Abstract
Although dependent on the integrity of a central pacemaker in the suprachiasmatic nucleus of the hypothalamus (SCN), endogenous daily (circadian) rhythms are expressed in a wide variety of peripheral organs. The pathways by which the pacemaker controls the periphery are unclear. Here, we used parabiosis between intact and SCN-lesioned mice to show that nonneural (behavioral or bloodborne) signals are adequate to maintain circadian rhythms of clock gene expression in liver and kidney, but not in heart, spleen, or skeletal muscle. These results indicate that the SCN regulates expression of circadian oscillations in different peripheral organs by diverse pathways.
Keywords: suprachiasmatic, parabiosis, entrainment
A wide variety of physiological events and behaviors exhibit pronounced endogenous daily (circadian) rhythms. Experimental destruction of the suprachiasmatic nucleus of the hypothalamus (SCN) results in arrhythmicity that is reversed by neurotransplantation of this structure. Persistence of rhythmicity depends on the operation of transcriptional-translational feedback loops among the products of critical genes, including Per1, Per2, and Bmal1 within the SCN. Circadian oscillations of Per1, Per2, and Bmal1 also occur in a variety of peripheral organs. Evidence has accumulated for both neural and humoral control of peripheral rhythms. For example, serum shock initiates oscillations of mPer1 expression in cultured hepatocytes and HeLa cells (1), and implants of fibroblasts that receive no innervation adopt the circadian phase of the host (2). Endocrine signals, including glucocorticoids, angiotensin II, and retinoic acid can shift the phase of peripheral clock gene expression (3–5). In addition, metabolites such as glucose may directly entrain peripheral oscillators or act indirectly to induce endocrine signals that regulate circadian rhythms of gene expression (6). On the other hand, the autonomic innervation of peripheral organs provides a potential pathway for entrainment (7–9). For example, SCN lesions that compromise catecholamine rhythms eliminate oscillations of clock gene expression in mouse liver (10).
Neural and endocrine pathways for peripheral entrainment are not mutually exclusive. Furthermore, different organs may vary in their dependence on one or another entraining signal. Such variation is not without precedent. Whereas behavioral rhythms appear to depend on humoral outputs of the SCN (11), endocrine rhythms may rely on axonal projections (12).
The technique of parabiosis offers unique advantages for investigation of the importance of blood-borne cues in the control of a variety of physiologic systems. This approach has been exploited in the study of cockroach circadian rhythms (13). Despite its useful application to study of metabolic signals in mice (14, 15), the effect on peripheral circadian rhythms of establishing vascular exchange without neural communication has not previously been investigated in vertebrates. We now report that parabiotic linkage of SCN-lesioned mice to intact partners reinstates circadian rhythmicity in some, but not other, peripheral organs.
Methods
Four groups of adult male C57/Bl6 mice were used in this experiment. Nonparabiosed controls were either unoperated (n = 11) or subjected to electrolytic SCN lesions under ketamine-xylazine anesthesia (n = 13). Lesions were produced by using tungsten rods (0.02 mm in diameter, A-M Systems, Everett, WA), insulated except at the cross-sectional tip with epoxylite resin. Electrodes were lowered 5.4 mm below the dural surface, 0.3 mm anterior to bregma, and at 0.2 mm on either side of the midline. A current of 1 mA was passed for 15 sec. Some animals were placed in running wheel cages in constant darkness to verify locomotor arrhythmicity for several weeks after the lesion. In all cases, mice were included only if subsequent histological analysis verified the presence of complete SCN lesions. Parabiosed controls consisted of two nonlesioned mice that were prepared as described (15). Experimental parabiotic pairs consisted of one SCN-lesioned mouse and one intact mouse. The position of the SCN-lesioned animal (left or right) was varied randomly in experimental pairs. This report is based on eight intact pairs and nine pairs comprised of intact and SCN-lesioned partners.
Animals were maintained in 12 h-light:12 h-dark with food and water available ad libitum from the time of parabiosis surgery until the last 3 days of the experiment. At that time, all mice were transferred to constant darkness (DD). Animals were killed by rapid decapitation on the third day of DD, at either zeitgeber time (ZT)1 or ZT13 (ZT12 = time of lights out in the previous light:dark cycle). Organs were rapidly removed and immediately frozen on dry ice. Brains were sectioned at 11 μm, mounted on microscope slides, and Nissl-stained to evaluate SCN lesions. Liver, kidney, spleen, heart, testis, and skeletal muscle (taken from the hind leg) were homogenized for RNA isolation by using Ultraspec reagent (Biotex Laboratories, Friendswood, TX).
Northern blots were used to measure the abundance of mPer1, mPer2, and mBmal1 relative to mGAPDH by using 32P-labeled cDNA probes as described (16). Briefly, RNA extracted from paired mice was run in adjacent lanes of the Northern gels. Equal numbers of samples collected at ZT1 and ZT13 were run on the same gels, and, for each organ, samples from all mice were run on two gels that were probed, exposed, and developed simultaneously. Blots were hybridized by using ULTRAhyb buffer (Ambion, Austin, TX) and exposed to Kodak BioMax MS film (Rochester, NY) at -80°C. Blots were stripped and sequentially reprobed for each clock transcript. Quantitative densitometry was used to express ratios between each transcript and GAPDH. The Kruskal–Wallis test was used to evaluate effects of time of day (ZT1 versus ZT13) and surgery (parabiosis and SCN lesion). Outcomes were considered statistically significant at P < 0.05.
Results
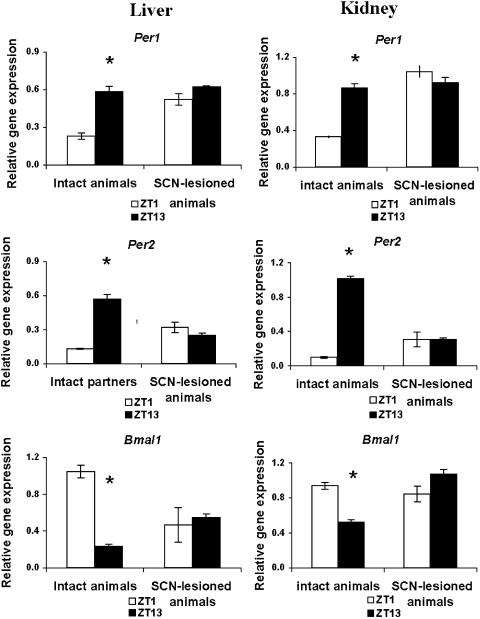
As expected, intact mice that were not parabiosed to other animals showed profound rhythms in relative mPer1, mPer2, and mBmal1 expression in both kidney and liver (Fig. 1). mPer expression was elevated at ZT13, and mBmal1 was elevated at ZT1. SCN lesions eliminated the ZT1 versus ZT13 differences in expression of all three genes (Fig. 1). Nonparabiosed SCN-lesioned mice showed tonically elevated mPer1 expression in both organs. In kidney, mPer2 expression was elevated at both ZT1 and ZT13 in SCN-lesioned animals, whereas in liver, mPer2 levels were intermediate between intact ZT1 and ZT13 values. In both organs, Bmal1 expression was arrhythmic at levels intermediate between the ZT1 and ZT13 values of intact mice. Parabiosis surgery did not, by itself, alter the rhythm of mPer1, mPer2, or mBmal1 expression in either liver or kidney: intact mice linked to intact partners routinely showed 3- to 5-fold differences between ZT1 and ZT13 relative gene expression, with normal phasing (Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 1.
SCN lesions eliminate circadian rhythms of peripheral gene expression in liver and kidney of control (nonparabiotic) mice. Mean (±SEM) expression of mPer1, mPer2, and mBmal1 (relative to GAPDH) in liver and kidney of intact or SCN-lesioned mice killed on the third day of constant darkness at ZT1 (white bars) or ZT13 (black bars) is shown. ZT1 values differed significantly from those at ZT13 in intact, but not in SCN-lesioned, mice (*, P < 0.05).
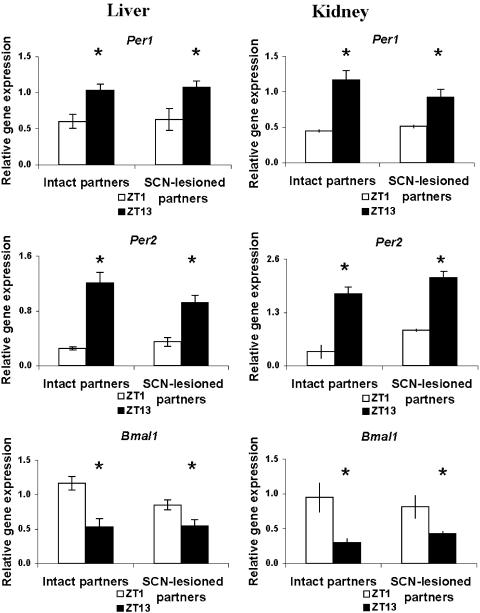
When SCN-lesioned mice were parabiotically linked to intact mice, the livers and kidneys of both partners showed ZT1 versus ZT13 differences in relative mPer1, mPer2, and mBmal1 expression (Fig. 2). The phase of expression was identical to that of intact animals, with mBmal1 expression peaking during subjective day and mPer expression peaking during subjective night. The amplitude of the circadian variation was most robust in the case of mPer2; while statistically significant and normally phased, the two-fold difference between ZT1 and ZT13 levels of relative mPer1 and Bmal1 expression in both intact and SCN-lesioned partners tended to be less than that of intact controls.
Fig. 2.
Parabiosis of SCN-lesioned mice to intact partners reinstates circadian rhythms of relative mPer1, mPer2, and mBmal1gene expression in liver and kidney. The nonlesioned (intact partners) and the brain damaged (SCN-lesioned partners) mice showed similar ZT1 versus ZT13 differences in mRNA values. Symbols are as in Fig. 1.
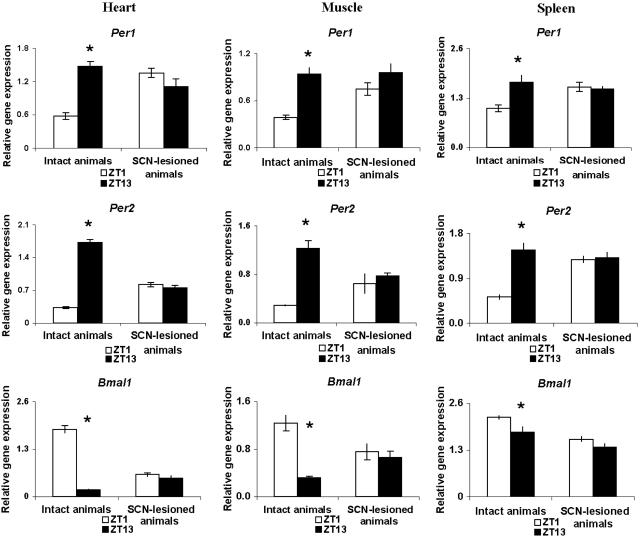
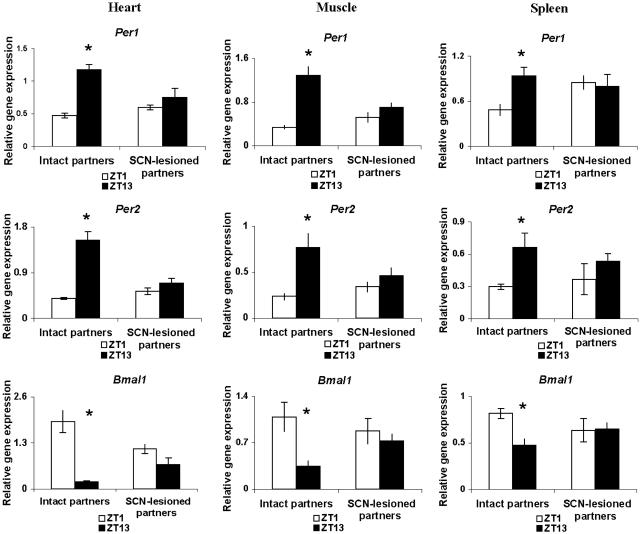
Circadian rhythms of mPer1, mPer2, and mBmal1 in the heart and skeletal muscle of intact mice resembled those in liver and kidney, whereas those in spleen tended to be of lower amplitude (Fig. 3). Parabiosis surgery, by itself, had little effect on circadian rhythms of gene expression in these organs (Fig. 6, which is published as supporting information on the PNAS web site). SCN lesions eliminated the circadian oscillations in the heart, skeletal muscle, and spleen of nonparabiosed mice (Fig. 3). Parabiosis to intact animals did not induce circadian rhythms of expression of any of the three clock genes studied in these organs of SCN-lesioned animals (Fig. 4), even though these rhythms had been reinstated in the kidney and liver of the same animals.
Fig. 3.
SCN lesions eliminate circadian rhythms of peripheral gene expression in heart, skeletal muscle, and spleen of control (nonparabiotic) mice. Symbols are as in Fig. 1.
Fig. 4.
Parabiosis of SCN-lesioned mice to intact partners fails to reinstate circadian rhythms of relative mPer1, mPer2, and mBmal1 gene expression in heart, spleen, and skeletal muscle. The nonlesioned animals (intact partners) show ZT1 and ZT13 patterns similar to those of mice in intact–intact pairs, whereas the brain-damaged mice (SCN-lesioned partners) showed no significant rhythms in mRNA values. Symbols are as in Fig. 1.
In agreement with some earlier studies (16–18), but at variance with others (19, 21, 22), we observed significant circadian variations in relative expression of mPer1, mPer2, and mBmal1 in the testis (Fig. 7, which is published as supporting information on the PNAS web site). As in hamsters (16), mRNA for all three clock genes rose at the same phase. The effects of SCN lesions on these rhythms were inconsistent between genes, however: differences between subjective day and subjective night were eliminated for mPer2 and mBmal1, but not for mPer1. Furthermore, parabiosis surgery eliminated these day-night differences in intact animals. Thus, we could not interpret the effects of parabiosis on rhythmicity of clock gene expression in the testis of SCN-lesioned mice.
Discussion
Our results indicate that neural input is not required for the persistence of circadian rhythms of mPer1, mPer2, or mBmal1 expression in liver and kidney. They do not establish the identity of signals that ensure rhythmicity in parabiotically paired animals. A strong possibility is the existence of a blood-borne cue, but it is not clear whether such a chemical signal is endocrine in nature. This hypothesis could be tested by examining the ability of brain-intact partners that have been subjected to endocrine extirpations (e.g., hypophysectomy) to restore peripheral circadian rhythms in parabiosed SCN-lesioned mice. Our results do not rule out the alternative possibility that blood-borne cues of non-endocrine origin, such as nutrients or metabolic products whose concentrations vary with circadian time, are responsible for the restoration of rhythmicity. For example, scheduled feeding can exert a striking effect on clock gene expression in mouse peripheral organs (6). Thus, it is possible that the rhythms we observed in liver and kidney of parabiotically linked, SCN-lesioned mice are driven by the meal pattern imposed by the brain-intact partner. The intact animal also may impose activity and body temperature fluctuations on the SCN-lesioned mouse that could substitute for an intact pacemaker, even in the absence of blood exchange.
In contrast to the liver and kidney, circadian rhythms of clock gene expression were not maintained in spleen, heart, or skeletal muscle of the same SCN-lesioned mice parabiosed to intact partners. This finding suggests that these organs differ from kidney and liver in their dependence on specific signals that are responsible for enforcing rhythmicity. Heart, muscle, and spleen may depend on different blood-borne signals that do not cross as readily between parabionts, or require higher concentrations of the same signals, than liver and kidney. Parabiotic pairs exchange 1–4% of their blood supply each minute, but only relatively stable molecules reach equilibrium. Until a specific signal is identified, it is not possible to estimate whether transfer is above or below a threshold value for particular organs. Another possibility is that heart, muscle, and spleen rely more heavily on neural signals than do liver and kidney. This result would resemble the differential dependence of endocrine versus behavioral rhythms on neural versus humoral outputs of the SCN (11, 20). Our results indicate that circadian rhythms of mPer1, mPer2, or Bmal1 expression in testis may be under still another means of central control.
It is also possible that parabiosis can reinstate circadian rhythms of clock gene expression in heart, muscle, and spleen of SCN-lesioned animals, but with an altered phase such that they were not detected with sampling at only ZT1 and ZT13. This possibility seems unlikely, in that robust differences between ZT1 and ZT13 values of mPer1, mPer2, and mBmal1 were apparent in these organs in intact mice. We note, however, that the amplitude of clock gene rhythms was somewhat lower in parabiotic pairs in which both partners were intact than in unoperated mice. This result may indicate that parabiosis blunts the amplitude or shifts the phase of some peripheral rhythms. Such effects may have masked the reestablishment of circadian rhythms in heart, spleen, and skeletal muscle. More frequent sampling of parabiotic pairs is required to address this question.
Parabiotically linked cockroaches sustained behavioral circadian rhythms in headless recipients, but it was argued that the rhythmic transfer of stress signals that do not normally provide internal entraining cues may have been responsible (13). The chronic and apparently stress-free nature of the parabiotic mouse preparation makes it unlikely that such artifacts can explain the effects reported here, and this would not explain the tissue-specific nature of the response in parabiosed lesioned mice. Further experimentation using the parabiotic preparation promises to elucidate the nature of the behavioral and chemical signals by which the SCN sustains circadian rhythms in specific peripheral organs.
Supplementary Material
Acknowledgments
We thank Karen Swiecanski for conscientious animal care. This work was supported by National Institute of Mental Health Grant R01 059166.
Abbreviations: SCN, suprachiasmatic nucleus of the hypothalamus; ZT, zeitgeber time.
References
- 1.Balsalobre, A., Damiola, F. & Schibler, U. (1998) Cell 93, 929-937. [DOI] [PubMed] [Google Scholar]
- 2.Pando, M. P., Morse, D., Cermakian, N. & Sassone-Corsi, P. (2002) Cell 110, 107-117. [DOI] [PubMed] [Google Scholar]
- 3.Balsalobre, A., Brown, S. A., Marcacci, L., Tronche, F., Kellendonk, C., Reichardt, H. M., Schutz, G. & Schibler, U. (2000) Science 289, 2344-2347. [DOI] [PubMed] [Google Scholar]
- 4.Nonaka, H., Emoto, N., Ikeda, K., Fukuya, H., Rohman, M. S., Raharjo, S. B., Yagita, K., Okamura, H. & Yokoyama, M. (2001) Circulation 104, 1746-1748. [DOI] [PubMed] [Google Scholar]
- 5.McNamara, P., Seo, S. P., Rudic, R. D., Sehgal, A., Chakravarti, D. & FitzGerald, G. A. (2001) Cell 105, 877-889. [DOI] [PubMed] [Google Scholar]
- 6.Le Minh, N., Damiola, F., Tronche, F., Schutz, G. & Schibler, U. (2001) EMBO J. 20, 7128-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueyama, T., Krout, K. E., Nguyen, X. V., Karpitskiy, V., Kollert, A., Mettenleiter, T. C. & Loewy, A. D. (1999) Nat. Neurosci. 2, 1051-1053. [DOI] [PubMed] [Google Scholar]
- 8.Bartness, T. J., Song, C. K. & Demas, G. E. (2001) J. Biol. Rhythms 16, 196-204. [DOI] [PubMed] [Google Scholar]
- 9.Buijs, R. M., la Fleur, S. E., Wortel, J., Van Heyningen, C., Zuiddam, L., Mettenleiter, T. C., Kalsbeek, A., Nagai, K. & Niijima, A. (2003) J. Comp. Neurol. 464, 36-48. [DOI] [PubMed] [Google Scholar]
- 10.Terazono, H., Mutoh, T., Yamaguchi, S., Kobayashi, M., Akiyama, M., Udo, R., Ohdo, S., Okamura, H. & Shibata, S. (2003) Proc. Natl. Acad. Sci. USA 100, 6795-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver, R., LeSauter, J., Tresco, P. A. & Lehman, M. N. (1996) Nature 382, 810-813. [DOI] [PubMed] [Google Scholar]
- 12.de la Iglesia, H. O., Meyer, J. & Schwartz, W. J. (2003) J. Neurosci. 23, 7412-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cymborowski, B. & Brady, J. (1972) Nat. New Biol. 236, 221-222. [DOI] [PubMed] [Google Scholar]
- 14.Finerty, J. (1952) Physiol. Rev. 32, 277-302. [DOI] [PubMed] [Google Scholar]
- 15.Harris, R. B. S., Zhou, J., Weigle, D. S. & Kuijper, J. L. (1997) Am. J. Physiol. 272, R1800-R1808. [DOI] [PubMed] [Google Scholar]
- 16.Tong, Y., Guo, H., Brewer, J. M., Lee, H., Lehman, M. N. & Bittman, E. L. (2004) J. Biol. Rhythms 19, 113-125. [DOI] [PubMed] [Google Scholar]
- 17.Zylka, M. J., Shearman, L. P., Weaver, D. R. & Reppert, S. M. (1998) Neuron 20, 1103-1110. [DOI] [PubMed] [Google Scholar]
- 18.Bittman, E. L., Doherty, L., Huang, L. & Paroskie, A. (2003) Am. J. Physiol. 285, R561-R569. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez, J. D., Chen, D., Storer, E. & Sehgal, A. (2003) Biol. Reprod. 69, 81-91. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Bernstein, E. L., Jetton, A. E., Matsumoto, S. I., Markuns, J. F., Lehman, M. N. & Bittman, E. L. (1999) Endocrinology 140, 207-218. [DOI] [PubMed] [Google Scholar]
- 21.Morse, D., Cermakian, N., Brancorsini, S., Parvinen, M. & Sassone-Corsi, P. (2003) Mol. Endocrinol. 17, 141-151. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto, Y. & Sancar, A. (1999) Brain Res. Mol. Brain Res. 71, 238-243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.