Abstract
Nuclear proteins play key roles in the fundamental regulation of genome instability, the phases of organ development, and physiological responsiveness through gene expression. Although nuclear proteins have been shown to account for approximately one-fourth of total proteins in yeast, no efficient method to identify novel nuclear proteins has been applied to plants. In this study, a trial to isolate nuclear proteins in rice was attempted, and several novel nuclear proteins showing a variety of subnuclear localizations were identified. The nuclear transportation trap (NTT) system, which is a modified two-hybrid system, isolated many nuclear proteins from rice (Oryza sativa) NTT cDNA libraries. Nuclear localization of the isolated proteins was confirmed by transient introduction of green fluorescent protein fusion constructs for a subset of protein genes into onion (Allium cepa) cells. The majority of these proteins, including novel proteins and proteins initially categorized as cytoplasmic proteins, were revealed to be localized in the nucleus. Detailed characterization of unknown proteins revealed various subnuclear localizations, indicating their possible association with chromatin and the nuclear matrix with a foci or speckle-like distribution. Some also showed dual distribution in the nucleus and cytoplasm. In the novel protein fraction, a protein was further identified for its chromatin-associated localization in a specific organ of rice by immunostaining. Thus, a variety of novel nuclear architectural proteins with chromatin or matrix associating abilities, which are important in nuclear organization by influencing certain organ developments or cell responsiveness, can be isolated using the NTT method. Because nuclear proteins other than transcription regulators have rarely been characterized in plants, such as matrix proteins and development-specific chromatin proteins, their identification and subsequent characterization could provide important information for genome-wide regulatory mechanisms controlled by nuclear organization.
INTRODUCTION
Dynamic nuclear organization, orchestrated by a complex network of nuclear proteins, is fundamental to an understanding of cellular development and physiology. In yeast, 27% of all proteins are localized in the nucleus (Kumar et al., 2002). This indicates that an unexpectedly large number of proteins function in the nucleus. By comparison, the number of nuclear proteins predicted by in silico analysis on the basis of genomic and known nuclear protein data comprise as much as 18% of all the proteins in yeast (Cokol et al., 2000). Nuclear proteome analysis has not yet been reported for other organisms, although the importance of such analyses for understanding nuclear organization and architecture, which coordinates gene expression and DNA replication, is clear. In contrast with yeast and mammals, a limited number of analyses of plant nuclear proteins have been performed. Some plant components of intranuclear compartments were reported to differ greatly from those of other organisms. Only a few plant nuclear matrix proteins have been characterized, and they have no obvious homology with known nuclear matrix proteins in yeast and mammals (Gindullis and Meier, 1999; Gindullis et al., 1999). Although a recent proteome analysis for the Arabidopsis thaliana nuclear matrix identified several nuclear matrix proteins with similarity to animal nuclear proteins, most of them were nucleolar or ribosomal components found in mammals (Calikowski et al., 2003). Furthermore, no homolog of the component of intermediate filament-like nuclear lamins has been found in the Arabidopsis and Oryza genomes, despite their essential role as fundamental nuclear components in animals. Biochemical, cell biological, and genetic studies, in relation to plant nuclear events, awaits the efficient isolation of a variety of nuclear proteins, including nuclear matrix/scaffold proteins and intermediate filament proteins.
Although genome sequencing of Arabidopsis (Arabidopsis Genome Initiative, 2000) and Oryza (Goff et al., 2002; Yu et al., 2002) has been nearly completed, a functional analysis of genes, their products, and interactions is only starting. More efficient methods of functional genomics, proteome analysis, and transcriptome analysis are necessary because plants have a relatively large number of genes and gene products compared with animals with a similar genome size. For example, Arabidopsis has 25,000 genes in its 125-Mb genome (Arabidopsis Genome Initiative, 2000), whereas Drosophila has 13,600 genes in its 180-Mb genome (Adams et al., 2000). Rice (Oryza sativa) has 40,000 to 50,000 genes in its 430-Mb genome (Goff et al., 2002; Yu et al., 2002), whereas Fugu rubripes (Japanese puffer fish) has 28,000 genes in its 365-Mb genome (Aparicio et al., 2002). Subcellular localization of gene products is a useful indicator for predicting their function, especially for previously uncharacterized genes in any organism. However, conventional proteomics approaches using isolated intracellular organelles to identify several proteins require much labor, time, and cost. A cell biological approach based on tagging proteins with an epitope or green fluorescent protein (GFP) followed by cytological detection of intracellular localization is a powerful alternative method for proteome analysis (Ross-MacDonald et al., 1997; Ding et al., 2000). A recent study using this method successfully identified subcellular localization of all 6100 yeast proteins (localizome; Kumar et al., 2002) and specifically localized approximately half of the uncharacterized proteins in yeast. Two methods have been used for genome-wide epitope tagging. One was direct cloning of PCR-amplified open reading frames into a yeast tagging and expression vector, and the other was random tagging by transposon insertion. The cDNA-tagging and random genome-tagging methods were very powerful for organisms with a small genome composed of a limited number of genes and gene-rich genome regions. A few trials to detect subcellular localization using GFP-fused plant cDNAs successfully identified specific subcellular proteins, including nuclear proteins in tobacco (Nicotiana tabacum) cells (Escobar et al., 2003; Van Damme et al., 2004). However, those methods still have limitations for isolating nuclear protein genes efficiently.
The nuclear transportation trap (NTT) system is a modified yeast two-hybrid method that can specifically trap proteins carrying a nuclear localization signal (NLS) in yeast (Ueki et al., 1998). The NTT system uses the transactivator NES-LexAD, which consists of three domains, the nuclear export signal (NES) of HIV Rev, the LexA DNA binding domain, and the GAL4 transactivation domain (GAL4AD), and has potential ability to induce LEU2 reporter gene expression. If a cDNA inserted into the cloning site downstream of this transactivator gene encodes the NLS, the expressed fusion protein can be imported into the nucleus. It then activates the LEU2 reporter gene and confers Leu prototrophy to the yeast host strain EGY48L. The NES signal prevents diffusible translocation of the fusion protein into the nucleus. Using this system, Ueki et al. (1998) successfully screened several novel nuclear proteins from the human fetal brain cDNA library. The NTT system offers several advantages for screening nuclear proteins. Firstly, simple direct screening of a cDNA library needs neither nuclear protein purification and sequencing nor construction of all tagged proteins. Secondly, the method can easily be combined with other molecular biological approaches to obtain epitope-tagged cDNA clones.
In this study, we applied the NTT system to rice and isolated 523 unique candidate genes encoding nuclear proteins in 2157 positive clones from three different NTT libraries. Subsequent localization analysis of GFP-fused proteins proved the isolated proteins were various novel nuclear proteins that had been categorized as hypothetical proteins or even nonnuclear proteins in the databases. These fusion proteins showed various nuclear localization patterns, such as colocalization with chromatin, accumulation in part of the nuclear matrix, and nuclear focus formation in diverse patterns in onion (Allium cepa) cells. Additional antibody analysis for one of these proteins also showed the same nuclear distribution and organ-specific expression in rice. A fraction of the proteins may be incorporated into the functional and structural link from the chromatin to the nuclear envelope via some of these components in plants. It is anticipated that the study of such protein components will shed light on the signal transfer machinery in relation to nuclear architectural control in the plant nuclei.
RESULTS
Construction of Rice NTT cDNA Libraries and Clone Screening
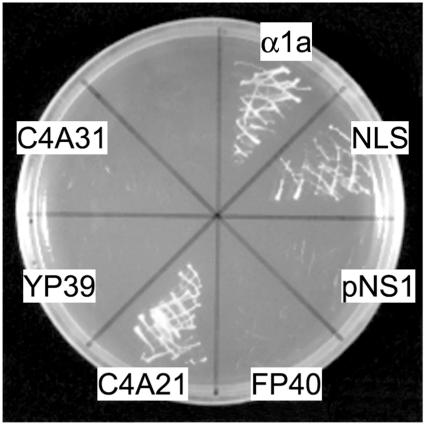
First, we examined the feasibility of the NTT selection system for plant nuclear proteins using rice importin α (importin-α1a; Jiang et al., 1998; Shoji et al., 1998) and four randomly picked cDNA clones from the cDNA libraries described below. These genes were ligated with the NTT vector that harbors a HIS3 gene as a selection marker and then introduced into the yeast strain EGY48L. We confirmed their nuclear transportation by checking their growth on a Leu-/His- selection plate. The yeast cells transformed with either importin-α1a or pNS-C4A21, which encodes an SC35-like splicing factor, grew well on the selection plate as shown in Figure 1. On the other hand, none of the remaining three clones, pNS-FP40, pNS-C4A31, or pNS-YP39, supported growth of the yeast cells (Figure 1). The pNS-FP40 construct encodes p68-like DEAD-box RNA helicase with a deletion for the predicted NLS on its N-terminal region. The pNS-C4A31 encodes a probable acidic endochitinase. The pNS-YP39 plasmid encodes a neoxanthin cleavage enzyme-like protein. These three proteins were predicted to localize in cytoplasm. These findings indicated that the NTT system can be successfully applied to the selection of plant nuclear proteins.
Figure 1.
NTT Selection against Plant Proteins.
Yeast strains containing rice cDNAs in the NTT vector, pNS1, were incubated on a Leu-/His- selection medium plate for 3 d at 30°C. α1a, pNS1 carrying the rice importin-α1a (Shoji et al., 1998); NLS, pNS-NLS carrying the SV40 large T-antigen NLS as a positive control (Ueki et al., 1998); pNS1, NTT vector with no insert as a negative control; FP40, pNS-FP40 encoding a p68-like DEAD-box RNA helicase with truncation of the N terminus NLS (DDBJ/GenBank/EMBL accession number AB110202); C4A21, pNS-C4A21 encoding an SC35-like splicing factor with N-terminal truncation (accession number AB110200); YP39, pNS-YP39 encoding a neoxanthin cleavage enzyme-like protein with C-terminal truncation (accession number AB110203); C4A31, pNS-C4A31 encoding a probable acidic endochitinase with N-terminal truncation (accession number AB1102001).
We used three rice cDNA libraries to identify nuclear protein genes. The libraries were constructed from RNA samples isolated from panicles of gamete generating stages (GGP library), from spikelets at early embryogenesis stages (EEP library), and from regenerating calli 4 d after regeneration treatment (C4 library). The libraries were first constructed as conventional plasmid or λ phage cDNA libraries and then converted into NTT libraries as described in Methods. Three different libraries were used based on two considerations. One was identification of as many nuclear proteins as possible, and the other was a comparison of nuclear protein contents among the three different samples. The three NTT libraries were then subjected to screening of nuclear transportable proteins. Approximately 0.05% of transformed clones representing His prototrophy were able to survive in the successive NTT selection on the His-/Leu- selection plate. In this screen, 2157 positive clones were obtained from the three different NTT cDNA libraries. After eliminating clones that showed very slow growth or had no, or a very short, insert, the sequence of the inserts in the remaining 1423 cDNA clones was determined. Sequence comparison among the 1423 clones identified 684 independent clones from the three different libraries (Table 1).
Table 1.
Clone Numbers in Each Analysis Step
| NTT cDNA Library
|
||||
|---|---|---|---|---|
| Clones Analyzed | GGP | EEP | C4 | Total |
| Positive clone | 868 | 677 | 612 | 2157 |
| Sequenced clone | 503 | 483 | 437 | 1423 |
| Independent clone | 206 | 268 | 210 | 684 |
Classification of the NTT Screened Proteins
Homology searches based on DNA sequences and predicted amino acid sequences of the 684 independent clones were performed against the DDBJ/EMBL/GenBank and the Swiss-Prot/PIR/DAD/PDB databases using the FASTA and BLAST programs (see Methods). We classified the 684 clones into five groups according to the homology search results (Table 2). The largest group with 177 clones (25.9%) was a nuclear-specific protein group. The clones in this group were classified further into eight subgroups based on their functional characteristics (Table 2). The largest subgroup was a transcriptional regulator subgroup, which comprised 107 clones (60.1% of this group). The second largest subgroup was an RNA processing protein group that comprised 27 clones (15.3%), such as splicing factors. The third largest subgroup of 14 clones (7.9%) encoded histones or other chromatin proteins. By contrast, a basal transcription factor, such as TFIIB, subgroup comprised a small population with only three clones (1.7%).
Table 2.
Classification of Independent Clones
| Independent Clone Number
|
||||||
|---|---|---|---|---|---|---|
| Group | Subgroup | GGP Library | EEP Library | C4 Library | Cumulative | Gene Number |
| Nuclear protein | 47 (22.8) | 70 (26.1) | 60 (28.6) | 177 (25.9) | 136 (26.0) | |
| Transcriptional regulator | 27 (57.4) | 39 (55.7) | 41 (68.3) | 107 (60.1) | 77 (56.6) | |
| RNA processing | 9 (19.1) | 9 (12.9) | 9 (15.0) | 27 (15.3) | 20 (14.7) | |
| Chromatin protein | 4 (8.5) | 6 (8.6) | 4 (6.7) | 14 (7.9) | 13 (9.6) | |
| Nuclear architecture | 4 (8.5) | 6 (8.6) | 0 (0) | 10 (5.6) | 9 (6.6) | |
| DNA replication, modification | 1 (2.1) | 1 (1.4) | 2 (3.3) | 4 (2.3) | 4 (2.9) | |
| Basal transcription factor | 1 (2.1) | 1 (1.4) | 1 (1.7) | 3 (1.7) | 3 (2.2) | |
| Nuclear transport | 0 (0) | 1 (1.4) | 0 (0) | 1 (0.6) | 1 (0.7) | |
| Others | 1 (2.1) | 7 (10.0) | 3 (5.0) | 11 (6.2) | 9 (6.6) | |
| Nuclear relating proteina | 20 (9.7) | 23 (8.6) | 33 (15.7) | 76 (11.1) | 49 (9.4) | |
| Unknown protein (with homology) | 70 (34.0) | 87 (32.5) | 60 (28.6) | 217 (31.7) | 176 (33.7) | |
| Unknown protein (without homology)b | 10 (4.9) | 12 (4.5) | 7 (3.3) | 29 (4.2) | 27 (5.2) | |
| Known as nonnuclear protein | 59 (28.6) | 76 (28.4) | 50 (23.8) | 185 (27.0) | 135 (25.8) | |
| Total | 206 | 268 | 210 | 684 | 523 | |
Scores in parentheses represent percentage. Subgroup clone numbers are underlined. Out-of-frame fused clones are not eliminated because their acceptance was reported in yeast (Stahl et al., 1995; Fromont-Racine et al., 1997).
Proteins of this group are cytoplasmic proteins, which are also detected in the nucleus.
This group may also contain clones derived from genomic DNA.
The second group with 76 clones (11.1%) was a nuclear-related protein group corresponding to proteins located both in the nucleus and the cytoplasm, such as ribosomal proteins and 14-3-3 family proteins. Clones classified into the third and the fourth groups (35.9% of the total independent clones) encoded proteins with unknown functions. Among them, 217 clones (31.7% of the total independent clones) classified into the third group showed homology with previously recorded proteins in the databases that lacked any functional annotation. Twenty-nine clones (4.2% of the total independent clones), classified into the fourth group, showed no significant homology to any sequences in the databases. The remaining 185 clones (27.0% of the total independent clones) in the fifth group showed significant amino acid sequence similarity to previously characterized nonnuclear proteins.
Composition ratio of individual groups or subgroup proteins in each library was comparable among the three different cDNA libraries. This consistency among libraries suggests that the nuclear protein trapping efficiency of this system was reliable. Overall, the 684 independent clones isolated from the three different libraries were derived from 523 unique genes as shown in Table 2. Sequence, accession numbers, BLAST search data, and other clone characters for 684 independent clones are available in the supplemental data online.
Analysis of Subcellular Localization of the Nonnuclear Group Proteins
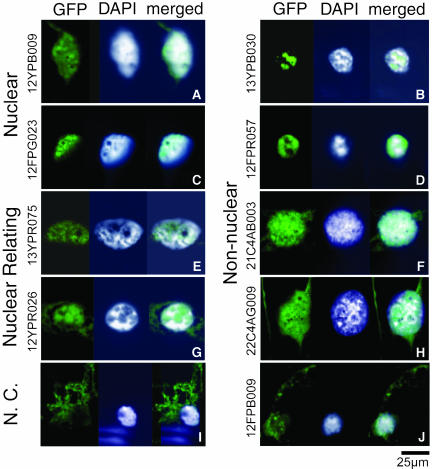
The clones classified into the nonnuclear protein group might possibly be false positives for various reasons, such as LEU2 gene activation by host cell modification. However, different clones encoding the same protein were identified reproducibly in many cases. These results indicated that the proteins classified into this group might have characteristics of nuclear proteins as well, although they were previously annotated as nonnuclear proteins. To test this idea, we examined the localization of 10 of these proteins after expression of GFP-tagged open reading frames in A. cepa (onion) epidermal cells. GFP fluorescence was detected in the nucleus at least for five proteins, as shown in Figures 2B, 2D, 2F, 2H, and 2J and Table 3. An example is ankyrin (13YPB030) fusion, which is a structural protein most often associated with the inside of the plasma membrane (Bennett, 1979). The GFP:ankyrin accumulated in a restricted part of the inner nuclear matrix region (Figure 2B), which was less stained with 4′,6-diamidino-2-phenylindole (DAPI) in the nucleus. Another example is HSP18, which has been generally thought to be a cytoplasmic protein (Helm et al., 1997). The GFP:HSP18 (21C4AB003) fusion localized in the nucleus (Figure 2F), and the GFP fluorescence corresponded well to the DAPI staining. In this case, a weak GFP fluorescence was also observed in the cytoplasm (Figure 2F). As a positive control, we also generated GFP-fusion constructs with subsets of the nuclear-specific group and the nuclear-related group proteins. In every case, the fusion proteins from these two groups were localized in the nucleus (Figures 2A, 2C, 2E, and 2G). These results suggest that a significant portion of the clones that had been initially classified into the nonnuclear group encode proteins that can be imported into the nucleus in plants.
Figure 2.
Nuclear Localization of GFP-Fused Proteins in the Allium Epidermal Cells.
The NTT clones introduced into onion cells are indicated vertically at the left. The image of the GFP fusion protein is shown at the left, and the nucleus stained with the DNA binding dye DAPI is in the middle, and the merged image of GFP with DAPI signals is at the right. The GFP figures represent localization of the following proteins fused to GFP.
(A) An uncharacterized zinc finger protein (protein ID BAA33204.1).
(C) A putative ADP-ribosylation factor GAP-like zinc finger–containing protein, ZiGA4 (protein ID BAB90399.1).
(E) A proteasome α subunit protein (protein ID AAB51521.1).
(G) A putative member of the 14-3-3 family proteins (DDBJ/EMBL/GenBank accession number AU032196, absent in the protein databases).
(I) A chlorophyll a/b binding protein used as a negative control (N.C.; protein ID AAC15992.1).
(B), (D), (F), (H), and (J) Nonnuclear group proteins whose fusion proteins were detected in the nucleus. Details of these clones are shown in Table 3. Bar = 25 μm. Note that the A. cepa epidermal cell has a variety of shapes, including various grooves and invaginations (Collings et al., 2000).
Table 3.
List of Nonnuclear and Unknown Group Proteins Used for GFP Fusion Protein Localization Analysis
| Group | Clone Name | DDBJ Accession Number | Clone Statusa | Hit Protein by Homology Search (Protein ID) | Predicted NLS by PSORT Searchb | NLS Score | MW of Fusion Proteinc | Nuclear Transport Efficiency | Corresponding Photograph |
|---|---|---|---|---|---|---|---|---|---|
| Nonnuclear proteins | 12FPB009 | AB110164 | Full | O. sativa putative fructokinase II protein (AAL26573.1) | n.f. | 0.0 | 62,934 | + | Figure 2J |
| 12FPR057 | AB110168 | Full | O. sativa protein similar to Zea mays caffeoyl-CoA 3-O-methyltransferase (BAA81774.1) | n.f. | 0.0 | 57,199 | ++ | Figure 2D | |
| 12YPR001 | AB110170 | Full | O. sativa putative thiamine biosynthesis protein (BAC45141.1) | n.f. | 0.0 | 66,344 | − | n.s.d | |
| 13YPB030 | AB110173 | Full | O. sativa putative ankyrin protein (AAG46181.1) | n.f. | 0.0 | 58,360 | ++ | Figure 2B | |
| 13YPG038 | AB110177 | Full | O. sativa katanin p60 subunit A 1-like protein (BAB90151.1) | Position 78-85 (RGKGFVDS) | 0.1 | 74,015 | − | n.s. | |
| 13YPG044 | AB110178 | Full | O. sativa putative nascent polypeptide associated complex α chain protein (BAB90246.1) | n.f. | 0.0 | 50,742 | − | n.s. | |
| 13YPR038 | AB110180 | Full | O. sativa Rubisco activase large isoform precursor protein (BAA97583.1) | n.f. | 0.0 | 76,859 | − | n.s. | |
| 14FPG037 | AB110186 | Full | O. sativa putative dihydroflavonol reductase protein (AAD24584.1) | n.f. | 0.0 | 65,110 | − | n.s. | |
| 21C4AB003 | AB110191 | Full | O. sativa class I low-molecular-weight heat shock protein 17.9 protein (AAK54445.1) | n.f. | 0.0 | 48,703 | + | Figure 2F | |
| 22C4AG009 | AB110193 | Full | O. sativa Asp aminotransferase protein (BAA03504.1) | n.f. | 0.0 | 73,960 | + | Figure 2H | |
| Unknown proteins | 12FPG037 | AB110165 | Full | O. sativa protein (BAA93034.1) | n.f. | 0.0 | 56,082 | +/− | n.s. |
| 12FPG084 | AB110166 | Partial | Arabidopsis F28N24.14 protein (AAF88121.1) | n.f. | 0.0 | 56,599 | +/− | n.s. | |
| 12FPR024 | AB110167 | Full | Arabidopsis dehydrogenase-like protein (BAB01322.1) | Position 111-118 (KGGAFVEA) | 0.1 | 61,787 | + | Figure 3H | |
| 12YPG019 | AB110169 | Full | Arabidopsis AT3g53990 protein (AAG40033.1) | n.f. | 0.0 | 49,223 | + | n.s. | |
| 13YPB021 | AB110171 | Full | O. sativa unknown protein (AAK14411.1) | n.f. | 0.0 | 44,266 | ++ | n.s. | |
| 13YPB022 | AB110172 | Full | Arabidopsis ERD15 protein (BAA06384.1) | n.f. | 0.0 | 49,239 | ++ | Figure 3F | |
| 13YPB053 | AB110174 | Partial | Arabidopsis putative protein F9K21.210 (CAB75492.1) | n.f. | 0.0 | 57,224 | ++ | n. s. | |
| 13YPG010 | AB110175 | Full | Arabidopsis putative protein T25B15-70 (CAC07921.1) | n.f. | 0.0 | 49,538 | + | Figure 3K | |
| 13YPG014 | AB110176 | Full | Arabidopsis putative protein F17A8.180 (CAB39650.1) | Position 3-6 (RKRK) | 0.3 | 54,315 | ++ | Figure 3C | |
| 13YPR033 | AB110179 | Full | Arabidopsis F2J10.8 protein (AAF76441.1) | n.f. | 0.0 | 78,095 | + | n.s. | |
| 13YPR054 | AB110181 | Partial | Arabidopsis putative dehydrogenase protein (AAF14837.1) | n.f. | 0.0 | 57,430 | − | n.s. | |
| 14FPB004 | AB110182 | Full | O. sativa YY2 protein (BAA23618.1) | n.f. | 0.0 | 72,412 | +/− | n.s. | |
| 14FPB010 | AB110183 | Full | Arabidopsis putative protein AT4g25840 (CAB39605.1) | n. f. | 0.0 | 56,046 | ++ | Figure 3D | |
| 14FPB077 | AB110184 | Full | Arabidopsis putative protein T30N20 130 (CAB96841.1) | n.f. | 0.0 | 51,163 | ++ | Figure 3E | |
| 14FPB137 | AB110185 | Full | Arabidopsis unknown protein F11B9.13 (AAG50959.1) | n.f. | 0.0 | 57,062 | + | n.s. | |
| 15FPR003 | AB110187 | Partial | Arabidopsis F8M12.21 protein (AAC33943.1) | n.f. | 0.0 | 62,124 | + | n.s. | |
| 16FPG004 | AB110188 | Full | Arabidopsis protein F14013.20 (BAB03019.1) | Position 248-251 (KKRK) | 0.3 | 57,715 | ++ | Figure 3A | |
| 17FPG011 | AB110189 | Full | Arabidopsis aluminum-induced protein-like protein (BAB11312.1) | n.f. | 0.0 | 56,779 | + | Figure 3J | |
| 17YPR003 | AB110190 | Full | Arabidopsis protein MCK7.9 (BAA96913.1) | n.f. | 0.0 | 66,059 | + | n.s. | |
| 21C4BG001 | AB110192 | Full | Arabidopsis protein MIJ24.6 (BAB08888.1) | Position 352-355 (RKKH) | 0.7 | 71,032 | + | Figure 3L | |
| 22C4AG010 | AB110194 | Partial | O. sativa B1153F04.12 protein (BAB62605.1) | n.f. | 0.0 | 45,826 | + | Figure 3I | |
| 23C4AG017 | AB110195 | Full | O. sativa P0038C05.1 protein (BAB19328.1) | n.f. | 0.0 | 39,018 | ++ | Figure 3G | |
| 23C4AG047 | AB110196 | Full | Arabidopsis protein K5F14.2 (BAB09934.1) | Position 84-87 (KRPR) | 0.3 | 56,461 | ++ | Figure 3B | |
| 23C4AG056 | AB110197 | Partial | Arabidopsis unknown protein AT4g37210 (AAK44004.1) | n.f. | 0.0 | 65,177 | ++ | n.s. | |
| 23C4AR004 | AB110198 | Full | Arabidopsis protein MTE24.1 (BAB01311.1) | n.f. | 0.0 | 46,905 | + | n.s. | |
| 23C4BG006 | AB110199 | Partial | Arabidopsis putative protein F14L2-150 (CAB88542.1) | n.f. | 0.0 | 48,195 | ++ | n.s. |
“Full” and “partial” indicate probable full-length and partial open reading frame encoding clones, respectively.
Numbers are calculated NLS position in each full-length protein. n.f., not found by PSORT search.
MW, molecular weight.
n.s., data not shown.
Novel Plant Nuclear Proteins in the Unknown Group
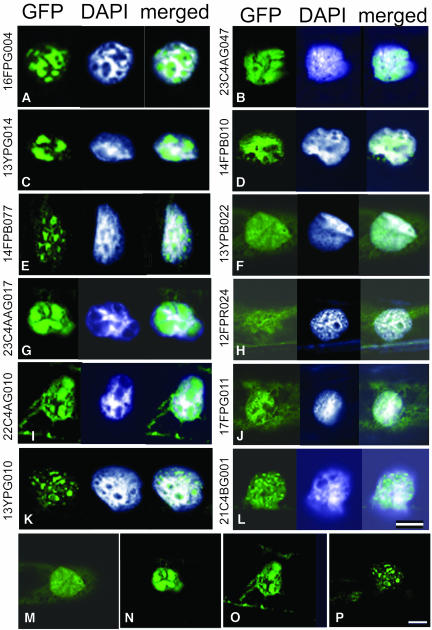
Nuclear localization of the unknown group proteins was the next issue to be clarified. Among the 26 unknown proteins fused with GFP, five clones were predicted to carry NLS. Including these five clones, 22 proteins showed nuclear localization upon introduction into the onion cells (Figure 3, Table 3). The localization was categorized into two major patterns of distribution. One was localization in the probable inner nuclear matrix or the chromatin sparse region as shown in Figures 3A, 3C, 3E, 3G, 3I, and 3K, and the other was DNA/chromatin colocalization as in Figures 3B, 3D, 3F, 3H, 3J, and 3L. The merged image of the GFP-fused proteins with DAPI-stained DNA/chromatin illustrates the distribution of the proteins in the nuclei. Three proteins out of the remaining four clones showed various localization patterns from nucleus to cytoplasm and are indicated by “+/−” for nuclear transportation efficiency in Table 3. However, the degree of localization preferentiality between the nucleus and cytoplasm was different among three proteins (data not shown). Only one protein, marked by “−” in Table 3, showed no nuclear localization in epidermal cells. If the three nuclear/cytoplasmic proteins were taken into account, 96% of unknown proteins examined here were revealed to be nuclear localizing proteins. These results showed that nuclear proteins were efficiently enriched by the NTT system and were easily obtained as cDNAs.
Figure 3.
Localization of the GFP-Fused Proteins of the Unknown Protein Group in Allium Epidermal Cells.
The images of the GFP fusion proteins are shown at the left, the nuclei stained with DAPI are in the middle, and the merged images with these two are at the right. The GFP figures represent localization of several unknown proteins fused to GFP. For detailed characteristics of these proteins, see Table 3. Bar = 25 μm.
(A), (C), (E), (G), (I), and (K) Proteins localized in an inner nuclear matrix region.
(B), (D), (F), (H), (J), and (L) Proteins colocalized with DNA/chromatin.
(M) to (P) Figures reduced to show whole cytoplasmic area of the cells in (F), (G), (I), and (K), respectively.
Various Distribution Patterns of GFP-Fused Proteins in the Plant Nuclei
More detailed observation of the GFP fusion protein signals seen in Figures 2 and 3 revealed diverse subnuclear localization patterns. The localization could be classified into three major patterns. The first pattern was accumulation in an inner nuclear matrix-like region (Figures 2B, 2D, 2G, 3A, 3C, 3E, 3G, 3I, and 3K). This pattern could be further separated into two subgroups. In one subgroup, several large GFP speckles occupied the majority of the nucleus but were excluded from the chromatin space marked by intense DAPI staining. Examples of this subgroup are 13YPB030, 12FPR057, 12YPR026, 16FPG004, 13YPG014, 23C4AG017, and 22C4AG010 (Figures 2B, 2D, 2G, 3A, 3C, 3G, and 3I, respectively). In the other subgroup, many smaller speckles or foci were distributed in interchromatin regions, as seen in the cases of 14FPB077 and 13YPG010 (Figures 3E and 3K, respectively).
The second pattern was an overlapping distribution with DAPI staining (Figures 2A, 2F, 2H, 3B, 3D, 3F, 3H, 3J, and 3L). In this group, GFP signals were uniformly spread in the chromatin region, and preferential distribution was not evident. The third pattern was speckles or foci distribution on a part of the nuclei or the whole nuclei. In this group, speckles/foci of various sizes and intensities were observed in 12FPG023, 13YPR075, 12FPB009, and 21C4BG001 (Figures 2C, 2E, 2J, and 3L, respectively). The 21C4BG001 fusion protein formed several small discrete foci in the entire nucleus and also in the cytoplasm at a lower density (Figure 3L). Repeat experiments showed a similar and reproducible localization pattern of GFP for each protein in the onion cells. No additional GFP signals in the cytoplasm or other cellular structures was seen apart from the signals shown in Figures 2 and 3. Diffusible distribution in the nucleus was not observed even for the small fusion proteins (in particular those smaller than 50 kD; Table 3) of 13YPB022, 23C4AG017, 22C4AG010, and 13YPG010 (Figures 3M to 3P, respectively). This suggested positive transportation of these proteins into the nucleus. Although these localization data were obtained using overexpression and GFP-fusion strategies in onion cells, they strongly suggest that rice nuclear proteins with various functions were isolated in this study. Protein distribution in the subnuclear compartments provides an insight into possible protein function. Using the distribution patterns of these proteins, we next attempted to discern the functional roles of several proteins that had not been previously characterized in plants.
Subnuclear Localization of the Nuclear Matrix Proteins
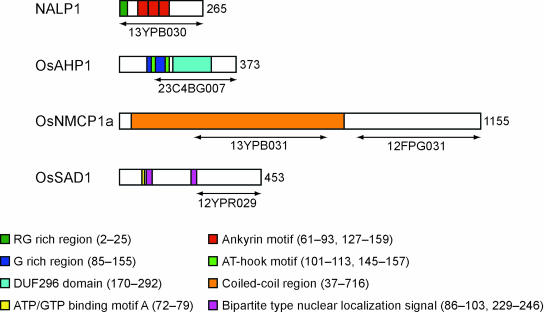
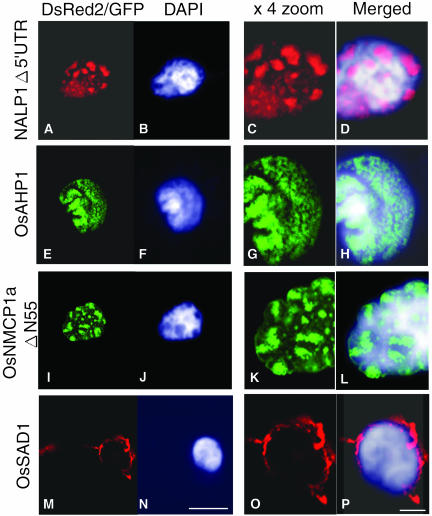
Isolation of housekeeping and stage-specific structural proteins fundamental to the plant nucleus was one of the objectives of our application of the NTT system. In fact, the NTT system appeared to work as an efficient tool to isolate such proteins, particularly in the case of proteins grouped in the unknown or nonnuclear categories. Four candidate proteins were selected for more detailed analysis. Two of them, named NALP1 and OsAHP1, may represent functional connections between the nuclear matrix components and the chromatin. The other two, named OsNMCP1 and OsSAD1, may connect the nuclear matrix and the nuclear envelope. Full-length cDNA clones corresponding to these proteins were isolated from the original cDNA libraries and characterized. The structural features of these four proteins are shown in Figure 4. The NALP1 (nuclear ankyrin like protein 1) is a homolog of ankyrin, which is a structural protein localized on the inside of the plasma membrane (Bennett, 1979). The NALP1 cDNA clone found in the NTT library contained a long 5′ untranslated region (UTR) and an N terminus Gly/Arg region (Figure 4). To see the distribution of NALP1 in more detail, we introduced three modified NALP1 constructs fused with GFP or DsRed2 into onion cells. The first construct contained a GFP-fused full-length cDNA (13YPB030) (Figure 4), and the localization of the resulting fusion protein in the nucleus is shown in Figure 2B. The protein expressed from the second construct, which lacked the 5′ UTR (Δ5′UTR) and was fused to DsRed2, showed a similar nuclear localization (Figures 5A, 5C, and 5D). The third NALP1 fusion protein lacked the N terminus Gly/Arg region but showed the same nuclear localization pattern as the first two versions (data not shown). No change in the speckled nature of the NALP1 protein distribution pattern in a probable inner nuclear matrix region with low DNA/chromatin content was seen among the three constructs. These results demonstrate that the restricted localization of the NALP1 protein can be attained by the portion beyond the Gly/Arg region to the C terminus.
Figure 4.
Primary Structures of Four Candidate Rice Nuclear Proteins.
Full-length cDNAs were isolated from the original cDNA libraries and used for GFP-fusion experiments. The numbers to the right of each protein indicate the number of amino acid residues of the predicted proteins. The regions cloned by the NTT method are shown on the bottom of each protein structure by arrowed bars. The motifs and characteristic regions are shown for each protein and described on the bottom of the figure. The DDBJ/GenBank/EMBL accession numbers are as follows: NALP1 (AB110173), OsAHP1 (AB110206), OsNMCP1a (AB110204), and OsSAD1 (AB110207).
Figure 5.
Subnuclear Localization of the GFP- or DsRed2-Fused Nucleus Associating Proteins in Allium Epidermal Cells.
Green and red show the GFP and the DsRed2 fusion proteins, respectively.
(B), (F), (J), and (N) are counterstained DAPI images of (A), (E), (I), and (M) nuclei, respectively.
(C), (G), (K), and (O) are expanded images of (A), (E), (I), and (M), respectively.
(D), (H), (L), and (P) are merged and enlarged images of each corresponding DsRed2/GFP with DAPI signals.
Bars in (N) and (P) = 20 and 5 μm and represent the length of the leftmost two and rightmost two figures, respectively.
The OsAHP1 is a probable HMG I/Y-type chromatin protein containing an AT-Hook motif (Figure 4). This protein motif has been shown to bind specific nucleotide sequences, including matrix attachment regions (Khadake and Rao, 1997). The OsAHP1 showed chromatin-associated localization over the entire nucleus in onion cells (Figures 5E to 5H). The protein was also shown to be distributed in a similar manner to the rice nuclei as shown in Figure 6B. OsNMCP1 shares 33.5% identity (33.5%) with the carrot (Daucus carota) nuclear matrix protein NMCP1 (Masuda et al., 1997) but has no similarity with any known nuclear matrix proteins in mammals and yeast. OsNMCP1 is predicted to have a coiled-coil structure (Figure 4). To examine the nuclear localization of OsNMCP1a, we used the GFP fusion of truncated OsNMCP1a in which 55 amino acids were deleted at the N terminus because no GFP fluorescence was observed when full-length OsNMCP1a was used (data not shown). The fluorescence was observed in the inner nuclear matrix region as aggregated foci or speckles, as well as in the nuclear peripheral region (Figures 5I, 5K, and 5L). The last clone, OsSAD1, has low homology (26.7% identity) with the C-terminal region of human SAD1. The UNC84, a SAD1 homolog of Caenorhabditis elegans, localizes at the nuclear envelope region (Malone et al., 1999). The OsSAD1 localized in the specified region of the nuclear envelope, in the outer nuclear matrix region, and partly in the endoplasmic reticulum as shown in Figures 5M, 5O, and 5P. These should provide useful information for specification of these protein functions in the dynamics of nuclear architecture.
Figure 6.
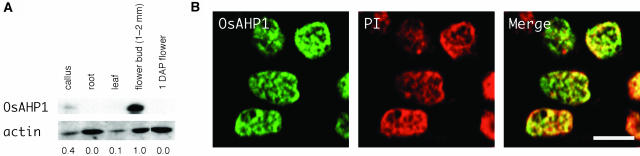
Tissue Specificity and Subnuclear Localization of OsAHP1 Protein in Rice.
(A) Immunoblotting of the protein extracts from several rice tissues reacted with anti-OsAHP1 (top) and actin (bottom) antibodies. Fifty micrograms of total crude proteins extracted from each of the indicated tissues were loaded. Values at the bottom of each lane represent the relative expression of OsAHP1 normalized by the level of actin expression. DAP, days after pollination.
(B) Immunostaining of the secondary parietal cells from the anther wall with an antibody for OsAHP1 followed by a fluorescent-labeled secondary antibody. The left panel (green) represents stained rice nuclei with the antibody, the middle panel (red) shows PI staining for chromatin, and the right panel (yellow) shows a merged figure of the antibody-stained OsAHP1 and the PI stain. Bar = 5 μm.
Organ- and Stage-Specific Expression of OsAHP1 in Rice
Organ-specific expression of nuclear proteins is an important issue for characterizing protein functions. We examined some nuclear protein genes for their organ/tissue specificity and obtained several candidate genes showing specific expression. One of them, an uncharacterized, but most probable, chromatin gene, OsAHP1, was further examined for expression and nuclear localization in rice using a polyclonal antibody against OsAHP1. Protein gel blot analysis on several rice organs or tissues (callus, root, leaf, 1- to 2-mm bud of a young panicle, and flowers 1 d after pollination) showed that the OsAHP1 protein was expressed most abundantly in the early developing flowers, lesser in the callus tissues, and not at all in the other organs examined (Figure 6A). The flowers of 1 to 2 mm long contained pollen mother and egg mother cells that had not entered into meiosis, three immature or four complete anther cell wall layers, anther filaments, and developing integuments and were all encased by palea and lemma. The antibody detected proteins mostly in the nuclei of these tissues at this stage but not in later stages (data not shown). This showed organ- and stage-specific expression of OsAHP1.
We next observed subnuclear localization of the OsAHP1 in a flower bud of 1 to 2 mm. In the secondary parietal cells of an anther wall, clear immunofluorescence antibody staining was detected over the whole nuclei but with uneven distribution (Figure 6B). Propidium iodide (PI) staining represents the chromatin region in the same nuclei. Both staining patterns of anti-OsAHP1 and PI were overlapped in the merged nuclei, though some regions were brighter with either stain (Figure 6B). The protein associating pattern with the chromatin observed in rice was very similar to the distribution pattern of the OsAHP1-GFP fusion protein seen in the onion nuclei. This result indicates that the subnuclear expression of OsAHP1-GFP transient protein detected in onion cells mimics the expression pattern of native OsAHP1 protein in rice cells.
DISCUSSION
In this study, we have exploited a two-stage proteomics approach to identify plant nuclear proteins. More than 500 candidate nuclear protein genes were identified that could be grouped into five classes, including many novel nuclear protein genes. Comparison of the composition of the classified nuclear proteins among three different organs and developmental stages showed no significant differences, as shown in Table 2. One exception was a nuclear architectural protein that was more abundant in the EEP and GGP libraries from the reproductive and embryogenesis organs but was not detected in the C4 library from regenerating calli composed mainly of vegetative cells. More than 75% of the identified clones were isolated only once in one of the three libraries, and this could be attributable to a shortage of cDNA clones analyzed. However, it is possible that various nuclear protein genes with specific characteristics in each organ could be trapped in this study, as shown in the later subsection. In addition, isolation and expression of the organ-specific nuclear protein OsAHP1 was revealed in this study (Figure 6). Thus, the NTT system combined with other specific methods could provide an efficient approach to identifying and characterizing nuclear proteins, which could in turn be used for further nucleus organization studies.
NLS and Nuclear Localization Ability of Isolated Proteins
Several new type plant NLSs have been reported in the individual nuclear protein studies (Robatzek and Somssich, 2001; Borrell et al., 2002). However, a systematic study about the general characteristics of plant NLSs has not yet been performed. The successful isolation of human and rice nuclear proteins by the NTT system shows that the general characteristics of NLS are common at least in part of the proteins in mammals, yeast, and higher plants. General NLS is known to consist of basic-rich residues, which are recognized by importin α and thus makes it possible to assign proteins that contain this type of NLS to nuclear proteins by in silico analysis. The NTT system provided another way to trap other types of NLS as revealed both in humans and rice. For example, β-catenin, which is imported by an importin α/β independent manner (Yokoya et al., 1999), was successfully identified by the NTT system (Ueki et al., 1998). In plants, non-basic-rich–type NLSs have not been characterized. Here, we could identify many NTT positive nuclear proteins that did not encode for the classical NLSs (Table 3).
Classification of 523 unique candidate nuclear proteins showed that 26% of these proteins (135 proteins) belong to the nonnuclear protein group. However, among them, five proteins out of 10 fused with GFP localized in the onion nuclei (Figure 2). Furthermore, ∼80% of the clones of this group reproducibly formed positive colonies by reintroduction of the NTT selected plasmids into yeast cells (data not shown). Therefore, it is indicated that at least 50% of the genes (∼70 genes) in the nonnuclear protein group encode nuclear transportable proteins. Based on this consideration, the recovery of plant nuclear proteins by NTT screening (nuclear-specific, nuclear-related, and nonnuclear groups) should approach 80%. Our localization analysis of GFP-fusion proteins (Figure 3) also indicate that approximately 90% of the proteins (at most 96% from 195 proteins) classified into the unknown protein group have the competence for nuclear import. Consequently, the ratio of nuclear localizing proteins both in the known and the unknown protein groups was ∼85% of the total isolated clones overall.
In the transformation experiments using onion cells, we observed proteins showing various patterns of localization associated with chromatin, the matrix-related region with a foci or speckle-structure, the nuclear membrane in the nucleus, and the cytoplasm of onion cells. There still remains a possibility that the localization pattern of the GFP-fusion proteins in onion cells was different from their original nature in rice cells because the GFP-fusion protein genes were driven by the 35S constitutive promoter, and the GFP-fusion itself might disrupt the binding ability of the fused proteins. Furthermore, in some cases, the lack of interactive protein that is originally present in rice cells might cause mislocalization of the introduced protein to an unusual subnuclear partition in onion cells.
However, repeated transformation experiments in onion epidermal cells showed a similar distribution pattern to the GFP-fusion proteins in the nucleus or cytoplasm, irrespective of their signal intensity (data not shown); this at least indicated nuclear localization ability of most proteins used in this study. In addition, localization of one of these proteins, OsAHP1, in the rice nuclei was confirmed by immunostaining and corresponded well to that detected in onion cells for OsAHP1-GFP (Figures 5E to 5H and 6B). In the case of bilateral protein localization in the nucleus and/or cytoplasm, the distribution pattern varied depending on the introduced cells (Table 3). Further examination using native conditions of rice cells is required to see whether they are shuttling proteins that move between the nuclei and the cytoplasm. In yeast, 7% of all proteins localize in both the nucleus and the cytoplasm. Moreover, such shuttling proteins include many proteins relating to transcription (∼35%) and cell cycle/mitosis (∼12%; Kumar et al., 2002). The Cdc25 protein, for example, is retained in the cytoplasm during an interphase by binding with 14-3-3 proteins and moves to the nucleus in an M phase (Kumagai and Dunphy, 1999). A recent study further proved that the 14-3-3 proteins also translocate to the nucleus and participate in nucleocytoplasmic transportation (Brunet et al., 2002). Consistent with these data, several NTT positive clones encoding 14-3-3 proteins were isolated in this study, and one of these proteins showed bilateral distribution in the nucleus and the cytoplasm (Figure 2G). These data suggest that the basal localization patterns of GFP-fusion proteins could reflect those expressed in rice cells. Our results thus indicate that the NTT system could also be used to obtain certain types of nuclear proteins showing specific localizations similar to nuclear-to-cytoplasmic shuttling proteins.
Novel Insights for Nuclear Matrix Components in Plants
Although nuclear matrix proteins play important roles in nuclear organization, plant nuclear matrix proteins have rarely been identified (Gindullis and Meier, 1999; Gindullis et al., 1999). For this reason, the search for plant nuclear matrix components was one of our interests. Our two-tiered strategy identified candidate proteins showing a variety of distribution patterns within the nuclear matrix region (Figure 5).
Ankyrin is generally found as a structural protein inside the plasma membrane (Bennett, 1979), but localization analysis of the rice ankyrin homolog NALP1 showed accumulation in a part of the inner nuclear matrix region or chromatin-poor space (Figures 2B and 5A to 5D). This localization pattern resembled the distribution of other nuclear architectural proteins, such as SATB1 (Shutao et al., 2003). NALP1 comprises three ankyrin repeats and a Gly/Arg-rich region at its N terminus and has no other known domain or motif (Figure 4). The nuclear localization of the deletion mutant of the Gly/Arg-rich region indicated that the Gly/Arg rich region was not important for nuclear localization. This region may function to associate with nuclear RNAs because a Gly/Arg-rich motif has been reported as an RNA binding motif, termed the RGG box (Kiledjian and Dreyfuss, 1992). The Arabidopsis counterpart of NALP1, AtAnkyrin3 (PIR accession number T04436), which is annotated as a homolog of ankyrin3, the human ankyrin3 (G), showed the second highest homology with the ankyrin repeat region of the NALP1. The NALP1 and the AtAnkyrin3 are proteins of 28 and 27 kD, respectively. These molecular weights are much smaller than the original human ankyrin3 (G) protein with molecular masses of 480 and 270 kD and are derived from alternatively spliced transcripts (Kordeli et al., 1995). Therefore, the NALP1 and the AtAnkyrin3 might be a novel type of plant-specific protein containing the ankyrin motif. Successful isolation of two additional plant type clones possessing ankyrin motifs (see supplemental data online) might also support this possibility.
The OsAHP1, a probable HMG I/Y–related chromatin protein containing an AT-Hook motif, is localized in a chromatin region similar to the wheat (Triticum aestivum) MAR binding protein AHM1 (Morisawa et al., 2000). Detailed analysis of the subnuclear localization of the fusion protein showed uneven distribution overlapping the DAPI stain (Figures 5E to 5H), suggesting an association of OsAHP1 with some specific regions of the chromatin structure. The proteins associating with the HMG I/Y–type proteins in the nuclear matrix have not been characterized well. Further analysis of the OsAHP1 is necessary to verify its chromatin association and/or organization by nuclear matrix components.
The OsNMCP1a does not contain any obvious known domains or motifs. However, it probably forms a coiled-coil structure, as indicated in Figure 4. We also isolated another coiled-coil protein, OsNMCP1b, which showed 27.6 and 28.4% amino acid identity to OsNMCP1a and the carrot NMCP1, respectively. The GFP or DsRed2 fusions of OsNMCP1a and OsNMCP1b showed clear nuclear localization (data not shown for OsNMCP1b). Although the previous NTT study concluded that NTT clones encoding coiled-coil proteins could be false positives (Ueki et al., 1998), nuclear localization of the GFP-fused OsNMCP1a showed that at least some of these proteins actually localize in the nucleus and function as nuclear proteins. The OsNMCP1aΔN55:GFP localized in the inner nuclear matrix and also on the nuclear envelope and/or the peripheral region (Figures 5I to 5L). Although we cannot exclude the possibility that the N-terminal deletion affected its subnuclear localization, our data showed that it possesses an ability to associate with the nuclear envelope as does carrot NMCP1 (Masuda et al., 1997). OsNMCP1a seems to contain a minimum of two NLSs because the NTT screening isolated two nonoverlapping clones, which cover amino acid positions 232 to 651 and 771 to 1160, respectively (Figure 4).
The OsSAD1 was annotated to have an ATP/GTP binding site motif A (P-loop) and two bipartite NLSs (Figure 4; Hagan and Yanagida, 1995). No other domain or motif was predicted. OsSAD1 contains one or more additional functional NLSs because our NTT clone used here, 12YPR029, did not contain the predicted bipartite NLSs on the N-terminal half (Figure 4). The DsRed2:OsSAD1 fusion protein localized at the nuclear periphery and part of the endoplasmic reticulum (Figures 5M to 5P). Although the SAD1 of Schizosaccharomyces pombe associates with a spindle pole body and is essential for both spindle formation and function, moderate overexpression of the SAD1 leads to association with the nuclear periphery (Hagan and Yanagida, 1995). The UNC-84, a SAD1 homolog of C. elegans, also localizes at the nuclear periphery (Malone et al., 1999). If the OsSAD1 is the rice counterpart of the SAD1/UNC-84 protein, localization of the rice protein during mitosis would be informative because higher plants lack centrosomes. We anticipate that further analysis of other nuclear matrix proteins identified in our NTT screening would provide a new insight into the organization of the plant nucleus.
Organ-Specific Expression of Nuclear Proteins
It was expected that organ- or tissue-specific nuclear proteins, especially transcription regulatory factors with roles in specific tissue function or development, could be isolated in this study. In fact, the homologs of ZmMADS1 and mEmBP-1, which are respectively expressed specifically in early stages of embryogenesis and the immature ear in maize (Zea mays) (Carlini et al., 1999; Heuer et al., 2001), were isolated from the EEP and GGP libraries generated from the corresponding organs in rice. In our preliminary experiment, several candidate clones showing organ- or stage-specific expression were detected (data not shown) not only in transcription regulators that were the most abundant in the known protein group (Table 2) but also in other protein groups. Besides the transcription regulators, we identified the chromatin-associating protein OsAHP1, which showed organ- and stage-specific expression in an early developing flower (Figure 6). From the expression pattern of OsAHP1 in the multiple tissues of the floral organ at the restricted stage, this chromatin protein may play a role in promoting the development of the whole reproductive organ, including the supporting tissues, or it may reorganize the nucleus prerequested for active respiration of flowering tissues. Some other proteins of probable matrix- and chromatin-associating characters could also be expected to show tissue-specific functions for some tissues. Although organ-specific expression of proteins is the next step of our study, these results suggest another way to use the NTT-isolated proteins for unraveling their functional characteristics in regulation of nuclear organization for organ development and physiological events.
Application and Adaptation of the NTT System to Nuclear Protein Analysis
Before our study, NTT screening was applied only once to the analysis of the human cDNA library (Ueki et al., 1998). Large-scale isolation and analysis of nuclear proteins using the three rice cDNA libraries will serve as another index for further application of the NTT method, especially for plant cell biologists. The outstanding advantages of the NTT system are savings in cost, time, and labor for large-scale nuclear protein isolation. In contrast with a biochemical approach that necessitates a large amount of proteins, a cDNA library for the NTT screening can be constructed with little effort from a small amount of sample. However, some points that have been observed and indicated in this study should be mentioned to ensure more efficient application of the NTT method.
The first is the efficiency of positive clone formation. Unlike the results for a human cDNA library where the efficiency of the NTT method resulted in isolation of 2% positive clones (Ueki et al., 1998), only 0.05% of the transformants were isolated as nuclear-trapped transformants in this study. We plated yeast cells transformed with the NTT libraries directly on selective plates, without any preincubation in nonselective medium, to minimize growth of false positive clones. The control experiment using SV40-NLS resulted in only 1% of clones surviving direct culturing on selective plates. By contrast, all clones (100%) were recovered after a two-step selection of successive culturing initially on a nonselective plate and then on a selective plate (data not shown). Difference of positive clone numbers between direct and two-step selection corresponded well to efficiencies of positive clone isolation obtained from the studies of rice and human cDNA libraries, respectively. The two-step selection may give scores more positive clones in the NTT experiment in human compared with that in rice.
The second is bias of the NTT selection. The most redundant cDNA clone obtained in our experiments encoded a zinc finger protein (Song et al., 1998), which covered 13.7 and 7.2% of the total sequenced clones of the GGP library and the EEP library, respectively. These zinc finger proteins may have high nuclear localizing activity because they were isolated by one-step direct selection. Meanwhile, the clones coding basal transcription factors and major chromatin proteins, such as histones, covered a very small fraction. The smaller-sized proteins, such as histones, might be eliminated at the cDNA library construction step because of the use of size-fractionated cDNAs of 1.0 to 3.5 kb. Some other clones might also be eliminated by either restorative or toxic effects of the proteins in the host yeast cells. Usage of shorter cDNA clones (0.5 to 1.0 kb in length) or partial cDNAs lacking toxic region(s) may be able to rescue these kinds of clones. Besides the above two points, an additional efficient way to apply cDNAs for use in the NTT system would be usage of a gateway system for constructing NTT cDNA libraries and then GFP-fusion clones and glutathione S-transferase (GST)- or HIS-tag clones. Combinational approaches using these convenient systems should provide many novel discoveries for nuclear proteins in plants.
Our study provides a new strategy for the isolation of various plant nuclear proteins that are difficult to predict as nuclear proteins using previously applied approaches. Currently, examination of plant nuclear organization is in its infancy. It is necessary to obtain and compile more data using various approaches to clarify nuclear protein characteristics, nuclear architecture, and complex nuclear functions. Toward this end, the NTT system should serve as an effective system to supplement conventional proteomics and in silico analysis.
METHODS
cDNA Library Construction
Plasmid cDNA libraries of whole rice panicles at meiotic stages and an early embryogenesis stage were constructed and amplified as described in the pZL1 vector instruction manual provided by its manufacturer (Invitrogen Life Technologies, Carlsbad, CA). The cDNA inserts were excised from plasmids by digesting with EcoRI and NotI, and then they were size fractionated by electrophoresis. The fractionated cDNA inserts with a size range of 1.0 to 3.5 kb were integrated into the vector of NTT system (pNS1 vector; Ueki et al., 1998). A λ phage cDNA library of a rice (Oryza sativa) callus, which had undergone regeneration for 4 d, was constructed and amplified (Uni-ZAP XR; Stratagene, La Jolla, CA). The cDNA library was converted to the NTT library as described above except that EcoRI and XhoI were used instead of EcoRI and NotI. Amplification of the NTT cDNA libraries was performed by introducing them into DH10B cells of Escherichia coli by electroporation. We ultimately obtained three cDNA libraries with ∼1 × 106, 5 × 105, and 1 × 105 independent clones derived from the panicle containing reproductive organs in meiotic stages, from the flowers with early embryogenic cells after pollination, and from calli with regenerating cells after 4 d of regeneration treatment, respectively.
NTT Assay and Sequencing of Inserted cDNAs
Yeast cells (EGY48L) were transformed with the NTT cDNA libraries following the method of Thompson et al. (1998). Transformants were incubated on a synthetic dropout (SD) medium plate lacking Leu and His (Leu-/His-) at 30°C for 3 to 7 d to detect LEU2 reporter gene expression. Rice importin-α1a (Shoji et al., 1998) and the NLS of SV40 large T-antigen (Ueki et al., 1998) were used as positive controls for the NTT assay. Positive clones were cultured in liquid SD medium (Leu-/His-) at 30°C for 24 h, and the plasmids were prepared using a Zymoprep Yeast Plasmid Miniprep kit (Zymo Research, Orange, CA). The plasmids obtained were further amplified by reintroduction (transforming) into E. coli DH10B. The cDNA clones were then sequenced using an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems Japan, Tokyo, Japan) according to the instruction manual using the following primers: NSF3, 5′-CGATGATGAAGATACCCCACCAAA-3′; NSF4, 5′-GCGTTTGGAATCACTACAGGGATG-3′; NSR4, 5′-TTCGTTTTAAAACCTAAGAGTCAC-3′.
Classification of Clones
The ClustalW program (Thompson et al., 1994) was used to distinguish redundant clones and independent clones. Sequenced clones were clustered into 686 kinds of clones. A homology search based on the DNA sequences and predicted amino acid sequences was performed against DDBJ/EMBL/GenBank and Swiss-Prot/PIR/DAD/PDB databases, respectively, using the FASTA or the BLAST program by accessing the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/searches-e.html). The threshold score of the unknown group with no homology to any sequences was defined as <150 by the FASTA optimal score. Classification into each protein group was based mainly on annotations of the highest hit gene or protein, but annotations of the second or the third ones were also applied when annotations of the first one were unavailable. Alternatively, PubMed searches (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) were used to find publications with nuclear localization data of the hit proteins, especially in the case of the nuclear-related protein and the nonnuclear protein groups. The NLSs were searched using the PSORT program by accessing the PSORT Web server (http://psort.ims.u-tokyo.ac.jp/). Coiled-coil regions were predicted using the COILS program as well as by accessing http://www.ch.embnet.org/index.html.
Microscopic Analysis of GFP-Fusion Proteins
A plasmid carrying 35SC4PPDK-sGFP(S65T) (Chiu et al., 1996) was modified for application to the NTT positive cDNA insert in two ways. First, an endogenous EcoRI site was deleted and novel SmaI and EcoRI sites were inserted just downstream of the sGFP(S65T) gene to construct pBlue35SC4PPDK-sGFP(S65T)SE2, and second, endogenous EcoRI and XhoI sites were deleted and novel EcoRI and XhoI sites were inserted to construct pBlue35SC4PPDK-sGFP(S65T)EX2. Two more vectors were also constructed. These vectors have DsRed2 in the position where the sGFP(S65T) is located in pBlue35SC4PPDK-sGFP(S65T)SE2 and pBlue35SC4PPDK-sGFP(S65T)EX2 and were named pBlue35SC4PPDK-Red2SE2 and pBlue35SC4PPDK-Red2EX2, respectively. The plasmids of the GFP gene fusions were introduced into onion (Allium cepa) epidermal cells by a particle bombardment (PDS-1000/He; Bio-Rad, Hercules, CA) using 1.0- or 1.6-μm gold particles at 1100 psi. After incubation in MS medium for 24 h, the onion cells were subjected to fluorescence microscopy analysis to detect GFP fusion proteins using a confocal laser scanning microscope (Fluoview FV300; Olympus Optical, Tokyo, Japan) or a fluorescence microscope (Axioskop 20; Carl Zeiss, Jena, Germany). Images were taken by a CCD camera (SenSys 0400; Photometrics, Tucson, AZ) and were processed with Adobe Photoshop 6.0 software (Mountain View, CA).
Antibody Production and Immunological Assays
For antibody production, the same fragment of OsAHP1 as used in the GFP-fusion protein construction was used to generate GST-fusion protein. The OsAHP1 fragment was ligated to the pENTR gateway vector (Invitrogen) and introduced into the pDEST15 gateway vector harboring GST tags by a clonase (Invitrogen) reaction. The crude proteins containing 6x His-tagged OsAHP1 were extracted from E. coli, BL21(DE3), 4 h after induction. The cells were homogenized in the Novagen BugBuster protein extraction reagent (EMD Biosciences, San Diego, CA), and the soluble fraction was collected by removing insoluble materials according to the methods described by the manufacturer. The collected fraction containing the GST-fused OsAHP1 protein was purified on a GST column following the procedures of the manufacturer. Immunization comprised of four injections of 10 mg of protein into rabbits every 2 weeks. Immune serum was extracted 52 d after the first injection and used for the immunological assay. For immunoblotting, proteins were extracted from the leaf, root, flowers either before or after flowering, and the regenerating callus of rice. Each tissue was crushed in liquid nitrogen, and the total crude extracts of the proteins were prepared following the method of Kikuchi et al. (1994) with slight modification. The tissue that was precipitated using trichloroacetic acid was washed with acetone and chilled at −20°C. To 50 mg of each of the tissues, 100 μL of 9 M urea solution containing 2% Triton X-100, 1% dithiothreitol, 20 μL of 10% lithium dodecyl sulfate, and 3 μL of 1 M Tris was added. The extracted proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The blotted membrane was treated with a 5% ECL blocking agent (Amersham Biosciences, Piscataway, NJ) and incubated with rabbit antiserum against OsAHP1 diluted to 1:20,000 in PBS supplemented with 0.1% Tween-20. Thereafter, it was incubated with a horseradish peroxidase–conjugated goat antibody to rabbit IgG (Amersham Biosciences) diluted to 1:200,000. For the control experiment, rabbit anti-actin (20-33 peptide) antibodies (Sigma-Aldrich, St. Louis, MO) diluted to 1:10,000 was used as the primary antibody. Finally, the blot was treated with ECL plus protein gel blot detection substrate (Amersham Biosciences) and exposed to an x-ray film.
Immunolocalization of the OsAHP1 was examined in paraffin-embedded sections of young buds. The buds were fixed with 4% paraformaldehyde in PMEG buffer (50 mM Pipes, 5 mM MgSO4, 10 mM EGTA, 4% [v/v] glycerol, and 0.2% dimethyl sulfoxide, pH 6.8) at 4°C overnight. The fixed samples were dehydrated in a graded ethanol series, replaced with xylene, and embedded in Paraplast Plus (Oxford Labware, St. Louis, MO). Microtome sections, 7 μm thick, were placed on glass slides coated with Vectabond (Vector Laboratories, Burlingame, CA). After treatment in 10 mM citrate buffer, pH 6.0, with microwave, they were treated with 3% BSA in PBS supplemented with 0.01% Triton X-100. The sections were labeled with anti-OsAHP1 rabbit antiserum diluted to a concentration of 1:1,600 in solution 1 of Can Get Signal (Toyobo, Osaka, Japan) at 4°C overnight and then with Alexa fluor 488-conjugated anti-rabbit IgG antibody (Molecular Probes, Eugene, OR) at 5 μg/mL in solution 2 of Can Get Signal at room temperature for 1 h. They were treated with 100 μg/mL of RNase A, stained with 10 μg/mL of PI, and mounted with 50% glycerol in PBS. The specimens were observed under a confocal microscope.
The cDNA sequences described in this article have been deposited with DNA Data Bank of Japan under accession numbers BP184341-BP185094 and AB110164-AB110207. Accession numbers for individual clones can be seen in the supplemental data online, although most are ESTs.
Supplementary Material
Acknowledgments
We express our gratitude to the following persons who kindly provided us with plasmids used in this study. pNS1 vector and pNS-NLS positive control were gifts from Kenji Nagahari at the Helix Research Institute, and rice importin-α1a was from Naoki Yamamoto at Ochanomizu University. We are grateful to Eric Richards (Washington University) for his kind discussion and critical reading of the manuscript. We also thank Kanae Sasaga, Akemi Ishii, and Mitsugu Eiguchi for their excellent technical assistance and other staff at the Plant Genetics Laboratory for their kind support and discussion of this study. This study was supported partly by a Grant-in-Aid for the Japan Society of the Promotion of Science Fellows (Grant 13002103-00) by the Ministry of Education, Culture, Sports, Science, and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nori Kurata (nkurata@lab.nig.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028456.
References
- Adams, M.D., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., et al. (2002). Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–813. [DOI] [PubMed] [Google Scholar]
- Bennett, V. (1979). Immunoreactive forms of human erythrocyte ankyrin are present in diverse cells and tissues. Nature 281, 597–599. [DOI] [PubMed] [Google Scholar]
- Borrell, A., Cutanda, M.C., Lumbreras, V., Pujal, J., Goday, A., Culianez-Macia, F.A., and Pages, M. (2002). Arabidopsis thaliana Atrab28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol. 50, 249–259. [DOI] [PubMed] [Google Scholar]
- Brunet, A., Kanai, F., Stehn, J., Xu, J., Sarbassova, D., Frangioni, J.V., Dalal, S.N., DeCaprio, J.A., Greenberg, M.E., and Yaffe, M.B. (2002). 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156, 817–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calikowski, T.T., Meulia, T., and Meier, I. (2003). A proteomic study of the Arabidopsis nuclear matrix. J. Cell. Biochem. 90, 361–378. [DOI] [PubMed] [Google Scholar]
- Carlini, L.E., Ketudat, M., Parsons, R.L., Prabhakar, S., Schmidt, R.J., and Guiltinan, M.J. (1999). The maize EmBP-1 orthologue differentially regulates Opaque2-dependent gene expression in yeast and cultured maize endosperm cells. Plant Mol. Biol. 41, 339–349. [DOI] [PubMed] [Google Scholar]
- Chiu, W.-I., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings, D.A., Carter, C.N., Rink, J.C., Scott, A.C., Wyatt, S.E., and Allen, N.S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12, 2425–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D.-Q., Tomita, Y., Yamamoto, A., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (2000). Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5, 169–190. [DOI] [PubMed] [Google Scholar]
- Escobar, N.M., Haupt, S., Thow, G., Boevink, P., Chapman, S., and Oparka, K. (2003). High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell 15, 1507–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.-C., and Legrain, P. (1997). Toward functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gindullis, F., and Meier, I. (1999). Matrix attachment region binding protein MFP1 is localized in discrete domains at the nuclear envelope. Plant Cell 11, 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindullis, F., Peffer, N., and Meier, I. (1999). MAF1, a novel plant protein interacting with matrix attachment region binding protein MFP1, is located at the nuclear envelope. Plant Cell 11, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Hagan, I., and Yanagida, M. (1995). The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm, K.W., Lee, G.J., and Vierling, E. (1997). Expression and native structure of cytosolic class II small heat-shock proteins. Plant Physiol. 114, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer, S., Hansen, S., Bantin, J., Brettschneider, R., Kranz, E., Lörz, H., and Dresselhaus, T. (2001). The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol. 127, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.-J., Imamoto, N., Matsuki, R., Yoneda, Y., and Yamamoto, N. (1998). Functional characterization of a plant importin α homologue. J. Biol. Chem. 273, 24083–24087. [DOI] [PubMed] [Google Scholar]
- Khadake, J.R., and Rao, M.R.S. (1997). Preferential condensation of SAR-DNA by histone H1 and its SPKK containing octapeptide repeat motif. FEBS Lett. 400, 193–196. [DOI] [PubMed] [Google Scholar]
- Kikuchi, H., Fujinawa, T., Kuribayashi, F., Nakanishi, A., Imajoh-Ohmi, S., Goto, M., and Kanegasaki, S. (1994). Induction of essential components of the superoxide generating system in human monoblastic leukemia U937 cells. J. Biochem. 116, 742–746. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M., and Dreyfuss, G. (1992). Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 11, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli, E., Lambert, S., and Bennett, V. (1995). AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of ranvier. J. Biol. Chem. 270, 2352–2359. [DOI] [PubMed] [Google Scholar]
- Kumagai, A., and Dunphy, W.G. (1999). Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., et al. (2002). Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C.J., Fixsen, W.D., Horvitz, H.R., and Han, M. (1999). UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126, 3171–3181. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Xu, Z.-J., Takahashi, S., Ito, A., Ono, M., Nomura, K., and Inoue, M. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long -helical domain. Exp. Cell Res. 232, 173–181. [DOI] [PubMed] [Google Scholar]
- Morisawa, G., Han-Yama, A., Moda, I., Tamai, A., Iwabuchi, M., and Meshi, T. (2000). AHM1, a novel type of nuclear matrix–localized, MAR binding protein with a single AT hook and a J domain–homologous region. Plant Cell 12, 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., and Somssich, I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. [DOI] [PubMed] [Google Scholar]
- Ross-MacDonald, P., Sheehan, A., Roeder, G.S., and Snyder, M. (1997). A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, K., Iwasaki, T., Matsuki, R., Miyao, M., and Yamamoto, N. (1998). Cloning of a cDNA encoding an importin-α and down-regulation of the gene by light in rice leaves. Gene 212, 279–286. [DOI] [PubMed] [Google Scholar]
- Shutao, C., Han, H.-J., and Kohwi-Shigematsu, T. (2003). Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51. [DOI] [PubMed] [Google Scholar]
- Song, J., Yamamoto, K., Shomura, A., Itadani, H., Zhong, H.S., Yano, M., and Sasaki, T. (1998). Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Res. 5, 95–101. [DOI] [PubMed] [Google Scholar]
- Stahl, G., Bidou, L., Rousset, J.P., and Cassan, M. (1995). Versatile vectors to study recoding: Conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.R., Register, E., Curotto, J., Kurtz, M., and Kelly, R. (1998). An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14, 565–571. [DOI] [PubMed] [Google Scholar]
- Ueki, N., Oda, T., Kondo, M., Yano, K., Noguchi, T., and Muramatsu, M. (1998). Selection system for genes encoding nuclear-targeted proteins. Nat. Biotechnol. 16, 1338–1342. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., Bouget, F.Y., Van Poucke, K., Inzé, D., and Geelen, D. (2004). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40, 386–398. [DOI] [PubMed] [Google Scholar]
- Yokoya, F., Imamoto, N., Tachibana, T., and Yoneda, Y. (1999). β-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell 10, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.