Abstract
Venom gland transcriptomes and proteomes of six Micrurus taxa (M. corallinus, M. lemniscatus carvalhoi, M. lemniscatus lemniscatus, M. paraensis, M. spixii spixii, and M. surinamensis) were investigated, providing the most comprehensive, quantitative data on Micrurus venom composition to date, and more than tripling the number of Micrurus venom protein sequences previously available. The six venomes differ dramatically. All are dominated by 2–6 toxin classes that account for 91–99% of the toxin transcripts. The M. s. spixii venome is compositionally the simplest. In it, three-finger toxins (3FTxs) and phospholipases A2 (PLA2s) comprise >99% of the toxin transcripts, which include only four additional toxin families at levels ≥0.1%. Micrurus l. lemniscatus venom is the most complex, with at least 17 toxin families. However, in each venome, multiple structural subclasses of 3FTXs and PLA2s are present. These almost certainly differ in pharmacology as well. All venoms also contain phospholipase B and vascular endothelial growth factors. Minor components (0.1–2.0%) are found in all venoms except that of M. s. spixii. Other toxin families are present in all six venoms at trace levels (<0.005%). Minor and trace venom components differ in each venom. Numerous novel toxin chemistries include 3FTxs with previously unknown 8- and 10-cysteine arrangements, resulting in new 3D structures and target specificities. 9-cysteine toxins raise the possibility of covalent, homodimeric 3FTxs or heterodimeric toxins with unknown pharmacologies. Probable muscarinic sequences may be reptile-specific homologs that promote hypotension via vascular mAChRs. The first complete sequences are presented for 3FTxs putatively responsible for liberating glutamate from rat brain synaptosomes. Micrurus C-type lectin-like proteins may have 6–9 cysteine residues and may be monomers, or homo- or heterodimers of unknown pharmacology. Novel KSPIs, 3× longer than any seen previously, appear to have arisen in three species by gene duplication and fusion. Four species have transcripts homologous to the nociceptive toxin, (MitTx) α-subunit, but all six species had homologs to the β-subunit. The first non-neurotoxic, non-catalytic elapid phospholipase A2s are reported. All are probably myonecrotic. Phylogenetic analysis indicates that the six taxa diverged 15–35 million years ago and that they split from their last common ancestor with Old World elapines nearly 55 million years ago. Given their early diversification, many cryptic micrurine taxa are anticipated.
Keywords: coralsnakes, Micrurus, venom gland transcriptomes, proteomes, 3FTx, phospholipase A2, molecular models, novel toxins
1. Introduction
The New World coralsnakes constitute a taxonomic complex of more than 70 species, traditionally divided into three genera (Micruroides, Leptomicrurus, and Micrurus) pertaining to the Family Elapidae. Because of their fossorial, semi-fossorial, or in a few cases, aquatic habits, coralsnakes are less often encountered by humans than are pit vipers occupying the same habitats. Coralsnakes produce smaller quantities of venom than pit vipers and other elapids of comparable length and they are more difficult to handle and to extract. Add to that the extreme difficulty of maintaining them in captivity for venom production, and it is not difficult to understand why venom chemistry of coralsnakes has lagged well behind that of viperids and larger elapids. Despite nearly 80 years of Micrurus venom research, fewer than 150 papers characterize Micrurus venoms or specific toxin constituents. Venoms of less than one-fourth of the recognized taxa have ever been examined in even the most superficial way. We have a modest understanding of the biochemical composition of only about five species, and pharmacologically, we know even less. To date, the coralsnake venom literature contains only two transcriptomic studies (Micrurus fulvius and M. tener) [1,2].
Accordingly, we characterized the venoms of six Brazilian Micrurus species (M. corallinus, M. l. carvalhoi, M. l. lemniscatus, M. paraensis, M. s. spixii, and M. surinamensis) that display great morphological and ecological diversity. Partial characterizations exist for M. corallinus, M. surinamensis, and M. l. lemniscatus venoms, but little has been reported for M. s. spixii and M. l. carvalhoi, and there have been no reports regarding M. paraensis. For each species, a venom gland transcriptome was sequenced using Illumina technology and venom peptides were identified with LC-MS.
2. Results and Discussion
2.1. Specimen Collection
Of the roughly 30 species of Micrurus that occur in the Amazon Basin, five are restricted to tropical Amazonian forests, while others, like Micrurus paraensis and M. s. spixii, can exist even in areas of contact with Amazonian cerrado (short-tree forest) [3,4,5,6]. The M. lemniscatus complex has two semi-aquatic taxa that are found in the Amazon Basin (M. l. lemniscatus and M. l. helleri) and a third (M. l. carvalhoi) that has the largest distribution of any coralsnake, from Rio Grande do Sul to the Northeast and the eastern Amazon, where it occupies primary and secondary cerrado, parts of the caatinga (a shrubby desert region in northeastern Brazil) and Atlantic forest [5]. Micrurus surinamensis exhibits similar adaptability, occurring not only in Amazonian rivers and their tributaries, but also at more northern latitudes, in the cerrado, and in areas of gallery forest along the Rio Araguaia [7]. Micrurus corallinus is associated with Atlantic forest, including zones of contact with cerrado [5].
Habitat utilization is very poorly known for most coralsnake species, and even in regions that appear more or less uniform (Amazonian forest or cerrado), coralsnakes may be present or absent, apparently depending upon less obvious habitat characteristics such as leaf litter. Ultimately, mineral content or soil pH, which govern plant species composition, may cause subtle differences in coralsnake habitats, thereby influencing distributions. Even semi-aquatic and aquatic species may be affected by such factors [5].
2.2. Transcriptomics and Proteomics
2.2.1. Characterization of Transcriptomes
The six transcriptomes yielded 1,051,787 contigs and the percentages of all reads assembled varied from 87.8 to 96.1%. Mean contig length ranged from 494 bp to 569 bp. Various other statistics are also available in Table S1.
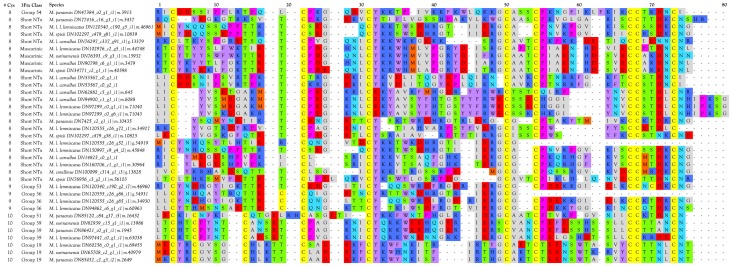
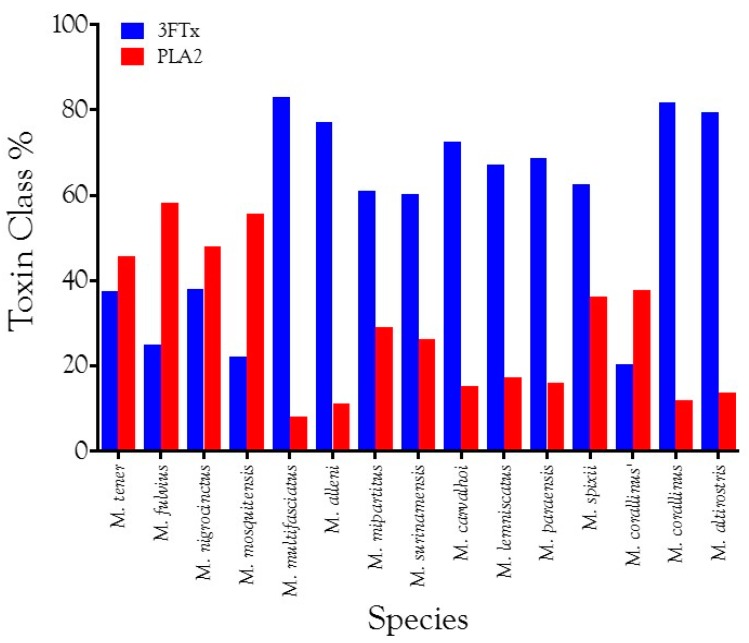
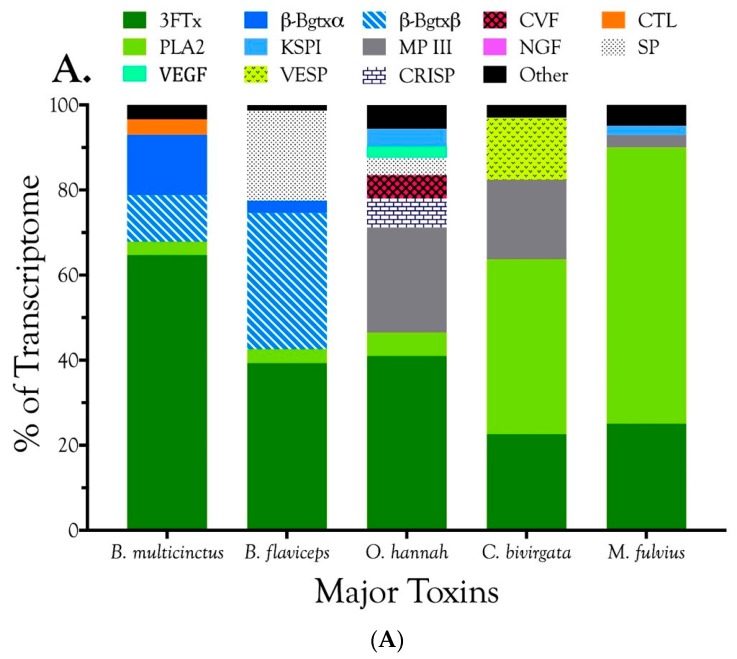
Venom gland transcriptomes of the six Micrurus species varied dramatically in composition. All transcriptomes were dominated by 2–6 toxin classes that accounted for 91.4–99.0% of the transcripts (Figure 1A; Table S2). The M. s. spixii transcriptome was compositionally the simplest. Three-finger toxins (3FTx) and phospholipases A2 (PLA2) amounted to just over 99.0% of the transcriptome, which comprised only four additional toxin families at levels of ≥0.1% (Figure 1A; Table S2). Other toxin families are present in all six venoms at trace levels (<0.005%). Venom of M. l. lemniscatus was the most complex, with at least 17 toxin families (Figure 1; Table S2). However, in each venome, multiple structural subclasses of 3FTXs and PLA2s are present. These variants have very different 3D structures and almost certainly differ in pharmacology as well.
Figure 1.
These Brazilian Micrurus venoms all contain three-finger toxins (3FTxs) and phospholipases A2 (PLA2s), but but they vary greatly in the relative proportions and subclasses thereof, and in the types and amounts of minor toxins as well. (A) Major toxins comprising ≥2% of the toxin portion a given transcriptome. The “other” portion of each venom (black) was comprised of minor components; (B) Minor toxins representing between 0.1% and 2.0% of the toxin transcriptome. Each venom contained still other toxins at trace levels, each amounting to less than 0.1% of the transcriptome. Micrurus s. spixii possesses the simplest venom, with 3FTxs and PLA2s accounting for just over 99% of the transcriptome, and comprising only six major and minor toxin classes.
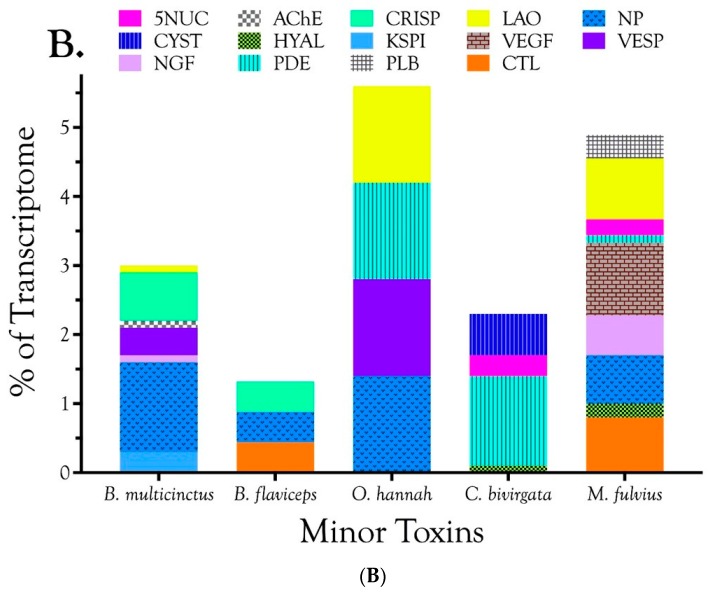
In addition to 3FTxs and PLA2s, all venoms also contained a putative nociceptive toxin (NOCI) subunit β (but not necessarily α), phospholipase B (PLB), and short vascular endothelial growth factors (VEGF-Fs) [8]. Minor components (0.1–2.0%) were found in venoms of all species except that of M. s. spixii (Table S2). The most abundant venom components after 3FTxs and PLA2s differed from venom to venom. In both M. l. carvalhoi and M. l. lemniscatus venoms, the next most abundant components were Kunitz serine protease inhibitors (KSPIs), a highly diversified venom protein family (Figure 1A; Table S2). In M. corallinus venom, C-type natriuretic peptide (CNP) was the next most abundant component, representing more than 29.5% of the transcriptome, followed by a C-type lectin-like protein (CTL) (3.4%), galactose-binding lectin (GBL) (3.4%), and a metalloprotease (MP) (2.1%). GBLs are normally extremely minor venom constituents (~0.1%) [9,10,11]. Micrurus paraensis venom had only one other major component beside 3FTxs and PLA2s. That was the nociceptive toxin β subunit (8.9%), homologous to MitTxβ from the venom of M. tener; however, interestingly, M. paraensis does not appear to produce the α subunit. At least from the western Amazon basin, Micrurus surinamensis venom is generally devoid of enzymatic components [12], but our transcriptome from this species contained 7.9% nerve growth factor (NGF), to which arginine esterase activity has been ascribed [13,14]. This is remarkable because this protease generally comprises no more than 0.2–0.7% of most venoms [10,11,15]. However, in the venom sample used for proteomics, this transcript was not even identified. Different specimens were used for the transcriptome and proteome, but both were collected in the vicinity of Altamira, Pará at localities separated by only 50 km. In a specimen from Estreito, Maranhão, about 800 km distant, NGF was present at a level of 0.4%. This is but one example of geographic and individual variation in this species. CNP was the remaining major component of the M. surinamensis transcriptome, at 2.7% (Figure 1; Table S2).
2.2.2. Characterization of Proteomes
The proteomes showed reasonable qualitative agreement with the transcriptomes, in that all but the most minor components of the transcriptomes were generally represented in the proteomes (Table S2). However, in contrast to our recent studies of Protobothrops venoms [10,11], the quantitative correspondence between transcriptomes and proteomes was extremely poor (Table S2). In large part, this was because the transcriptomes and venom samples came from different snakes, but this does not provide an entirely satisfactory justification.
3FTxs proved especially problematic in that their estimated abundances tended to be drastically lower in the proteomes than the transcriptomes (Table S2). We can offer no adequate explanation for this discrepancy. The lower representation of 3FTxs inflated values for all other venom proteins. Given the disparities between transcriptome and proteome, we ran a 4–12% SDS-DTT Bis-Tris polyacrylamide gel in MES buffer (pH 8.3), using M. l. carvalhoi venom (62.5, 125, 187.5 µg/well) to determine whether the transcriptomic or the proteomic result was accurate (Figure S1). Mass spectrometry was used to confirm what protein families were present in each band on the gel. 3FTxs were present in all of the lower bands on the gel (Fractions 10–16) and in some of the upper bands as well (3–6). Clearly, the venom is amply supplied with 3FTxs, as the transcriptome implies. While two of the digestions were enzymatic, the formic acid cleavage essentially cannot fail, so it seems unlikely that generation of peptides suitable for mass spectrometry was the source of the problem. A more likely explanation has to do with identification of the peptides sequenced by mass spectrometry. Since protein identification relies on interpretation of the spectra, the tremendous diversity of micrurine 3FTxs and the use of different specimens for transcriptomes and proteomes may have partially confounded peptide identification.
In 1995, using traditional Edman degradation, Aird and Silva, Jr. began sequencing toxins from M. surinamensis collected in the vicinity of Letícia, Colombia. Funding ran out before the sequences did and the partial sequences were never published, except for the first 37 residues of a short neurotoxin [16]. In 2008, Olamendi-Portugal et al. [17], using a combination of Edman degradation and mass spectrometry, completed the sequences of six toxins, from venom of specimens captured at Iquitos, Loreto, Peru. Both sets of sequences were identical, to the extent of our sequences. However, in this study, our two venom samples came from Pará and Maranhão, Brazil, at the eastern end of the M. surinamensis range. We found good matches for three of the sequences (MS5, MS4, and MS2) of Olamendi-Portugal et al. [17], with percent identities ranging from 89.1–97.7%. However, the best matches to their sequences MS1, MS3, and MS11 were very poor, displaying percent identities of only 44.9–50.1%. No BLAST hit for our Letícia, Colombia short neurotoxin sequence was found in the M. surinamensis transcriptome from Pará, Brazil.
2.2.3. 3-Finger Toxins (3FTx)
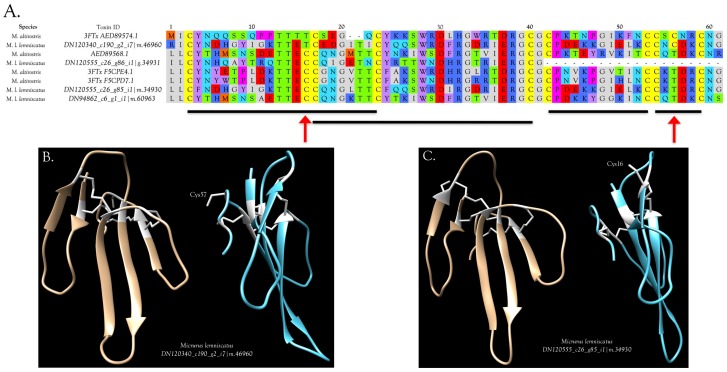
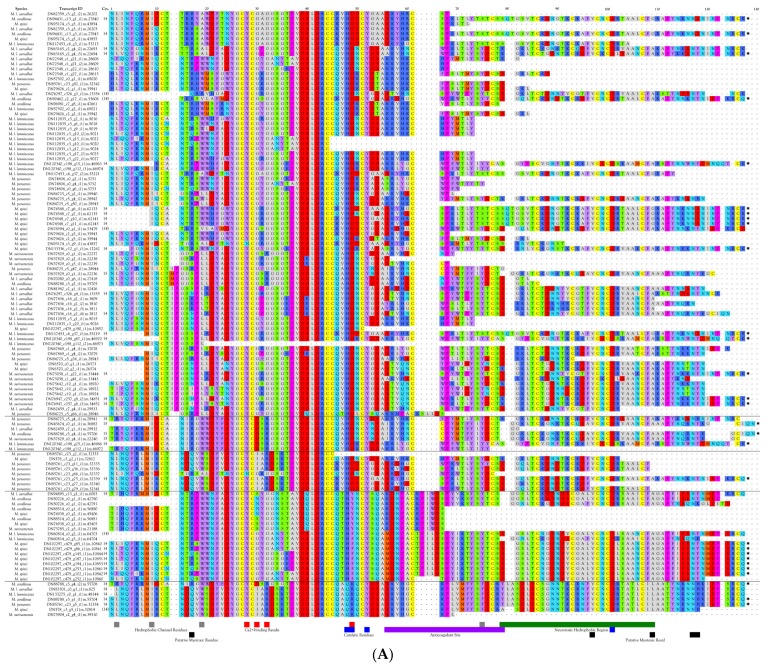
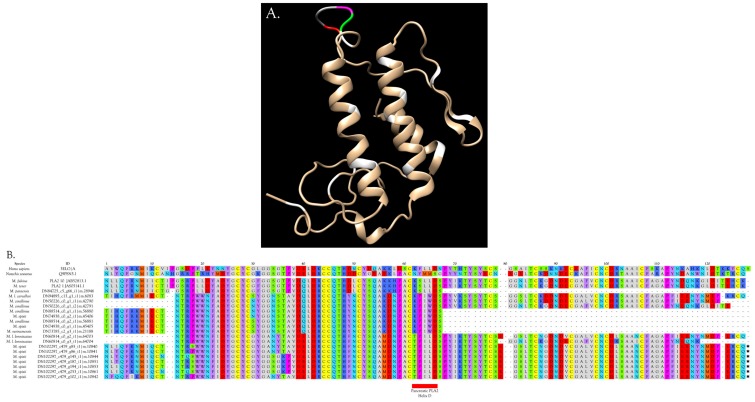
Three-finger toxins are significant components of all six venoms examined in this study, comprising from 20.3 to 72.6% of the transcriptomes (Figure 1, Figure 2 and Figure S2; Table S2). 3FTxs having 8, 9, and 10 cysteines are present (Figure 2 and Figure S2), although interestingly, among New World elapids, 9-Cys toxins have been found only in the venoms of M. l. lemniscatus and M. altirostris to date (Figure 3 and Figure S2). Two of the 9-Cys toxins, one from each species, have the extra Cys in position 56 (Figure 3A,B and Figure S2). A BLAST search failed to locate any Old World elapid 3FTx with this disulfide bond arrangement in the top 100 hits [18,19]. The remaining six toxins have the extra Cys residue in position 16 (Figure 3A,C), giving the two subclasses of 3FTx very different 3D profiles49.
Figure 2.
Micrurus venoms are rich in 3FTxs displaying an astonishing variety of primary structures. Micrurine 3FTxs may possess 8, 9, or 10 Cys residues with different disulfide patterns in each group. Pharmacologies are almost entirely unknown. Some probably target nicotinic acetylcholine receptors of reptilian neuromuscular junctions, but their potential targets in mammals are unknown. Most 3FTxs have 21-residue signal peptides. In the interest of creating a figure of manageable size for the journal format, all signal peptides have been deleted here, and only sequences originating in this study have been included. Full sequences of all 184 micrurine 3FTxs can be found in Figure S2.
Figure 3.
Primary and 3D structures of 9-Cys 3FTxs from Micrurus venoms include two structural subclasses. (A) To date, micrurine 3FTxs with 9 cysteines have been found only in venoms of Micrurus l. lemniscatus and M. altirostris; however, among these, the extra cysteine can appear in either of two positions, indicated by red arrows. Putative conserved disulfide bonds are indicated with black bars. Signal peptides, 21 residues in length, were almost invariant (MKTLL LTLVV VTIVC LDFGH T). M. l. lemniscatus toxin DN120340 had an L/Q substitution in position 4 and a V/L substitution in position 14; (B) Front and side views of the ribbon model of M. l. lemniscatus DN120340; (C) Front and side views of the ribbon model of M. l. lemniscatus DN120555. SWISS-MODEL was used to select the best templates for the Micrurus toxins (Laticauda semifasciata, erabutoxin, 2era.1.A for M. l. lemnsicatus DN120340 and Naja atra cobrotoxin (1coe.1.A) for M. l. lemniscatus DN120555) and to construct a preliminary model. Then models were refined and energy minimizations were performed with UCSF Chimera [21]. Disulfide bonds are shown in white.
This arrangement apparently has arisen independently in only two Old World elapids, Walterinnesia aegyptia (C1IC48.1) [20] and Micropechis ikaheka (AHZ08817.1, Paiva, O.K. unpublished). The existence of 9-Cys toxins suggests the possibility of covalent homodimeric 3FTxs. Alternatively, they could form heterodimers with some other toxin class that also has a free Cys, or it may be that the Cys is actually free and is involved in target binding.
3FTxs with 10 Cysteine Residues
Micrurine 3FTx toxins with 10 Cys residues (19 sequences) have the disulfide bond arrangement of γ-bungarotoxin, rather than that of α- or κ-bungarotoxin (Figure 4A and Figure S2); thus, close homologs of these two better-known 3FTx subclasses do not appear to be present in Micrurus venoms. γ-bungarotoxin is a neuronal nAChR antagonist with unique specificity [22,23]. 10-Cys 3FTxs fall into a subclass of elapid 3FTxs originally known as “weak neurotoxins”, under the assumption that they target nAChRs ineffectively. Later they were dubbed “non-conventional” toxins [24]. While some of the Old World elapid 3FTxs eventually proved to be neuronal toxins, rather than antagonists of neuromuscular junction nAChRs, it finally became clear that many “non-conventional” toxins do not target nAChRs at all [25,26,27].
Figure 4.
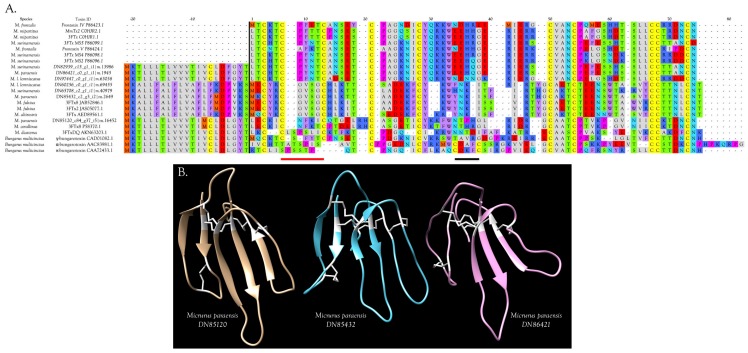
Micrurine 3FTxs with 10 cysteine residues include several subclasses. (A) 3FTxs with 10 cysteines show the disulfide bond pattern of γ-bungarotoxin (red bar), but not of α- and κ-bungarotoxins (black bar). Thus, homologs of the latter two 3FTx subclasses probably are not found in New World coralsnake venoms. Residues to the left of the black vertical line constitute signal peptides; (B) Ribbon models of three different subclasses of 10-Cys 3FTxs from the venom of M. paraensis: DN85120 (tan), DN85432 (blue), and DN86421 (violet), from (A). 3D representations were made with UCSF Chimera [21] based on models generated with SWISS-MODEL, using the structure of candoxin (1jgk.1.A) as a template for DN85120, bucandin (1ijc.1.A) as a template for DN85432, both from Bungarus candidus venom, and Bungarus multicinctus γ-bungarotoxin (1mr6.1.A) for DN86421. Disulfide bonds are shown in white.
Micrurine 10-Cys 3FTxs fall into at least three sub-subclasses, represented by M. paraensis: toxins DN85432, DN86421, and DN85120 (Figure 4A,B and Figure S2). Homologs of DN85432 have radically different signal peptides rich in phenylalanine and commence with the N-terminal sequence MECYR, suggesting unusual pharmacology (Figure 4A). Homologs of DN86421 and DN 85120 have similar signal peptides. The former have N-terminal sequences commencing with LTCK/HT, while the latter have LECKI, with many other internal sequence differences also (Figure 4A).
3FTxs and GABA Receptors
Rosso et al. [28] reported that micrurotoxins 1 and 2, from the venom of Costa Rican Micrurus mipartitus, bind to GABAA receptors at subnanomolar concentrations. Nicotinic acetylcholine receptors (nAChR) were not affected by these toxins. The two micrurotoxins, which differ by a single Arg/His substitution at position 33, bind to a benzodiazepine-like binding site at the α+/β− subunit interface, allosterically enhancing receptor susceptibility to agonist, which potentiates receptor opening and desensitization [28]. Unfortunately, because no structures were available, it is not possible to know whether any homologs of the micrurotoxins are found in our transcriptomes.
3FTxs with 8 Cysteine Residues
The overwhelming majority of micrurine 3FTxs with eight cysteines follow the classic α-neurotoxin/cardiotoxin disulfide bond pattern (3–24, 17–41, 43–54, 55–60) [29,30,31]. Numbering varies slightly with the toxin or alignment in question. While most micrurine 8-Cys 3FTxs terminate one or two residues beyond the most C-terminal Cys, some of the toxins that we sequenced have C-terminal extensions of 2–14 amino acids (Figure 2 and Figure S2). Many of these probably target nicotinic acetylcholine receptors of reptilian or fish neuromuscular junctions, but in the absence of pharmacological studies, little more can be said.
Interestingly, two toxins from Micrurus fulvius and Micrurus tener introduce an entirely new structure in which the Cys54-Cys60 disulfide is absent and a new disulfide has apparently formed between Cys6 and Cys11 (Figure 5A). The magnitude of the resulting structural variation is readily apparent when comparing ribbon models of the two subclasses (Figure 5B). In addition, the signal peptides of these two classes show numerous differences (Figure 5A).
Figure 5.
3FTxs bearing 8 cysteine residues show two very different disulfide bond patterns. (A) The classic short α-neurotoxin/cardiotoxin pattern, represented above by Naja nigricollis Cardiotoxin 1 and by Micrurus fulvius 3FTxs 9a & b, is employed by all but two micrurine 3FTxs (M. tener 3FTx1 and M. fulvius 3FTx5a). The novel disulfide in the latter two toxins is indicated with a red bar. Conserved disulfides are denoted with black bars. Signal peptides are shown to the left of the black vertical line; (B) Ribbon models of the two subclasses of 8-Cys 3FTxs from the venom of M. fulvius: 3FTx 5a (JAB52850.1) (tan) and 3FTx 9a (JAS05068.1) (blue) from (A). Disulfide bonds are shown in white. SWISS-MODEL selected bucandin (1f94.1.A) from Bungarus candidus venom as the best template for M. fulvius 3FTx 5a, and 3FTx 3b (4rud.1.B) from M. fulvius venom as the best template for M. fulvius 3FTx 9a. Models were refined and energy minimizations were performed using UCSF Chimera [21].
Muscarinic Toxins (MTx)
Toxins that agonize or antagonize muscarinic acetylcholine receptors with far greater specificity than small, organic ligands [32,33,34,35,36], and that bind with low nanomolar affinity, were first discovered in the venoms of mambas (Dendroaspis sp.) [37,38,39]. Muscarinic toxins, 3FTxs with eight cysteines, account for about 2% of crude Dendroaspis angusticeps venom, by mass [40]. Paradoxically, mamba venoms contain both muscarinic acetylcholine receptor (mAChR) agonists and antagonists, which, to the best of our knowledge, has never been explained.
mAChRs have been classified into five types (M1–M5) (Table 1), with the M3 type predominating in epithelial and smooth muscle cells [41]. M3-mAChRs mediate endothelium-dependent vasorelaxation in coronary arterial circulation [42], and activation of M3 and M5 mAChRs in the vasculature causes vasodilation [43] (Table 1). Neither m1- nor m4-toxin binds to M2, M3, or M5 receptors [33,35]. In contrast, MT2 acts as a partial agonist at M3-mAChRs [35]. Jolkkonen et al. [44] opined that MTα and MTβ, two muscarinic toxins from Dendroaspis polylepis venom, are probably mAChR agonists, based on their capacity to contract guinea pig ileum; thus, these toxins would be expected to induce vasodilation and hypotension. Ryberg et al. [45] have shown that through its actions on muscarinic receptors, acetylcholine causes vasodilation of rat carotid and submandibular arteries and vasoconstriction of jugular and submandibular veins. Acetylcholine, can cause vasodilation or vasoconstriction, depending upon the mAChR class to which it binds. Mamba muscarinic toxins may act as either agonists or antagonists, but it appears that in a well coordinated strategy, they simultaneously agonize vasodilatory muscarinic receptors and antagonize vasoconstrictive receptors, causing vasodilation of all vascular beds, profound hypotension, and circulatory shock [46] (Table 1).
Table 1.
Pharmacology of mammalian muscarinic acetylcholine receptors (mAChRs) compared with actions of mamba muscarinic toxins. mAChRs (blue) comprise five classes (M1-M5) with broad peripheral and central tissue distributions. Ryberg et al. [45] have shown that through its actions on muscarinic receptors, acetylcholine causes vasodilation of rat carotid and submandibular arteries and vasoconstriction of jugular and submandibular veins. The response depends upon the mAChR class to which it binds. Mamba (Dendroaspis) muscarinic toxins (red) may act as either agonists or antagonists of mAChRs, but it appears that they agonize vasodilatory mAChRs and antagonize vasoconstrictive receptors. The net result is profound hypotension [46,47]. References for mAChR and ligand pharmacology, and for toxin effects are as follows: M1 [45,48]; M2 [49,50,51,52]; M3 [42,53,54,55]; M4 [45,56]; M5 [57]; m1-toxin/MT7 [33,58,59]; m2-toxin [60]; MT2/MTα/MTβ [35,61]; m4-toxin/MT3/MT2/MT7 [35,59,62]; MTα/MTβ [44].
| mAChR Class | Tissue Target | Agent Action | Organism | Effect | Reference | Toxin | Reference | Action | Probable Effect |
|---|---|---|---|---|---|---|---|---|---|
| M1 | Rostral ventrolateral medulla | Agonist | Humans | Vasoconstriction, Tachycardia | Medina et al., 1997 | m1-Toxin; MT7 | Max et al., 1993; Jolkkonen, 1996; Liang et al., 1996 | Antagonist | Hypotension |
| M1 | Submandibular & jugular vein endothelium | Agonist | Rats | Vasoconstriction | Ryberg et al., 2008 | ||||
| M2 | Cerebral ventricles | Antagonist | SH rats | Hypotension | Brezenoff et al., 1988 | m2-toxin | Carsi et al., 1999 | Antagonist | Hypotension |
| M2 | Cerebral ventricles | Agonist | Rats | Hypertension, Tachycardia | Pazos et al., 1986; Kubo, 1998; Ozkutlu et al., 1993 | ||||
| M2 | Cardiac Atrium | Mice | Negative Inotropy | Nishimaru et al., 2000 | |||||
| M3 | Aorta | Agonist | Mice | Vasodilation | Khurana et al, 2004 | MT2; MTα; MTβ | Bradley, 2000; Jolkkonen et al., 1995 | Agonist | Hypotension |
| M3 | Resistance vessels | Agonist | Mice | Vasodilation | Bruning et al., 1994; Gericke et al., 2011 | ||||
| M3 | Mesenteric vessels | Agonist | Rat | Vasodilation | Hendriks et al., 1992 | ||||
| M3 | Coronary arteries | Agonist | Mice | Vasodilation | Lamping et al., 2004 | ||||
| M4 | Submandibular & jugular vein endothelium | Agonist | Rats | Vasoconstriction | Ryberg et al., 2008 | m4-toxin; MT3; MT2; MT7 | Jolkkonen et al., 1994; Liang et al., 1996; Bradley, 2000 | Antagonist | Hypotension |
| M4 | Spinal Cord | Binding only | Rats | Not determined | Höglund & Baghdoyan, 1997 | ||||
| M5 | Cerebral blood vessels | Agonist | Mice | Vasodilation | Yamada et al., 2001 | MTα; MTβ | Jolkkonen et al., 1995 | Agonist | Hypotension |
In overall structure, MTs resemble traditional short α-neurotoxins that antagonize nAChRs [63]. Servent et al. [64] outlined structural attributes that characterize D. angusticeps muscarinic toxins, based upon assays in mammals. These include four disulfide bonds, an N-terminal LTCV sequence, a C-terminal TDKCNX sequence, the sequence CP(D/A)GQN(L/V)CFK in the region connecting loops I and II, and the sequence GC(A/V)ATCP between loops II and III.
The six Micrurus transcriptomes were searched for muscarinic toxins 1–4. Micrurus l. carvalhoi, M. corallinus, M. l. lemniscatus, M. s. spixii, and M. surinamensis, as well as M. altirostris, M. fulvius, and M. tener, possess toxins with substantial similarity to Muscarinic Toxin 1 (MT1) from the venom of Dendroaspis angusticeps (Figure 6). However, of the attributes listed by Servent et al. [64], micrurine muscarinic-like toxins share only the disulfide pattern, but so do many toxins with entirely different pharmacologies. Moreover, there are also enough non-synonymous amino acid substitutions in the Micrurus sequences to raise doubts as to whether these really are muscarinic antagonists. Their 3D structures and surface hydrophobicities vary considerably. Even if they are muscarinic antagonists, Jolkkonen et al. [36] noted that minor differences in the amino acid sequences of the various muscarinic toxins from mamba venoms confer very different pharmacologies upon the toxins. In particular, micrurine toxins have 3-5 tyrosine and tryptophan residues in the N-terminal 11 residues that are not present in MT1 (Figure 6). On the other hand, the 21-residue MT1 signal peptide is identical to those of four Micrurus MTs (Figure 6).
Figure 6.
To date, all Micrurus venoms contain 3FTxs with significant similarities to muscarinic toxins from Dendroaspis angusticeps venom; however, these toxins also display numerous differences. It seems unlikely that these toxins could exhibit the same pharmacology in mammals as the mamba toxins, but they may antagonize mAChRs in reptiles, fishes, and onychophorans, the natural prey of coralsnakes. Aligned sequences of mamba muscarinic antagonists and putative muscarinic toxins from Micrurus venoms. Signal peptides are shown to the left of the black vertical line.
All of the Micrurus species investigated here had an MT2-like sequence, except for M. paraensis (Figure 6); however, these are the same sequences that resemble MT1. Nonetheless, in order to align the micrurine sequences with MT2, it is not necessary to gap the sequences as when comparing them with MT1 (Figure 6). That of M. corallinus is 7 residues shorter than a normal MT2. All eight micrurine toxins share their cysteine positions with MT2, and all except the toxins from M. fulvius and M. tener, which have a P39A substitution, share P39 and P68 also. None of the micrurine toxins have MT2 P75. All except the M. corallinus, M. fulvius, and M. tener sequences share all of the acidic residues, except for the C-terminal aspartate, which oddly, only the M. corallinus toxin has. Furthermore, all Micrurus MT-like toxins have R48, and all but the odd toxin from M. corallinus also have F46, K47, W49, and H50. All micrurine toxins except for that of M. corallinus possess N43 and N72, and all of them have N85. All eight micrurine MT-like toxins (five from this study plus those from M. altirostris, M. fulvius, and M. tener) share the same 21-residue signal peptide with MT2, except that three of our coralsnake toxins have either of two valine to methionine substitutions (Figure 6). Alignments with Dendroaspis angusticeps muscarinic toxins MT3 and MT4 present the same sorts of identities seen with MT1 and MT2.
da Silva et al. [65] isolated a 7048 Da protein from the venom of M. l. lemniscatus, which they dubbed MT-Mlα, because it displaced [3H]QNB from two types of binding site in rat hippocampus. It also antagonized accumulation of phosphate induced by carbachol, but with ~15-fold less potency than the M1 antagonist, pirenzipine. MT-Mlα [65] had the N-terminal sequence LICFICFSPTAH, which is unlike the sequences of any Dendroaspis muscarinic toxins. It also cannot be aligned with any sequence from our M. l. lemniscatus transcriptome. While we do not doubt their pharmacology, their sequence seems questionable. Only the 10-Cys 3FTxs have two cysteines separated by three residues (Figure 2 and Figure S2). While some of these have a tyrosine in position 4, none of them has a hydrophobic residue in position 5.
Until appropriate pharmacological assays are done with these toxins, we will not know whether they actually are muscarinic toxins, or whether they have some entirely different target. Given their interesting combination of identities and differences with the mamba toxins, it may be that these toxins are genuine muscarinic toxins, but adapted to incapacitate reptiles or fish, rather than mammals or birds. Mammalian muscarinic receptors are glycoproteins of 460–590 amino acids, depending upon the type. They show considerable sequence similarity from one mammal species to another (within types) [66]; however, BLAST searches of the NCBI nr database using the human M1 muscarinic receptor sequence as a query yielded identity scores with reptilian muscarinic receptors (M1–M5) of only 45–72%. Presumably glycosylation patterns may also differ between mammals and reptiles. While glycosylation apparently does not affect receptor function, toxins that target reptilian muscarinic receptors would be expected to differ significantly from those targeting mammalian receptors.
3FTxs that Impact Glutamate Receptors
Montandon [67] investigated the neurotoxicity of M. l. carvalhoi venom using rat cerebrocortical synaptosomes. She concluded that it was primarily caused by liberation of l-glutamate by venom phospholipases A2, as a result of phospholipid hydrolysis and membrane disruption; however, she also determined that a 3FTx with a mass of 6675 Da was responsible for at least some of the neurotransmitter liberation. The action of this 3FTx appeared at least partially dependent upon the action of voltage-gated calcium channels, since it was inhibited when calcium was replaced with strontium.
Freire Donato investigated possible pharmacological targets responsible for neurotoxicity of crude Micrurus l. carvalhoi venom using an MTT viability assay in neonatal rat hippocampal neurons [68]. Neurotoxicity involved NMDA and nicotinic receptors. In addition, the venom, a reverse phase fraction thereof, and two purified 3FTxs liberated l-glutamate (Glu) from adult male rat cerebrocortical synaptosomes. Ca2+ is required for Glu liberation. Voltage-sensitive calcium channels (VSCCs) (Types N, P, and L) participated, and Glu release was reduced by 10 μM ω-conotoxin GVIA, 30 nM ω-agatoxin IVA, or 3 μM nifedipine; however, even when applied together, the three antagonists were unable to block all Glu liberation. Both NMDA and AMPA receptors were involved in neurotransmitter release, since it was abolished by combined application of the ionotropic glutamate receptor antagonists, MK-801 and CNQX (10 µM each), but not by either administered alone. Toxin Ml_7294_NTX (1 μM) released approximately 2.3 nmol Glu/mg of synaptosomal protein in 35 min. In comparison, 33 mM KCl liberated approximately 8 nmol/mg in the same period, indicating that the toxin is approximately 104 times more potent than KCl on a molar basis. A possible mechanism of action is as follows. These toxins may activate pre-synaptic AMPA/kainate receptors, when the membrane is at rest. Upon activation, these receptors would depolarize the membrane, activating VSCCs (Types N, P, and L), and NMDA receptors. Subsequent entry of calcium via both calcium channels and NMDA receptors would result in exacerbated neurotransmitter liberation, as observed.
Freire Donato determined that the N-terminal sequence of the 3FTx from M. l. carvalhoi venom responsible for glutamate release was: LICYVSMCGAKMTCPCEGNNLCEYYAVPVF. This sequence aligns modestly well with three of our sequences from the same taxon, except that it appears to contain two spurious cysteine residues (underlined) that have no equivalent in any other 3FTx sequences. However, if these are deleted, the alignment with our sequences is excellent (Figure 7). The 3FTx responsible for glutamate liberation from rat cerebrocortical synaptosomes appears to be a 66-residue, 8-Cys toxin. Identical sequences are found in all of our transcriptomes except for that of M. paraensis. These probably represent the first complete sequences of glutamate-liberating 3FTxs from any snake venom. Venoms of M. l. carvalhoi and M. l. lemniscatus each have two other sequences with very high similarity (Figure 7).
Figure 7.
Freire Donato [68] determined the N-terminal amino acid sequence of a toxin from the venom of M. l. carvalhoi that provokes the release of glutamate from rat cortical synaptosomes. While that sequence contained two cysteine residues that appear spurious, when those are deleted, her sequence aligns well with sequences from five of the six Micrurus transcriptomes. These appear to represent the first complete sequences of snake toxins that provoke glutamate release.
Cardiotoxins/Cytotoxins/β-Cardiotoxin
Based on electrophysiological evidence, Vital Brazil [69] concluded that cardiotoxins are present in M. fulvius venom. In a chromatographic survey of 11 Micrurus venoms, Silva Jr. et al. [70] found that molecules with the chromatographic behavior of Naja kaouthia cardiotoxin 1 were present only in the venoms of M. albicinctus, M. Braziliensis, and M. frontalis; however, they could not positively identify the toxins, which were most likely other 3FTxs. To date, no transcriptomic studies have provided any evidence for the presence of Naja-like cardiotoxins in any Micrurus venom, including even the deep transcriptomic study of M. fulvius venom glands [1]. BLAST searches of our transcriptomes produced no cardiotoxin/cytotoxin hits, and BLAST searches of the NCBI nr and tsa_nr databases, restricted to Micrurus, located only neurotoxin sequences. These “hits” resulted from the near identity of the signal peptide sequences. It seems most likely that the toxins responsible for the activity described by Vital Brazil are probably PLA2s, and we do not anticipate finding typical cardiotoxins/cytotoxins in Micrurus venoms.
β-cardiotoxin was discovered in the venom of Ophiophagus hannah a decade ago [71]. This 63-residue 3FTx binds to β1 and β2 adrenergic receptors to promote dose-dependent bradycardia, rather than the tachycardia that typifies the activity of conventional cardiotoxins. BLAST searches of our six transcriptomes and all Micrurus sequences from the NCBI nr database yielded no venom proteins with similarity greater than 50%. Micrurus paraensis yielded no hit at all. As of this writing, it seems probable that Micrurus venoms do not contain β-cardiotoxin-like proteins either.
Fasciculins
Kumar et al. [72] reported that M. fulvius venom possesses anti-cholinesterase activity comparable to that of various Dendroaspis and Naja venoms. This is difficult to interpret since Dendroaspis venoms possess fasciculins, which no Naja venoms are known to have and because the responsible component was retained by a 100 kDa-cutoff dialysis membrane, whereas a 7 kDa protein such as a fasciculin should have been lost. To date, no fasciculins have been reported in any Micrurus venom, and BLAST searches of our transcriptomes failed to produce even low-similarity hits. Possibly some Micrurus venoms contain an unrelated anti-cholinesterase protein, but this seems unlikely.
Calcicludine- or Calciseptine-like Toxins
Mamba venoms also contain potent antagonists of mammalian L-type calcium channels (calciseptine, calcicludine, FS2, etc.) [73,74]. However, BLAST searches of our transcriptomes and previously published toxin sequences from Micrurus venoms venoms failed to yield any candidates. We conclude that Micrurus venoms probably do not contain mamba-like, L-type calcium channel antagonists.
2.2.4. 5′-Nucleotidase (5NUC)
5NUCs dephosphorylate purine mononucleotides to release free nucleosides (principally adenosine), which contribute to prey hypotension [46]. All eight Micrurus species examined to date have 5′-nucleotidases in their venoms (Figure S3). These are present at levels ranging from ~0–0.05% (Table S2). At 573–588 residues, with no signal peptides, micrurine venom 5NUCs are slightly shorter than their crotalid counterparts. Micrurine 5′-nucleotidases bear two different C-termini, one 14 residues longer than the other. Interestingly, the longer variant also appears in 5NUCs of the pitvipers, Protobothrops elegans and Protobothrops flavoviridis [11], and Gloydius brevicaudus (Ogawa and Yanoshita, unpublished, BAG82602.1. Micrurine 5NUCS are structurally almost invariant, in addition to their close identity with the crotalid enzymes (Figure S3).
2.2.5. Acetylcholinesterase (AChE)
Micrurine acetylcholinesterases (AChEs) are modestly large proteins, containing approximately 585 amino acids. However, they are not expressed at biologically significant levels as they are in some Old World elapines. Based upon the hypertensive action of acetylcholine at M2 and M4 muscarinic receptors (Table 1), Aird [46] concluded that AChE might have a secondary role of promoting hypotension. That hypothesis now appears more unlikely, in light of subsequent research. Given that MT2 toxin, an M3 receptor agonist, comprises ~50% of the muscarinic toxins in Dendroaspis angusticeps venom, mambas seem to be targeting primarily vascular M3 muscarinic receptors, not only with MT2, but also with venom acetylcholine, to produce profound hypotension [47,75,76], an effect intensified by dendrotoxins, which liberate neurotransmitter [77], and fasciculins, which inhibit prey AChE so as to prevent transmitter degradation [78,79,80]. All of the foregoing implies that the primary function of AChE in the venoms of those elapids that produce it, is to starve neuromuscular junctions of transmitter, in concert with postsynaptic antagonists of nicotinic AChRs.
Among our Micrurus transcriptomes, AChE is most abundant in M. l. lemniscatus venom, where AChE transcripts total only 0.05% of the transcriptome (Table S2). Micrurine AChEs are highly variable in primary structure (Figure S4). Some, such as M. s. spixii transcript DN97541_c0_g1_i1|m.57626, are predicted to have 29-residue signal peptides, while others, such as M. tener AChE (JAS05178.1), are expected to have signal peptides of only 23 residues, with shorter N-termini.
2.2.6. Aminopeptidases A and N (APA and APN)
Both APA and APN convert kallidin to bradykinin [81,82]. Cerebral microvascular aminopeptidase A inactivates the vasoconstrictive peptides, angiotensins I and II, by cleaving the amino-terminal Asp-Arg bond [83]. APN does not cleave mediators of hypotension and plasma extravasation, such as bradykinin and Substance P [84,85,86]. Thus, it is highly likely that the role of micrurine aminopeptidases A and N (M) is to promote hypotension, in concert with other venom constituents [46].
All of the transcriptomes except that of M. surinamensis had an APA sequence (Figure S5). The Micrurus sequences comprise 958 residues and are largely identical to the APA from Gloydius brevicaudus venom [87]. Because they are membrane-anchored proteins, they are predicted to lack signal peptides. Micrurus s. spixii had the highest APA transcript abundance, but that constituted only 0.09% of the transcriptome (Table S2).
Aminopeptidase N (EC 3.4.11.2), also known as Aminopeptidase M [84] presented a similar picture. All of the transcriptomes except for that of M. paraensis contained an APN transcript, as did that of M. fulvius (Figure S6). All of our transcripts were incomplete, but they were nearly identical to the M. fulvius sequence, and all Micrurus sequences were very similar to the Gloydius brevicaudus sequence (BAG82599.1; Yuko Ogawa and Ryohei Yanoshita, 2007, unpublished). Among micrurine APNs, that of M. surinamensis was the most abundant at 0.01% of its transcriptome (Table S2).
2.2.7. C-Type Lectin-like Proteins (CTLs)
Fry and Wüster [88] proposed that C-type lectin-like proteins were recruited into venoms twice. Galactose-binding lectins are widespread in venoms and appear to have been basal to the Colubroidea tree. However, heterotetrameric toxins that lack mannose-binding capacity [89,90], seem to have been a strictly viperid invention. Viperid CTLs are all multimeric proteins composed of heterodimers that act upon platelets [91]. Some of them inhibit platelet aggregation, while others agglutinate, or aggregate and activate platelets. Viperid CTLs often bind to more than one type of platelet receptor [91]. However, with the discovery of bungarine, micrurine, and acanthophiine lectins [92,93], it is clear that recruitment of C-type lectin-like proteins is more complicated than Fry and Wüster suggested.
CTLs have been reported previously in M. corallinus, M. fulvius, and M. tener venoms [1,2,94], although there have been no pharmacological characterizations to date, nor any assessment of structural variability within this toxin family. Micrurine CTL transcripts encode proteins of 149–150 amino acids, with putative signal peptides of 20 residues (Figure S7). These New World elapid CTLs exhibit tremendous sequence variation, and they may comprise 8–10 structural subclasses. CTLs from Micrurus venoms may have 6, 7, 8, or 9 Cys residues, while viperid CTLs normally have 8. This situation raises the possibility of heterodimeric toxins, similar to those of viperid venoms, but these toxins could also be complexed with a non-CTL moiety, or the extra Cys could simply be free. Regardless, the Cys patterns suggest a minimum of 3–4 structural subclasses, though whether they differ pharmacologically cannot be surmised at present. Nine toxins also bear C-terminal extensions of 2–6 amino acids (Figure S7). However, of the 40 sequences presented here, 31 bear an EPN sequence (residues 113–115) that is characteristic of mannose binding lectins [95] (Figure S7). In addition, three micrurine CTLs bear a KPS sequence in these positions. Five possess an ATS sequence, and one M. l. lemniscatus toxin has a QPD sequence, indicating that the latter may be a galactose-binding lectin (GBL) [95]. Nonetheless, its Cys pattern is different from those of putative micrurine GBLs, discussed below. Among our samples, CTLs ranged in abundance from trace quantities in M. s. spixii to 1.87% in M. paraensis and 3.35% in M. corallinus. These significant quantities suggest a CTL role of some importance, at least in the latter two species.
2.2.8. Cysteine-Rich Secretory Proteins (CRISPs)
CRISPs are apparently not abundant components of any snake venoms, but they are widely distributed taxonomically [96]. Interestingly, they have also apparently been recruited into all varanid lizard venoms, in which they constitute the second most abundant venom constituent (Ivan Koludarov, personal communication). The CRISPs, ablomin (Gloydius blomhoffii), triflin (Protobothrops flavoviridis), and latisemin (Laticauda semifasciata) are L-type Ca2+ channel antagonists of depolarization-induced arterial smooth muscle contraction [97]; thus, they promote vasodilation and hypotension. Patagonin, a CRISP isolated from the venom of the colubrid, Philodryas patagoniensis, damaged murine skeletal muscle [98].
All six Micrurus transcriptomes contained CRISP sequences of 238–239 residues (Figure S8). The sequences from M. l. carvalhoi, M. l. lemniscatus, and M. surinamensis are identical over the first 201 residues, at which point they terminate. We do not know whether they are complete or truncated. The sequences of M. corallinus and M. s. spixii differ somewhat from the three foregoing, but that of M. paraensis is radically different, being more similar to the CRISP from Ovophis okinavensis venom [10]. Micrurine CRISPs range from trace levels in most species to 0.24% of the M. l. lemniscatus transcriptome (Table S2).
2.2.9. Cysteine-Rich Secretory Proteins with EGF Domains (CRISP-EGF)
All of the Micrurus transcriptomes possessed from two to five CRISP-EGF sequences. These are very minor venom components, ranging from trace quantities to a maximum of 0.13% in M. surinamensis venom. Snake venom CRISP-EGF transcripts apparently encode proteins of 362 amino acids with 28-residue signal peptides. Structurally they appear to comprise three subclasses, one of which includes CRISP-EGF transcripts from Micrurus fulvius and from crotalid venoms and colubrid tissues [1,10,99] (Figure S9).
The second and third subclasses consist only of micrurine sequences from the present study. The second subclass is most closely related to an uncharacterized protein from the Protobothrops mucrosquamatus genome (XP_015675368) and a CRISP with an EGF domain from the Thamnophis sirtalis genome (XP_013930246.1) (Figure S9). Both of these latter proteins exceed 960 amino acids, whereas the micrurine sequences appear to contain fewer than 400 residues, although we cannot be absolutely certain of this since we did not isolate a stop codon. However, all of the subgroup 2 sequences terminate with the same EIGCV sequence, which is very unlikely if they are all incomplete.
Unfortunately, the third subclass is represented only by two pairs of incomplete sequences. The first pair (M. surinamensis DN51727_c0_g2_i1_m.16805 and M. paraensis DN13321_c0_g1_i1|m.25776), is most closely related to Protobothrops flavoviridis fibulin-1-like protein (XP_015686848.1; 97% identity over the 90-residue M. surinamensis sequence). The second pair of sequences (M. s. spixii DN99876_c0_g3_i1|m.54981 and M. surinamensis DN81846_c0_g1_i1|m.38354) is most closely related to Protobothrops mucrosquamatus fibulin-1 (XP_015670949.1; 92% identity over the 94-residue M. surinamensis sequence) (Figure S9). All four of these are expressed at essentially trace levels (<0.0002%), so they may be contaminating tissue proteins. Human fibulins include Cys-rich, Ca2+-binding, extracellular matrix and plasma glycoproteins of 566-683 amino acids [100]. Epidermal growth factor [101] and extracellular matrix proteins containing EGF domains [102] possess a six-Cys motif that is repeated nine times in fibulins [101]. According to Tran et al. [103], fibulin-1 is the major fibulin in human blood, occurring at a concentration of 30–40 μg/mL, 1000× the concentration of fibulin-2. Tran et al. found that this 340-kDa polypeptide consistently co-purified with fibrinogen. Fibulin-1 is incorporated into clots in vitro and has been detected in thrombi in vivo. In the presence of fibrinogen, platelets bind to human fibulin-1 [104]. Accordingly, there is a conceivable role for fibulin-like proteins in envenomation.
The aggregating proteoglycans (aggrecan, versican, neurocan, and brevican) are important components of many extracellular matrices [105]. Their N-terminal globular domains bind hyaluronan and their C-termini contain C-type lectin domains. In the presence of Ca2+, the lectin domains of aggrecan and versican bind to the Ca2+-binding EGF repeats of fibulin-1, with KDs in the low nM range [105]. Versican is highly expressed in arteries, veins, and capillaries [106]. Perhaps, given the capacity of fibulin-1 to bind fibrinogen and platelets, Micrurus venom fibulin homologs promote clot formation, their smaller size notwithstanding. Possibly, fibulin-1-like proteins complement the actions of prey serine proteases. Eagle [106] found that a mixture of unspecified Micrurus venoms was capable of weakly clotting fresh, citrated horse plasma, due to apparent prothrombin activation, and not to direct cleavage of fibrinogen. Crotalid thrombin-like enzymes promote formation of small clots that stimulate prey anticoagulation cascades to destroy the clots, with the result that prey blood is quickly rendered incoagulable. However, serine proteases are minor components of micrurine venoms, and to date, there is no evidence that they are thrombin-like. It also remains to be seen whether micrurine fibulin-1-like proteins bind and activate platelets. Perhaps, given that they are ~40% smaller than human fibulin-1, in concert with prey serine proteases, they promote defective clot formation so as to render the prey’s blood incoagulable, but given their expression levels and their probable non-enzymatic character, they probably contribute little to envenomation.
2.2.10. Cystatin (CYST)
Cystatins have not been reported previously as snake venom constituents and we make no claim that they are. Cystatins participate in cell survival, proliferation, and differentiation, cell signaling, and immunomodulation [107], roles that do not seem as pertinent to envenomation as their well-known capacity to inhibit cysteine proteases. However, they are present in the Micrurus venoms examined here in only trace quantities (0–0.02% of the transcriptomes). These transcripts encode 136-residue proteins with 24-residue signal peptides and three cysteine residues; thus, the mature proteins are slightly smaller than phospholipases A2. Mammalian cystatin families 1 and 2 contain proteins of 100–120 amino acid residues. Those in family 1 are synthesized without signal peptides or disulfide bonds, and are normally intracellular. Those in family 2 have signal peptides and disulfide bonds, and are secreted proteins [108].
We isolated cystatin transcripts from all six species (Figure S10); however, no cystatin sequences have been reported from any other Micrurus species to date. The six cystatin sequences presented here are largely invariant, owing to their low expression levels. There are only 7 variable positions in the 112-residue proteins. Their cysteine residues occur in the same positions as do mammalian Family 2 inhibitors, with which they share only 45–55% sequence similarity. However, it seems probable that these are secreted proteins, as venom proteins have to be, unless they are secreted in microsomes [109].
2.2.11. Dipeptidyl Peptidase IV (DPP IV)
DPP IV was first discovered in snake venoms by Silva Jr. and Aird [110] in a study of coralsnake venom lethality. High levels of DPP IV are found in blood capillaries [111,112], suggesting that its function is to control hypertension, because of its ability to catabolize Substance P [113], neuropeptide Y, and peptide YY [114,115]. Aird [46] offered a hypothetical explanation for the presence of DPP IV in venoms, suggesting that its role was to counteract a hypertensive response on the part of envenomated prey by destroying hypertensive peptidyl hormones released to offset venom-induced hypotension.
Silva Jr. and Aird [110] found DPP-IV activity in all venoms assayed (16 Micrurus, Bungarus multicinctus, Naja naja, and Bothrops moojeni), except for those of M. albicinctus and M. l. carvalhoi. It was subsequently found in a broad taxonomic array of venoms [116] and DPP IV transcripts have now been reported in various crotalid, viperid, and elapid venoms [10,11,117,118]. All six transcriptomes contained DPP IV transcripts, and ironically, given its generally low enzyme titers, M. surinamensis had two. One of these contained a 26-residue insert not seen in any other DPP IV transcript reported to date (Figure S11). This variant is remarkable given the generally low variability seen in snake venom DPP IVs. Even crotalid DPP IVs vary relatively little from micrurine sequences; however, DPP IV is a trace level component in all six venoms examined here.
2.2.12. Galactose-Binding Lectins (GBL)
Galactose-binding lectins, a subset of C-type lectins, were first reported by Gartner et al. [119] from the venom of Bothrops jararaca. These Ca2+-dependent lectins induce platelet aggregation [9,120] and are potent mitogens [121,122]. In all previous studies, GBLs have been found at very low levels [120], and their known sequences display very little sequence variation [10]. Accordingly, Aird et al. [10] postulated that given their mitogenic properties, the primary function of GBLs may be to upregulate venom synthesis in the venom gland when venom supplies become depleted. However, with the present findings, that view may have to be revised somewhat. While several species in the present study also have very low GBL titers (0–0.19%) (Table S2), M. paraensis (1.93%) and M. corallinus (3.35%) have by far the highest levels reported to date, suggesting that in these two species at least, GBL hemagglutinating and edematogenic properties [122] may play a significant role in envenomation.
Moreover, the collection of GBL sequences yielded by this study presents a picture of greater sequence diversity than hitherto seen (Figure S12). Micrurus GBLs comprise two groups, each having 158 residues, including a predicted 23-residue signal peptide. One group commences with an unusual CCC sequence shared with various Australian elapids, while the other group has a YTC sequence. The former group has 10 Cys residues while the latter group has 8, presumably arranged in disulfide bonds. While the two groups are generally similar, displaying the same invariant and variable positions, several other positions appear to differ consistently. At position 33 (Figure S12), all but one of the CCC toxins have proline, while all of the YTC toxins have serine. At position 58, the CCC toxins have phenylalanine, histidine or leucine, in order of abundance, whereas the YTC toxins all have histidine. Lastly, at position 81, all the CCC toxins have serine or threonine, whereas all the YTC toxins have serine or phenylalanine (Figure S12). The significance of these differences, if any, is unknown.
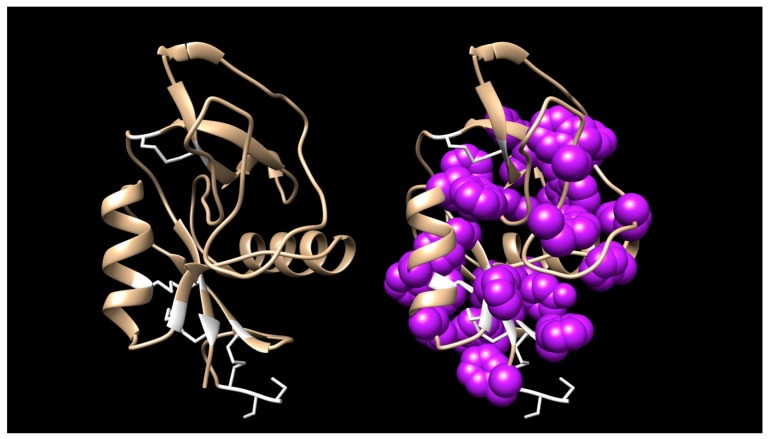
Of particular interest is the large number of invariant aromatic residues, including W7, Y15, W24, Y55, Y59, W67, W79, W81, F/Y89, W92, Y115, W118, and F/Y129 (numbering as per Figure S12). In addition, 3 other positions appear overwhelming aromatic with only occasional non-aromatic substitutions at F13, F18, F/Y30, and F/Y135. Many GBLs also have additional aromatic residues that do not appear in all homologs, e.g., F52(I,L), Y57/S, F/Y77(R/G), Y/F100(L/S), W101(Q/N/K/R), W110(V/L), F112(L/V/A/N/S), and F127(R/N) (Figure S12). While phenylalanine and tyrosine are interchangeable at several of these apparently obligatory aromatic positions, tryptophan is never substituted, suggesting structural, as well as chemical roles. We are unaware of any other toxin family with such a high aromatic amino acid content, an attribute that fairly well begs for a structural/functional investigation (Figure 8).
Figure 8.
The galactose-binding lectin from Micrurus corallinus venom (DN79899_c0_g1_i1|m30755) showing the molecule’s secondary structure (left) and the same model with the aromatic residues shown in a space-filling representation. Aromatic residues tend to be clustered in the core, but most tyrosines have their hydroxyl groups exposed on the surface and several of the tryptophans are also partially exposed. The functional reason for this extremely high aromatic content is unknown. The model was made with SWISS-MODEL using a GBL from Protobothrops mucrosquamatus venom as a template. The model was then further refined and energy minimization was performed using Chimera [21].
2.2.13. Guanylyl Cyclase (QC)
Guanylyl cyclase is believed to serve only the function of cyclizing N-terminal glutamine residues of specific venom proteins to prevent their rapid degradation in host tissues. The acidic subunit of crotoxin, from the venom of Crotalus durissus terrificus, consists of three chains derived from an acidic phospholipase A2 [123]. The B and C chains are blocked with pyroglutamate [123,124]. Thus, GC is not a snake venom constituent in the usual sense, although it is usually present in trace quantities. All six coralsnake transcriptomes possess identical 368-residue transcripts with putative 23-residue signal peptides that comprise from 0–0.02% of their respective transcriptomes. Even the GC transcript of the habu, Protobothrops flavoviridis, differs from the micrurine GCs at only 5 residues (Figure S13).
2.2.14. Hyaluronidase (HYAL)
Hyaluronidase is generally a minor enzymatic component of snake venoms. Venom hyaluronidase has been deemed a “spreading factor” because its degradation of extracellular matrix enables venom hydrolases and non-enzymatic constituents to attack additional targets [125,126]. As such, hyaluronidase probably serves primarily to digest the prey. HYALs have been recorded to date in 10 Micrurus venoms (Figure S14), including all six of the species we studied. Micrurine hyaluronidases comprise 465 amino acids and lack signal peptides. In our transcriptomes, hyaluronidases accounted for between ~0% (M. s. spixii and M. l. lemniscatus) and 0.27% (M. surinamensis) of the transcriptomes. Consistent with the finding of Aird et al. [11] that the most abundant venom proteins evolve most rapidly, hyaluronidases show strikingly little sequence variation. Most Micrurus species appear to have only one hyaluronidase transcript, but M. l. carvalhoi has three (Figure S14).
2.2.15. Kunitz Serine Protease Inhibitors (KSPI)
Kunitz serine protease inhibitors have been recognized for decades as a family of short, structurally similar toxins (59–61 residues with three disulfides) with diverse functions ranging from non-neurotoxic inhibitors of trypsin or chymotrypsin to neurotoxic ligands of potassium and calcium channels, such as the dendrotoxins [74,127,128,129,130,131]. Dufton [132] noted that relatively few amino acid substitutions are necessary to turn a protease inhibitor into a dendrotoxin. The present study confirms an earlier sequence from M. tener and shows that KSPIs also include a handful of homologous, 232-residue toxins, in several Micrurus venoms (Figure S15). The latter should not be confused with SERPINs, which are nearly twice the size of the long KSPIs.
These long KSPIs have atypical, 20-residue signal peptides commencing with MRREKS (Figures S15 and S16), instead of the usual 24-residue signal peptide commencing with MSSGGL [127]. Long KSPIs have an invariant, acidic N-terminus, ESPPD (Figures S15 and S16). Their N-termini show considerable homology to typical, short KSPIs, with a strongly aromatic (WWY) sequence (residues 48–50) and six cysteines in their usual locations. Four hydrophilic residues (NQNN, NENN, NKNN, or NANN) are found at positions 68–71 (Figures S15 and S16). Typically, after the last cysteine, KSPIs terminate with two or three residues, one of which is aliphatic.
In contrast, the long KSPIs continue with an 11-residue span that is largely aliphatic, mimicking a signal peptide (Figure S16). The next 40 residues contain 6 prolines and two cysteines. The two cysteines are nine residues apart, as is the first pair of cysteines in the short KSPIs. Positions 139–141 show another aromatic triplet (WYF), like that mentioned above. The next two pairs of cysteines are separated by 8 and 5 residues, exactly as their counterparts in the short toxins. The long KSPIs also have a block of hydrophilic residues (positions 159–162: NKNN), mimicking a similar block in their own N-termini and in the short toxins (positions 68–71) (N*NN, in which the asterisk may be Q, R, K, D, E, or A) (Figure S16). Because such a variety of sequence segments align perfectly with the N-termini of the short toxins, it appears that the long KSPIs were formed partly due to a gene duplication, in which a short toxin was duplicated and grafted onto its own C-terminus (Figure 9). There is even some suggestion of a second duplication, with what appears to be another pseudo-signal peptide (residues 109–125); however, in the second putative duplication, sequence homology degrades very significantly (Figure S16).
Figure 9.
Four of the species examined here have sequences that are strikingly similar to the α subunit of the nociceptive toxin (MitTx) from Micrurus tener venom, which even the closely related M. fulvius lacks. Nonetheless, the South American Micrurus toxins have five substitutions (indicated by triangles), four of which are strongly non-synonymous. Signal peptides 24 residues long were essentially invariant (MSSGG LLLLL GLLTL CAELT PVSS). Only M. l. lemniscatus toxin DN110178 had a G/R substitution in position 5.
Because the N-termini of these toxins show great similarity to the short toxins, it seems probable that these long toxins are functional KSPIs; however, their specificity is unknown. It is likely that the long KSPIs do not target them same prey proteins as the short toxins. Their presence in eight Micrurus species may offer a clue as to they prey specificity. On the other hand, it is difficult to argue strongly for any great importance in envenomation in view of their expression levels: M. surinamensis, 0.006% of the transcriptome; M. s. spixii, 0.007%; M. paraensis, 0.0003%. As a percentage of the total KSPI level in these venoms, these toxins represent: M. surinamensis, 0.65% of the transcriptome: M. s. spixii, 1.1%; M. paraensis, 0.04%. If long KSPIs are non-enzymatic, that means they have stoichiometric relationships with their targets. If so, their contribution to envenomation is presumably minor, but that still begs the question of their function, a question that pertains to most snake venom KSPIs. Beyond the six cysteines, they share very few residues with α-dendrotoxin; thus, they probably do not block K+ channels, unless the differences reflect differences in mammalian and reptilian K+ channels.
2.2.16. L-Amino Acid Oxidase (LAO)
The flavin-enzyme, L-amino acid oxidase (LAO), is responsible for the bright yellow color of many solenoglyph venoms, in which LAO content can approach 10% [10]. It is a more minor constituent of Micrurus venoms, ranging from essentially 0% (M. s. spixii venom) to 0.84% (M. corallinus). Snake venom L-amino acid oxidase (EC 1.4.3.2) generates H2O2 from the conversion of L-amino acids to keto acids. While anticoagulant and apoptotic roles have been demonstrated for LAO, Aird [46] argued that LAO’s nearly ubiquitous distribution in snake venoms implies an important role in envenomation and suggested that LAO’s most important function is to promote hypotension by augmenting nitric oxide production via stimulation of soluble guanylate cyclase. Micrurine LAOs are large proteins, with 499 residues and predicted 18-residue signal peptides (Figure S17). They exhibit relatively little sequence variation, except at specific residues, where the same sorts of amino acid substitutions are encountered across species.
2.2.17. Metalloproteases, SVMP Type III (P-III)
Snake venom metalloproteases (SVMPs) are classified according to their domain organization [133,134,135]. P-I SVMPs are the simplest class of enzymes, containing only a metalloproteinase catalytic domain. P-II SVMPs contain a metalloproteinase domain linked to a disintegrin domain. P-III SVMPs are the most structurally complex, containing catalytic, disintegrin-like, and cysteine-rich domains [133]. Many venoms contain multiple metalloprotease forms; however, elapid venoms tend to be depauperate in SVMPs relative to viperid venoms (~70% less than viperid venoms) [136,137]. All P-III SVMPs are assumed to be catalytically active. The majority of these cause hemorrhage, but some specifically inhibit platelet aggregation, cleave von Willebrand factor, or activate prothrombin or Factor X [135].
Micrurus venoms appear to contain predominantly one Type III metalloprotease, comprising 614 residues, including a 20-residue signal peptide (Figure S18). One M. l. lemniscatus transcript (lemniscatus DN101949_c32_g1_i4|m.13925) yields a mature protein with a mass of 66,657 D and a predicted pI of 8.37. Except for other micrurine MPs, this protein is most similar to atragin, from the venom of Naja atra [138]. The metalloprotease content of the venoms we examined varies from essentially 0% in M. s. spixii to 2.1% in M. corallinus. The latter suggests a significant function in envenomation. Nonetheless, the pharmacological and physiological effects provoked by micrurine metalloproteases are unknown.
2.2.18. Natriuretic Peptides (NP)
Atrial and brain natriuretic peptides had been known for several years when C-type natriuretic peptide was discovered in porcine brain by Sudoh et al. [138]. Both atrial and brain natriuretic peptides resemble ribbons, folded across themselves to form a loop, pinned with a single disulfide bond and having two tails [138]. C-type natriuretic peptides terminate with the second cysteine so that they have only a single tail. Nonetheless, their pharmacological spectrum is similar to those of atrial and brain natriuretic peptides [138].
The first snake venom natriuretic peptide was discovered two years later in the venom of the eastern green mamba (Dendroaspis angusticeps) (DNP) [139]. Natriuretic peptides have subsequently been discovered in a variety of elapid and crotalid venoms [95,140,141,142,143].
Natriuretic peptide transcripts were found in all six of the Micrurus species we investigated. They have also been reported from M. fulvius and M. altirostris. Like ANP and BNP, micrurine NPs have 25-residue signal peptides (Figure S19). After removal of the signal peptide, another 61 residues must also be cleaved from the N-terminus, assuming an N-terminal tail length equivalent to that of DNP, bearing an invariant glutamate residue. Since human CNP lacks a C-terminal tail, it is unclear whether these micrurine peptides have tails, and if so, how long they are. One of the M. surinamensis NPs bears what appears to be a premature stop codon in position 134 (Figure S19). That transcript may be a misassembly or a pseudogene, since it also has an extra cysteine not seen in any other transcript. On the other hand, even truncated at that point, it still has 8 residues beyond the C-terminus of DNP.
No other NP transcript bears a stop codon, even though 2 M. l. carvalhoi transcripts are 195 and 267 residues in length. These two transcripts encode one and two additional NPs, respectively, in a manner resembling that of bradykinin-potentiating peptides in crotalid venoms [140,144] (Figure S19). Thus, it is impossible to say with absolute certainty how long these transcripts should be, or the natriuretic peptides they encode. Nonetheless, in M. corallinus and M. s. spixii venoms we repeatedly isolated a peptide -DRIGNVSGMGCNHVRT-, which corresponds to residues 73–89 (Figure S19). Interestingly, this peptide spans the first and second NP sequence repeats.
Immediately upstream from the N-terminal glutamate residue, most NPs display an -GPAK- or -GLAK- sequence. One group of three NPs from venoms of M. l. carvalhoi, M. corallinus, and M. l. lemniscatus, deviates sharply from this pattern, having a -GLDT- sequence. The mature NP sequences are likewise very unusual. Between the cysteines, two of the hydrophilic residues have been replaced with hydrophobic aliphatic residues (Q95L, D98V). The nearly invariant M106 has also been replaced with L. Functionally the effect of these changes is unknown, but these toxins should be much less hydrophilic than most NPs.
2.2.19. Nerve Growth Factor (NGF)
Micrurine NGF transcripts encode 244 amino acids plus 18-residue signal peptides (Figure S20). They are highly similar sequences, displaying numerous blocks of invariant residues. Most of the variable positions show specific sets of substitutions, synonymous and non-synonymous, that occur without regard to species. The only strongly divergent sequences include a set of six transcripts from M. l. lemniscatus, which vary at numerous positions. Another group of four transcripts from M. l. lemniscatus and M. corallinus are also significantly different from all others; however, the significance of these variations, if any, is unknown (Figure S20). NGFs function as arginine esterases, so they probably contribute to venom hypotensive activity via nitric oxide liberation and histamine release [14,15,46,145,146]. Mouse salivary NGFs activate plasminogen, their only known action upon a biologically important, non-neural substrate [147,148], but it is not clear whether snake venom NGFs can also do this. If so, they would hinder blood clotting, consistent with envenomation strategies [46].
2.2.20. Nociceptive Toxins (NOCI)
Bohlen et al. [149] reported a non-covalent, heterodimeric toxin (MitTx) from the venom of M. tener that specifically targets ASIC1 acid-sensing ion channels to induce intense, persistent pain. The toxin consists of a Kunitz serine protease inhibitor homolog and a non-catalytic phospholipase. The latter lacks the essential active site histidine and aspartate residues. Interestingly, M. tener has a series of PLA2 toxins with this same modification.
Four of the six Brazilian coralsnakes we examined (except M. l. carvalhoi and M. paraensis) have sequences that are very similar to MitTxα, the KSPI subunit, but there are no other sequences in the NCBI nr database that provide a match of better than ~52%. Even M. fulvius does not have an equivalent toxin (Figure 9). MitTxα homologs have predicted 24-residue signal peptides, leaving 60-residue mature toxins. The Brazilian sequences differ from the MitTxα sequence at five positions, but only one of the substitutions (Q15R) is reasonably conservative (Figure 9). The others include F13H, D22G, S23A, and F37N. Accordingly, it is impossible to say whether these toxins are actually functional MitTxα homologs.
For MitTxβ, SignalP also predicts a 24-residue signal peptide, resulting in a 125-residue mature protein. All six Brazilian species also possess toxins that differ at only 7 positions from M. tener MitTxβ (Figure 10). These include P2S, R21Q, A24S, V34I, N39K, V60E, and T109A, numbering from the start of the mature protein; however, only the substitutions at positions 21, 34 and 39 are reasonably conservative. Nonetheless, all of the Brazilian Micrurus proteins are also non-catalytic. Like MitTxβ they possess an unusual QAQKQ sequence between the fifth and sixth cysteine residues that is not shared by any other Micrurus PLA2s. Micrurus tener has at least two other PLA2s that are nearly identical to MitTxβ, with conservative substitutions at V27M and Q125R. Without pharmacological studies, it is impossible to say whether all of these toxins are actually nociceptive, like MitTx; however, their common structural similarities and their distinctness from all related toxins, particularly micrurine PLA2s, suggest that they probably are.
Figure 10.
All six Micrurus species examined produced a close homolog of MitTxβ, the nociceptive toxin β subunit from Micrurus tener, including M. paraensis and M. l. carvalhoi, which apparently do not produce the α-subunit. The South American Micrurus sequences are basically identical, and differ from that of MitTxβ at only six positions (indicated by triangles). The three M. tener PLA2 sequences had 21-residue signal peptides while MitTxβ and putative homologs had 24-residue signal peptides. 21-residue signal peptides had the sequence MNPAH LLVLA AVCVS LLGASS. MitTxβ and putative homologs had a three-residue N-terminal extension, MDK.
In summary, it is impossible to say at present whether the South American species examined here possess functional equivalents to MitTx, and if so, which subunit arose first. It seems probable that M. l. carvalhoi and M. paraensis do not have them, given the apparent lack of an MitTxα sequence, but perhaps these species employ another KSPI in this role. Or perhaps these South American MitTx homologs have other functions, as yet undiscovered.
2.2.21. Phosphodiesterase (PDE)
Snake venom phosphodiesterase contributes to prey hypotension by liberating purine nucleotides (especially ATP) from prey nucleic acids. The nucleotides can be subsequently dephosphorylated by venom and prey 5′-nucleotidases to release free nucleosides. Among nucleosides, adenosine plays an especially significant role in envenomation [46]. All of the Micrurus venoms investigated here apparently have a single phosphodiesterase gene (850–852 amino acids), except for M. surinamensis, which, barring a transcript misassembly, may have two (Figure S21). The transcript in question shows an inserted glycine at position 700, and a glutamate residue at position 698, where all other sequences have leucine. Signal P predicts signal peptides of 18 residues, but they are probably actually 22 residues long, given the unbroken 14-residue run of aliphatic amino acids. PDE sequences are highly conserved, given the similarity of Micrurus sequences to Protobothrops sequences (Old World crotalids) (Figure S21). PDEs are expressed at trace levels in five of the Micrurus species and at 0.02% in M. corallinus. Their low variability and low expression levels apparently provide additional support for the principle that the most abundant venom proteins evolve most rapidly [11].
2.2.22. Phospholipases A2 (PLA2)
Overview and Diversity of PLA2s
Kocholaty et al. [150] found that Micrurus fulvius venom exhibited higher PLA2 activity than venoms of several species of Bungarus and Naja, and Ophiophagus hannah. Ramsey et al. [151] fractionated M. fulvius venom on CM-Cellulose and found PLA2 activity in every fraction. Possani et al. [152] fractionated the venom of M. fulvius microgalbineus in the process of purifying a PLA2, the first Micrurus toxin to be isolated and characterized from any species. Their gel filtration profile of the crude venom on Sephadex G-50 and subsequent subfractionation of the main peak by cation exchange chromatography also revealed PLA2 activity in every fraction, suggesting that M. fulvius venom is preponderantly PLA2. This was confirmed when Margres et al. [71] produced the first coralsnake venom gland transcriptome and found that PLA2 transcripts comprised 63.4% of the transcriptome. By proteomic means, Vergara et al. [153] obtained similar results. Not only is M. fulvius venom composed largely of PLA2, but 54 of the 63 complete Micrurus PLA2 sequences in the public domain come from that species. Likewise, the M. nigrocinctus proteome appears to be largely PLA2s (48.0%) [154].
However, venoms of other coralsnakes are not so dominated by PLA2s. Both M. surinamensis (Letícia, Colombia) [12,155] and M. l. carvalhoi venoms [155] show relatively little PLA2 activity. However, activity does not necessarily reflect the amount of PLA2 present. Silva Jr. and Aird [70] reported that in M. f. fulvius venom, the enzymatic activity level was much lower than expected, relative to the amount of PLA2 present. At that time, non-neurotoxic, non-catalytic, myotoxic PLA2s were known [156,157,158], but only later did it become clear just how much variation in catalytic activity exists among catalytic phospholipases [159]. For instance, Kopper et al. [160] reported an 8.9-fold variation in PLA2 activity/mg of venom protein among 13 specimens of M. tener, suggesting that interspecific comparisons based on pooled samples may be more reliable than those based on single specimens.
Micrurus fulvius venom caused direct and irreversible cardiac depression in cats, in addition to respiratory insufficiency, but exerted little effect on conductivity of rat phrenic nerve [161]. Myoglobin was not released despite large amounts of free hemoglobin; hence the authors concluded that M. fulvius PLA2s act like cardiotoxins. Myotoxic, catalytic PLA2s had been known from crotalines for at least 9 years [162,163]; however, myotoxic elapid PLA2s were not reported until 1975 [164]. The first non-catalytic, myotoxic PLA2 would not be discovered until 1984 [156,157,158] in the venom of the pit viper, Agkistrodon piscivorus.
Assays for phospholipase activity depend heavily upon the types of phospholipases present, the phospholipid substrate used, the assay temperature, the organizational state of the phospholipids, the presence or absence of detergents, and other factors [165,166,167]. Aird and Silva Jr. [12] detected PLA2 activity against phosphatidylcholine in venoms of 11 coralsnake taxa. Echoing the earlier finding of Kocholaty [150], all but M. surinamensis venom exhibited higher PLA2 activity than the two outgroup species, Bothrops moojeni and Naja kaouthia; however, such data provide little indication of what types of PLA2 are present. Moreover, as Silva Jr. et al. [70] discovered, in some cases PLA2 activity correlated well with PLA2 content as revealed by chromatographic profiles (M. surinamensis and M. l. lemniscatus, the least and most active samples, respectively), but in other cases it did not. Chromatographic profiles suggested very high PLA2 content in both M. fulvius and Naja kaouthia venoms, but both were relatively inactive in the phosphatidylcholine assay system. Undoubtedly, the activity differences reflect the diversity of pharmacological functions of different PLA2s. de Roodt et al. [168] reported that myotoxicity was present only in Micrurus venoms with the highest PLA2 activity (M. fulvius, M. nigrocinctus, M. pyrrhocryptus), but that all six venoms examined, including additionally M. altirostris, M. baliocoryphus, and M. surinamensis venoms, manifested PLA2 activity. Olamendi-Portugal et al. [17] detected peptides corresponding to PLA2s in M. surinamensis venom, but the pharmacological types could not be determined from the short sequences available.
Because they are not confounded by assay conditions and variations in catalytic rate, etc., transcriptomic studies promise to resolve at least the questions regarding compositional differences between venoms. High-throughput techniques (e.g., Illumina technology) [71] yield much more quantitative data than was possible with earlier molecular biological techniques [169]. Technological advances are occurring at such a rapid rate that current Illumina technology produces roughly 5 − 10x the number of reads as in the Margres study [1], and very soon, the current technology will likely be supplanted by technologies offering vastly longer reads.
Fernández et al. [170] compared Micrurus species for which venom gland transcriptomes are available and reported generalized north-south clines in PLA2 and 3FTx content. They opined that North American species are generally richer in PLA2 and poorer in 3FTxs, while South American species show a reverse trend. However, with the addition of the present data, the suggested clines break down, at least in part because the geographic rule does not consider the influences of either phylogeny or dietary ecology (Figure 11). The venoms examined in this study had PLA2 levels ranging from 15.3–37.7% (Table S2), with three of them below 17.2%. Populations of M. corallinus from southeastern Brazil do not necessarily follow the geographic trend trend suggested by Fernández et al. [170] (Figure 11).
Figure 11.
When Micrurus species for which transcriptomic data are available, are arranged in an approximately northwestern to southeastern sequence, the gradient of high PLA2 and low 3FTx concentrations in the North to high 3FTx and low PLA2 concentration in the South previously suggested by Fernández et al. [170], no longer appears as obvious.
For the sake of simplicity, the discussion below approaches micrurine phospholipases A2 on the basis of biological activity, but the result is more simplistic than simple, because nature disdains such classifications. Many micrurine PLA2s possess more than one biological activity (myotoxicity, cytotoxicity, neurotoxicity, anticoagulation, pro-inflammation, etc.), as has been shown for crotalid neurotoxic PLA2s [171].
Micrurine PLA2s have transcripts that encode 115–120 residues plus 27-residue signal peptides. These PLA2 signal peptides terminate in paired hydroxylated amino acids, usually -SS-, but occasionally -ST- (Figure 12A and Figure S22). All of them possess a Cys residue in position -15, but they exhibit several N-terminal sequence patterns. Most of the signal peptides commence with MNPAH- or MILAH-, although a small number have MLIFLW-. The latter usually have one or two extra cysteines in position -19 or positions -18 and -19. There appears to be little relationship between signal peptide structure and expressed protein structure.
Figure 12.
Structures of 121 new micrurine phospholipases A2, illustrating their great structural diversity. For a comparison of all known micrurine PLA2s, see Figure S22. (A) Of 121 PLA2s with partial or complete structures, 202 are apparently catalytic, having the requisite H48, D49, Y52, and D101 in their active sites. The remaining 42 are apparently non-catalytic. Positions indicated in red across the bottom designate residues involved in Ca2+-binding [172]. Positions indicated in blue are catalytic residues. The anticoagulant site [173] and the neurotoxic hydrophobic region [172,174] are indicated by purple and green bars, respectively. Residues proposed by Alape-Girón et al. [175] to be involved in myotoxicity of elapid PLA2s are indicated below with black cells. Residues participating in the hydrophobic channel, via which substrate enters the catalytic site are indicated in gray [176]. PLA2s from the two North American species, M. fulvius and M. tener, form a structural cluster that is relatively distinctive from the much more variable South American sequences, which probably also represent much greater pharmacological diversity. Numbers of cysteine residues are given for sequences that are complete. Numbers of cysteines in parentheses are for sequences truncated at the N-terminal end, for which the cysteines can be reliably predicted, owing to the relative invariability of the N-termini. Asterisks indicate stop codons.Micrurine venoms include 42 presumably non-catalytic PLA2s that appear to be myotoxic. These comprise four loose subclasses, shown here separated by horizontal lines. The first two groups possess all four catalytic residues, but apparently have a disrupted Ca2+-binding site (D or I in lieu of G30, and R in place of G32). Toxins in the third group have H48, but D49 has been replaced by Y49 or F49, and G30 with N or S. The fourth subclass, comprising 9 PLA2s from all six Brazilian species and M. tener, has replaced H48 with Q and D49 with K. G30 has been replaced with D, I, or V. Thus, it seems improbable that any of these can bind Ca2+; (B) Space-filling models of catalytic residues, Ca2+-binding residues, and residues forming the hydrophobic channels of four micrurine PLA2s. See Part A of this figure for the roles and locations of individual residues. The PLA2 from M. altirostris (upper right) is a catalytic toxin, possessing all of the essential residues. The other three are presumably non-catalytic. In the M. s. spixii toxin (lower right), the catalytic D47 has been replaced with F47. While the usual G28 and G30 are present in the hydrophobic channel (not visible in this projection), a bulky W17 has replaced L17, presumably disrupting the channel. In the M. corallinus toxin in the lower left, the catalytic H46 has been replaced by Q46. K47 has been substituted for D47, replacing a negative charge involved in Ca2+-binding with a positive charge. R30 has replaced G30 in the Ca2+-binding loop and W17 has replaced L17 in the hydrophobic channel. Lastly, in the M. corallinus PLA2 in the upper left, R2 and W17 occlude the hydrophobic channel, while R30 is present in what should have been the Ca2+-binding loop. SWISS-MODEL employed a cardiotoxic PLA2 with a D-loop from Ophiophagus hannah (1gp7.1.A) as a template for M. spixii DN102297. M. altirostris AED89576 was modelled after the subunits of a homotetrameric PLA2 from Micropechis ikaheka (1pow.1.ABCD). The model of M. corallinus DN88788-55704 was based upon the 3D structure of M. tener MitTxβ (4ntw.1.C), while that for M. corallinus DN88788-55706 was modeled after a PLA2 isoform from Naja naja sagittifera (1xxw.1.A). The latter is one subunit of a noncovalent heterodimer.
The present study yielded 121 new micrurine PLA2 sequences, partial and complete (Figure 12A). More significantly, all six Micrurus transcriptomes in this study possessed non-catalytic, presumably monomeric PLA2s, the first reported for elapid venoms. These toxins fall into four structural subgroups, depending upon which of the catalytic and Ca2+-binding residues have been substituted non-synonymously (Figure 12A,B). There were 42 such sequences, including two from M. tener venom.
Most micrurine PLA2s have the expected 14 cysteine residues (Figure 12A); however, because of mutations some have only 13. However, more than a dozen PLA2s have 15 Cys residues. Seven of these have an extra Cys at position 67, a variant structure not reported previously (Figure 12A). In the case of the nigroxins, the extra Cys occurs at position 59 [175]. PLA2 #27 from M. fulvius has a 15th Cys residue at position 102 (Figure 12A), but since no other PLA2 displays this variant, this is assumed just to be an unusual mutation. Whether this extra cysteine has any functional significance is unknown. Three toxins have an extra Cys at position 121. One of these, M. surinamensis DN37829_c0_g1_i1|m.22236, has extra Cys residues at both positions 67 and 121. Since it also has an incomplete C-terminus, we cannot rule out the possibility of a 16th cysteine at position 126. Nonetheless, it is probable that most of the 15-Cys toxins form an intermolecular disulfide bond with either another phospholipase, or with a non-homologous protein. A covalent homodimer seems most likely since the micrurine 15-Cys toxins do not align with the PLA2 subunit of β-bungarotoxin, which has only 13 cysteines. Nonetheless, non-PLA2 toxins with odd numbers of Cys residues, and which therefore could potentially bind covalently to 15-Cys PLA2s, include the long KSPIs (13 Cys)(Figure S16), serine proteases (most have 15)(Figure S25), serpins (33 Cys)(Figure S26), NGFs (7 Cys, some have 8)(Figure S20), MPs (39 Cys)(Figure S18), and CTLs (6, 7, 8, 9 Cys)(Figure S7). Of these, the long KSPIs seem unlikely, both because of their low abundance and their distributions. SPs, serpins, MPs, and NGFs seem unlikely by reason of their large sizes. The most likely candidates would be CTLs with either 7 or 9 cysteines, although such toxins would be a compositional novelty; however, such a toxin class might target and activate blood platelets very effectively.
In the vast majority of PLA2s, catalytic or non-catalytic, C11 is followed by T12 and then 7–8 other hydrophilic and/or basic residues (Figure 12A and Figure S22). However, 34 PLA2s follow T12 with an insert of IPG or MPG. The function of this structural novelty is unknown. Whether it serves to render these PLA2s resistant to degradation by prey proteases, after the manner of crotaline bradykinin-potentiating peptides, or whether it presents key residues to be recognized by molecular targets in prey tissues is unknown. PLA2s of this type are present in all six transcriptomes that we examined, and in the transcriptomes of M. fulvius, M. tener, and M. altirostris (Figure S22; Table S2). Those with the MPG sequence are extremely minor, ranging from 0.005–0.02% of the PLA2 content in their respective transcriptomes. The IPG forms are more abundant, representing from 0.6% of all PLA2s in the M. paraensis transcriptome to 86.2% in M. surinamensis. Our M. corallinus transcriptome did not have an IPG PLA2 (Figure 12A).
Myotoxic PLA2s
Gutiérrez et al. [177] found that six of seven Micrurus venoms they investigated (M. alleni, M. frontalis, M. n. mosquitensis, M. n. nigrocinctus, M. dumerilii carinicauda, M. surinamensis) induced rapid and qualitatively similar myonecrosis. Myonecrosis provoked by M. nigrocinctus venom was especially profound, while M. surinamensis exhibited much less activity. Only M. mipartitus (probably M. multifasciatus) venom was non-myotoxic. Gutiérrez et al. [178] later demonstrated that M. nigrocinctus venom released massive amounts of myoglobin, creatine, and creatine kinase, but caused relatively little hemolysis, unless calcium were present in the media. Whether phospholipolytic or cytotoxic in nature, the observed hypercontraction of myofilaments suggested that at least some micrurine PLA2s are myotoxic and that they destroy muscle largely via massive calcium influx [179], which also poisons mitochondria. The former authors also suggested that cardiotoxin-like molecules are probably also present in M. nigrocinctus venom, a conclusion also reached by Vital Brazil for M. fulvius venom [69]; however, this has yet to be confirmed in any Micrurus venom to date.
Arroyo et al. [180] purified a PLA2 from M. nigrocinctus venom and reported that after intramuscular (i.m.) injection, serum creatine kinase levels spiked within 1.5 h. They concluded that the toxin’s primary target is the plasma membrane. Thereafter, myofilaments were clumped, sarcoplasmic reticulum was disrupted, and some mitochondria were damaged. Goularte et al. [181] proposed that the M. nigrocinctus myotoxin initially binds presynaptically to the neuromuscular junction of mouse phrenic nerve-diaphragm preparations, followed by sub- and postsynaptic effects. They concluded that the latter were the most important cause of the neuromuscular blockade. A catalytic phospholipase A2 from M. s. spixii venom appears to present a similar picture, although there is insufficient information to draw any firm conclusions [182]. Carvalho et al. [183] demonstrated that in cultured rat primary hippocampal cells, two PLA2s isolated from Micrurus l. lemniscatus venom induced cell death, exhibiting elements of apoptosis, autophagy, and necrosis.
Lind and Eaker [184] noted that the Lys-Lys-Lys sequence (positions 100–102) in notexin is also encountered in porcine pancreatic PLA2 and in the myotoxic notechis II-5. They suggested that these residues might be essential for neurotoxicity, myotoxicity, or both. Alape-Girón et al. [175] systematically compared the sequences of the myotoxic nigroxins A and B from M. nigrocinctus venom with elapid PLA2s known to cause myonecrosis upon i.m. injection. These included Naja mossambica CM-I, CM-II, and CM-III [185], Oxyuranus scutellatus OS1, which binds in calcium-independent fashion to the muscle (M-type) PLA2 receptor [186], Notechis scutatus notexin and II-5, [187,188], and compared them with the β-bungarotoxin A chain and two pancreatic PLA2s known not to be myotoxic. They identified a series of amino acid residues that were highly conserved in the myotoxic PLA2s (Arg15, Val97, Ala109, Asn117, and an aromatic residue in position 118, Figure 12A) (residue numbers vary with the sequences compared and the resulting insertion of sequence gaps).
Gutiérrez and Ownby [189] noted that some myotoxic PLA2s exhibit myotoxicity even if injected intravenously, whereas others require i.m. injection to exert this effect. They proposed that the systemic myotoxins are sufficiently specific for muscle that they will find their way to cell membranes of skeletal muscle cells regardless of the route of injection. Lemnitoxin appears to illustrate this principle [190]. Other PLA2s, that are less specific for muscle than for some other target, e.g., neurotoxic PLA2s, bind to the more specific targets first, inducing myotoxicity secondarily, or if injected directly into muscle [189]. However, it should be emphasized that given the myriad structural variations of PLA2s, all of our attempts at classification are overly simplistic and are bound to prove unsatisfactory. Gutiérrez et al. [191] have discussed the complexities of such an undertaking in regard to myotoxicity, and Sampaio et al. [192] have more recently shown that secondary pharmacological effects continue to be discovered even for toxins as well studied as crotoxin. Moreover, in some cases, different PLA2s achieve the same pharmacological effect by different mechanisms [173].
Nuñez et al. [193] showed that a C-terminal 13-amino acid peptide from the K49 myotoxin of Agkistrodon p. piscivorus was able to lyse cultured skeletal muscle cells, indicating that it contained the structural elements necessary for myotoxicity; however, crotalid PLA2s have longer C-termini than micrurine PLA2s [159], so clearly, crotalids and micrurines accomplish myotoxicity by different means [190]. Of the 42 non-catalytic PLA2s, 32 retain H48, but 18 have replaced D49 with Y or F (Figure 12A). The remaining 9 PLA2s have replaced residues 48–50 with QKQ and are found in venoms of all six of the Brazilian species and that of M. tener (Figure 12A). Surprisingly, no non-catalytic sequence has been reported from M. fulvius to date, so far as we are aware. Two other groups of apparently non-catalytic PLA2s have the four catalytic residues, but apparently have a disrupted Ca2+-binding site (residues 30–32) (Figure 12A). At present, it does not appear possible to draw many conclusions regarding structure and function of micrurine PLA2s, owing to the lack of pharmacological studies; however, most micrurine PLA2s display all of the residues identified by Alape-Girón et al. [175] as being essential for myotoxicity (Figure 12A).
Hemorrhagic PLA2s
Francis et al. discovered a novel, toxic Type I phospholipase in the venom of the Australian tiger snake (Notechis scutatus) [194,195]. These toxins cause transient hypotension, but acute organ damage due to hemorrhage; hence, they were denominated “hemorrhagic toxins.” Structurally what makes them unusual is that they retain Helix D, a loop of amino acids characteristic of pancreatic PLA2s, that sits atop the two large parallel α-helices (Figure 13A). This extra loop causes them to migrate on SDS-PAGE with apparent molecular weights of 18–23 kDa. From M. frontalis venom, Francis et al. [196] isolated basic, 21–23 kDa PLA2s that cross-reacted strongly with rabbit polyclonal antibodies raised against the hemorrhagic PLA2 from Notechis s. scutatus venom, and concluded that most or all of the PLA2s in M. frontalis venom are of this type. However, to date, no sequence has been published for any of the M. frontalis hemorrhagic PLA2s. Based on the presence of a Helix D-like structure, a probable hemorrhagic phospholipase sequence from venom of M. fulvius was published in 2013, although the authors did not identify it as such [1]. The present study identified 18 new micrurine hemorrhagic phospholipases in all six Brazilian species. These (and two sequences from M. fulvius and M. tener) show great sequence similarity to Notechis HTe and to human pancreatic PLA2 (Figure 13B). Francis et al. [196] reported that the hemorrhagic phospholipases from M. frontalis venom lacked phospholipase activity, and now we can explain why. Of the 20 hemorrhagic PLA2s reported here, 17 have Tyr50 in the active site instead of Asp50; thus, they cannot bind the Ca2+ ion required for catalysis. The sequences from M. fulvius, M. tener, and M. l. carvalhoi do have Asp50 and probably are catalytic (Figure 13B).
Figure 13.
This study identified 18 putative micrurine hemorrhagic phospholipases (HPLA2s) in all six Brazilian Micrurus species examined. All have an extra D loop (colored residues) that sits atop the two main helices, like human pancreatic PLA2 and Notechis scutatus HT. These D loops all have one of three sequences: KSLLD, KPIWD, or TPILD. (A) A TPILD toxin from M. s. spixii (DN102297_c479_g187_i1|m.10851) modeled after a cardiotoxic PLA2 with a D-loop, from Ophiophagus hannah (1gp7.1.A); (B) All of the HPLA2s are non-catalytic, except for the KSLLD forms from M. l. carvalhoi, M. fulvius, and M. tener, which do have the requisite catalytic and Ca2+-binding residues in the active site, and key residues that form the hydrophobic channel.
The structural requirements for Helix D are not entirely clear, beyond the fact that it comprises five residues. Micrurus hemorrhagic phospholipases (HP) display one of three sequences: KSLLD, KPIWD, or TPILD. The third and fourth positions of HPs appear to always require a hydrophobic amino acid, either aliphatic, aromatic, or methionine, although the latter has only been found in the hemorrhagic toxin (HT) from Notechis [195], to date (Figure 13). Like the human pancreatic PLA2, all Micrurus HPs have aspartate in the fifth position, although Notechis HT has serine. The Brazilian Micrurus HPs all have proline in the second position, except for M. l. carvalhoi, which along with M. fulvius and M. tener, has serine, a strongly non-synonymous substitution. In the first position, most Micrurus HPs have lysine, as in pancreatic PLA2; however, a nearly equal number of toxins have threonine, and HT has asparagine. Thus, it appears that the first position simply requires a hydrophilic residue (Figure 13).
Intravascular hemolysis, manifested by hemoglobinuria and resulting in anemia, has been reported in dogs bitten by M. fulvius [197,198,199]. Arce-Bejarano et al. [200] confirmed that catalytically active PLA2s are responsible for the hemolysis and identified one of the three active fractions as being identical to PLA2 2b reported by Margres et al. [1]. In vitro, dog and mouse erythrocytes were found to be highly susceptible to the PLA2s, while those of rabbits and humans were unaffected. The difference appeared related to a high ratio of phosphatidylcholine/sphingomyelin in erythrocyte plasma membranes. Mechanical stress, after incubation with venom, significantly increased hemolysis, suggesting that in vivo shear stresses associated with circulation probably contribute to cell lysis [200].
Neurotoxic PLA2s
Lambeau et al. [201] suggested that the diversity of pathophysiological effects evoked by snake venom PLA2s probably reflects the occurrence of specific receptors for them. They found that neuronal PLA2 receptors in brain display high affinity for toxic snake venom PLA2s, but not for non-toxic PLA2s. A second type of receptor, the muscle type, binds the hemorrhagic snake venom PLA2s with high affinity, as well as pancreatic and human inflammatory PLA2s, but not the neurotoxic snake venom PLA2s. It is likely that relatively small structural differences may account for these differing affinities.
A possible illustration of this principle is seen in MiDCA1, a presynaptic, monomeric phospholipase A2 from M. dumerillii carinicauda venom that produces a neuromuscular blockade in vertebrate nerve-muscle preparations [202]. MiDCA1 causes a spontaneous release of transmitter at neuromuscular junctions, followed by a depression of transmitter release, a pattern also observed in M. laticollaris venom [203]. The initial increase results from a direct effect of MiDCA1 on presynaptic potassium channels, and neurotoxicity is Ca2+-dependent, indicating that catalytic activity is essential for neurotoxicity [202].
MiDCA1 possesses an intact Ca2+-binding loop and an intact active site (Figure 12 and Figure S22). However, the authors noted that while MiDCA1 possesses a number of highly conserved amino acid residues that are generally considered necessary for myotoxicity (R17, V97, A109, N117, and Y118; numbering after Figure 12 and Figure S22) [175], it shows little myotoxicity. They surmised that its lack of myotoxicity might be due to its relatively low enzymatic activity [202]. However, MiDCA1 has E17 in place of the essential R17. In addition, MiDCA1 has V20 instead of W20 in the hydrophobic channel that is essential to admit the substrate, although L2, I9, and Y75 are present (Figure 12 and Figure S22). It is possible that active site inaccessibility could also contribute to low catalytic activity.
Neurotoxic PLA2s are significant venom components in the venoms of some species. Alapé-Giron et al. [204,205] reported that not only are PLA2s significant components of M. nigrocinctus venom, but that a number of these are antigenically related to notexin, the monomeric, neurotoxic PLA2 from Notechis scutatus venom. Concurrently, Goularte et al. [181,206] reported significant neurotoxicity in M. nigrocinctus venom. Oliveira et al. [207] reported on the neurotoxic effects (behavioral, EEG and histopathologic) of intrahippocampal injection of four neurotoxic PLA2s from M. l. lemniscatus venom. These toxins were convulsive, but not epileptogenic at low doses (2.1 µg/rat). Higher doses (1–2.9 µg/rat) caused massive neurodegeneration of the dorsal hippocampus within 7 days. Vergara et al. [153] reported that PLA2-containing fractions (14 different PLA2 variants) accounted for 58.3% of M. fulvius venom protein. Other species, such as M. pyrrhocryptus, manifest low PLA2 activity, with no myotoxicity or presynaptic neurotoxicity [208]. Bénard-Valle et al. [209] reported a PLA2 with very high catalytic activity, but no discernible toxicity. They suggested that its primary role might be digestion of prey. Unfortunately, these studies presented no sequences that can be linked to the pharmacology they reported.
Kini and Iwanaga [172] studied presynaptic neurotoxic venom PLA2s, which display a bewildering structural diversity, from monomers to heteropentamers. They argued that neurotoxic PLA2s have to be able to interact with membranes, and that a hydrophobic site must be essential. They proposed a region of about 30 amino acids from residues 79–109 (Figure 12 and Figure S22) as the likely binding site. Guanidination or carbamylation of basic residues in Naja nigricollis PLA2 had earlier been shown to reduce toxicity by nearly 90% without affecting catalytic activity [210,211,212]. This suggested that basic residues are probably involved in targeting the PLA2 to the presynaptic terminus.
To explain variable susceptibilities of different tissues to different PLA2s, Kini and Evans [213] proposed the existence of specific target sites on plasma membranes of target cell types. They suggested that these are recognized by specific pharmacological sites on PLA2s that are complementary to one another in terms of charges, hydrophobicity and van der Waal’s forces. The higher the affinity between the enzyme’s pharmacological site and its target, the more specific is the pharmacological activity of the PLA2. Target sites are usually proteins, and various target proteins have now been identified for different presynaptic PLA2 toxins. Binding constants for PLA2-target protein interactions are 4–6 orders of magnitude stronger than those for PLA2-lipid interactions [214].
Comparisons with notexin show that levels of acidic, basic, and hydrophobic residues in this region are generally similar among micrurine PLA2s, although many of the latter have one or two additional basic residues at the C-terminus, and their exact positioning differs somewhat from those of notexin (Figure 12 and Figure S22). This may reflect the preference of most Micrurus for reptilian prey, but these toxins may also be active against other vertebrate motor nerve terminals as well.
Anticoagulant PLA2s
Salazar et al. [215] reported that both M. isozonus and M. tener venoms inhibit ADP-induced platelet aggregation and concluded that inhibition could be due to the presence of disintegrins, ADPases, or to proteolytic digestion of ADP receptors. However, a more likely explanation is that inhibition was caused by Type I PLA2s, which are well known from elapid venoms [216,217,218,219,220,221], and that are able to inhibit aggregation induced by ADP, collagen, and epinephrine. Depending upon the species and the PLA2 in question, this inhibition may involve catalytic or non-catalytic mechanisms.
While antiplatelet activity is anticoagulant in a broad sense, antiplatelet activities should not be confused with PLA2-mediated “anticoagulant” activity, which refers to inhibition of the external tenase and/or prothrombinase complexes [173]. According to Kini, the anticoagulant structural region embraces residues 56–79 in Type I PLA2s (Figure 12, Figure 14 and Figure S22). Strongly anticoagulant enzymes possess more basic residues in this region, while weakly anticoagulant enzymes possess more neutral or acidic residues. Naja nigricollis CM-IV, a strong anticoagulant, binds coagulation factor FXa and prevents formation of the prothrombinase complex [173]. While there have not yet been comparable studies of anticoagulant pharmacologies of micrurine PLA2s, it seems likely that like CM-IV, some of the catalytic PLA2s will also inhibit the prothrombinase complex by a mixture of enzymatic and non-enzymatic mechanisms.
Figure 14.
Weakly anticoagulant PLA2s differ from strongly anticoagulant enzymes, such as CM-IV, at five amino acid residues, when tested against mammalian blood. Micrurus PLA2s lack the residues in CM-IV that mimic coagulation Factor Va light chain and tissue factor [176]. The vast majority of micrurine PLA2s have 4–6 basic residues in the anticoagulant site, whereas Naja nigricollis CM-IV has only three. In positions 55–60, where CM-IV has only a solitary Lys, most Micrurus PLA2s have four His, Lys, or Arg residues (positions 57–61 in Figure 12 and Figure S22). It is impossible to predict what effect this might have on specific binding to vertebrate coagulation factors, but this may be a modification for reptiles or fish. Many Micrurus PLA2s are probably at least weakly anticoagulant.
Kini and Evans [222] concluded that weakly anticoagulant PLA2s differ from strongly anticoagulant enzymes, such as CM-IV, at five amino acid residues. Micrurine position numbers, in parentheses, correspond to those in Figure 14.
negatively charged Glu-54 (53) is replaced by neutral residues;
positively charged Lys-55 (54) is replaced by negatively charged Glu;
uncharged Gly-57 (56) is replaced by negatively charged Glu;
positively charged Lys-75 (69) is replaced by Ser or Thr;
positively charged Lys-77 (71) is replaced by negatively charged Glu or Asp
The Micrurus PLA2s do not offer simple comparisons.
Half of micrurine PLA2s (49.7%) have Asp in position 54 (position 53 in Figure 14), like CM-IV. Gly or Ala occupy this position in 37.8%, while 9.8% have a hydroxylated amino acid, and 2.6% have Asn. See Figure 12, Figure 14, and Figure S22 for all PLA2s;
No micrurine PLA2s have Lys in position 53 (position 54 in Figure 14). All have Glu (41.8%), Gln (8.9%), Asp (6.8%), or Ala (13.1%), Thr (29.5%), like the weak anticoagulants;
Most Micrurus PLA2s (53.2%) have Glu in position 57 instead of Gly (Figure 14). 30% have Lys; 4.2% have Ser, and 0.4% have Tyr. An additional 11% have an aliphatic/hydrophobic residue (Ile, Ala, Met, or Val);
No micrurine PLA2s have Lys in position 75 (position 69 in Figure 14). Overwhelmingly they have Ser (80.4%). An additional 14.2% have Thr and 5.4% have Ile;
Only 5% of micrurine PLA2s have Lys in position 77 (71 in Figure 14), whereas 50% have Asp. Other substitutions include Thr (27.7%), Tyr (4.0%), Glu (10.4%), and Asn (3.0%).
Micrurus PLA2s appear to lack residues in CM-IV that mimic coagulation Factor Va light chain and tissue factor [173]. If this were not sufficiently complicated, the vast majority of micrurine PLA2s have 4–6 basic residues in the anticoagulant site, whereas Naja nigricollis CM-IV has only three (Figure 12 and Figure 14). In fact, in positions 54–60, where CM-IV has only a solitary Lys, most Micrurus PLA2s have four His, Lys, or Arg residues (Figure 14; positions 57–61 in Figure 12A). It is impossible to predict what effect this might have on specific binding to vertebrate coagulation factors, but perhaps this is a modification for reptilian prey. Many Micrurus PLA2s may be weakly anticoagulant against mammalian platelets, but they should be tested against fish or reptile platelets before any conclusions are drawn.
Pro-Inflammatory PLA2s
Some micrurine PLA2s also exhibit pro-inflammatory activity. Intraplantar injection of lemnitoxin (M. l. lemniscatus) in rats induced dose- and time-dependent edema of rapid onset [195]. It was detectable after only 5 min and remained undiminished for 6 h, disappearing after 24 h. Doses of 100–500 µg of lemnitoxin provoked substantial degranulation of mast cells, and the release of histamine and serotonin resulted increased vascular permeability [190].
2.2.23. Phospholipase B (PLB)
First discovered in the venom of the Australian elapid, Pseudechis porphyriacus, by Doery and Pearson [223], phospholipase B (PLB) apparently contributes to generalized hydrolysis of cell membrane lipids. A single PLB gene is present in venom glands of all six species examined and in those of M. fulvius as well (Figure S23). They range from 547–551 amino acids in length and mRNA transcripts lack a signal peptide. Six invariant Cys residues suggest three disulfide bonds; however, in a Trp-rich segment (positions 18–24) the M. l. lemniscatus and M. s. spixii toxins have an extra Cys, while the M. l. carvalhoi and M. surinamensis toxins have two more (Figure S23). Beyond this segment, the PLBs show remarkably little structural variation. They are not major toxins, ranging in abundance from 0.01% in the venom of M. l. carvalhoi to 0.59% in that of M. corallinus.
2.2.24. Ribonuclease (RNAse A)
RNAse is essentially absent in all of the transcriptomes, except for that of M. surinamensis, where it comprises 0.7% of the toxin transcriptome (Figure S24; Table S2). Micrurine RNAse transcripts have 141 residues and apparently lack signal peptides. The sequences are largely invariant, showing amino acid substitutions, all non-synonymous, at only 3 positions (V/D60; R/G63; F/S85) (Figure S24). Oddly, 3 of our 6 M. l. carvalhoi transcripts and both transcripts from M. l. lemniscatus had a stop codon at position 118 rather than the usual position 142. As with nearly everything else in Micrurus venoms, the functional significance of this truncation is unknown. The loss of the C-terminal 24 residues suggests that that these have become pseudogenes, since the next 17 residues after the stop codon are the same as in full-length RNAse transcripts (Figure S24). However, we cannot exclude the possibility that these apparently dysfunctional enzymes have adopted another function instead.
2.2.25. Serine Proteases (SP)
A single serine protease transcript of 265 residues was found in each of the six Micrurus venoms examined in the present study (Figure S25). The signal peptide accounts for the first 18 residues. These align well with the transcript published from M. tener venom. Two additional serine protease transcripts from the transcriptome of M. fulvius do not appear to be closely related to the other seven. Given the great similarity of the micrurine serine proteases presented here, the M. fulvius sequences may represent contaminating blood or tissue serine proteases. When the M. paraensis sequence was used as a query string, the most similar sequence was a trypsin-like serine protease from Hydrophis hardwickii venom [224]. Harobin is anti-coagulant, and has both fibrinolytic and fibrinogenolytic activities. It exhibits a preference for the Bβ-chain of fibrinogen, but eventually cleaves the Aα-chain and the γ-chain after prolonged incubation. In vitro, harobin is hypotensive, cleaving high molecular weight kininogen so as to release bradykinin, and also angiotensin-2 [224]. While the presence of this serine protease is consistent with snake envenomation strategies elaborated by [46], it is a very minor component in all venoms examined (≤0.1%), and it probably contributes relatively little to envenomation sequelae.
2.2.26. Serine Protease Inhibitors (Serpins)
Serpins constitute a protein superfamily, comprising 16 clades [225]. While named for their ability to inhibit serine proteases of the chymotrypsin family, some are also capable of inhibiting members of other protease families, while others lack inhibitory activity altogether [225]. Micrurine serpins consist of 501-504 residues plus a 19-residue signal peptide; thus, they are roughly twice the size of the proteases they inhibit. They are approximately 6x longer than KSPIs (Figure S26).
In Micrurus venom glands, these transcripts are almost invariant, showing amino acid substitutions at only about 24 positions. Interestingly, while protease inhibitors generally have 1:1 stoichiometry with their targets, serpins are able to irreversibly disrupt protease structure [226], detach, and move to another target molecule to repeat the inhibition [227]. If serpins truly are venom constituents, they are extremely minor, ranging from 0–0.04% of the transcriptomes examined in this study. In various Micrurus venoms, KSPIs are much more abundant than serpins, but if they can function in a pseudo-enzymatic manner, absolute concentration would be less significant.
To the best of our knowledge, no studies of snake venom serpins have been undertaken, but serpins are constituents of parasitoid wasp venoms and the saliva of ticks and mosquitos [228]. Their employment by hematophagous arthropods probably holds the key to their presence in snake venoms. Serpins isolated from saliva of Aedes mosquitos inhibit Factor Xa by an atypical mechanism involving reversible inhibition with a 1:1 stoichiometry [229,230]. Ticks rely primarily upon serpins to render host blood incoagulable. A serpin from the saliva of Ixodes ricinus, called Iris, acts as an anticoagulant, by interfering with platelet aggregation and by delaying fibrinolysis [231]. A second serpin, Irs-2, is capable of inhibiting thrombin and Cathepsin G [232]. Other tick salivary serpins have similar activity profiles [233,234,235]. We suggest, therefore, that the most probable role for snake venom serpins is anticoagulation.
2.2.27. Vascular Endothelial Growth Factors (VEGF)
VEGFs are widely distributed in snake venoms, and comprise long (VEGF-C) and short VEGF (VEGF-F) subclasses. VEGF-Cs consistently have mRNA transcripts comprising 421 residues, with a signal peptide of 16 residues. Seven micrurine VEGF-C sequences have been determined to date, including six from this study. Interestingly, they are essentially structurally invariant, with the only deviation being a conservative I to F substitution at position 97 (Figure S27). No VEGF-C sequence has been reported from Micrurus tener, which does have a VEGF-F. VEGF-Cs, known primarily for their role in lymphangiogenesis [236], have not been reported previously as venom components, and the low expression levels seen here (0–0.02% of the venom gland transcriptomes) do not argue that they are; however, their capacity to induce increased vascular permeability [237] could predispose them to co-option for that purpose.
VEGF-Fs (211–235 residues) are structurally more complex, with at least three major structural variants, although at individual amino acid positions, like the VEGF-Cs, they are extremely conservative. Variable positions include either E or Q at position 5, G or D at position 12, S, P, or A at position 110, and T or I at position 113 (Figure S28). Following an invariant E116, strange things begin to happen. Position 117 may be N or K, and in transcripts from M. corallinus, M. fulvius, and M. surinamensis, it is missing altogether (Figure S28). Toxins with K at this position then have an extremely basic, 24-residue insert that is absent in the N117 or -117 toxins (Figure S28). These toxins have 14 K or R residues from positions 118–137. All sequences are invariant from residues 142–185. Position 185 is R in all known sequences, except for the M. corallinus transcript mentioned above, in which K is present. The majority of VEGF-Fs have a hydrophilic 25-residue C-terminal sequence, commencing with a short hydrophobic segment, YLHLL; however, three toxins terminate with a short, basic sequence, CEKPRR.
2.2.28. Vespryns (VESP)
Pung et al. [238] isolated a novel 12-kDa toxin from the venom of the king cobra that acts centrally to induce hypolocomotion and pain in mammalian prey. Named ohanin, it became the first snake venom vespryn. Additional vespryns have subsequently been sequenced from various elapids [239] and crotalids [2,10,240,241], and from the viperid, Echis coloratus [242]. Crotalid vespryns are 28–32 residues longer at their N-termini than the elapid toxins.
All six Brazilian Micrurus venoms possess a single vespryn sequence, except for the venoms of M. paraensis and M. l. carvalhoi, in which there were two (Figure S29). Their sequences are nearly identical to that of ohanin. Micrurine vespryn mRNA transcripts comprise 190 residues and lack signal peptides. No vespryns have been reported from M. fulvius venom, although a search of the NCBI nr database produced two sequences in M. fulvius that share several 3–4-residue sequence segments with the Ophiophagus toxin (NCBI GAEP01000222.1 and GAEP01001746.1). Nonetheless, their sequence similarities to other micrurine vespryns are so limited that functional identity seems unlikely. Interestingly, the closely related M. tener does have a legitimate vespryn sequence. Vespryn transcripts have also been reported in the venom gland transcriptomes of M. altirostris, where they comprised 0.2% of the transcriptome [95]. Moreover, vespryn peptides have been sequenced from M. nigrocinctus venom, comprising 3.8% of the proteome [154]. Vespryns appear at only trace levels in most of the venoms examined here, although they comprised 0.22% of the M. s. spixii transcriptome and 1.66% in M. corallinus.
2.2.29. Waprins (WAPR)
Waprins have been reported to have anti-bacterial [243] and protease inhibitory activity [244]. It is unclear whether either of these activities is germane to envenomation, although a waprin-like sequence fused to a KSPI domain was identified in the venom gland transcriptome of Sistrurus catenatus edwardsi [245]. This suggests that protease-inhibitory activity may be the key. Waprin-like sequences are present in all six Micrurus venoms examined in this study, but they have not been reported in venoms of M. fulvius [1], M. tschudii [246], M. clarki [247], M. alleni, or M. mosquitensis [170]. Micrurine waprin-like sequences are more similar to the 135-residue colubrid opisthoglyph and crotalid waprin-like sequences than to those of Old World elapids (78–80 residues) (Figure S30). Colubrid and crotalid waprin-like proteins have 23-residue signal peptides; however, none of our sequences have complete signal peptides and the sequence from M. tener is so different at the N-terminus that no definite conclusions can be drawn regarding the lengths of other micrurine toxins. However, it appears that all micrurine waprin-like sequences have an N-terminal Glu, as in the colubrid and crotalid sequences (Figure S30). Waprin-like toxins are minor components of micrurine venoms, ranging from essentially 0 to 0.44% in M. l. lemniscatus venom.
2.2.30. Phylogenetic Conclusions
We used all venom and tissue protein transcripts (4650 protein families) with the Ophiophagus hannah genome as an outgroup to estimate relationships among the Micrurus species examined here (Figure 15). The data suggest that these six taxa diverged from one another 15–35 million years ago, and from the last common ancestor with Old World elapines about 55 million years ago. Given this early divergence, we expect to find numerous cryptic species among the micrurines. It is apparent that M. corallinus and M. paraensis are closely related (Figure 15). This is unsurprising, given that both are monad-banded species. Nor is the close relationship between M. l. lemniscatus and M. l. carvalhoi unexpected, given their former subspecific status [248,249]. Micrurus surinamensis is more closely related to M. l. lemniscatus and M. l. carvalhoi than to the monad-banded species and M. s. spixii. Most interesting is the distant relationship of M. s. spixii to the other five species (Figure 15). Slowinski et al. [249] proposed that M. s. spixii is more closely related to the triad-banded species of the M. frontalis group, from the cerrado of central Brazil. While these data do not suggest whether it is closer to M. corallinus and M. paraensis or to M. l. lemniscatus, M. l. carvalhoi, and M. surinamensis, the latter seems far more likely based upon morphological data and it will be interesting to see whether high-throughput transcriptomes from other species will eventually confirm this supposition.
Figure 15.
A phylogenetic tree based upon all venom and tissue proteins (4650 protein families) in the six transcriptomes suggests that these Micrurus species diverged 15–35 million years ago. The New World elapids split from the last common ancestor with Old World elapines nearly 55 million years ago. The tree indicates close relationships between two monad-banded species, M. corallinus and M. paraensis, and between M. l. lemniscatus and M. l. carvalhoi. Micrurus surinamensis is closely allied to M. l. lemniscatus and M. l. carvalhoi. Micrurus s. spixii is not particularly close to any of the other species, supporting the assertion of Slowinski [249] that it is more closely related to the triad-banded species of the M. frontalis complex in the Brazilian cerrado. The Ophiophagus hannah genome [250] provided data for the outgroup to root the tree. Blue bars indicate the 95% confidence intervals for the nodes. Taxonomic abbreviations: para (M. paraensis), cora (M. corallinus), suri (M. surinamensis), carv (M. l. carvalhoi), lemn (M. l. lemniscatus), spix (M. s. spixii), and hann (O. hannah).
Micrurine transcriptomes developed during the present study were compared with those of three Old World ophiophagus species, Bungarus flaviceps, Bungarus multicinctus, and Calliophis bivirgata and with the genome of Ophiophagus hannah [250,251,252,253]. The former two transcriptomes were developed without the use of next-generation sequencing technology. As a result, they lack resolution with regard to minor venom components. Nonetheless, comparisons were instructive (Figure 16). Contrary to our expectations, venoms of different snake-eating taxa shared few attributes (Figure 16A). In all of these venoms, 3FTxs were major components. PLA2s are major components in M. fulvius and Calliophis, but relatively minor components in the other three Old World species (Figure 16A). Both kraits possess β-bungarotoxin, a krait invention, but the other three lack it. Metalloproteases are significant constituents of Calliophis and Ophiophagus venoms, but minor in M. fulvius and missing in the kraits (Figure 16A). Cytotoxins/cardiotoxins, generally considered a cobra specialty, are absent in Ophiophagus venom. The minor toxin classes present an even more varied picture, if that be possible (Figure 16B).
Figure 16.
Transcriptomes of Old World elapids most closely related to Micrurus show highly divergent venomes in terms of both major (A) and minor (B) venom constituents. 3FTxs are major components of all of these venoms; however, phospholipase content varies significantly. Phospholipases A2 are a much more significant proportion of Calliophis and Micrurus venoms than of Bungarus or Ophiophagus venoms, even when β-bungarotoxin is considered. Venoms of Bungarus multicinctus and Bungarus flaviceps show significant quantities of β-bungarotoxin, although with strongly unequal quantities of the α and β subunits. Ophiophagus hannah and Calliophisbivirgata venoms possess large quantities of metalloproteases which are absent in kraits and minor components in most coralsnake venoms. Minor constituents are even more diverse.
3. Conclusions
The present study approximately triples the amount of New World coralsnake venom protein data in the public domain. Despite the fact that all of these taxa eat snakes and amphisbaenians (M. surinamensis and M. l. lemniscatus also take fish) the six venoms profiled here vary dramatically in composition, indicating that there is more than one way to efficiently kill the same type of prey organism. Coralsnakes experiment continually with venom composition, both qualitatively and quantitatively. Micrurines may well prove to have the most diversified venoms of any snakes, offering a treasure trove of ligands with novel and potentially useful pharmacologies and these six venomes differ dramatically. In each, 2–6 toxin classes account for 91–99% of toxin transcripts. Other toxin families are present in all six venoms at trace levels (<0.005%). In addition to 3FTxs and PLA2s, all venoms also contain nociceptive toxin (MitTx) subunit β, phospholipase B, and vascular endothelial growth factors. Minor components (0.1–2.0%) are found in all venoms except that of M. s. spixii. The most abundant venom components after 3FTxs and PLA2s differ from venom to venom. Numerous novel toxin chemistries include 3FTxs with previously unknown 8- and 10-cysteine arrangements, suggesting new 3D structures and target specificities. 9-cysteine toxins raise the possibility of covalent, homodimeric 3FTxs or heterodimeric toxins with unknown subunit compositions and pharmacologies.
4. Materials and Methods
4.1. Collection of Snakes and Venom Samples
All specimens except for that of M. corallinus, from Rio de Janeiro, Brazil, were captured in the municipality of Altamira, Pará, Brazil (Table S3). Prior to venom extraction, snakes were kept at low temperature (±4 °C) to reduce their physical activity. Individual venom samples were obtained by placing each fang into a microcapillary tube (70 µL). Glands were manually massaged and then the venom was transferred to individual Eppendorf tubes (2.0 mL) and stored in liquid nitrogen until lyophilization.
Given the tremendous geographic range of M. surinamensis, venom samples from a second specimen captured at Altamira and from two other specimens collected at Estreito, Maranhão, Brazil, were subjected to proteomic comparisons only in an effort to assess intraspecific variation in this species.
4.2. Collection of Tissue Samples
Four specimens of each species from the same locality were used for sample collection. Tissues were collected immediately after administration of a lethal dose of B-Euthanasia (sodium pentobarbital). Samples of visceral organs (heart, liver, kidneys, testes, and skeletal muscle) were extracted, transferred to individual cryotubes (2.0 mL), and immediately frozen in liquid nitrogen. Two aliquots of each tissue sample were also preserved in 100% ethanol until use for DNA amplification and sequencing. Venom glands were extracted under a stereoscopic microscope to clear all attached muscle tissue. Glands were then subdivided into three or four smaller pieces and transferred immediately to individual cryotubes with RNAlater (Qiagen, Venlo, The Netherlands). They were maintained at 4 °C until RNA extraction and analyses. Specimens were collected under permit from the Brazilian Institute of the Environment and Renewable Natural Resources (Instituto Brazileiro do Meio Ambiente e dos Recursos Naturais Renováveis) (IBAMA), 02001.001848/2006-75, under ACCTMB 473/2014 and 647/2015.
4.3. Transcriptomics
4.3.1. Removal of Venom Glands
Specimens were anesthetized with sodium pentobarbital injected into the dorsolateral musculature. They were considered fully anesthetized when all three of the following conditions were met: lack of righting reflex, lack of withdrawal reflex when the tail was tapped repeatedly with a finger, lack of response when the venter was stroked with a finger. The supralabial scales and skin were separated from the underlying tissues with a scalpel. Then the venom duct was severed with a fine scissors just behind the maxilla. Using a scissors, the gland and underlying muscle were cut away, placed in a pre-labeled cryotube containing RNAlater® (Sigma®, St. Louis, MO, USA), (1 mL for up to 100 mg tissue). Tissue samples were maintained in RNAlater up to three days at 37 °C, or up to one week at 25 °C, and then transferred to a −80 °C freezer until RNA extraction.
4.3.2. Isolation of Total mRNA from Venom Glands and Muscle
RNA was isolated using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Venom gland samples (50–100 mg) were suspended in 1 mL of Trizol reagent, incubated on ice for 10 min, and then homogenized with a TissueRuptor (Qiagen). Probes used for homogenization had been previously treated with DEPC (Diethylpyrocarbonate—Sigma®). For phase separation, 0.2 mL of chloroform were added per mL of Trizol. Samples were shaken vigorously by hand for 15 s and incubated for 15 min on ice. Samples were then centrifuged at 12,000× g for 15 min at 4 °C. Each sample was separated into a lower red phenol-chloroform phase, an interphase, and a colorless, upper aqueous phase. Aqueous phases were carefully removed by angling the tubes at 45° and pipetting the aqueous solutions into new tubes. RNA was isolated by adding 0.5 mL of 100% isopropanol to each collected aqueous solution. Samples were vortexed gently, incubated at room temperature for 10 min, and then centrifuged at 12,000× g for 10 min at 4 °C. Supernatants were discarded, leaving gel-like or white pellets that contained the RNA. Pellets were washed with 1 mL of 75% ethanol per sample and then centrifuged at 7500× g for 5 min at 4 °C. Washes were discarded and pellets were air dried for 5–10 min. Pellets were subsequently suspended in RNase-free water. RNA quantification was carried out with a Thermo Scientific NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the 260/280 nm absorbance ratio was used to assess RNA purity. A ratio of ~2.0 was generally accepted for RNA. RNA sample integrity was also evaluated using denaturant agarose gel electrophoresis. Qiagen RNeasy Micro Kits (Qiagen, Venlo, The Netherlands) were used to clean and concentrate RNA samples. Lyophilized samples were shipped from Brazil to OIST Graduate University for further processing.
4.3.3. mRNA Isolation and First and Second Strand cDNA Synthesis
With very minor modifications mRNA purification and cDNA synthesis essentially followed the protocols in Aird et al. [11]. Detailed methods are elaborated in the Supplementary Information and RIN numbers are provided in (Supplementary Table S3).
4.3.4. Library Sequencing
Size selection was performed on amplified libraries using 12.0/12.5% double solid phase reversible immobilization [254] and 300- to 700-bp fragments were selected. Libraries were pooled so that all contained the same amount of cDNA. Pooled libraries were then size-selected by gel extraction on a Minelute column (Qiagen) to achieve an optimal insert size for the Illumina Hiseq PE150 (~500 bp). Size distribution of the library was confirmed using an Agilent 2100 BioAnalyzer (Agilent Technologies, Waldbronn, Germany), and library concentration was determined using qPCR. Pooled and sized libraries were sent to Beijing Genomics Institute (Shenzhen, China) for sequencing.
4.4. Proteomics
Protein reduction and carboxyamidomethylation methods and protein cleavage techniques are found in the Supplementary Materials and Methods.
4.4.1. Protein Sequencing by Liquid Chromatography-Mass Spectrometry
All samples were analyzed using a Thermo Scientific Q-Exactive Plus Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The mass spectrometer was equipped with an HPLC (Dionex Ultimate 3000 nanoRSLC), an autosampler (HTC PAL, CTC Analytics) and a nanoelectrospray ion source. Each sample was injected in a volume of 5 μL, separated on a Zorbax 300SB C18 column (0.3 × 150 mm; Agilent, Agilent Technologies, Waldbronn, Germany) at 40 °C. A one-hour gradient was employed (1%B to 32%B in 45 min, 32%B to 45%B in 15 min, with a final wash at 75%B for 5 min and reequilibration at 1%B for 10 min.). Solvent A was 2% acetonitrile in 0.1% formic acid, and solvent B was 98% acetonitrile and 0.1% formic acid. A flow rate of 3.0 μL/min was used for peptide separation. The temperature of the heated capillary was 300 °C, and 1.9 kV spray voltage was applied to all samples. Mass spectrometer settings were: full MS scan range 350 to 1500 m/z, with a mass resolution of 70,000,30 μs scan time, and AGC set to 1 × e6 ions, and fragmentation MS2 of the 20 most intense ions.
4.4.2. Protein Identification
Thermo RAW data was directly analyzed on Mascot (version 2.4, Thermo Fisher Scientific, Waltham, MA, USA) and Proteome Discoverer (version 1.4, using Sequest HT, Thermo Fisher Scientific, Waltham, MA, USA). Search parameters for both algorithms were: a maximum of two missed cleavages, with precursor and fragment mass tolerance set to 20 ppm and 0.01 Da, respectively. Carboxyamidomethylation of cysteine was set as a fixed modification, while methionine oxidation, asparagine and glutamine deamidation, and N-terminal acetylation were set as variable modifications for tryptic and chymotryptic digests. Asparagine and glutamine deamidation and formylation of peptide N-termini were set as variable modifications for formic acid digests. Reagents used for sequencing (trypsin, R and K; chymotrypsin, F, L, W, and Y; formic acid, D and M) were specified in each case. For relative quantification, we used the peak areas of the three most abundant, unique peptides per protein.
A database of our transcript data was constructed using TransDecoder [255], with default parameters and PFAM database lookup. These relaxed settings were chosen to avoid the loss of small, naturally occurring peptides in the venoms. The common Repository of Adventitious Proteins—cRAP [256] was merged with this constructed database for searching.
4.4.3. Sequence and Structural Analyses
Sequences were aligned and compared using Geneious v 8.1.8 [257]. Signal peptides were predicted using SignalP 4.0 [258]. Three-dimensional molecular models were created using SWISS-MODEL [259], which matches a novel amino acid sequence to homologous sequences for which solved crystal or NMR structures are available. Overhanging N- or C-terminal residues not modeled by SWISS-MODEL were added with UCSF Chimera, with which disulfide bonds were formed and energy minimizations were executed [21].
4.5. Bioinformatics
Transcriptome Assembly and Quantitation
Sequencing was performed by BGI (Shenzhen, China) on a HiSeq 4000 in PE 250 mode, with adaptor filtering as part of the service. Libraries had 48.3 ± 7.9 S.D. million read pairs comprising 14.5 ± 2.4 S.D. Reference transcriptomes were assembled using Trinity (2.1.1) [260] with default parameters after trimming with Trimmomatic, with the SLIDINGWINDOW:4:30 MINLEN:65 parameters and Nextera adaptor sequences [261]. Reads were then re-mapped to the assembly using the RSEM (v1.2.13) pipeline, with bowtie2 as the mapper [262,263], to determine Fragments per kilobase mapped (FPKM).
4.6. Phylogenetics
In order to generate a phylogenetic tree, predicted proteins were first grouped using OrthoMCL v2.0.9 [264] with default parameters percentMatchCutoff = 50; evalueExponentCutoff = −5 in the config file. Clusters with fewer than 4 sequences were discarded, yielding 4560 families. For each cluster, only one sequence was retained from each species, and duplicates were removed. Translated proteins (using bioPython v.1.64 [265] were aligned with Muscle v3.8.31 [266]. Finally, a phylogenetic tree was constructed using RAxML v.7.3.5 [267], running 100 bootstrap replicates: raxml -T 24 -m PROTGAMMAWAG -s {input.phy} -n outRaxml -x 12345 -p 12345 -q {input.partitions} -# 100 -f a. Divergence times were acquired via MEGA7 RelTime [268], using a Poisson model: megacc -a reltime_ml_protein.mao -d concat.fasta -t micrurus.nexus -g groups.txt -c Calibration.txt -o timeTree_Calibration. Some parts of the code were written with Snakemake [269] in order to automate it. All code is available on a github repository [270].
Acknowledgments
We thank Okinawa Institute of Science and Technology Graduate University for its generous support of the Ecology and Evolution Unit.
Supplementary Materials
The following are available online at www.mdpi.com/2072-6651/9/6/187/s1.
Table S1. Transcriptome sequencing statistics.
Table S2. Venom protein composition for six Brazilian Micrurus species.
Table S3. Specimen collection data.
Figure S1. SDS-DTT 4–12% gradient gel (Bis-Tris pH 8.3) confirmed that the abundance of 3FTx in Micrurus l. carvalhoi venom suggested by transcriptomics (72.6%) (Table S2) is likely accurate, while the abundance suggested by mass spectrometry (2.3%) (Table 2) cannot possibly be correct. (A) The gradient gel itself is shown. Molecular weight markers are shown in the left and right lanes. Venom was loaded in the three center wells: 62.5 µg (left); 125 µg (middle); 187.5 µg (right). Fraction numbers are provided to the right of the most concentrated venom sample. (B) Proteins identified in the fractions, after in-gel digestion with trypsin, followed by mass spectrometry. Fraction numbers correspond to those in panel A. 3FTxs were found in Fractions 10–16, where they were significant components. 3FTxs were also found in Fractions 3–6.
Figure S2. Micrurus venoms are rich in 3FTxs displaying an astonishing variety of primary structures. Micrurine 3FTxs may possess 8, 9, or 10 Cys residues with different disulfide patterns in each group. Pharmacologies are almost entirely unknown. Some probably target nicotinic acetylcholine receptors of reptilian neuromuscular junctions, but their potential targets in mammals are unknown. Most 3FTx have 21-residue signal peptides, shown to the left of the black vertical line; however, Group 28 is unique in that it has 24-residue signal peptides.
Figure S3. 5′-Nucleotidases dephosphorylate purine mononucleotides so as to release free nucleosides (principally adenosine), which contribute to prey hypotension [46]. All eight Micrurus species examined to date have 5′-nucleotidases in their venoms. These are present at levels ranging from ~0–0.05% (Table S2). With 573–588 residues and no signal peptides, micrurine venom 5NUCs are slightly shorter than their crotalid counterparts. They bear two different C-termini, one 14 residues longer than the other. Interestingly, the longer variant also appears in 5NUCs of the pitvipers, Protobothrops elegans and Protobothrops flavoviridis [11], and Gloydius brevicaudus [271]. Micrurine 5NUCS are structurally almost invariant, in addition to their close identity with the crotalid enzymes.
Figure S4. Micrurine acetylcholinesterases (AChEs) are modestly large proteins, containing approximately 585 amino acids. However, they are not expressed at biologically significant levels in Micrurus venoms, as they are in some Old World elapid taxa. The primary function of AChE in the venoms of those elapids that produce it, is presumably to starve neuromuscular junctions of transmitter, in concert with postsynaptic antagonists of nicotinic AChRs. Among the Micrurus transcriptomes, AChE is most abundant in M. l. lemniscatus venom, where AChE transcripts total only 0.05% of the transcriptome (Table S2). Micrurine AChEs are highly variable in primary structure. Some, such as M. s. spixii transcript DN97541_c0_g1_i1|m.57626, are predicted to have 29-residue signal peptides, while others, such as M. tener AChE (JAS05178.1), are expected to have signal peptides of only 23 residues, with shorter N-termini.
Figure S5. Aminopeptidase A (APA) converts kallidin to bradykinin [82,87], a potent inducer of hypotension. All of the transcriptomes except that of M. surinamensis had an APA sequence. The Micrurus sequences comprise 958 residues and are largely identical to the APA from Gloydius brevicaudus [87]. Micrurus s. spixii had the highest APA transcript abundance, but that constituted only 0.09% of the transcriptome (Table S2).
Figure S6. Like APA, Aminopeptidase N (APN) also converts kallidin to bradykinin [82,87]. All of our transcriptomes except for that of M. paraensis contained an APN transcript, as did that of M. fulvius. All were also incomplete, but were nearly identical to the M. fulvius sequence insofar as comparisons could be made. Micrurus sequences were also very similar to the Gloydius brevicaudus sequence (BAG82599.1; Yuko Ogawa and Ryohei Yanoshita, 2007, unpublished). Among micrurine APNs, that of M. surinamensis was the most abundant at 0.01% of its transcriptome (Table S2).
Figure S7. New World elapid C-type lectin-like proteins (CTLs) exhibit tremendous sequence variation, and may comprise as many as 8-10 structural subclasses. Micrurine CTLs may have 6, 7, 8, or 9 Cys residues, and the 7-Cys toxins present two different patterns, raising the possibility of heterodimeric CTLs. However, there have been no pharmacological characterizations of any Micrurus CTLs to date. Among our samples, CTLs ranged in abundance from trace quantities in M. s. spixii to 1.87% in M. paraensis and 3.35% in M. corallinus. These significant quantities suggest a role of some importance, at least in the latter two species. Signal peptide sequences are shown to the left of the heavy black vertical line.
Figure S8. Various crotalid CRISPs act as L-type Ca2+ channel antagonists of depolarization-induced arterial smooth muscle contraction [97]; thus, they promote vasodilation and hypotension. All six Micrurus transcriptomes contained CRISP sequences of 221 residues plus 19-residue signal peptides (left of black line). The sequences from M. l. carvalhoi, M. l. lemniscatus, and M. surinamensis are identical over the first 201 residues, at which point they terminate. We do not know whether they are complete or truncated. The sequences of M. corallinus and M. s. spixii differ somewhat from the three foregoing, but that of M. paraensis is radically different, being more similar to the CRISP from Ovophis okinavensis venom [10]. Micrurine CRISPs range from trace levels in most species to 0.24% of the M. l. lemniscatus transcriptome (Table S2).
Figure S9. Micrurine CRISP-EGFs appear to comprise three protein subclasses, one of which includes CRISP-EGF transcripts from Micrurus fulvius and crotalid venoms and from colubrid tissues. This first subclass has 28-residue signal peptides. The second and third subclasses consist only of micrurine sequences from the present study. The second subclass is most closely related to an uncharacterized protein from the Protobothrops mucrosquamatus genome and a CRISP with an EGF domain from the Thamnophis sirtalis genome. Both of these proteins exceed 960 amino acids. The third subclass is represented only by two pairs of incomplete sequences, both of which appear related to fibulins. All four of these are expressed at trace levels (<0.0002%); thus, they may be contaminating tissue proteins.
Figure S10. Cystatins have not been reported previously as snake venom constituents and we make no claim that they are. However, they are present in the Micrurus venoms examined here, although in only trace quantities (0–0.02% of the transcriptomes). Micrurine cystatins possess 24-residue signal peptides and have their Cys residues in the same positions as mammalian cystatins, with which they share only 45-55% of their amino acids. However, it appears that they probably are secreted proteins.
Figure S11. All six Micrurus transcriptomes contained a DPP IV transcript and that of M. surinamensis contained two. DPP IVs comprise 752–779 amino acids, but because they are membrane-bound in microsomes [117], they have no signal peptides. One sequence from M. surinamensis has a 27-residue insert that is lacking in the other homologs.
Figure S12. Aird et al. [10] postulated that the primary function of galactose-binding lectins (GBLs) may be to upregulate venom synthesis in the venom gland when venom supplies become depleted. However, with the present findings, that view may have to be revised somewhat. While several species in the present study also have typically low GBL titers (0–0.19%)(Table S2), M. paraensis (1.93%) and M. corallinus (3.35%) have the highest levels reported in any snake venom to date, suggesting that in these two species, GBL hemagglutinating and edematogenic properties [122] may play a significant role in envenomation. Signal peptides are shown to the left of the vertical black line.
Figure S13. Guanylyl cyclase (GC) is believed to serve only the function of cyclizing N-terminal glutamine residues of specific venom proteins to prevent their rapid degradation in the host. For this reason, GC is not a snake venom constituent in the usual sense, although it is usually present in trace quantities. All six coral snake transcriptomes possessed identical 368-residue transcripts with a putative 23-residue signal peptide (left of black vertical line) that comprised from 0–0.02% of their respective transcriptomes. Even the GC transcript of the habu, Protobothrops flavoviridis, differed from the micrurine GCs at only 5 residues.
Figure S14. Hyaluronidases are generally minor enzymatic components of snake venoms. They have been recorded to date in 10 Micrurus venoms, including all six species that we studied. Micrurine hyaluronidases comprise 465 amino acids and lack signal peptides. In our transcriptomes, hyaluronidases accounted for between ~0% (M. s. spixii and M. l. lemniscatus) and 0.27% (M. surinamensis) of the transcriptomes. Consistent with the finding of Aird et al. (2015) that the most abundant venom proteins evolve most rapidly, hyaluronidases show strikingly little sequence variation. Most Micrurus species apparently have only one hyaluronidase transcript, but oddly, M. l. carvalhoi had three.
Figure S15. In addition to typical elapid Kunitz serine protease inhibitors (short KSPIs, 59–61 residues), some Micrurus venoms contain novel KSPIs (long KSPIs) 232 residues in length. The physiological targets of both classes are unknown. The long KSPIs should not be confused with SERPINs, which are nearly twice again as long (Figure S26). Signal peptides of either 20 or 24 residues are shown to the left of the black vertical line.
Figure S16. Four Micrurus species possess long KSPIs (232 residues) that appear to have arisen through gene duplication of a short KSPI, with the duplicated portion being grafted onto the C-terminus of a short KSPI. The long KSPIs have 13 Cys residues, but the disulfide pattern is unknown; however, the second group of six cysteines has the same spacing as the first six (See also Figure S15). In this figure, the C-terminal half of the sequences has been cut and re-aligned with the N-terminal half to illustrate the Cys pattern match.
Figure S17. The flavin-enzyme, L-amino acid oxidase (LAO), is responsible for the bright yellow color of many solenoglyph venoms, in which LAO content can approach 10% [10]. Snake venom L-amino acid oxidase (EC 1.4.3.2) generates H2O2 from the conversion of L-amino acids to keto acids. It is a more minor constituent of Micrurus venoms, ranging from essentially 0% (M. s. spixii) to 0.84% (M. corallinus). Micrurine LAOs exhibit relatively little sequence variation, except at specific residues, where the same types of amino acid substitutions are encountered across species.
Figure S18. Most Micrurus venoms appear to contain only one Type III metalloprotease, comprising 614 residues, including a 20-residue signal peptide (left of the vertical black line). One M. l. lemniscatus transcript (lemniscatus DN101949_c32_g1_i4|m.13925) yields a mature protein with a mass of 66,657 D and a predicted pI of 8.37. This protein is most similar to atragin, from the venom of Naja atra [137]. The metalloprotease content of the venoms we examined varies from essentially 0% in M. s. spixii to 2.1% in M. corallinus. The latter suggests a significant function in envenomation. Nonetheless, the pharmacological and physiological effects provoked by micrurine metalloproteases are unknown.
Figure S19. Natriuretic peptide transcripts were found in all six of the Micrurus species we investigated. They have also been reported in venom gland transcriptomes of M. fulvius and M. altirostris as well. Interestingly, two of our M. l. carvalhoi transcripts show evidence of gene duplication. One of them contains at least three copies of an NP. The sequence of human CNP is shown across the bottom for comparison. NP transcripts have 25-residue signal peptides (left of the vertical black line), but we cannot draw any conclusions regarding the total length of the transcripts. Solid brown lines separate the repeats.
Figure S20. Micrurine nerve growth factor (NGF) transcripts encode 227 amino acids plus 18-residue signal peptides. They are highly similar sequences, displaying numerous blocks of invariant residues. Most variable positions show specific sets of substitutions, synonymous and non-synonymous, that occur without regard to species. The only strongly divergent sequences include a set of six transcripts from M. l. lemniscatus, which vary at numerous positions. Another group of four transcripts from M. l. lemniscatus and M. corallinus are also significantly different from all others; however, the significance of these variations, if any, is unknown.
Figure S21. All of the Micrurus venoms investigated here apparently have a single phosphodiesterase gene, except for M. surinamensis, which barring a transcript misassembly, may have two. The transcript in question shows an inserted glycine at position 700, and a glutamate residue at position 698, where all other sequences have leucine. Signal P predicts signal peptides of 18 residues, but they are probably actually 22 residues long, given the unbroken 14-residue run of aliphatic amino acids. PDE sequences are highly conserved, given the similarity of Micrurus sequences to Protobothrops sequences (Old World crotalids). PDEs are expressed at trace levels in five of the Micrurus species and at 0.02% in M. corallinus.
Figure S22. Structures of micrurine phospholipases A2, illustrating their great structural diversity. Of 244 PLA2s with partial or complete structures, 202 are apparently catalytic, having the requisite H48, D49, Y52, and D101 in their active sites. The remaining 42 are apparently non-catalytic. Positions indicated in red across the bottom designate residues involved in Ca2+-binding [172]. Positions indicated in blue are catalytic residues. The anticoagulant site [173] and the neurotoxic hydrophobic region [172,174] are indicated by purple and green bars, respectively. Residues proposed by Alape-Girón et al. [175] to be involved in myotoxicity of elapid PLA2s are indicated below with black cells. Residues participating in the hydrophobic channel, via which substrate enters the catalytic site are indicated in gray [176]. PLA2s from the two North American species, M. fulvius and M. tener, form a structural cluster that is relatively distinctive from the much more variable South American sequences, which probably also represent much greater pharmacological diversity. SignalP predicted signal peptides of 21 residues; however, elapid PLA2s sequenced by Edman degradation commence with residue 28, confirming that the signal peptides are actually 27 residues in length. Numbers of cysteine residues are given for sequences that are complete. Numbers of cysteines in parentheses are for sequences truncated at the N-terminal end, for which the cysteines can be reliably predicted, owing to the relative invariability of the N-termini. Asterisks indicate stop codons.
Figure S23. A phospholipase B (PLB) transcript was found in each of the six Micrurus species examined. PLB is believed to contribute to envenomation by hydrolyzing membrane phospholipids [224].
Figure S24. RNAse is essentially absent in all of the transcriptomes, except for that of M. surinamensis, where it comprises 0.7% of the transcriptome. Micrurine RNAse transcripts have 141 residues and apparently lack signal peptides. The sequences are largely invariant, showing amino acid substitutions, all non-synonymous, at only 3 positions (V/D60; R/G63; F/S85). Oddly, 3 M. l. carvalhoi transcripts and both transcripts from M. l. lemniscatus have stop codons at position 118. The functional significance of this truncation is unknown. The loss of the C-terminal 24 residues suggests that that these have become pseudogenes, since the next 17 residues after the stop codon are the same as in full-length RNAse transcripts. However, we cannot exclude the possibility that these apparently dysfunctional enzymes have adopted another function instead.
Figure S25. A single serine protease transcript of 265 residues was found in each of the six Micrurus venoms examined in the present study. The signal peptide accounts for the first 18 residues. Except for their signal peptides, these protease sequences align well with the transcripts published from M. tener venom. Two serine protease transcripts from the transcriptome of M. fulvius do not appear closely related to the other seven. Given the great similarity of the micrurine serine proteases presented here, the M. fulvius sequences may represent contaminating blood or tissue serine proteases. The functions of micrurine serine proteases are unknown.
Figure S26. Serpins consist of 501–504 residues plus a 19-residue signal peptide (left of vertical black line). They are approximately 6x longer than KSPIs (Figure S15). In Micrurus venom glands, these transcripts are almost invariant, showing amino acid substitutions at only about 24 positions. Serpins constitute 0–0.04% of the transcriptomes examined in this study. Their probable role in envenomation is to contribute to anticoagulation.
Figure S27. Micrurine long VEGFs (VEGF-Cs) are nearly structurally invariant. VEGF-C mRNA transcripts comprise 399 residues, with a signal peptide of uncertain length. SignalP v4.1 predicts 16-residue signal peptides (dashed vertical line), but this would leave a long, hydrophobic N-terminus. While that is possible, a 24-residue signal peptide would seem more reasonable (solid vertical black line). No VEGF-C has been reported from Micrurus tener venom, which does have a short VEGF.
Figure S28. Short VEGFs (VEGF-Fs) are conservative in amino acid composition, but display at least three major structural variants. Position 142 may be N or K, or missing altogether in one transcript from M. corallinus. Toxins with K142 then have an extremely basic 24-residue insert, with 14 K or R residues from positions 142–162. The majority of VEGF-Fs have a hydrophilic 25-residue C-terminal sequence, commencing with a short hydrophobic segment, YLHLL. However, three toxins terminate with a short hydrophilic sequence, CEKPRR. It is unclear whether sequences 9–13 are truncated or complete sequences. VEGF-Fs were present in all six of our transcriptomes. They have 26-residue signal peptides (vertical black line).
Figure S29. Ohanin, the first known vespryn, which was isolated from the venom of the king cobra (Ophiophagus hannah), produces hypolocomotion and hyperalgesia in mice [239]. Its effects in snakes may be similar, but at present, no pharmacological data are available. All six Brazilian Micrurus venoms possessed single vespryn sequences, except for the venoms of M. paraensis and M. l. carvalhoi, in which there were two. Micrurus vespryns are nearly identical to ohanin and display limited sequence variation. Micrurine vespryn transcripts comprise 190 residues and lack signal peptides. Vespryns appear at trace levels in most of the venoms examined here, although they comprised 0.22% of the M. s. spixii transcriptome and 1.66% in M. corallinus.
Figure S30. Micrurine waprin-like sequences are more similar to opisthoglyph colubrid and crotalid waprin-like sequences than to those of Old World elapids. Waprin-like sequences have been reported from all Micrurus venoms except that of M. fulvius. Colubrid and crotalid waprin-like proteins have 23-residue signal peptides (left of vertical black line). None of our sequences have complete signal peptides and the sequence from M. tener is so different at the N-terminus that no definite conclusions can be drawn regarding it length in Micrurus. However, it appears that all micrurine waprin-like sequences have an N-terminal Q, as in the colubrid and crotalid sequences.
Appendix A
Appendix A.1. First Strand cDNA Synthesis
RNA was isolated using the Trizol method and was purified with Qiagen RNeasy Micro Kits. One μg of total RNA was used for first strand cDNA synthesis. ERCC ExFold RNA Spike-In Mix (Life Technologies, Carlsbad, CA, USA) was included in the cDNA synthesis reaction according to the manufacturer’s protocol. To the RNA and spike-in mix, 1 μL of 10 μM poly T_START oligo.
(5′-AATTGCAGTGGTATCAACGCAGAGCGGCCGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTVN) was added. The 10-μL reaction was incubated at 65 °C for 3 min, and then chilled on ice. To this, a 10-μL mixture, containing 4 μL of 5x first strand synthesis buffer (Invitrogen, Carlsbad, CA, USA), 1 μL of 10 mM dNTP (Promega, Madison, WI, USA), 2 μL of 0.1 M DTT (Invitrogen), 1 μL of 12 μM template-switching RNA oligo. (5′-AAGCAGUGGUAUCAACGCAGAGUACAUGGG), 1 μL RNase inhibitor (Qiagen), and 1 μL Superscript II Reverse Transcriptase (Invitrogen) was added to each sample. Reactions were incubated at 42 °C for 60 min and the enzyme was heat-inactivated at 65 °C for 15 min. 80 μL MilliQ water were added to each cDNA reaction.
Appendix A.2. Second Strand cDNA Synthesis
Second strand cDNA was synthesized with 9 PCR cycles. The 50-μL PCR reaction consisted of 1x Phusion HF buffer (Thermo Scientific, Waltham, MA, USA), 200 μM dNTP (Promega), 0.5 μM START primer (5′-CGCCAGGGTTTTCCCAGTCACGACAATTGCAGTGGTATCAACGCAGA), 0.5 μM TS_long primer (5′-CTTGTAGGTTAAGTGGAGAGCTAACAATTTCACACAGGAAAGCAGTGGTATCAACGC), 0.5 μL of 2 U/μL Phusion DNA polymerase (Thermo Scientific), and 5 μL diluted cDNA. Eight 50-μL PCR reactions were set up for each cDNA sample. PCR was carried out with the following conditions: initial denaturation at 98 °C for 30 s, with 9 cycles of denaturation at 98 °C for 10 s, 68 °C for 6 min, followed by final extension at 72 °C for 10 min. PCR reactions were concentrated with an Amicon Ultra-0.5 column (Millipore, Billerica, MA, USA).
Concentrated PCR products were purified by solid phase reversible immobilization using Dynabeads MyOne Carboxylic Acid (Invitrogen). 18% PEG was used for purification. 1 ng of ds-cDNA was used library preparation and sequencing, using a Nextera XT DNA sample kit (Illumina, San Diego, CA, USA) according to manufacturer’s directions. Twelve cycles of PCR were used for library amplification.
Appendix A.3. Size Selection of the Pooled cDNA Libraries
Libraries were pooled so that all six contained the same amount of cDNA. Pooled libraries were then size-selected by gel extraction to achieve an optimal insert size for the Illumina Hiseq PE150 (~500 bp). Size distribution of the library was confirmed using an Agilent 2100 BioAnalyzer (Figure A1).
Figure A1.
Size selection of pooled cDNA libraries confirmed that they had an average length of ~500 bp.
Appendix A.4. Reduction and Alkylation of Snake Venoms Prior to Enzymatic Digestion
Crude venoms were gently vortexed for several seconds to suspend microsomes. Reduction was accomplished by adding 36 µL 8 M urea, 2 µL venom (125 µg/µL), 2 µL 500 mM DTT in ultrapure water, and 10 µL 500 mM of ammonium bicarbonate to 200 µL PCR tubes. Tubes were incubated at 60 °C for 45 min in an MJ PTC 200 Peltier thermocycler. Then, 4 µL 500 mM sodium iodoacetate were added to each tube and mixed gently. Tubes were incubated at 37 °C for 30 min in the dark. Afterward, an additional 2 µL 500 mM DTT were added to quench the alkylation reaction. Sample desalting was done by pipetting the reaction from the PCR tubes into 10K Nanosep centrifugal filters (Pall Corp., New York, NY, USA) and centrifuging them at 18,407× g for 25 min. Samples were rehydrated with 50 µL of 50 mM ammonium bicarbonate and were again centrifuged to further desalt the samples.
Appendix A.5. Tryptic Hydrolyses
After the second centrifugation, 10 µL 8 M urea were added to the membrane and the membrane surface was wetted with the urea by rotating the tube manually. Then 90 µL 50 mM ammonium bicarbonate were added to the membrane and the filter was vortexed gently to re-suspend desalted proteins. Solutions were pipetted from the filter and placed in clean 200 µL PCR tubes. Finally, 10 µg of trypsin (Pierce 90055) dissolved in 10 µL 1 mM HCl were added to each tube. Tubes were incubated 24 h at 37 °C. Then, samples were pipetted back into their original 10 K Nanosep filters and centrifuged again at 18,407× g for 30 min to collect the de-proteinated filtrate which was frozen at −30 °C until analysis by mass spectrometry.
Appendix A.6. Chymotryptic Hydrolyses
The procedure for chymotrypsin (Pierce 90056) was nearly identical to that for trypsin, except that after wetting the Nanosep filter with 10 µL 8 M urea, 10 µL 50 mM ammonium bicarbonate, 5 µL 200 mM CaCl2, and 65 µL ultrapure water were added to the membrane. All subsequent procedures were identical to those for trypsin.
Appendix A.7. Formic Acid Hydrolyses
Crude venoms were gently vortexed for few seconds. Digestion of proteins was done by mixing 2 µL venom, 4 µL 500 mM DTT in ultrapure water and 94 µL 2% formic acid (Wako 063-04192) in 200 µL PCR tubes. Tubes were incubated for 2 h at 99 °C and then frozen at −30 °C until analysis by mass spectrometry.
Appendix A.8. Digestion and Extraction of Peptides from SDS PAGE Gels
Gel bands were excised in order of decreasing molecular weight. Each fraction was reduced at 56 °C with 10 mM dithiothreitol in 50 mM ammonium bicarbonate for 10 min, and then alkylated with 55 mM iodoacetamide in 50 mM ammonium bicarbonate for 20 min in the dark. Samples were washed with 50 mM ammonium bicarbonate in 50% acetonitrile, followed by acetonitrile. After washing, each fraction was digested with trypsin overnight at 37 °C. Peptides were extracted from excised gel bands using 5% formic acid and 50% acetonitrile in water. After extraction, peptides were concentrated with a centrifugal vacuum concentrator and then resuspended in 0.1% formic acid in water.
Author Contributions
N.J.d.S. oversaw the collection of specimens and identified them V.A.S. and M.P.d.C.T prepared and shipped the RNA samples to OIST. L.Q. made the cDNA libraries. A.V.-B. preformed the mass spectrometric analysis of venompeptides and identified the proteins they represented. M.L.G. and A.S.M. handled all bioinformatic analyses. S.D.A. performed the venom digestions, made the sequence alignments, analyzed the sequences, created the figures, and wrote the Introduction and most of the Results and Discussion. L.Q., A.V.-B., M.L.G., and A.S.M. wrote pertinent sections of the Materials and Methods. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Data Accessibility
Transcriptomic sequences have been deposited in the DNA Databank of Japan (DDBJ) under the project ID: PRJDB5628. BioSample IDs are: SAMD00076719-SAMD00076724, and raw data may be accessed under ID: DRA005678 (DRX083388-DRX083393).
References
- 1.Margres M.J., Aronow K., Loyacano J., Rokyta D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013;14:531. doi: 10.1186/1471-2164-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rokyta D.R., Margres M.J., Calvin K. Post-transcriptional Mechanisms Contribute Little to Phenotypic Variation in Snake Venoms. G3. 2015;5:2375–2382. doi: 10.1534/g3.115.020578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunha O., Nascimento F.P. Ofídios da Amazônia XIV—As espécies de Micrurus, Bothrops, Lachesis e Crotalus do sul do Pará e oeste do Maranhão, incluindo áreas de cerrado deste estado (Ophidia: Elapidae e Viperidae) Bol. Mus. Para. Emílio Goeldi Zool. 1982;112:1–58. [Google Scholar]
- 4.Da Silva N.J., Jr. Novas ocorrências de Micrurus braziliensis Roze, 1967 (Serpentes: Elapidae) em áreas de tensão ambiental no Centro-Oeste Brazileiro. Estudos. 2007;34:931–956. [Google Scholar]
- 5.Rodrigues de Almeida P.C., Costa Prudente A.L., Curcio F.F., Urbano Rodrigues M.T. Biologia e História Natural. In: Silva N.J. Jr., editor. As Cobras Corais do Brasil: Biologia, Taxonomia, Veneno e Envenenamentos. Editora PUC Goiás; Goiânia, Brazil: 2016. [Google Scholar]
- 6.Da Silva N.J., Jr. Capítulo 3: Características Morfológicas das Cobras-Corais do Novo Mundo. In: da Silva N.J. Jr., editor. As Cobras-Corais do Brazil: Biologia, Taxonomia, Venenos e Envenenamentos. Editora PUC Goiás; Goiânia, Brazil: 2016. [Google Scholar]
- 7.Da Silva N.J., Jr., Pessoa H.L.R. Micrurus surinamensis (Surinam Coralsnake) geographic distribution. Herpetol. Rev. 2008;39:372. [Google Scholar]
- 8.Suto K., Yamazaki Y., Morita T., Mizuno H. Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: Insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1. J. Biol. Chem. 2005;280:2126–2131. doi: 10.1074/jbc.M411395200. [DOI] [PubMed] [Google Scholar]
- 9.Gartner T.K., Ogilvie M.L. Isolation and characterization of three Ca2+-dependent beta-galactoside-specific lectins from snake venoms. Biochem. J. 1984;224:301–307. doi: 10.1042/bj2240301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aird S.D., Watanabe Y., Villar-Briones A., Roy M.C., Terada K., Mikheyev A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis) BMC Genom. 2013;14:790. doi: 10.1186/1471-2164-14-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aird S.D., Aggarwal S., Villar-Briones A., Tin M.M.-Y., Terada K., Mikheyev A.S. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genom. 2015;2015:647. doi: 10.1186/s12864-015-1832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aird S.D., da Silva N.J., Jr. Comparative enzymatic composition of Brazilian coral snake (Micrurus) venoms. Comp. Biochem. Physiol. B. 1991;99:287–294. doi: 10.1016/0305-0491(91)90043-D. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Polo J.R., Bomar H., Beck C., Hall K. Nerve growth factor from Crotalus adamanteus snake venom. J. Biol. Chem. 1978;253:6140–6148. [PubMed] [Google Scholar]
- 14.Siigur E., Neuman T., Järve V., Tara A., Siigur J. Isolation and characterization of nerve growth factor from Vipera lebetina (snake) venom. Comp. Biochem. Physiol. B. 1985;81:211–215. doi: 10.1016/0305-0491(85)90185-3. [DOI] [PubMed] [Google Scholar]
- 15.Kostiza T., Meier J. Nerve growth factors from snake venoms: Chemical properties, mode of action and biological significance. Toxicon. 1996;34:787–806. doi: 10.1016/0041-0101(96)00023-2. [DOI] [PubMed] [Google Scholar]
- 16.Silva N.J., Jr., Griffin P.R., Aird S.D. Primary structure of a short postsynaptic neurotoxin from the venom of Micrurus surinamensis surinamensis. Toxicon. 1993;31:108. [Google Scholar]
- 17.Olamendi-Portugal T., Batista C.V.F., Restano-Cassulini R., Pando V., Villa-Hernandez O., Zavaleta-Martínez-Vargas A., Salas-Arruz M.C., Rodríguez de la Vega R.C., Becerril B., Possani L.D. Proteomic analysis of the venom from the fish eating coral snake Micrurus surinamensis: Novel toxins, their function and phylogeny. Proteomics. 2008;8:1919–1932. doi: 10.1002/pmic.200700668. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S.F., Wootton J.C., Gertz E.M., Agarwala R., Morgulis A., Schäffer A.A., Yu Y.-K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samejima Y., Aoki-Tomomatsu Y., Yanagisawa M., Mebs D. Amino acid sequence of two neurotoxins from the venom of the Egyptian black snake (Walterinnesia aegyptia) Toxicon. 1997;35:151–157. doi: 10.1016/S0041-0101(96)00138-9. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Aird S.D., Womble G.C., Yates J.R., 3rd, Griffin P.R. Primary structure of γ-bungarotoxin, a new postsynaptic neurotoxin from venom of Bungarus multicinctus. Toxicon. 1999;37:609–625. doi: 10.1016/S0041-0101(98)00199-8. [DOI] [PubMed] [Google Scholar]
- 23.Chang L.S., Lin J. cDNA sequence analysis of a novel neurotoxin homolog from Taiwan banded krait. Biochem. Mol. Biol. Int. 1997;43:347–354. doi: 10.1080/15216549700204131. [DOI] [PubMed] [Google Scholar]
- 24.Nirthanan S., Gopalakrishnakone P., Gwee M.C.E., Khoo H.E., Kini R.M. Non-conventional toxins from Elapid venoms. Toxicon. 2003;41:397–407. doi: 10.1016/S0041-0101(02)00388-4. [DOI] [PubMed] [Google Scholar]
- 25.Utkin Y.N., Kukhtina V.V., Kryukova E.V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V.I. “Weak toxin” from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001;276:15810–15815. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 26.Rzhevskiĭ D.I., Murashev A.N., Kukhtina V.V., Tsetlin V.I., Utkin I.N. The weak neurotoxin from Naja kaouthia Cobra venom decreases the arterial blood pressure in rats. Bioorg. Khim. 2001;27:221–223. doi: 10.1023/a:1011337522449. [DOI] [PubMed] [Google Scholar]
- 27.Servent D., Winckler-Dietrich V., Hu H.-Y., Kessler P., Drevet P., Bertrand D., Ménez A. Only snake curaremimetic toxins with a fifth disulfide bond have high affinity for the neuronal α7 nicotinic receptor. J. Biol. Chem. 1997;272:24279–24286. doi: 10.1074/jbc.272.39.24279. [DOI] [PubMed] [Google Scholar]
- 28.Rosso J.-P., Schwarz J.R., Diaz-Bustamante M., Céard B., Gutiérrez J.M., Kneussel M., Pongs O., Bosmans F., Bougis P.E. MmTX1 and MmTX2 from coral snake venom potently modulate GABAA receptor activity. Proc. Natl. Acad. Sci. USA. 2015;112:E891–E900. doi: 10.1073/pnas.1415488112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang C.C., Yang H.J., Chiu R.H. The position of disulfide bonds in cobrotoxin. Biochim. Biophys. Acta. 1970;214:355–363. doi: 10.1016/0005-2795(70)90013-9. [DOI] [PubMed] [Google Scholar]
- 30.Endo Y., Sato S., Ishii S., Tamiya N. The disulphide bonds of erabutoxin a, a neurotoxic protein of a sea-snake (Laticauda semifasciata) venom. Biochem. J. 1971;122:463–467. doi: 10.1042/bj1220463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fryklund L., Eaker D. Complete covalent structure of a cardiotoxin from the venom of Naja nigricollis (African black-necked spitting cobra) Biochemistry. 1975;14:2865–2871. doi: 10.1021/bi00684a012. [DOI] [PubMed] [Google Scholar]
- 32.Dörje F., Wess J., Lambrecht G., Tacke R., Mutschler E., Brann M.R. Antagonist binding profiles of five cloned human muscarinic receptor subtypes. J. Pharmacol. Exp. Ther. 1991;256:727–733. [PubMed] [Google Scholar]
- 33.Max S.I., Liang J.S., Potter L.T. Purification and properties of m1-toxin, a specific antagonist of m1 muscarinic receptors. J. Neurosci. 1993;13:4293–4300. doi: 10.1523/JNEUROSCI.13-10-04293.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerusalinsky D., Kornisiuk E., Alfaro P., Quillfeldt J., Alonso M., Verde E.R., Cerveñansky C., Harvey A. Muscarinic toxin selective for m4 receptors impairs memory in the rat. Neuroreport. 1998;9:1407–1411. doi: 10.1097/00001756-199805110-00029. [DOI] [PubMed] [Google Scholar]
- 35.Bradley K.N. Muscarinic toxins from the green mamba. Pharmacol. Ther. 2000;85:87–109. doi: 10.1016/S0163-7258(99)00064-9. [DOI] [PubMed] [Google Scholar]
- 36.Jolkkonen M., Oras A., Toomela T., Karlsson E., Järv J., Åkerman K.E.O. Kinetic evidence for different mechanisms of interaction of black mamba toxins MTα and MTβ with muscarinic receptors. Toxicon. 2001;39:377–382. doi: 10.1016/S0041-0101(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 37.Adem A., Asblom A., Johansson G., Mbugua P.M., Karlsson E. Toxins from the venom of the green mamba Dendroaspis angusticeps that inhibit the binding of quinuclidinyl benzilate to muscarinic acetylcholine receptors. Biochim. Biophys. Acta. 1988;968:340–345. doi: 10.1016/0167-4889(88)90025-0. [DOI] [PubMed] [Google Scholar]
- 38.Ducancel F., Rowan E.G., Cassar E., Harvey A.L., Ménez A., Boulain J.C. Amino acid sequence of a muscarinic toxin deduced from the cDNA nucleotide sequence. Toxicon. 1991;29:516–520. doi: 10.1016/0041-0101(91)90026-N. [DOI] [PubMed] [Google Scholar]
- 39.Karlsson E., Risinger C., Jolkkonen M., Wernstedt C., Adem A. Amino acid sequence of a snake venom toxin that binds to the muscarinic acetylcholine receptor. Toxicon. 1991;29:521–526. doi: 10.1016/0041-0101(91)90027-O. [DOI] [PubMed] [Google Scholar]
- 40.Karlsson E., Jolkkonen M., Satyapan N., Adem A., Kumlin E., Hellman U., Wernstedt C. Protein toxins that bind to muscarinic acetylcholine receptors. Ann. N. Y. Acad. Sci. 1994;710:153–161. doi: 10.1111/j.1749-6632.1994.tb26623.x. [DOI] [PubMed] [Google Scholar]
- 41.Walch L., Brink C., Norel X. The muscarinic receptor subtypes in human blood vessels. Therapie. 2001;56:223–226. [PubMed] [Google Scholar]
- 42.Lamping K.G., Wess J., Cui Y., Nuno D.W., Faraci F.M. Muscarinic (M) receptors in coronary circulation: Gene-Targeted mice define the role of M2 and M3 receptors in response to acetylcholine. Arterioscler. Thromb. Vasc. Biol. 2004;24:1253–1258. doi: 10.1161/01.ATV.0000130661.82773.ca. [DOI] [PubMed] [Google Scholar]
- 43.Harvey R.D. Handbook of Experimental Pharmacology. Springer; Berlin, Germany: 2011. Muscarinic Receptor Agonists and Antagonists: Effects on Cardiovascular Function; pp. 299–316. [DOI] [PubMed] [Google Scholar]
- 44.Jolkkonen M., Van Giersbergen P.L., Hellman U., Wernstedt C., Oras A., Satyapan N., Adem A., Karlsson E. Muscarinic toxins from the black mamba Dendroaspis polylepis. Eur. J. Biochem. 1995;234:579–585. doi: 10.1111/j.1432-1033.1995.579_b.x. [DOI] [PubMed] [Google Scholar]
- 45.Ryberg A.T., Selberg H., Soukup O., Gradin K., Tobin G. Cholinergic submandibular effects and muscarinic receptor expression in blood vessels of the rat. Arch. Oral Biol. 2008;53:605–616. doi: 10.1016/j.archoralbio.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Aird S.D. Ophidian envenomation strategies and the role of purines. Toxicon. 2002;40:335–393. doi: 10.1016/S0041-0101(01)00232-X. [DOI] [PubMed] [Google Scholar]
- 47.Wangai J., Ng’ang’a J.N., Mungai S., Muthaura C.N., Thairu K., Telang B.V. Mechanisms of centrally induced vasodepressor response in cats after lateral ventricular administration of whole venom of Dendroaspis angusticeps-Part I. Agressologie. 1982;23:301–304. [PubMed] [Google Scholar]
- 48.Medina A., Bodick N., Goldberger A.L., Mac Mahon M., Lipsitz L.A. Effects of central muscarinic-1 receptor stimulation on blood pressure regulation. Hypertension. 1997;29:828–834. doi: 10.1161/01.HYP.29.3.828. [DOI] [PubMed] [Google Scholar]
- 49.Brezenoff H.E., Vargas H., Xiao Y.F. Blockade of brain M2 muscarinic receptors lowers blood pressure in spontaneously hypertensive rats. Pharmacology. 1988;36:101–105. doi: 10.1159/000138365. [DOI] [PubMed] [Google Scholar]
- 50.Pazos A., Wiederhold K.H., Palacios J.M. Central pressor effects induced by muscarinic receptor agonists: Evidence for a predominant role of the M2 receptor subtype. Eur. J. Pharmacol. 1986;125:63–70. doi: 10.1016/0014-2999(86)90083-X. [DOI] [PubMed] [Google Scholar]
- 51.Kubo T. Cholinergic mechanism and blood pressure regulation in the central nervous system. Brain Res. Bull. 1998;46:475–481. doi: 10.1016/S0361-9230(98)00041-0. [DOI] [PubMed] [Google Scholar]
- 52.Ozkutlu U., Onat F., Aslan A.N., Oktay S. Central muscarinic M2 cholinoceptors involved in cholinergic hypertension. Eur. J. Pharmacol. 1993;250:349–354. doi: 10.1016/0014-2999(93)90020-I. [DOI] [PubMed] [Google Scholar]
- 53.Bruning T.A., Hendriks M.G., Chang P.C., Kuypers E.A., van Zwieten P.A. In vivo characterization of vasodilating muscarinic-receptor subtypes in humans. Circ. Res. 1994;74:912–919. doi: 10.1161/01.RES.74.5.912. [DOI] [PubMed] [Google Scholar]
- 54.Hendriks M.G., Pfaffendorf M., Van Zwieten P.A. Characterization of the muscarinic receptor subtype mediating vasodilation in the rat perfused mesenteric vascular bed preparation. J. Auton. Pharmacol. 1992;12:411–420. doi: 10.1111/j.1474-8673.1992.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 55.Gericke A., Sniatecki J.J., Mayer V.G.A., Goloborodko E., Patzak A., Wess J., Pfeiffer N. Role of M1, M3, and M5 muscarinic acetylcholine receptors in cholinergic dilation of small arteries studied with gene-targeted mice. Am. J. Physiol. Heart Circ. Physiol. 2011;300:H1602–H1608. doi: 10.1152/ajpheart.00982.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Höglund A.U., Baghdoyan H.A. M2, M3 and M4, but not M1, muscarinic receptor subtypes are present in rat spinal cord. J. Pharmacol. Exp. Ther. 1997;281:470–477. [PubMed] [Google Scholar]
- 57.Yamada M., Lamping K.G., Duttaroy A., Zhang W., Cui Y., Bymaster F.P., McKinzie D.L., Felder C.C., Deng C.X., Faraci F.M., et al. Cholinergic dilation of cerebral blood vessels is abolished in M(5) muscarinic acetylcholine receptor knockout mice. Proc. Natl. Acad. Sci. USA. 2001;98:14096–14101. doi: 10.1073/pnas.251542998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolkkonen M. Ph.D. Thesis. University of Uppsala; Uppsala, Sweden: 1996. Muscarinic Toxins from Dendroaspis (mamba) Venoms. Peptides Selective for Subtypes of Acetylcholine Muscarinic Receptors. [Google Scholar]
- 59.Liang J.S., Carsi-Gabrenas J., Krajewski J.L., McCafferty J.M., Purkerson S.L., Santiago M.P., Strauss W.L., Valentine H.H., Potter L.T. Anti-muscarinic toxins from Dendroaspis angusticeps. Toxicon. 1996;34:1257–1267. doi: 10.1016/S0041-0101(96)00109-2. [DOI] [PubMed] [Google Scholar]
- 60.Carsi J.M., Valentine H.H., Potter L.T. m2-toxin: A selective ligand for M2 muscarinic receptors. Mol. Pharmacol. 1999;56:933–937. doi: 10.1124/mol.56.5.933. [DOI] [PubMed] [Google Scholar]
- 61.Jolkkonen M., Adem A., Hellman U., Wernstedt C., Karlsson E. A snake toxin against muscarinic acetylcholine receptors: Amino acid sequence, subtype specificity and effect on guinea-pig ileum. Toxicon. 1995;33:399–410. doi: 10.1016/0041-0101(94)00102-E. [DOI] [PubMed] [Google Scholar]
- 62.Jolkkonen M., van Giersbergen P.L., Hellman U., Wernstedt C., Karlsson E. A toxin from the green mamba Dendroaspis angusticeps: Amino acid sequence and selectivity for muscarinic m4 receptors. FEBS Lett. 1994;352:91–94. doi: 10.1016/0014-5793(94)00933-3. [DOI] [PubMed] [Google Scholar]
- 63.Ségalas I., Roumestand C., Zinn-Justin S., Gilquin B., Ménez R., Ménez A., Toma F. Solution structure of a green mamba toxin that activates muscarinic acetylcholine receptors, as studied by nuclear magnetic resonance and molecular modeling. Biochemistry. 1995;34:1248–1260. doi: 10.1021/bi00004a019. [DOI] [PubMed] [Google Scholar]
- 64.Servent D., Blanchet G., Mourier G., Marquer C., Marcon E., Fruchart-Gaillard C. Muscarinic toxins. Toxicon. 2011;58:455–463. doi: 10.1016/j.toxicon.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Da Silva D.C., de Medeiros W.A.A., Batista I.D.F.C., Pimenta D.C., Lebrun I., Abdalla F.M.F., Sandoval M.R.L. Characterization of a new muscarinic toxin from the venom of the Brazilian coral snake Micrurus lemniscatus in rat hippocampus. Life Sci. 2011;89:931–938. doi: 10.1016/j.lfs.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 66.Dhein S., van Koppen C.J., Brodde O.E. Muscarinic receptors in the mammalian heart. Pharmacol. Res. 2001;44:161–182. doi: 10.1006/phrs.2001.0835. [DOI] [PubMed] [Google Scholar]
- 67.Montandon A.C. Master’s Thesis. Universidade Federal de Minas Gerais: Belo Horizonte; Minas Gerais, Brazil: 2013. Avaliação da liberação de L-glutamato por sinaptosomas cérebro-corticais de rato pela ação da peçonha da serpente coral Sul-Americana, Micrurus lemniscatus (Linnaeus, 1758), e por algumas de suas fraçOões semi-purificadas. [Google Scholar]
- 68.Freire Donato M. Ph.D. Thesis. Federal University of Minas Gerais, Belo Horizonte; Minas Gerais, Brazil: 2014. Peçonha da serpente Micrurus l. lemniscatus (Roze, 1967) Caracterização Parcial das Propriedades Bioquímica e Farmacológicas: Neurotoxicidade e Atividade pré Sináptica de Toxinas Três-Dígitos. [Google Scholar]
- 69.Vital Brazil O. Coral snake venoms: Mode of action and pathophysiology of experimental envenomation. Rev. Inst. Med. Trop. Sao Paulo. 1987;29:119–126. doi: 10.1590/S0036-46651987000300001. [DOI] [PubMed] [Google Scholar]
- 70.Da Silva N.J., Jr., Griffin P.R., Aird S.D. Comparative chromatography of Brazilian coral snake (Micrurus) venoms. Comp. Biochem. Physiol. B. 1991;100:117–126. doi: 10.1016/0305-0491(91)90093-S. [DOI] [PubMed] [Google Scholar]
- 71.Rajagopalan N., Pung Y.F., Zhu Y.Z., Wong P.T.H., Kumar P.P., Kini R.M. Beta-cardiotoxin: A new three-finger toxin from Ophiophagus hannah (king cobra) venom with beta-blocker activity. FASEB J. 2007;21:3685–3695. doi: 10.1096/fj.07-8658com. [DOI] [PubMed] [Google Scholar]
- 72.Kumar V., Rejent T.A., Elliott W.B. Anticholinesterase activity of elapid venoms. Toxicon. 1973;11:131–138. doi: 10.1016/0041-0101(73)90074-3. [DOI] [PubMed] [Google Scholar]
- 73.De Weille J.R., Schweitz H., Maes P., Tartar A., Lazdunski M. Calciseptine, a peptide isolated from black mamba venom, is a specific blocker of the L-type calcium channel. Proc. Natl. Acad. Sci. USA. 1991;88:2437–2440. doi: 10.1073/pnas.88.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schweitz H., Heurteaux C., Bois P., Moinier D., Romey G., Lazdunski M. Calcicludine, a venom peptide of the Kunitz-type protease inhibitor family, is a potent blocker of high-threshold Ca2+ channels with a high affinity for L-type channels in cerebellar granule neurons. Proc. Natl. Acad. Sci. USA. 1994;91:878–882. doi: 10.1073/pnas.91.3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raina R.K., Ng’ang’a J.N., Njoroge D.K., Telang B.V. Further studies on the mechanism of vasodepressor response in cats after intravenous administration of venom from the snake Dendroaspis jamesoni (Jameson’s mamba) Toxicon. 1977;15:561–570. doi: 10.1016/0041-0101(77)90107-6. [DOI] [PubMed] [Google Scholar]
- 76.Wangai J., Ng’ang’a J.N., Njoroge D., Thairu K., Telang B.V. Centrally acting hypotensive fraction in the venom of Dendroaspis angusticeps. Experientia. 1978;34:874–876. doi: 10.1007/BF01939676. [DOI] [PubMed] [Google Scholar]
- 77.Harvey A.L., Karlsson E. Dendrotoxin from the venom of the green mamba, Dendroaspis angusticeps. A neurotoxin that enhances acetylcholine release at neuromuscular junction. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1980;312:1–6. doi: 10.1007/BF00502565. [DOI] [PubMed] [Google Scholar]
- 78.Wangai J., Thairu K., Bharaj B.S., Telang B.V. Identification and isolation of three acetylcholinesterase inactivating fractions in the venom of Dendroaspis angusticeps. Acta Physiol. Acad. Sci. Hung. 1982;60:83–88. [PubMed] [Google Scholar]
- 79.Rodríguez-Ithurralde D., Silveira R., Barbeito L., Dajas F. Fasciculin, a powerful anticholinesterase polypeptide from Dendroaspis angusticeps venom. Neurochem. Int. 1983;5:267–274. doi: 10.1016/0197-0186(83)90028-1. [DOI] [PubMed] [Google Scholar]
- 80.Karlsson E., Mbugua P.M., Rodriguez-Ithurralde D. Fasciculins, anticholinesterase toxins from the venom of the green mamba Dendroaspis angusticeps. J. Physiol. Paris. 1984;79:232–240. [PubMed] [Google Scholar]
- 81.Orawski A.T., Susz J.P., Simmons W.H. Metabolism of bradykinin by multiple coexisting membrane-bound peptidases in lung: Techniques for investigating the role of each peptidase using specific inhibitors. Adv. Exp. Med. Biol. 1989;247:355–364. doi: 10.1007/978-1-4615-9546-5_59. [DOI] [PubMed] [Google Scholar]
- 82.Kokkonen J.O., Kuoppala A., Saarinen J., Lindstedt K.A., Kovanen P.T. Kallidin- and Bradykinin-Degrading Pathways in Human Heart : Degradation of Kallidin by Aminopeptidase M Like Activity and Bradykinin by Neutral Endopeptidase. Circulation. 1999;99:1984–1990. doi: 10.1161/01.CIR.99.15.1984. [DOI] [PubMed] [Google Scholar]
- 83.Bausback H.H., Churchill L., Ward P.E. Angiotensin metabolism by cerebral microvascular aminopeptidase A. Biochem. Pharmacol. 1988;37:155–160. doi: 10.1016/0006-2952(88)90712-5. [DOI] [PubMed] [Google Scholar]
- 84.Ward P.E., Benter I.F., Dick L., Wilk S. Metabolism of vasoactive peptides by plasma and purified renal aminopeptidase M. Biochem. Pharmacol. 1990;40:1725–1732. doi: 10.1016/0006-2952(90)90348-O. [DOI] [PubMed] [Google Scholar]
- 85.Holzer P., Guth P.H. Neuropeptide control of rat gastric mucosal blood flow. Increase by calcitonin gene-related peptide and vasoactive intestinal polypeptide, but not substance P and neurokinin A. Circ. Res. 1991;68:100–105. doi: 10.1161/01.RES.68.1.100. [DOI] [PubMed] [Google Scholar]
- 86.Lu B., Figini M., Emanueli C., Geppetti P., Grady E.F., Gerard N.P., Ansell J., Payan D.G., Gerard C., Bunnett N. The control of microvascular permeability and blood pressure by neutral endopeptidase. Nat. Med. 1997;3:904–907. doi: 10.1038/nm0897-904. [DOI] [PubMed] [Google Scholar]
- 87.Ogawa Y., Murayama N., Fujita Y., Yanoshita R. Characterization and cDNA cloning of aminopeptidase A from the venom of Gloydius blomhoffi brevicaudus. Toxicon. 2007;49:1172–1181. doi: 10.1016/j.toxicon.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 88.Fry B.G., Wüster W. Assembling an arsenal: Origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 2004;21:870–883. doi: 10.1093/molbev/msh091. [DOI] [PubMed] [Google Scholar]
- 89.Drickamer K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999;9:585–590. doi: 10.1016/S0959-440X(99)00009-3. [DOI] [PubMed] [Google Scholar]
- 90.Morita T. C-type lectin-related proteins from snake venoms. Curr. Drug Targets Cardiovasc. Haematol. Disord. 2004;4:357–373. doi: 10.2174/1568006043335916. [DOI] [PubMed] [Google Scholar]
- 91.Lu Q., Clemetson J.M., Clemetson K.J. Snake venoms and hemostasis. J. Thromb. Haemost. 2005;3:1791–1799. doi: 10.1111/j.1538-7836.2005.01358.x. [DOI] [PubMed] [Google Scholar]
- 92.Zha H.G., Lee W.H., Zhang Y. Cloning of cDNAs encoding C-type lectins from Elapidae snakes Bungarus fasciatus and Bungarus multicinctus. Toxicon. 2001;39:1887–1892. doi: 10.1016/S0041-0101(01)00172-6. [DOI] [PubMed] [Google Scholar]
- 93.Earl S.T.H., Robson J., Trabi M., de Jersey J., Masci P.P., Lavin M.F. Characterisation of a mannose-binding C-type lectin from Oxyuranus scutellatus snake venom. Biochimie. 2011;93:519–527. doi: 10.1016/j.biochi.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 94.Corrêa-Netto C., Junqueira-de-Azevedo Ide L., Silva D.A., Ho P.L., Leitao-de-Araujo M., Alves M.L., Sanz L., Foguel D., Zingali R.B., Calvete J.J. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J. Proteom. 2011;74:1795–1809. doi: 10.1016/j.jprot.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 95.Drickamer K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature. 1992;360:183–186. doi: 10.1038/360183a0. [DOI] [PubMed] [Google Scholar]
- 96.Yamazaki Y., Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–231. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 97.Yamazaki Y., Koike H., Sugiyama Y., Motoyoshi K., Wada T., Hishinuma S., Mita M., Morita T. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 2002;269:2708–2715. doi: 10.1046/j.1432-1033.2002.02940.x. [DOI] [PubMed] [Google Scholar]
- 98.Peichoto M.E., Mackessy S.P., Teibler P., Tavares F.L., Burckhardt P.L., Breno M.C., Acosta O., Santoro M.L. Purification and characterization of a cysteine-rich secretory protein from Philodryas patagoniensis snake venom. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2009;150:79–84. doi: 10.1016/j.cbpc.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 99.Rokyta D.R., Lemmon A.R., Margres M.J., Aronow K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus) BMC Genom. 2012;13:312. doi: 10.1186/1471-2164-13-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Argraves W.S., Tran H., Burgess W.H., Dickerson K. Fibulin is an extracellular matrix and plasma glycoprotein with repeated domain structure. J. Cell Biol. 1990;111:3155–3164. doi: 10.1083/jcb.111.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott J., Urdea M., Quiroga M., Sanchez-Pescador R., Fong N., Selby M., Rutter W.J., Bell G.I. Structure of a mouse submaxillary messenger RNA encoding epidermal growth factor and seven related proteins. Science. 1983;221:236–240. doi: 10.1126/science.6602382. [DOI] [PubMed] [Google Scholar]
- 102.Engel J., Jürgen E. EGF-like domains in extracellular matrix proteins: Localized signals for growth and differentiation? FEBS Lett. 1989;251:1–7. doi: 10.1016/0014-5793(89)81417-6. [DOI] [PubMed] [Google Scholar]
- 103.Tran H., Tanaka A., Litvinovich S.V., Medved L.V., Haudenschild C.C., Argraves W.S. The interaction of fibulin-1 with fibrinogen. A potential role in hemostasis and thrombosis. J. Biol. Chem. 1995;270:19458–19464. doi: 10.1074/jbc.270.33.19458. [DOI] [PubMed] [Google Scholar]
- 104.Godyna S., Diaz-Ricart M., Argraves W.S. Fibulin-1 mediates platelet adhesion via a bridge of fibrinogen. Blood. 1996;88:2569–2577. [PubMed] [Google Scholar]
- 105.Aspberg A., Adam S., Kostka G., Timpl R., Heinegård D. Fibulin-1 is a ligand for the C-type lectin domains of aggrecan and versican. J. Biol. Chem. 1999;274:20444–20449. doi: 10.1074/jbc.274.29.20444. [DOI] [PubMed] [Google Scholar]
- 106.Eagle H. The coagulation of blood by snake venoms and its physiologic significance. J. Exp. Med. 1937;65:613–639. doi: 10.1084/jem.65.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keppler D. Towards novel anti-cancer strategies based on cystatin function. Cancer Lett. 2006;235:159–176. doi: 10.1016/j.canlet.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 108.Abrahamson M. Cystatins. Methods Enzymol. 1994;244:685–700. doi: 10.1042/bss0700179. [DOI] [PubMed] [Google Scholar]
- 109.Ogawa Y., Kanai-Azuma M., Akimoto Y., Kawakami H., Yanoshita R. Exosome-like vesicles in Gloydius blomhoffii blomhoffii venom. Toxicon. 2008;51:984–993. doi: 10.1016/j.toxicon.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 110.Da Silva N.J., Jr., Aird S.D. Prey specificity, comparative lethality and compositional differences of coral snake venoms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001;128:425–456. doi: 10.1016/S1532-0456(00)00215-5. [DOI] [PubMed] [Google Scholar]
- 111.Heymann E., Mentlein R. Has dipeptidyl peptidase IV an effect on blood pressure and coagulation? Klin. Wochenschr. 1984;62:2–10. doi: 10.1007/BF01725186. [DOI] [PubMed] [Google Scholar]
- 112.Werner J.A., Schünke M., Tillmann B. Histochemical visualization of lymphatic capillaries in the rat: A comparison of methods demonstrated at the posterior pharyngeal surface. Arch. Histol. Jpn. 1987;50:505–514. doi: 10.1679/aohc.50.505. [DOI] [PubMed] [Google Scholar]
- 113.Ahmad S., Wang L., Ward P.E. Dipeptidyl(amino)peptidase IV and aminopeptidase M metabolize circulating substance P in vivo. J. Pharmacol. Exp. Ther. 1992;260:1257–1261. [PubMed] [Google Scholar]
- 114.Zukowska-Grojec Z., Haass M., Bayorh M.A. Neuropeptide Y and peptide YY mediate nonadrenergic vasoconstriction and modulate sympathetic responses in rats. Regul. Pept. 1986;15:99–110. doi: 10.1016/0167-0115(86)90080-7. [DOI] [PubMed] [Google Scholar]
- 115.Mentlein R., Dahms P., Grandt D., Krüger R. Proteolytic processing of neuropeptide Y and peptide YY by dipeptidyl peptidase IV. Regul. Pept. 1993;49:133–144. doi: 10.1016/0167-0115(93)90435-B. [DOI] [PubMed] [Google Scholar]
- 116.Aird S.D. Snake venom dipeptidyl peptidase IV: Taxonomic distribution and quantitative variation. Comp. Biochem. Physiol. B. 2008;150:222–228. doi: 10.1016/j.cbpb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 117.Ogawa Y., Yuko O., Yoshie M., Nobuhiro M., Ryohei Y. Characterization and cDNA cloning of dipeptidyl peptidase IV from the venom of Gloydius blomhoffi brevicaudus. Comp. Biochem. Physiol. B. 2006;145:35–42. doi: 10.1016/j.cbpb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 118.St. Pierre L., Birrell G.W., Earl S.T., Wallis T.P., Gorman J.J., de Jersey J., Masci P.P., Lavin M.F. Diversity of toxic components from the venom of the evolutionarily distinct black whip snake, Demansia vestigiata. J. Proteome Res. 2007;6:3093–3107. doi: 10.1021/pr0701613. [DOI] [PubMed] [Google Scholar]
- 119.Gartner T.K., Stocker K., Williams D.C. Thrombolectin: A lectin isolated from Bothrops atrox venom. FEBS Lett. 1980;117:13–16. doi: 10.1016/0014-5793(80)80902-1. [DOI] [PubMed] [Google Scholar]
- 120.Ogilvie M.L., Dockter M.E., Wenz L., Gartner T.K. Isolation and characterization of lactose-binding lectins from the venoms of the snakes Lachesis muta and Dendroaspis jamesonii. J. Biochem. 1986;100:1425–1431. doi: 10.1093/oxfordjournals.jbchem.a121848. [DOI] [PubMed] [Google Scholar]
- 121.Mastro A.M., Hurley D.J., Winning R.K., Filipowski R., Ogilvie M.L., Gartner T.K. Mitogenic activity of snake venom lectins. Cell Tissue Kinet. 1986;19:557–566. doi: 10.1111/j.1365-2184.1986.tb00756.x. [DOI] [PubMed] [Google Scholar]
- 122.Lomonte B., Rojas G., Gutiérrez J.M., Ramírez G. Isolation of a galactose-binding lectin from the venom of the snake Bothrops godmani (Godmann’s pit viper) Toxicon. 1990;28:75–81. doi: 10.1016/0041-0101(90)90008-U. [DOI] [PubMed] [Google Scholar]
- 123.Aird S.D., Kaiser I.I., Lewis R.V., Kruggel W.G. Rattlesnake presynaptic neurotoxins: Primary structure and evolutionary origin of the acidic subunit. Biochemistry. 1985;24:7054–7058. doi: 10.1021/bi00346a005. [DOI] [PubMed] [Google Scholar]
- 124.Aird S.D., Yates J.R., 3rd, Martino P.A., Shabanowitz J., Hunt D.F., Kaiser I.I. The amino acid sequence of the acidic subunit B-chain of crotoxin. Biochim. Biophys. Acta. 1990;1040:217–224. doi: 10.1016/0167-4838(90)90079-U. [DOI] [PubMed] [Google Scholar]
- 125.Tu A.T., Hendon R.R. Characterization of lizard venom hyaluronidase and evidence for its action as a spreading factor. Comp. Biochem. Physiol. B. 1983;76:377–383. doi: 10.1016/0305-0491(83)90086-X. [DOI] [PubMed] [Google Scholar]
- 126.Girish K.S., Mohanakumari H.P., Nagaraju S., Vishwanath B.S., Kemparaju K. Hyaluronidase and protease activities from Indian snake venoms: Neutralization by Mimosa pudica root extract. Fitoterapia. 2004;75:378–380. doi: 10.1016/j.fitote.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 127.Zupunski V., Kordis D., Gubensek F. Adaptive evolution in the snake venom Kunitz/BPTI protein family. FEBS Lett. 2003;547:131–136. doi: 10.1016/S0014-5793(03)00693-8. [DOI] [PubMed] [Google Scholar]
- 128.Sigle R., Hackett M., Aird S.D. Primary structures of four trypsin inhibitor E homologs from venom of Dendroaspis angusticeps: Structure-function comparisons with other dendrotoxin homologs. Toxicon. 2002;40:297–308. doi: 10.1016/S0041-0101(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 129.Tytgat J., Jan T., Isabel V., Chris U., Van Beeumen J. New polypeptide components purified from mamba venom. FEBS Lett. 2001;491:217–221. doi: 10.1016/S0014-5793(01)02201-3. [DOI] [PubMed] [Google Scholar]
- 130.Strydom D.J., Joubert F.J. The amino acid sequence of a weak trypsin inhibitor B from Dendroaspis polylepis polylepis (black mamba) venom. Hoppe Seylers Z. Physiol. Chem. 1981;362:1377–1384. doi: 10.1515/bchm2.1981.362.2.1377. [DOI] [PubMed] [Google Scholar]
- 131.Harvey A.L. Twenty years of dendrotoxins. Toxicon. 2001;39:15–26. doi: 10.1016/S0041-0101(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 132.Dufton M.J. Proteinase inhibitors and dendrotoxins. Sequence classification, structural prediction and structure/activity. Eur. J. Biochem. 1985;153:647–654. doi: 10.1111/j.1432-1033.1985.tb09349.x. [DOI] [PubMed] [Google Scholar]
- 133.Fox J.W., Serrano S.M.T. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005;45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 134.Fox J.W., Serrano S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBS J. 2008;275:3016–3030. doi: 10.1111/j.1742-4658.2008.06466.x. [DOI] [PubMed] [Google Scholar]
- 135.Takeda S., Takeya H., Iwanaga S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim. Biophys. Acta. 2012;1824:164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 136.Li S., Wang J., Zhang X., Ren Y., Wang N., Zhao K., Chen X., Zhao C., Li X., Shao J., et al. Proteomic characterization of two snake venoms: Naja naja atra and Agkistrodon halys. Biochem. J. 2004;384:119–127. doi: 10.1042/BJ20040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guan H.-H., Goh K.-S., Davamani F., Wu P.-L., Huang Y.-W., Jeyakanthan J., Wu W.-G., Chen C.-J. Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J. Struct. Biol. 2010;169:294–303. doi: 10.1016/j.jsb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sudoh T., Minamino N., Kangawa K., Matsuo H. C-type natriuretic peptide (CNP): A new member of natriuretic peptide family identified in porcine brain. Biochem. Biophys. Res. Commun. 1990;168:863–870. doi: 10.1016/0006-291X(90)92401-K. [DOI] [PubMed] [Google Scholar]
- 139.Schweitz H., Vigne P., Moinier D., Frelin C., Lazdunski M. A new member of the natriuretic peptide family is present in the venom of the green mamba (Dendroaspis angusticeps) J. Biol. Chem. 1992;267:13928–13932. [PubMed] [Google Scholar]
- 140.Murayama N., Hayashi M.A., Ohi H., Ferreira L.A., Hermann V.V., Saito H., Fujita Y., Higuchi S., Fernandes B.L., Yamane T., et al. Cloning and sequence analysis of a Bothrops jararaca cDNA encoding a precursor of seven bradykinin-potentiating peptides and a C-type natriuretic peptide. Proc. Natl. Acad. Sci. USA. 1997;94:1189–1193. doi: 10.1073/pnas.94.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ho P.L., Soares M.B., Maack T., Gimenez I., Puorto G., Furtado M.F., Raw I. Cloning of an unusual natriuretic peptide from the South American coral snake Micrurus corallinus. Eur. J. Biochem. 1997;250:144–149. doi: 10.1111/j.1432-1033.1997.00144.x. [DOI] [PubMed] [Google Scholar]
- 142.Fry B.G., Wickramaratana J.C., Lemme S., Beuve A., Garbers D., Hodgson W.C., Alewood P. Novel natriuretic peptides from the venom of the inland taipan (Oxyuranus microlepidotus): Isolation, chemical and biological characterisation. Biochem. Biophys. Res. Commun. 2005;327:1011–1015. doi: 10.1016/j.bbrc.2004.11.171. [DOI] [PubMed] [Google Scholar]
- 143.St Pierre L., Flight S., Masci P.P., Hanchard K.J., Lewis R.J., Alewood P.F., de Jersey J., Lavin M.F. Cloning and characterisation of natriuretic peptides from the venom glands of Australian elapids. Biochimie. 2006;88:1923–1931. doi: 10.1016/j.biochi.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 144.Higuchi S., Murayama N., Saguchi K.-I., Ohi H., Fujita Y., da Silva N.J., Jr., de Siqueira R.J.B., Lahlou S., Aird S.D. A novel peptide from the ACEI/BPP-CNP precursor in the venom of Crotalus durissus collilineatus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2006;144:107–121. doi: 10.1016/j.cbpc.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 145.Kostiza T., Dahinden C.A., Rihs S., Otten U., Meier J. Nerve growth factor from the venom of the Chinese cobra Naja naja atra: Purification and description of non-neuronal activities. Toxicon. 1995;33:1249–1261. doi: 10.1016/0041-0101(95)00086-2. [DOI] [PubMed] [Google Scholar]
- 146.Guo L.Y., Zhu J.F., Wu X.F., Zhou Y.C. Cloning of a cDNA encoding a nerve growth factor precursor from the Agkistrodon halys Pallas. Toxicon. 1999;37:465–470. doi: 10.1016/S0041-0101(98)00177-9. [DOI] [PubMed] [Google Scholar]
- 147.Orenstein N.S., Dvorak H.F., Blanchard M.H., Young M. Nerve growth factor: A protease that can activate plasminogen. Proc. Natl. Acad. Sci. USA. 1978;75:5497–5500. doi: 10.1073/pnas.75.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Young M. Proteolytic activity of nerve growth factor: A case of autocatalytic activation. Biochemistry. 1979;18:3050–3055. doi: 10.1021/bi00581a022. [DOI] [PubMed] [Google Scholar]
- 149.Bohlen C.J., Chesler A.T., Sharif-Naeini R., Medzihradszky K.F., Zhou S., King D., Sanchez E.E., Burlingame A.L., Basbaum A.I., Julius D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kocholaty W.F., Ledford E.B., Daly J.G., Billings T.A. Toxicity and some enzymatic properties and activities in the venoms of Crotalidae, Elapidae and Viperidae. Toxicon. 1971;9:131–138. doi: 10.1016/0041-0101(71)90006-7. [DOI] [PubMed] [Google Scholar]
- 151.Ramsey H.W., Snyder G.K., Kitchen H., Taylor W.J. Fractionation of coral snake venom. Toxicon. 1972;10:67–72. doi: 10.1016/0041-0101(72)90092-X. [DOI] [PubMed] [Google Scholar]
- 152.Possani L.D., Alagon A.C., Fletcher P.L., Jr., Varela M.J., Julia J.Z. Purification and characterization of a phospholipase A2 from the venom of the coral snake, Micrurus fulvius microgalbineus (Brown and Smith) Biochem. J. 1979;179:603–606. doi: 10.1042/bj1790603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vergara I., Pedraza-Escalona M., Paniagua D., Restano-Cassulini R., Zamudio F., Batista C.V.F., Possani L.D., Alagón A. Eastern coral snake Micrurus fulvius venom toxicity in mice is mainly determined by neurotoxic phospholipases A2. J. Proteom. 2014;105:295–306. doi: 10.1016/j.jprot.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 154.Fernandez J., Alape-Giron A., Angulo Y., Sanz L., Gutierrez J.M., Calvete J.J., Lomonte B. Venomic and antivenomic analyses of the Central American coral snake, Micrurus nigrocinctus (Elapidae) J. Proteome Res. 2011;10:1816–1827. doi: 10.1021/pr101091a. [DOI] [PubMed] [Google Scholar]
- 155.Cecchini A.L., Marcussi S., Silveira L.B., Borja-Oliveira C.R., Rodrigues-Simioni L., Amara S., Stabeli R.G., Giglio J.R., Arantes E.C., Soares A.M. Biological and enzymatic activities of Micrurus sp. (Coral) snake venoms. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005;140:125–134. doi: 10.1016/j.cbpb.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 156.Maraganore J.M., Merutka G., Cho W., Welches W., Kézdy F.J., Heinrikson R.L. A new class of phospholipases A2 with lysine in place of aspartate 49. Functional consequences for calcium and substrate binding. J. Biol. Chem. 1984;259:13839–13843. [PubMed] [Google Scholar]
- 157.Lomonte B., Gutiérrez J.M. A new muscle damaging toxin, myotoxin II, from the venom of the snake Bothrops asper (terciopelo) Toxicon. 1989;27:725–733. doi: 10.1016/0041-0101(89)90039-1. [DOI] [PubMed] [Google Scholar]
- 158.Gutiérrez J.M., Lomonte B., Cerdas L. Isolation and partial characterization of a myotoxin from the venom of the snake Bothrops nummifer. Toxicon. 1986;24:885–894. doi: 10.1016/0041-0101(86)90089-9. [DOI] [PubMed] [Google Scholar]
- 159.Montecucco C., Gutiérrez J.M., Lomonte B. Cellular pathology induced by snake venom phospholipase A2 myotoxins and neurotoxins: Common aspects of their mechanisms of action. Cell. Mol. Life Sci. 2008;65:2897–2912. doi: 10.1007/s00018-008-8113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kopper R.A., Harper G.R., Zimmerman S., Hook J. Comparison of total protein and phospholipase A(2) levels in individual coralsnake venoms. Toxicon. 2013;76:59–62. doi: 10.1016/j.toxicon.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 161.Weis R., McIsaac R.J. Cardiovascular and muscular effects of venom from coral snake, Micrurus fulvius. Toxicon. 1971;9:219–228. doi: 10.1016/0041-0101(71)90073-0. [DOI] [PubMed] [Google Scholar]
- 162.Maeno H., Mitsuhashi S., Okonogi T., Hoshi S., Homma M. Studies on Habu snake venom. (V). Myolysis caused by phospholipase A in Habu snake venom. Jpn. J. Exp. Med. 1962;32:55–64. [PubMed] [Google Scholar]
- 163.Kurashige S., Hara Y., Kawakami M., Mitsuhashi S. Studies on Habu snake venom. VII. Heat-stable myolytic factor and development of its activity by addition of phospholipase A. Jpn. J. Microbiol. 1966;10:23–31. doi: 10.1111/j.1348-0421.1966.tb00288.x. [DOI] [PubMed] [Google Scholar]
- 164.Harris J.B., Johnson M.A., Karlsson E. Pathological responses of rat skeletal muscle to a single subcutaneous injection of a toxin isolated from the venom of the Australian tiger snake, Notechis scutatus scutatus. Clin. Exp. Pharmacol. Physiol. 1975;2:383–404. doi: 10.1111/j.1440-1681.1975.tb01846.x. [DOI] [Google Scholar]
- 165.Kensil C.R., Dennis E.A. Action of cobra venom phospholipase A2 on the gel and liquid crystalline states of dimyristoyl and dipalmitoyl phosphatidylcholine vesicles. J. Biol. Chem. 1979;254:5843–5848. [PubMed] [Google Scholar]
- 166.Napias C., Heilbronn E. Phospholipase A2 activity and substrate specificity of snake venom presynaptic toxins. Biochemistry. 1980;19:1146–1151. doi: 10.1021/bi00547a017. [DOI] [PubMed] [Google Scholar]
- 167.Radvanyi F., Saliou B., Bon C., Strong P.N. The interaction between the presynaptic phospholipase neurotoxins beta-bungarotoxin and crotoxin and mixed detergent-phosphatidylcholine micelles. A comparison with non-neurotoxic snake venom phospholipases A2. J. Biol. Chem. 1987;262:8966–8974. [PubMed] [Google Scholar]
- 168.de Roodt A.R., Lago N.R., Stock R.P. Myotoxicity and nephrotoxicity by Micrurus venoms in experimental envenomation. Toxicon. 2012;59:356–364. doi: 10.1016/j.toxicon.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 169.Leao L.I., Ho P.L., Junqueira-de-Azevedo Ide L. Transcriptomic basis for an antiserum against Micrurus corallinus (coral snake) venom. BMC Genom. 2009;10:112. doi: 10.1186/1471-2164-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fernández J., Vargas-Vargas N., Pla D., Sasa M., Rey-Suárez P., Sanz L., Gutiérrez J.M., Calvete J.J., Lomonte B. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in New World elapids. Toxicon. 2015;107:217–233. doi: 10.1016/j.toxicon.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 171.Chen Y.-H., Wang Y.-M., Hseu M.-J., Tsai I.-H. Molecular evolution and structure-function relationships of crotoxin-like and asparagine-6-containing phospholipases A2 in pit viper venoms. Biochem. J. 2004;381:25–34. doi: 10.1042/BJ20040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kini R.M., Iwanaga S. Structure-function relationships of phospholipases. I: Prediction of presynaptic neurotoxicity. Toxicon. 1986;24:527–541. doi: 10.1016/0041-0101(86)90173-X. [DOI] [PubMed] [Google Scholar]
- 173.Kini R.M. Structure–function relationships and mechanism of anticoagulant phospholipase A2 enzymes from snake venoms. Toxicon. 2005;45:1147–1161. doi: 10.1016/j.toxicon.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 174.Kini R.M., Iwanaga S. Structure-function relationships of phospholipases. II: Charge density distribution and the myotoxicity of presynaptically neurotoxic phospholipases. Toxicon. 1986;24:895–905. doi: 10.1016/0041-0101(86)90090-5. [DOI] [PubMed] [Google Scholar]
- 175.Alape-Girón A., Persson B., Cederlund E., Flores-Díaz M., Gutiérrez J.M., Thelestam M., Bergman T., Jörnvall H. Elapid venom toxins: Multiple recruitments of ancient scaffolds. Eur. J. Biochem. 1999;259:225–234. doi: 10.1046/j.1432-1327.1999.00021.x. [DOI] [PubMed] [Google Scholar]
- 176.White S.P., Scott D.L., Otwinowski Z., Gelb M.H., Sigler P.B. Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue. Science. 1990;250:1560–1563. doi: 10.1126/science.2274787. [DOI] [PubMed] [Google Scholar]
- 177.Gutiérrez J., JoséMaría G., Bruno L., Elsa P., Luis C., Ermila R. Local effects induced by coral snake venoms: Evidence of myonecrosis after experimental inoculations of venoms from five species. Toxicon. 1983;21:777–783. doi: 10.1016/0041-0101(83)90066-1. [DOI] [PubMed] [Google Scholar]
- 178.Gutiérrez J.M., Arroyo O., Chaves F., Lomonte B., Cerdas L. Pathogenesis of myonecrosis induced by coral snake (Micrurus nigrocinctus) venom in mice. Br. J. Exp. Pathol. 1986;67:1–12. [PMC free article] [PubMed] [Google Scholar]
- 179.Wrogemann K., Pena S.D. Mitochondrial calcium overload: A general mechanism for cell-necrosis in muscle diseases. Lancet. 1976;1:672–674. doi: 10.1016/S0140-6736(76)92781-1. [DOI] [PubMed] [Google Scholar]
- 180.Arroyo O., Rosso J.P., Vargas O., Gutierrez J.M., Cerdas L. Skeletal muscle necrosis induced by a phospholipase A2 isolated from the venom of the coral snake Micrurus nigrocinctus nigrocinctus. Comp. Biochem. Physiol. B. 1987;87:949–952. doi: 10.1016/0305-0491(87)90417-2. [DOI] [PubMed] [Google Scholar]
- 181.Goularte F.C., da Cruz-Höfling M.A., Corrado A.P., Rodrigues-Simioni L. Electrophysiological and ultrastructural analysis of the neuromuscular blockade and miotoxicity induced by the Micrurus nigrocinctus snake venom. Acta Physiol. Pharmacol. Ther. Latinoam. 1999;49:290–296. [PubMed] [Google Scholar]
- 182.Terra A.L., Moreira-Dill L.S., Simoes-Silva R., Monteiro J.R., Cavalcante W.L., Gallacci M., Barros N.B., Nicolete R., Teles C.B., Medeiros P.S., et al. Biological characterization of the Amazon coral Micrurus spixii snake venom: Isolation of a new neurotoxic phospholipase A2. Toxicon. 2015;103:1–11. doi: 10.1016/j.toxicon.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 183.Carvalho N.D., Garcia R.C., Ferreira A.K., Batista D.R., Cassola A.C., Maria D., Lebrun I., Carneiro S.M., Afeche S.C., Marcourakis T., et al. Neurotoxicity of coral snake phospholipases A2 in cultured rat hippocampal neurons. Brain Res. 2014;1552:1–16. doi: 10.1016/j.brainres.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 184.Lind P., Peter L., David E. Amino acid sequence of a lethal myotoxic phospholipase A2 from the venom of the common sea snake (Enhydrina schistosa) Toxicon. 1981;19:11–24. doi: 10.1016/0041-0101(81)90113-6. [DOI] [PubMed] [Google Scholar]
- 185.Lin W.W., Chang P.L., Lee C.Y., Joubert F.J. Pharmacological study on phospholipases A2 isolated from Naja mossambica mossambica venom. Proc. Natl. Sci. Counc. Repub. China B. 1987;11:155–163. [PubMed] [Google Scholar]
- 186.Lambeau G., Schmid-Alliana A., Lazdunski M., Barhanin J. Identification and purification of a very high affinity binding protein for toxic phospholipases A2 in skeletal muscle. J. Biol. Chem. 1990;265:9526–9532. [PubMed] [Google Scholar]
- 187.Halpert J., Eaker D. Isolation and amino acid sequence of a neurotoxic phospholipase A from the venom of the Australian tiger snake Notechis scutatus scutatus. J. Biol. Chem. 1976;251:7343–7347. [PubMed] [Google Scholar]
- 188.Dixon R.W., Harris J.B. Myotoxic activity of the toxic phospholipase, notexin, from the venom of the Australian tiger snake. J. Neuropathol. Exp. Neurol. 1996;55:1230–1237. doi: 10.1097/00005072-199612000-00006. [DOI] [PubMed] [Google Scholar]
- 189.Gutiérrez J.M., Ownby C.L. Skeletal muscle degeneration induced by venom phospholipases A2: Insights into the mechanisms of local and systemic myotoxicity. Toxicon. 2003;42:915–931. doi: 10.1016/j.toxicon.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 190.Casais-e-Silva L.L., Teixeira C.F.P., Lebrun I., Lomonte B., Alape-Girón A., Gutiérrez J.M. Lemnitoxin, the major component of Micrurus lemniscatus coral snake venom, is a myotoxic and pro-inflammatory phospholipase A2. Toxicol. Lett. 2016;257:60–71. doi: 10.1016/j.toxlet.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 191.Gutiérrez J.M., Alberto Ponce-Soto L., Marangoni S., Lomonte B. Systemic and local myotoxicity induced by snake venom group II phospholipases A2: Comparison between crotoxin, crotoxin B and a Lys49 PLA2 homologue. Toxicon. 2008;51:80–92. doi: 10.1016/j.toxicon.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 192.Sampaio S.C., Hyslop S., Fontes M.R.M., Prado-Franceschi J., Zambelli V.O., Magro A.J., Brigatte P., Gutierrez V.P., Cury Y. Crotoxin: Novel activities for a classic beta-neurotoxin. Toxicon. 2010;55:1045–1060. doi: 10.1016/j.toxicon.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 193.Nunez C.E., Angulo Y., Lomonte B. Identification of the myotoxic site of the Lys49 phospholipase A2 from Agkistrodon piscivorus piscivorus snake venom: Synthetic C-terminal peptides from Lys49, but not from Asp49 myotoxins, exert membrane-damaging activities. Toxicon. 2001;39:1587–1594. doi: 10.1016/S0041-0101(01)00141-6. [DOI] [PubMed] [Google Scholar]
- 194.Francis B., Brian F., Williams E.S., Corrine S., Kaiser I.I. Proteins isolated from the venom of the common tiger snake (Notechis scutatus scutatus) promote hypotension and hemorrhage. Toxicon. 1993;31:447–458. doi: 10.1016/0041-0101(93)90180-Q. [DOI] [PubMed] [Google Scholar]
- 195.Francis B., Coffield J.A., Simpson L.L., Kaiser I.I. Amino Acid Sequence of a New Type of Toxic Phospholipase A2 from the Venom of the Australian Tiger Snake (Notechis scutatus scutatus) Arch. Biochem. Biophys. 1995;318:481–488. doi: 10.1006/abbi.1995.1257. [DOI] [PubMed] [Google Scholar]
- 196.Francis B.R., da Silva Junior N.J., Seebart C., Casais e Silva L.L., Schmidt J.J., Kaiser I.I. Toxins isolated from the venom of the Brazilian coral snake (Micrurus frontalis frontalis) include hemorrhagic type phospholipases A2 and postsynaptic neurotoxins. Toxicon. 1997;35:1193–1203. doi: 10.1016/S0041-0101(97)00031-7. [DOI] [PubMed] [Google Scholar]
- 197.Marks S.L., Mannella C., Schaer M. Coral snake envenomation in the dog: Report of four cases and review of the literature. J. Am. Anim. Hosp. Assoc. 1990;26:629–634. [Google Scholar]
- 198.Peterson M.E. Snake bite: Pit vipers. Clin. Tech. Small Anim. Pract. 2006;21:174–182. doi: 10.1053/j.ctsap.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 199.Pérez M.L., Fox K., Schaer M. A retrospective evaluation of coral snake envenomation in dogs and cats: 20 cases (1996–2011) J. Vet. Emerg. Crit. Care. 2012;22:682–689. doi: 10.1111/j.1476-4431.2012.00806.x. [DOI] [PubMed] [Google Scholar]
- 200.Arce-Bejarano R., Lomonte B., Gutiérrez J.M. Intravascular hemolysis induced by the venom of the Eastern coral snake, Micrurus fulvius, in a mouse model: Identification of directly hemolytic phospholipases A2. Toxicon. 2014;90:26–35. doi: 10.1016/j.toxicon.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 201.Lambeau G., Ancian P., Nicolas J.P., Beiboer S.H., Moinier D., Verheij H., Lazdunski M. Structural elements of secretory phospholipases A2 involved in the binding to M-type receptors. J. Biol. Chem. 1995;270:5534–5540. doi: 10.1074/jbc.270.10.5534. [DOI] [PubMed] [Google Scholar]
- 202.Belo C.A., Toyama M.H., Toyama Dde O., Marangoni S., Moreno F.B., Cavada B.S., Fontana M.D., Hyslop S., Carneiro E.M., Boschero A.C. Determination of the amino acid sequence of a new phospholipase A(2) (MIDCA1) isolated from Micrurus dumerilii carinicauda venom. Protein J. 2005;24:147–153. doi: 10.1007/s10930-005-7838-1. [DOI] [PubMed] [Google Scholar]
- 203.Carbajal-Saucedo A., Floriano R.S., Dal Belo C.A., Olvera-Rodriguez A., Alagon A., Rodrigues-Simioni L. Neuromuscular activity of Micrurus laticollaris (Squamata: Elapidae) venom in vitro. Toxins. 2014;6:359–370. doi: 10.3390/toxins6010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Alape-Girón A., Alberto A.-G., Bruno L., Bjorn G., Da Silva N.J., Monica T. Electrophoretic and immunochemical studies of Micrurus snake venoms. Toxicon. 1994;32:713–723. doi: 10.1016/0041-0101(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 205.Alape-Girón A., Stiles B., Schmidt J., Girón-Cortes M., Thelestam M., Jörnvall H., Bergman T. Characterization of multiple nicotinic acetylcholine receptor-binding proteins and phospholipases A2 from the venom of the coral snake Micrurus nigrocinctus. FEBS Lett. 1996;380:29–32. doi: 10.1016/0014-5793(95)01543-4. [DOI] [PubMed] [Google Scholar]
- 206.Goularte F.C., Cruz-Höfling M.A., Cogo J.C., Gutiérrez J.M., Rodrigues-Simioni L. The ability of specific antivenom and low temperature to inhibit the myotoxicity and neuromuscular block induced by Micrurus nigrocinctus venom. Toxicon. 1995;33:679–689. doi: 10.1016/0041-0101(94)00178-B. [DOI] [PubMed] [Google Scholar]
- 207.Oliveira D.A., Harasawa C., Seibert C.S., Casais e Silva L.L., Pimenta D.C., Lebrun I., Sandoval M.R. Phospholipases A2 isolated from Micrurus lemniscatus coral snake venom: Behavioral, electroencephalographic, and neuropathological aspects. Brain Res. Bull. 2008;75:629–639. doi: 10.1016/j.brainresbull.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 208.Camargo T.M., de Roodt A.R., da Cruz-Höfling M.A., Rodrigues-Simioni L. The neuromuscular activity of Micrurus pyrrhocryptus venom and its neutralization by commercial and specific coral snake antivenoms. J. Venom Res. 2011;2:24–31. [PMC free article] [PubMed] [Google Scholar]
- 209.Bénard-Valle M., Carbajal-Saucedo A., de Roodt A., Lopez-Vera E., Alagon A. Biochemical characterization of the venom of the coral snake Micrurus tener and comparative biological activities in the mouse and a reptile model. Toxicon. 2013;77:6–15. doi: 10.1016/j.toxicon.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 210.Condrea E., Rapuano B.E., Fletcher J.E., Yang C.C., Rosenberg P. Effects of arginine modification of Naja nigricollis and Naja naja atra snake venom phospholipases A2 on enzymatic activity, lethality and anticoagulant action. Toxicon. 1981;19:721–725. doi: 10.1016/0041-0101(81)90109-4. [DOI] [PubMed] [Google Scholar]
- 211.Rosenberg P., Condrea E., Rapuano B.E., Soons K.R., Yang C.C. Dissociation of pharmacological and enzymatic activities of snake venom phospholipases A2 by modification of carboxylate groups. Biochem. Pharmacol. 1983;32:3525–3530. doi: 10.1016/0006-2952(83)90298-8. [DOI] [PubMed] [Google Scholar]
- 212.Rosenberg P., Condrea E., Fletcher J.E., Rapuano B.E. Dissociation between enzymatic activity and pharmacological properties of snake venom phospholipases A2. Toxicon. 1983;21:371–375. doi: 10.1016/0041-0101(83)90232-5. [DOI] [PubMed] [Google Scholar]
- 213.Kini R.M., Evans H.J. A model to explain the pharmacological effects of snake venom phospholipases A2. Toxicon. 1989;27:613–635. doi: 10.1016/0041-0101(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 214.Kini R.M. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 215.Salazar A.M., Vivas J., Sánchez E.E., Rodríguez-Acosta A., Ibarra C., Gil A., Carvajal Z., Girón M.E., Estrella A., Navarrete L.F., et al. Hemostatic and toxinological diversities in venom of Micrurus tener tener, Micrurus fulvius fulvius and Micrurus isozonus coral snakes. Toxicon. 2011;58:35–45. doi: 10.1016/j.toxicon.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Evans H.J., Franson R., Qureshi G.D., Moo-Penn W.F. Isolation of anticoagulant proteins from cobra venom (Naja nigricollis). Identity with phospholipases A2. J. Biol. Chem. 1980;255:3793–3797. [PubMed] [Google Scholar]
- 217.Yuan Y., Jackson S.P., Mitchell C.A., Salem H.H. Purification and characterisation of a snake venom phospholipase A2: A potent inhibitor of platelet aggregation. Thromb. Res. 1993;70:471–481. doi: 10.1016/0049-3848(93)90089-7. [DOI] [PubMed] [Google Scholar]
- 218.Rudrammaji L.M., Machiah K.D., Kantha T.P., Gowda T.V. Role of catalytic function in the antiplatelet activity of phospholipase A2 cobra (Naja naja naja) venom. Mol. Cell. Biochem. 2001;219:39–44. doi: 10.1023/A:1011002606505. [DOI] [PubMed] [Google Scholar]
- 219.Sundell I.B., Theakston R.D., Kamiguti A.S., Harris R.J., Treweeke A.T., Laing G.D., Fox J.W., Warrell D.A., Zuzel M. The inhibition of platelet aggregation and blood coagulation by Micropechis ikaheka venom. Br. J. Haematol. 2001;114:852–860. doi: 10.1046/j.1365-2141.2001.03045.x. [DOI] [PubMed] [Google Scholar]
- 220.Yang W.-L., Peng L.-S., Zhong X.-F., Wei J.-W., Jiang X.-Y., Ye L.-T., Zou L., Tu H.-B., Wu W.-Y., Xu A.-L. Functional expression and characterization of a recombinant phospholipase A2 from sea snake Lapemis hardwickii as a soluble protein in E. coli. Toxicon. 2003;41:713–721. doi: 10.1016/S0041-0101(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 221.Satish S., Tejaswini J., Krishnakantha T.P., Gowda T.V. Purification of a Class B1 platelet aggregation inhibitor phospholipase A2 from Indian cobra (Naja naja) venom. Biochimie. 2004;86:203–210. doi: 10.1016/j.biochi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 222.Kini R.M., Evans H.J. Structure-function relationships of phospholipases. The anticoagulant region of phospholipases A2. J. Biol. Chem. 1987;262:14402–14407. [PubMed] [Google Scholar]
- 223.Doery H.M., Pearson J.E. Phospholipase B in snake venoms and bee venom. Biochem. J. 1964;92:599–602. doi: 10.1042/bj0920599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.He J., Chen S., Gu J. Identification and characterization of Harobin, a novel fibrino(geno)lytic serine protease from a sea snake (Lapemis hardwickii) FEBS Lett. 2007;581:2965–2973. doi: 10.1016/j.febslet.2007.05.047. [DOI] [PubMed] [Google Scholar]
- 225.Rau J.C., Beaulieu L.M., Huntington J.A., Church F.C. Serpins in thrombosis, hemostasis and fibrinolysis. J. Thromb. Haemost. 2007;5(Suppl. 1):102–115. doi: 10.1111/j.1538-7836.2007.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bock P.E., Olson S.T., Björk I. Inactivation of thrombin by antithrombin is accompanied by inactivation of regulatory exosite I. J. Biol. Chem. 1997;272:19837–19845. doi: 10.1074/jbc.272.32.19837. [DOI] [PubMed] [Google Scholar]
- 227.Huntington J.A., Read R.J., Carrell R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 228.Meekins D.A., Kanost M.R., Michel K. Serpins in arthropod biology. Semin. Cell Dev. Biol. 2017;62:105–119. doi: 10.1016/j.semcdb.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stark K.R., James A.A. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J. Biol. Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- 230.Calvo E., Mizurini D.M., Sá-Nunes A., Ribeiro J.M.C., Andersen J.F., Mans B.J., Monteiro R.Q., Kotsyfakis M., Francischetti I.M.B. Alboserpin, a factor Xa inhibitor from the mosquito vector of yellow fever, binds heparin and membrane phospholipids and exhibits antithrombotic activity. J. Biol. Chem. 2011;286:27998–28010. doi: 10.1074/jbc.M111.247924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Prevot P.-P., Adam B., Boudjeltia K.Z., Brossard M., Lins L., Cauchie P., Brasseur R., Vanhaeverbeek M., Vanhamme L., Godfroid E. Anti-hemostatic effects of a serpin from the saliva of the tick Ixodes ricinus. J. Biol. Chem. 2006;281:26361–26369. doi: 10.1074/jbc.M604197200. [DOI] [PubMed] [Google Scholar]
- 232.Chmelar J., Oliveira C.J., Rezacova P., Francischetti I.M.B., Kovarova Z., Pejler G., Kopacek P., Ribeiro J.M.C., Mares M., Kopecky J., et al. A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation. Blood. 2011;117:736–744. doi: 10.1182/blood-2010-06-293241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ibelli A.M.G., Kim T.K., Hill C.C., Lewis L.A., Bakshi M., Miller S., Porter L., Mulenga A. A blood meal-induced Ixodes scapularis tick saliva serpin inhibits trypsin and thrombin, and interferes with platelet aggregation and blood clotting. Int. J. Parasitol. 2014;44:369–379. doi: 10.1016/j.ijpara.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tirloni L., Kim T.K., Coutinho M.L., Ali A., Seixas A., Termignoni C., Mulenga A., da Silva Vaz I., Jr. The putative role of Rhipicephalus microplus salivary serpins in the tick-host relationship. Insect Biochem. Mol. Biol. 2016;71:12–28. doi: 10.1016/j.ibmb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Xu T., Lew-Tabor A., Rodriguez-Valle M. Effective inhibition of thrombin by Rhipicephalus microplus serpin-15 (RmS-15) obtained in the yeast Pichia pastoris. Ticks Tick Borne Dis. 2016;7:180–187. doi: 10.1016/j.ttbdis.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 236.Roy H., Bhardwaj S., Ylä-Herttuala S. Biology of vascular endothelial growth factors. FEBS Lett. 2006;580:2879–2887. doi: 10.1016/j.febslet.2006.03.087. [DOI] [PubMed] [Google Scholar]
- 237.Karkkainen M.J., Haiko P., Sainio K., Partanen J., Taipale J., Petrova T.V., Jeltsch M., Jackson D.G., Talikka M., Rauvala H., et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat. Immunol. 2004;5:74–80. doi: 10.1038/ni1013. [DOI] [PubMed] [Google Scholar]
- 238.Pung Y.F., Wong P.T.H., Kumar P.P., Hodgson W.C., Kini R.M. Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 2005;280:13137–13147. doi: 10.1074/jbc.M414137200. [DOI] [PubMed] [Google Scholar]
- 239.St Pierre L., Fischer H., Adams D.J., Schenning M., Lavidis N., de Jersey J., Masci P.P., Lavin M.F. Distinct activities of novel neurotoxins from Australian venomous snakes for nicotinic acetylcholine receptors. Cell. Mol. Life Sci. 2007;64:2829–2840. doi: 10.1007/s00018-007-7352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Junqueira-de-Azevedo I.L.M. Lachesis muta (Viperidae) cDNAs Reveal Diverging Pit Viper Molecules and Scaffolds Typical of Cobra (Elapidae) Venoms: Implications for Snake Toxin Repertoire Evolution. Genetics. 2006;173:877–889. doi: 10.1534/genetics.106.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rokyta D.R., Wray K.P., Lemmon A.R., Lemmon E.M., Caudle S.B. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 242.Hargreaves A.D., Swain M.T., Logan D.W., Mulley J.F. Testing the Toxicofera: Comparative reptile transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon. 2014;92:140–156. doi: 10.1016/j.toxicon.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 243.Tomee J.F., Koëter G.H., Hiemstra P.S., Kauffman H.F. Secretory leukoprotease inhibitor: A native antimicrobial protein presenting a new therapeutic option? Thorax. 1998;53:114–116. doi: 10.1136/thx.53.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhu J., Nathan C., Jin W., Sim D., Ashcroft G.S., Wahl S.M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C.D., et al. Conversion of proepithelin to epithelins: Roles of SLPI and elastase in host defense and wound repair. Cell. 2002;111:867–878. doi: 10.1016/S0092-8674(02)01141-8. [DOI] [PubMed] [Google Scholar]
- 245.Pahari S., Mackessy S.P., Manjunatha Kini R. The venom gland transcriptome of the Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): Towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea) BMC Mol. Biol. 2007;8:115. doi: 10.1186/1471-2199-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sanz L., Pla D., Pérez A., Rodríguez Y., Zavaleta A., Salas M., Lomonte B., Calvete J.J. Venomic Analysis of the Poorly Studied Desert Coral Snake, Micrurus tschudii tschudii, Supports the 3FTx/PLA₂ Dichotomy across Micrurus Venoms. Toxins. 2016;8:178. doi: 10.3390/toxins8060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lomonte B., Sasa M., Rey-Suárez P., Bryan W., Gutiérrez J.M. Venom of the Coral Snake Micrurus clarki: Proteomic Profile, Toxicity, Immunological Cross-Neutralization, and Characterization of a Three-Finger Toxin. Toxins. 2016;8:138. doi: 10.3390/toxins8050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Da Silva N.J., Jr., Sites J.W., Jr. Phylogeny of South American triad coral snakes (Elapidae: Micrurus) based on molecular characters. Herpetologica. 2001;57:1–22. [Google Scholar]
- 249.Slowinski J.B. A Phylogenetic Analysis of the New World Coral Snakes (Elapidae: Leptomicrurus, Micruroides, and Micrurus) Based on Allozymic and Morphological Characters. J. Herpetol. 1995;29:325–338. doi: 10.2307/1564981. [DOI] [Google Scholar]
- 250.Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J., et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Siang A.S., Doley R., Vonk F.J., Kini R.M. Transcriptomic analysis of the venom gland of the red-headed krait (Bungarus flaviceps) using expressed sequence tags. BMC Mol. Biol. 2010;11:24. doi: 10.1186/1471-2199-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jiang Y., Li Y., Lee W., Xu X., Zhang Y., Zhao R., Zhang Y., Wang W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011;12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tan C.H., Fung S.Y., Yap M.K.K., Leong P.K., Liew J.L., Tan N.H. Unveiling the elusive and exotic: Venomics of the Malayan blue coral snake (Calliophis bivirgata flaviceps) J. Proteom. 2016;132:1–12. doi: 10.1016/j.jprot.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 254.Tin M.M.-Y., Economo E.P., Mikheyev A.S. Sequencing degraded DNA from non-destructively sampled museum specimens for RAD-tagging and low-coverage shotgun phylogenetics. PLoS ONE. 2014;9:e96793. doi: 10.1371/journal.pone.0096793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Transdecoder. [(accessed on 5 June 2017)]; Available online: http://transdecoder.sf.net, version r20131110.
- 256.Common Repository of Adventitious Proteins. [(accessed on 5 June 2017)]; Available online: http://www.thegpm.org/crap/
- 257.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 259.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 260.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Li L. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.bioPython v1.64. [(accessed on 7 June 2017)]; Available online: http://biopython.org/wiki/Download.
- 266.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 268.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Köster J., Rahmann S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 270.Github Repository. [(accessed on 5 June 2017)]; Available online: https://github.com/migrau/phylopipe.
- 271.Ogawa Y., Murayama N., Yanoshita R. Molecular cloning and characterization of ecto-5′-nucleotidase from the venoms of Gloydius blomhoffi. Toxicon. 2009;54:408–412. doi: 10.1016/j.toxicon.2009.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptomic sequences have been deposited in the DNA Databank of Japan (DDBJ) under the project ID: PRJDB5628. BioSample IDs are: SAMD00076719-SAMD00076724, and raw data may be accessed under ID: DRA005678 (DRX083388-DRX083393).