Abstract
More than half a century after the discovery of transposable elements, the number of genetically defined autonomous elements that have been isolated and characterized molecularly in any one species remains surprisingly small. Because of its rich genetic history, maize (Zea mays) is, by far, the plant with the largest number of such elements. Yet, even in maize, a maximum of only two autonomous elements have been characterized in any transposon superfamily. This article describes the isolation and molecular and genetic characterization of Mx (for mobile element induced by x-rays), a third autonomous member of the hAT transposon superfamily in maize. Mx is 3731 bp long, ends in 13-bp terminal inverted repeats (TIRs), and causes an 8-bp duplication of the target site. Mx and rMx (for responder to Mx), its 571-bp nonautonomous partner, define a classical family of interacting transposable elements. Surprisingly, the TIRs of Mx and rMx are only 73% identical, and the subterminal sequences are even less so, suggesting that Mx and rMx may represent diverging transposable elements still capable of mobilization by the same transposase. Sequences that are closer to the ends of either Mx or rMx are present in the maize genome. Mx is predicted to encode a 674–amino acid protein that is homologous to the Ac transposase. Although Mx and Ac are closely related, they do not interact. Other data suggest that maize may possess at least five families of hAT transposons that do not interact with each other. The possible origin of noninteracting transposon families within the same superfamily is discussed.
INTRODUCTION
Interrelationships between transposable elements in maize (Zea mays) have been established traditionally by complementation tests between a nonautonomous element residing at a reporter allele and an autonomous element residing elsewhere. If the elements complement to produce a mutable or unstable reporter phenotype, they are considered to belong to the same system (McClintock, 1956b) or family (Fedoroff, 1983). If they do not, they are assigned to different families. The application of this deceptively simple, yet effective, test has led to the grouping of transposons causing a wide range of mutable phenotypes in maize into several families (Peterson, 1988), of which the best characterized are Ac-Ds, Spm/En-dSpm/I, and MuDR-Mu1.
The subsequent DNA sequence analysis of genetically active transposons has permitted their independent classification into larger groups, termed superfamilies, based generally on three criteria: the sequence of the transposon terminal inverted repeats (TIRs), the homology of their putative transposases, and the size of the host target site duplication (TSD). The five main superfamilies of DNA transposons recognized in plants are hAT, CACTA, Mutator, PIF/Harbinger, and Tc1/mariner, each defined by one or more genetically characterized founding members (Feschotte et al., 2002a). The numerous transposon-related sequences uncovered by the genome sequencing projects in Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000) and rice (Oryza sativa; Goff et al., 2002) have been placed into one or another of these five large categories. Although the biological activity of these sequences has not been examined, most are expected to be immobile. In fact, miniature inverted-repeat transposable elements—the predominant transposons in or near genes—are so stable that they have been exploited as genetic markers in both maize and rice (Feschotte et al., 2002b).
The situation in maize differs from that in Arabidopsis and rice in that many genetically defined transposable element families were identified decades before the establishment of a genome sequencing project. The maize inbred lines B73 and Mo17 that have been chosen for sequencing by the public (Cone et al., 2002) and private (Palaisa et al., 2003) sectors, respectively, are not likely to carry active transposable elements, as they are known to lack even the more common ones, such as Uq (Cormack et al., 1988). However, active transposons are certainly present in wild populations and even in some inbred lines (e.g., van der Walt and Brink, 1969; Montanelli et al., 1984; Peterson and Salamini, 1986; Cormack et al., 1988). The extreme natural diversity in maize and its long history as a model organism for genetic research have contributed to the identification, genetic characterization, and preservation of many systems of mutability (Peterson, 1988; Neuffer et al., 1997; http://w3.ag.uiuc.edu/maize-coop/mgc-info.html), only a fraction of which have been characterized molecularly. For example, the autonomous transposons Dt (Rhoades, 1938), Fcu (Gonella and Peterson, 1977), Mrh, and Mut (Rhoades and Dempsey, 1982) have yet to be isolated. In some instances, a nonautonomous element of the family has been isolated and sequenced, allowing its tentative assignment to a superfamily based on TIR homology and TSD size. By those criteria, rDt (Brown et al., 1989) and rMut (Dennis et al., 1988) would be members of the hAT superfamily, and rMrh (Shepherd et al., 1989) would be a member of the Mutator superfamily. Further molecular characterization of these families awaits the isolation of their respective autonomous transposons.
In spite of the paucity of autonomous transposons isolated in plants (Feschotte et al., 2002b; Kunze and Weil, 2002), it is clear that several distinct transposon families belonging to the same superfamily can coexist in one species. Thus, in maize, the hAT superfamily includes Ac-Ds and Bg-rBg (Hartings et al., 1991b); the CACTA superfamily includes Spm/En-dSpm/I and Shooter (Panavas et al., 1999), and the Mutator superfamily includes MuDR (Walbot and Rudenko, 2002) and Jittery (Xu et al., 2004). This means that each superfamily contains groups of related transposons that do not interact with each other, raising the issue of how related autonomous transposons in the same species can evolve different specificities.
This article describes the isolation and characterization of Mx (for mobile element induced by x-rays), a new autonomous transposon of the hAT superfamily in maize. This transposon is responsible for the instability of bz-x3m, a mutation from an X-irradiated stock (Mottinger, 1973) that carries a nonautonomous element termed rMx (for responder to Mx) (Mottinger, 1992). Mx is more closely related to Ac and the Tam3 element of Antirrhinum majus (Hehl et al., 1991) than to Bg or the Tag1 element of Arabidopsis (Liu and Crawford, 1998), but cannot trans-activate Ds excision nor contribute to Ac's dosage effect. Surprisingly, the TIRs of Mx and rMx are only 73% identical, and the subterminal sequences are even more different, suggesting that Mx and rMx may represent diverging transposable elements that can still interact with each other. The possible origin of noninteracting transposon families within the same superfamily is discussed.
RESULTS
Isolation and Characterization of rMx, the Nonautonomous Element of the Mx-rMx System
The bz-x3m mutation was acquired from John Mottinger in 1982 and introduced into the genetic background of the W22 inbred by backcrossing repeatedly to a W22 sh bz-R wx tester line and selecting bz-m spotted seed each generation. After the fourth backcross, a Sh bz-x3m wx stock was established by self-pollinating the heterozygous BC4 plants and identifying progeny plants that produced mostly spotted seed and did not segregate sh in their selfed ears. These plants were the foundation of the W22 bz-x3m stock used in the work described below. The W22 bz-x3m stock produces a finely spotted kernel phenotype (Figures 1A and 1B), similar to that described earlier by Mottinger (1973) (1992), except that the frequency of somatic reversion appears to be lower in the W22 version. Both stocks behave similarly, though, in that the overall somatic reversion level (kernel spot size and number) is higher when they are used as female, rather than male, parents in crosses to a bz-R tester. As in the original mutant, some kernels are uniformly bronze and lack obvious Bz′ revertant sectors, but most do not breed true. In the presence of the R-r transcription factor carried by most color-converted W22 lines (Brink, 1956), the somatic instability of bz-x3m is also evident as red sectors in otherwise bronze anthers (Figure 1C).
Figure 1.
bz-x3m Phenotypes.
(A) Ear of a W22 bz-x3m homozygote, showing the finely spotted kernel phenotype arising from late somatic reversions in the aleurone layer. A few kernels show larger sectors produced from earlier reversion events.
(B) Close-up of (A), showing the small size of the majority of revertant sectors in the aleurone.
(C) Spotted and red anthers in a W22 bz-x3m homozygote. Most reversions occur late in development and result in very finely spotted anthers. However, reversions can also occur earlier, as indicated by the two florets with exclusively red anthers.
Mottinger (1992) established that the mutability of bz-x3m was because of an ∼0.5-kb nonautonomous element at bz, which he designated rMx, and a closely linked autonomous element, which he designated Mx. To characterize this transposable element system more fully, the rMx insertion in bz was PCR amplified with bz primers and sequenced. rMx is 571 bp long, has 13-bp imperfect TIR, contains 15 copies of the hexanucleotide CCCGAA or its reverse complement within the subterminal 170 bp at either end, and produces an 8-bp TSD. The rMx insertion site (GTGGAGGA) is located in the bz second exon, close to the 3′ end of the gene. The sequences of the 13-bp imperfect TIRs are 5′-TAGCACTGGGCAT-3′ at the 5′ end and 5′-ATGCCCAGTCCTA-3′ at the 3′ end (the imperfect noncomplementary bases are underlined). Sequences homologous to rMx are present in ∼15 copies in a W22 background (data not shown).
BLASTX analysis of the GenBank sequence databases using the rMx sequence as query established that rMx had no significant similarity to any known protein. By contrast, BLASTN analysis revealed that the terminal 180 bp at either end of rMx were highly similar to the ends of the Tz86 transposon. Tz86 is a maize endogenous transposon identified in a sh1 mutable allele recovered after virus infection (Mottinger et al., 1984). Although Tz86 has not been sequenced, 250 bp of each Tz86-sh1 junction are available from GenBank (M10174 and M10175). Figure 2 shows the sequence alignment of the Tz86 and rMx ends. Tz86 and rMx are 77% identical from position 1 to 181 at the 5′ end (140/181) and 81% identical from position 402 to 571 at the 3′ end (137/170). The high identity of their ends suggests that rMx and Tz86 may belong to the same transposon superfamily. However, the internal part of rMx (from position 182 to 401) shows little similarity to the rest of the Tz86 sequence.
Figure 2.
DNA Sequence Comparison between rMx and Tz86.
The entire 571-bp rMx element from bz-x3m was compared with the Tz86 ends. The Tz86 sequence is a 439-nucleotide composite, consisting of 207 nucleotides from the 5′ end (GenBank accession M10174) and 232 nucleotides from the 3′ end (GenBank accession M10175). Sequences were aligned using the program MultiAlin (http://prodes.toulouse.inra.fr/multalin/multalin.html). The graphical output highlights identical nucleotides as white letters in black boxes and purine transitions as white letters in gray boxes (Box-Shader 3.21, http://www.ch.embnet.org/software/BOX_form.html).
bz-x3m is a mutable allele that reverts occasionally to Bz′ (full purple) in the germ line (Mottinger, 1992). In testcrosses using the W22 bz-x3m stock as female parent, the frequency of Bz′ germinal revertants is ∼0.5%. To investigate whether rMx produces typical transposon footprints upon excision, the transposon empty sites of eight independent Bz′ revertants were PCR amplified and sequenced. The results are presented in Table 1. The Bz′ germinal revertants had footprints of 8+0, 8+3, 8+6, and 8+9 (i.e., nucleotide additions in multiples of three), in agreement with the requirement for restoration of the correct reading frame in Bz′ functional derivatives. Mutations of bz-x3m to a stable bronze form are harder to identify because of the incomplete penetrance of the bz-x3m mutation, even in self-pollinated ears (Mottinger, 1992). However, one such derivative was evident as a large premeiotic sector of plump, bronze seeds in the testcross ear of a Sh bz-x3m/sh bz-R heterozygote. In this nonfunctional bz-s derivative, rMx had not excised, but had, instead, caused a deletion of 214 bp of bz sequences immediately proximal to the insertion (Table 1). Such adjacent deletions are also generated by other plant transposable elements, such as Mu1 (Taylor and Walbot, 1985) and Ac (Dooner, 1985), and probably arise from abortive excision events.
Table 1.
rMx Germinal Excision Footprints in bz-x3m
| Selections | Sequence of Empty Site | Excision Pattern | ||
|---|---|---|---|---|
| bz-x3m | GTGGAGGA | rMx | GTGGAGGA | |
| Bz′-1 | GTGGAGGA | 8+0 | ||
| Bz′-2 | GTGGAGGA | 8+0 | ||
| Bz′-3 | GTGGA | c | GAGGA | 8+3 |
| Bz′-4 | GTGGAG | c | TGGAGGA | 8+6 |
| Bz′-5 | GTGGAG | c | TGGAGGA | 8+6 |
| Bz′-6 | GTGGAG | c | TGGAGGA | 8+6 |
| Bz′-7 | GTGGAGG | tcc | TGGAGGA | 8+9 |
| Bz′-8 | GTGGAGG | tcc | TGGAGGA | 8+9 |
| bz-s1 | 214 bp deletion | rMx | GTGGAGGA | Adjacent deletion |
Separation and Resynthesis of Mx and rMx
To isolate and characterize the autonomous Mx element molecularly, it is first necessary to derive stocks in which Mx and bz-x3m have been separated so that Mx can be scored genetically on the basis of its ability to trans-activate the excision of rMx from bz-x3m. Mottinger (1992) was able to separate the nonautonomous bz-x3m mutant from Mx and, thus, to map the location of the autonomous element in his stocks. Not surprisingly for a mobile element, Mx did not map to a single locus in the maize genome. In addition to a location tightly linked to bz, Mx could occupy other locations: one was distal to c1 on the same chromosome and another one was unlinked to bz. The genetic tests described below show that, in the W22 bz-x3m stock, Mx is present in a single copy that maps distal to the sh locus.
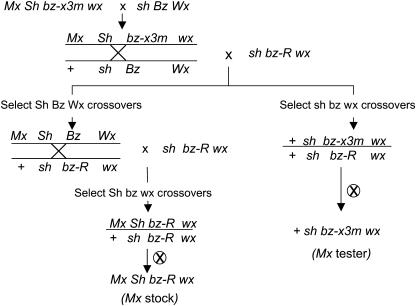
Figure 3 diagrams the crossing scheme used to separate Mx and rMx. For ease in presentation, Mx is shown on the bz-x3m parental chromosome at the location deduced from the experiment itself. Hence, the genotype of the W22 bz-x3m stock is represented as Mx Sh bz-x3m wx. That stock was crossed to a sh Bz Wx stock, and the resulting Mx Sh bz-x3m wx/+ sh Bz Wx F1 heterozygote was testcrossed to sh bz-R wx to select reciprocal recombinants between sh and bz. None of the sh bz wx crossovers (Figure 3, right) had spots and all of them bred true (putative + sh bz-x3m wx/+ sh bz-R wx), suggesting that Mx was located distal to sh on 9S. These recombinants were confirmed by DNA gel blots to carry rMx in bz (data not shown), so they should serve as bona fide Mx testers. This inference was confirmed by further analysis of the reciprocal Sh Bz Wx recombinants.
Figure 3.
Genetic Scheme to Separate Mx and rMx in the W22 bz-x3m Stock.
See text for details.
The Sh Bz Wx crossovers (putative Mx Sh Bz Wx/+ sh bz-R wx at the left of Figure 3) were backcrossed to sh bz-R wx, and Sh bz wx second-cycle recombinants were obtained. The Sh bz wx recombinants should be Mx Sh bz-R wx/+ sh bz-R wx in genotype because Mx was inferred to be distal to Sh. The putative genotypic assignment of these recombinants was verified by crossing them to the Mx tester line described above (+ sh bz-x3m wx) and establishing that they could reactivate spotting (i.e., rMx somatic excision) in the Mx tester. At the same time, the W22 sh bz-R wx parental line was confirmed to lack Mx activity because it could not reactivate spotting in the Mx tester. Neither could several other W22 lines tested, indicating that Mx is not present in the W22 inbred. Although the original bz-x3m stock appeared to have an Mx element distal to sh and a second one between sh and bz (Mottinger, 1992), the derived W22 bz-x3m stock lacks the latter. Most likely, the second Mx element transposed and was lost during the introgression of the Sh bz-x3m chromosome segment into a W22 background.
Isolation and Characterization of Mx, the Autonomous Element of the Mx-rMx System
Stepwise Isolation of Progressively Larger Members of the Mx Family by Primer Walking
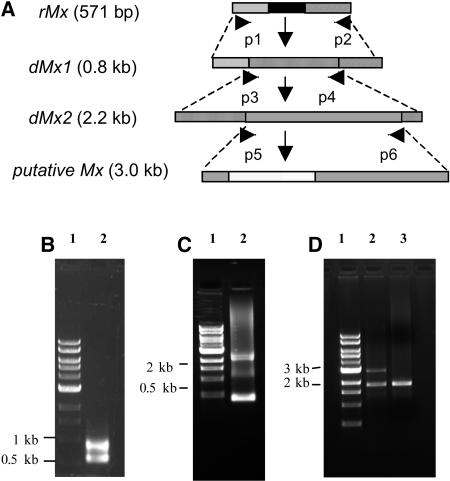
Hybridization of DNA gel blots with the short rMx probe failed to identify any bands that cosegregated with the Mx element in the Mx Sh bz-R wx and + sh bz-x3m wx recombinants of Figure 3. To generate a more suitable probe for the isolation of Mx, a three-step PCR procedure was developed based on the fact that all known autonomous maize transposons have defective relatives in the genome that have undergone internal deletions of varying lengths (Kunze and Weil, 2002; Walbot and Rudenko, 2002). The procedure, diagrammed in Figure 4A and henceforth referred to as primer walking, allowed the amplification of progressively larger members of the Mx family.
Figure 4.
Primer Walking to Isolate Mx.
(A) Sequential PCR steps to amplify progressively larger members of the Mx family. p1 to p6 stand for specific primers used in the amplification reactions. Boxes with same shading indicate identical or highly similar sequences.
(B) Ethidium bromide (EtBr)–stained agarose gel of PCR amplification products with primers p1 and p2. Lane 1, 1-kb ladder of molecular size markers; lane 2, PCR products from Mx bz-x3m, showing the 0.8- and 0.5-kb bands, corresponding to dMx1 and rMx, respectively.
(C) EtBr-stained agarose gel of PCR amplification products with primers p3 and p4. Lane 1, 1-kb ladder of molecular size markers; lane 2, PCR products from Mx bz-x3m, showing the 2.2-kb band corresponding to dMx2.
(D) EtBr-stained agarose gel of PCR amplification products with primers p5 and p6. Lane 1, 1-kb ladder of molecular size markers; lane 2, PCR products from Mx bz-x3m, showing the 3.0-kb band corresponding to the putative Mx element; lane 3, PCR products from W22 bz-R.
First, using primers from rMx sequences just three nucleotides away from either end (p1 and p2 in Figure 4A), a 0.8-kb band, designated dMx1, and the expected 0.5-kb rMx band were PCR amplified from total genomic DNA of the W22 Mx bz-x3m stock (Figure 4B) and sequenced. The ends of dMx1 (166 bp) are highly similar (81%) to the rMx ends and identical to the Tz86 ends in the database. However, the dMx1 internal region is novel and shows no similarity to that of rMx. Second, using primers based on this internal region (p3 and p4 in Figure 4A), a 2.2-kb fragment, designated dMx2, was PCR amplified (Figure 4C) and sequenced. BLASTX analysis predicts that dMx2 encodes an incomplete protein with homology to the Ac transposase. Therefore, the dMx2 fragment is derived from a member of the hAT superfamily of transposons. Third, using primers based on the dMx2 internal region (p5 and p6 in Figure 4A), a 3.0-kb fragment, designated putative Mx, was PCR amplified (Figure 4D) and sequenced. Consistent with the notion that this fragment comes from Mx, the putative Mx band was only amplified from the W22 Mx bz-x3m stock, but not from the W22 bz-R line. Analysis of the putative Mx sequence revealed that this fragment also encodes a putative Ac-like transposase.
Isolation of Mx
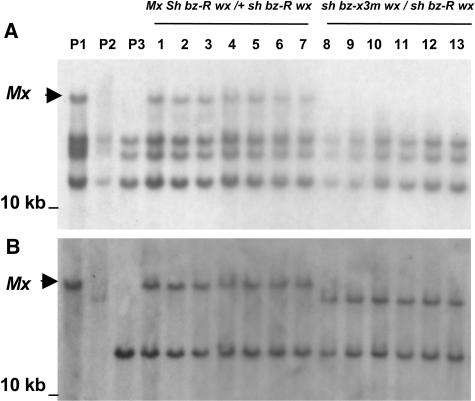
A DNA gel blot of the recombinant individuals shown in Figure 3 was then hybridized sequentially with probes made from the PCR amplification products described above. A probe from the 2.2-kb dMx2 fragment (Figures 4A and 4C) revealed the presence of a unique EcoRI band in the W22 Mx bz-x3m parent and the Mx Sh bz-R wx/+ sh bz-R wx recombinants (Figure 5A, lanes P1 and 1 to 7), but not in the + sh bz-x3m wx/+ sh bz-R wx reciprocal recombinants or the sh Bz Wx and sh bz-R wx parents (Figure 5A, lanes 8 to 13, P2, and P3). Thus, the unique band cosegregates with Mx activity. Similar unique bands were also detected in gel blots of DNAs digested with either SalI, SacI, or BglII (data not shown).
Figure 5.
DNA Gel Blot Analysis of Recombinants from the W22 Mx bz-x3m Stock.
Reciprocal recombinants from Mx Sh bz-x3m/+ sh Bz heterozygotes were obtained by the scheme diagrammed in Figure 3. Genomic DNA was digested with EcoRI, blotted onto Nylon membranes, and hybridized sequentially to radiolabeled probes from dMx2 (A) (Figure 4A) and tmx (B), the Mx flanking region. P1, Mx Sh bz-x3m wx parent; P2, sh Bz Wx parent; P3, sh bz-R wx testcross parent; lanes 1 to 7, Mx Sh bz-R wx/+ sh bz-R wx recombinants; lanes 8 to 13, + sh bz-x3m wx/+ sh bz-R wx recombinants. The arrowhead identifies the position of Mx.
The missing ends of the putative Mx element (Figure 4A) and the chromosomal region flanking it (tmx) were amplified by inverse PCR (IPCR) using a gel-purified, dMx2-hybridizing, SalI fragment from the Mx bz-x3m line as template and primers located near the ends of the putative Mx sequence. Rehybridization of the recombinants' DNA gel blot with the tmx probe (Figure 5B) supports the premise that tmx corresponds to the region flanking Mx. tmx detects a single polymorphic EcoRI band in the three parents used in the crossing scheme of Figure 3. The band present in the P1 Mx Sh bz-x3m parent is also present in all the Mx Sh bz-R wx/+ sh bz-R wx recombinants (lanes 1 to 7), but is absent in the + sh bz-x3m wx/+ sh bz-R wx recombinants (lanes 8 to 13). These recombinants have, instead, the band that is present in the P2 sh Bz Wx parent. Finally, the band present in the P3 sh bz-R wx testcross parent is shared, as expected, with all of the recombinants. Using tmx as a probe on recombinant inbred membranes (Burr et al., 1988), the putative Mx was mapped to a location 2 centimorgan (cM) distal to c1 and 6 cM distal to sh1 on 9S (data not shown). The tmx polymorphism between the P2 and P3 parents can be explained by linkage drag during the derivation of those lines, which are known to carry different c1 alleles: P2 carries a C1-s (super-C1) allele, whereas P3 carries a regular C1 allele. The DNA gel blot data is thus consistent with the genetic crossover results shown in Figure 3.
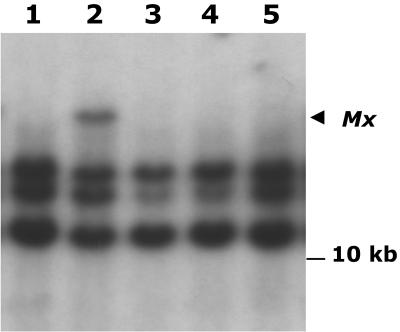
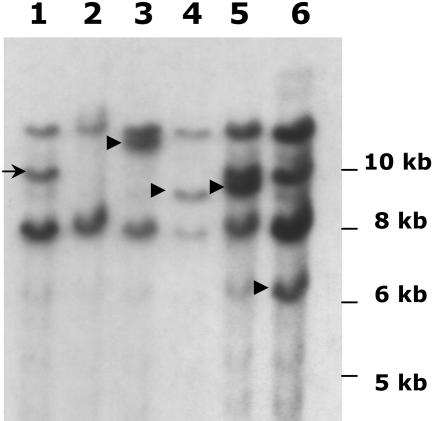
The above hybridization data show that the putative Mx element, isolated by a combination of PCR primer walking and IPCR, cosegregates with Mx activity and resides at a chromosomal location (tmx) consistent with the mapped position of Mx in the W22 Mx bz-x3m stock. That this element is in fact the autonomous Mx transposon was confirmed by the following genetic test. Excision losses of Mx from the W22 Mx bz-x3m stock were sought in a screen for exceptional nonspotted seeds among the seed progeny of a cross to sh bz-R wx. Several uniformly bronze exceptions were obtained among 144 spotted seeds. Three of these exceptions bred true, producing no spotted seeds in their self-progenies. Crosses of these three exceptions to the Mx bz-R stock (Figure 3, left) yielded spotted seeds, indicating that the bz-x3m allele was intact and that somatic excision of rMx from bz could be reactivated. Crosses of the exceptions to the Mx tester + sh bz-x3m (Figure 3, right) yielded only bronze seeds, indicating that Mx had been lost from the genome. DNA gel blot analysis of the exceptions (Figure 6) showed they had lost the EcoRI band corresponding to the putative Mx element (arrowhead, cf. Figure 5), thus verifying the latter's identity as the true Mx. Using primers based on the tmx sequence, the Mx excision site in these exceptions was amplified and sequenced. Examination of the sequence revealed that Mx had excised and left typical transposon footprints at the empty site (Table 2). The demonstration that the somatic excision of rMx from bz-x3m can be activated in a stock carrying the putative Mx element, but not in derivatives lacking it, establishes unambiguously that that element is Mx.
Figure 6.
DNA Gel Blot Analysis of Exceptions from Mx bz-x3m That Have Lost Mx.
Genomic DNA was digested with EcoRI, blotted onto nylon membranes, and hybridized to a radiolabeled probe from dMx2. Lane 1, the Mx tester line + sh bz-x3m wx; lane 2, Mx Sh bz-x3m; lanes 3 to 5, three + Sh bz-x3m derivatives from Mx Sh bz-x3m. The arrowhead identifies the position of Mx.
Table 2.
Analysis of Mx Excision Footprints
| Selections | Sequence of Empty Site | Excision Pattern | ||
|---|---|---|---|---|
| Mx-TSD | CACTACAC | Mx | CACTACAC | |
| Mx-xis1a | CACTACAC | gtg | CACTACAC | 8+9 |
| Mx-xis2a | CACTACA | g | CTACAC | 8+6 |
| Mx-xis3a | CACTACA | gt | ACTACAC | 8+8 |
| Mx-xis4b | CACTACAC | 8+0 | ||
| Mx-xis5b | CACTACA | gt | ACTACAC | 8+8 |
Mx excisions xis1, xis2, and xis3 were identified as nonspotted exceptions in a Mx- bz-x3m × bz-R cross.
Mx excisions xis4 and xis5 were identified by DNA blots among the spotted individuals in the same cross.
Mx Is a Member of the hAT Transposon Superfamily
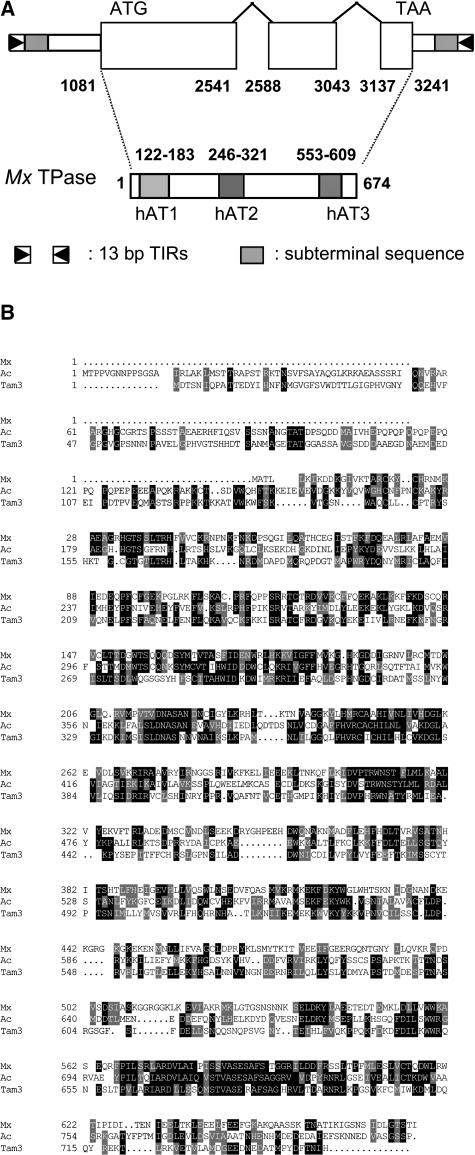
A complete Mx transposon was amplified using tmx primers and sequenced. Figure 7A diagrams the overall structure of the element. Mx is 3731 bp long, ends in 13-bp imperfect TIRs identical to those of Tz86, contains 17 subterminal direct repeats of the hexanucleotide CCCGAA or its reverse complement, and causes an 8-bp duplication of the target site (5′-CACTACAC-3′). The 180-bp terminal sequences at the 5′ and 3′ ends, respectively, are only 76 and 78% similar to those of rMx, but are identical with those of dMx1 and Tz86. Thus, Mx and Tz86 may be the same element, based on the limited Tz86 sequence available. The Mx transcript is predicted to consist of three exons encoding a 674–amino acid protein that is homologous to the Ac transposase. Figure 7B shows the sequence alignment of the proteins encoded by Mx and Ac. Like the Ac transposase, the protein encoded by Mx contains three domains, designated hAT1, hAT2, and hAT3, which are highly conserved among members of the hAT transposon superfamily (Kunze and Weil, 2002). This large superfamily has been named after the autonomous elements hobo in Drosophila melanogaster, Ac in maize, and Tam3 in Antirrhinum (Calvi et al., 1991) and includes transposons from plants, animals, and fungi.
Figure 7.
Mx Structure and Sequence Comparison.
(A) Structure of Mx, showing the location of the 13-bp TIRs, the subterminal hexanucleotide repeats, and the exons and introns of its predicted protein, Mx TPase. This protein is homologous to the transposases encoded by Ac and other members of the hAT transposon superfamily, which share three conserved domains, indicated as hAT1, hAT2, and hAT3.
(B) Amino acid sequence alignment of the putative transposases encoded by Mx, Ac, and Tam3. The latter two are the main representatives of autonomous hAT elements from monocots and dicots, respectively. The sequences were aligned with MultiAlin, and the aligned sequences were decorated with BoxShade 3.21. Identity and similarity of amino acid residues are indicated by black and gray shading, respectively.
Transposition of Mx
To confirm that Mx could reinsert in the genome, as suggested by earlier mapping data (Mottinger, 1992), the spotted seed progeny from the testcross of a Mx Sh bz-x3m/+ sh bz-R heterozygote was screened for new Mx-hybridizing bands by DNA gel blots. Out of 95 individuals analyzed, two had novel bands, but retained Mx at its original location (Figure 8, lanes 5 and 6). This outcome—the joint recovery of transposon copies at both donor and receptor sites—is also common in Ac transposition (Chen et al., 1987). As indicated below, the alternative outcome—loss of the transposon copy at the donor site—was also uncovered, suggesting that Ac and Mx may transpose by the same mechanism (Brink and Nilan, 1952; Greenblatt and Brink, 1962). Mx transposants that have lost Mx from its original position were found serendipitously during the reprobing with an Mx probe of a blot containing the DNA of a small number of spotted progeny from a cross between Mx Sh bz-x3m and sh bz-R. The pattern of Mx-hybridizing bands in these individuals is shown in Figure 8 (lanes 3 and 4), and the sequence of their Mx excision sites is given in Table 2 (xis4 and xis5). Thus, as with Ac, transposition of Mx may or may not result in a net increase in the original copy number of the transposon (Chen et al., 1987, 1992).
Figure 8.
DNA Gel Blot Analysis of Mx Transposition.
Genomic DNA was digested with NcoI, blotted onto Nylon membranes, and hybridized to an internal Mx probe extending from position 761 to 1710 in the nucleotide sequence. Lane 1, bz-x3m stock (Mx Sh bz-x3m); lane 2, Mx tester line (+ sh bz-x3m); lanes 3 to 6, derivatives of the bz-x3m stock with transposed Mx elements. Arrow, original Mx element; arrowheads, transposed Mx elements.
Mx Homologous Sequences in Maize and Its Relatives
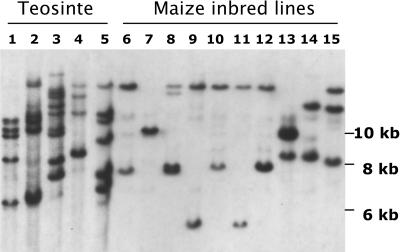
An rMx probe, which shares homology with Mx only at its ends, detects multiple bands in W22 (data not shown). To examine the relative abundance of Mx homologous sequences in maize and its closest relatives, a blot containing DNA from several maize inbred lines and teosinte accessions was hybridized with an Mx internal sequence. The probe sequence, which extends from position 760 to 1710 and includes part of the Mx first exon (Figure 7A), is completely missing in the two dMx elements isolated in this study (Figure 4A), so it will not detect these or other similarly deleted defective Mx elements. Figure 9 shows the hybridization result. The 10 different Corn Belt inbreds examined are as follows: A188, A636, B73, BSSS53, 4Co63, H99, M14, Mo17, W22, and W23 (lanes 6 to 15). The five teosinte plants examined came from two different collections of Z. mays ssp mexicana (lanes 1 and 2, from Estado, Mexico, and lane 5, from Michoacán) and one collection of Z. mays ssp parviglumis (lanes 3 and 4, from Michoacán). Mx internal sequences are present in low copy number (one to four) in the maize inbreds and in somewhat higher copy number (five to nine) in the teosinte plants, which are known to be highly heterozygous. The potential Mx activity of those sequences in the inbreds in A636, B73, 4Co63, H99, M14, and Mo17 was tested by crossing each of them to an Mx tester line (see Figure 3) and scoring for bz-m kernels in the F2 progeny. Eight F2 ears were scored for each inbred and no bz-m seed were found. This test would have detected an active Mx element anywhere in the genome, except at locations very closely linked (<1 cM) to bz. The failure to detect Mx activity makes it highly unlikely that any of the Mx-hybridizing sequences in the tested inbreds possess Mx activity.
Figure 9.
DNA Gel Blot Analysis of Mx Abundance in Maize Inbreds and Wild Relatives.
Genomic DNA was digested with NcoI, blotted onto Nylon membranes, and hybridized to the same internal Mx probe as in Figure 8. Lanes 1 and 2, Z. mays ssp mexicana, collected in Estado, Mexico; lanes 3 and 4, Z. mays ssp parviglumis, collected in Michoacán; lane 5, Z. mays ssp mexicana, collected in Michocán; lane 6, A188; lane 7, A636; lane 8, B73; lane 9, BSSS53; lane 10, 4Co63; lane 11, H99; lane 12, M14; lane 13, Mo17; lane 14, W22; lane 15, W23.
Mx Dosage Effect
Mottinger (1992) found that increasing the dosage of a 9S chromosome arm carrying bz-x3m and two Mx elements resulted in an increase in the frequency of spots per kernel and in the frequency of kernels with larger spots, which are indicative of earlier rMx excision events. In that experiment, dosage of the autonomous element and the reporter allele were varied jointly. Because Mx is present in single copy in the W22 Mx Sh bz-x3m stock and absent in the derived + sh bz-x3m stock (Figure 3, right), it is possible to vary the dosage of Mx, while holding the dosage of the bz-x3m reporter allele constant, by crossing the two stocks reciprocally. In this reciprocal cross, kernels borne on the Mx Sh bz-x3m parent will have two doses of Mx, whereas those borne on the + sh bz-x3m parent will have a single dose of Mx. All will have three doses of the bz-x3m reporter. The number of spots in 75 kernels from each cross was counted and the average number of spots per kernel calculated. The averages were 3.7 in one-Mx kernels and 4.2 in two-Mx kernels. The distributions of kernel spot number in the two crosses were broad (standard deviations approaching means) and virtually identical. The spot size was small and remarkably uniform in all kernels. Thus, increasing the dosage of the Mx element located 2 cM distal to c1 in 9S from one to two does not appear to have an obvious effect on the excision of rMx from bz-X3m in a W22 background.
Transposons of the Mx and Ac Systems Do Not Cross-React
The availability of bz reporter alleles for Mx and Ac makes it readily feasible to test whether these two hAT transposon systems can cross-react with each other. To determine whether Mx could trans-activate excision of Ds elements from bz, a bz-m2(D1) stock, carrying a Ds deletion derivative of Ac in the second exon (Dooner et al., 1986), and a Bz-wm stock, carrying a Ds1 element in the promoter (Schiefelbein et al., 1988), were pollinated with Mx bz-R as test and with bz-R wx-m7(Ac) as positive control. To determine, reciprocally, whether Ac could trans-activate excision of rMx from bz-x3m, the + sh bz-x3m stock was pollinated with bz-R wx-m7(Ac) as test and with Mx bz-R as positive control. None of the kernels from any of the test crosses were spotted, whereas most of the kernels from the control crosses were, indicating that Mx could not substitute for Ac nor Ac for Mx. Furthermore, ears from reciprocal crosses between bz-m2(Ac) and bz-R displayed the characteristic inverse dosage effect of Ac, independent of the Mx constitution of the bz-R stock, indicating that Mx did not interfere with Ac's dosage effect. Thus, Ac-Ds and Mx-rMx are two active hAT transposon systems in the same species that have evolved different specificities and do not interact with each other.
DISCUSSION
Somatic instability of the bz-x3m mutation, expressed as a spotted kernel phenotype, was ascribed by Mottinger (1992) to a two-element system consisting of rMx, a nonautonomous transposon at bz, and Mx, an autonomous transposon located nearby. Mottinger (1973) had earlier established by conventional genetic tests that the factor responsible for the instability of bz-x3m did not correspond to the autonomous element of any of the major families then known. A W22 version of bz-x3m, developed by repeated backcrosses of the original mutant to that inbred and selection for spotted kernels, was used in this work to isolate rMx and Mx. This stock carries a single active Mx element at a position 6 cM distal to sh1 on 9S. Recombinant stocks carrying only bz-x3m or Mx were derived and used as Mx tester and source, respectively, in subsequent genetic tests of transposon interactions and to authenticate Mx candidates isolated molecularly.
The nonautonomous rMx element is 571 bp long, ends in 13-bp TIRs with a single internal mismatch, and is flanked by an 8-bp TSD. rMx retains no vestiges of a transposase (TPase)-coding sequence, but its terminal sequences resemble those of Tz86, a maize transposon whose ends only are known. The 181 nucleotides at the 5′ ends of rMx and Tz86 are 77% identical, and the 170 nucleotides at the respective 3′ ends are 81% identical. Tz86 has been described as a 3.6-kb transposon that generates a 10-bp TSD and lacks a discernible TIR (Dellaporta et al., 1984). A reexamination of the Tz86-sh1 junction sequences in light of the alignment between the Tz86 and rMx ends (Figure 2) leads to the revised conclusion that, like rMx, Tz86 has a 13-bp imperfect TIR (TCACAGTGGGCAT at the 5′ end, and ATGCCCAGCGTGA at the 3′ end, noncomplementary bases underlined) and produces an 8-bp, rather than a 10-bp, TSD (GGCTGATG). The 13-bp TIRs of Tz86 and rMx differ from each other by 3 and 4 bp, respectively, at the 5′ and 3′ ends. The internal part of rMx shows little similarity to the rest of the available Tz86 sequence.
There are ∼15 copies of rMx homologous sequences in the W22 inbred. Like most other transposons, rMx can excise germinally, leaving either no footprints or typical transposon footprints upon excision, and can create adjacent deletions of host DNA, probably from abortive transposition events. The germinal excision frequency of rMx in W22 is estimated to be at least 0.5%, based on the reversion of bz-x3m to Bz′. However, this is clearly an underestimate because rMx sits in the second exon of the bz gene and many excisions will likely produce a stable bronze phenotype. Such stable bz derivatives are difficult to identify because of the incomplete penetrance of the bz-x3m mutation. In bz-m2(Ac), a completely penetrant mutable allele that harbors an Ac element in the second exon of bz, stable bz derivatives outnumber Bz′ revertants by a factor of 4 to 1 (McClintock, 1956a; Dooner and Belachew, 1989).
Because an rMx probe failed to reveal any bands that cosegregated with Mx, an iterative PCR approach was developed to isolate Mx. This approach, termed primer walking and diagrammed in Figure 4, is based upon the fact that all known autonomous maize transposons have defective relatives in the genome that have suffered internal deletions of variable sizes. The approach consists of amplifying internal sequences from progressively larger members of a family with primers based on unique sequences from the last element isolated (i.e., sequences that are not shared with the smaller elements isolated earlier). A 3.7-kb element was eventually isolated and confirmed to be Mx on the basis of the following cosegregation and excision evidence. First, it was present in the parental Mx bz-x3m and recombinant Mx bz-R stocks, but not in the parental + bz-R or recombinant + bz-x3m stocks. Second, and more importantly, it was lost in Mx− derivatives from Mx bz-x3m homozygotes, where it was replaced by characteristic excision footprints at the prior site of insertion.
Mx displays features typical of other class II transposons in maize. Sequences homologous to the ends of Mx are present in multiple copies in the maize genome, whereas sequences homologous to the central part of Mx are present in low copy number (one to four) in Corn Belt inbred lines and in somewhat higher copy number in different teosinte accessions. However, none of the inbreds tested possessed Mx activity. Sequence analysis of Mx revealed it to be a member of the hAT superfamily of transposons, which also includes Ac. The transposons in this group are categorized primarily on the basis of sequence similarities between their TIRs, amino acid homology among the element-encoded TPases, and the formation of 8-bp TSDs (Kunze and Weil, 2002). Like Ac, Mx contains several copies of a hexanucleotide motif in its subterminal regions, although the hexanucleotide sequences differ. The Mx hexanucleotide repeat is also present in rMx, although in fewer copies (15 versus 17). The Ac TIRs and subterminal repeats have been shown to bind the Ac TPase in vitro (Kunze and Starlinger, 1989; Becker and Kunze, 1997) and most likely play a role in determining the specificity of transposon interactions.
Mx also resembles Ac in its transposition properties: transposed copies of the element can be recovered either with or without a copy of the element at the donor site. The frequency of germinal excision and loss of Mx from its location 6 cM distal to sh1 in 9S is in the low percentage (3/144), similar to that of Ac from bz-m2(Ac) (Dooner and Belachew, 1989). Ac transposes preferentially to linked sites (Greenblatt, 1984; Dooner and Belachew, 1989) and Mx probably does, too, based on the fact that stocks carrying Mx elements at three different linked locations in 9S have been derived from the original bz-x3m mutant stock (Mottinger, 1992; this article). However, Mx may lack Ac's typical negative dosage effect. Ac normally displays a negative dosage effect in which increasing copies of the element result in a developmental delay and reduced frequency of transpositions of either Ac or Ds (McClintock, 1952; Dellaporta and Moreno, 1994), although a few cases of Ac elements with either positive or no dosage effect have been described (Heinlein and Starlinger, 1991; Brutnell et al., 1997). In the original bz-X3m line, which had two Mx elements in 9S, Mottinger (1992) reported a positive dosage effect of Mx on rMx excision, whereas in our W22 derived line, which has a single Mx element at a different location, we do not see an obvious effect when Mx dosage is increased from one to two. The absence of negative dosage effect of Mx on the somatic excision of rMx may mean that the Mx TPase protein does not form inactive aggregates as readily as the Ac TPase (Heinlein et al., 1994).
The combination of three copies of bz-x3m and either one or two copies of Mx, results in a very fine spotting pattern, indicative of infrequent and late somatic excisions (Figure 1). Interestingly, the 13-bp 5′ and 3′ TIRs of Mx and rMx are only 73% identical (19/26). This is the highest degree of divergence found between the TIRs of an autonomous and a nonautonomous member of a transposon family in plants and suggests that Mx and rMx could be elements that are diverging from each other, yet can still interact so that the genetic complementation test places them in the same family. Analysis of the extensive collection of maize sequences in the Genome Survey Sequence section of GenBank supports this view. Searches of that database using as queries the 5′ and 3′ terminal 180 nucleotides of Mx and rMx, which contain the elements' TIRs and all the subterminal hexanucleotide repeats, yielded sequences that shared TIRs with rMx and were more similar to the ends of rMx than of Mx (81 to 96% versus 57 to 72% identity at the 5′ end and 85 to 96% versus 63 to 72% identity at the 3′ end; see Supplemental Table 1 online) and sequences that, conversely, shared TIRs with Mx and were more similar to the ends of Mx than of rMx (82 to 99% versus 63 to 71% identity at the 5′ end and 89 to 98% versus 64 to 75% identity at the 3′ end; see Supplemental Table 1 online). This indicates that maize has several defective transposons, such as dMx1, that are more closely related to Mx than rMx, its genetically defined nonautonomous partner, and whose terminal sequences may be better substrates for the Mx TPase than those of rMx. In Ac, both the TIRs and the subterminal repeats bind to the TPase and are required for Ac excision (Feldmar and Kunze, 1991; Becker and Kunze, 1996; Weil and Kunze, 2000). By analogy, both the TIRs and the hexanucleotide repeats at either end are probably involved in the transposition of Mx and rMx. Perhaps the rather low somatic and germinal excision of rMx from bz-x3m is because of an inefficient interaction between the Mx TPase and the rMx TIRs. If so, a reporter allele harboring a defective transposon of the dMx1 type might be expected to produce a more highly mutable phenotype than bz-x3m in the presence of Mx.
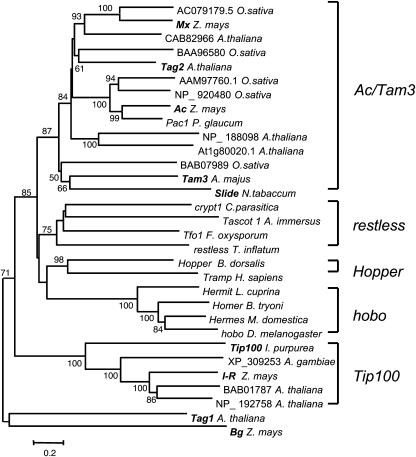
The evolution of the hAT transposon superfamily has been studied by many investigators (Kempken and Windhofer, 2001; Rubin et al., 2001; Robertson, 2002). These studies have revealed that the hAT superfamily is very ancient, probably predating the plant–fungi–animal split 1000 million years ago. To examine the relationship of Mx to other hAT transposons, a similar analysis was performed with 31 hAT TPases from plants, animals, and fungi (see Methods for the basis of selecting sequences). The amino acid sequences were aligned by ClustalX, and the phylogenetic tree shown in Figure 10 was constructed using neighbor joining in MEGA version 2.1 (Kumar et al., 2001). The phylogenetic analysis allows the following main conclusions.
Figure 10.
Phylogenetic Tree of hAT Transposases.
Sequences were aligned using ClustalX, and the tree was constructed using neighbor joining in MEGA version 2.1. Each sequence is identified by either its transposon name or GenBank accession number and the name of its source species. Plant autonomous elements are indicated in bold. Numbers above the branches indicate the percentage of 1000 bootstrap replications in which that branch was present. The well-supported clades are indicated by brackets and given the name of the first described element(s) in that clade.
First, and as noted by others (Kempken and Windhofer, 2001; Robertson, 2002), the plant hAT transposons fall into different clades. One clade contains the genetically defined elements Ac (Pohlman et al., 1984); (Muller-Neumann et al., 1984), Tam3 (Hehl et al., 1991), and Slide (Grappin et al., 1996) from maize, Antirrhinum, and tobacco (Nicotiana tabacum), respectively, plus several Arabidopsis and rice homologs. Another clade includes Tip100 from morning glory (Ipomoea tricolor; Ishikawa et al., 2002), I-R from maize (W. Eggleston, personal communication), and a transposon from the mosquito Anopheles gambia. The more distantly related elements Bg from maize (Hartings et al., 1991a) and Tag1 from Arabidopsis (Liu and Crawford, 1998) do not belong to either of the above clades and may form yet a third, albeit weakly supported, group. Hence, it is not surprising that Ac and Bg do not cross-mobilize each other's defective elements (Hartings et al., 1991b).
Second, the Mx element falls in the same clade as Ac and is closely related to it. Yet, although Ac and Mx are much closer to each other, they still do not interact: Mx cannot trans-activate Ds excision nor contribute to Ac's dosage effect. Thus, modern maize has several families of active and noninteracting transposable elements belonging to the same hAT superfamily. If one adds to the above three, Dt (Rhoades, 1938) and I-R (Eggleston et al., 1995), which have been demonstrated not to interact genetically with Ac and whose partial molecular characterization classifies them as hAT elements (Kunze and Weil, 2002; Robertson, 2002), then there may be at least five families of noninteracting hAT elements of different degree of relatedness in a single species. This observation raises the interesting question of how transposons of the same superfamily can evolve different specificities within one species.
Transposons of the Tc1/mariner superfamily appear to have spread frequently by horizontal transfer during evolution (Hartl et al., 1997; Robertson, 2002). Lampe et al. (2001) have explained the coexistence of distinct autonomous mariner elements in the same genome by a model that invokes transposon amplification, inactivation of most copies by a variety of mechanisms leading to stabilization of the transposon copy number, accumulation of neutral or functionally different mutations in active copies, and horizontal transfer of a divergent functional copy to another species. After horizontal transfer, any mutation that increased the element's activity would be of benefit to the element, as long as it did not affect the fitness of the host. One such mutation would be a cis-acting suppressor mutation in the TIR. If this process was repeated several times, new elements would be expected to diverge from the original to the point that they would no longer interact if they came together again in the same organism. In this model, mutations in the TPase and the TIRs that lead to novel transposon specificities occur in separate organisms.
Unlike the situation with Tc1/mariner elements, there is currently no compelling evidence that transposons of the hAT superfamily have undergone horizontal transfer in evolution. In fact, Rubin et al. (2001) concluded that, in contrast with what had been proposed earlier (Calvi et al., 1991), there was no evidence for transkingdom horizontal transfer of hAT elements and that the question of intrakingdom horizontal transfer could not be resolved from the available data. Therefore, a different model for the evolution of transposon specificities may need to be invoked to explain the cooccurrence of different hAT autonomous elements in the same species.
The following model assumes that hAT TPases resemble the Ac TPase in possessing a dimerization domain (hAT3), which is essential for enzyme activity and also plays a role in the formation of inactive aggregates, although an additional, as yet uncharacterized, domain is also required for Ac multimer formation (Essers et al., 2000; Kunze and Weil, 2002). A mutation in the TPase dimerization domain would allow the new TPase to escape inactivation if the new monomer failed to heterodimerize with the original, more abundant TPase monomers (or did so poorly), but was still able to form active homodimers. Consequently, the new TPase would contribute disproportionately to total activity because it would escape the aggregation inactivation mechanism of the original TPase. However, this will not affect the relative numbers of old and new transposons until the latter's TIRs change, likely in two steps, to favor their recognition by the new TPase. First, a chance mutation in the TIR of one of the more numerous original elements, followed by the homogenization of both TIRs by a conversion-like mechanism, would produce an element that would serve as a better substrate for the new TPase. Thereafter, the relative copy number of that element will increase in individuals that also harbor the element encoding the new TPase. This differential amplification will enhance the probability that the original TPase gene carried in one of the elements with modified TIRs will be replaced with a new TPase gene by mechanisms such as ectopic gene conversion or template switching during the occasional gap repair that follows hAT element excision (Yan et al., 1999). The resulting new element and its progenitor will begin to evolve independently of each other because their oligomerization inactivation mechanisms would not cross-react. Mx and rMx, with their unusually divergent TIRs, may represent transposons in the process of evolving different specificities. If so, an autonomous element with rMx-like ends and encoding a TPase closely related to, but different from, that of Mx may be present in maize.
METHODS
Plant Materials
The bz-x3m mutation was obtained from John Mottinger, backcrossed four times to a W22 stock carrying the bz-R allele, and selfed twice to derive the bz-x3m stock used in this work. All other stocks were also in a W22 background. bz-R is the stable reference allele for the bz locus. The mutable alleles bz-m2(D1), Bz-wm, and wx-m7(Ac) were obtained from B. McClintock (McClintock, 1962, 1964). A188, A636, B73, BSSS53, 4Co63, H99, M14, Mo17, and W23 are standard maize (Zea mays) inbred lines from various public breeding programs (Gerdes et al., 1993). The teosinte accessions were from collections made by Jerry Kermicle in various Mexican states.
PCR and Sequencing
PCR was performed according to the protocol of QiaTaq (Qiagen, Valencia, CA). The fragment containing rMx was amplified from genomic DNA of the bz-x3m mutant using the pair of bz primers Bz-C (5′-CTCAACACGTTCCCAGGC-3′) and Bz-3R (5′-AAACCTCTGAACAGCAAGACGACC-3′). These primers are located 591 bp upstream and 172 bp downstream, respectively, of the rMx insertion site. Germinal rMx excision sites were amplified using the genomic DNAs of Bz′/bz-R revertants as templates and primers Bz-C and Bz-3R. The sequence corresponding to primer Bz-C is deleted from the bz-R allele (Ralston et al., 1988), so only the bz fragment from the Bz′ revertant allele is amplified in the reaction. The sequences of the six oligonucleotide primers used for the primer walking method diagrammed in Figure 4 are as follows: p1, 5′-CACTGGGCATAAAACCCGAGCCC-3′; p2, 5′-CTGGGCATTCGGGTTAGCCCGAA-3′; p3, 5′-CGGGCTTAATCGGGTAGCAACACC-3′; p4, 5′-CCGTTCTACCCGATTTCGTTGCAG-3′; p5, 5′-GTACACCTGGCCGGATCCGTTCAA-3′; p6, 5′-CCAGTAGACTTGCTGCTCACTGGT-3′.
The Mx flanking fragment (tmx) was isolated by IPCR, as optimized in our lab (Cowperthwaite et al., 2002), using genomic DNA from the bz-x3m mutant as template. The primers used in the IPCR are as follows: dMx2-5′R, 5′- AAGCCGGATGCTCCAGACGGCCAC-3′, and dMx2-3′F, 5′- GTGTTGTACGCTTCTGCTAGTGTA-3′. The Mx element was amplified from the W22 bz-x3m stock using primers based on tmx. These primers are tMx-5′-1F (5′-TCGACGTCCGTGATCAACGCCGTT-3′) and tMx-3′-2R (5′-GGCGAAAAAGAACAGTGGGCGCAC-3′). The Mx germinal excision sites (empty sites) were amplified from the tmx′ Sh bz-x3m/+ sh bz-R heterozygotes using primers tMx-5′-1F and tMx-3′-2R. These PCR products were then run on 8% PAGE gels to detect size differences between the amplified alleles. The PCR products were cloned into pGEM-T easy (Promega, Madison, WI) and transformed into DH5α competent cells. Plasmids were purified with a Qiagen spin miniprep kit. Eight randomly selected clones from each empty site amplification were sequenced to characterize the footprints of Mx excision. DNA sequencing was performed in an ABI 377 sequencer (Perkin-Elmer, Torrance, CA) following the manufacturer's instructions. The sequences of Mx and rMx have been submitted to and are being processed by GenBank.
Nucleic Acid Extraction and Hybridization
Genomic DNA from seedlings and mature leaves was prepared by a urea extraction procedure (Greene et al., 1994). A total of 15 to 20 μg of genomic DNA was digested with various restriction enzymes (New England Biolabs, Beverly, MA), resolved on a 0.8% agarose gel in Tris acetate EDTA buffer, and transferred a to Hybond N+ membrane according to the manufacturer's protocol (Amersham Biosciences, Buckinghamshire, UK). Radioactive probes were prepared with a Ready-to-Go labeling kit (Amersham Biosciences). Membrane hybridization and washing conditions followed the recommendations of the manufacturer. The fragment for the dMx2 probe was released from the dMx2-pGEM-T easy clone by EcoRI digestion. The primers used for generating the Mx internal probe, with their Mx location indicated in parentheses, are Mx-5′-2F (760 to 783) and Mx-3′-4R (1686 to 1710). The fragment for the tmx probe (∼280 bp long) was amplified from genomic DNA of the W22 bz-R stock, using primers tMx-5′-1F and tMx-3′-1R (5′-GCGCTAAACTAATGCGGAAAGAGG-3′), cloned into pGEM-T easy, and sequenced.
Chromosome Mapping
Recombinant inbred (RI) lines from a cross between T232 and CM37 were used in chromosome mapping (Burr et al., 1988). Gels were blotted onto Hybond N+ membranes and hybridized with a radiolabeled Mx-adjacent fragment (tmx). Several restriction enzymes were tested on the parental genomic DNA, and EcoRI gave the most distinct restriction fragment length polymorphisms. The segregation of restriction fragment length polymorphisms in RI populations was scored and compared with the RI database at Brookhaven National Lab.
Phylogenetic Analysis
Initial BlastX searches of the GenBank nonredundant databases were performed using the Mx sequence as query and limiting the searches to viridiplantae, fungi, and metazoa, respectively. Hits containing an open reading frame longer than 400 amino acids were chosen for further analysis. Some hAT transposons of the more distant Tip100 group (Robertson, 2002), which were not recovered initially, were also incorporated in the phylogenetic analysis. The putative Slide transposase sequence was deduced from the homology of its predicted open reading frames to other hAT transposases in the database. To reduce the redundancy within the data, sequences with a high degree of similarity to another sequence from the same species were eliminated so that only one of them was included in the comparison. The full-length amino acid sequences were aligned by ClustalX (Thompson et al., 1997), and a phylogenetic tree was constructed using neighbor joining in MEGA version 2.1 (http://www.megasoftware.net/), with 1000 bootstrap replicates and the pairwise-deletion option for handling gaps.
Sequence data for Mx, rMx, dMx1, and dMx2 have been deposited with the EMBL/GenBank data libraries under accession numbers AY753670, AY753671, AY786440, and AY786441, respectively.
Supplementary Material
Acknowledgments
We thank members of the Dooner lab, particularly Yubin Li, for comments on the manuscript, John Mottinger for seeds of the bz-x3m mutation, Bill Eggleston for the unpublished sequence of I-R, Brandon Gaut and Chunguang Du for help with the phylogenetic analysis, and Ben Burr for the analysis of the RI mapping data. This research was supported by a Busch-Waksman Predoctoral Fellowship from Rutgers University to Z.X. and National Science Foundation Grant MCB 99-04646 and Waksman Institute start-up funds to H.K.D.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hugo K. Dooner (dooner@waksman.rutgers.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027797.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Becker, H.A., and Kunze, R. (1996). Binding sites for maize nuclear proteins in the subterminal regions of the transposable element Activator. Mol. Gen. Genet. 251, 428–435. [DOI] [PubMed] [Google Scholar]
- Becker, H.A., and Kunze, R. (1997). Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol. Gen. Genet. 254, 219–230. [DOI] [PubMed] [Google Scholar]
- Brink, R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41, 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R.A., and Nilan, R.A. (1952). The relation between light variegated and medium variegated pericarp in maize. Genetics 37, 519–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.J., Mattes, M.G., O'Reilly, C., and Shepherd, N.S. (1989). Molecular characterization of rDt, a maize transposon of the Dotted controlling element system. Mol. Gen. Genet. 215, 239–244. [DOI] [PubMed] [Google Scholar]
- Brutnell, T.P., May, B.P., and Dellaporta, S.L. (1997). The Ac-st2 element of maize exhibits a positive dosage effect and epigenetic regulation. Genetics 147, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, B., Burr, F.A., Thompson, K.H., Albertson, M.C., and Stuber, C.W. (1988). Gene mapping with recombinant inbreds in maize. Genetics 118, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi, B.R., Hong, T.J., Findley, S.D., and Gelbart, W.M. (1991). Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell 66, 465–471. [DOI] [PubMed] [Google Scholar]
- Chen, J., Greenblatt, I.M., and Dellaporta, S.L. (1987). Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117, 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Greenblatt, I.M., and Dellaporta, S.L. (1992). Molecular analysis of Ac transposition and DNA replication. Genetics 130, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K.C., et al. (2002). Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiol. 130, 1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack, J.B., Cox, D.F., and Peterson, P.A. (1988). Presence of the transposable element Uq in maize breeding material. Crop Sci. 28, 941–944. [Google Scholar]
- Cowperthwaite, M., Park, W., Xu, Z., Yan, X., Maurais, S.C., and Dooner, H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14, 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Chomet, P.S., Mottinger, J.P., Wood, J.A., Yu, S.M., and Hicks, J.B. (1984). Endogenous transposable elements associated with virus infection in maize. Cold Spring Harb. Symp. Quant. Biol. 49, 321–328. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., and Moreno, M.A. (1994). Gene tagging with Ac/Ds elements in maize. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer Verlag), pp. 219–233.
- Dennis, E.S., Finnegan, E.J., Taylor, B.H., Peterson, T.A., Walker, A.R., and Peacock, W.J. (1988). Maize transposable elements: Structure, function, and regulation. In Plant Transposable Elements, O.E. Nelson, ed (New York: Plenum Press), pp. 101–113.
- Dooner, H.K. (1985). A deletion adjacent to the Ac insertion site in a stable bz derivative from bz-m2(Ac). In Plant Genetics, M. Freeling, ed (New York: Alan R. Liss), pp. 561–573.
- Dooner, H.K., and Belachew, A. (1989). Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., English, J., Ralston, E., and Weck, E. (1986). A single genetic unit specifies two transposition functions in the maize element Activator. Science 234, 210–211. [DOI] [PubMed] [Google Scholar]
- Eggleston, W.B., Alleman, M., and Kermicle, J.L. (1995). Molecular organization and germinal instability of R-stippled maize. Genetics 141, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, L., Adolphs, R.H., and Kunze, R. (2000). A highly conserved domain of the maize Activator transposase is involved in dimerization. Plant Cell 12, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N. (1983). Controlling elements in maize. In Mobile Genetic Elements, J.A. Shapiro, ed (New York: Academic Press), pp. 1–63.
- Feldmar, S., and Kunze, R. (1991). The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 10, 4003–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., Jiang, N., and Wessler, S.R. (2002. a). Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., Zhang, Y., and Wessler, S.R. (2002. b). Miniature inverted repeat transposable elements and their relationship to established DNA transposons. In Moble DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 1147–1158.
- Gerdes, J.T., Behr, C.F., Coors, J.G., and Tracy, W.F. (1993). Compilation of North American Maize Breeding Germplasm. (Madison, WI: Crop Science Society of America).
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Gonella, J.A., and Peterson, P.A. (1977). Controlling elements in a tribal maize from Colombia: Fcu, a two-unit system. Genetics 85, 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grappin, P., Audeon, C., Chupeau, M.C., and Grandbastien, M.A. (1996). Molecular and functional characterization of Slide, an Ac-like autonomous transposable element from tobacco. Mol. Gen. Genet. 252, 386–397. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I.M. (1984). A chromosome replication pattern deduced from pericarp phenotypes resulting from movement of the transposable element Modulator in maize. Genetics 108, 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, I.M., and Brink, R.A. (1962). Twin mutations in medium variegated pericarp maize. Genetics 47, 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, B., Walko, R., and Hake, S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138, 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings, H., Rossi, V., Lazzaroni, N., Thompson, R.D., Salamini, F., and Motto, M. (1991. a). Nucleotide sequence of the Bg transposable element of Zea mays L. Maydica 36, 355–359. [Google Scholar]
- Hartings, H., Spilmont, C., Lazzaroni, N., Rossi, V., Salamini, F., Thompson, R.D., and Motto, M. (1991. b). Molecular analysis of the Bg-rbg transposable element system of Zea mays L. Mol. Gen. Genet. 227, 91–96. [DOI] [PubMed] [Google Scholar]
- Hartl, D.L., Lohe, A.R., and Lozovskaya, E.R. (1997). Modern thoughts on an ancyent marinere: Function, evolution, regulation. Annu. Rev. Genet. 31, 337–358. [DOI] [PubMed] [Google Scholar]
- Hehl, R., Nacken, W.K., Krause, A., Saedler, H., and Sommer, H. (1991). Structural analysis of Tam3, a transposable element from Antirrhinum majus, reveals homologies to the Ac element from maize. Plant Mol. Biol. 16, 369–371. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., Brattig, T., and Kunze, R. (1994). In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and Petunia protoplasts. Plant J. 5, 705–714. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., and Starlinger, P. (1991). Variegation patterns caused by transposable element Ac. Maydica 36, 309–316. [Google Scholar]
- Ishikawa, N., Johzuka-Hisatomi, Y., Sugita, K., Ebinuma, H., and Iida, S. (2002). The transposon Tip100 from the common morning glory is an autonomous element that can transpose in tobacco plants. Mol. Genet. Genomics 266, 732–739. [DOI] [PubMed] [Google Scholar]
- Kempken, F., and Windhofer, F. (2001). The hAT family: A versatile transposon group common to plants, fungi, animals, and man. Chromosoma 110, 1–9. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). Molecular Evolutionary Genetics Analysis (MEGA) Software. (Tempe, AZ: Arizona State University).
- Kunze, R., and Starlinger, P. (1989). The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8, 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., and Weil, C.F. (2002). The hAT and CACTA superfamilies of plant transposons. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 565–610.
- Lampe, D.J., Walden, K.K., and Robertson, H.M. (2001). Loss of transposase-DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol. Biol. Evol. 18, 954–961. [DOI] [PubMed] [Google Scholar]
- Liu, D., and Crawford, N.M. (1998). Characterization of the putative transposase mRNA of Tag1, which is ubiquitously expressed in Arabidopsis and can be induced by Agrobacterium-mediated transformation with dTag1 DNA. Genetics 149, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1952). Chromosome organization and gene expression. Cold Spring Harb. Symp. Quant. Biol. 16, 13–47. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1956. a). Mutation in maize. Carnegie Inst. Wash. Yrbk. 55, 323–332. [Google Scholar]
- McClintock, B. (1956. b). Intranuclear systems controlling gene action and mutation. Brookhaven Symp. Biol. 8, 58–74. [PubMed] [Google Scholar]
- McClintock, B. (1962). Topographical relations between elements of control systems in maize. Carnegie Inst. Wash. Yrbk. 61, 448–461. [Google Scholar]
- McClintock, B. (1964). Aspects of gene regulation in maize. Carnegie Inst. Wash. Yrbk. 63, 592–602. [Google Scholar]
- Montanelli, C., Di Fonzo, N., Marotta, R., Soave, C., and Salamini, F. (1984). Occurrence and behavior of the components of the o2-m(r)-Bg system of maize controlling elements. Mol. Gen. Genet. 197, 209–218. [Google Scholar]
- Mottinger, J. (1973). Unstable mutants of bronze induced by premeiotic X-ray treatment in maize. Theor. Appl. Genet. 43, 190–195. [DOI] [PubMed] [Google Scholar]
- Mottinger, J.P. (1992). Studies on the Mx transposable element system in maize recovered from X-irradiated stocks. Mol. Gen. Genet. 236, 96–104. [DOI] [PubMed] [Google Scholar]
- Mottinger, J.P., Dellaporta, S.L., and Keller, P.B. (1984). Stable and unstable mutations in Aberrant Ratio stocks of maize. Genetics 106, 751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Neumann, M., Yoder, J., and Starlinger, P. (1984). The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198, 19–24. [Google Scholar]
- Neuffer, M.G., Coe, E.H., and Wessler, S. (1997). The Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Palaisa, K.A., Morgante, M., Williams, M., and Rafalski, A. (2003). Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas, T., Weir, J., and Walker, E.L. (1999). The structure and paramutagenicity of the R-marbled haplotype of Zea mays. Genetics 153, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, P.A. (1988). The mobile element systems in maize. In Plant Transposable Elements, O.E. Nelson, ed (New York: Plenum Press), pp. 43–68.
- Peterson, P.A., and Salamini, F. (1986). A search for active mobile elements in the Iowa Stiff Stalk Synthetic maize population and some derivatives. Maydica 31, 163–172. [Google Scholar]
- Pohlman, R., Fedoroff, N., and Messing, J. (1984). The nucleotide sequence of the maize controlling element Activator. Cell 37, 635–644. [DOI] [PubMed] [Google Scholar]
- Ralston, E.J., English, J., and Dooner, H.K. (1988). Sequence of three bronze alleles of maize and correlation with the genetic fine structure. Genetics 119, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M. (1938). Effect of the Dt gene on the mutability of the a1 allele in maize. Genetics 23, 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M., and Dempsey, E. (1982). The induction of mutable systems in plants with the high-loss mechanism. Maize Genet. Coop. News Lett. 56, 21–26. [Google Scholar]
- Robertson, H.M. (2002). Evolution of DNA transposons in eukaryotes. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 1093–1110.
- Rubin, E., Lithwick, G., and Levy, A.A. (2001). Structure and evolution of the hAT transposon superfamily. Genetics 158, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J.W., Furtek, D.B., Dooner, H.K., and Nelson, O.E., Jr. (1988). Two mutations in a maize bronze1 allele caused by transposable elements of the Ac-Ds family alter the quantity and quality of the gene product. Genetics 120, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, N.S., Rhoades, M.M., and Dempsey, E. (1989). Genetic and molecular characterization of a-mrh-Mrh, a new mutable system of Zea mays. Dev. Genet. 10, 507–519. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P., and Walbot, V. (1985). A deletion adjacent to the maize transposable element Mu1 accompanies loss of Adh1 expression. EMBO J. 4, 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt, W.J., and Brink, R.A. (1969). Geographic distribution of paramutable and paramutagenic R alleles in maize. Genetics 61, 677–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V., and Rudenko, G.N. (2002). MuDR/Mu transposable elements of maize. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 533–564.
- Weil, C.F., and Kunze, R. (2000). Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 26, 187–190. [DOI] [PubMed] [Google Scholar]
- Xu, Z., Yan, X., Maurais, S., Fu, H., O'Brien, D.G., Mottinger, J., and Dooner, H.K. (2004). Jittery, a Mutator distant relative with a paradoxical mobile behavior: Excision without reinsertion. Plant Cell 16, 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., Martinez-Ferez, I.M., Kavchok, S., and Dooner, H.K. (1999). Origination of Ds elements from Ac elements in maize: Evidence for rare repair synthesis at the site of Ac excision. Genetics 152, 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.