Abstract
Many studies in both plant and animal systems have shown that matrix attachment regions (MARs) can increase expression of transgenes in whole organisms or cells in culture. Because histochemical assays often indicate variegated transgene expression, a question arises: Do MARs increase transgene expression by increasing the percentage of cells expressing the transgene (likelihood), by increasing the level of expression in expressing cells (magnitude), or both? To address this question, we used flow cytometry to measure green fluorescent protein (GFP) expression in individual tobacco (Nicotiana tabacum) cells from lines transformed by Agrobacterium tumefaciens. We conclude that MAR-mediated overall increases in transgene expression involve both likelihood and magnitude. On average, cell lines transformed with the Rb7 MAR-containing vector expressed GFP at levels 2.0- to 3.7-fold higher than controls. MAR lines had fewer nonexpressing cells than control lines (10% versus 45%), and the magnitude of GFP expression in expressing cells was greater in MAR lines by 1.9- to 2.9-fold. We also show that flow cytometry measurements on cells from isogenic lines are consistent with those from populations of independently transformed cell lines. By obviating the need to establish isogenic lines, this use of flow cytometry could greatly simplify the evaluation of MARs or other sequence elements that affect transgene expression.
INTRODUCTION
In recent years, gene transfer into a wide variety of plant species has become almost common practice, but lack of predictable and reliable transgene expression has plagued efforts to fully exploit gene transfer technology for plant improvement and basic research. Initially, major differences in transgene expression between independent primary transformants were attributed to transgene silencing caused by integration of transgenes into or near heterochromatin regions, a phenomenon similar to position effects that have been well characterized in Drosophila melanogaster (fruitfly) (reviewed in Weiler and Wakimoto, 1995). We now know that gene silencing is more complex, and although we are still far from understanding variability in the silencing of transgenes, great progress has been made in understanding transcriptional gene silencing, posttranscriptional gene silencing, and the interplay between the two (Carrington and Ambros, 2003; Grewal and Moazed, 2003; Matzke and Matzke, 2003; Schramke and Allshire, 2003).
It is clear that to derive maximum benefits from the use of transgenic plants for biotechnology and basic research, gene silencing must be understood, and methods to combat gene silencing must be devised. One approach that has shown promise in combating gene silencing involves the use of matrix attachment regions (MARs) to flank transgenes (Allen et al., 2000). MARs are AT-rich DNA sequences that bind to the nuclear matrix in vitro and are thought to organize chromosomal DNA into loop domains (Bonifer et al., 1990; Bode et al., 1996; Martelli et al., 2002). Early experiments in animal cells indicated that MARs increased transgene expression and decreased transformant-to-transformant variability in transgene expression (Stief et al., 1989; Bonifer et al., 1990; Phi-Van et al., 1990). Subsequent work in plant systems also demonstrated an increase in transgene expression and/or a reduction in variability (Breyne et al., 1992; Allen et al., 1993, 1996; Mlynarova et al., 1994, 1995; Han et al., 1997; Liu and Tabe, 1998; Ülker et al., 1999; Vain et al., 1999; Cheng et al., 2001; Mendu et al., 2001; Fukuda and Nishikawa, 2003; Mankin et al., 2003; Butaye et al., 2004; van der Geest et al., 2004). The observations of increases in expression and/or reductions in variability have not, however, been consistent. Some reports indicate that MARs have no effect on transgene expression (van Leeuwen et al., 2001; De Bolle et al., 2003; Sidorenko et al., 2003), and in at least two cases, a decrease in transgene expression has been reported (Breyne et al., 1992; Holmes-Davis and Comai, 2002).
Direct comparison of the results of the different studies are difficult to make because various plant species, MARs, transformation methods, promoters, and reporter genes have been used. Both cultured cells and intact plants have served as experimental material, and a wide variety of tissues in various stages of development have been sampled. In nearly all cases, histochemical approaches and/or extractive approaches have been used to assess activity of reporter genes. Histochemical approaches are not quantitative, but they do reveal cell-to-cell epigenetic variation in reporter gene expression. Approaches in which the product of the reporter gene is extracted from tissues and then measured are quantitative, but they obscure epigenetic cell-to-cell differences. Attempts have been made using the firefly (Photinus pyralis) luciferase reporter gene to simultaneously quantitate transgene expression and spatial variation in transgene expression in whole tobacco (Nicotiana tabacum) plants (van Leeuwen et al., 2001). In these studies, however, transgene expression was not measured at the resolution of individual cells.
In assays for transgene expression in both cultured cells and in whole plants, we and others have noticed that the expression is often variegated (Allen et al., 1993; Ülker et al., 1999; van Leeuwen et al., 2001; Reddy et al., 2003; Bastar et al., 2004), that is, some cells appear to express the transgene and others do not. This raises the possibility that the MAR-mediated elevation in transgene expression measured by extractive procedures is due to epigenetic changes in the distribution of expressing cells within a population. That is, increases in transgene expression may reflect a greater proportion of cells expressing the transgene rather than an increase in the level of expression in expressing cells.
To test this possibility, we have used flow cytometry to measure green fluorescent protein (GFP) expression in individual, cultured tobacco cells from clonal lineages of cells transformed by Agrobacterium tumefaciens. Binary vectors used for transformation carried a GFP reporter gene flanked on both sides by the tobacco Rb7 MAR, which we have previously used (Allen et al., 1996; Ülker et al., 1999; Mendu et al., 2001; Mankin et al., 2003). GFP levels were measured in protoplasts generated from the various transgenic lines and compared with levels in protoplasts generated from lines carrying control constructs without MARs. We discovered that the cell lines carrying constructs including the Rb7 MAR exhibited both a higher percentage of cells expressing GFP and a higher level of GFP expression in the expressing cells. These effects were observed not only by compiling the results from many individual cell lines but also by making measurements on large pools of transformed cell lines.
RESULTS
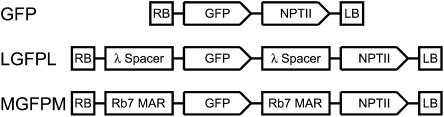
To test the effect of MARs on GFP expression in A. tumefaciens–transformed tobacco cells in culture, we used a T-DNA construct in which the GFP cassette was flanked on both sides by the Rb7 MAR. This construct is denoted MGFPM (Figure 1). We compared results for this construct with results for two control constructs. In the first control (GFP), the GFP cassette is inserted into the T-DNA alone. In the second control, the GFP cassette is flanked by a 1.2-kb region of phage λ DNA that does not bind to tobacco nuclear matrices (N. Mendu and S. Spiker, unpublished results). This control (LGFPL) allows us to discriminate sequence-dependent MAR effects from effects that may be due to MARs acting simply as spacer elements to distance the GFP cassette from neighboring T-DNA and host genomic DNA. The soluble-modified, red-shifted GFP with its enhanced fluorescence (Davis and Vierstra, 1998) was used because similar GFP variants were found to be more suitable for flow cytrometry (reviewed in Galbraith et al., 1999b).
Figure 1.
Schematic Diagrams of T-DNA Regions of Transformation Vectors.
The right and left borders of the T-DNA are indicated with RB and LB, respectively. In the control (GFP) construct, the soluble-modified, red-shifted GFP gene is driven by the 35S promoter of Cauliflower mosaic virus with the nos polyadenylation region. The NPTII (neomycin phosphotransferase selectable marker gene) is driven by the nos promoter with the g7T polyadenylation region. An additional control construct (LGFPL) contains direct repeats of 1195 bp of λ phage spacer DNA. MGFPM contains direct repeats of the 1167-bp Rb7 MAR. Diagram schematics are not to scale.
Improved Flow Cytometry Resolution by Debris Discrimination
Because tobacco suspension cells grow in filamentous structures that cannot be analyzed by flow cytometry, it is necessary to disrupt the cell walls that hold these filaments together. A gentle digestion with cellulases and pectolyases releases individual cells as protoplasts, which can pass one at a time through the flow cell of the cytometer to measure GFP expression in individual cells. The protoplasting procedure creates some debris, such as free nuclei and some protoplasts with compromised plasma membrane integrity. GFP diffuses out of free nuclei and compromised protoplasts. Thus, GFP-expressing protoplasts with damaged membranes would be counted as nonexpressing protoplasts. Because this would lead to an underestimate of GFP in the cell population, including these entities in our analyses would yield spurious results.
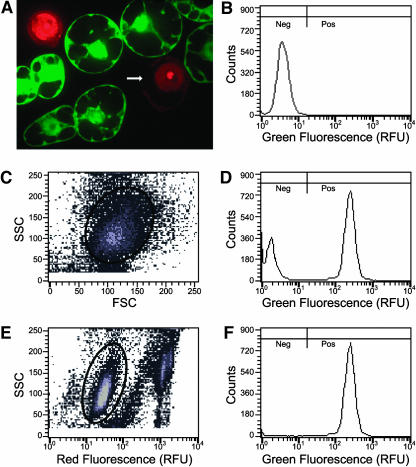
To illustrate these phenomena, a cell line was identified that had high levels of uniform, nonvariegated, GFP fluorescence before protoplasting. As determined by microscopy, all cells in this cell line had levels of GFP fluorescence that were indistinguishable from one another (data not shown). Protoplasts were prepared from this cell line, stained with propidium iodide, and subjected to confocal microscopy (Figure 2A). Clearly, GFP fluorescence is not uniform for all protoplasts. Protoplasts with compromised plasma membrane integrity allow GFP to diffuse out of the cell and propidium iodide to diffuse in and stain the nucleus. Despite having defects in plasma membrane integrity, these protoplasts may still be able to maintain their size and general shape. Spurious flow cytometry data from these protoplasts and from free nuclei can be excluded from analysis by measuring and accounting for the high levels of red fluorescence associated with propidium iodide staining.
Figure 2.
Propidium Iodide Staining of Protoplast Preparations Improves Flow Cytometry Resolution.
(A) A 1-μm thick confocal section of a propidium iodide–stained protoplast preparation of a tobacco cell line uniformly expressing GFP after stable transformation with MGFPM. Intact protoplasts exclude propidium iodide (PI), whereas free nuclei and nuclei in protoplasts with compromised plasma membrane integrity stain brightly. GFP is absent from protoplasts in which nuclei stain brightly with PI. The arrow indicates a missing portion of the plasma membrane.
(B) Green fluorescence histogram of wild-type protoplasts. Fluorescence was measured in relative fluorescence units (RFU). Because in wild-type cells green fluorescence intensity is below 17 RFU, transformed cells with green fluorescence below 17 RFU are considered GFP negative cells (Neg), and cells with green fluorescence above 17 RFU are considered GFP positive cells (Pos).
(C) MGFPM stably transformed protoplasts stained with PI subjected to biparametric analysis of side light scatter (SSC) versus forward light scatter (FSC). Gated events inside the oval are plotted in (D).
(D) Green fluorescence histogram of gated MGFPM protoplasts from (C).
(E) The same data from the same MGFPM stably transformed protoplasts stained with PI were subjected to biparametric analysis of side light scatter versus relative red fluorescence. Gated events inside the oval are plotted in (F). These gated events have background levels of PI fluorescence similar to that of wild-type cells (data not shown).
(F) Green fluorescence histogram of gated MGFPM protoplasts from (E). Note that after exclusion of protoplasts and debris that have high levels of PI fluorescence, the mode consistent with wild-type fluorescence was lost.
Intrinsic physical parameters such as forward light scatter (a measure of particle size) and side light scatter (a measure of cytoplasmic granularity) (Shapiro, 1995) can be used to discriminate between clusters of light scatter values from cells and debris such as damaged cells, which often have reduced light scatter properties (McGann et al., 1988). Some of the data from debris in our protoplast preparations can thus be excluded by gating, that is, including only data points consistent with the light-scattering properties of protoplasts, as defined by an oval outline in Figure 2C. When gating based only on light scattering is used, it appears as though a substantial proportion of the cells (32% in this example) have low levels of fluorescence equivalent to those of wild-type cells (cf. Figure 2D with 2B). Because microscopic evaluation of the cells before protoplasting showed that all cells displayed uniform GFP fluorescence, it appears that the proportion of cells that have no GFP fluorescence can be overestimated if the protoplasts with compromised membranes and residual nuclei are not gated away (Figure 2D). This problem can be alleviated by gating data based on both side light scatter (to exclude data from debris that is not consistent with light scatter properties of protoplasts) and from red fluorescence associated with propidium iodide staining (to exclude data from free nuclei and protoplasts with compromised membranes that have allowed GFP to diffuse out of the cell and propidium iodide to diffuse in and stain nuclei). When the same data as shown in Figures 2C and 2D were gated based on side light scatter and red fluorescence (Figure 2E), only 0.9% of the cells had fluorescence properties consistent with wild-type cells (Figure 2F). With this approach, the error associated with false-negative cells can be greatly reduced and the ability to quantitate the proportion of cells with no detectable GFP fluorescence greatly improved. This gating approach was used in all analyses that follow.
Rb7 MAR Increases the Likelihood and Magnitude of GFP Expression in Individual Cell Lines
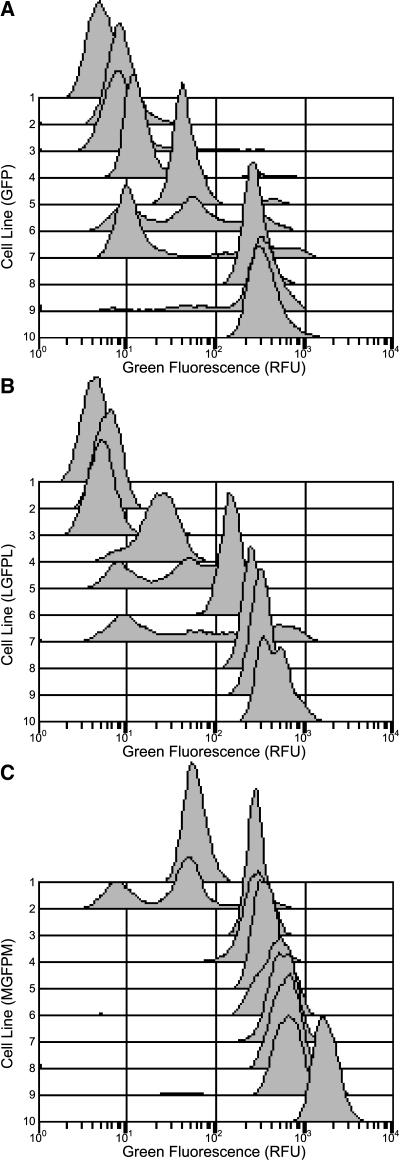
Three independent replicates of A. tumefaciens–mediated transformation experiments were conducted to assess the effect of MARs on GFP expression in NT-1 cells in culture. Each replicate contained three T-DNA construct treatments to generate ∼30 cell lines per T-DNA construct for ∼270 cell lines for all three replicates. Average GFP expression across all lines for each of the treatments does not by itself adequately describe variation between lines and within lines. To illustrate this variation, a sample of results from flow cytometric analysis is displayed in Figure 3. The data used to generate histograms in this figure include green fluorescence values for 15,000 protoplasts from each cell line that were present in a gated region (as in Figure 2E). The cell lines were ordered from lowest to highest total fluorescence for each of the treatments, and fluorescence profiles from every third cell line are presented. From this figure, it can be seen that green fluorescence varies more than two orders of magnitude. Control cell lines (Figures 3A and 3B) tend to have many cells with green fluorescence in the same low range as wild-type cells (below 17 relative fluorescence units [RFU]). Cell lines containing the Rb7 MAR (Figure 3C) have very few cells with low levels of green fluorescence and many cells with higher levels of green fluorescence.
Figure 3.
Green Fluorescence Histograms of Individual Transgenic Cell Lines.
Histograms from each treatment are ordered by their average green fluorescence. For simplicity, only every third histogram, ranked by average green fluorescence (lowest fluorescence, line 1; highest fluorescence, line 10) is displayed. Data shown from replicate 2 only. Green fluorescence histograms from cell lines transformed with the GFP (A), LGFPL (B), and MGFPM (C) T-DNA vectors. Note in (C), the cell line with the second lowest average green fluorescence (line 2) has a mode of cells expressing in the 300 RFU region, but the mode is partially obscured by the histograms plotted below it.
Variegation (variation within lines) of green fluorescence is evident in Figure 3. Whereas some lines have cells with either a low mode of green fluorescence (∼10 RFU and in the range of autofluorescence of wild-type cells) or a high mode of green fluorescence (∼300 RFU or higher), other cell lines have cells that display multiple modes of green fluorescence (e.g., lines 5, 6, and 7 in Figure 3A). In many of these lines, an intermediate mode of green fluorescence occurs (∼50 RFU). In some lines, the range of green fluorescence is so wide that modes are less distinct (e.g., line 7 in Figure 3B). Even in lines that have only high levels of green fluorescence, variegation occurs. For example, in the highest expressing of the LGFPL lines (line 10 in Figure 3B), the green fluorescence profile has an obvious shoulder.
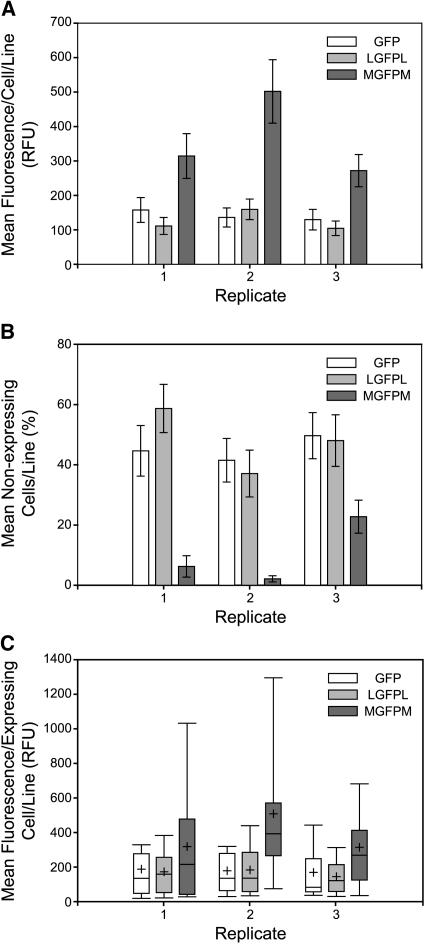
Figure 3 illustrates the cell-to-cell and line-to-line variation in green fluorescence that would be obscured if only summary statistics were presented. However, summary statistics are necessary to quantitate the effects that MARs have on GFP expression in the transformed cell lines. Presented in Figure 4A are the average green fluorescence values per cell in all cell lines transformed with a given construct in each of the replicate experiments. Cells that have green fluorescence properties characteristic of wild-type cells (below 17 RFU) are termed GFP negative cells, whereas cells that have levels of green fluorescence greater than wild-type cells are termed GFP positive cells. Data from all cells, both GFP negative and GFP positive, are included in Figure 4A. This type of analysis is analogous to transgene expression studies in which the product of the reporter gene is extracted from a large population of cells. The resulting quantitative measurements ignore cell-to-cell variation within cell lines. In the data shown in Figure 4A, expression levels of the two controls (LGFPL and GFP alone) are not significantly different in each replicate (P > 0.05, see Methods). Cell lines transformed with the MAR construct (MGFPM) had significantly higher GFP fluorescence in each replicate (P < 0.05) when compared with lines carrying GFP alone. Fold increases of 1.99, 3.69, and 2.11 were observed in the three replicates.
Figure 4.
Summary of GFP Expression in Individual Transgenic Cell Lines for Three Replicate Experiments.
Tobacco protoplasts were analyzed by flow cytometry, and results from ∼30 cell lines each of GFP, LGFPL, and MGFPM T-DNA vector transformations are shown for each replicate.
(A) Mean GFP fluorescence per cell in cell lines transformed by each of the vectors (data from both GFP positive cells and GFP negative cells are included). Error bars denote se.
(B) Mean percentage of GFP negative cells in cell lines transformed by each of the vectors. Error bars denote se.
(C) Box plots of GFP fluorescence per GFP positive cell in cell lines transformed by each of the vectors (data from GFP negative cells is not included). The boundaries of the boxes indicate the 25th and 75th percentiles. The whiskers above and below the box indicate the 10th and the 90th percentiles. The median and the mean are represented by a solid line within the box and a plus sign, respectively.
Increases in GFP expression resulting from the use of the Rb7 MAR in transgenic experiments could be accounted for by two possible scenarios, which are not mutually exclusive: (1) a greater percentage of expressing cells in lines containing MGFPM (likelihood) and/or (2) higher levels of expression in individual cells in lines containing MGFPM (magnitude). Flow cytometric analysis, unlike extractive procedures, allows for the quantification of subpopulations of GFP expression needed to investigate these two possibilities.
To address the first possibility, the percentage of GFP negative cells was determined for each cell line. The averages of these percentages for all cell lines of each treatment in each of the three replicates are shown in Figure 4B. The two control lines (LGFPL and GFP alone) are not statistically different in each replicate (P > 0.05). Cell lines transformed with the MAR-containing construct (MGFPM) had significantly lower percentages of negative cells than controls (P < 0.05) in replicates 1 and 2 (replicate 1, GFP 45% and MGFPM 6%; replicate 2, GFP 41% and MGFPM 2%). In replicate three, cell lines transformed with the MAR-containing construct had borderline significant (P = 0.064) lower percentages of negative cells than controls (GFP 50% and MGFPM 23%). These results support the hypothesis that the presence of Rb7 MAR increases the likelihood of GFP expression by decreasing the percentage of nonexpressing cells.
MARs may also increase overall levels of GFP expression by increasing the magnitude of expression in the expressing cells. To address this possibility, the average green fluorescence per expressing cell in all cell lines transformed with a given construct in each of the replicate experiments is plotted in Figure 4C. Strikingly, the range of green fluorescence above the 75th percentile for cell lines transformed with MGFPM extends to higher values than the controls. There are more cell lines with higher average levels of green fluorescence (GFP positive cells only) in the MAR treatments than in controls. When the means are compared, the two control lines (LGFPL and GFP alone) are not statistically different in each replicate (P > 0.05). In replicates two and three, cell lines transformed with MGFPM had significantly higher green fluorescence than control lines (P < 0.05) with fold increases of 2.85 and 1.87, respectively. In replicate one, expressing cells in cell lines transformed with MGFPM had a higher average level of green fluorescence than those transformed with control constructs, but the difference was not statistically significant (P = 0.23). Even though our criteria for statistical significance was not met in all replicates, these results support the hypothesis that the Rb7 MAR can increase the magnitude of GFP expression in expressing cells.
GFP Expression Is Stable over Time with and without Continued Selection
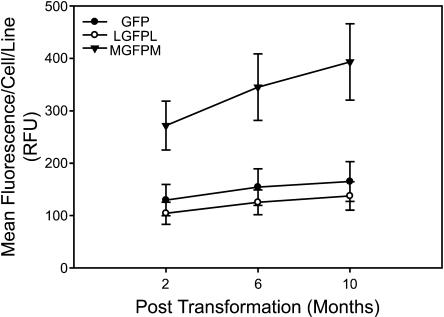
To evaluate possible changes in GFP expression over time in liquid culture, cell lines were maintained with kanamycin selection after the first flow cytometry analysis was conducted. After 4 and 8 additional months (with weekly passages), green fluorescence data were collected. No statistically significant changes in mean GFP expression were observed for any of the treatments (Figure 5). Similarly, no statistically significant changes in the percentages of GFP positive cells or the levels of GFP expression in positive cells were observed for any of the treatments (data not shown). Additionally, a subset of cell lines in each treatment was cultured without kanamycin selection over the final 4 months of the time course. Again, no changes in mean GFP expression levels or percentages of expressing cells were observed for any treatments (data not shown).
Figure 5.
GFP Expression Is Stable over Time in Individual Transgenic Cell Lines.
Cell lines were maintained on selection for 10 months, and green fluorescence was measured by flow cytometry. Data from all cells (expressing and nonexpressing) are included. Error bars denote se.
Rb7 MAR Increases the Likelihood and Magnitude of GFP Expression in Population Cell Cultures
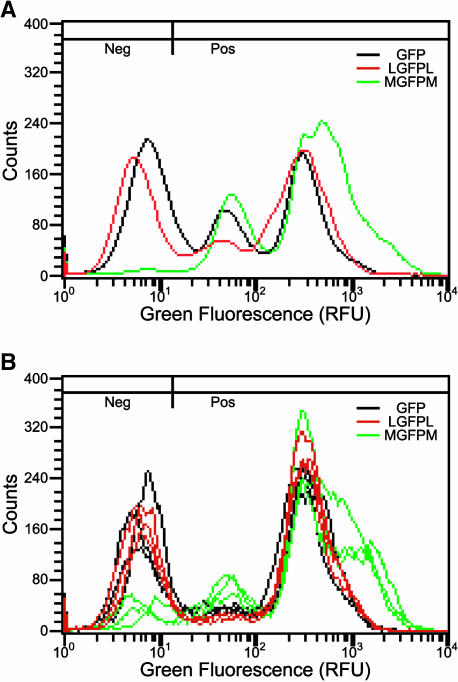
Plots of histograms and summary statistics in Figures 3 and 4 indicate that including the Rb7 MAR in T-DNA vectors increases both the likelihood and the magnitude of GFP expression. The graphical representations in Figure 6 support these conclusions. By averaging all of the histograms from each treatment in the second replicate as shown in Figure 6A, overall differences between treatments can be observed. Rb7 MAR-containing lines on average have more expressing cells, and these cells express at higher levels compared with control lines. Note that MAR-containing lines and control lines have approximately the same proportion of GFP positive cells expressing at ∼300 RFU and below, but the MAR-containing lines have a much higher proportion of cells expressing above 300 RFU. This shoulder at the higher end of the profile could account for nearly all of the MAR-mediated reduction in the percentage of nonexpressing cells.
Figure 6.
Comparison of GFP Expression in Averaged Individual Cell Lines and Population Cell Cultures.
(A) Average green fluorescence histograms of all 30 individual cell lines for each treatment from the second replicate experiment (10 of which are shown in Figure 3).
(B) Green fluorescence histograms from population cultures established from the same A. tumefaciens cocultivation transformation plates used to isolate individual lines summarized in (A). Three independent cultures were established for each T-DNA vector. Parameters for histograms are the same as in Figure 2B.
In traditional approaches to determining the effect of specific DNA elements (e.g., promoters, enhancers, and MARs), many separate independently transformed cell lines are created, and the averages of transgene expression in the lines are measured. This approach can be quite laborious. Because flow cytometry allows detection of GFP expression in individual cells, it is possible to pool large populations of cells derived from many different transformation events and still retain an underlying distribution of GFP expression that would be lost if cell lines were pooled and assayed with a traditional, extractive approach. This pooling strategy allows for the assessment of the effect of any specific DNA element on transgene expression across a wide range of A. tumefaciens–mediated transformation events. Instead of isolating and propagating individual cell lines, aliquots of entire A. tumefaciens and tobacco cell culture cocultivations (representing numerous independent transformants) were used to initiate liquid cultures under kanamycin selection. Cells from these pooled lines were subjected to the same flow cytometry analysis as outlined previously for individual cell lines. The results are shown in Figure 6B. Strikingly, MGFPM population culture lines have fewer GFP negative cells (7.7%) than the GFP and LGFPL lines, which collectively average 33.9% GFP negative cells. This observation is very similar to the results in individual cell lines (Figures 4B and 6A). Another distinguishing feature is that population culture lines containing MGFPM have shoulders on the high end of the profile. The presence of this subset of cells expressing GFP at a higher level than control constructs is consistent with our observations in individual cell lines (Figures 4C and 6A).
Variegated GFP Expression Arises from Cells with the Same Genotype
In experiments involving the generation of individual cell lines that have been transformed with foreign DNA, it is desirable to use methods of DNA transformation and tissue propagation that eliminate or reduce the frequency of chimeric cell lines. Even though care is taken to ensure that each cell line arises from a single transformation event, it is possible that multiple transformation events could occur next to each other on culture plates and not be distinguished from a single event. If individual cell lines were composed of chimeric populations, conclusions about epigenetic variation in transgene expression could be compromised.
In our experiments, cell lines are originally identified as microcalli that appear as small colonies on a lawn of nontransformed tobacco NT-1 cells. It is likely that each microcallus results from a single transformation event as the microcalli are well separated on the culture plates. Nevertheless, the possibility that the microcalli are chimeric exists. The microcalli are picked and subcultured on solid media containing kanamycin before transfer to liquid media. During cell line propagation on solid medium containing kanamycin, sectors of GFP expression were sometimes observed (Figure 7A, left column). Sectors were observed in all three rounds of solid selection (data not shown). Approximately one-third of the calli displayed sectors of GFP fluorescence on the second plate of solid selection, whereas approximately one-tenth of the calli displayed sectors of GFP fluorescence on the third plate of solid selection. To minimize the possibility of carrying chimeric lines into liquid culture, only small subsets of cells (pieces ∼1 mm in diameter) were used to initiate new callus. Therefore, our flow cytometry results are indicative of the variation that exists during the last solid-state propagation in which the entire callus was used to initiate liquid cell cultures.
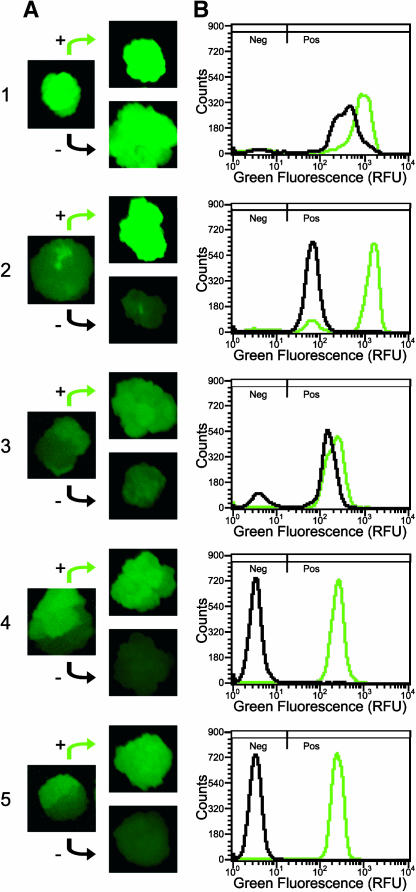
Figure 7.
Isolation of Callus Sectors Expressing GFP at Different Levels and Flow Cytometric Analysis.
(A) A subset of calli display sectored GFP phenotype (left column). Tissue from these sectors was subcultured on fresh solid medium (right column). The plus sign denotes tissue from the sector with the higher level of green fluorescence, whereas the minus sign denotes tissue from the sector with the lower level of green fluorescence. Sectored calli from each T-DNA vector were chosen for further analysis: sample 1 from GFP T-DNA; sample 2 from MGFPM T-DNA; samples 3, 4, and 5 from LGFPL T-DNA.
(B) GFP fluorescence histograms of liquid cultures initiated from sectors in the right column of (A). The black histogram is from the (−) sector, and the green histogram is from the (+) sector. Parameters for histograms are the same as in Figure 2B.
To determine whether sectors arose from a mixture of transformation events or are descendents of an epigenetic change after a single transformation event, tissues from sectors of single calli were independently propagated as shown in the right column of Figure 7A. The calli in the left column of Figure 7A result from enlargement of the original microcalli picked from the transformation plates. Thus, the calli in the right column of Figure 7A have not undergone the additional round of subculturing that was performed for all other previously mentioned individual NT-1 lines (see Methods). Calli from the sectors were used to initiate liquid suspension cultures and subjected to flow cytometry analysis (Figure 7B). In all cases, cultures derived from sectors, which by visual inspection had higher levels of GFP fluorescence, were demonstrated by flow cytometry to have a higher percentage of positive cells and/or higher levels of fluorescence in positive cells.
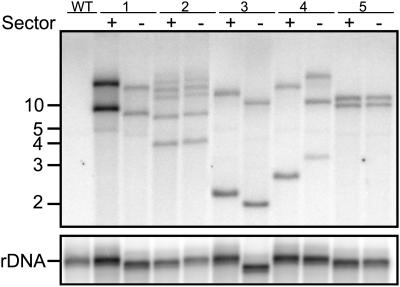
To determine if the sectors have the same genotype, DNA from the liquid suspension cultures arising from the sectors was isolated and subjected to DNA gel blot analysis (Figure 8). A 458-bp region common to all of the T-DNA constructs tested was used to probe genomic DNA cut with a methylation-insensitive restriction enzyme, HindIII. Because this enzyme cuts only once in the T-DNA, the resulting fragments will have distinct sizes based on the next closest genomic HindIII site (unless a tandem T-DNA repeat flanks the right border). In four out of five sector pairs, the banding patterns match exactly (Figure 8, sector pairs 1, 2, 3, and 5), indicating a single transformation event. Sector pair 4 clearly has a different banding pattern, suggesting this cell line may be composed of lineages representing two distinct transformation events. In any case, four out of five sector pairs demonstrate banding patterns consistent with identical genotypes. This is most likely an overestimate of the frequency of chimerism in populations used to generate data in Figures 3, 4, 5, and 6A because we investigated chimerism only in lines that displayed sectoring. Furthermore, the sectored calli were subjected to one fewer round of subculturing than the lines used to investigate the effects of MARs on transgene expression. Based on the observations that only one of five cell lines with obvious sectoring was chimeric and that of the cell lines used to generate data concerning the effect of MARs on GFP expression only one in 10 had obvious sectoring, we would estimate that only one in 50 (2%) of our cell lines was chimeric.
Figure 8.
Genotyping Sectors by DNA Gel Blot Analysis.
DNA was isolated from liquid cultures initiated from callus sectors (Figure 6A, right column). Blots of 15 μg of DNA digested with the methylation-insensitive restriction enzyme HindIII (which cuts only once within the T-DNA) and separated by electrophoresis were probed with a portion of the right T-DNA border common to all vectors. Each banding pattern represents a transgenic fingerprint for each sector. The blot was reprobed with rDNA to account for differences in DNA loading and differences in DNA mobility attributed to possible differences in salt concentrations. Numbers to the left are approximate DNA size markers in kilobases.
DISCUSSION
Whereas methods for using flow cytometry based on GFP expression in plants have been available for several years (Galbraith et al., 1995; Sheen et al., 1995), few applications have been reported. These examples include abscisic acid signal transduction in rice (Oryza sativa) (Desikan et al., 1999; Hagenbeek et al., 2000; Gampala et al., 2001; Hagenbeek and Rock, 2001), root development in Arabidopsis thaliana (Birnbaum et al., 2003), flow cytometry protocol development and mosaic observations in tobacco leaves (Bohanec et al., 2002; Bastar et al., 2004), and nuclear targeting in tobacco (Grebenok et al., 1997). Only the last four cases contained examples of flow cytometry analysis of stably transformed plants, and of these only the last three studies include attempts to quantify GFP expression. One of the true powers of flow cytometry is analysis of heterogeneity in a cell population because properties are measured on an individual cell basis rather than as a population average (Yanpaisan et al., 1999). The data presented here represent an application of flow cytometry to characterize epigenetic variation that can exist in transgenic plant cell populations.
An important factor in our ability to quantitate GFP expression variegation in stably transformed cell lines is to accurately estimate the number of cells that have wild-type levels of fluorescence. Because of the nature of the protoplasting procedure and the fact that in this study several samples had to be prepared simultaneously and rapidly, nuclei and protoplasts with compromised plasma membrane integrity frequently occurred in our preparations. Because GFP diffuses out of compromised protoplasts but they may have physical properties otherwise nearly indistinguishable from intact protoplasts, the number of cells with wild-type levels of fluorescence can be overestimated. To correct for this phenomenon, we used an approach common in animal systems by which cells are stained with propidium iodide to exclude necrotic cells from analysis, for example, Migliaccio et al. (2000). Under conditions where debris is a problem, adding this step to existing plant cell flow cytometry protocols would enhance our ability to discriminate between expressing and nonexpressing subpopulations of cells.
We have previously characterized the effects of flanking tobacco Rb7 MAR sequences on transgene expression in NT-1 tobacco suspension culture cells and whole tobacco plants (Allen et al., 1996; Ülker et al., 1999). In cultured cells, we observed a 60-fold increase in β-glucuronidase (GUS) expression by direct DNA transformation, but only a moderate twofold to threefold increase in whole plants. In this study, we chose A. tumefaciens–mediated transformation and observed a twofold to fourfold increase in GFP expression in NT-1 tobacco suspension culture cells. There are several differences between our past and present experiments with MARs in NT-1 cells. The lower fold effects we have observed in this study may be due to any one or combination of these differences. A. tumefaciens–mediated transformation results in less complex integration patterns than does direct DNA transformation (Pawlowski and Somers, 1996; DeBuck et al., 1998). In this work, the reporter gene and selectable marker reside on the same vector, whereas they were on separate vectors in our previous work. We have used a different reporter gene and an assay that does not depend upon enzymatic activity. Variation in mean expression values of control treatments may also account for fold differences. That is, when expression values for control constructs approach zero, the fold difference can dramatically increase as Cheng et al. (2001) noted when they determined the effect of the Rb7 MAR on GUS and GFP expression in rice.
The MAR-mediated increases in GFP expression that we observed were in part due to a greater proportion of cells that express GFP (i.e., cells in which GFP was not silenced). Additionally, some of the cells that express GFP express it at higher levels in MAR-containing lines than in control lines. Even in cell lines demonstrated not to be chimeric, we often observed epigenetic differences in the levels of GFP expression as evidenced by more than one mode of GFP expression level (Figure 7). The results presented in Figure 3 demonstrate that variegation within cell lines may be obscured by traditional extractive methods of quantitating-reporter gene expression.
In work we report here, we were able to assess epigenetic variation in transgene expression within isogenic cell lines. Because Brouwer et al. (2002) used traditional methods of assessing transgene expression in their studies of the effect of MARs on LUC expression in maize (Zea mays) cells in culture, they were not able to assess epigenetic variation within a line. They did however examine variation across a large number of transgenic lines. The authors noted fewer nonexpressing lines among lines that contained either the yeast (Saccharomyces cerevisiae) ARS1 MAR or the maize Adh1 5′ MAR than among control lines. Based on these observations, they suggested that MARs increase the likelihood that an integrated gene will be expressed but not the level of expression (Brouwer et al., 2002). Mankin et al. (2003) observed a similar MAR-induced reduction in the frequency of low expressing tobacco cell lines, but also noted that when these low expressing cell lines were removed from the analysis, MAR lines still showed an increase in GUS expression. However, it was not possible for Mankin and coworkers to determine if these increases in expression were due to greater proportions of expressing cells, cells expressing at higher levels, or both. Our data are in agreement with those of Brouwer et al. (2002) and Mankin et al. (2003), in that when MARs are incorporated into transgene constructs, we see an increase in the likelihood of expression. However, in contrast with Brouwer et al. (2002), we observe that the expressing cells in lines transformed with the MAR-containing construct express at higher levels than the expressing cells in lines transformed with control constructs.
We set out to determine whether flow cytometry could be used to assess the effect of MARs on transgene expression in cultures derived from large pools of transformed cells and whether conclusions drawn from work with pools of transformed lines would parallel those drawn from work with individual cell lines. We found that flow cytometry of population cultures transformed with MAR-containing constructs resulted in distinctive profiles of GFP expression that were not present in control population cultures. These profiles are consistent with our observations on individual cell lines. In both cases, presence of the Rb7 MAR increased the percentage of expressing cells as well as the levels of expression in a subpopulation of expressing cells. Our results indicate that population cultures studied by flow cytometry may be a useful tool to quickly and easily evaluate the effect of various sequence elements on transgene expression.
Whereas flow cytometry analysis allowed us to quantify differing modes of GFP expression within cell lines, it is possible that these differences could be attributed to chimeric cell populations. A few of the cell lines exhibited sectors that differed in GFP expression in calli cultured on solid media. DNA gel blot analysis demonstrated that in most cases the sectoring was due to epigenetic changes after an original transformation event rather than genetic chimerism. For example, in sector pair 5 (Figure 7), there was a complete loss of GFP expression, and this loss of expression was not accompanied by the loss of the T-DNA insert tested in DNA gel blots (Figure 8). However, in one of the five sectored lines we tested, the original microcallus was most likely composed of cells from more than one transformation event. This observation stresses the need for additional rounds of subculturing to increase the probability of fixing a single genotype in each cell line. Our results in individual cell lines were based on one additional round of solid media subculture, which is expected to further minimize the frequency of chimeric cell lines.
In this study, we have not investigated the molecular mechanism by which the Rb7 MAR increases GFP expression. However, we have observed that cells with the same genotype can include both GFP negative and GFP positive cells and that the Rb7 MAR reduces the frequency of GFP negative, nonexpressing cells. These results are consistent with previous suggestions (Allen et al., 2000; Mlynarova et al., 2003) that MARs can reduce the severity of gene silencing. A large portion of the MAR effect can now be understood as an effect on the frequency of silencing events within a clone of transgenic cells.
Although it is generally accepted that MARs function only upon stable integration into the genome, minor increases in transient expression have been reported for the Rb7 MAR, suggesting a possible classical enhancer effect (Allen et al., 1996; Cheng et al., 2001). Additionally, MARs may act to facilitate the activity of nearby enhancers as van der Geest and Hall (1997) reported for the β-phaseolin 5′ MAR. Whereas the mechanism by which the Rb7 MAR influences transgene expression is not yet clear, the fact that we observe higher levels of GFP expression in GFP positive cells compared with controls suggests a possible role in enhancer-like activity. Another possibility is raised by the existence of additional modes of higher GFP expression that occur in MAR-containing lines but not in control lines (Figure 6). It is possible that when a threshold of transgene expression is reached, silencing is triggered in control cell lines (Dougherty and Parks, 1995; Que et al., 1997), whereas in MAR-containing lines, MARs may act to suppress the triggering of silencing above the threshold.
In summary, we have used flow cytometry to measure GFP expression in individual plant cells. Our results indicate that the Rb7 MAR increases the proportion of cells expressing the transgene, presumably by reducing gene silencing, while also increasing the levels of expression in a subpopulation of expressing cells. Our observations on the effect of the Rb7 MAR on transgene expression are consistent between individual isogenic cell lines and large pools of transformed cell lines. Thus, the use of flow cytometry to assess gene expression in pools of transformed cell lines may provide a tool to quickly evaluate the effect of various DNA elements on transgene expression of many transformation events.
METHODS
Recombinant Plasmids
Standard cloning techniques (Sambrook et al., 1989) were used to create three binary vectors with T-DNA regions containing GFP described in Figure 1. The first control construct (pGFP) was kindly provided by Luke Mankin and originally described as pLMNC-b47 (Mankin, 2000). This binary vector (pGFP) contains the GFP cassette from psmRS-GFP (Davis and Vierstra, 1998) as well as the nptII kanamycin resistance gene cassette and binary vector backbone from pGPTV-KAN (Becker et al., 1992). To generate an additional control construct (pLGFPL), the GFP cassette from psmRS-GFP (Davis and Vierstra, 1998) was cloned into pLMNC-b73 (Mankin, 2000), replacing the GUS cassette and resulting in GFP flanked by direct repeats of a 1.2-kb region of phage λ DNA (GenBank accession number J02459, nucleotides 21231 to 22425). Similarly, the MAR-containing construct (pMGFPM) was made by cloning the same GFP cassette into pLMNC-b72 (Mankin, 2000), replacing the GUS cassette and resulting in GFP flanked by direct repeats of the 1.2-kb Nicotiana tabacum Rb7 MAR fragment from pRb7-6 (Hall et al., 1991). All T-DNA constructs contained the same orientation of gene cassettes and the same vector backbone sequences.
Plant Cell Transformation and Propagation
T-DNA vectors were electroporated into an Agrobacterium tumefaciens strain (UIA143/pMP90) described by Callaway et al. (1996). To examine the effect of the Rb7 MAR on GFP expression in stable transformants, NT-1 cells from tobacco suspension cultures were maintained and transformed as described by An (1985) and Allen et al. (1993). These cells contain essentially no chlorophyll. Transformants were selected on solid NT-1 medium (An, 1985) supplemented with 200 mg/L of Timintin (PhytoTechnology Laboratories, Shawnee Mission, KS) and 50 mg/L of kanamycin (Sigma-Aldrich, St. Louis, MO). Microcalli were clearly visible after 2 weeks, and 72 putative transformants per construct were isolated by transferring the microcalli to a second plate of fresh medium. After 2 weeks, a small randomly selected piece of the resulting callus tissue (∼1 mm in diameter) was transferred to a final third plate of fresh medium. After 2 additional weeks, the entire callus of 30 randomly selected calli was dispersed in 5 mL of fresh liquid medium supplemented with 200 mg/L of Timintin and 50 mg/L of kanamycin. Liquid cultures were grown at 27°C under constant light conditions on an orbital shaker (125 rpm) and transferred weekly by adding 0.5 mL to 5 mL of fresh medium.
During the growth on the second fresh medium plate previously mentioned, many calli exhibited a sectored GFP phenotype. To investigate the possibility that the sectored calli were chimeric (the result of multiple transformation events), calli of a subset of these were further investigated. While under far-blue light (described below), small pieces of calli (∼1 mm in diameter) representing different sectors were transferred to a third plate of fresh medium. After 3 weeks, liquid cultures were initiated as mentioned previously. DNA gel blots of DNA from cultures arising from the different sectors were used to determine if sectors arose from separate transformation events.
Population-based liquid cultures were initiated by resuspending the original A. tumefaciens and NT-1 cocultivation plate with 5 mL of media supplemented with 200 mg/L of Timintin and 50 mg/L of kanamycin. Of this 5 mL, 400 μL were plated on solid medium to isolate individual cell lines as described previously, and the remaining 4.6 mL became the population culture.
GFP Imaging
Confocal sections of protoplasts were obtained with a Perkin-Elmer UltraVIEW live cell imager (Perkin-Elmer, Boston, MA). NT-1 calli were imaged every 4 d throughout the course of solid medium selection. A Nikon Coolpix 990 (370-nm filter; Tokyo, Japan) was attached to a tripod and positioned vertically over Petri plates containing NT-1 callus. Two Dark Reader (Model DL-09B) hand lamps (Clare Chemical Research, Dolores, CO) were used to excite GFP fluorophors with far-blue light (∼420 to 500 nm). Images of the calli were processed in Adobe Photoshop (San Jose, CA) to remove signal from red and blue channels.
Sample Preparation, Flow Cytometry, and Analysis
Existing techniques in flow cytometry and protoplast preparation methods (Galbraith et al., 1995, 1999a, 1999b) were adapted and modified for our specific application. Protoplasts were obtained from 4-d-old NT-1 liquid cultures by the following procedure. The protoplasting solution was composed of 0.4 M mannitol, 10 mM Mes, pH 5.7, 1 mg/mL of pectolyase Y-23, and 10 mg/mL of Cellulase RS (Karlan Research Products, Santa Rosa, CA). Three hundred microliters of pelleted cells were suspended in a 0.4 M mannitol, 10 mM Mes, pH 5.7 solution, pelleted at 450g for 3 min, and resuspended in 500 μL of protoplasting solution. Incubation for 45 min at 28°C on a rotating shaker (125 rpm) yielded >75% liberated protoplasts. The remaining steps were performed at 4°C. Protoplasts were centrifuged for 3 min at 100g, washed in a 0.4 M mannitol, 10 mM Mes, pH 5.7, solution, and resuspended in 1 mL of a 0.4 M mannitol, 10 mM Mes, pH 5.7, solution. Samples were then passed through a 50-μm mesh and stained with 5 μL of 1 mg/mL of propidium iodide before flow cytometric analysis.
Protoplasts were stored on ice a maximum of 2 h until flow cytometry was conducted. The flow cytometer FACSCalibur (Becton-Dickinson, San Jose, CA) was operated at a laser wavelength of 488 nm with a power output of 15 mW, using a sort-sense flow cell equipped with a 100-nm orifice. Forward angle light scatter signals, side angle light scatter signals, and fluorescence emission signals corresponding to wavelengths of 520 to 530 nm (green) and 564 to 606 nm (yellow/orange) were collected at a maximum flow rate of 1000 cells/s with a total accumulation of 15,000 viable cells based on populations of minimal propidium iodide staining. To collect data on 15,000 viable cells, ∼30,000 total events were needed depending on the effectiveness of the protoplasting procedure for each sample. The forward light scatter detector was operated at a gain setting of 9.9 and a high voltage of E-1V. The side light scatter detector high voltage was set to 319 V with no gain adjustments. The photomultiplier tube (PMT) high voltages were 370 to 450 V (green channel) based on calibration beads (discussed below) and 447 V (yellow/orange channel) with no gain adjustments for either. Uniparametric and biparametric displays were generated with CellQuest 3.3 (Becton-Dickinson).
To ensure that degradation of laser intensity and other instrument-specific fluctuations did not affect our results taken over several time points, Flow Check YG 6.0 microspheres (Polysciences, Warrington, PA) were used to calibrate the flow cytometer before each use. Three distinct intensities of green fluorescence provided three calibration points to adjust the PMT voltage of the green channel. Because the fluorescence intensities of the microspheres in one set may not match those of a second set from the same manufacturer, the PMT voltage of the green channel was adjusted to match calibration points for the original set, and then with the same instrument settings, microspheres from the second set were analyzed to determine the new calibration points for this second set.
Because neither green fluorescence intensity data nor percentage of GFP positive cells were normally distributed as determined by the Kolmogorov-Smirnov test (Chakravarti et al., 1967), a nonparametric Mann-Whitney test was used to compare treatments and calculate P values (Snedecor and Cochran, 1989).
DNA Extraction and DNA Gel Blot Analysis
DNA was isolated from frozen tissue collected from transgenic tobacco cells using a modified CTAB extraction procedure (Murray and Thompson, 1980) as described by Johnson et al. (1995). Fifteen micrograms of genomic DNA was digested with HindIII (Promega, Madison, WI), a methylation-insensitive restriction enzyme, for 12 h at 37°C in a reaction volume of 50 μL containing 30 units of restriction enzyme according to the instructions of the manufacturer. The fragments were then separated by electrophoresis for 16 h in a 1% agarose gel in Tris acetate EDTA and blotted onto MagnaGraph nylon membranes (Osmonics, Westborough, MA) by overnight capillary transfer. Membranes were prehybridized overnight at 65°C with hybridization buffer from an AlkPhos Direct kit (Amersham Biosciences, Piscataway, NJ) and hybridized overnight in the same buffer at 65°C with a 458-bp PCR-amplified fragment immediately internal to the right border of all T-DNAs tested. The primers used were T-DNA RB probe forward (5′-CCAATTCGCCCTATAGTGAGTCGT-3′) and T-DNA RB probe reverse (5′-CATGCACATACAAATGGACGAACGG-3′). After PCR amplification, the probe was 32P-labeled by random-primed synthesis with Rediprime II (Amersham Biosciences). Membranes were washed five times, with the final two washes being the most stringent, consisting of 0.5× SSC and 0.1% SDS at 65°C for 20 min each. Hybridized membranes were then placed in phosphor screens for varying amounts of time and read on a Storm 840 phosphor imager (Amersham Biosciences). Similarly, membranes were probed with a 256-bp PCR-amplified fragment from the 18S rRNA coding region of NT-1 genomic DNA with primers 18S rDNA forward (5′-ACCAGACTCATAGAGCCCGGTATT-3′) and 18S rDNA reverse (5′-TCATGATAACTCGACGGATCGCAC-3′).
Acknowledgments
We thank the Arabidopsis Biological Resource Center and Richard Vierstra for kindly providing psmRS-GFP and Luke Mankin for kindly providing pLMNC-b47, pLMNC-b72, and pLMNC-b73. We thank Janet Dow at the North Carolina State University College of Veterinary Medicine Flow Cytometry Facility. We also thank Kirk Francis, Niki Robertson, and Arthur Weissinger for helpful discussions and Jeff Thorne and Amy Anderson for statistical consultation. Christopher Halweg was supported by a Tri-Agency fellowship (National Science Foundation/USDA/Department of Energy) and a Graduate Assistance in Areas of National Need fellowship (Department of Education).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steven Spiker (steven_spiker@ncsu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028100.
References
- Allen, G.C., Hall, G., Jr., Michalowski, S., Newman, W., Spiker, S., Weissinger, A.K., and Thompson, W.F. (1996). High-level transgene expression in plant cells: Effects of a strong scaffold attachment region from tobacco. Plant Cell 8, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G.C., Hall, G.E., Jr., Childs, L.C., Weissinger, A.K., Spiker, S., and Thompson, W.F. (1993). Scaffold attachment regions increase reporter gene expression in stably transformed plant cells. Plant Cell 5, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G.C., Spiker, S., and Thompson, W.F. (2000). Use of matrix attachment regions (MARs) to minimize transgene silencing. Plant Mol. Biol. 43, 361–376. [DOI] [PubMed] [Google Scholar]
- An, G.H. (1985). High-efficiency transformation of cultured tobacco cells. Plant Physiol. 79, 568–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastar, M.T., Luthar, Z., Skof, S., and Bohanec, B. (2004). Quantitative determination of mosaic GFP gene expression in tobacco. Plant Cell Rep. 22, 939–944. [DOI] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Bode, J., Stengert-Iber, M., Kay, V., Schlake, T., and Dietz-Pfeilstetter, A. (1996). Scaffold/matrix-attached regions: Topological switches with multiple regulatory functions. Crit. Rev. Eukaryot. Gene Expr. 6, 115–138. [DOI] [PubMed] [Google Scholar]
- Bohanec, B., Luthar, Z., and Rudolf, K. (2002). A protocol for quantitative analysis of green fluorescent protein-transformed plants, using multiparameter flow cytometry with cluster analysis. Acta Biol. Cracov. Ser. Bot. 44, 145–153. [Google Scholar]
- Bonifer, C., Vidal, M., Grosveld, F., and Sippel, A.E. (1990). Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 9, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne, P., Vanmontagu, M., Depicker, A., and Gheysen, G. (1992). Characterization of a plant scaffold attachment region in a DNA fragment that normalizes transgene expression in tobacco. Plant Cell 4, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, C., Bruce, W., Maddock, S., Avramova, Z., and Bowen, B. (2002). Suppression of transgene silencing by matrix attachment regions in maize: A dual role for the maize 5′ ADH1 matrix attachment region. Plant Cell 14, 2251–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye, K.M., Goderis, I.J., Wouters, P.F., Pues, J.M., Delaure, S.L., Broekaert, W.F., Depicker, A., Cammue, B.P., and De Bolle, M.F. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39, 440–449. [DOI] [PubMed] [Google Scholar]
- Callaway, A., Liu, W., Andrianov, V., Stenzler, L., Zhao, J., Wettlaufer, S., Jayakumar, P., and Howell, S.H. (1996). Characterization of cauliflower mosaic virus (CaMV) resistance in virus-resistant ecotypes of Arabidopsis. Mol. Plant-Microbe Interact. 9, 810–818. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chakravarti, I.M., Laha, R.G., and Roy, J. (1967). Handbook of Methods of Applied Statistics. (New York: Wiley).
- Cheng, Z.Q., Targolli, J., and Wu, R. (2001). Tobacco matrix attachment region sequence increased transgene expression levels in rice plants. Mol. Breed. 7, 317–327. [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- De Bolle, M.F.C., Butaye, K.M.J., Coucke, W.J.W., Goderis, I.J.W.M., Wouters, P.F.J., van Boxel, N., Broekaert, W.F., and Cammue, B.P.A. (2003). Analysis of the influence of promoter elements and a matrix attachment region on the inter-individual variation of transgene expression in populations of Arabidopsis thaliana. Plant Sci. 165, 169–179. [Google Scholar]
- De Buck, S., Jacobs, A., Van Montagu, M., and Depicker, A. (1998). Agrobacterium tumefaciens transformation and cotransformation frequencies of Arabidopsis thaliana root explants and tobacco protoplasts. Mol. Plant-Microbe Interact. 11, 449–457. [DOI] [PubMed] [Google Scholar]
- Desikan, R., Hagenbeek, D., Neill, S.J., and Rock, C.D. (1999). Flow cytometry and surface plasmon resonance analyses demonstrate that the monoclonal antibody JIM19 interacts with a rice cell surface component involved in abscisic acid signalling in protoplasts. FEBS Lett. 456, 257–262. [DOI] [PubMed] [Google Scholar]
- Dougherty, W.G., and Parks, T.D. (1995). Transgenes and gene suppression: Telling us something new? Curr. Opin. Cell Biol. 7, 399–405. [DOI] [PubMed] [Google Scholar]
- Fukuda, Y., and Nishikawa, S. (2003). Matrix attachment regions enhance transcription of a downstream transgene and the accessibility of its promoter region to micrococcal nuclease. Plant Mol. Biol. 51, 665–675. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Anderson, M.T., and Herzenberg, L.A. (1999. a). Flow cytometric analysis and FACS sorting of cells based on GFP accumulation. Methods Cell Biol. 58, 315–341. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Herzenberg, L.A., and Anderson, M.T. (1999. b). Flow cytometric analysis of transgene expression in higher plants: Green fluorescent protein. Methods Enzymol. 302, 296–315. [DOI] [PubMed] [Google Scholar]
- Galbraith, D.W., Lambert, G.M., Grebenok, R.J., and Sheen, J. (1995). Flow cytometric analysis of transgene expression in higher plants: Green-fluorescent protein. Methods Cell Biol. 50, 3–14. [DOI] [PubMed] [Google Scholar]
- Gampala, S.S., Hagenbeek, D., and Rock, C.D. (2001). Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J. Biol. Chem. 276, 9855–9860. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J., Lambert, G.M., and Galbraith, D.W. (1997). Characterization of the targeted nuclear accumulation of GFP within the cells of transgenic plants. Plant J. 12, 685–696. [Google Scholar]
- Grewal, S.I., and Moazed, D. (2003). Heterochromatin and epigenetic control of gene expression. Science 301, 798–802. [DOI] [PubMed] [Google Scholar]
- Hagenbeek, D., Quatrano, R.S., and Rock, C.D. (2000). Trivalent ions activate abscisic acid-inducible promoters through an ABI1-dependent pathway in rice protoplasts. Plant Physiol. 123, 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbeek, D., and Rock, C.D. (2001). Quantitative analysis by flow cytometry of abscisic acid-inducible gene expression in transiently transformed rice protoplasts. Cytometry 45, 170–179. [DOI] [PubMed] [Google Scholar]
- Hall, G., Jr., Allen, G.C., Loer, D.S., Thompson, W.F., and Spiker, S. (1991). Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc. Natl. Acad. Sci. USA 88, 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, K.H., Ma, C.P., and Strauss, S.H. (1997). Matrix attachment regions (MARs) enhance transformation frequency and transgene expression in poplar. Transgenic Res. 6, 415–420. [Google Scholar]
- Holmes-Davis, R., and Comai, L. (2002). The matrix attachment regions (MARs) associated with the Heat Shock Cognate 80 gene (HSC80) of tomato represent specific regulatory elements. Mol. Genet. Genomics 266, 891–898. [DOI] [PubMed] [Google Scholar]
- Johnson, E., Miklas, P.N., Stavely, J.R., and Martinezcruzado, J.C. (1995). Coupling-phase and repulsion-phase Rapds for marker assisted selection of Pi-181996 rust resistance in common bean. Theor. Appl. Genet. 90, 659–664. [DOI] [PubMed] [Google Scholar]
- Liu, J.W., and Tabe, L.M. (1998). The influences of two plant nuclear matrix attachment regions (MARs) on gene expression in transgenic plants. Plant Cell Physiol. 39, 115–123. [DOI] [PubMed] [Google Scholar]
- Mankin, S.L. (2000). Chromatin Elements and Transgene Expression in Tobacco Cells. PhD dissertation (Raleigh, NC: North Carolina State University).
- Mankin, S.L., Allen, G.C., Phelan, T., Spiker, S., and Thompson, W.F. (2003). Elevation of transgene expression level by flanking matrix attachment regions (MAR) is promoter dependent: A study of the interactions of six promoters with the RB7 3′ MAR. Transgenic Res. 12, 3–12. [DOI] [PubMed] [Google Scholar]
- Martelli, A.M., Falcieri, E., Zweyer, M., Bortul, R., Tabellini, G., Cappellini, A., Cocco, L., and Manzoli, L. (2002). The controversial nuclear matrix: A balanced point of view. Histol. Histopathol. 17, 1193–1205. [DOI] [PubMed] [Google Scholar]
- Matzke, M., and Matzke, A.J. (2003). RNAi extends its reach. Science 301, 1060–1061. [DOI] [PubMed] [Google Scholar]
- McGann, L.E., Walterson, M.L., and Hogg, L.M. (1988). Light scattering and cell volumes in osmotically stressed and frozen-thawed cells. Cytometry 9, 33–38. [DOI] [PubMed] [Google Scholar]
- Mendu, N., Massel, M., and Spiker, S. (2001). Increasing loop domain size does not diminish effects of matrix attachment regions on transgene expression in tobacco cells culture. FEBS Lett. 496, 66–67. [DOI] [PubMed] [Google Scholar]
- Migliaccio, A.R., Bengra, C., Ling, J., Pi, W., Li, C., Zeng, S., Keskintepe, M., Whitney, B., Sanchez, M., Migliaccio, G., and Tuan, D. (2000). Stable and unstable transgene integration sites in the human genome: Extinction of the green fluorescent protein transgene in K562 cells. Gene 256, 197–214. [DOI] [PubMed] [Google Scholar]
- Mlynarova, L., Hricova, A., Loonen, A., and Nap, J.P. (2003). The presence of a chromatin boundary appears to shield a transgene in tobacco from RNA silencing. Plant Cell 15, 2203–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., Jansen, R.C., Conner, A.J., Stiekema, W.J., and Nap, J.P. (1995). The MAR-mediated reduction in position effect can be uncoupled from copy number-dependent expression in transgenic plants. Plant Cell 7, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarova, L., Loonen, A., Heldens, J., Jansen, R.C., Keizer, P., Stiekema, W.J., and Nap, J.P. (1994). Reduced position effect in mature transgenic plants conferred by the chicken lysozyme matrix-associated region. Plant Cell 6, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski, W.P., and Somers, D.A. (1996). Transgene inheritance in plants genetically engineered by microprojectile bombardment. Mol. Biotechnol. 6, 17–30. [DOI] [PubMed] [Google Scholar]
- Phi-Van, L., von Kries, J.P., Ostertag, W., and Strätling, W.H. (1990). The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol. Cell. Biol. 10, 2302–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que, Q., Wang, H.Y., English, J.J., and Jorgensen, R.A. (1997). The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, M.S., Dinkins, R.D., and Collins, G.B. (2003). Gene silencing in transgenic soybean plants transformed via particle bombardment. Plant Cell Rep. 21, 676–683. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schramke, V., and Allshire, R. (2003). Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Shapiro, H.M. (1995). Practical Flow Cytometry. (New York: Wiley-Liss).
- Sheen, J., Hwang, S., Niwa, Y., Kobayashi, H., and Galbraith, D.W. (1995). Green-fluorescent protein as a new vital marker in plant cells. Plant J. 8, 777–784. [DOI] [PubMed] [Google Scholar]
- Sidorenko, L., Bruce, W., Maddock, S., Tagliani, L., Li, X., Daniels, M., and Peterson, T. (2003). Functional analysis of two matrix attachment region (MAR) elements in transgenic maize plants. Transgenic Res. 12, 137–154. [DOI] [PubMed] [Google Scholar]
- Snedecor, G.W., and Cochran, W.G. (1989). Statistical Methods. (Ames, IA: Iowa State University Press).
- Stief, A., Winter, D.M., Strätling, W.H., and Sippel, A.E. (1989). A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341, 343–345. [DOI] [PubMed] [Google Scholar]
- Ülker, B., Allen, G.C., Thompson, W.F., Spiker, S., and Weissinger, A.K. (1999). A tobacco matrix attachment region reduces the loss of transgene expression in the progeny of transgenic tobacco plants. Plant J. 18, 253–263. [Google Scholar]
- Vain, P., Worland, B., Kohli, A., Snape, J.W., Christou, P., Allen, G.C., and Thompson, W.F. (1999). Matrix attachment regions increase transgene expression levels and stability in transgenic rice plants and their progeny. Plant J. 18, 233–242. [Google Scholar]
- van der Geest, A., Welter, M.E., Woosley, A.T., Pareddy, D.R., Pavelko, S., Skokut, M., and Ainley, W.M. (2004). A short synthetic MAR positively affects transgene expression in rice and Arabidopsis. Plant Biotechnol. J. 2, 13–26. [DOI] [PubMed] [Google Scholar]
- van der Geest, A.H., and Hall, T.C. (1997). The beta-phaseolin 5′ matrix attachment region acts as an enhancer facilitator. Plant Mol. Biol. 33, 553–557. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, W., Mlynarova, L., Nap, J.P., van der Plas, L.H., and van der Krol, A.R. (2001). The effect of MAR elements on variation in spatial and temporal regulation of transgene expression. Plant Mol. Biol. 47, 543–554. [DOI] [PubMed] [Google Scholar]
- Weiler, K.S., and Wakimoto, B.T. (1995). Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29, 577–605. [DOI] [PubMed] [Google Scholar]
- Yanpaisan, W., King, N.J.C., and Doran, P.M. (1999). Flow cytometry of plant cells with applications in large-scale bioprocessing. Biotechnol. Adv. 17, 3–27. [DOI] [PubMed] [Google Scholar]