Abstract
To study genome evolution and diversity in barley (Hordeum vulgare), we have sequenced and compared more than 300 kb of sequence spanning the Rph7 leaf rust disease resistance gene in two barley cultivars. Colinearity was restricted to five genic and two intergenic regions representing <35% of the two sequences. In each interval separating the seven conserved regions, the number and type of repetitive elements were completely different between the two homologous sequences, and a single gene was absent in one cultivar. In both cultivars, the nonconserved regions consisted of ∼53% repetitive sequences mainly represented by long-terminal repeat retrotransposons that have inserted <1 million years ago. PCR-based analysis of intergenic regions at the Rph7 locus and at three other independent loci in 41 H. vulgare lines indicated large haplotype variability in the cultivated barley gene pool. Together, our data indicate rapid and recent divergence at homologous loci in the genome of H. vulgare, possibly providing the molecular mechanism for the generation of high diversity in the barley gene pool. Finally, comparative analysis of the gene composition in barley, wheat (Triticum aestivum), rice (Oryza sativa), and sorghum (Sorghum bicolor) suggested massive gene movements at the Rph7 locus in the Triticeae lineage.
INTRODUCTION
Comparative genetic mapping in grasses has shown that the gene order is well conserved along the grass chromosomes, despite large differences in genome size, chromosome number, ploidy level, and content of repetitive elements (Gale and Devos, 1998; Bennetzen, 2000; Keller and Feuillet, 2000; Gaut, 2002). Recently, comparative studies of large stretches of BAC DNA sequences at orthologous loci in barley (Hordeum vulgare), sorghum (Sorghum bicolor), wheat (Triticum aestivum), maize (Zea mays), and rice (Oryza sativa) have shown numerous exceptions to colinearity at the molecular level, revealing a mosaic gene conservation in the grass genomes that was overlooked by genetic mapping. These studies have shown that rearrangements caused by insertion/deletion of transposable elements, duplication, insertion, deletion, and inversion of genes as well as gene movements have shaped the different grass genomes since their divergence from a common ancestor 50 to 70 million years ago (Mya) (for recent reviews, see Feuillet and Keller, 2002; Bennetzen and Ma, 2003). Comparative genomic analysis also provided insight into the dynamics of genome evolution by revealing some of the mechanisms underlying the rearrangements such as the rapid turnover of long-terminal repeat (LTR) retrotransposons by unequal and illegitimate recombination or gene amplification followed by gene movements at hotspot regions for sequence divergence (Bennetzen, 2002; Li and Gill, 2002; Ramakrishna et al., 2002; Song et al., 2002). Finally, these studies have revealed differences in the dynamics of genome evolution in different lineages of the grass family and have demonstrated the importance of whole or partial genome duplications during the evolution of these genomes (Gaut et al., 2000; Ilic et al., 2003; Paterson et al., 2003, 2004; Guyot and Keller, 2004).
To date, most of the studies have compared orthologous loci between members of the grass subfamilies Andropogoneae (maize and sorghum), Pooideae (barley and wheat), and Erhartoideae (rice). Comparisons between and/or within these families have provided information on rearrangements that have occurred in a time frame of 50 to 60 to 10 to 14 million years (Bennetzen and Ramakrishna, 2002; Feuillet and Keller, 2002; Ramakrishna et al., 2002; Song et al., 2002; Bennetzen and Ma, 2003; Gu et al., 2003; Ilic et al., 2003). More recently, comparisons have been performed between the homoeologous genomes of wheat that have radiated 2.5 to 4.5 Mya (Huang et al., 2002). Large sequences were compared at orthologous storage protein loci in the homoeologous A and Am genomes of T. monococcum and T. durum (Wicker et al., 2003) and in the A, B, and D genomes of T. durum and Aegilops tauschii (Gu et al., 2004; Kong et al., 2004). In both cases, conservation was mainly restricted to the gene space, and rearrangements, including gene duplications and rapid divergence of intergenic regions through the movement of retroelements, were observed. Thus, even in recently diverged species, rapid and dynamic genome evolution limits the possibilities to assess the type, rate, and precise mechanisms of microrearrangements in grass genomes (Wicker et al., 2003). Intraspecific comparisons were expected to help address these questions. Very recently, Fu and Dooner (2002) and Song and Messing (2003) have compared BAC sequences from different inbred lines of maize. Surprisingly, there was as much rearrangement between two homologous regions in maize as between orthologous loci in two different species. Dramatic differences were not only found in the composition and length of retroelement blocks in the intergenic regions but also in the gene space where several genes were missing. Fu and Dooner (2002) suggested that gene deletions might have resulted from the retrotransposon invasion that occurred in the last 2 to 3 million years in the maize lineage. Moreover, they postulated that independent events of retrotransposon invasion in different individuals of the population of modern maize progenitors are at the origin of the high intergenic sequence variability found in maize. These findings raised many questions. Is this exceptional haplotype variation restricted to the maize genome history, and does it only concern particular regions of the genome, such as those carrying genes that are not essential for plant development (e.g., pigmentation genes)?
We have recently sequenced 212 kb at the leaf rust resistance locus Rph7 in the susceptible barley cultivar Morex (Brunner et al., 2003). Here, we have isolated a physical contig of 350 kb from the homologous region in the resistant cultivar Cebada Capa and have compared 226 kb of this contig with 126 kb of sequence from Morex. No conservation in the size and composition of the intergenic regions and the absence of one gene in Cebada Capa indicate rapid and recent genome divergence in the barley genome. In addition, PCR-based haplotype analyses at the Rph7 locus and at three other independent loci revealed large variability in the intergenic regions in the cultivated H. vulgare gene pool. Finally, comparative analysis of the gene composition at the Rph7 locus in four grass genomes indicates gene movements specifically in the Triticeae lineage.
RESULTS
Establishment of a 350-kb Contig at the Rph7 Locus in Cepada Capa
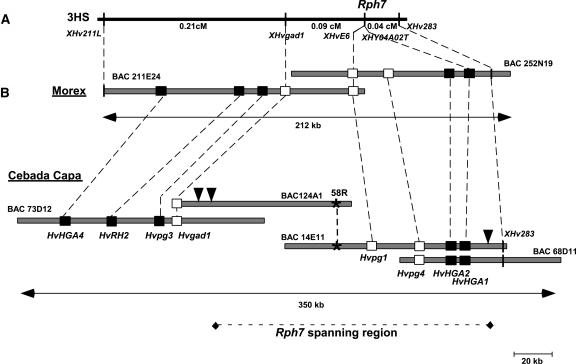
We have recently sequenced 212 kb at the Rph7 locus in the leaf rust susceptible barley cultivar Morex (Brunner et al., 2003). Ten putative genes were identified, whereof five are candidates for Rph7. None of them showed characteristics of known disease resistance (R) genes, suggesting that Rph7 is either a new type of R gene or that it is absent in cv Morex. To test these hypotheses, we have constructed and screened a pooled BAC library of the Rph7 donor line, cv Cebada Capa (Isidore et al., 2005). Screening was performed by PCR with primers corresponding to the Hvpg1 and Hvpg4 genes, which cosegregate with Rph7, as well as to the Hvgad1 gene, which is located proximal to Rph7 (Figure 1). In total, 10 independent BAC clones were isolated. Hybridization of NotI fingerprinted BAC DNA with probes derived from BAC-end sequences as well as probes corresponding to the HvHGA4, Hvrh2, Hvpg3, HvHGA1, and HvHGA2 genes (Figure 1B; data not shown) showed that the 10 BAC clones form a single physical contig of 350 kb represented by BACs 73D12, 124A1, 14E11, and 68D11 (Figure 1B). Clones 124A1 and 14E11, which span the Rph7 region between the flanking markers Hvgad1 and Hv283 (Figure 1A), were chosen for sequencing. Both BACs were sequenced to a coverage of 11X, which resulted in seven contigs for BAC 124A1 and four contigs for BAC 14E11. The gaps were closed by PCR with primers designed at the ends of the contigs. This resulted in two contiguous sequences of 12,258 bp (AY642925) and 184,425 bp (AY642926) separated by a region consisting of LTR retrotransposons of the BARE-1 family. This region could not be assembled to completion because of very high sequence identity between the BARE-1 sequences. However, some single nucleotide differences and different target site duplications (TSDs) suggested that at least four different BARE-1 sequences are present in this interval, which is ∼30 kb long. The 12,258- and 184,425-bp sequences were completely annotated (AY642925-26) using a combination of analytical tools and BLAST searches against the databases.
Figure 1.
Establishment of a 350-kb Physical Contig at the Rph7 Locus in the Resistant Cultivar Cebada Capa.
(A) Genetic map of the Rph7 locus on chromosome 3HS (Brunner et al., 2003).
(B) Comparison of the two physical maps in the susceptible cultivar Morex (Brunner et al., 2003) and the resistant cultivar Cebada Capa. Boxes represent the genes located at the Rph7 locus. White boxes correspond to the three genes (Hvgad1, Hvpg1, and Hvpg4) that were used for screening the Cebada Capa BAC library. The asterisk represents a BAC-end probe that was derived from BAC 58F1 and was used to establish the connection between BACs 124A1 and 14E11. The region spanning the Rph7 gene between the distal and proximal recombination break points (indicated with black vertical arrowheads) is shown as a dotted line.
Comparison of Homologous Contigs in Morex and Cebada Capa Reveals Dramatic and Recent Genome Rearrangements at the Rph7 Locus
The sequences between Hvgad1 and Hv283 (Figure 1) were compared in the two cultivars Morex and Cebada Capa. This region represents 126.6 kb of sequence in Morex (Brunner et al., 2003; position 85,000 to 211,664 in AF521177) and ∼226 kb in Cebada Capa, indicating a large size (∼100 kb) difference between the two regions. Seven conserved regions (CR1-7) ranging from 5 to 12.7 kb were identified by dot plot analysis (see Supplemental Figure 1 online). In total, 53 kb representing 42 and 23% of the sequence in Morex and Cebada Capa, respectively, is conserved with an average of 96% identity. Five conserved regions (CR1 and CR3-6) correspond to sequences containing the Hvgad1, Hvpg1, Hvpg4, HvHGA1, and HvHGA2 genes, whereas CR2 (8.3 kb) and CR7 (5 kb) are intergenic regions (see Supplemental Figure 1 online). Coding regions account for 16.6 kb (31%) of the conserved 53 kb stretch, whereas the rest (69%) mainly corresponds to 3′ and 5′ untranslated regions (UTRs) with an average size of 2.7 kb.
Large variations in the size of the intervals separating the seven conserved regions are found between the two sequences. In the CR4-CR5 interval, ∼22 kb of additional sequence is found in Morex compared with Cebada Capa. In the remaining intervals, additional sequence (100 kb) is found in the intergenic regions of Cebada Capa (see Supplemental Figures 1 and 2 online). The additional sequences correspond to repetitive elements (40%) and to other, putatively nonrepetitive, sequences (60%). Consequently, although there are more repetitive elements in Cebada Capa (one CACTA transposon, six solo LTRs, and three complete LTR retrotransposons) (see Supplemental Figure 2 online) compared with Morex (one solo LTR and two complete LTR retrotransposons), the proportion of repetitive sequence in Cebada Capa (50%) is very similar to the one previously found in Morex (55%; Brunner et al., 2003).
To study the structure and evolution of the nonconserved intergenic regions, the seven conserved regions (CR1-CR7) were assembled into single DNA stretches of 53 kb as if they would represent an ancestral sequence present in both cultivars (Figure 2A). The position and type of additional sequences that were identified in Morex and Cebada Capa were then compared (Figure 2A). Repetitive elements were found in all intervals except in the region between CR4 and CR5 (Figure 2A). Most belong to different families, and those belonging to the same family did not correspond to conserved elements. For example, in the CR1-CR2 interval, a BARE-1 retrotransposon is found at the same position in both sequences. However, the presence of different TSDs demonstrates that they do not correspond to the same element. In both cultivars, different types of transposable elements, partial elements as well as duplicated and inverted sequences were identified in the intergenic region between the Hvgad1 and Hvpg1 genes (Figure 2A; see Supplemental Figure 2 online). By contrast, only complete retrotransposons and a solo LTR were identified in the other intergenic regions. Recent retrotransposition of BARE-1 elements in the Hvgad1-Hvpg1 interval in both cultivars is suggested by the complete identity (100%) of the LTR of BARE-1_211E24-1 in Morex (Brunner et al., 2003) and the very high similarity observed among the different BARE-1 sequences in the 30-kb region in Cebada Capa. In Cebada Capa, sequence expansion in this interval also resulted from two duplications of ∼21 and 9 kb flanking the solo LTR SLB2 and the CACTA transposon Caspar_124A1-1 (see Supplemental Figure 2 online). Together, these data suggest that the Hvgad1-Hvpg1 intergenic region was subjected to more rearrangements than the other regions and that despite the difference in sequence composition, this characteristic property is conserved.
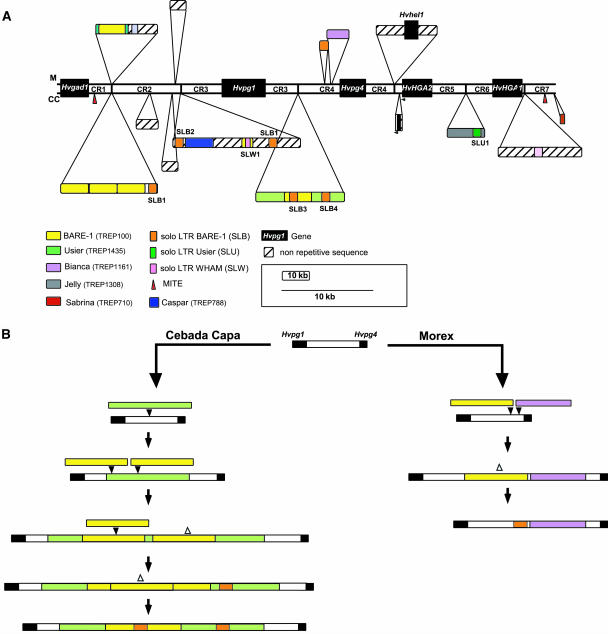
Figure 2.
Sequence Rearrangements in the Intergenic Regions in Morex and Cebada Capa.
(A) Schematic representation of 53 kb of DNA sequence corresponding to the assembly of the seven conserved regions (CR1-7) in Morex and Cebada Capa. The top line (M) represents the 53 kb of sequence from Morex and the bottom one (CC) the sequence from Cebada Capa. Filled boxes with color codes given in the figure represent transposable elements that have been inserted in each sequence. Hatched boxes represent nonrepetitive sequences that also differ from the common sequence stretch. Red triangles represent miniature inverted-repeat transposable elements. Genes are represented as black boxes and identified by name. The size scale for the conserved regions (CR1-CR7) is indicated as a line and is three times larger than the one (indicated as a box) used for the nonconserved repetitive and nonrepetitive sequences. Two small arrowheads indicate the duplicated region of 804 bp found near the 3′ end of HvHGA2 in Cebada Capa.
(B) Model for the evolution of the Hvpg1-Hvpg4 intergenic region (CR3-CR4) in Cebada Capa (left) and Morex (right). Color codes are the same as in (A). Filled arrowheads indicate the insertion of an element, whereas empty ones indicate unequal recombination between LTR of the same elements leading to solo LTRs with identical TSDs.
The 7.7-kb region between Hvpg1 and Hvpg4 (CR3-CR4) is highly conserved (98% identity). This allowed us to identify the exact insertion positions for the different retroelements and to estimate the sequence and timing of insertions and rearrangements that have shaped this region (Figure 2B). A BARE-1 solo LTR and a Bianca retrotransposon were found in this interval in Morex (Brunner et al., 2003), whereas in Cebada Capa, two full-length LTR retrotransposons (Usier_14E11-1 and BARE-1_124A1-1) and two BARE-1 solo LTRs (SLB3 and 4) were identified (Figure 2A). We have determined the number of substitutions in the LTR sequences of the complete retrotransposons. Assuming that at the time of insertion both LTRs of a retrotransposon are 100% identical and using a mutation rate of 6.5 × 10−9 substitutions per synonymous site per year (Gaut et al., 1996; SanMiguel et al., 1998), the approximate age of the complete elements was estimated (Table 1). This analysis indicates that Bianca_252N19-1 was inserted less than 1 Mya (0.92) in the progenitor of Morex, whereas in Cebada Capa, the oldest element Usier_14E11-1 was inserted ∼2 Mya. Subsequent insertions and intraelement unequal recombination of three BARE-1 retrotransposons followed the insertion of Usier_14E11-1 in Cebada Capa (Figure 2B). Our estimates indicate that the complete BARE-1_124A1-1 element inserted ∼1.30 Mya consistent with its location within Usier_14E11-1. The presence of the SLB3 and four solo LTRs in BARE-1_124A1-1 indicates retrotransposition and intraelement recombination within the last million years (Figure 2B). We have also estimated the age of the conserved 7.7-kb ancestral intergenic sequence using the same molecular clock. Our estimate indicates a divergence time of 1.15 million years (±0.15 million years; data not shown), which is younger than the insertion time of the Usier_14E11-1 retroelement (2.15 million years, ±0.3 million years). A similar discrepancy was found in the HvHGA2-HvHGA1 interval (data not shown). These discrepancies between the age of the retroelements and the region in which they are inserted support previous suggestions (SanMiguel et al., 1998; Ma et al., 2004; Ma and Bennetzen, 2004) that LTR retroelements evolve at least two times faster than the genes and the UTR regions. If we divide the insertion time estimates of all retroelements in this region by a factor 2, our data suggest high retroelement activity within the last 500,000 years in this region of the barley genome.
Table 1.
Estimated Times of Retrotransposon Insertion at the Rph7 Locus
| Elements | K (±sd)a | Time Mya (±sd) |
|---|---|---|
| BARE 1_211E24-1 (Morex) | 0 | 0 |
| Bianca_252N19-1 (Morex) | 0.012 (0.008) | 0.92 (0.6) |
| BARE1_14E11-1 (Cebada Capa) | 0.017 (0.003) | 1.30 (0.23) |
| Usier_14E11-1 (Cebada Capa) | 0.028 (0.004) | 2.15 (0.30) |
| Jelly_14E11-1 (Cebada Capa) | 0.022 (0.006) | 1.69 (0.46) |
K, estimated number of substitutions per nucleotide site and its standard deviation. K is based on the γ-K2P model for LTR (Kimura, 1980). The average substitution rate of 6.5 × 10−9 substitutions per synonymous site per year that was estimated for the adh1 and adh2 genes in maize (Gaut et al., 1996) was applied to estimate the divergence time.
No repetitive elements were detected in the CR4-CR5 interval, but a large insertion of 22.2 kb containing the Hvhel1 gene is found in Morex (Figure 2A). This demonstrates that differences between the two barley sequences are not restricted to the repetitive sequence blocks but are also found in the gene composition. In the homologous interval in Cebada Capa, a small sequence of 804 bp corresponding to a duplication of 519 bp of the 3′ end and 3′UTR of HvHGA2 and 285 bp of additional sequence was found (Figure 2A). In Morex, the HvHGA1 and HvHGA2 genes are separated by an intergenic region of 6.4 kb, whereof 5.6 kb is conserved in Cebada Capa (with 95% identity). An insertion of 15.4 kb that mostly (13.6 kb) corresponds to a complete LTR retroelement (Jelly_14E11-1) interrupted by a Usier solo LTR (SLU1) occurred in Cebada Capa (Figure 2A). Jelly_14E11-1 was estimated to have inserted 1.7 Mya, confirming that at the Rph7 locus, the retroelements are not older than 2 million years. Finally, in the CR6-CR7 interval, 29 kb of additional sequence is found in Cebada Capa compared with Morex. Sequence analysis did not clearly identify repetitive patterns or elements, but short homologies by BLASTX with the hypothetical protein pTREP15 and putative rice transposases or reverse transcriptase indicates the presence of an unidentified element or remaining traces of an older element. In the noncoding CR7 region, (TA) microsatellites and XI_AF521177-1 elements are conserved at slightly different positions relative to each other in the two regions. Together, our data show a lack of conservation in the length and composition of the intergenic regions between the two barley Rph7 homologous regions, indicating rapid and recent sequence divergence in the barley genome. Moreover, the fact that no additional genes were identified in the sequence of the resistant cultivar Cebada Capa indicates that Rph7 is likely a new type of disease R gene.
To examine the effect of such a high level of sequence divergence on recombination in the Rph7 region, we have analyzed the recombination breakpoints in Cebada Capa and Morex with the assumption that the sequence in Morex is highly similar to the one of Bowman, the parent used for genetic linkage analysis (Brunner et al., 2003). Using a combination of restiction fragment length polymorphism hybridizations and restriction fragment analysis on the BAC sequences, we have identified one recombination break point in a 1.8-kb sequence next to XHv283 in the conserved region CR7 and two recombination events in the CR1-CR2 interval (Figure 1). In this interval, ∼7.5 kb of the CR1 and CR2 regions as well as sequences from the BARE-1 retroelements are available for homologous recombination. These data indicate that despite overall low sequence conservation between the two haplotypes, there are enough conserved regions that can serve as target sequences for homologous recombination and that recombination is not completely blocked at this locus.
High Variability Is Found in Intergenic Regions at the Rph7 Locus in the Cultivated Barley Gene Pool
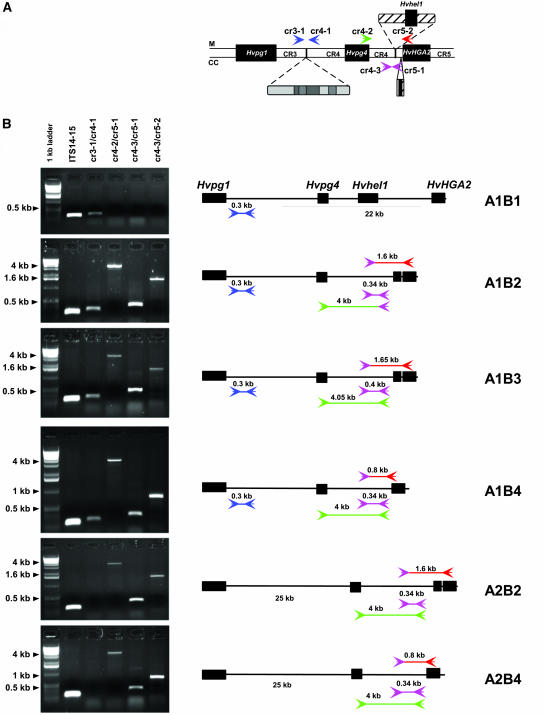
To investigate whether the variability observed in the intergenic regions of Morex and Cebada Capa specifically reflects the genetic distance between the two lines or if there is generally high variability in the cultivated barley gene pool, primers were designed in the CR3, CR4, and CR5 regions (Figure 3A) and used to amplify genomic DNA from 39 additional cultivated H. vulgare lines. PCR amplification across the CR3-CR4 interval (primers cr3-1/cr4-1) produced a 300-bp fragment in Morex, whereas no amplification product was obtained from Cebada Capa because of the insertion of 25 kb of repetitive sequence (Figure 3B). The analysis of the 39 lines showed amplification of the 300-bp fragment in 22 lines and no amplification in 17 lines (Table 2), indicating the presence of at least two haplotypes represented by Morex (A1) and Cebada Capa (A2) at this locus in the cultivated barley gene pool. A similar strategy was used for amplification in the CR4-CR5 interval. In this case, two sense primers were designed at the 5′ end of the Hvpg4 gene (cr4-2) and on the left border of the insertion point for the 22-kb sequence in Morex (cr4-3), and two reverse primers were designed on the right side of the insertion point of the Morex 22kb sequence (cr5-1) and on the 3′ end of HvHGA2 (cr5-2) (Figure 3A). In Morex, none of the primer combinations resulted in a PCR product because of the 22-kb insertion in this interval. In Cebada Capa, amplification with cr4-3/cr5-1 resulted in a 340-bp fragment, PCR with cr4-2/cr5-1 gave a 4-kb fragment, and the cr4-3/cr5-2 combination lead to the amplification of a 1.6-kb fragment (Figure 3B). Amplification with these three different primer combinations resulted in four different types of products in the 39 lines (Figure 3B). Eight lines, including near-isogenic lines carrying different Rph genes in the background of Bowman, gave the same pattern as in Morex (i.e., no product; B1 haplotype) (Table 2). Sixteen lines produced the same fragments as in Cebada Capa (B2 haplotype) (Table 2). Among these lines, 14 that originate from Ethiopia, Eritrea, Uruguay, and Argentina carry the Rph7 resistance allele. Two other lines (Quinn and Magnif104) showed a 60-bp size difference with all primer combinations, indicating a small insertion between primers cr4-3 and cr5-1 (Table 2, B3 haplotype). Finally, 13 lines (B4 haplotype) amplified a 0.8-kb fragment with the primers cr4-3/cr5-2 instead of the expected 1.6-kb fragment, whereas the other fragments had the same size as in Cebada Capa. Cloning and sequencing of the 0.8-kb fragment demonstrated that the size difference results from the absence of the duplicated 804 bp found at the 3′ end of the HvHGA2 gene in Cebada Capa. In summary, at least two haplotypes (A1 and A2) can be found for the CR3-CR4 interval and four (B1, B2, B3, and B4) for the CR4-CR5 interval (Figure 3B). Analysis of the haplotype combinations for both intervals in the 41 lines indicated that six (A1B1, A1B2, A1B3, A1B4, A2B2, and A2B4) out of the eight possible combinations were found in the cultivated barley gene pool (Figure 3B, Table 2). Interestingly, all the lines containing the Rph7 resistance allele had the same haplotype (A2B2, Table 2). The line Sudan also had the A2B2 haplotype, but it is not known to carry the Rph7 resistance allele. In artificial leaf rust infection tests, Sudan was resistant to the AvrRph7 isolate, although the resistance phenotype was different than that observed with Cebada Capa and the 14 lines with the A2B2 haplotype (data not shown). These results suggest that Sudan may carry a different resistance allele of Rph7 and that all lines carrying Rph7 likely originate from a single donor line.
Figure 3.
Haplotype Combinations Found in the Hvpg1-Hvpg4-HvHGA2 Intergenic Regions in 41 Cultivated Barley Lines.
(A) Schematic representation of the Hvpg1-Hvpg4 and Hvpg4-HvHGA2 intervals in Morex (M) and Cebada Capa (CC) showing the position, orientation, and names of the primers used in the analysis.
(B) PCR products amplified by four primer combinations and schematic representation of the six haplotype combinations identified in the 41 barley lines. The ITS14-15 primer combination was used as a positive control for PCR amplification. PCR products are represented as lines between the primers with their size above it.
Table 2.
Haplotype Combinations in the CR3-CR4 and CR4-CR5 Intervals at the Rph7 Locus and Haplotypes at the Vrn2, Vrn1, and MWG838-MWG010 Loci in 41 Cultivated Barley Lines
|
Rph7
|
||||||
|---|---|---|---|---|---|---|
| Cultivars | R Gene | CR3-CR4 | CR4-CR5 | Vrn2 | Vrn1 | MWG838-MWG010 |
| Morex | A1 | B1 | C1 | D1 | E1 | |
| Bowman | A1 | B1 | C2 | D1 | E1 | |
| Bowman*7/Hor2596 | Rph9 | A1 | B1 | C2 | D1 | E1 |
| Clipper BC8/5*Bowman | Rph10 | A1 | B1 | C2 | D1 | E1 |
| Bowman*5/Clipper BC67 | Rph11 | A1 | B1 | C2 | D1 | E1 |
| Bowman*6/PI531849 | Rph13 | A1 | B1 | C2 | D1 | E2 |
| Bowman*4/PI584760 | Rph14 | A1 | B1 | C2 | D1 | E1 |
| Sundance | A1 | B1 | C2 | D1 | E1 | |
| Franka | —— | A1 | B1 | C1 | D2 | E2 |
| Tunisian | A1 | B2 | C2 | D1 | E1 | |
| Egypt 4 | Rph8 | A1 | B2 | C2 | D1 | E1 |
| Quinn | Rph2,5 | A1 | B3 | C2 | D3 | E1 |
| Magnif104 | Rph5 | A1 | B3 | C2 | D1 | E1 |
| Ribari | Rph3 | A1 | B4 | C2 | D1 | E1 |
| Gold | Rph4 | A1 | B4 | C2 | D1 | E1 |
| CI1243 | Rph9 | A1 | B4 | C2 | D1 | E2 |
| Triumph | Rph12 | A1 | B4 | C1 | D1 | E1 |
| Elisa | A1 | B4 | C2 | D1 | E1 | |
| JCJ-188 | A1 | B4 | C1 | D1 | E1 | |
| L94 | A1 | B4 | C2 | D1 | E2 | |
| Meltan | A1 | B4 | C1 | D1 | E1 | |
| Michka | A1 | B4 | C1 | D1 | E1 | |
| Pallas | —— | A1 | B4 | C2 | D1 | E1 |
| Cebada Capa | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Ab1122 | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Cebada Forrajera | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Dabat | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Debra Sina | Rph7 | A2 | B2 | C2 | D1 | E1 |
| A2210 | Rph7 | A2 | B2 | C2 | D1 | E1 |
| A2211 | Rph7 | A2 | B2 | C2 | D1 | E1 |
| A2212 | Rph7 | A2 | B2 | C2 | D1 | E1 |
| La Estanzuela | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Hor4445 | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Hanka | Rph7 | A2 | B2 | C1 | D1 | E1 |
| Heris | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Ellinor | Rph7 | A2 | B2 | C1 | D1 | E1 |
| Bowman*8/Cebada Capa | Rph7 | A2 | B2 | C2 | D1 | E1 |
| Sudan | Rph1 | A2 | B2 | C2 | D1 | E1 |
| Peruvian | Rph2 | A2 | B4 | C2 | D1 | E1 |
| Bolivia | Rph2,6 | A2 | B4 | C2 | D1 | E1 |
| Lechtaler | Rph4 | A2 | B4 | C2 | D2 | E1 |
The presence of known leaf rust disease resistance genes in the cultivars is indicated in the second column. Cultivars belonging to one of the six haplotype combinations (AxBx) found at the Rph7 locus are separated from each other by single lines.
To test whether the variability observed at the Rph7 locus does not specifically reflect high variability at a fungal disease resistance locus, we have scanned all barley BAC sequences that do not originate from a disease resistance locus and are available from the databases for intergenic regions amenable to PCR amplification. Six intergenic regions that can result in PCR products ranging from 0.5 to 3.9 kb in Morex were identified from BAC sequences originating from the Hordein (AY268139), Waxy1 (WX1) (AF474373), Vrn1 (Wg644) (AY013246), and Vrn2 (AY485643) loci. Six primer pairs were designed based on the open reading frames predicted in the annotations and BLASTN analysis against the EST database. In addition, one primer pair was designed in an intergenic region of 1.4 kb originating from a BAC located in the telomeric region of chromosome 3HL between markers MWG838 and MWG010 (N. Stein, personal communication). Out of six primer combinations that amplified the expected product from Morex, three showed the presence of at least two haplotypes in the corresponding intergenic regions on the set of 41 cultivated barley lines. Two to three different haplotypes were found at the orthologous wheat Vrn2 and Vrn1 loci on chromosome 4H and 5H, respectively, and two were found at the MWG838-MWG010 locus on 3HL (Table 2; see Supplemental Figure 3 online). The cultivars that belong to a particular haplotype were different at each independent locus (Table 2). From these data, we conclude that the variability found at the Rph7 locus is not specific for this locus but is indicative for a large variability in intergenic regions of the genome from cultivated barley.
Evolution of the Gene Composition at Rph7 Orthologous Loci in Wheat, Barley, Rice, and Sorghum
The structure and transcriptional orientation of the genes identified in Morex and Cebada Capa are highly conserved. The Hvpg4, Hvgad1, HvHGA2, and HvHGA1 alleles share nucleotide sequence identity ranging from 98 to 99.6% over the entire sequence. The Hvpg1 alleles are also highly conserved (98% identity at the nucleotide sequence level) except for a deletion of 198 bp at the C terminus of the Cebada Capa allele, which results in the lack of 66 amino acids corresponding to two terminal repeats of a Gly-rich repeat domain. We had previously shown that microcolinearity between barley and rice at the Rph7 locus was restricted to the conservation of members of the HGA gene family on rice chromosome 1 and that genes orthologous to Hvpg1, Hvpg3, Hvpg4, and Hvgad1 are located on the homeologous group 3 chromosomes in wheat (Brunner et al., 2003). A BLASTN search with all the gene sequences against the recently created database of mapped wheat EST (http://wheat.pw.usda.gov/wEST/blast/) allowed us to identify the location of these orthologs in deletion bins on the wheat chromosomes. HvHGA1 (E value = 1e-126), HvHGA2 (E value = 1e-75) , Hvpg1 (E value = 1e-79), Hvpg3 (E value = 3e-22), and Hvpg4 (E value = 4e-50) orthologs were identified in the three most distal bins of chromosomes 3A, 3B, and 3D (3AS4-0.45-1.00, 3BS8-0.78-1.00 and 3BS1-0.33-0.57, and 3DS6-0.55-1.00), confirming the conservation of the Rph7 locus composition in barley and wheat (Figure 4). In addition, BLAST hits for Hvpg4 (E value = 4e-50) and Hvgad1 (E value = 3e-27) were found in bins of chromosomes 4AS, 4BL, and 4DL.
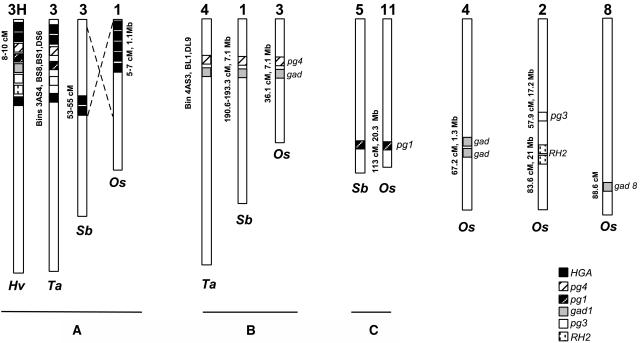
Figure 4.
Orthologous Relationships between the Genes Located at the Rph7 Locus in Barley, Wheat, Rice, and Sorghum.
The location of the genes on genetic maps and/or on physical maps is indicated on the left side of the chromosomes. Hv, Hordeum vulgare; Ta, Triticum aestivum; Sb, Sorghum bicolor; Os, Oryza sativa. The inversion identified by Klein et al. (2003) between rice chromosome 1 and sorghum chromosome 3 is indicated with dashed lines. A, B, and C indicate groups of synteny between at least two of the grass species investigated.
In rice, previous BLASTN and TBLASTX searches had indicated the presence of homologs for Hvrh2, Hvpg1, Hvhel1, Hvpg3, Hvpg4, and Hvgad1 in the rice genome, but the chromosomal location could not be assessed at that time (Brunner et al., 2003). The recent release of 12 rice pseudomolecules by TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) allowed us to identify the chromosomal location of genes homologous to Hvrh2, Hvpg3, hvgad1, Hvpg1, and Hvpg4 in rice. Using BLASTN, single hits were found for a pg1 gene (E value = 6e-48) on chromosome 11 and for a pg3 gene (E value = 2e-18) and two paralogs of rh2 (E value = 3e-28) on chromosome 2 (Figure 4). Four hits were identified with the Hvgad1 sequence: one on chromosome 3, 46 kb away from a homolog of Hvpg4 (E value = 3e-49), two for homologous genes located 13.5 kb from each other on chromosome 4, and the best hit (E value = 6e-94) was found with a gad gene (gad8) on chromosome 8 (Figure 4). Thus, these data indicate that putative orthologs of the genes found at the Rph7 locus in barley are present in rice but are found on five noncolinear chromosomes.
To investigate the origin of this large rearrangement, we have identified homologs for several of these genes and determined their chromosomal location in sorghum, a member of the Andropogoneae subfamily. Sorghum ESTs were identified for the HvHGA1, HvHGA2, Hvpg1, Hvpg4, and Hvgad1 genes and were used to screen six dimensional BAC DNA pools (Klein et al., 2003). The presence of sorghum genes homologous to the barley genes within each positive BAC was confirmed after hybridization with probes for the different barley genes (data not shown). In total, seven BACs were identified. Three (61E22, 63A11, and 52N1) contain a homolog of Hvpg1, two (76H6a and 67F2) carry homologous genes to HvHGA2 and HvHGA1, and single BACs have been identified with Hvpg4 (73H13) and Hvgad1 (62H18). The chromosomal location of each BAC was determined by integrating the BACs with the sorghum genetic map (Klein et al., 2000) or by BAC-based fluorescence in situ hybridization (FISH) analysis (data not shown). BACs 76H6 and 67F2 harboring the HGA1 and HGA2 genes were located on sorghum chromosome 3 (53 to 55 centimorgan [cM]) in a position colinear to rice chromosome 1 and to the Triticeae chromosome 3 (Figure 4). This indicates the conservation of an HGA gene family at colinear positions in rice, barley, wheat, and sorghum (group A, Figure 4). BACs 73H13 (pg4) and 62H18 (gad) mapped on sorghum chromosome 1 (190.6 to 193.3 cM) at a position orthologous to rice chromosome 3 (36.1 cM). Homologs of the pg4 and gad genes were also found in bins of chromosome group 4 in wheat (see above). Thus, these two genes are found in colinear regions (group B, Figure 4) on wheat chromosome group 4, rice chromosome 3, and sorghum chromosome 1. Finally, the three BACs carrying pg1 homologs mapped to sorghum chromosome 5 (Menz et al., 2002; Kim et al., 2004), which is syntenic to rice chromosome 11 where the rice pg1 ortholog was identified (group C, Figure 4). Together, these data indicate that the genes in sorghum are not found at a single locus colinear with the Triticeae group 3 but are located on different chromosomes at syntenic positions with rice. The fact that a similar gene distribution is found in members of two different subfamilies (Andropogoneae and Erhartoideae) suggests gene translocation events in the ancestral group 3 chromosomes during the evolution of the Triticeae genomes.
DISCUSSION
Intraspecific Comparison of Homologous Loci Reveals Mechanisms Underlying Rapid Genome Evolution in Barley
In this study, we have compared more than 300 kb of homologous sequence from two cultivated barley lines that differ in their resistance to leaf rust. Conservation was limited to five genic and two intergenic regions leaving up to 77% divergent sequence between the two loci. This high level of sequence divergence reflects a completely different composition within the intervals spacing the seven conserved regions. In these intervals, neither the type of repetitive elements nor their insertion positions were conserved between the two cultivars. Moreover, one putative helicase gene was absent in Cebada Capa compared with Morex. The difference in the gene composition was not as dramatic as the one recently described in comparative studies between maize inbreds. At the bronze bz locus, four genes were missing in one inbred line compared with the other (Fu and Dooner, 2002), whereas at the zein z1C-1 locus, 10 genes were affected by segmental duplications, insertions, and deletions that have occurred differentially in the two inbreds (Song and Messing, 2003). Additional comparisons at other loci in maize and barley are needed to determine whether the apparent higher level of gene rearrangement observed in maize compared with barley is genome specific as suggested by Song and Messing (2003) or rather locus specific.
Except for a small DNA stretch in the 7.7-kb Hvpg1-Hvpg4 intergenic region, which was previously annotated as a partial element (George_252N19-1) in Morex (Brunner et al., 2003), there was no evidence for conservation of any repetitive element between the two barley sequences. Although we cannot exclude that part of the conserved regions correspond to new, not yet identified repetitive elements, this absence of conservation is striking. It particularly contrasts with recent findings in comparative analysis between orthologous loci on wheat homoeologous genomes. At the low molecular weight GluA-3 locus, a single solo LTR of the retrotransposon Wilma was conserved between the A and Am genomes (Wicker et al., 2003), and at the high molecular weight Glu-1 loci on the A, B, and D genomes, partial sequences of Wilma and Sabrina LTR retrotransposons as well as a stowaway miniature inverted-repeat element were colinear (Gu et al., 2004; Kong et al., 2004). Similarly, a CACTA transposon, an LTR retrotransposon, and five fold-back elements were found at conserved positions on the A and Am genomes at the Lr10 orthologous resistant loci (E. Isidore and B. Keller, unpublished results). Several lines of evidence further support the hypothesis of a very dynamic barley genome with recent activity of transposable elements: (1) estimates of divergence time indicate retroelement movements not older than 1 million years, (2) the majority of the retroelements are complete, and one BARE-1 element has identical LTR, and (3) none of the repetitive elements are conserved. Evidence for an active barley genome was also found by Kalendar et al. (2000) who showed that BARE-1 retrotransposition has been induced in response to environmental stress during evolution of wild barley populations in Israel. Halterman and Wise (2004) have also recently shown that members of the Mla powdery mildew resistance gene family have been subjected to differential insertion of repetitive elements within and flanking the open reading frames in four barley cultivars. In the maize lineage, it is estimated that retrotransposon invasion occurred in the past 3 million years (SanMiguel et al., 1998; Gaut et al., 2000). Fu and Dooner (2002) have suggested that this invasion has occurred separately in different individuals of the population from which modern maize eventually evolved, resulting in large variability between allelic regions in inbreds. It is possible that similar events are responsible for the absence of conservation between the two barley cultivars. It is not yet known when retroelement invasion occurred in the barley or the wheat lineages and whether this occurred in one wave such as in maize. However, together with the recent comparative studies between wheat homoeologous genomes (Wicker et al., 2003; Gu et al., 2004; Kong et al., 2004) and between Mla alleles in barley (Halterman and Wise, 2004), our data suggest retroelement movements in the different lineages of modern wheat and barley in recent evolutionary times (1 to 3 million years). The complete lack of colinearity between repetitive elements found at barley and wheat orthologous loci (Ramakrishna et al., 2002; Gu et al., 2003) and the low amount of conserved elements between wheat homoeologous loci and barley homologous regions suggest a complete turnover of the repetitive sequences in the Triticeae genomes within a period of less than 4 to 5 million years. Wicker et al. (2003) have recently suggested less than 3 million years for the complete elimination of elements in the intergenic regions in wheat.
The average divergence time estimates for most of the genes present at the Rph7 locus (1.2 million years; data not shown) suggest a radiation time for wild barley lineages comparable to the maize lineages (1 to 2 million years) (Gaut and Clegg, 1993) and to the 0.5 to 1 million years estimated for the A genomes of wheat (Huang et al., 2002). This is in the same time frame as the estimated divergence time for the two rice subspecies O. sativa ssp indica and japonica (Bennetzen, 2000; Ma and Bennetzen, 2004). However, in contrast with the high level of variability observed between the progenitors of modern maize and barley cultivars, high conservation has been found between orthologous sequences in the two rice subspecies (Song et al., 2002; Han and Xue, 2003; Ma and Bennetzen, 2004). Thus, these data indicate large differences in the activity of the different grass genomes in the last 2 million years of evolution and support the idea that rice has a more stable genome than other grasses (Ilic et al., 2003). Finally, our data and the recent findings in maize (Fu and Dooner, 2002; Song and Messing, 2003) also demonstrate that intraspecific comparisons can be as informative as interspecific comparisons in revealing the mosaic organization of orthologous sequences in grass genomes.
Despite high sequence divergence, interesting features were conserved in intergenic regions of both cultivars. First, in the two sequences, some regions (e.g., Hvpg4-HvHGA2 interval) seem to be protected from transposable element invasion, whereas others, such as the Hvgad1-Hvpg1 intergenic regions, have complex patterns of sequence rearrangements and contain several partial sequences of repetitive elements. Mechanisms underlying the preferential insertion of repetitive elements at target sites in plant genomes are not yet well understood, and except for repeat sequences of knob DNA in maize (Ananiev et al., 1998), no particular sequences have been specifically associated yet with the insertion of repetitive elements. In this respect, the conservation of the insertion position for the BARE-1 retrotransposons in the CR1-CR2 interval is very interesting. Thus, intraspecific comparisons may be very helpful in identifying conserved regions with preferential insertion of transposable elements. A second interesting trend is the conservation of a ratio of ∼50% in the proportion of repetitive to nonrepetitive sequence in both regions. This is lower than the average 70% found at other loci in barley (Dubcovsky et al., 2001; Rostoks et al., 2002) and might therefore be locus specific, reflecting particular evolutionary constraints associated with the presence of essential genes in this gene-rich region.
Finally, this comparison has allowed us to determine that there are no additional candidate genes for Rph7 at the resistance locus in Cebada Capa, demonstrating that Rph7 belongs to a new type of disease resistance gene. Complementation experiments to identify Rph7 among the four candidate genes Hvpg1, Hvpg4, HvHGA1, and HvHGA2 are currently underway.
Large Haplotype Variability in the Cultivated Barley Gene Pool
Our analysis indicates high diversity at the Rph7 locus in the cultivated gene pool. The relative representation of the different haplotype combinations in 41 H. vulgare lines shows that the combinations observed in Morex and in Cebada Capa do not correspond to the predominant (A1B4) haplotype. In fact, the A1B1 combination of Morex was only found in three other lines, and the A2B2 combination of Cebada Capa was detected in the lines known to carry the Rph7 resistance allele. The presence of a single resistant haplotype for Rph7 in cultivars that originate from Ethiopia, Eritrea, Uruguay, and Argentina suggests a single origin of the Rph7 gene, very likely in the Fertile Crescent, the center of origin of barley (Badr et al., 2000). We are currently analyzing a larger set of 200 wild barley lines from Israel and the Fertile Crescent to determine to what extent the diversity observed in the cultivated gene pool originates from the wild relatives and to study the relative abundance of the Rph7 resistance haplotype in the wild gene pool to understand the origin of this important disease resistance gene.
So far, genetic diversity studies in cultivated barley have been performed with different DNA marker techniques and have given contradictory results depending on the locus or the marker type used (Graner et al., 2003). Here, we show that PCR-based analysis of intergenic regions can also be used to detect the genetic diversity present in the cultivated barley germplasm. Our results support and extend recent results of Halterman and Wise (2004), indicating variability in flanking regions of the Mla genes. Moreover, we show that variability in intergenic regions is not restricted to disease resistance loci, which are often considered as more variable than other loci, but is also found at three independent loci. Further BAC sequence comparisons between barley homologous regions should help to confirm these data and to determine whether variability is mainly based on transposable element activity in the intergenic regions or also involves gene loss and gene relocation, such as in maize, with a possible impact on gene dosage and recombination distribution along the chromosomes. Furthermore, expression studies of the genes found at the Rph7 locus in Morex and Cebada Capa will be performed to determine whether, similar to the finding of Song and Messing (2003) in maize, allelic gene expression is differentially regulated in noncolinear barley haplotypes.
A Complex History of Rearrangements Involving Gene Movements Is Responsible for the Gene Diversity at the Rph7 Locus in the Triticeae
The Rph7 locus is a diverse gene-rich locus and represents a very good model for studying complex gene rearrangements during the evolution of grass genomes. Our comparative analysis in barley, wheat, sorghum, and rice suggests that the gene composition observed in barley and wheat results from gene movements specifically in the Triticeae lineage. The orthologous relationships found in sorghum and rice indicate that the gene distribution in these species likely reflects an ancestral pattern. As the number of comparative studies between rice and other grass genomes increases, there are more examples of genes that are conserved in the different genomes but are found at nonorthologous positions on the chromosomes (Li and Gill, 2002; Song et al., 2002; Ilic et al., 2003). Recently, Guyot et al. (2004) have analyzed the conservation of more than 200 ESTs mapped in deletion bins on the short arm of wheat chromosome 1AS with the sequence of the 12 rice pseudomolecules. Less than 20% of the ESTs were found in colinear regions on rice chromosome 5S, whereas the remaining ESTs were distributed at nonorthologous loci on all other rice chromosomes. Song et al. (2002) have suggested a possible mechanism underlying gene movements based on gene amplification followed by illegitimate recombination. Interestingly, the gad and rh2 genes, which have been relocated on chromosome 3HS at the Rph7 locus in barley, belong to gene families in barley and rice and are duplicated on rice chromosomes 4 and 2.
Several BAC libraries are now available from wheat, and a BAC library from Brachypodium sylvaticum that represents an interesting intermediate between the rice and Triticeae genomes has been recently constructed (Foote et al., 2004). Further sequence comparisons at the Rph7 orthologous regions from sorghum, wheat, and Brachypodium should provide relevant information to support or reject the hypothesis of gene movements and to understand molecular mechanisms at the origin of the gene composition at the Rph7 locus in the Triticeae.
METHODS
Plant Material
The characteristics and the origin of the 41 barley (Hordeum vulgare) breeding lines used in the PCR-based haplotype analysis are described by Brunner et al. (2000). Among these lines, 14 are known to possess the Rph7 resistance allele (Table 2). The other lines either have other Rph genes (Weibull et al., 2003) or are not known to contain any leaf rust resistance gene (Table 2).
Shotgun Sequencing and Sequence Analysis
Screening of the Cebada Capa BAC library and identification of single BAC clones was performed as described by Isidore et al. (2005). PCR screening for the Hvgad1 gene was performed with the following primers: gad-1 (5′-CACACCACGCCTACTCCTAC-3′) and gad-2 (5′-ACGAAGGACGCCAGGTTCAG-3′). Preparation of BAC DNA for fingerprint analysis and BAC-end sequencing as well as shotgun libraries for sequencing the BAC clones 14E11 and 124A1 was made as previously described (Stein et al., 2000). A total of 3779 clones were sequenced on an ABI PRISM 377 automatic sequencer (Applied Biosystems, Foster City, CA) from both ends (average length, 788 bp). Base calling and quality of the shotgun sequences (predicted error rate 1/3 kb) were processed using PHRED (Ewing et al., 1998) and assembled using the PHRAP assembly engine (version 0.990319; provided by P. Green, http://www.phrap.org). The STADEN software package was used to finish the assembly (Bonfield et al., 1995). Gaps between the subcontigs were filled by PCR using 18- to 24-mer oligonucleotides designed at the contig ends. DNA sequences were analyzed using BLASTN, BLASTX, and TBLASTX algorithms (Altschul et al., 1997) against public DNA and protein sequence databases. Detailed sequence analysis was performed with the GCG sequence analysis aoftware package version 10.1 (Madison, WI) and by dot plot analysis (DOTTER; Sonnhammer and Durbin, 1995). Analysis of repetitive sequences and transposable elements was performed by BLASTN and BLASTX searches against public databases, the database for Triticeae repetitive DNA (Wicker et al., 2002), and a local database for repetitive DNA. For gene prediction, the RiceGAAS annotation system (Sakata et al., 2002) as well as comparison against the rice (Oryza sativa) full-length cDNA collection (Kikuchi et al., 2003) was used. BLASTN and TBLASTX searches against the 12 rice pseudomolecules recently released by TIGR (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) and against the mapped wheat (Triticum aestivum) EST database (http://wheat.pw.usda.gov/wEST/blast/) were performed to identify the position of genes orthologous to the barley genes in rice and wheat, respectively.
Identification and Mapping of Sorghum BACs Containing Genes Homologous to the Rph7 Locus
Sequences of the barley genes Hvgad1, HvHGA1, HvHGA2, Hvpg4, Hvpg1, and Hvpg3 were used to identify homologous sequences in the sorghum (Sorghum bicolor) EST collection (http://fungen.org/blast/blast.html). STS primers were designed against each sorghum EST using primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and used in PCR reactions to screen six-dimensional BAC DNA pools from the sorghum genotype IS3620C (Klein et al., 2003). Positive BACs were identified after electrophoresis on 4% agarose gels and visualization with SYBR Gold (Molecular Probes, Eugene, OR). BAC clones were subjected to low-pass sequence scanning to assess gene content and to aid in alignment to the rice genome sequence as described by Klein et al. (2003). BACs were fingerprinted using high information content fingerprinting (Klein et al., 2003) and integrated into the sorghum physical map with FPC V7.1 (Soderlund et al., 2000). A subset of the positive BACs (76H6, 67F2, 73H13, and 62H18) integrated into existing DNA contigs that had been previously linked to the sorghum genetic map (Klein et al., 2000). FISH was used to map the chromosomal location of BACs 52N1, 61E22, and 63A11, containing a homolog of Hvpg1. Cells for chromosome spreads were prepared from anthers of sorghum genotype BTx623, and BAC DNA used for FISH was isolated as described by Islam-Faridi et al. (2002). For BAC mapping, a probe cocktail containing the labeled BAC DNAs (52N1, 63A11, and 61E22) as well as two BACs developed previously as karyotype probes for sorghum chromosome 5 (Kim et al., 2002) were hybridized to sorghum pachytene spreads according to Hanson et al. (1996). To assess the location and relative intensity of FISH signals, blue (4′,6-diaminosino-2-2-phenylindole signal from chromosomal DNA), green (fluorescein isothiocyanate from BAC probes), and red (Cy3 from BAC probes) signals were measured from digital images using Optimas v 6.0 (Media Cybernetics, Carlsbad, CA). Image capture and data analysis were done as described (Islam-Faridi et al., 2002).
Dating of Retrotransposon Insertions
Dating of LTR retrotransposon insertions and divergence time estimations were performed based on the method described by SanMiguel et al. (1998). MEGA2 (Kumar et al., 2001) was used to calculate the number of transition and transversion mutations. Insertion dates were estimated using the Kimura two-parameter method (Kimura, 1980). A mutation rate of 6.5 × 10−9 substitutions per synonymous site per year, based on the adh1 and adh2 loci of grasses (Gaut et al., 1996), was applied.
Haplotype Analysis
PCR was performed on 50 ng of genomic DNA in 25 μL containing 0.5 units of Taq DNA-polymerase (Sigma-Aldrich, Buchs, Switzerland), 1× PCR buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, and 0.001% gelatin), 100 μM deoxynucleotide triphosphate, and 400 nM primers described in Supplemental Table 1 online. Amplifications were performed in a PTC-200 thermocycler (MJ Research, Bioconcept, Switzerland) as follows: 3 min at 94°C, 30 cycles of 45 s at 94°C, and 45 s at 53 to 63°C depending on the primer combination (see Supplemental Table 1 online) followed by 1 min (CR4-3/CR5-1 and CR3-1/CR4-1) or 2 min (CR4-3/CR5-2) at 72°C. The extension of the amplified products was achieved at 72°C for 5 min. PCR with the primers pairs CR4-2/CR5-1, 635P2_G4F/635P2_G5R, and 615K1_G2F/615K1_G3R was performed on 50 ng of genomic DNA with the TaKaRa ExTaq polymerase (TaKaRA, Dalian, Japan) according to the manufacturer's instructions. A control PCR was performed with the ITS14/ITS15 primers that amplify a 240-bp fragment corresponding to the rDNA internal transcribed spacer region, ITS1, as described by De Bustos et al. (2002). PCR products were separated by electrophoresis on 0.75 to 1.5% agarose gels and visualized under UV light after ethidium bromide staining.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY642925 and AY642926.
Supplementary Material
Acknowledgments
We thank Edith Schlagenhauf and Romain Guyot for their help in sequence assembly and analysis. This work was supported by Grants 3100-066840 and 3100-65114 from the Swiss National Science Foundation. This work was supported in part by National Science Foundation Plant Genome Research Grant DBI-0321578 (P.K.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Beat Keller (bkeller@botinst.unizh.ch) and Catherine Feuillet (catherine.feuillet@clermont.inra.fr).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028225.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Complex structure of knob DNA on maize chromosome 9. Retrotransposon invasion into heterochromatin. Genetics 149, 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr, A., Muller, K., Schafer-Pregl, R., El Rabey, H., Effgen, S., Ibrahim, H.H., Pozzi, C., Rohde, W., and Salamini, F. (2000). On the origin and domestication history of Barley (Hordeum vulgare). Mol. Biol. Evol. 17, 499–510. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2000). Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell 12, 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2002). Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115, 29–36. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Ma, J.X. (2003). The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr. Opin. Plant Biol. 6, 128–133. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Ramakrishna, W. (2002). Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48, 821–827. [DOI] [PubMed] [Google Scholar]
- Bonfield, J.K., Smith, K., and Staden, R. (1995). A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., Keller, B., and Feuillet, C. (2000). Molecular mapping of the Rph7.g leaf rust resistance gene in barley (Hordeum vulgare L.). Theor. Appl. Genet. 101, 783–788. [Google Scholar]
- Brunner, S., Keller, B., and Feuillet, C. (2003). A large rearrangement involving genes and low copy DNA interrupts the microcolinearity between rice and barley at the Rph7 locus. Genetics 164, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bustos, A., Loarce, Y., and Jouve, N. (2002). Species relationships between antifungal chitinase and nuclear rDNA (internal transcribed spacer) sequences in the genus Hordeum. Genome 45, 339–347. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., Ramakrishna, W., SanMiguel, P.J., Busso, C.S., Yan, L.L., Shiloff, B.A., and Bennetzen, J.L. (2001). Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol. 125, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., and Keller, B. (2002). Comparative genomics in the grass family: Molecular characterization of grass genome structure and evolution. Ann. Bot. 89, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote, T.N., Griffiths, S., Allouis, S., and Moore, G. (2004). Construction and analysis of a BAC library in the grass Brachypodium sylvaticum: Its use as a tool to bridge the gap between rice and wheat in elucidating gene content. Funct. Integr. Genomics 4, 26–33. [DOI] [PubMed] [Google Scholar]
- Fu, H.H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. (2002). Evolutionary dynamics of grass genomes. New Phytol. 154, 15–28. [Google Scholar]
- Gaut, B.S., and Clegg, M.T. (1993). Molecular evolution of the Adh1 locus in the genus Zea. Proc. Natl. Acad. Sci. USA 90, 5095–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., d'Ennequin, M.L., Peek, A.S., and Sawkins, M.C. (2000). Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. USA 97, 7008–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner, A., Bjornstad, A., Konishi, T., and Ordon, F. (2003). Molecular diversity of the barley genome. In Diversity in Barley (Hordeum vulgare), R.V. Bothmer, T.V. Hintum, H. Knüpffer, and K. Sato, eds (Amsterdam: Elsevier Science), pp. 122–141.
- Gu, Y.Q., Anderson, O.D., Londeore, C.F., Kong, X.Y., Chibbar, R.N., and Lazo, G.R. (2003). Structural organization of the barley D-hordein locus in comparison with its orthologous regions of wheat genomes. Genome 46, 1084–1097. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., Coleman-Derr, D., Kong, X.Y., and Anderson, O.D. (2004). Rapid genome evolution revealed by comparative sequence analysis of orthologous regions from four Triticeae genomes. Plant Physiol. 135, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot, R., and Keller, B. (2004). Ancestral genome duplication in rice. Genome 47, 610–614. [DOI] [PubMed] [Google Scholar]
- Guyot, R., Yahiaoui, N., Feuillet, C., and Keller, B. (2004). In silico comparative analysis reveals a mosaic conservation of genes within a novel colinear region in wheat chromosome 1AS and rice chromosome 5S. Funct. Integr. Genomics 4, 47–58. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A., and Wise, R.P. (2004). A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 38, 215–226. [DOI] [PubMed] [Google Scholar]
- Han, B., and Xue, Y. (2003). Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 6, 134–138. [DOI] [PubMed] [Google Scholar]
- Hanson, R.E., Islam-Faridi, M.N., Percival, E.A., Crane, C.F., Ji, Y., McKnight, T.D., Stelly, D.M., and Price, H.J. (1996). Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105, 55–61. [DOI] [PubMed] [Google Scholar]
- Huang, S.X., Sirikhachornkit, A., Faris, J.D., Su, X.J., Gill, B.S., Haselkorn, R., and Gornicki, P. (2002). Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48, 805–820. [DOI] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100, 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidore, E., Scherrer, B., Bellec, A., Budin, K., Faivre-Rampant, P., Waugh, R., Keller, B., Caboche, M., Feuillet, C., and Chalhoub, B. (2005). Direct targeting and isolation of genes of interest using improved pooled BAC libraries cloning and screening strategy. Funct. Integr. Genomics, in press. [DOI] [PubMed]
- Islam-Faridi, M.N., Childs, K.L., Klein, P.E., Hodnett, G., Menz, M.A., Klein, R.R., Rooney, W.L., Mullet, J.E., Stelly, D.M., and Price, H.J. (2002). A molecular cytogenetic map of sorghum chromosome 1: Fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar, R., Tanskanen, J., Immonen, S., Nevo, E., and Schulman, A.H. (2000). Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 97, 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, B., and Feuillet, C. (2000). Colinearity and gene density in grass genomes. Trends Plant Sci. 5, 246–251. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. [DOI] [PubMed] [Google Scholar]
- Kim, J.S., Childs, K.L., Faridi, N.I., Menz, M.A., Klein, R.R., Klein, P.E., Price, H.J., Mullet, J.E., and Stelly, D.M. (2002). Integrated karyotyping of sorghum by in situ hybridization of landed BACs. Genome 45, 402–412. [DOI] [PubMed] [Google Scholar]
- Kim, J.S., Klein, P.E., Klein, R.R., Price, H.J., Mullet, J.E., and Stelly, D.M. (Oct. 16, 2004). Chromosome identification and nomenclature of Sorghum bicolor. Genetics 10.1534/genetics.104.035980. [DOI] [PMC free article] [PubMed]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Klein, P.E., Klein, R.R., Cartinhour, S.W., Ulanch, P.E., Dong, J., Obert, J.A., Morishige, D.T., Schlueter, S.D., Childs, K.L., Ale, M., and Mullet, J.E. (2000). A high-throughput AFLP-based method for constructing integrated genetic and physical maps: Progress toward a sorghum genome map. Genome Res. 10, 789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P.E., Klein, R.R., Vrebalov, J., and Mullet, J.E. (2003). Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34, 605–621. [DOI] [PubMed] [Google Scholar]
- Kong, X.Y., Gu, Y.Q., You, F.M., Dubcovsky, J., and Anderson, O.D. (2004). Dynamics of the evolution of orthologous and paralogous portions of a complex locus region in two genomes of allopolyploid wheat. Plant Mol. Biol. 54, 55–69. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Li, W.L., and Gill, B.S. (2002). The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the Triticeae. Genetics 160, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Bennetzen, J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101, 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Devos, K.M., and Bennetzen, J.L. (2004). Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz, M.A., Klein, R.R., Mullet, J.E., Obert, J.A., Unruh, N.C., and Klein, P.E. (2002). A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol. Biol. 48, 483–499. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., and Chapman, B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101, 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Peterson, D.G., Estill, J.C., and Chapman, B.A. (2003). Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 13, 644–650. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. (2002). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks, N., et al. (2002). Genomic sequencing reveals gene content, genomic organization, and recombination relationships in barley. Funct. Integr. Genomics 2, 51–59. [DOI] [PubMed] [Google Scholar]
- Sakata, K., Nagamura, Y., Numa, H., Antonio, B.A., Nagasaki, H., Idonuma, A., Watanabe, W., Shimizu, Y., Horiuchi, I., Matsumoto, T., Sasaki, T., and Higo, K. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. (2000). Contigs built with fingerprints, markers, and FPC V4. Genome Res. 10, 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, V., and Messing, J. (2002). Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and Messing, J. (2003). Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100, 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., and Durbin, R. (1995). A Dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167, 1–10. [DOI] [PubMed] [Google Scholar]
- Stein, N., Feuillet, C., Wicker, T., Schlagenhauf, E., and Keller, B. (2000). Subgenome chromosome walking in wheat: A 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc. Natl. Acad. Sci. USA 97, 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibull, J., Walther, U., Sato, K., Habekuss, A., Kopahnke, D., and Proeseler, G. (2003). Diversity in resistance to biotic stress. In Diversity in Barley (Hordeum vulgare), R.V. Bothmer, T.V. Hintum, H. Knüpffer, and K. Sato, eds (Amsterdam: Elsevier Science), pp. 143–178.
- Wicker, T., Matthews, D.E., and Keller, B. (2002). TREP: A database for Triticeae repetitive elements. Trends Plant Sci. 7, 561–562. [Google Scholar]
- Wicker, T., Yahiaoui, N., Guyot, R., Schlagenhauf, E., Liu, Z.D., Dubcovsky, J., and Keller, B. (2003). Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15, 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.