Abstract
The Drosophila RNA‐binding protein Sex‐lethal (Sxl) is a potent post‐transcriptional regulator of gene expression that controls female development. It regulates the expression of key factors involved in sex‐specific differences in morphology, behavior, and dosage compensation. Functional Sxl protein is only expressed in female flies, where it binds to U‐rich RNA motifs present in its target mRNAs to regulate their fate. Sxl is a very versatile regulator that, by shuttling between the nucleus and the cytoplasm, can regulate almost all aspects of post‐transcriptional gene expression including RNA processing, nuclear export, and translation. For these functions, Sxl employs multiple interactions to either antagonize RNA‐processing factors or to recruit various coregulators, thus allowing it to establish a female‐specific gene expression pattern. Here, we summarize the current knowledge about Sxl function and review recent mechanistic and structural studies that further our understanding of how such a seemingly ‘simple’ RNA‐binding protein can exert this plethora of different functions.
Keywords: alternative splicing, post‐transcriptional regulation of gene expression, RNA‐binding protein, sex determination, sex‐lethal, translational control
Abbreviations
APA>, alternative polyadenylation
CPSF, cleavage and polyadenylation specificity factor
DSE, downstream sequence element
dsx, doublesex
eIFs, eukaryotic initiation factors
fru, fruitless
GSC, germline stem cell
how, held out wings
msl‐2, male‐specific lethal‐2
N, Notch
NMD, nonsense‐mediated mRNA decay
nos, nanos
ORFs, open reading frames
PABP, poly(A)‐binding protein
PIC, preinitiation complex
RBPs, RNA‐binding proteins
RNPs, ribonucleoprotein particles
RRMs, RNA‐recognition motifs
snf, sans fille
Sxl, sex‐lethal
tra, transformer
Unr, upstream of N‐ras
uORFs, upstream open reading frames
vir, virilizer
In eukaryotes post‐transcriptional regulation of gene expression plays a critical role in almost all cellular processes. Regulation is mainly achieved by RNA‐binding proteins (RBPs) that recognize specific RNA elements and form ribonucleoprotein particles (RNPs) to control RNA fate from synthesis to decay. In Drosophila melanogaster, female development is governed by a single RNA‐binding protein, Sex‐lethal (Sxl), which controls the expression of key factors that regulate sex‐specific traits. Not surprisingly, interfering with Sxl expression in female flies is lethal, as well as its forced expression in males, which has initially led to the identification of the gene locus 1.
The choice whether to be male or female is made early in Drosophila embryogenesis by an X‐chromosome‐counting mechanism that produces functional Sxl protein only in females. Sxl expression is established in the precellular embryo through four proteins encoded on the X‐chromosome: Runt, Scute, SisA, and Unpaired. Since female Drosophila carry two X‐chromosomes, the gene dose for these proteins exceeds the threshold required to activate Sxl expression via the ‘establishment’ promoter Pe (which otherwise stays silent in males that carry a single X‐chromosome). Even in the absence of Sxl protein, transcripts from the Pe promoter are spliced in a productive fashion, giving rise to mRNAs that encode functional Sxl protein. This protein is then required to instruct productive splicing of transcripts from the maintenance promoter (SxlPm) which is activated later in development in both sexes. This way, Sxl establishes an autoregulatory, positive feedback loop for permanent and self‐sustained expression that commits to female development only in flies that carry two X‐chromosomes [reviewed in 2].
Sxl‐dependent post‐transcriptional regulation is, however, not restricted to its own transcript. It recognizes and binds to regulatory RNA sequences in numerous other transcripts to control protein expression. Among the most important targets are the transformer (tra), Notch (N), male‐specific lethal‐2 (msl‐2), and nanos (nos) mRNAs which code for proteins that are involved in the regulation of female morphology and behavior, dosage compensation, and germline homeostasis 2, 3, 4, 5, 6, 7, 8.
Despite its many different functions, Sxl has a surprisingly simple architecture. The central part of the protein contains two RNA‐recognition motifs (RRMs) which bind to U‐rich and GU‐rich RNA sequences and which participate in protein–protein interactions. The flanking, Glycine‐rich N‐terminal domain can stabilize RNA interactions through oligomerization. In addition, it also participates in interactions with multiple different regulatory partners.
Sxl is ubiquitously expressed in females and while showing a mostly nuclear localization in somatic tissues 9, Sxl subcellular distribution in the germline is dynamic and more complex: in stem cells and during early differentiation, it accumulates in the cytoplasm, but redistributes to nuclei during later stages of development 10, 11.
Intriguingly, Sxl has functions in both the nucleus and the cytoplasm and acts on multiple, different levels to control splicing, polyadenylation, RNA export, and translation of its RNA targets. In this review, we will follow the path of the mRNA from synthesis and processing in the nucleus to translation in the cytoplasm, highlighting the regulatory functions of Sxl along the way. We will compare Sxl‐dependent regulation and discuss shared principles or differences in the mechanisms that it employs to control expression of its target mRNAs.
Regulation of RNA processing and export by Sxl
In the nucleus, Sxl has been found to associate with specific regions of nurse cell polytene chromosomes 11 which has been attributed to the binding of nascent transcripts to regulate their fate. Nuclear functions of Sxl involve the regulation of alternative splicing, alternative polyadenylation, and RNA export. Sxl‐mediated regulation of splicing contributes to suppression of dosage compensation and elicits a female‐specific splicing cascade to govern female development [which has been extensively reviewed elsewhere 2, 4, 6, 8, 12, 13, 14, 15]. Sxl appears to target numerous mRNAs to regulate their splicing, as Sxl‐binding sites are predicted in the vicinity of many alternative exons that exhibit sex‐specific splicing patterns 16. Here, we will focus on the well‐studied, major targets of Sxl (msl‐2, tra, and sxl mRNA) and briefly summarize the most important aspects of Sxl‐mediated regulation of alternative splicing.
Autoregulatory splicing to enforce Sxl expression in female flies
The Sxl gene produces several sex‐specific mRNA isoforms. In male flies, in the absence of Sxl protein, splicing generates mRNAs that include exon 3. This exon contains a premature termination codon and the translation of exon 3‐containing RNAs produces a truncated, nonfunctional Sxl protein. Moreover, premature termination codons often affect RNA stability by triggering nonsense‐mediated mRNA decay (NMD) which might explain the low levels of sxl mRNA that can be detected in male flies 17.
The utilization of ‘poison exons’ to control protein synthesis is not limited to Sxl, but in fact many other genes encoding for splicing regulators of the SR‐protein or hnRNP family contain similar termination cassette exons that, upon inclusion in the mature mRNA, result in the production of nonfunctional peptides and accelerated RNA decay by NMD 18, 19. These exons are thought to have a homeostatic function: when the level of a respective splicing regulator protein gets too high, it promotes inclusion of the poison exon in its own mRNA. This generates an autoregulatory, negative feedback loop that maintains protein concentrations within the physiological range. Sxl employs a similar splicing‐regulatory strategy. However, Sxl‐mediated alternative splicing creates a positive feedback loop: once produced, the Sxl protein represses inclusion of the poison cassette exon to reinforce and maintain its expression in female flies 20, 21.
But how does Sxl control splicing of the poison exon in its own mRNA? Regulation is complex and involves numerous interactions between Sxl and the splicing machinery. The Sxl primary transcript contains multiple Sxl‐binding sites that surround the poison exon, the most critical ones for regulation are, however, located quite far away from the splice sites (>200nt upstream and downstream of the poison exon) 22, 23.
Sxl can interact with components of both the U1 and U2 small nuclear ribonucleoproteins (snRNPs) that play key roles in the recognition of the 5′ and 3′ splice sites, defining the borders between exons and introns (Fig. 1A). An important role of the snRNP component Sans fille (Snf) in Sxl autoregulatory splicing was first established by genetic experiments, where snf mutants disrupted the Sxl‐positive feedback loop 10, 24, 25, 26. Snf shares homology with the mammalian U1A and U2B’’ spliceosomal proteins and is an integral component of both the U1 and the U2 snRNPs 27, 28, 29. Sxl and Snf interact directly via their RRM domains and, at least in the case of the U1 snRNP, incorporation of Snf into snRNPs and interaction with Sxl are not mutually exclusive 30. As Snf is also a component of U2 snRNPs it seems likely that Sxl might establish a similar interaction with this snRNP. Moreover, Sxl can also bind to another component of the U1 snRNP, the protein U1‐70K. Functional importance of this interaction is provided by the observation that skipping of the sxl poison exon becomes U1‐70K dependent when Snf and Sxl levels are low 30. Importantly, it was demonstrated that Sxl does not interfere with the recruitment of the U1 snRNP to the 5′ splice site at the end of the poison exon 31. Taken together, this has fuelled the hypothesis that Sxl interacts with the U1 and U2 snRNPs that are bound to the splice sites that flank the critical exon of sxl mRNA. By interfering with their function in splicing, a dead‐end complex is formed and the inclusion of the poison exon in the mature mRNA is suppressed in female flies 6, 26, 32.
Figure 1.
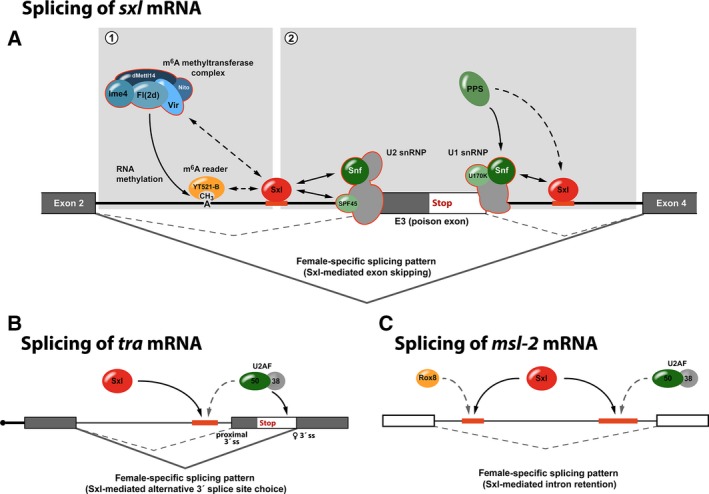
Sxl‐mediated regulation of alternative splicing. (A) Autoregulatory splicing of sxl mRNA. Sxl associates with regulatory sequences that flank the alternative, male‐specific exon E3 (poison exon) to suppress its inclusion in the mature transcript. Regulation not only requires individual proteins of the m6A methyltransferase complex and the reader protein YT521‐B (1) but also involves direct contacts of Sxl with components of the splicing machinery (2). (B) Sxl‐dependent regulation of alternative 3′ splice site usage in transformer mRNA. To prevent utilization of the proximal 3′ splice site (proximal 3′ss), Sxl competes with U2AF50 for binding of a U‐rich sequence (red), diverting splicing to the distal, female‐specific splice site (♀3′ss). (C) Sxl‐mediated intron retention in the 5′ UTR of msl‐2 mRNA. By competition for the same RNA‐binding sites, Sxl antagonizes Rox8/TIA and U2AF50 to prevent splicing.
In addition, via its N‐terminal domain, Sxl can also interact with SPF45, a protein with functions in DNA repair and splicing 33, 34. SPF45 is incorporated into the U2 snRNP and plays an important role in 3′ splice site selection and its activation during the second catalytic step of splicing 34. In female flies with reduced levels of Sxl, the deletion of the C‐terminal domain of SPF45 results in female lethality due to the inclusion of the poison exon 33. While interfering with Snf and U1‐70K is believed to stall splicing during the early stages 6, Sxl‐mediated regulation via SPF45 interferes with the second catalytic step of the splicing reaction in vitro 34. This might provide an additional layer of regulation and a fail‐safe mechanism that ensures exon skipping in female flies 30.
As splicing occurs cotranscriptionally, coupling between the transcription and splicing machineries can be observed—many factors involved in transcription and its regulation also influence alternative splicing [reviewed in 35]. A possible link between the transcription machinery and sxl alternative splicing is provided by the finding that the protein PPS (Protein Partner of Sans fille) is required for exon skipping. PPS bears homology to transcription factors and is loaded onto the Sxl primary transcript. It interacts directly with Snf and its knockout blocks Sxl autoregulation resulting in germline tumors 31. How PPS influences alternative splicing of the sxl mRNA remains to be uncovered.
Genetic analyses have revealed that the autoregulatory feedback loop of Sxl also depends on the presence of Virilizer (Vir) and Female‐lethal (2)d (Fl(2)d) 36, 37. The molecular function of both proteins long remained elusive until more recently when the mammalian orthologs were demonstrated to be subunits of the methyltransferase complex involved in N6‐methyladenosine (m6A) RNA modification 38, 39, 40. In Drosophila, this complex encompasses also the proteins Inducer of meiosis 4 (Ime4), dMettl14/CG7818, and Spenito (Nito) 41 (Fig. 1A). Similar to Vir and Fl(2)d, depletion of other members of the methyltransferase complex impairs productive splicing of sxl in female flies, resulting in a bias toward male sex and typical Sxl loss‐of‐function phenotypes 41, 42, 43. Moreover, depletion of YT521‐B, a protein that recognizes and binds to m6A via its YTH domain, phenocopies the Sxl‐splicing defect and its forced expression in males results in lethality which can be rescued by the removal of Ime4 41, 42. In sum, this supports the idea that m6A RNA modification plays a critical role in the regulation of sxl alternative splicing in female flies.
Intriguingly, m6A modifications appear to cluster around the Sxl‐binding sites that flank the alternative, male‐specific exon in sxl mRNA 42; and, as demonstrated by coimmunoprecipitation experiments, Sxl can interact with Vir and Fl(2)d 44. This raises the question whether Sxl plays a role in recruiting the methyltransferase complex to modify its own transcript and to allow for subsequent binding of YT521‐B (in this context it is interesting to note that the mammalian ortholog of Sxl, HuR, has been identified as a potential m6A‐binding protein 45, hinting that Sxl‐like proteins might also be more directly involved in m6A‐mediated gene regulation without the requirement for YTH domain‐containing reader proteins). As the human m6A reader protein YTHDC1 interacts with numerous splicing factors to control alternative RNA processing 46, its Drosophila ortholog YT521‐B might employ similar regulatory interactions—however, the details of how it contributes to exon skipping in sxl mRNA remain to be uncovered.
Autoregulatory splicing of Sxl is robust and tolerates the removal of some of the regulatory factors without compromising exon skipping. Hence, the molecular phenotypes of U1‐70K, SPF45, and YT521‐B depletion only become apparent in a genetically ‘sensitized’ fly background. Deletion of YT521‐B or the C‐terminal domain of SPF45 does not impact on the sxl splicing pattern in female flies when two functional copies of the Sxl gene are present. Upon removal of one Sxl copy, however, the autoregulatory feedback loop can no longer be established and the poison exon is retained in the sxl transcripts 33, 42. Similarly, exon skipping becomes U1‐70K dependent only when the levels of both Sxl and Snf are reduced 30. These findings are consistent with the hypothesis that multiple or redundant pathways operate in parallel to prevent exon inclusion in the mature transcript in females. Loss of one regulatory pathway can be tolerated, if the other regulatory circuits are still operational, providing robust and (almost) fail‐safe regulation of sxl splicing.
Sxl directs female development through regulated splicing of transformer mRNA
In Drosophila, most aspects of sexually dimorphic traits are governed by a splicing cascade that is initiated by the Sxl‐dependent alternative splicing of tra mRNA. Tra protein is itself a female‐specific splicing regulator that, once produced, controls splicing of doublesex (dsx) and fruitless (fru). This results in expression of the female versions of the transcription factors Dsx and Fru that control most aspects of female morphology and behavior 2, 4, 6, 8, 12, 13, 14.
Sxl‐mediated regulation of tra splicing involves usage of a pair of alternative 3′ splice sites at the end of the first intron (Fig. 1B). 3′ splice site recognition requires the heterodimeric splicing factor U2AF which in Drosophila consists of two subunits of 38 and 50 kDa. It recognizes 3′ splice site‐associated polypyrimidine tracts and stabilizes binding of the U2 snRNP during spliceosome assembly. U2AF has an ~ 100‐fold higher affinity for the pyrimidine tract of the proximal splice site 47 and hence its utilization is favored over the distal one. Splicing to the proximal site introduces a premature translation termination codon and results in the absence of functional Tra protein in male flies.
In female flies, Sxl diverts splicing to the distal 3′ splice site to produce an mRNA with an extended open reading frame that encodes functional Tra protein. Regulation involves competition between Sxl and U2AF for binding to the 3′ splice site pyrimidine tract. Sxl has a high affinity to the proximal polypyrimidine tract (which contains two closely spaced U stretches), but does not bind to the downstream one. Therefore, it competes with U2AF only on the proximal 3′ splice site, diverting U2AF to the downstream pyrimidine tract and thus activating the weaker, distal splice site 47, 48, 49.
Regulation of alternative splice site choice in tra is not complete. The female‐specific, short RNA isoform can be detected at approximately equimolar concentrations along with the non–sex‐specific longer version. As Sxl and U2AF directly compete for binding to the proximal pyrimidine tract, the extent of regulation appears to reflect the relative abundance and activity of the two proteins—and (at least in vitro) Sxl‐dependent regulation can be overcome by the addition of recombinant U2AF, shifting splicing back to the proximal splice site 47.
The N‐terminal domain of Sxl is important for regulation of tra splicing as N‐terminally truncated Sxl does not support splice site switching in male flies 50. An explanation for this phenomenon is offered by the finding that the Sxl N‐terminal domain provides cooperativity between individual Sxl molecules upon binding to RNA regions that contain two adjacent binding sites, each consisting of a short U‐run (as in the case of the proximal pyrimidine tract of tra) 51, 52. Deletion of the N‐terminal domain of Sxl would therefore diminish binding to the proximal splice site of tra mRNA. According to the competition model, this would shift the balance toward binding of U2AF, favoring splicing to the proximal 3′ splice site. The competition model has, however, been questioned by the finding that overexpression of the N‐terminal domain of Sxl in male flies also resulted in the deregulation of tra splicing, promoting usage of the distal splice site by a yet unidentified mechanism 53.
Intriguingly, also m6A RNA modification has been implicated in the regulation of tra mRNA 3′ splice site choice as the presence of Fl(2)d promoted the Sxl‐dependent, female‐specific splicing pattern. A dose dependency could be demonstrated and tra alternative splicing became Fl(2)d independent when sufficient Sxl was provided 36. But, alike to the splicing regulation of sxl mRNA, it is not understood how the m6A RNA modification mark contributes to alternative splicing of tra.
Regulation of msl‐2 alternative splicing
Female Drosophila carry two X‐chromosomes (XX), while male animals only have a single one (XY). Dosage compensation occurs by hypertranscription of the single male X‐chromosome to achieve a gene dose comparable to the output of the two female X‐chromosomes [reviewed in 3, 5, 54]. Not surprisingly, assembly of an active dosage compensation complex (DCC) in female flies results in lethality and hence has to be stringently controlled 55. This is achieved by Sxl‐dependent, post‐transcriptional regulation of the limiting component of the DCC, male‐specific lethal‐2 (msl‐2) 56, 57, 58. Intriguingly, msl‐2 repression involves multiple levels of regulation including alternative splicing, nuclear retention, and translational repression that will be covered in individual chapters of this review. In sum, the combination of regulatory mechanisms ensures a robust and fail‐safe shut‐off of Msl‐2 protein synthesis, effectively preventing the assembly of a functional DCC in female flies.
Alternative splicing of msl‐2 mRNA is regulated by Sxl and involves retention of a 5′ UTR intron (Fig. 1C), resulting in the expression of a female‐specific transcript that, in comparison to the male‐specific isoform, contains 133 additional nucleotides in its 5′ UTR 55, 59, 60. Regulation is mediated by two U‐rich stretches within the intron that are located close to the 5′‐ and 3′ splice sites 57, 61. Similar to the regulation of tra, Sxl exerts control over the 3′ splice site by antagonizing U2AF binding to the intronic pyrimidine tract. This is facilitated by the longer than usual distance between the polypyrimidine tract and the 3′ splice site, weakening the interaction of U2AF with the RNA and allowing efficient competition by Sxl 62. In addition, Sxl blocks splice site recognition at the other end of the intron by an analogous mechanism. The 5′ splice site of the msl‐2 5′ UTR intron is rather weak and its effective utilization depends on the protein TIA‐1 (Rox8), which promotes recruitment of the U1 snRNP 63, 64. Sxl competes with TIA‐1 for binding to the pyrimidine tract close to the 5′ splice site, preventing recruitment of the U1 snRNP to block splicing 61.
Sxl as a regulator of alternative polyadenylation
The enhancer of rudimentary (e(r)) locus produces sex‐specific mRNA isoforms by alternative polyadenylation (APA). In male flies, polyadenylation occurs at a proximal site, while in female flies two different polyadenylation sites are utilized, resulting in RNA isoforms that differ in their 3′ UTR length 65 (Fig. 2).
Figure 2.
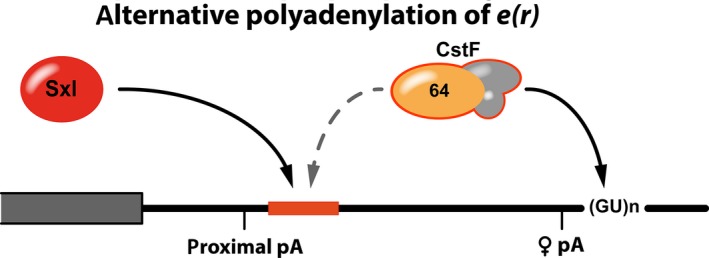
Sxl‐dependent alternative polyadenylation of enhancer of rudimentary mRNA. Activation of the proximal poly(A) site (proximal pA) requires the binding of CstF to a downstream RNA sequence (red). Sxl can compete with and displace CstF‐64, diverting it to a downstream site. This results in activation of a distal polyA‐site (♀ pA) and an extended 3′ UTR in female flies.
Three Sxl‐binding sites are predicted downstream of the proximal polyadenylation site but only the first one plays an important role in polyA‐site choice. In the absence of Sxl it serves as a downstream sequence element (DSE) for polyadenylation. For this, it is bound by a subunit of the cleavage stimulation factor, CstF‐64, which promotes binding of the cleavage and polyadenylation specificity factor (CPSF) to the hexanucleotide motif of the polyadenylation signal, thus promoting the utilization of the proximal polyadenylation site.
In female flies, Sxl competes with CstF‐64 for binding to the RNA sequence element, reducing the utilization of the proximal polyadenylation site. This results in activation of a downstream polyadenylation site and production of a mature transcript with a longer 3′ UTR that retains the Sxl‐binding motifs. Translation of RNAs with this longer, female‐specific UTR is markedly reduced in comparison to reporters that bear the short 3′ UTR of e(r) 65. It is tempting to speculate that, alike to msl‐2, Sxl might also employ two regulatory pathways for the control of E(r) expression: Sxl‐mediated regulation of APA results in retention of its binding sites in the mature mRNA that then might serve Sxl‐dependent translational control in the cytoplasm.
Intriguingly, regulation of e(r) APA is germline specific: despite the presence of Sxl in somatic tissues, only the short mRNA isoform can be detected 65. This suggests that additional factors might be involved in the Sxl‐dependent regulation of e(r) APA.
The role of Sxl in How‐dependent nuclear retention of msl‐2 mRNA
As described above, Sxl blocks the splicing of the 5′ UTR intron of msl‐2 mRNA in female flies. The retained intron harbors two Sxl‐binding sites that are important for translational repression in the cytoplasm (see below), as well as flanking sequences that contain two binding sites for the protein Held out wings (How) 66. How is a member of the STAR family of RNA‐binding proteins that have been implicated in various aspects of RNA metabolism, including splicing, RNA localization, RNA stability, and translation. Depletion of the protein from Drosophila cells, however, does neither affect Sxl‐dependent msl‐2 mRNA splicing, nor translational repression but rather results in nuclear retention of the RNA to prevent Msl‐2 protein synthesis in the cytoplasm 66. Sxl and How interact directly and both proteins can associate with msl‐2 mRNA independently of each other. Efficient nuclear retention of the msl‐2 mRNA, however, requires both factors and it has been proposed that they exert function by blocking binding of a yet unidentified factor required for RNA export 66.
Cytoplasmic Sxl as a regulator of translation
Besides its nuclear functions, Sxl can also operate in the cytoplasm to control mRNA translation. Sxl‐binding sites are common and the protein associates with many mRNA species and probably regulates translation of numerous transcripts including its own mRNA 16, 50, 55, 65, 67, 68. Here, we will focus on three mRNA targets which exemplarily highlight the diversity of Sxl‐mediated translational regulation. These transcripts encode proteins involved in dosage compensation (Msl‐2), germline maintenance (Nanos), and aspects of sexual dimorphism (Notch) 56, 57, 58, 68, 69.
Eukaryotic translation is complex and can be divided into three subsequent steps—translation initiation, elongation, and termination/recycling. In most cases translation initiation is the rate‐limiting step and target of many regulatory pathways [reviewed in 70]. It requires, besides the two ribosomal subunits, 12 eukaryotic initiation factors (eIFs) to assemble an elongation‐competent 80S ribosome on an initiation codon. Most mRNAs are translated by the scanning mechanism [reviewed in 71] that involves linear inspection of the mRNA sequence in the 5′ to 3′ direction by a ribosomal preinitiation complex (PIC) in order to identify a suitable initiation codon and to subsequently trigger ribosomal subunit joining and peptide synthesis. Recruitment of the PIC to the mRNA is stimulated by two RNA features that interact functionally: the 5′ cap structure and the poly(A) tail. A ‘closed‐loop’ structure is established through interaction of the cap‐binding complex eIF4F (consisting of eIF4e, 4G, and 4A) with the poly(A)‐binding protein (PABP) that are bound to the mRNA cap and poly(A) tail, respectively.
Initiation codon selection by the scanning PIC depends not only on complementarity with the anticodon of the initiator tRNA (Met‐tRNAi) but also on the sequence environment surrounding the potential initiation codon. Skipping of codons in a poor context in favor of translation initiation at a downstream site (a process termed ‘leaky scanning’) is commonly encountered in eukaryotes, as well as translation of open reading frames (ORFs) other than the annotated main ORF [reviewed in 72]. This also includes translation of upstream open reading frames (uORFs) that can affect and regulate the translation of downstream ORFs by a variety of mechanisms [reviewed in 73].
It was found that Sxl employs two different mechanisms to regulate translation initiation: 1) it can prevent recruitment of the PIC to the mRNA and 2) it can control the activity of a uORF to prevent scanning PICs from reaching the physiologically relevant main ORF 67, 74, 75.
Suppression of dosage compensation by Sxl‐mediated translational control of msl‐2 mRNA
msl‐2 mRNA carries six Sxl‐binding sites (denoted A‐ to F‐site) located in its 5′ leader and 3′ trailer sequences. Two of the binding sites (A‐ and B‐site) are located within the 5′ untranslated region (UTR) facultative intron, which is spliced out in male flies, but retained in females (see above); four additional Sxl‐binding motifs are found in the 3′ UTR 56, 57, 58. The 5′ Sxl‐binding sites are longer than their 3′ counterparts (U11 and U16 versus U9 and U7) and are also involved the regulation of alternative splicing and nuclear retention (see above). The two most 3′ Sxl‐binding sites (E‐ and F‐sites) are each flanked by a binding site for the protein Upstream of N‐ras (Unr) 76, 77. Translational repression does not require all of the Sxl‐binding sites and a construct that carries the 5′ B‐site and the 3′ E‐ and F‐sites is fully active in repression 75, 78.
Intriguingly, Sxl employs two regulatory mechanisms with mutually reinforcing blocks to translation initiation to establish tight control of Msl‐2 protein expression 74, 75, 78 (Fig. 3A). The regulatory pathway that operates via the 5′ UTR differs mechanistically from regulation via the 3′ UTR and targets a different step of translation initiation 74. Here, Sxl bound to the 5′ regulatory element activates a short upstream open reading frame to prevent scanning ribosomal subunits from reaching the initiation codon of the Msl‐2 coding region. Regulation cannot be simply explained by a roadblock model, where Sxl binds in the path of ribosomes to prevent scanning, as replacing Sxl by another high‐affinity RNA‐binding protein does not fully recapitulate regulation. Importantly, mapping of the RNA sequences that are minimally required for repression via this pathway allowed bioinformatic prediction of further Sxl target RNAs and suggested that this type of Sxl‐mediated regulation is not unique to msl‐2 mRNA 67. However, the exact mechanism of how Sxl controls initiation at upstream open reading frames to prevent downstream translation remains unknown, and attempts to identify molecular targets or potential corepressors that might be involved in 5′ regulation were so far unsuccessful.
Figure 3.
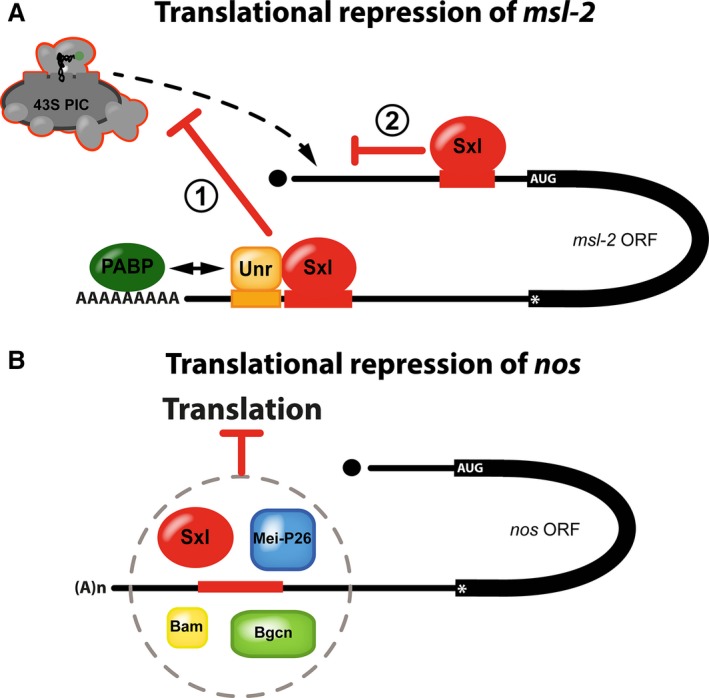
Sxl controls translation of its targets by multiple mechanisms. (A) Translational repression of msl‐2 mRNA involves two blocks to translation initiation. Bound to the 3′ UTR regulatory elements, Sxl recruits the corepressor protein UNR (1). Unr interacts directly with the polyA‐binding protein (PABP) to prevent recruitment of ribosomal preinitiation complexes (43S PIC) to the 5′ end of the RNA. Ribosomes escaping this first regulatory mechanism are then challenged by additional molecules of Sxl bound to the 5′ UTR that prevent scanning ribosomes from reaching the msl‐2 open reading frame (2) (modified from [84]). (B) Translational repression of nanos mRNA in the female germline depends on a U‐rich RNA element in the 3′ UTR of the RNA (shown in red). Besides Sxl, the proteins Mei‐P26, Bam, and Bgcn are required for regulation and all four proteins appear to form a complex to repress translation by a yet unknown mechanism.
Regulation via the 3′ Sxl‐binding sites of msl‐2 mRNA targets an earlier step of translation initiation by interfering with stable association of small ribosomal subunits with the RNA 75. In addition, this regulatory pathway critically requires the corepressor protein Upstream of N‐ras (Unr) and its depletion abolishes regulation via the 3′ UTR. Unr is an RNA‐binding protein itself that associates with many RNAs in a sex‐specific fashion which is mostly attributed to sex‐specific differences in the UTRs of its target genes 79. However, in the case of msl‐2, Unr stably associates with the mRNA only in the presence of Sxl 76, 77, 80. Hence, in male flies, where Sxl is absent, Unr does not bind the 3′ UTR of the msl‐2 transcript and cannot affect its translation.
Unr has five cold shock nucleic acid‐binding domains (CSDs) but only the first one is required for complex formation with Sxl and msl‐2 mRNA 81. It can interact with Sxl in solution even in the absence of nucleic acids; however, stable association can only be observed in the presence of RNA 76. Recent structural analyses of the Sxl‐Unr‐msl‐2 repressive complex have revealed the basis of the highly synergistic binding behavior. Association of Sxl with the more 5′ nucleotides of msl‐2 RNA occurs in a manner that is comparable to Sxl bound to a fragment of tra pre‐mRNA. In the presence of UNR, an extended stretch of msl‐2 mRNA is specifically recognized (compare Fig. 4A,B) 82, 83. Upon association with both Sxl and Unr, the msl‐2 region downstream of the canonical Sxl‐binding site becomes sandwiched between the two proteins and wraps around the side of RRM1 of Sxl to establish additional contacts (Fig. 4B). A set of ternary interactions is formed that resemble a ‘triple zipper’ where all three components of the complex participate in mutual interactions, explaining the extraordinary cooperativity in complex formation 83. This intimate association of Sxl, Unr, and msl‐2 mRNA and the synergistic binding behavior allows for the formation of a specific and highly stable repressor complex to interfere with translation.
Figure 4.
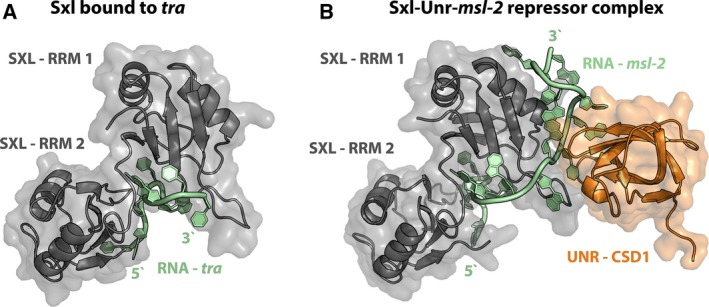
Structural insights into Sxl function. (A) Sxl bound to the proximal pyrimidine tract of tra mRNA [82; PDB: 1B7F]. The two RRMs of Sxl (gray) specifically recognize the U‐rich pyrimidine tract of tra mRNA (green). RNA contacts are mostly established with the beta‐sheet surfaces of the two RRMs. (B) Structure of the Sxl‐Unr repressor complex bound to a fragment of msl‐2 mRNA [83; PDB: 4QQB]. Binding of Sxl (gray) to the 3′ UTR regulatory elements of msl‐2 mRNA (green) occurs in a manner comparable to recognition of tra mRNA. Upon binding of Unr (cold shock domain 1, shown in orange), additional, ternary contacts are formed that involve the side of RRM1 of Sxl and allow specific recognition of an extended stretch of the RNA.
But how does translational regulation occur and which of the proteins acts as repressor? Insight comes from tethering experiments that exploit the specific interaction of a phage‐derived peptide with a well‐characterized RNA motif. By fusing the phage‐derived peptide to either Sxl or Unr, the individual proteins can be artificially recruited to reporter RNAs that carry target sites for the peptide in the 3′ UTR, circumventing formation of the entire repressor complex. Sxl shows no detectable activity upon tethering to a 3′ UTR 80. This suggests that either important functional interactions are disturbed upon tethering of the protein, or that it cannot act as a direct repressor of translation via the 3′ UTR‐binding sites and that its primary function in 3′ UTR‐mediated repression of msl‐2 lies rather in the assembly of the regulatory complex. In contrast, tethered Unr represses translation even in the absence of Sxl, demonstrating that it can act as a bona fide translational regulator. While complex assembly on msl‐2 involves mostly CSD1 of Unr, repressor activity requires a larger portion of the protein, including the two N‐terminal CSDs 81.
Regulation via the 3′ trailer of msl‐2 mRNA further requires the poly(A) tail of the RNA; and a direct protein–protein interaction between Drosophila Unr and the poly(A)‐binding protein (PABP) could be demonstrated 84. PABP plays an important role during translation initiation by mediating the functional interaction of the poly(A) tail and the 5′ cap structure of the RNA to form a ‘closed‐loop’ conformation. For this, poly(A) tail‐bound PABP interacts with the scaffolding protein, eIF4G, which in turn stabilizes cap‐bound eIF4E, establishing a physical interaction between the two RNA termini that stimulates translation initiation. However, ‘closed‐loop’ formation was unaffected in repressed msl‐2 ribonucleoproteins, demonstrating that regulation by Sxl and Unr targets ribosome binding after PABP‐mediated recruitment of eIF4G and 4E by a yet unknown mechanism 84.
Controlling cell fate by translational silencing of nos
In the female ovaries, the tip of each germarium houses a small number of germline stem cells (GSCs). Signaling by the surrounding tissue defines a stem cell niche that helps to maintain GSC fate by preventing differentiation. The GSCs cycle in mitosis and after cell division the two daughter cells adopt a different fate: one cell remains in the stem cell niche to replenish the pool of GSCs (self‐renewal), whereas the other cell moves out of the niche and differentiates into a cystoblast (CB). The CB then undergoes four synchronous cell divisions with incomplete cytokinesis to form a cyst of 16 interconnected cells, one of which will develop into the oocyte [reviewed in 7]. Changes in the fine balance between self‐renewal and differentiation result in female sterility, either by GSC loss (failure to self‐renew) or by failure to differentiate into oocytes.
Tissue homeostasis in the female germline requires Sxl and its ablation results in the lack of differentiation, overproliferation of immature CBs, and germline tumor formation 85, 86, 87, 88. Recently, nanos (nos) mRNA could be identified as a target of Sxl‐mediated translational repression in the female germline 69. The Nos protein is itself a translational repressor, that, together with its partner Pumilio (Pum), represses translation of differentiation genes to allow self‐renewal of GSCs 89, 90, 91. Hence, females mutant for nos exhibit a phenotype ‘opposite’ to Sxl and (besides other phenotypes) loose GSCs by differentiation, resulting in empty ovaries 92. In order to maintain germline homeostasis, Nos is expressed in GSCs where it is involved in the maintenance of stemness—in cells exiting the stem cell niche, however, nos needs to be transiently repressed by Sxl to trigger differentiation. It is interesting to note that nos mRNA is presumably not the only Sxl target in the female germline: inappropriate expression of Nos in the progeny of GSCs is necessary for tumor growth but does neither suffice to trigger malignant transformation nor to block differentiation 69, 93.
The nos transcript contains three Sxl‐binding motifs, but only a single one located in the 3′ UTR appears to be of relevance for Sxl‐mediated translational control (Fig. 3B) and its mutation mitigates regulation 69. Importantly, Sxl alone appears not to be sufficient for nos translational regulation: GSCs express Nos protein despite the presence of high levels of Sxl 7, 69. This suggests that—similar to repression via the 3′ UTR of msl‐2 that strictly depends on the co‐repressor Unr (see above) 76, 77, 80—other factors might be involved in the regulation of nos translation. However, unlike in msl‐2, the nos mRNA has no identifiable Unr‐binding site adjacent to the Sxl‐binding motif (own, unpublished observations). Therefore, it appears unlikely that a Sxl‐Unr repressor complex can be formed on nos RNA. But if Unr does not participate in the regulation of nos translation together with Sxl, which other corepressors are then involved in translational control?
Besides Sxl, three additional proteins are likely to participate directly in the repression of nos mRNA in the female germline: Bag of marbles (Bam), Benign gonial cell neoplasm (Bgcn), and Meiotic‐P26 (Mei‐P26) 85, 93, 94 (Fig. 3B). Mutants in either gene result in de‐repression of nos, causing germline differentiation defects to varying degrees 85, 95, 96, 97, 98, 99, 100, 101. Multiple interactions are reported between the individual proteins. Bgcn, which shows homology to DExD‐box ATP‐dependent RNA helicases but lacks critical amino acids important for ATPase and helicase function 100, binds the C‐terminal region of Bam 93, 94. Furthermore, Bgcn can interact with Mei‐P26, a protein of the TRIM‐NHL family of RNA‐binding proteins, which in turn can coimmunoprecipitate Sxl 94. Hence, all four proteins might form a complex to transiently repress nos translation and to initiate differentiation of stem cells into cystoblasts.
More recently also the deadenylase Twin (Drosophila CCR4) was implicated in GSC self‐renewal and differentiation 102. It interacts with Bam through N‐terminal Leucine‐rich repeats 102, but it remains unclear whether it plays a direct role in nos translational regulation. Depletion of Twin impacts on Bam expression levels 102, therefore, the Twin differentiation phenotype might be caused by reduced levels of Bam in the absence of Twin.
In sum, nos translational control by Sxl differs dramatically from repression of msl‐2 mRNA. Repression of msl‐2 mRNA is long‐lasting and needs to persist throughout the lifetime of females in order to prevent toxicity from DCC formation. In contrast, Sxl‐dependent repression of nos translation is transient and needs to be precisely modulated as it occurs only during differentiation from GSCs to early cysts. In later cysts Nos protein is again detectable and regulation switches to a Sxl‐independent mechanism that ensures spatial and temporal control of Nos protein expression in the developing embryo. Hence, Nos post‐transcriptional regulation is complex, involves several layers of regulation and different regulatory factors [reviewed in 103, 104].
Despite being intensively studied using in vitro systems, cultured cells, and flies 56, 57, 58, 77, 105, so far only a single corepressor protein has been identified to be required for msl‐2 mRNA translational regulation 76, 77, 80. In the case of nos translational control, at least three additional proteins (Bam, Bgcn, and Mei‐P26) appear to form a repressive complex with Sxl to directly contribute to regulation 69, 93, 94. However, it remains unclear how the regulatory complex assembles on the RNA (it contains several RNA‐binding proteins), how and at what stage it controls translation, and how it is disassembled to subsequently allow translation of Nos.
Regulation of female morphology by fine‐tuning Notch translation
Despite the overall body plan of male and female flies being largely similar, significant differences between the sexes can be observed not only in the gonads but also in other, nonreproductive organs. Some of these sexually dimorphic traits are independent of Tra and Dsx and hence of the female‐specific splicing cascade that is initiated and maintained by Sxl (see above). This includes, for example, the larger body size of females and the number of bristles on abdominal segment A5 68, 106.
To achieve sex‐specific patterning independent of Tra, Sxl negatively regulates the expression of Notch (N) 68, an essential transcription factor that controls cell fate and pattern formation during development [reviewed in 107]. Overexpression of Sxl in wing imaginal disks abolishes N expression and exacerbates the N wing phenotype—thickened veins and a notch in the wing margin. Conversely, reduction in Sxl expression results in increased N protein levels, suppresses certain N phenotypes, and even rescues lethality associated with some hypomorphic N alleles. Moreover, alteration of the Sxl expression level affects the number of bristles on the sternite of abdominal segment A5, which is under the control of the N signaling pathway 68, 108. This demonstrates that downregulation of Notch signaling by Sxl contributes to sexually dimorphic traits in flies.
Regulation of N by Sxl appears to be direct as Sxl immunoprecipitates N mRNA which, similar to msl‐2, contains several Sxl‐binding motifs (two in its 5′‐, four in the 3′ UTR) 68. Also, N mRNA harbors multiple upstream initiation codons—cis‐acting RNA elements that in msl‐2 mRNA are required for strong repression via the 5′ UTR 67. But again, there is no apparent Unr‐binding motif associated with any of the Sxl‐binding motifs (own, unpublished observations), suggesting that an Sxl‐Unr repressor complex cannot form on N mRNA.
Translation regulation of N mRNA by Sxl appears to differ from msl‐2 translational control by several means. In the case of msl‐2, translation of the mRNA has to be tightly repressed to shut‐off peptide synthesis and to prevent toxicity associated with formation of the DCC in female flies (see above). In contrast, Notch signaling is essential during development and, to maintain this fundamental signaling pathway, N expression must not be completely shut‐off by Sxl. Formation of a stable repressor complex for persistent and strong translational repression therefore appears unnecessary or even harmful. Instead of completely shutting off translation, it is conceivable that the role of Sxl rather lies in fine‐tuning of N protein synthesis to instruct development of female morphology.
By mutation of individual Sxl‐binding sites, it has been demonstrated that msl‐2 reporters defective in either 5′ or 3′ UTR‐mediated repression exhibit an attenuated response to Sxl. In contrast to wt msl‐2 mRNA that shows an almost on/off switch‐like response to Sxl, a more graded translational regulation is observed on these mutant reporters and low levels of DCC formation can be detected in female flies under physiological Sxl protein concentration 56, 58, 67, 74. This raises the question whether two mutually reinforcing, translation regulatory pathways necessarily have to operate on N mRNA (like on msl‐2 mRNA) or whether some of the Sxl‐binding motifs are nonfunctional in regulation to allow a more graded translational response. The lack of Unr‐binding motifs associated with the Sxl‐binding sites in N RNA favors the latter idea; however, the contribution of corepressors other than Unr cannot be ruled out.
Balancing Sxl expression—finding the right concentration for survival
Hypomorphic Sxl alleles result in Msl‐2 protein expression and assembly of the DCC in female flies 109, whereas overexpression of Sxl in female wing imaginal disks abolishes N expression 108. This implies that Sxl concentrations have to be maintained within a narrow physiological range: too little Sxl and msl‐2 mRNA will be de‐repressed, too much Sxl and N signaling will be shut off. Both scenarios would be deleterious for female flies. This raises the question of how Sxl expression is kept in check and maintained within adequate and physiological concentrations.
Both positive and negative feedback mechanisms exist that generate a regulatory circuit to maintain Sxl protein levels within homeostatic concentrations. Sxl directs splicing of its own mRNA in the productive, female pattern, generating positive feedback loop to promote its own synthesis (see above). At the same time Sxl can inhibit its own translation 50, providing negative feedback and balancing Sxl protein concentration. Also, Hrp48, an abundant RNA‐binding protein of the hnRNP A/B family, has been identified as a negative regulator of Sxl expression. It functions as homeotic factor to prevent accumulation of excessive levels of Sxl thus allowing proper N expression and preventing toxicity 108. It has been proposed that, in order to regulate Sxl expression, Hrp48 requires the Sxl protein itself, such that if Sxl levels become too low, regulation is alleviated 110. This would provide an additional regulatory mechanism to carefully balance Sxl concentrations and to prevent accumulation of excessive protein levels. Since Hrp48 does neither affect splicing and polyadenylation of the Sxl mRNA, nor its stability 108, it is likely that it exerts its function via translational control. Hence, it is tempting to speculate that Hrp48 might be yet another corepressor protein that acts together with Sxl.
Sxl as versatile regulator of partnerships and competition
In sum, Sxl acts as a negative regulator of RNA processing and translation. By blocking utilization of splice sites or polyadenylation elements, Sxl promotes exon skipping (sxl mRNA), intron retention (msl‐2 mRNA), alternative 3′ splice site use (tra mRNA), and alternative polyadenylation (e(r) mRNA). Despite this apparent similarity, regulation by Sxl differs drastically on the individual mRNAs in terms of cofactor requirement, and strength and duration of regulation. For some RNAs, regulation can be explained by a simple competition with processing factors (msl‐2 intron retention and tra alternative 3′ splice site usage), while other regulatory pathways are complex and involve numerous different regulatory partners (sxl exon skipping). In the latter case, regulation is strong with no detectable inclusion of the poison exon in females, while regulated alternative splicing of tra and msl‐2 and APA of e(r) is incomplete and significant amounts of the non–sex‐specific mRNA isoforms are still produced.
The same holds true for Sxl‐dependent translational repression in the cytoplasm. Translational control of msl‐2 mRNA is almost complete and persists throughout the lifetime of female Drosophila, requiring continuous regulation and the extraordinary stability of the Sxl‐Unr repressor complex. In contrast, regulation of nanos mRNA translation is strong, but spatially and temporally restricted. Only a single Sxl‐dependent regulatory mechanism appears to operate via a 3′ UTR RNA regulatory element, involving corepressors other than Unr. Finally, Sxl‐mediated regulation of Notch translation is rather a fine‐tuning of protein synthesis instead of an on/off switch, as full repression of Notch signaling has dire consequences.
It is striking to note, that several RNAs appear to be regulated at multiple levels by Sxl. To maintain physiological protein levels, Sxl controls both, splicing and translation, of its own mRNA creating homeotic feedback. Similarly, e(r) expression appears to be regulated by Sxl on the level of RNA processing and translation, adjusting protein levels in the germline. Finally, Msl‐2 protein synthesis is even controlled on three levels to block formation of an active DCC in female flies: in the nucleus, Sxl prevents splicing of an intron that contains regulatory elements to prevent nuclear export; in addition, RNA that escapes to the cytoplasm is translationally silenced. Employing multiple regulatory mechanisms that jointly act to repress Msl‐2 protein synthesis in female flies ensures tight and fail‐safe regulation which is critical for survival of females.
Taken together, Sxl operates as a very versatile regulator of gene expression to tailor peptide synthesis to the cellular requirements. But how can the difference in regulation of its RNA targets be explained? A simple model that takes into account the position and sequence of the Sxl‐binding sites does not suffice to predict the regulatory potential. Although N and msl‐2 mRNA both carry similar Sxl‐binding motifs (that closely match the consensus sequence) within their 5′ and 3′ UTRs 111, 112, regulation results either in an almost complete block of translation, or a rather modest repression. In another study, RNAs that harbor comparable regulatory features—a 5′ UTR Sxl‐binding motif associated with a uORF—exhibit varying degrees of regulation in response to Sxl, also ranging from a modest decrease in translation to an almost complete shut‐off 67. This suggests that the context of the Sxl‐binding sites, additional RNA regulatory elements, and/or the partners that are employed by Sxl strongly influence the outcome of regulation.
To achieve regulation, Sxl often acts as a remodeler of ribonucleoprotein composition, either by competing with and evicting factors that recognize similar RNA elements, or by recruitment of regulatory co‐factors to adjacent sequences, often conveying a sex‐specific function to them. Binding with high affinity to U‐ or GU‐rich sequences, Sxl efficiently antagonizes the binding of RNA processing factors, such as CstF‐64 and U2AF65. But Sxl is not a general inhibitor of these complexes. To promote splicing, U2AF shows a broad binding specificity to be able to recognize the large number of different polypyrimidine tracts that are present in the introns of pre‐mRNAs. In contrast, Sxl has a strong preference for U‐rich sequences interrupted by Gs. Competition between Sxl and U2AF, therefore, is thought to occur only on sequences that resemble Sxl‐binding sites, leaving unaffected the vast majority of polypyrimidine tracts that are recognized by U2AF 111, 113. A similar scenario can be envisioned for the regulation of alternative polyadenylation where Sxl and CstF compete: both proteins recognize similar sequences but show different binding site length requirements 111.
The best studied example for cooperation of Sxl with one of its regulatory partners is the intricate interaction with Unr in the translational repression of msl‐2 mRNA (Fig. 4B). Both proteins can independently bind various RNA targets to control gene expression—interaction of Unr with the 3′ UTR of msl‐2, however, requires Sxl and binding occurs synergistically 76, 77, 83. In contrast to Unr, binding of the 5′ UTR of msl‐2 mRNA by the coregulator How occurs independently of Sxl 66. However, Sxl plays at least an indirect role in its recruitment to msl‐2. The How response elements (HREs) are located within the facultative 5′ UTR intron of msl‐2, and Sxl‐mediated intron retention preserves them in the mature transcript. But intron retention is not sufficient for How to exert its function in regulating Msl‐2 expression which further requires Sxl 66.
In most other cases, it remains elusive how Sxl cooperates with its protein partners and regulates their activity to control mRNA processing and protein synthesis. For control of sxl alternative splicing, multiple interactions between Sxl and the splicing machinery are reported and regulation possibly involves several mechanisms. However, the molecular details of how Sxl prevents splicing still remain mostly enigmatic. Also, translational control of nanos requires proteins in addition to Sxl. Multiple interactions among the regulatory factors suggest formation of a higher order complex. This is thought to include at least three (putative) RNA‐binding proteins: Sxl, Mei‐P26, and Bgcn but it remains unclear whether all three proteins contribute to RNA recognition, and if so, if RNA binding is synergistic and involves RNA sequences in addition to the Sxl‐binding motif.
Finally, it appears as if not all regulatory partners of Sxl have yet been identified. YT521‐B and Hrp48 have been proposed to function together with Sxl in gene regulation 41, 42, 110. Moreover, despite ubiquitous expression of Sxl, regulation of e(r) APA is germline specific and potentially involves additional regulatory factors. Also, translational repression via the 5′ UTR of msl‐2 cannot simply be explained by Sxl impeding the progress of scanning ribosomes (roadblock model), implying the requirement for additional regulatory factors. In this context, interactions of Sxl with the translation machinery were detected but functional evidence is still missing 67. Moreover, interactions of the Glycine‐rich N‐terminal domain of Sxl with hnRNPL and Hrb87F, two abundant RNA‐binding proteins of the hnRNP family, are reported 52. But so far, no data are available that might hint at the molecular function of these proteins in Sxl‐dependent regulation of gene expression.
Functions of Sxl‐related proteins
Intriguingly, the function of Sxl as the master regulator of female development appears to be limited to the Drosophila clade. In other insects, the major sex‐determining switch gene is not Sxl, but transformer 6, 114, 115, 116. This suggests that the key function of Sxl in female development of D.melanogaster was evolutionarily acquired rather recently 117. The change in function also coincides with a gene duplication event that occurred early in Drosophilid evolution, presumably lessening evolutionary pressure on Sxl and allowing it to evolve more rapidly to adopt its function as a master regulator of female development 117, 118.
The ancestral, non–sex‐specific function of Sxl, however, still remains enigmatic. Insights might be provided by functional analysis of the paralog originating from the gene duplication event, sister of sex‐lethal (ssx) 117. Sxl and Ssx proteins share a high degree of identity between their central domains that encompass the two RRMs, the N‐ and C‐terminal domains are, however, quite distinct. Ssx is a nonessential gene and its knockout shows no apparent phenotype under standard laboratory conditions 117; but its knockdown increases sensitivity to infection by gram‐positive bacteria and suggests a function in immunity 119.
Sxl is related to the ELAV (embryonic lethal, abnormal visual system)/Hu RNA‐binding proteins, a phylogenetically highly conserved protein family with three members in Drosophila (ELAV, FNE, and RBP9) and four in mammals (HuB, HuC, HuD, and HuR). Many of them are expressed in the nervous system where they regulate neuronal differentiation and synaptic plasticity. Alike to Sxl, they shuttle between the cytoplasm and the nucleus to regulate various aspects of gene expression, including splicing, polyadenylation, RNA stability and localization, and translation [reviewed in 120, 121, 122].
Several aspects of regulation by ELAV/Hu proteins share striking similarities with Sxl‐mediated post‐transcriptional control of gene expression. The two Drosophila proteins ELAV and FNE establish an autoregulatory feedback loop by binding the 3′ UTR of their own transcripts. This prevents excessive protein accumulation which otherwise would result in lethality 123, 124, 125. In mammals, nuclear Hu proteins control the neuron‐specific splicing of the calcitonin/CGRP gene. By competing with TIA‐1/TIAR for binding to a U‐rich sequence, they promote exon skipping resulting in synthesis of the neurotransmitter CGRP instead of the peptide hormone calcitonin 126. Similarly, Hu proteins can promote exon skipping in the NF1 mRNA by interfering with TIA‐1 and U2AF binding 127, in much the same way as observed for Sxl. And finally, both human Hu proteins and Drosophila ELAV can regulate alternative polyadenylation 128, 129, however, in this case, a simple competition with CstF‐64 does not suffice to explain regulation.
The close relationship between Sxl and other ELAV proteins is further revealed by partially overlapping functions. They recognize and bind to U‐rich RNA sequences and, when coexpressed, Sxl can induce neuron‐specific alternative splicing of an ELAV target mRNA. Moreover, heterozygosity for RBP9 and Sxl results in female lethality. Importantly, this can be rescued by genetically interfering with DCC formation, suggesting that RBP9 can function in Msl‐2 repression 130.
Since the discovery of Sxl almost 40 years ago 1, Sxl‐based research has yielded invaluable insights into the mechanisms and fundamental principles of diverse aspects of post‐transcriptional gene regulation. But despite the impressive progress, still many important questions remain to be explored. In many cases our understanding of the mechanisms and molecular details of Sxl‐mediated regulation is still very limited. Moreover, Sxl‐binding sites are predicted in hundreds of different transcripts 16, 55, 67, but it remains unclear how many of them are directly controlled by Sxl. Yet, the presence of so many potential targets indicates that Sxl‐dependent regulation might be much more broadly employed to regulate gene expression in female flies than previously anticipated. If so, does regulation follow the same principles that were established on the known and well‐characterized targets, or are different Sxl‐mediated control mechanisms and coregulators just waiting to be identified? The next decades of research on Sxl will sure stay exciting!
Author contributions
RM, MG and JM drafted the manuscript.
Acknowledgements
We apologize to our colleagues whose work could not be cited due to space limitations. Moreover, we would like to acknowledge the following funding agencies for their financial support: POLONEZ1 (grant UMO‐2015/19/P/NZ1/02514 from the National Science Centre) to MG, Bavarian Research Network for Molecular Biosystems (BioSysNet), the German Research Foundation (DFG ME4238/1‐1 within the SFB960/2, TP B11), and the German Federal Ministry of Education and Research (BMBF, 01ZX1401D) to JM.
Edited by: Wilhelm Just
References
- 1. Cline TW (1978) Two closely linked mutations in Drosophila melanogaster that are lethal to opposite sexes and interact with daughterless. Genetics 90, 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salz HK and Erickson JW (2010) Sex determination in Drosophila: the view from the top. Fly (Austin) 4, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Graindorge A, Militti C and Gebauer F (2011) Posttranscriptional control of X‐chromosome dosage compensation. Wiley Interdiscip Rev RNA 2, 534–545. [DOI] [PubMed] [Google Scholar]
- 4. Penalva LO and Sanchez L (2003) RNA binding protein sex‐lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev 67, 343–359, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prestel M, Feller C and Becker PB (2010) Dosage compensation and the global re‐balancing of aneuploid genomes. Genome Biol 11, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salz HK (2011) Sex determination in insects: a binary decision based on alternative splicing. Curr Opin Genet Dev 21, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Slaidina M and Lehmann R (2014) Translational control in germline stem cell development. J Cell Biol 207, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venables JP, Tazi J and Juge F (2012) Regulated functional alternative splicing in Drosophila . Nucleic Acids Res 40, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bopp D, Bell LR, Cline TW and Schedl P (1991) Developmental distribution of female‐specific Sex‐lethal proteins in Drosophila melanogaster . Genes Dev 5, 403–415. [DOI] [PubMed] [Google Scholar]
- 10. Bopp D, Horabin JI, Lersch RA, Cline TW and Schedl P (1993) Expression of the Sex‐lethal gene is controlled at multiple levels during Drosophila oogenesis. Development 118, 797–812. [DOI] [PubMed] [Google Scholar]
- 11. Hinson S and Nagoshi RN (2002) The involvement of ovarian tumour in the intracellular localization of sex‐lethal protein. Insect Mol Biol 11, 241–248. [DOI] [PubMed] [Google Scholar]
- 12. Graveley BR (2002) Sex, AGility, and the regulation of alternative splicing. Cell 109, 409–412. [DOI] [PubMed] [Google Scholar]
- 13. Lopez AJ (1998) Alternative splicing of pre‐mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 14. Förch P and Valcarcel J (2003) Splicing regulation in Drosophila sex determination. Prog Mol Subcell Biol 31, 127–151. [DOI] [PubMed] [Google Scholar]
- 15. Schütt C and Nöthiger R (2000) Structure, function and evolution of sex‐determining systems in Dipteran insects. Development 127, 667–677. [DOI] [PubMed] [Google Scholar]
- 16. Robida MD, Rahn A and Singh R (2007) Genome‐wide identification of alternatively spliced mRNA targets of specific RNA‐binding proteins. PLoS One 2, e520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salz HK, Maine EM, Keyes LN, Samuels ME, Cline TW and Schedl P (1989) The Drosophila female‐specific sex‐determination gene, Sex‐lethal, has stage‐, tissue‐, and sex‐specific RNAs suggesting multiple modes of regulation. Genes Dev 3, 708–719. [DOI] [PubMed] [Google Scholar]
- 18. Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE and Ares M (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense‐mediated decay. Genes Dev 21, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lareau LF, Inada M, Green RE, Wengrod JC and Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929. [DOI] [PubMed] [Google Scholar]
- 20. Bell LR, Horabin JI, Schedl P and Cline TW (1991) Positive autoregulation of sex‐lethal by alternative splicing maintains the female determined state in Drosophila . Cell 65, 229–239. [DOI] [PubMed] [Google Scholar]
- 21. Sakamoto H, Inoue K, Higuchi I, Ono Y and Shimura Y (1992) Control of Drosophila sex‐lethal pre‐mRNA splicing by its own female‐specific product. Nucleic Acids Res 20, 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horabin JI and Schedl P (1993) Sex‐lethal autoregulation requires multiple cis‐acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol 13, 7734–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Horabin JI and Schedl P (1993) Regulated splicing of the Drosophila sex‐lethal male exon involves a blockage mechanism. Mol Cell Biol 13, 1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oliver B, Kim YJ and Baker BS (1993) Sex‐lethal, master and slave: a hierarchy of germ‐line sex determination in Drosophila . Development 119, 897–908. [DOI] [PubMed] [Google Scholar]
- 25. Albrecht EB and Salz HK (1993) The Drosophila sex determination gene snf is utilized for the establishment of the female‐specific splicing pattern of sex‐lethal. Genetics 134, 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salz HK and Flickinger TW (1996) Both loss‐of‐function and gain‐of‐function mutations in snf define a role for snRNP proteins in regulating sex‐lethal pre‐mRNA splicing in Drosophila development. Genetics 144, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuels M, Deshpande G and Schedl P (1998) Activities of the sex‐lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res 26, 2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flickinger TW and Salz HK (1994) The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev 8, 914–925. [DOI] [PubMed] [Google Scholar]
- 29. Harper DS, Fresco LD and Keene JD (1992) RNA binding specificity of a Drosophila snRNP protein that shares sequence homology with mammalian U1‐A and U2‐B” proteins. Nucleic Acids Res 20, 3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nagengast AA, Stitzinger SM, Tseng CH, Mount SM and Salz HK (2003) Sex‐lethal splicing autoregulation in vivo: interactions between SEX‐LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development 130, 463–471. [DOI] [PubMed] [Google Scholar]
- 31. Johnson ML, Nagengast AA and Salz HK (2010) PPS, a large multidomain protein, functions with sex‐lethal to regulate alternative splicing in Drosophila . PLoS Genet 6, e1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deshpande G, Samuels ME and Schedl PD (1996) Sex‐lethal interacts with splicing factors in vitro and in vivo. Mol Cell Biol 16, 5036–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaouki AS and Salz HK (2006) Drosophila SPF45: a bifunctional protein with roles in both splicing and DNA repair. PLoS Genet 2, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI and Valcarcel J (2002) Splicing regulation at the second catalytic step by Sex‐lethal involves 3′ splice site recognition by SPF45. Cell 109, 285–296. [DOI] [PubMed] [Google Scholar]
- 35. Naftelberg S, Schor IE, Ast G and Kornblihtt AR (2015) Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 84, 165–198. [DOI] [PubMed] [Google Scholar]
- 36. Granadino B, Penalva LO and Sanchez L (1996) The gene fl(2)d is needed for the sex‐specific splicing of transformer pre‐mRNA but not for double‐sex pre‐mRNA in Drosophila melanogaster . Mol Gen Genet 253, 26–31. [DOI] [PubMed] [Google Scholar]
- 37. Hilfiker A, Amrein H, Dubendorfer A, Schneiter R and Nöthiger R (1995) The gene Virilizer is required for female‐specific splicing controlled by Sxl, the master gene for sexual development in Drosophila . Development 121, 4017–4026. [DOI] [PubMed] [Google Scholar]
- 38. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS et al (2014) Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Res 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X et al (2014) A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter‐Ovanesyan D, Habib N, Cacchiarelli D et al (2014) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 8, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade‐Navarro MA, Poeck B, Helm M et al (2016) m6A modulates neuronal functions and sex determination in Drosophila . Nature 540, 242–247. [DOI] [PubMed] [Google Scholar]
- 42. Haussmann IU, Bodi Z, Sanchez‐Moran E, Mongan NP, Archer N, Fray RG and Soller M (2016) m6A potentiates Sxl alternative pre‐mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304. [DOI] [PubMed] [Google Scholar]
- 43. Yan D and Perrimon N (2015) spenito is required for sex determination in Drosophila melanogaster . Proc Natl Acad Sci USA 112, 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ortega A, Niksic M, Bachi A, Wilm M, Sanchez L, Hastie N and Valcarcel J (2003) Biochemical function of female‐lethal (2)D/Wilms’ tumor suppressor‐1‐associated proteins in alternative pre‐mRNA splicing. J Biol Chem 278, 3040–3047. [DOI] [PubMed] [Google Scholar]
- 45. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob‐Hirsch J, Amariglio N, Kupiec M et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- 46. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY et al (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- 47. Valcarcel J, Singh R, Zamore PD and Green MR (1993) The protein sex‐lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre‐mRNA. Nature 362, 171–175. [DOI] [PubMed] [Google Scholar]
- 48. Inoue K, Hoshijima K, Sakamoto H and Shimura Y (1990) Binding of the Drosophila sex‐lethal gene product to the alternative splice site of transformer primary transcript. Nature 344, 461–463. [DOI] [PubMed] [Google Scholar]
- 49. Sosnowski BA, Belote JM and McKeown M (1989) Sex‐specific alternative splicing of RNA from the transformer gene results from sequence‐dependent splice site blockage. Cell 58, 449–459. [DOI] [PubMed] [Google Scholar]
- 50. Yanowitz JL, Deshpande G, Calhoun G and Schedl PD (1999) An N‐terminal truncation uncouples the sex‐transforming and dosage compensation functions of sex‐lethal. Mol Cell Biol 19, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J and Bell LR (1994) The Sex‐lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev 8, 2072–2085. [DOI] [PubMed] [Google Scholar]
- 52. Wang J, Dong Z and Bell LR (1997) Sex‐lethal interactions with protein and RNA. Roles of glycine‐rich and RNA binding domains. J Biol Chem 272, 22227–22235. [DOI] [PubMed] [Google Scholar]
- 53. Deshpande G, Calhoun G and Schedl PD (1999) The N‐terminal domain of Sxl protein disrupts Sxl autoregulation in females and promotes female‐specific splicing of tra in males. Development 126, 2841–2853. [DOI] [PubMed] [Google Scholar]
- 54. Conrad T and Akhtar A (2012) Dosage compensation in Drosophila melanogaster: epigenetic fine‐tuning of chromosome‐wide transcription. Nat Rev Genet 13, 123–134. [DOI] [PubMed] [Google Scholar]
- 55. Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V and Kuroda MI (1995) Expression of msl‐2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila . Cell 81, 867–877. [DOI] [PubMed] [Google Scholar]
- 56. Bashaw GJ and Baker BS (1997) The regulation of the Drosophila msl‐2 gene reveals a function for sex‐lethal in translational control. Cell 89, 789–798. [DOI] [PubMed] [Google Scholar]
- 57. Gebauer F, Merendino L, Hentze MW and Valcarcel J (1998) The Drosophila splicing regulator sex‐lethal directly inhibits translation of male‐specific‐lethal 2 mRNA. RNA 4, 142–150. [PMC free article] [PubMed] [Google Scholar]
- 58. Kelley RL, Wang J, Bell L and Kuroda MI (1997) Sex lethal controls dosage compensation in Drosophila by a non‐splicing mechanism. Nature 387, 195–199. [DOI] [PubMed] [Google Scholar]
- 59. Bashaw GJ and Baker BS (1995) The msl‐2 dosage compensation gene of Drosophila encodes a putative DNA‐binding protein whose expression is sex specifically regulated by Sex‐lethal. Development 121, 3245–3258. [DOI] [PubMed] [Google Scholar]
- 60. Zhou S, Yang Y, Scott MJ, Pannuti A, Fehr KC, Eisen A, Koonin EV, Fouts DL, Wrightsman R, Manning JE et al (1995) Male‐specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex‐specific regulation and encodes a protein with a RING finger and a metallothionein‐like cysteine cluster. EMBO J 14, 2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Förch P, Merendino L, Martinez C and Valcarcel J (2001) Modulation of msl‐2 5′ splice site recognition by Sex‐lethal. RNA 7, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Merendino L, Guth S, Bilbao D, Martinez C and Valcarcel J (1999) Inhibition of msl‐2 splicing by Sex‐lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402, 838–841. [DOI] [PubMed] [Google Scholar]
- 63. Del Gatto‐Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stevenin J and Breathnach R (2000) The RNA‐binding protein TIA‐1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol 20, 6287–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Förch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P and Valcarcel J (2000) The apoptosis‐promoting factor TIA‐1 is a regulator of alternative pre‐mRNA splicing. Mol Cell 6, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 65. Gawande B, Robida MD, Rahn A and Singh R (2006) Drosophila sex‐lethal protein mediates polyadenylation switching in the female germline. EMBO J 25, 1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Graindorge A, Carre C and Gebauer F (2013) Sex‐lethal promotes nuclear retention of msl2 mRNA via interactions with the STAR protein HOW. Genes Dev 27, 1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Medenbach J, Seiler M and Hentze MW (2011) Translational control via protein‐regulated upstream open reading frames. Cell 145, 902–913. [DOI] [PubMed] [Google Scholar]
- 68. Penn JK and Schedl P (2007) The master switch gene sex‐lethal promotes female development by negatively regulating the N‐signaling pathway. Dev Cell 12, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chau J, Kulnane LS and Salz HK (2012) Sex‐lethal enables germline stem cell differentiation by down‐regulating Nanos protein levels during Drosophila oogenesis. Proc Natl Acad Sci USA 109, 9465–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sonenberg N and Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hinnebusch AG (2014) The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83, 779–812. [DOI] [PubMed] [Google Scholar]
- 72. Ingolia NT (2016) Ribosome footprint profiling of translation throughout the genome. Cell 165, 22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hinnebusch AG, Ivanov IP and Sonenberg N (2016) Translational control by 5′‐untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Beckmann K, Grskovic M, Gebauer F and Hentze MW (2005) A dual inhibitory mechanism restricts msl‐2 mRNA translation for dosage compensation in Drosophila . Cell 122, 529–540. [DOI] [PubMed] [Google Scholar]
- 75. Gebauer F, Grskovic M and Hentze MW (2003) Drosophila sex‐lethal inhibits the stable association of the 40S ribosomal subunit with msl‐2 mRNA. Mol Cell 11, 1397–1404. [DOI] [PubMed] [Google Scholar]
- 76. Abaza I, Coll O, Patalano S and Gebauer F (2006) Drosophila UNR is required for translational repression of male‐specific lethal 2 mRNA during regulation of X‐chromosome dosage compensation. Genes Dev 20, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Duncan K, Grskovic M, Strein C, Beckmann K, Niggeweg R, Abaza I, Gebauer F, Wilm M and Hentze MW (2006) Sex‐lethal imparts a sex‐specific function to UNR by recruiting it to the msl‐2 mRNA 3′ UTR: translational repression for dosage compensation. Genes Dev 20, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gebauer F, Corona DF, Preiss T, Becker PB and Hentze MW (1999) Translational control of dosage compensation in Drosophila by sex‐lethal: cooperative silencing via the 5′ and 3′ UTRs of msl‐2 mRNA is independent of the poly(A) tail. EMBO J 18, 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mihailovic M, Wurth L, Zambelli F, Abaza I, Militti C, Mancuso FM, Roma G, Pavesi G and Gebauer F (2012) Widespread generation of alternative UTRs contributes to sex‐specific RNA binding by UNR. RNA 18, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grskovic M, Hentze MW and Gebauer F (2003) A co‐repressor assembly nucleated by sex‐lethal in the 3′UTR mediates translational control of Drosophila msl‐2 mRNA. EMBO J 22, 5571–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Abaza I and Gebauer F (2008) Functional domains of Drosophila UNR in translational control. RNA 14, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y and Yokoyama S (1999) Structural basis for recognition of the tra mRNA precursor by the sex‐lethal protein. Nature 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 83. Hennig J, Militti C, Popowicz GM, Wang I, Sonntag M, Geerlof A, Gabel F, Gebauer F and Sattler M (2014) Structural basis for the assembly of the Sxl‐Unr translation regulatory complex. Nature 515, 287–290. [DOI] [PubMed] [Google Scholar]
- 84. Duncan KE, Strein C and Hentze MW (2009) The SXL‐UNR corepressor complex uses a PABP‐mediated mechanism to inhibit ribosome recruitment to msl‐2 mRNA. Mol Cell 36, 571–582. [DOI] [PubMed] [Google Scholar]
- 85. Chau J, Kulnane LS and Salz HK (2009) Sex‐lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics 182, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schüpbach T (1985) Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex‐lethal in Drosophila melanogaster . Genetics 109, 529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hashiyama K, Hayashi Y and Kobayashi S (2011) Drosophila sex lethal gene initiates female development in germline progenitors. Science 333, 885–888. [DOI] [PubMed] [Google Scholar]
- 88. Salz HK, Cline TW and Schedl P (1987) Functional changes associated with structural alterations induced by mobilization of a P element inserted in the sex‐lethal gene of Drosophila . Genetics 117, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Harris RE, Pargett M, Sutcliffe C, Umulis D and Ashe HL (2011) Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell 20, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Joly W, Chartier A, Rojas‐Rios P, Busseau I and Simonelig M (2013) The CCR4 deadenylase acts with Nanos and Pumilio in the fine‐tuning of Mei‐P26 expression to promote germline stem cell self‐renewal. Stem Cell Reports 1, 411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Deshpande G, Calhoun G, Yanowitz JL and Schedl PD (1999) Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99, 271–281. [DOI] [PubMed] [Google Scholar]
- 92. Forbes A and Lehmann R (1998) Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690. [DOI] [PubMed] [Google Scholar]
- 93. Li Y, Minor NT, Park JK, McKearin DM and Maines JZ (2009) Bam and Bgcn antagonize Nanos‐dependent germ‐line stem cell maintenance. Proc Natl Acad Sci USA 106, 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li Y, Zhang Q, Carreira‐Rosario A, Maines JZ, McKearin DM and Buszczak M (2013) Mei‐p26 cooperates with Bam, Bgcn and Sxl to promote early germline development in the Drosophila ovary. PLoS One 8, e58301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Page SL, McKim KS, Deneen B, Van Hook TL and Hawley RS (2000) Genetic studies of mei‐P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila . Genetics 155, 1757–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM and Knoblich JA (2008) Mei‐P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ohlstein B and McKearin D (1997) Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development 124, 3651–3662. [DOI] [PubMed] [Google Scholar]
- 98. McKearin D and Ohlstein B (1995) A role for the Drosophila bag‐of‐marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121, 2937–2947. [DOI] [PubMed] [Google Scholar]
- 99. McKearin DM and Spradling AC (1990) bag‐of‐marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev 4, 2242–2251. [DOI] [PubMed] [Google Scholar]
- 100. Ohlstein B, Lavoie CA, Vef O, Gateff E and McKearin DM (2000) The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH‐box proteins and interacts genetically with bag‐of‐marbles. Genetics 155, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wei G, Oliver B, Pauli D and Mahowald AP (1994) Evidence for sex transformation of germline cells in ovarian tumor mutants of Drosophila . Dev Biol 161, 318–320. [DOI] [PubMed] [Google Scholar]
- 102. Fu Z, Geng C, Wang H, Yang Z, Weng C, Li H, Deng L, Liu L, Liu N, Ni J et al (2015) Twin promotes the maintenance and differentiation of germline stem cell lineage through modulation of multiple pathways. Cell Rep 13, 1366–1379. [DOI] [PubMed] [Google Scholar]
- 103. Lasko P (2011) Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip Rev RNA 2, 408–416. [DOI] [PubMed] [Google Scholar]
- 104. Andrews S, Snowflack DR, Clark IE and Gavis ER (2011) Multiple mechanisms collaborate to repress nanos translation in the Drosophila ovary and embryo. RNA 17, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Gebauer F and Hentze MW (2007) Studying translational control in Drosophila cell‐free systems. Methods Enzymol 429, 23–33. [DOI] [PubMed] [Google Scholar]
- 106. Cline TW and Meyer BJ (1996) Vive la difference: males vs females in flies vs worms. Annu Rev Genet 30, 637–702. [DOI] [PubMed] [Google Scholar]
- 107. Ehebauer M, Hayward P and Martinez‐Arias A (2006) Notch signaling pathway. Sci STKE 2006, cm7. [DOI] [PubMed] [Google Scholar]
- 108. Suissa Y, Kalifa Y, Dinur T, Graham P, Deshpande G, Schedl P and Gerlitz O (2010) Hrp48 attenuates Sxl expression to allow for proper notch expression and signaling in wing development. Proc Natl Acad Sci USA 107, 6930–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hilfiker A, Yang Y, Hayes DH, Beard CA, Manning JE and Lucchesi JC (1994) Dosage compensation in Drosophila: the X‐chromosomal binding of MSL‐1 and MLE is dependent on Sxl activity. EMBO J 13, 3542–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Suissa Y, Ordan E, Deshpande G and Gerlitz O (2011) Males and females: creating differences while maintaining the similarities. Fly (Austin) 5, 25–28. [DOI] [PubMed] [Google Scholar]
- 111. Singh R, Valcarcel J and Green MR (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract‐binding proteins. Science 268, 1173–1176. [DOI] [PubMed] [Google Scholar]
- 112. Sakashita E and Sakamoto H (1994) Characterization of RNA binding specificity of the Drosophila sex‐lethal protein by in vitro ligand selection. Nucleic Acids Res 22, 4082–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Singh R, Banerjee H and Green MR (2000) Differential recognition of the polypyrimidine‐tract by the general splicing factor U2AF65 and the splicing repressor sex‐lethal. RNA 6, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hediger M, Henggeler C, Meier N, Perez R, Saccone G and Bopp D (2010) Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex‐determining pathway in the housefly. Genetics 184, 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Verhulst EC, Beukeboom LW and van de Zande L (2010) Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328, 620–623. [DOI] [PubMed] [Google Scholar]
- 116. Pane A, Salvemini M, Delli Bovi P, Polito C and Saccone G (2002) The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129, 3715–3725. [DOI] [PubMed] [Google Scholar]
- 117. Cline TW, Dorsett M, Sun S, Harrison MM, Dines J, Sefton L and Megna L (2010) Evolution of the Drosophila feminizing switch gene sex‐lethal. Genetics 186, 1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Traut W, Niimi T, Ikeo K and Sahara K (2006) Phylogeny of the sex‐determining gene sex‐lethal in insects. Genome 49, 254–262. [DOI] [PubMed] [Google Scholar]
- 119. Ayres JS, Freitag N and Schneider DS (2008) Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178, 1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Colombrita C, Silani V and Ratti A (2013) ELAV proteins along evolution: back to the nucleus? Mol Cell Neurosci 56, 447–455. [DOI] [PubMed] [Google Scholar]
- 121. Hinman MN and Lou H (2008) Diverse molecular functions of Hu proteins. Cell Mol Life Sci 65, 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Simone LE and Keene JD (2013) Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev 23, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Samson ML (1998) Evidence for 3′ untranslated region‐dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA‐binding protein ELAV. Genetics 150, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Borgeson CD and Samson ML (2005) Shared RNA‐binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res 33, 6372–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Samson ML and Chalvet F (2003) Found in neurons, a third member of the Drosophila elav gene family, encodes a neuronal protein and interacts with elav. Mech Dev 120, 373–383. [DOI] [PubMed] [Google Scholar]
- 126. Zhu H, Hasman RA, Barron VA, Luo G and Lou H (2006) A nuclear function of Hu proteins as neuron‐specific alternative RNA processing regulators. Mol Biol Cell 17, 5105–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhu H, Hinman MN, Hasman RA, Mehta P and Lou H (2008) Regulation of neuron‐specific alternative splicing of neurofibromatosis type 1 pre‐mRNA. Mol Cell Biol 28, 1240–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhu H, Zhou HL, Hasman RA and Lou H (2007) Hu proteins regulate polyadenylation by blocking sites containing U‐rich sequences. J Biol Chem 282, 2203–2210. [DOI] [PubMed] [Google Scholar]
- 129. Soller M and White K (2003) ELAV inhibits 3′‐end processing to promote neural splicing of ewg pre‐mRNA. Genes Dev 17, 2526–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zaharieva E, Haussmann IU, Brauer U and Soller M (2015) Concentration and localization of coexpressed ELAV/Hu proteins control specificity of mRNA processing. Mol Cell Biol 35, 3104–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]


