Abstract
Objective
To analyse the proportions of protein identity between Zika virus and dengue, Japanese encephalitis, yellow fever, West Nile and chikungunya viruses as well as polymorphism between different Zika virus strains.
Methods
We used published protein sequences for the Zika virus and obtained protein sequences for the other viruses from the National Center for Biotechnology Information (NCBI) protein database or the NCBI virus variation resource. We used BLASTP to find regions of identity between viruses. We quantified the identity between the Zika virus and each of the other viruses, as well as within-Zika virus polymorphism for all amino acid k-mers across the proteome, with k ranging from 6 to 100. We assessed accessibility of protein fragments by calculating the solvent accessible surface area for the envelope and nonstructural-1 (NS1) proteins.
Findings
In total, we identified 294 Zika virus protein fragments with both low proportion of identity with other viruses and low levels of polymorphisms among Zika virus strains. The list includes protein fragments from all Zika virus proteins, except NS3. NS4A has the highest number (190 k-mers) of protein fragments on the list.
Conclusion
We provide a candidate list of protein fragments that could be used when developing a sensitive and specific serological test to detect previous Zika virus infections.
Résumé
Objectif
Analyser les pourcentages de similarité protéique entre le virus Zika et les virus de la dengue, de l'encéphalite japonaise, de la fièvre jaune, du Nil occidental et du chikungunya, ainsi que le polymorphisme entre différentes souches du virus Zika.
Méthodes
Nous avons utilisé les séquences protéiques publiées du virus Zika et avons obtenu les séquences protéiques des autres virus dans la banque protéique du National Center for Biotechnology Information (NCBI) ou dans la base de données Virus Variation du NCBI. Nous avons utilisé BLASTP pour identifier les régions de similarité entre les virus. Nous avons quantifié la similarité entre le virus Zika et chacun des autres virus ainsi que le polymorphisme du virus Zika pour tous les k-mers d'acides aminés, dans tout le protéome, avec k allant de 6 à 100. Nous avons étudié l'accessibilité des fragments protéiques en calculant la surface accessible au solvant pour les protéines d'enveloppe et non structurale-1 (NS1).
Résultats
Au total, nous avons identifié 294 fragments protéiques du virus Zika qui présentent à la fois un faible degré de similarité avec les autres virus et un faible degré de polymorphisme entre les souches du virus Zika. Notre liste comprend des fragments protéiques issus de toutes les protéines du virus Zika, à l'exception de la protéine NS3. Le plus grand nombre de fragments protéiques de notre liste (190 k-mers) correspond à la protéine NS4A.
Conclusion
Nous proposons une liste de fragments protéiques candidats, qui pourraient être utilisés pour concevoir un test sérologique sensible et spécifique pour dépister les infections antérieures par le virus Zika.
Resumen
Objective
Analizar las proporciones de identidad proteica entre el virus de Zika y los virus del dengue, la encefalitis japonesa, la fiebre amarilla, el Nilo Occidental y el chikungunya, así como el polimorfismo entre las distintas cepas del virus de Zika.
Métodos
Se utilizaron secuencias de proteínas publicadas para el virus de Zika y secuencias de proteínas obtenidas para los otros virus de la base de datos de proteínas del Centro Nacional para la Información Biotecnológica (NCBI) o la fuente de información sobre la variación de virus del NCBI. Se utilizó el programa BLASTP para encontrar regiones de identidad entre los virus. Se cuantificó la identidad entre el virus de Zika y cada uno de los otros virus, así como el polimorfismo del virus de Zika para todos los k-mers de aminoácidos a través del proteoma, con una variación de k de 6 a 100. Se evaluó la accesibilidad de los fragmentos proteicos calculando la superficie accesible solvente para las proteínas de envoltura y no estructurales 1 (NS1).
Resultados
En total, se identificaron 294 fragmentos proteicos del virus de Zika con una proporción escasa de identidad con otros virus y con niveles bajos de polimorfismos entre las distintas cepas del virus de Zika. En la lista se incluyen fragmentos proteicos de todas las proteínas del virus de Zika, salvo la NS3. La NS4A cuenta con el mayor número (190 k-mers) de fragmentos proteicos de la lista.
Conclusión
Se proporcionó una lista de posibles fragmentos proteicos que podrían utilizarse para desarrollar una prueba serológica sensible y específica para detectar infecciones del virus de Zika anteriores.
ملخص
الغرض
تحليل مقدار تماثل البروتين بين فيروس زيكا وحمى الضنك، والتهاب الدماغ الياباني، وحمى الصفراء، وفيروس غرب النيل، وشيكونغونيا فضلاً عن تعددية الأشكال بين سلالات فيروس زيكا.
الطريقة
استخدمنا تسلسلات البروتين المنشورة لفيروس زيكا وحصلنا على تسلسلات البروتين لغيره من الفيروسات من قاعدة البيانات الخاصة بالبروتين للمركز الوطني لمعلومات التقانة الحيوية (NCBI) أو مورد اختلاف الفيروسات لمركز NCBI. كما استخدمنا أداة BLASTP للعثور على مناطق التماثل بين الفيروسات. وقمنا بإجراء تحديد كمي لمقدار التماثل بين فيروس زيكا وكلٍ من الفيروسات الأخرى فضلاً عن تعددية الأشكال في فيروس زيكا نفسه لكل ميرات k للأحماض الأمينية السائدة عبر البروتيوم، حيث تتراوح k من 6 إلى 100. وقمنا بتقييم إمكانية الوصول إلى شظايا البروتين من خلال حساب المساحة السطحية التي يمكن للمذيبات الوصول من خلالها للبروتينات المغلفة والبروتين اللابنيوي-1 (NS1).
النتائج
إجمالاً، قمنا بتحديد 294 شظية من شظايا البروتين الخاص بفيروس زيكا مع انخفاض نسبة التماثل مع الفيروسات الأخرى وانخفاض مستويات تعدد الأشكال بين سلالات فيروس زيكا. وتتضمن القائمة شظايا البروتين من جميع بروتينات فيروس زيكا باستثناء بروتين NS3. ويتمتع بروتين NS4A بالرقم الأكبر (190 من ميرات k ) من شظايا البروتين الواردة في القائمة.
الاستنتاج
قمنا بإنشاء قائمة مرشحين لشظايا البروتين والتي يمكن استخدامها عند تطوير اختبار مصلي حساس ومحدد لاكتشاف الحالات السابقة للإصابة بفيروس زيكا.
摘要
目的
旨在分析寨卡病毒、登革热、流行性乙型脑炎、黄热病、西尼罗河以及基孔肯雅热病毒之间的蛋白质识别率以及不同寨卡病毒株之间的多态性。
方法
我们使用已公布的寨卡病毒蛋白质序列,并从国家生物技术信息中心 (NCBI) 蛋白质数据库或国家生物技术信息中心 (NCBI) 病毒变异资源中获取了其他病毒的蛋白质序列。 我们使用 BLASTP 来找出病毒之间的识别区域。 我们量化了寨卡病毒和其他各种病毒之间的蛋白质识别以及寨卡病毒内部多态性,以识别蛋白质组中的所有氨基酸 k-mer,其中 k 的变化范围为 6 到 100。通过计算外膜蛋白和非结构蛋白 1 (NS1) 的溶剂可及表面,我们对蛋白质片段的可及性进行了评估。
结果
我们共识别出 294 个寨卡病毒蛋白质片段,相较于其他病毒,其识别率较低,且寨卡病毒株之间的多态性程度较低。 上述清单包括所有寨卡病毒蛋白质的蛋白质片段,非结构蛋白 3 (NS3) 除外。 清单中,非结构蛋白 4A (NS4A) 的蛋白质片段数目(190 个 k-mer)最高。
结论
我们提供了一份蛋白质片段补充目录,可在开发敏感的特殊血清学测试时使用,以检测之前的寨卡病毒感染情况。
Резюме
Цель
Проанализировать пропорции белковой идентичности между вирусом Зика и вирусами лихорадки денге, японского энцефалита, желтой лихорадки, лихорадки Западного Нила и лихорадки чикунгунья, а также полиморфизм между различными штаммами вируса Зика.
Методы
Мы использовали опубликованные последовательности белка для вируса Зика и получили последовательности белка для других вирусов из базы данных Национального центра биотехнологической информации (NCBI) или ресурса вирусных вариаций NCBI. Мы использовали программу BLASTP, чтобы найти области идентичности между вирусами. Мы провели количественную оценку идентичности между вирусом Зика и каждым из других вирусов, а также оценку полиморфизма между различными штаммами вируса Зика для всех k-меров аминокислот всего протеома, где k находится в пределах от 6 до 100. Мы оценили доступности фрагментов белка путем расчета доступной для растворителя области поверхности для белков оболочки и неструктурного белка-1 (NS1).
Результаты
В целом мы идентифицировали 294 фрагмента белка вируса Зика с низкой долей идентичности с другими вирусами и низкими уровнями полиморфизма среди штаммов вируса Зика. Этот список включает белковые фрагменты от всех белков вируса Зика, за исключением NS3. В этом списке NS4A имеет самое большое количество (190 k-меров) фрагментов белка.
Вывод
Мы подготовили список белковых фрагментов-кандидатов, которые можно использовать при разработке чувствительного и специфического серологического теста для выявления ранее обнаруженных инфекций, вызываемых вирусом Зика.
Introduction
Monitoring the geographic and the demographic distribution of people infected with Zika virus is important for informing decision-makers and researchers during the ongoing epidemic. Health officials also need further knowledge about the associations between Zika virus infection and its sequelae, such as microcephaly and Guillain–Barré syndrome. However, the absence of a sensitive and specific serological test for detecting prior Zika virus infection impedes research. According to the World Health Organization’s Target product profiles for better diagnostic tests for Zika virus infection,1 such a test must be able to differentiate between chikungunya, dengue and Zika viruses, since these mosquito-borne arboviruses can be co-circulating and can cause similar symptoms.2
Dengue and Zika viruses belong to the virus family Flaviviridae, while chikungunya virus belongs to the Togaviridae family. Although they belong to different virus families, Zika and chikungunya viruses share some similarities in envelope protein folding and membrane fusion mechanisms.3
Active Zika virus infections can be detected by nucleic acid-based diagnostic tools.4,5 However, developing serological diagnostic tests to detect previous Zika virus infections has been challenging, because of cross-reactivity between antibodies against different arboviruses.6–12 Hence, current serological assays, such as enzyme-linked immunosorbent assay (ELISA) and plaque reduction neutralization tests, may not be able to distinguish if a person has been infected with Zika virus or another flavivirus or if a person has received a previous yellow fever or Japanese encephalitis vaccination.13,14 A study has shown that neutralizing monoclonal antibodies generated against recombinant fragments of the envelope protein of dengue virus serotype 2 tend to be cross-reactive among flaviviruses, while nonneutralizing antibodies seem to be virus specific.15
We hypothesize that immunogenic protein regions with sequence dissimilarity may exist across arthropod-borne viruses (arboviruses) and that antibodies targeting these regions may be less likely to be cross-reactive. Identifying such regions could aid the development of specific microarray-based serological tests, such as a peptide microarray, to detect Zika virus and/or other related viruses. A peptide microarray is a high-throughput method for detecting interactions between peptides and antibodies and is composed of multiple spots of peptides on a solid surface.16 We also hypothesize that protein regions that are more conserved among different strains of the Zika virus are more likely to contribute to the sensitivity of the peptide microarray. Thus, to identify Zika virus conserved protein fragments that are variable among other virus species, we analysed proportions of protein sequence identity across virus species and protein polymorphism among different strains of Zika virus. We analysed the flaviviruses Zika, dengue, West Nile, Japanese encephalitis and yellow fever, and the alphavirus chikungunya.
Methods
We used publicly available proteomic sequencing data (Table 1). For the Zika virus, we used data set A from Faria et al.17 We downloaded the protein sequences of Japanese encephalitis virus, yellow fever virus and chikungunya virus from the National Center for Biotechnology Information (NCBI) protein database and the sequences for dengue virus serotypes 1–4 and West Nile virus from NCBI virus variation resource.18
Table 1. Proteomic sequencing data used to compare identity between viruses and within viruses.
| Species | Collection date | WHO Region | No. of samples |
|---|---|---|---|
| ZIKV | 1947–2015 | African, Americas, Western Pacific | 34 |
| DENV1 | 01/01/2010– 06/01/2016 | African, Americas, European, South-East Asia, Western Pacific | 171 |
| DENV2 | 01/01/2010– 06/01/2016 | Americas, Eastern Mediterranean, South-East Asia, Western Pacific | 158 |
| DENV3 | 01/01/2010– 06/01/2016 | Americas, Eastern Mediterranean, South-East Asia, Western Pacific | 62 |
| DENV4 | 01/01/2010– 06/01/2016 | Americas, South-East Asia, Western Pacific | 58 |
| WNV | 01/01/2008–06/01/2016 | Americas, European, South-East Asia, | 44 |
| JEV | 1951–2012 | South-East Asia, Western Pacific | 19 |
| YFV | 1981–2016 | African, Americas, Western Pacific | 31 |
| CHIKV | 1953-2015 | African, Americas, European, South-East Asia, Western Pacific | 212 |
CHIKV: chikungunya virus; DENV1−4: dengue virus serotype 1; JEV: Japanese encephalitis virus; WHO: World Health Organization; WNV: West Nile virus; YFV; yellow fever virus; ZIKV: Zika virus.
Note: For ZIKV, we used data set A from Faria et al.17 We downloaded the protein sequences of JEV, YFV and CHIKV from the National Center of Biotechnology Information (NCBI) protein database and sequences for DENV serotypes 1–4 and WNV from NCBI virus variation resource.18
We used BLASTP19 to find regions of identity between arboviruses, applying a default Expect (E)-value threshold of 10, that is the expected number of hits of the observed similarity, by chance, is fewer than 10. The results are robust and we obtained the same results when E-value thresholds were 5 or 50. When comparing the chikungunya and the Zika viruses, we used an E-value threshold of 1000, because chikungunya does not belong to the Flaviviridae family and we could not identify any regions of similarity when using an E-value threshold of 10. For all protein fragments across the proteome, we calculated the proportion of shared amino acids between virus species and polymorphism among different Zika virus strains. We analysed protein fragments of different lengths, so called k-mers (where k is the amino acid length of the protein fragment), with k equal to 6 or ranging from 10 to 100. We used a sliding window approach, where we moved the window one amino acid at a time along the proteome to include every possible k-mer. To be conservative, we identified protein fragment identity between species by the maximum identity among all the pairs of strains for each window considered. For analysing the identity with dengue virus, we used the highest identity between the Zika virus and all four serotypes of the dengue virus for each window considered. To assess if protein identity between the Zika virus and each of the dengue serotype was significantly associated with polymorphism within each dengue virus serotype, we calculated P-values by using Pearson's correlation test.
To identify polymorphisms within viruses, we used both the average pairwise difference and the proportion of polymorphic sites. Average pairwise difference is calculated by averaging the proportions of differences in peptide sequences from all pairs of the virus strains. We chose to plot the proportion of polymorphic sites in the figures because it is less sensitive to population structure and/or sampling bias.
To identify potential protein fragments that could be used for diagnostic tests, we selected k-mers with low proportion of identity between the Zika virus and other arboviruses as well as low polymorphism between different strains of Zika virus as lead candidate protein fragments. The rationale for this approach was that fragments with low between-species identity and low within-species polymorphism are most likely to have both the required specificity and sensitivity for such tests. We chose k-mers in the bottom quintile of values of identity and polymorphism for each k-mer length.
Insights into protein structures are critical for assessing the possible antigenicity of peptides, because buried peptides are less likely to be antigenic.20 To determine if any of the fragments are exposed or buried in the two Zika virus proteins with available protein structures, the envelope protein and the non-structural (NS) protein 1, we calculated the solvent accessible surface area for each amino acid. We used the published structures of dimeric NS1 (protein data bank identification, PDB ID: 5GS6)21 and the envelope protein in the biological assembly of the mature virus (PDB ID: 5IRE).22 To calculate the solvent accessible surface area, we used the linear combinations of pairwise overlaps method23 and used 10 Å2 as the upper limit for buried residues, as this value corresponds to half the surface area of a single water molecule. The regions at the C-terminal end of the dengue virus envelope protein interact with the viral lipid membrane24 and are unlikely to be exposed. Due to the high structural similarity of the envelope proteins between dengue and Zika viruses, we assume that the region from residue 404 to the C-terminus in Zika virus envelope protein is also buried. For the lead candidate list, we excluded the k-mers without any continuous exposed peptides longer than five amino acids in the two proteins, because exposed peptides are more likely to be antigenic. The threshold of five amino acids was chosen because 99.7% of experimentally determined antigenic B-cell epitopes for flaviviruses found in Virus Pathogen Database and Analysis Resource database are longer than five amino acids.25 We obtained the list of theses epitopes through the database’s web site at http://www.viprbrc.org/.
Results
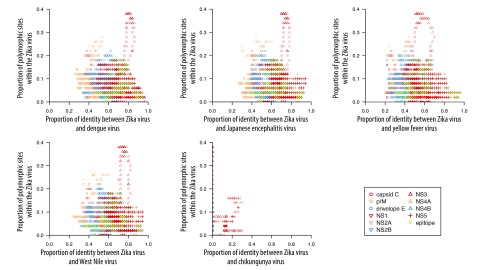
On average, Zika virus shares 55.6% amino acid sequence identity with dengue virus, 46.0% with yellow fever virus, 56.1% with Japanese encephalitis virus, 57.0% with West Nile virus and 1.3% with chikungunya virus. The identity between Zika virus and other viruses and Zika virus polymorphism for all k-mers are available from the corresponding author. As an example, Fig. 1 and Fig. 2 show the identity between Zika virus and other viruses investigated and polymorphisms within the Zika virus for all 50-mer peptides.
Fig. 1.
Zika virus polymorphism versus identity between Zika virus and other arboviruses, 50-mers across the Zika virus proteome
E: envelope; NS; non-structural; prM; precursor membrane.
Notes: 50-mers across the Zika virus proteome were analysed, using a sliding window approach. Fifty-mers containing known epitopes in non-Zika virus flaviviruses are shown in green. Polymorphic sites are the sites that vary among different strains of Zika virus.
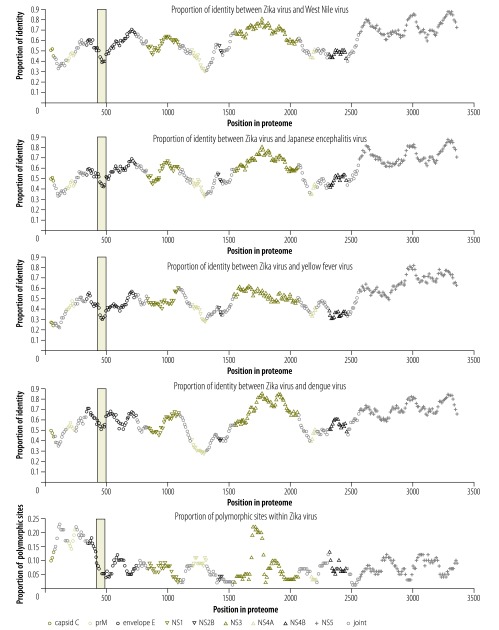
Fig. 2.
Sliding-window identity between Zika virus and other flaviviruses and within-Zika virus polymorphism
E: envelope; NS; non-structural; prM; precursor membrane.
Notes: The window size is 50 amino acids and step size is 5 amino acids. The light green shaded area (position 430–500) shows low between-species identity and low within-Zika virus polymorphism.
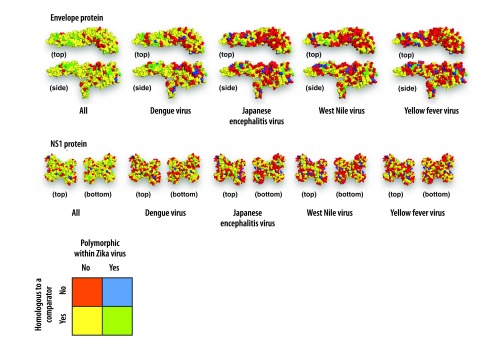
Fig. 3 shows protein fragments mapped to the corresponding envelope or NS1 proteins. The exposed areas of the proteins show regions with both low identity with other flaviviruses and low Zika virus polymorphism.
Fig. 3.
Mapping per-site identity and polymorphism onto the structures of Zika virus envelope protein and nonstructural protein 1 dimer
NS1: nonstructural protein 1.
Notes: Sites polymorphic within Zika virus or with high identity between Zika virus and other viruses could compromise respectively sensitivity and specificity of a serological test. Thus the most useful sites are expected to be those that lack identity with other viruses and lack polymorphism within Zika virus, shown in red. Identity between a comparator virus and Zika virus is shown in yellow; polymorphism within Zika virus is shown in blue; and sites with both properties are shown in green. The side view of the envelope protein shows the two transmembrane helices on the bottom. These helices likely remain buried within the lipid bilayer envelope and hence are unavailable for interactions with antibodies. In the structures including all viruses, sites are shown as homologous if they are homologous between Zika virus and any of the flaviviruses. Homologous regions are larger for dengue virus because sites are shown as homologous if they are homologous between Zika virus and any of the four dengue virus serotypes.
The lead candidate list for developing a specific and sensitive microarray-based serological test contains 294 protein fragments. These fragments have low similarity between viruses, low polymorphism within the Zika virus and continuous exposed peptides longer than five amino acids (Table 2; available at: http://www.who.int/bulletin/volumes/95/7/16-182105). The list excluded 10.9% (36/330) of k-mers containing previously identified B-cell epitopes for other flaviviruses than Zika, because they are likely to be cross-reactive. Protein fragments from all Zika virus proteins, except NS3, are present in the list. NS4A has the highest number (190 k-mers) of candidate protein fragments (Table 3).
Table 2. The lead candidate list of Zika virus protein fragments with low proportion of identity with other flaviviruses and low polymorphism.
| Position in proteome, aa |
Protein | Average pairwise difference | Polymorphic sites, % | k-mera | Homology with other flaviviruses, % |
Peptide sequence | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | DENV | JEV | YFV | WNV | |||||
| 26 | 95 | capsid C | 0.0078 | 0.0429 | 70 | 0.4857 | 0.4857 | 0.2286 | 0.5000 | PFGGLKRLPAGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRII |
| 28 | 97 | capsid C | 0.0019 | 0.0286 | 70 | 0.5000 | 0.4714 | 0.2286 | 0.5000 | GGLKRLPAGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINA |
| 32 | 101 | capsid C | 0.0077 | 0.0429 | 70 | 0.5000 | 0.4714 | 0.2571 | 0.4857 | RLPAGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARKEK |
| 33 | 102 | capsid C | 0.0077 | 0.0429 | 70 | 0.4857 | 0.4714 | 0.2571 | 0.4857 | LPAGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARKEKK |
| 34 | 103 | capsid C | 0.0077 | 0.0429 | 70 | 0.4857 | 0.4857 | 0.2571 | 0.5000 | PAGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARKEKKR |
| 35 | 104 | capsid C | 0.0077 | 0.0429 | 70 | 0.4857 | 0.4857 | 0.2571 | 0.5000 | AGLLLGHGPIRMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARKEKKRR |
| 45 | 94 | capsid C | 0.0027 | 0.0400 | 50 | 0.4800 | 0.4600 | 0.2600 | 0.4800 | RMVLAILAFLRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRI |
| 54 | 93 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2500 | 0.4250 | LRFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLR |
| 55 | 94 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2500 | 0.4250 | RFTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRI |
| 56 | 95 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2500 | 0.4250 | FTAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRII |
| 57 | 96 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2250 | 0.4250 | TAIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIIN |
| 58 | 97 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.3750 | 0.2000 | 0.4000 | AIKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINA |
| 59 | 98 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.3750 | 0.2250 | 0.4000 | IKPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINAR |
| 60 | 99 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2500 | 0.3750 | KPSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARK |
| 61 | 100 | capsid C | 0.0033 | 0.0500 | 40 | 0.4750 | 0.4000 | 0.2250 | 0.3750 | PSLGLINRWGSVGKKEAMEIIKKFKKDLAAMLRIINARKE |
| 87 | 92 | capsid C | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | DLAAML |
| 131 | 136 | pr | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.0000 | 0.1667 | AYYMYL |
| 132 | 137 | pr | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.0000 | 0.1667 | YYMYLD |
| 133 | 138 | pr | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.0000 | 0.1667 | YMYLDR |
| 231 | 240 | membrane | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | SQTWLESREY |
| 411 | 450 | envelope | 0.0044 | 0.0750 | 40 | 0.4500 | 0.4250 | 0.2250 | 0.3750 | CSKKMTGKSIQPENLEYRIMLSVHGSQHSGMIGHETDENR |
| 412 | 451 | envelope | 0.0044 | 0.0750 | 40 | 0.4250 | 0.4250 | 0.2000 | 0.3500 | SKKMTGKSIQPENLEYRIMLSVHGSQHSGMIGHETDENRA |
| 413 | 452 | envelope | 0.0044 | 0.0750 | 40 | 0.4250 | 0.4250 | 0.2250 | 0.3750 | KKMTGKSIQPENLEYRIMLSVHGSQHSGMIGHETDENRAK |
| 414 | 453 | envelope | 0.0044 | 0.0750 | 40 | 0.4250 | 0.4250 | 0.2000 | 0.3750 | KMTGKSIQPENLEYRIMLSVHGSQHSGMIGHETDENRAKV |
| 415 | 454 | envelope | 0.0044 | 0.0750 | 40 | 0.4500 | 0.4000 | 0.2000 | 0.3500 | MTGKSIQPENLEYRIMLSVHGSQHSGMIGHETDENRAKVE |
| 419 | 448 | envelope | 0.0020 | 0.0333 | 30 | 0.4667 | 0.4000 | 0.2000 | 0.3667 | SIQPENLEYRIMLSVHGSQHSGMIGHETDE |
| 420 | 449 | envelope | 0.0020 | 0.0333 | 30 | 0.4667 | 0.4000 | 0.2000 | 0.3667 | IQPENLEYRIMLSVHGSQHSGMIGHETDEN |
| 421 | 450 | envelope | 0.0020 | 0.0333 | 30 | 0.4667 | 0.3667 | 0.2000 | 0.3333 | QPENLEYRIMLSVHGSQHSGMIGHETDENR |
| 422 | 451 | envelope | 0.0020 | 0.0333 | 30 | 0.4667 | 0.3667 | 0.2000 | 0.3000 | PENLEYRIMLSVHGSQHSGMIGHETDENRA |
| 436 | 445 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | SQHSGMIGHE |
| 438 | 447 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | HSGMIGHETD |
| 439 | 448 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | SGMIGHETDE |
| 440 | 449 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.2000 | 0.2000 | 0.2000 | GMIGHETDEN |
| 441 | 450 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.2000 | 0.2000 | 0.2000 | MIGHETDENR |
| 442 | 451 | envelope | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.2000 | IGHETDENRA |
| 626 | 665 | envelope | 0.0029 | 0.0500 | 40 | 0.4000 | 0.4750 | 0.3000 | 0.4750 | KVPAQMAVDMQTLTPVGRLITANPVITESTENSKMMLELD |
| 627 | 666 | envelope | 0.0029 | 0.0500 | 40 | 0.4000 | 0.4750 | 0.3000 | 0.4750 | VPAQMAVDMQTLTPVGRLITANPVITESTENSKMMLELDP |
| 629 | 658 | envelope | 0.0020 | 0.0333 | 30 | 0.4333 | 0.4667 | 0.3000 | 0.4333 | AQMAVDMQTLTPVGRLITANPVITESTENS |
| 913 | 918 | NS1 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.0000 | 0.0000 | 0.0000 | FVRAAK |
| 913 | 922 | NS1 | 0.0000 | 0.0000 | 10 | 0.4000 | 0.1000 | 0.2000 | 0.2000 | FVRAAKTNNS |
| 914 | 919 | NS1 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.0000 | 0.0000 | 0.0000 | VRAAKT |
| 915 | 920 | NS1 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | RAAKTN |
| 1294 | 1343 | NS2A | 0.0043 | 0.0600 | 50 | 0.3600 | 0.3600 | 0.3000 | 0.3200 | LAILAALTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPFVMAL |
| 1295 | 1344 | NS2A | 0.0043 | 0.0600 | 50 | 0.3800 | 0.3600 | 0.2800 | 0.3200 | AILAALTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPFVMALG |
| 1299 | 1318 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.3500 | 0.2500 | 0.3500 | ALTPLARGTLLVAWRAGLAT |
| 1299 | 1338 | NS2A | 0.0018 | 0.0250 | 40 | 0.3500 | 0.3250 | 0.2500 | 0.3250 | ALTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLP |
| 1299 | 1348 | NS2A | 0.0043 | 0.0600 | 50 | 0.3600 | 0.3400 | 0.3000 | 0.3000 | ALTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPFVMALGLTAV |
| 1300 | 1319 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.3500 | 0.3000 | 0.3500 | LTPLARGTLLVAWRAGLATC |
| 1300 | 1339 | NS2A | 0.0018 | 0.0250 | 40 | 0.3500 | 0.3250 | 0.2500 | 0.3250 | LTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPF |
| 1300 | 1349 | NS2A | 0.0043 | 0.0600 | 50 | 0.3600 | 0.3400 | 0.3000 | 0.3000 | LTPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPFVMALGLTAVR |
| 1301 | 1320 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.3500 | 0.2500 | 0.3500 | TPLARGTLLVAWRAGLATCG |
| 1301 | 1340 | NS2A | 0.0018 | 0.0250 | 40 | 0.3750 | 0.3000 | 0.2500 | 0.3000 | TPLARGTLLVAWRAGLATCGGFMLLSLKGKGSVKKNLPFV |
| 1302 | 1311 | NS2A | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | PLARGTLLVA |
| 1302 | 1321 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.3000 | 0.2000 | 0.3000 | PLARGTLLVAWRAGLATCGG |
| 1303 | 1322 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.2500 | 0.1500 | 0.2500 | LARGTLLVAWRAGLATCGGF |
| 1304 | 1323 | NS2A | 0.0000 | 0.0000 | 20 | 0.3500 | 0.2500 | 0.1500 | 0.2500 | ARGTLLVAWRAGLATCGGFM |
| 1305 | 1324 | NS2A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3000 | 0.1500 | 0.3000 | RGTLLVAWRAGLATCGGFML |
| 1309 | 1318 | NS2A | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | LVAWRAGLAT |
| 1310 | 1319 | NS2A | 0.0000 | 0.0000 | 10 | 0.4000 | 0.2000 | 0.2000 | 0.2000 | VAWRAGLATC |
| 1311 | 1320 | NS2A | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | AWRAGLATCG |
| 1312 | 1321 | NS2A | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | WRAGLATCGG |
| 1313 | 1322 | NS2A | 0.0000 | 0.0000 | 10 | 0.2000 | 0.3000 | 0.2000 | 0.3000 | RAGLATCGGF |
| 1314 | 1323 | NS2A | 0.0000 | 0.0000 | 10 | 0.1000 | 0.2000 | 0.2000 | 0.2000 | AGLATCGGFM |
| 1315 | 1324 | NS2A | 0.0000 | 0.0000 | 10 | 0.2000 | 0.3000 | 0.2000 | 0.3000 | GLATCGGFML |
| 1317 | 1322 | NS2A | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.1667 | 0.1667 | ATCGGF |
| 1318 | 1323 | NS2A | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.1667 | 0.1667 | TCGGFM |
| 1326 | 1331 | NS2A | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.1667 | 0.1667 | SLKGKG |
| 1328 | 1333 | NS2A | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.1667 | 0.1667 | KGKGSV |
| 1328 | 1337 | NS2A | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | KGKGSVKKNL |
| 1354 | 1363 | NS2A | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | INVVGLLLLT |
| 1459 | 1468 | NS2B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | GPPMREIILK |
| 1460 | 1469 | NS2B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.2000 | 0.2000 | 0.2000 | PPMREIILKV |
| 1461 | 1470 | NS2B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.2000 | 0.2000 | 0.3000 | PMREIILKVV |
| 1462 | 1467 | NS2B | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | MREIIL |
| 1462 | 1471 | NS2B | 0.0000 | 0.0000 | 10 | 0.4000 | 0.1000 | 0.1000 | 0.2000 | MREIILKVVL |
| 1463 | 1472 | NS2B | 0.0000 | 0.0000 | 10 | 0.4000 | 0.1000 | 0.1000 | 0.2000 | REIILKVVLM |
| 1474 | 1493 | NS2B | 0.0000 | 0.0000 | 20 | 0.4000 | 0.2500 | 0.2000 | 0.3500 | ICGMNPIAIPFAAGAWYVYV |
| 1475 | 1494 | NS2B | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3000 | 0.2500 | 0.3000 | CGMNPIAIPFAAGAWYVYVK |
| 1476 | 1495 | NS2B | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3500 | 0.2500 | 0.3000 | GMNPIAIPFAAGAWYVYVKT |
| 1477 | 1496 | NS2B | 0.0000 | 0.0000 | 20 | 0.3500 | 0.3500 | 0.2500 | 0.3000 | MNPIAIPFAAGAWYVYVKTG |
| 1478 | 1497 | NS2B | 0.0000 | 0.0000 | 20 | 0.3500 | 0.4000 | 0.2500 | 0.3500 | NPIAIPFAAGAWYVYVKTGK |
| 1483 | 1492 | NS2B | 0.0000 | 0.0000 | 10 | 0.4000 | 0.2000 | 0.2000 | 0.3000 | PFAAGAWYVY |
| 1484 | 1493 | NS2B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.2000 | 0.2000 | 0.2000 | FAAGAWYVYV |
| 2116 | 2215 | NS4A | 0.0091 | 0.0600 | 100 | 0.3900 | 0.3500 | 0.3500 | 0.4400 | GAAFGVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEI |
| 2117 | 2216 | NS4A | 0.0091 | 0.0600 | 100 | 0.3900 | 0.3500 | 0.3400 | 0.4400 | AAFGVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIE |
| 2118 | 2217 | NS4A | 0.0084 | 0.0500 | 100 | 0.4000 | 0.3500 | 0.3400 | 0.4400 | AFGVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEP |
| 2119 | 2218 | NS4A | 0.0084 | 0.0500 | 100 | 0.4000 | 0.3400 | 0.3300 | 0.4400 | FGVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPA |
| 2120 | 2179 | NS4A | 0.0057 | 0.0500 | 60 | 0.4333 | 0.3333 | 0.3500 | 0.3833 | GVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTV |
| 2120 | 2189 | NS4A | 0.0049 | 0.0429 | 70 | 0.3857 | 0.3286 | 0.3286 | 0.4286 | GVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMR |
| 2120 | 2199 | NS4A | 0.0052 | 0.0500 | 80 | 0.4125 | 0.3875 | 0.3375 | 0.4625 | GVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFG |
| 2120 | 2209 | NS4A | 0.0046 | 0.0444 | 90 | 0.3889 | 0.3556 | 0.3333 | 0.4667 | GVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWL |
| 2120 | 2219 | NS4A | 0.0041 | 0.0400 | 100 | 0.3900 | 0.3400 | 0.3300 | 0.4400 | GVMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPAR |
| 2121 | 2180 | NS4A | 0.0057 | 0.0500 | 60 | 0.4333 | 0.3333 | 0.3500 | 0.3833 | VMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVS |
| 2121 | 2190 | NS4A | 0.0049 | 0.0429 | 70 | 0.3857 | 0.3286 | 0.3286 | 0.4143 | VMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRN |
| 2121 | 2200 | NS4A | 0.0052 | 0.0500 | 80 | 0.4125 | 0.3875 | 0.3500 | 0.4500 | VMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGM |
| 2121 | 2210 | NS4A | 0.0046 | 0.0444 | 90 | 0.3889 | 0.3556 | 0.3444 | 0.4556 | VMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLM |
| 2121 | 2220 | NS4A | 0.0041 | 0.0400 | 100 | 0.4000 | 0.3500 | 0.3400 | 0.4400 | VMEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARI |
| 2122 | 2181 | NS4A | 0.0057 | 0.0500 | 60 | 0.4333 | 0.3333 | 0.3500 | 0.4000 | MEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSL |
| 2122 | 2191 | NS4A | 0.0049 | 0.0429 | 70 | 0.4000 | 0.3429 | 0.3429 | 0.4286 | MEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNK |
| 2122 | 2201 | NS4A | 0.0052 | 0.0500 | 80 | 0.4125 | 0.3875 | 0.3500 | 0.4625 | MEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMV |
| 2122 | 2211 | NS4A | 0.0046 | 0.0444 | 90 | 0.4000 | 0.3667 | 0.3444 | 0.4667 | MEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMW |
| 2122 | 2221 | NS4A | 0.0041 | 0.0400 | 100 | 0.4100 | 0.3600 | 0.3400 | 0.4500 | MEALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIA |
| 2123 | 2182 | NS4A | 0.0057 | 0.0500 | 60 | 0.4500 | 0.3500 | 0.3667 | 0.4167 | EALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLG |
| 2123 | 2192 | NS4A | 0.0049 | 0.0429 | 70 | 0.4143 | 0.3571 | 0.3571 | 0.4429 | EALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKG |
| 2123 | 2202 | NS4A | 0.0052 | 0.0500 | 80 | 0.4250 | 0.3875 | 0.3625 | 0.4750 | EALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVT |
| 2123 | 2212 | NS4A | 0.0046 | 0.0444 | 90 | 0.4000 | 0.3667 | 0.3556 | 0.4667 | EALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWL |
| 2123 | 2222 | NS4A | 0.0041 | 0.0400 | 100 | 0.4100 | 0.3600 | 0.3400 | 0.4500 | EALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIAC |
| 2124 | 2183 | NS4A | 0.0024 | 0.0333 | 60 | 0.4500 | 0.3333 | 0.3667 | 0.4000 | ALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGI |
| 2124 | 2193 | NS4A | 0.0020 | 0.0286 | 70 | 0.4286 | 0.3571 | 0.3714 | 0.4429 | ALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGI |
| 2124 | 2203 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3875 | 0.3625 | 0.4750 | ALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTL |
| 2124 | 2213 | NS4A | 0.0024 | 0.0333 | 90 | 0.4000 | 0.3556 | 0.3556 | 0.4556 | ALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLS |
| 2124 | 2223 | NS4A | 0.0021 | 0.0300 | 100 | 0.4100 | 0.3500 | 0.3400 | 0.4400 | ALGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACV |
| 2125 | 2184 | NS4A | 0.0024 | 0.0333 | 60 | 0.4500 | 0.3500 | 0.3667 | 0.4167 | LGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIF |
| 2125 | 2194 | NS4A | 0.0020 | 0.0286 | 70 | 0.4429 | 0.3714 | 0.3714 | 0.4571 | LGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIG |
| 2125 | 2204 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3875 | 0.3625 | 0.4875 | LGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLG |
| 2125 | 2214 | NS4A | 0.0024 | 0.0333 | 90 | 0.4111 | 0.3667 | 0.3556 | 0.4667 | LGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSE |
| 2125 | 2224 | NS4A | 0.0021 | 0.0300 | 100 | 0.4100 | 0.3600 | 0.3400 | 0.4500 | LGTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVL |
| 2126 | 2185 | NS4A | 0.0024 | 0.0333 | 60 | 0.4500 | 0.3333 | 0.3500 | 0.4167 | GTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFF |
| 2126 | 2195 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3714 | 0.3571 | 0.4571 | GTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGK |
| 2126 | 2205 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3750 | 0.3500 | 0.4875 | GTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGA |
| 2126 | 2215 | NS4A | 0.0024 | 0.0333 | 90 | 0.4222 | 0.3556 | 0.3444 | 0.4556 | GTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEI |
| 2126 | 2225 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3500 | 0.3300 | 0.4400 | GTLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLI |
| 2127 | 2186 | NS4A | 0.0024 | 0.0333 | 60 | 0.4333 | 0.3167 | 0.3500 | 0.4000 | TLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFV |
| 2127 | 2196 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3714 | 0.3714 | 0.4429 | TLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKM |
| 2127 | 2206 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3625 | 0.3500 | 0.4750 | TLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGAS |
| 2127 | 2216 | NS4A | 0.0024 | 0.0333 | 90 | 0.4222 | 0.3444 | 0.3444 | 0.4444 | TLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIE |
| 2127 | 2226 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3500 | 0.3300 | 0.4300 | TLPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIV |
| 2128 | 2187 | NS4A | 0.0024 | 0.0333 | 60 | 0.4333 | 0.3167 | 0.3500 | 0.4167 | LPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVL |
| 2128 | 2197 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3857 | 0.3714 | 0.4571 | LPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMG |
| 2128 | 2207 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3625 | 0.3500 | 0.4750 | LPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASA |
| 2128 | 2217 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3556 | 0.4444 | LPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEP |
| 2128 | 2227 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3500 | 0.3300 | 0.4300 | LPGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2129 | 2188 | NS4A | 0.0024 | 0.0333 | 60 | 0.4333 | 0.3333 | 0.3500 | 0.4333 | PGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLM |
| 2129 | 2198 | NS4A | 0.0020 | 0.0286 | 70 | 0.4429 | 0.3857 | 0.3571 | 0.4571 | PGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGF |
| 2129 | 2208 | NS4A | 0.0027 | 0.0375 | 80 | 0.4125 | 0.3625 | 0.3375 | 0.4750 | PGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAW |
| 2129 | 2218 | NS4A | 0.0024 | 0.0333 | 90 | 0.4222 | 0.3444 | 0.3444 | 0.4444 | PGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPA |
| 2129 | 2228 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3500 | 0.3300 | 0.4300 | PGHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2130 | 2179 | NS4A | 0.0029 | 0.0400 | 50 | 0.4600 | 0.3200 | 0.3600 | 0.3600 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTV |
| 2130 | 2189 | NS4A | 0.0024 | 0.0333 | 60 | 0.4167 | 0.3167 | 0.3333 | 0.4167 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMR |
| 2130 | 2199 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3857 | 0.3429 | 0.4571 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFG |
| 2130 | 2209 | NS4A | 0.0027 | 0.0375 | 80 | 0.4125 | 0.3500 | 0.3375 | 0.4625 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWL |
| 2130 | 2219 | NS4A | 0.0024 | 0.0333 | 90 | 0.4111 | 0.3333 | 0.3333 | 0.4333 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPAR |
| 2130 | 2229 | NS4A | 0.0021 | 0.0300 | 100 | 0.4100 | 0.3500 | 0.3200 | 0.4300 | GHMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2131 | 2180 | NS4A | 0.0029 | 0.0400 | 50 | 0.4600 | 0.3200 | 0.3600 | 0.3800 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVS |
| 2131 | 2190 | NS4A | 0.0024 | 0.0333 | 60 | 0.4167 | 0.3167 | 0.3333 | 0.4167 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRN |
| 2131 | 2200 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3857 | 0.3571 | 0.4571 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGM |
| 2131 | 2210 | NS4A | 0.0027 | 0.0375 | 80 | 0.4125 | 0.3500 | 0.3500 | 0.4625 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLM |
| 2131 | 2220 | NS4A | 0.0024 | 0.0333 | 90 | 0.4222 | 0.3444 | 0.3444 | 0.4444 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARI |
| 2131 | 2230 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3600 | 0.3300 | 0.4400 | HMTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLL |
| 2132 | 2181 | NS4A | 0.0029 | 0.0400 | 50 | 0.4400 | 0.3000 | 0.3600 | 0.3800 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSL |
| 2132 | 2191 | NS4A | 0.0024 | 0.0333 | 60 | 0.4333 | 0.3167 | 0.3500 | 0.4167 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNK |
| 2132 | 2201 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3714 | 0.3571 | 0.4571 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMV |
| 2132 | 2211 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3500 | 0.3500 | 0.4625 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMW |
| 2132 | 2221 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3444 | 0.4444 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIA |
| 2132 | 2231 | NS4A | 0.0021 | 0.0300 | 100 | 0.4200 | 0.3500 | 0.3300 | 0.4300 | MTERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2133 | 2192 | NS4A | 0.0024 | 0.0333 | 60 | 0.4500 | 0.3333 | 0.3667 | 0.4333 | TERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKG |
| 2133 | 2202 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3714 | 0.3714 | 0.4714 | TERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVT |
| 2133 | 2212 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3500 | 0.3625 | 0.4625 | TERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWL |
| 2133 | 2222 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3444 | 0.4444 | TERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIAC |
| 2133 | 2232 | NS4A | 0.0021 | 0.0300 | 100 | 0.4300 | 0.3600 | 0.3400 | 0.4300 | TERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2134 | 2203 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3857 | 0.3714 | 0.4857 | ERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTL |
| 2134 | 2213 | NS4A | 0.0027 | 0.0375 | 80 | 0.4250 | 0.3500 | 0.3625 | 0.4625 | ERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLS |
| 2134 | 2223 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3444 | 0.4444 | ERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACV |
| 2134 | 2233 | NS4A | 0.0021 | 0.0300 | 100 | 0.4300 | 0.3700 | 0.3500 | 0.4400 | ERFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2135 | 2204 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3714 | 0.3714 | 0.5000 | RFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLG |
| 2135 | 2214 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3500 | 0.3625 | 0.4750 | RFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSE |
| 2135 | 2224 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3444 | 0.4556 | RFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVL |
| 2135 | 2234 | NS4A | 0.0021 | 0.0300 | 100 | 0.4400 | 0.3700 | 0.3500 | 0.4500 | RFQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVL |
| 2136 | 2215 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3500 | 0.3625 | 0.4750 | FQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEI |
| 2136 | 2225 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3444 | 0.4556 | FQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLI |
| 2136 | 2235 | NS4A | 0.0021 | 0.0300 | 100 | 0.4400 | 0.3800 | 0.3500 | 0.4600 | FQEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLI |
| 2137 | 2216 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3500 | 0.3625 | 0.4750 | QEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIE |
| 2137 | 2226 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3556 | 0.3444 | 0.4556 | QEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIV |
| 2137 | 2236 | NS4A | 0.0021 | 0.0300 | 100 | 0.4500 | 0.3900 | 0.3600 | 0.4700 | QEAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIP |
| 2138 | 2217 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3500 | 0.3750 | 0.4750 | EAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEP |
| 2138 | 2227 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3556 | 0.3444 | 0.4556 | EAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2138 | 2237 | NS4A | 0.0021 | 0.0300 | 100 | 0.4600 | 0.4000 | 0.3700 | 0.4800 | EAIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPE |
| 2139 | 2208 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3571 | 0.3571 | 0.5000 | AIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAW |
| 2139 | 2218 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3375 | 0.3625 | 0.4625 | AIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPA |
| 2139 | 2228 | NS4A | 0.0024 | 0.0333 | 90 | 0.4444 | 0.3444 | 0.3444 | 0.4444 | AIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2139 | 2238 | NS4A | 0.0021 | 0.0300 | 100 | 0.4700 | 0.4000 | 0.3700 | 0.4800 | AIDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEP |
| 2140 | 2189 | NS4A | 0.0029 | 0.0400 | 50 | 0.4600 | 0.3000 | 0.3600 | 0.4400 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMR |
| 2140 | 2199 | NS4A | 0.0036 | 0.0500 | 60 | 0.4833 | 0.3833 | 0.3667 | 0.4833 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFG |
| 2140 | 2209 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3429 | 0.3571 | 0.4857 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWL |
| 2140 | 2219 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3250 | 0.3500 | 0.4500 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPAR |
| 2140 | 2229 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3333 | 0.4444 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2140 | 2239 | NS4A | 0.0021 | 0.0300 | 100 | 0.4700 | 0.4000 | 0.3600 | 0.4800 | IDNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPE |
| 2141 | 2190 | NS4A | 0.0029 | 0.0400 | 50 | 0.4600 | 0.3000 | 0.3600 | 0.4400 | DNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRN |
| 2141 | 2210 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3429 | 0.3714 | 0.4857 | DNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLM |
| 2141 | 2220 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3375 | 0.3625 | 0.4625 | DNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARI |
| 2141 | 2230 | NS4A | 0.0024 | 0.0333 | 90 | 0.4444 | 0.3556 | 0.3444 | 0.4556 | DNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLL |
| 2141 | 2240 | NS4A | 0.0021 | 0.0300 | 100 | 0.4800 | 0.4100 | 0.3600 | 0.4900 | DNLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPEK |
| 2142 | 2191 | NS4A | 0.0029 | 0.0400 | 50 | 0.4600 | 0.3000 | 0.3600 | 0.4400 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNK |
| 2142 | 2201 | NS4A | 0.0036 | 0.0500 | 60 | 0.4667 | 0.3667 | 0.3667 | 0.4833 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMV |
| 2142 | 2211 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3429 | 0.3571 | 0.4857 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMW |
| 2142 | 2221 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3375 | 0.3500 | 0.4625 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIA |
| 2142 | 2231 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3444 | 0.3333 | 0.4444 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2142 | 2241 | NS4A | 0.0021 | 0.0300 | 100 | 0.4800 | 0.4100 | 0.3600 | 0.4900 | NLAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPEKQ |
| 2143 | 2212 | NS4A | 0.0031 | 0.0429 | 70 | 0.4286 | 0.3429 | 0.3714 | 0.4857 | LAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWL |
| 2143 | 2222 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3375 | 0.3500 | 0.4625 | LAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIAC |
| 2143 | 2232 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3556 | 0.3444 | 0.4444 | LAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2143 | 2242 | NS4A | 0.0021 | 0.0300 | 100 | 0.4800 | 0.4200 | 0.3700 | 0.5000 | LAVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPEKQR |
| 2144 | 2213 | NS4A | 0.0031 | 0.0429 | 70 | 0.4286 | 0.3429 | 0.3714 | 0.4857 | AVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLS |
| 2144 | 2223 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3375 | 0.3500 | 0.4625 | AVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACV |
| 2144 | 2233 | NS4A | 0.0024 | 0.0333 | 90 | 0.4333 | 0.3667 | 0.3556 | 0.4556 | AVLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2145 | 2214 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3571 | 0.3714 | 0.5000 | VLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSE |
| 2145 | 2224 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3500 | 0.3500 | 0.4750 | VLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVL |
| 2145 | 2234 | NS4A | 0.0024 | 0.0333 | 90 | 0.4444 | 0.3778 | 0.3556 | 0.4667 | VLMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVL |
| 2146 | 2215 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3571 | 0.3571 | 0.4857 | LMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEI |
| 2146 | 2225 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3500 | 0.3375 | 0.4625 | LMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLI |
| 2146 | 2235 | NS4A | 0.0024 | 0.0333 | 90 | 0.4556 | 0.3889 | 0.3444 | 0.4667 | LMRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLI |
| 2147 | 2216 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3571 | 0.3429 | 0.4857 | MRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIE |
| 2147 | 2226 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3625 | 0.3250 | 0.4625 | MRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIV |
| 2147 | 2236 | NS4A | 0.0024 | 0.0333 | 90 | 0.4556 | 0.4000 | 0.3556 | 0.4778 | MRAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIP |
| 2148 | 2217 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3571 | 0.3571 | 0.4857 | RAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEP |
| 2148 | 2227 | NS4A | 0.0027 | 0.0375 | 80 | 0.4375 | 0.3625 | 0.3250 | 0.4625 | RAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2148 | 2237 | NS4A | 0.0024 | 0.0333 | 90 | 0.4667 | 0.4111 | 0.3667 | 0.4889 | RAETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPE |
| 2149 | 2218 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3571 | 0.3571 | 0.4857 | AETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPA |
| 2149 | 2228 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3625 | 0.3375 | 0.4625 | AETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2149 | 2238 | NS4A | 0.0024 | 0.0333 | 90 | 0.4778 | 0.4222 | 0.3778 | 0.5000 | AETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEP |
| 2150 | 2219 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3429 | 0.3571 | 0.4714 | ETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPAR |
| 2150 | 2229 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3625 | 0.3375 | 0.4625 | ETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2150 | 2239 | NS4A | 0.0024 | 0.0333 | 90 | 0.4889 | 0.4222 | 0.3778 | 0.5000 | ETGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPE |
| 2151 | 2220 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3429 | 0.3571 | 0.4714 | TGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARI |
| 2151 | 2230 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3625 | 0.3375 | 0.4625 | TGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLL |
| 2151 | 2240 | NS4A | 0.0024 | 0.0333 | 90 | 0.4889 | 0.4222 | 0.3667 | 0.5000 | TGSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIPEPEK |
| 2152 | 2221 | NS4A | 0.0031 | 0.0429 | 70 | 0.4714 | 0.3571 | 0.3571 | 0.4857 | GSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIA |
| 2152 | 2231 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3625 | 0.3375 | 0.4625 | GSRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2153 | 2222 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3429 | 0.3429 | 0.4714 | SRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIAC |
| 2153 | 2232 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3625 | 0.3375 | 0.4500 | SRPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2154 | 2223 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3429 | 0.3286 | 0.4714 | RPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACV |
| 2154 | 2233 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3750 | 0.3375 | 0.4625 | RPYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2155 | 2224 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3571 | 0.3143 | 0.4714 | PYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVL |
| 2155 | 2234 | NS4A | 0.0027 | 0.0375 | 80 | 0.4500 | 0.3875 | 0.3250 | 0.4625 | PYKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVL |
| 2156 | 2225 | NS4A | 0.0031 | 0.0429 | 70 | 0.4571 | 0.3571 | 0.3143 | 0.4714 | YKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLI |
| 2156 | 2235 | NS4A | 0.0027 | 0.0375 | 80 | 0.4625 | 0.4000 | 0.3375 | 0.4750 | YKAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLI |
| 2157 | 2226 | NS4A | 0.0031 | 0.0429 | 70 | 0.4429 | 0.3714 | 0.3000 | 0.4714 | KAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIV |
| 2157 | 2236 | NS4A | 0.0027 | 0.0375 | 80 | 0.4625 | 0.4125 | 0.3375 | 0.4875 | KAAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLIP |
| 2158 | 2227 | NS4A | 0.0020 | 0.0286 | 70 | 0.4429 | 0.3714 | 0.3000 | 0.4714 | AAAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2159 | 2228 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3714 | 0.3143 | 0.4714 | AAAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2160 | 2219 | NS4A | 0.0024 | 0.0333 | 60 | 0.4500 | 0.3500 | 0.3167 | 0.4833 | AAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPAR |
| 2160 | 2229 | NS4A | 0.0020 | 0.0286 | 70 | 0.4429 | 0.3714 | 0.3000 | 0.4714 | AAQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2161 | 2230 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3857 | 0.3143 | 0.4857 | AQLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLL |
| 2162 | 2231 | NS4A | 0.0020 | 0.0286 | 70 | 0.4571 | 0.3857 | 0.3143 | 0.4857 | QLPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2163 | 2232 | NS4A | 0.0010 | 0.0143 | 70 | 0.4714 | 0.4000 | 0.3286 | 0.4857 | LPETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2164 | 2233 | NS4A | 0.0010 | 0.0143 | 70 | 0.4571 | 0.4000 | 0.3429 | 0.4857 | PETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2165 | 2234 | NS4A | 0.0010 | 0.0143 | 70 | 0.4571 | 0.4000 | 0.3286 | 0.4857 | ETLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVL |
| 2166 | 2235 | NS4A | 0.0010 | 0.0143 | 70 | 0.4571 | 0.4143 | 0.3286 | 0.5000 | TLETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLI |
| 2168 | 2227 | NS4A | 0.0012 | 0.0167 | 60 | 0.4333 | 0.3667 | 0.3000 | 0.4833 | ETIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2169 | 2228 | NS4A | 0.0012 | 0.0167 | 60 | 0.4333 | 0.3500 | 0.3167 | 0.4833 | TIMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2170 | 2229 | NS4A | 0.0012 | 0.0167 | 60 | 0.4167 | 0.3500 | 0.3167 | 0.4833 | IMLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2171 | 2230 | NS4A | 0.0012 | 0.0167 | 60 | 0.4333 | 0.3500 | 0.3333 | 0.4833 | MLLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLL |
| 2172 | 2231 | NS4A | 0.0012 | 0.0167 | 60 | 0.4167 | 0.3500 | 0.3167 | 0.4833 | LLGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2173 | 2232 | NS4A | 0.0012 | 0.0167 | 60 | 0.4167 | 0.3500 | 0.3167 | 0.4667 | LGLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2174 | 2233 | NS4A | 0.0012 | 0.0167 | 60 | 0.4167 | 0.3667 | 0.3333 | 0.4833 | GLLGTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2177 | 2226 | NS4A | 0.0014 | 0.0200 | 50 | 0.4000 | 0.3600 | 0.3000 | 0.4800 | GTVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIV |
| 2178 | 2227 | NS4A | 0.0014 | 0.0200 | 50 | 0.3800 | 0.3600 | 0.2800 | 0.4800 | TVSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2179 | 2228 | NS4A | 0.0014 | 0.0200 | 50 | 0.4000 | 0.3600 | 0.3000 | 0.4800 | VSLGIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2182 | 2231 | NS4A | 0.0014 | 0.0200 | 50 | 0.4200 | 0.4000 | 0.3200 | 0.4800 | GIFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2183 | 2232 | NS4A | 0.0014 | 0.0200 | 50 | 0.4200 | 0.4000 | 0.3200 | 0.4600 | IFFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2184 | 2233 | NS4A | 0.0014 | 0.0200 | 50 | 0.4000 | 0.4200 | 0.3400 | 0.4800 | FFVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVV |
| 2185 | 2234 | NS4A | 0.0014 | 0.0200 | 50 | 0.4000 | 0.4200 | 0.3400 | 0.4800 | FVLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVL |
| 2186 | 2235 | NS4A | 0.0014 | 0.0200 | 50 | 0.4200 | 0.4400 | 0.3600 | 0.4800 | VLMRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLVVLI |
| 2188 | 2227 | NS4A | 0.0018 | 0.0250 | 40 | 0.4000 | 0.4000 | 0.3250 | 0.4500 | MRNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVV |
| 2189 | 2228 | NS4A | 0.0018 | 0.0250 | 40 | 0.4000 | 0.3750 | 0.3250 | 0.4250 | RNKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVF |
| 2190 | 2229 | NS4A | 0.0018 | 0.0250 | 40 | 0.4000 | 0.4000 | 0.3250 | 0.4500 | NKGIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFL |
| 2192 | 2231 | NS4A | 0.0018 | 0.0250 | 40 | 0.4000 | 0.4000 | 0.3250 | 0.4500 | GIGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLL |
| 2193 | 2232 | NS4A | 0.0018 | 0.0250 | 40 | 0.4000 | 0.4000 | 0.3250 | 0.4250 | IGKMGFGMVTLGASAWLMWLSEIEPARIACVLIVVFLLLV |
| 2203 | 2222 | NS4A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.2500 | 0.3000 | 0.3500 | LGASAWLMWLSEIEPARIAC |
| 2207 | 2226 | NS4A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3000 | 0.3000 | 0.2500 | AWLMWLSEIEPARIACVLIV |
| 2208 | 2227 | NS4A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3000 | 0.3000 | 0.2500 | WLMWLSEIEPARIACVLIVV |
| 2210 | 2229 | NS4A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3500 | 0.3000 | 0.3000 | MWLSEIEPARIACVLIVVFL |
| 2212 | 2231 | NS4A | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3500 | 0.3000 | 0.3000 | LSEIEPARIACVLIVVFLLL |
| 2316 | 2335 | NS4B | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3500 | 0.3000 | 0.4000 | TPAVQHAVTTSYNNYSLMAM |
| 2317 | 2326 | NS4B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | PAVQHAVTTS |
| 2318 | 2323 | NS4B | 0.0000 | 0.0000 | 6 | 0.1667 | 0.1667 | 0.1667 | 0.1667 | AVQHAV |
| 2318 | 2327 | NS4B | 0.0000 | 0.0000 | 10 | 0.2000 | 0.3000 | 0.2000 | 0.3000 | AVQHAVTTSY |
| 2318 | 2337 | NS4B | 0.0000 | 0.0000 | 20 | 0.3500 | 0.2500 | 0.3000 | 0.3000 | AVQHAVTTSYNNYSLMAMAT |
| 2319 | 2328 | NS4B | 0.0000 | 0.0000 | 10 | 0.2000 | 0.3000 | 0.2000 | 0.3000 | VQHAVTTSYN |
| 2319 | 2338 | NS4B | 0.0000 | 0.0000 | 20 | 0.4000 | 0.3000 | 0.3000 | 0.3500 | VQHAVTTSYNNYSLMAMATQ |
| 2418 | 2427 | NS4B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.3000 | 0.2000 | 0.3000 | VVTDIDTMTI |
| 2419 | 2428 | NS4B | 0.0000 | 0.0000 | 10 | 0.3000 | 0.2000 | 0.2000 | 0.2000 | VTDIDTMTID |
| 2422 | 2427 | NS4B | 0.0000 | 0.0000 | 6 | 0.1667 | 0.0000 | 0.1667 | 0.0000 | IDTMTI |
| 2423 | 2428 | NS4B | 0.0000 | 0.0000 | 6 | 0.3333 | 0.0000 | 0.0000 | 0.0000 | DTMTID |
| 2453 | 2458 | NS4B | 0.0000 | 0.0000 | 6 | 0.3333 | 0.0000 | 0.1667 | 0.0000 | TAWGWG |
| 2453 | 2462 | NS4B | 0.0000 | 0.0000 | 10 | 0.4000 | 0.3000 | 0.2000 | 0.3000 | TAWGWGEAGA |
| 2454 | 2459 | NS4B | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | AWGWGE |
| 2703 | 2708 | NS5 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | YTSTMM |
| 2704 | 2709 | NS5 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | TSTMME |
| 2705 | 2710 | NS5 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.1667 | 0.1667 | 0.1667 | STMMET |
| 3403 | 3412 | NS5 | 0.0000 | 0.0000 | 10 | 0.3000 | 0.0000 | 0.2000 | 0.0000 | STQVRYLGEE |
| 3404 | 3413 | NS5 | 0.0000 | 0.0000 | 10 | 0.3000 | 0.0000 | 0.2000 | 0.0000 | TQVRYLGEEG |
| 3405 | 3414 | NS5 | 0.0000 | 0.0000 | 10 | 0.4000 | 0.0000 | 0.2000 | 0.0000 | QVRYLGEEGS |
| 3408 | 3413 | NS5 | 0.0000 | 0.0000 | 6 | 0.3333 | 0.0000 | 0.1667 | 0.0000 | YLGEEG |
aa: amino acid; DENV: dengue virus; JEV: Japanese encephalitis virus; NS: non-structural; pr: precursor; WNV: West Nile virus; YFV: yellow fever virus.
a K-mer is the protein fragment’s length in amino acids.
Note: None of the peptides had any homology with chikungunya virus.
Table 3. The number of Zika virus protein fragments selected as lead candidates for developing a serological test.
| Protein | No. of protein fragments |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6-mer | 10-mer | 20-mer | 30-mer | 40-mer | 50-mer | 60-mer | 70-mer | 80-mer | 90-mer | 100-mer | Total | |
| Capsid C | 1 | 0 | 0 | 0 | 8 | 1 | 0 | 6 | 0 | 0 | 0 | 16 |
| prM | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| E | 0 | 6 | 0 | 5 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 18 |
| NS1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| NS2A | 4 | 10 | 7 | 0 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 28 |
| NS2B | 1 | 7 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| NS3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS4A | 0 | 0 | 5 | 0 | 5 | 14 | 24 | 44 | 38 | 32 | 28 | 190 |
| NS4B | 5 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 |
| NS5 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| Total | 21 | 34 | 20 | 5 | 23 | 19 | 24 | 50 | 38 | 32 | 28 | 294 |
E: envelope; NS; non-structural; prM; precursor membrane.
Note: mer refers to the amino acids length of the protein fragment. Candidate proteins were selected based on identity between Zika virus and other viruses, within-Zika virus polymorphism, and protein structure.
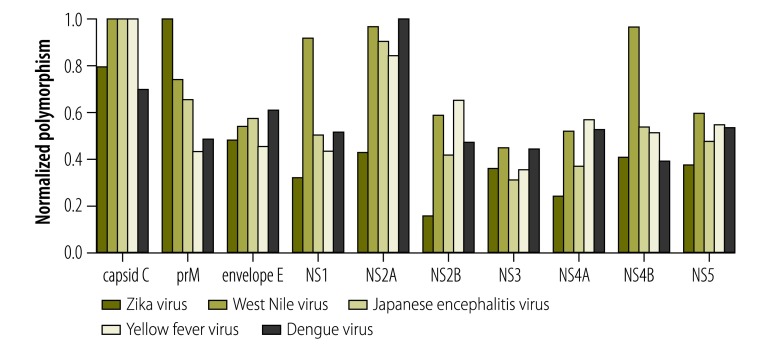
As Zika virus infection is associated with birth defects that are not seen in other flavivirus infections, we compared identity and polymorphism of proteins between flaviviruses. Overall, the level of identity between Zika virus and other flaviviruses is similar to the level of identity seen when comparing other flaviviruses with each other (available from the corresponding author). In contrast, one region (amino acid positions 430–500 in the proteome) in the envelope protein shows both low identity between Zika virus and other flaviviruses and low polymorphism within Zika virus (Fig. 2) and the relative polymorphism of NS2A and NS2B is on average 53.6% and 69.5% lower in Zika virus than in other flaviviruses, respectively (Fig. 4).
Fig. 4.
Normalized within-species polymorphism for each gene of each virus
E: envelope; NS; non-structural; prM; precursor membrane.
Notes: The proportion of polymorphic sites of each gene is normalized by the highest proportion of polymorphic sites for each virus.
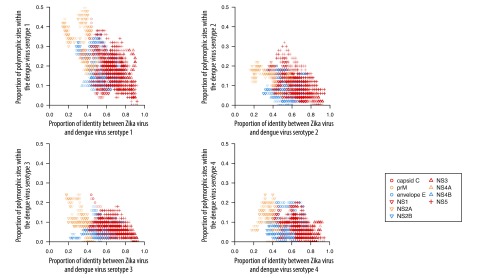
Protein identity between dengue and Zika viruses is negatively associated with polymorphism within the dengue virus proteins (P-values < 0.01 for all dengue serotypes; Fig. 5). This result can be explained by so-called negative selection, i.e. protein regions under stronger selective constraints tend to be more conserved and have higher identity between species and lower polymorphism within species.26 We did not observe a similar association for within-Zika virus polymorphism, which might be due to fewer strains analysed and/or smaller effective size of the global Zika virus population from which sequences were sampled, resulting in lower selection efficiency.
Fig. 5.
Dengue virus polymorphism versus identity with Zika virus
E: envelope; NS; non-structural; prM; precursor membrane.
Notes: 50-mers across the Dengue virus proteome were analysed, using a sliding window approach. Dengue virus polymorphism is negatively associated with the identity between dengue virus and Zika virus.
Discussion
Here we identified regions within the Zika virus proteome that have low identity with other viruses and low within-species polymorphism. These regions may be used to develop new serological diagnostic tests to detect Zika virus infection. However, for some of the identified regions, their antigenic properties are unknown and, therefore, these regions would first need to be evaluated for such properties. The regions identified as antigenic could then be used for developing a peptide microarray, where a collection of identified peptides are displayed on a surface. Antibodies generated during a previous Zika virus infection will then be able to bind to these displayed peptides. The read-out of the microarray is the fluorescent signal generated by fluorescence-coupled secondary antibodies that have bound to the serum antibody–peptide complexes. An advantage of assessing multiple peptides simultaneously in one test is that individual peptides do not need to generate a strong signal, since the intensities of signals of all different antibody–peptide complexes can be incorporated into a composite signal. Through statistical modelling the signal generated can be used to distinguish Zika virus infection and other infections.16 Microarrays also have a greater potential to identify prior virus infections than neutralization-based assays, because microarrays can detect a broader range of antibodies than only antibodies that neutralize the virus and protect against infections. Peptide microarrays have been used to differentiate between serological responses to closely related bacterial pathogens16 and to detect previous viral infections.27
The computational selection strategy used here represents a targeted approach, which reduces the number of potential candidate peptides. These peptides could be used for creating a peptide–antibody signature for a given viral infection. Once the signature is identified, a diagnostic test employing only the most important peptides contributing to that signature can be designed and produced. While our computational analysis of k-mers focused on linear epitopes, specific and sensitive linear epitopes together may be sufficient to distinguish different arboviruses. Moreover, depending on how a serological diagnostic test is produced, some of the longer k-mers might fold with sufficient similarity to their native folding to present conformational epitopes.
Our analysis showed that NS1 protein polymorphism is low. Therefore, using peptides from the NS1 protein for diagnostic test might result in a high-sensitivity test for detecting antibodies against Zika virus from different geographical locations. On the contrary, the identity of NS1 protein across flaviviruses is not particularly low compared to other proteins (third highest among 10 proteins), suggesting that NS1 is not the top candidate protein for low cross-reactivity. Recently, Euroimmun AG (Lübeck, Germany) developed a Zika virus ELISA for immunoglobulins (Ig)M and IgG, based on the NS1 protein. Preliminary results show that the test is Zika virus specific.28,29 However, the small sample size, the fact that the samples were not from regions with endemic dengue and the lack of samples from patients with different stages of infection weaken the conclusion.28,29 Moreover, because each diagnostic test has its advantages and disadvantages, having multiple approaches available is helpful for providing an accurate diagnosis. A sensitive and specific diagnostic test detecting several arbovirus infections simultaneously would be valuable,1 so that only one assay is required to diagnose active and previous flavivirus infection(s). While we designed the sequence analysis for specificity and sensitivity of detection of Zika virus infection, the same type of analysis could be used for identifying specific and sensitive markers for each arbovirus. By including specific and sensitive markers from all arboviruses in the same peptide microarray, the microarray has the potential to detect several arbovirus infections simultaneously.
To further dissect the molecular mechanism leading to the Zika virus sequelae not seen with other flaviviruses, the protein fragments presented in the candidate list may be useful. The low polymorphisms in NS2A and NS2B proteins might be good candidates to start investigating the possible molecular link between Zika virus and microcephaly and Guillain–Barré syndrome.
Peptide-sequence identity is unlikely to fully predict cross-reactivity due to other factors, such as glycosylation. Nonetheless, this analysis based on publicly available sequences provides a step towards the development of a serological test that can distinguish previous Zika virus and co-circulating arbovirus infections.1
Acknowledgements
We thank Xiaowu Liang and Taj Azarian. David Camerini is also affiliated with the Center for Virus Research, University of California, Irvine, USA.
Funding:
This study was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number U54GM088558 and MOE2012-T3-1-008, 15/1/82/27/001, H16/99/b0/013 for the Singapore side.
Competing interests:
HC, YHG, RGH, PJB and SMS declare no conflict of interest. DC has dual employment at UC Irvine and Antigen Discovery Inc. ML has received consulting fees from Pfizer and Affinivax and Antigen Discovery and grants through Harvard TH Chan School of Public Health from Pfizer and PATH Vaccine Solutions.
References
- 1.Target product profiles for better diagnostic tests for Zika virus infection. Geneva: World Health Organization; 2016. Available from: http://www.who.int/csr/research-and-development/zika-tpp.pdfhttp://[cited 2017 Feb 7].
- 2.Ioos S, Mallet HP, Leparc Goffart I, Gauthier V, Cardoso T, Herida M. Current Zika virus epidemiology and recent epidemics. Med Mal Infect. 2014. July;44(7):302–7. 10.1016/j.medmal.2014.04.008 [DOI] [PubMed] [Google Scholar]
- 3.Strauss JH, Strauss EG. Virus evolution: how does an enveloped virus make a regular structure? Cell. 2001. April 6;105(1):5–8. 10.1016/S0092-8674(01)00291-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016. May 19;165(5):1255–66. 10.1016/j.cell.2016.04.059 [DOI] [PubMed] [Google Scholar]
- 5.Charrel RN, Leparc-Goffart I, Pas S, de Lamballerie X, Koopmans M, Reusken C. Background review for diagnostic test development for Zika virus infection. Bull World Health Organ. 2016. August 1;94(8):574–584D. 10.2471/BLT.16.171207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allwinn R, Doerr HW, Emmerich P, Schmitz H, Preiser W. Cross-reactivity in flavivirus serology: new implications of an old finding? Med Microbiol Immunol. 2002. March;190(4):199–202. 10.1007/s00430-001-0107-9 [DOI] [PubMed] [Google Scholar]
- 7.Koraka P, Zeller H, Niedrig M, Osterhaus AD, Groen J. Reactivity of serum samples from patients with a flavivirus infection measured by immunofluorescence assay and ELISA. Microbes Infect. 2002. October;4(12):1209–15. 10.1016/S1286-4579(02)01647-7 [DOI] [PubMed] [Google Scholar]
- 8.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008. August;14(8):1232–9. 10.3201/eid1408.080287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tappe D, Rissland J, Gabriel M, Emmerich P, Gunther S, Held G, et al. First case of laboratory-confirmed Zika virus infection imported into Europe, November 2013. Euro Surveill. 2014. January 30;19(4):20685. 10.2807/1560-7917.ES2014.19.4.20685 [DOI] [PubMed] [Google Scholar]
- 10.Vorou R. Letter to the editor: diagnostic challenges to be considered regarding Zika virus in the context of the presence of the vector Aedes albopictus in Europe. Euro Surveill. 2016;21(10):30161. 10.2807/1560-7917.ES.2016.21.10.30161 [DOI] [PubMed] [Google Scholar]
- 11.Zammarchi L, Stella G, Mantella A, Bartolozzi D, Tappe D, Günther S, et al. Zika virus infections imported to Italy: clinical, immunological and virological findings, and public health implications. J Clin Virol. 2015. February;63:32–5. 10.1016/j.jcv.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 12.Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016. August 4;536(7614):48–53. 10.1038/nature18938 [DOI] [PubMed] [Google Scholar]
- 13.Revised diagnostic testing for Zika, chikungunya, and dengue viruses in US public health laboratories. Atlanta: Centers for Disease Control and Prevention; 2016. Available from: https://www.cdc.gov/zika/pdfs/denvchikvzikv-testing-algorithm.pdf [cited 2016 Jun 21].
- 14.Haug CJ, Kieny MP, Murgue B. The Zika challenge. N Engl J Med. 2016. May 12;374(19):1801–3. 10.1056/NEJMp1603734 [DOI] [PubMed] [Google Scholar]
- 15.Megret F, Hugnot JP, Falconar A, Gentry MK, Morens DM, Murray JM, et al. Use of recombinant fusion proteins and monoclonal antibodies to define linear and discontinuous antigenic sites on the dengue virus envelope glycoprotein. Virology. 1992. April;187(2):480–91. 10.1016/0042-6822(92)90450-4 [DOI] [PubMed] [Google Scholar]
- 16.Felgner PL, Kayala MA, Vigil A, Burk C, Nakajima-Sasaki R, Pablo J, et al. A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc Natl Acad Sci U S A. 2009. August 11;106(32):13499–504. 10.1073/pnas.0812080106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faria NR, Azevedo RdoS, Kraemer MU, Souza R, Cunha MS, Hill SC, et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science. 2016. April 15;352(6283):345–9. 10.1126/science.aaf5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brister JR, Bao Y, Zhdanov SA, Ostapchuck Y, Chetvernin V, Kiryutin B, et al. Virus Variation Resource–recent updates and future directions. Nucleic Acids Res. 2014. January;42(Database issue):D660–5. 10.1093/nar/gkt1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009. December 15;10(1):421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Regenmortel MH, Pellequer JL. Predicting antigenic determinants in proteins: looking for unidimensional solutions to a three-dimensional problem? Pept Res. 1994. Jul-Aug;7(4):224–8. [PubMed] [Google Scholar]
- 21.Xu X, Song H, Qi J, Liu Y, Wang H, Su C, et al. Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure. EMBO J. 2016. October 17;35(20):2170–8. 10.15252/embj.201695290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, et al. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016. April 22;352(6284):467–70. 10.1126/science.aaf5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiser J, Shenkin PS, Still WC. Approximate atomic surfaces from linear combinations of pairwise overlaps (LCPO). J Comput Chem. 1999. 20(2):217–30. [DOI] [Google Scholar]
- 24.Marzinek JK, Holdbrook DA, Huber RG, Verma C, Bond PJ. Pushing the envelope: dengue viral membrane coaxed into shape by molecular simulations. Structure. 2016. August 2;24(8):1410–20. 10.1016/j.str.2016.05.014 [DOI] [PubMed] [Google Scholar]
- 25.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012. January;40(Database issue):D593–8. 10.1093/nar/gkr859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Zhang Y, Lei X, Zhang Z. Natural selection of protein structural and functional properties: a single nucleotide polymorphism perspective. Genome Biol. 2008. April 8;9(4):R69. 10.1186/gb-2008-9-4-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015. June 5;348(6239):aaa0698. 10.1126/science.aaa0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Press release. Lübeck: EUROIMMUN; 2016. Available from: https://www.euroimmun.com/index.php?eID=dumpFile&t=f&f=3013&token=6cc0bb2eb7c10f484ae50ad91e69cd6424088529 [cited 2017 Mar 17].
- 29.Huzly D, Hanselmann I, Schmidt-Chanasit J, Panning M. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill. 2016. April 21;21(16):30203. 10.2807/1560-7917.ES.2016.21.16.30203 [DOI] [PubMed] [Google Scholar]