Anna Śmietańska for valuable suggestions and editorial work. This research was supported by Jagiellonian University Medical College grant no. K/ZDS/006363 and by the KNOW system.
References
- 1.Devlin TM. Textbook of biochemistry with clinical correlations. 7. New York: Wiley; 2011. [Google Scholar]
- 2.Sillitoe I, Lewis TE, Cuff AL, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees J, Lehtinen S, Studer R, Thornton JM, Orengo CA (2015) CATH: comprehensive structural and functional annotations for genome sequences Nucleic Acids Res. doi:10.1093/nar/gku947 [DOI] [PMC free article] [PubMed]
- 3.Skolnick J, Gao M, Zhou H. How special is the biochemical function of native proteins? F1000Res. 2016;2016:5. doi: 10.12688/f1000research.7374.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis IW, Arendall WB, 3rd, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14(2):265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Banach M, Prymula K, Jurkowski W, Konieczny L, Roterman I. Fuzzy oil drop model to interpret the structure of antifreeze proteins and their mutants. J Mol Model. 2012;18:229–237. doi: 10.1007/s00894-011-1033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banach M, Konieczny L, Roterman I. Use of the “fuzzy oil drop” model to identify the complexation area in protein homodimers. In: Roterman-Konieczna I, editor. Protein folding in silico. Oxford: Woodhead; 2012. pp. 95–122. [Google Scholar]
- 7.Banach M, Konieczny L, Roterman I. Ligand-binding-site recognition. In: Roterman-Konieczna I, editor. Protein folding in silico. Oxford: Woodhead; 2012. pp. 78–94. [Google Scholar]
- 8.Banach M, Konieczny L, Roterman I. Can the structure of hydrophobic core determine the complexation site? In: Roterman-Konieczna I, editor. Identification of ligand binding site and protein-protein interaction area. Berlin: Springer; 2013. pp. 41–54. [Google Scholar]
- 9.Prymula K, Jadczyk T, Roterman I. Catalytic residues in hydrolases: analysis of methods designed for ligand-binding site prediction. J Comput Aided Mol Des. 2011;25:117–133. doi: 10.1007/s10822-010-9402-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auerbach G, Ostendorp R, Prade L, Korndörfer I, Dams T, Huber R, Jaenicke R. Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization. Structure. 1998;6:769–781. doi: 10.1016/S0969-2126(98)00078-1. [DOI] [PubMed] [Google Scholar]
- 11.Tougard P, Bizebard T, Ritco-Vonsovici M, Minard P, Desmadril M. Structure of a circularly permuted phosphoglycerate kinase. Acta Crystallogr D Biol Crystallogr. 2002;58:2018–2023. doi: 10.1107/S0907444902015548. [DOI] [PubMed] [Google Scholar]
- 12.Williams JC, Zeelen JP, Neubauer G, Vriend G, Backmann J, Michels PA, Lambeir AM, Wierenga RK. Structural and mutagenesis studies of leishmania triosephosphate isomerase: a point mutation can convert a mesophilic enzyme into a superstable enzyme without losing catalytic power. Protein Eng. 1999;12:243–250. doi: 10.1093/protein/12.3.243. [DOI] [PubMed] [Google Scholar]
- 13.Cook WJ, Senkovich O, Aleem K, Chattopadhyay D. Crystal structure of Cryptosporidium parvum pyruvate kinase. PLoS One. 2012;7:e46875. doi: 10.1371/journal.pone.0046875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart PJ, Balbirnie MM, Ogihara NL, Nersissian AM, Weiss MS, Valentine JS, Eisenberg D. A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry. 1999;38:2167–2178. doi: 10.1021/bi982284u. [DOI] [PubMed] [Google Scholar]
- 15.Naismith JH, Habash J, Harrop S, Helliwell JR, Hunter WN, Wan TC, Weisgerber S, Kalb AJ, Yariv J. Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution. Acta Crystallogr D Biol Crystallogr. 1993;49:561–571. doi: 10.1107/S0907444993006390. [DOI] [PubMed] [Google Scholar]
- 16.Cowan SW, Newcomer ME, Jones TA. Crystallographic refinement of human serum retinol binding protein at 2A resolution. Proteins. 1990;8:44–61. doi: 10.1002/prot.340080108. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn P, Tarentino AL, Plummer TH, Jr, Van Roey P. Crystal structure of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F at 2.2-A resolution. Biochemistry. 1994;33:11699–11706. doi: 10.1021/bi00205a005. [DOI] [PubMed] [Google Scholar]
- 18.Banner DW, Bloomer A, Petsko GA, Phillips DC, Wilson IA. Atomic coordinates for triose phosphate isomerase from chicken muscle. Biochem Biophys Res Commun. 1976;72:146–155. doi: 10.1016/0006-291X(76)90972-4. [DOI] [PubMed] [Google Scholar]
- 19.Lederer F, Glatigny A, Bethge PH, Bellamy HD, Matthew FS. Improvement of the 2.5 A resolution model of cytochrome b562 by redetermining the primary structure and using molecular graphics. J Mol Biol. 1981;148:427–448. doi: 10.1016/0022-2836(81)90185-6. [DOI] [PubMed] [Google Scholar]
- 20.Saul FA, Poljak RJ. Crystal structure of human immunoglobulin fragment Fab New refined at 2.0 A resolution. Proteins. 1992;14:363–371. doi: 10.1002/prot.340140305. [DOI] [PubMed] [Google Scholar]
- 21.Abad-Zapatero C, Griffith JP, Sussman JL, Rossmann MG. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J Mol Biol. 1987;198:445–467. doi: 10.1016/0022-2836(87)90293-2. [DOI] [PubMed] [Google Scholar]
- 22.Timkovich R, Dickerson RE. Refined crystal structure of dogfish M4 apo-lactate dehydrogenase. J Biol Chem. 1976;251:4033–4046. doi: 10.2210/pdb155c/pdb. [DOI] [PubMed] [Google Scholar]
- 23.Camara-Artigas A, Williams JC, Allen JP. Structure of cytochrome c2 from Rhodospirillum centenum. Acta Crystallogr D Biol Crystallogr. 2001;57:1498–1505. doi: 10.1107/S0907444901010423. [DOI] [PubMed] [Google Scholar]
- 24.Bhatia G, Finzel BC, Kraut J (1984) Electron transport protein (cytochrome) from Rhodospirillum rubrum PDB Depositions Authors: of 2C2C. http://www.rcsb.org/pdb/explore/explore.do?structureId=2c2c
- 25.Takano T. Methods and applications in crystallographic computing. Oxford: Clarendon; 1984. p. 262. [Google Scholar]
- 26.Yu LJ, Unno M, Kimura Y, Yanagimoto K, Oh-oka H, Wang-otomo ZY. Structure analysis and characterization of the cytochrome c-554 from thermophilic green sulfur photosynthetic bacterium Chlorobaculum tepidum. Photosynth Res. 2013;118:249–258. doi: 10.1007/s11120-013-9922-2. [DOI] [PubMed] [Google Scholar]
- 27.Konieczny L, Bryliński M, Roterman I. Gauss-function-based model of hydrophobicity density in proteins. In Silico Biol. 2006;6:15–22. [PubMed] [Google Scholar]
- 28.Kalinowska B, Banach M, Konieczny L, Roterman I. Application of divergence entropy to characterize the structure of the hydrophobic core in DNA interacting proteins. Entropy. 2015;17(3):1477–1507. doi: 10.3390/e17031477. [DOI] [Google Scholar]
- 29.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/S0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 30.Levitt MA. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976;104:59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- 31.Roterman I, Banach M, Kalinowska B, Konieczny L. Influence of the aqueous environment on protein structure—a plausible hypothesis concerning the mechanism of amyloidogenesis. Entropy. 2016;18(10):351. doi: 10.3390/e18100351. [DOI] [Google Scholar]
- 32.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22:79–86. doi: 10.1214/aoms/1177729694. [DOI] [Google Scholar]
- 33.Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Lees JG, Lewis TE, Studer RA, Rentzsch R. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41:D490–D498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Beer TAP, Berka K, Thornton JM, Laskowski RA. PDBsum additions. Nucleic Acids Res. 2014;42:D292–D296. doi: 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banach M, Prudhomme N, Carpentier M, Duprat E, Papandreou N, Kalinowska B, Chomilier J, Roterman I. Contribution to the prediction of the fold code: application to immunoglobulin and flavodoxin cases. PLoS One. 2015;10(4):e0125098. doi: 10.1371/journal.pone.0125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banach M, Konieczny L, Roterman I. The fuzzy oil drop model, based on hydrophobicity density distribution, generalizes the influence of water environment on protein structure and function. J Theor Biol. 2014;359:6–17. doi: 10.1016/j.jtbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Sałapa K, Kalinowska B, Jadczyk T, Roterman I. Measurement of hydrophobicity distribution in proteins – complete redundant protein data bank. Bio-Algorithms and Med-Systems. 2012;8(2):195–206. [Google Scholar]
- 38.Lin MM, Zewail AH. Hydrophobic forces and the length limit of foldable protein domains. Proc Natl Acad Sci U S A. 2012;109(25):9851–9856. doi: 10.1073/pnas.1207382109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levinthal C. Are there pathways for protein folding? J Chem Phys. 1968;65:44–45. [Google Scholar]
- 40.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/S0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 41.Agashe VR, Shastry MC, Udgaonkar JB. Initial hydrophobic collapse in the folding of barstar. Nature. 1995;377(6551):754–757. doi: 10.1038/377754a0. [DOI] [PubMed] [Google Scholar]
- 42.Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282(5389):740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 43.Burke MG, Woscholski R, Yaliraki SN. Differential hydrophobicity drives self-assembly in Huntington's disease. Proc Natl Acad Sci U S A. 2003;100(24):13928–13933. doi: 10.1073/pnas.1936025100. [DOI] [PMC free article] [PubMed] [Google Scholar]