Abstract
The genus Streptocarpus comprises species with diverse body plans. Caulescent species produce leaves from a conventional shoot apical meristem (SAM), whereas acaulescent species lack a conventional SAM and produce only a single leaf (the unifoliate form) or clusters of leaves from the base of more mature leaves (the rosulate form). These distinct morphologies reflect fundamental differences in the role of the SAM and the process of leaf specification. A subfamily of KNOTTED-like homeobox (KNOX) genes are known to be important in regulating meristem function and leaf development in model species with conventional morphologies. To test the involvement of KNOX genes in Streptocarpus evolution, two parologous KNOX genes (SSTM1 and SSTM2) were isolated from species with different growth forms. Their phylogenetic analysis suggested a gene duplication before the subgeneric split of Streptocarpus and resolved species relationships, supporting multiple evolutionary origins of the rosulate and unifoliate morphologies. In S. saxorum, a caulescent species with a conventional SAM, KNOX proteins were expressed in the SAM and transiently downregulated in incipient leaf primordia. The ability of acaulescent species to initiate leaves from existing leaves was found to correlate with SSTM1 expression and KNOX protein accumulation in leaves and to reflect genetic differences at two loci. Neither locus corresponded to SSTM1, suggesting that cis-acting differences in SSTM1 regulation were not responsible for evolution of the rosulate and unifoliate forms. However, the involvement of KNOX proteins in leaf formation in rosulate species suggests that they have played an indirect role in the development of morphological diversity in Streptocarpus.
INTRODUCTION
Morphological variation is a prerequisite of evolution, yet its underlying genetic basis is poorly understood. Related species are often distinguished by relatively minor variations in organ size and form. In Drosophila, subtle morphological differences have been attributed to changes in expression of regulatory transcription factor genes (e.g., Stern, 1998; Sucena et al., 2003). Variation in the activity of transcription factor genes also underlies some of the artificially selected morphological changes that accompanied the domestication of maize (Zea mays) (Doebley et al., 1998; Doebley and Lukens, 1998; Wang et al., 1999). In contrast with closely related species, higher order taxa tend to differ more dramatically in body plan, for example, in the number, identity, or relative positions of organs. Such evolutionary innovations may have also involved transcription factor genes because the changes sometimes correlate with altered transcription factor expression and can be mimicked by mutations in transcription factor genes in both plants and animals (Doebley and Lukens, 1998; Carroll, 2000).
One genus of plants that encompasses species with different body plans is Streptocarpus, the Cape primrose. Streptocarpus consists of ∼140 African and Madagascan species that fall into two main clades on the basis of internal transcribed spacer (ITS) sequence phylogenies and chromosome numbers, and are broadly consistent with classical taxonomic subdivisions (Hilliard and Burtt, 1971; Möller and Cronk, 2001). Clade I consists of caulescent species that form stems and leaves conventionally from a shoot apical meristem (SAM; Figure 1B). Clade I corresponds to subgenus Streptocarpella with the genus Saintpaulia (African violets) nested within it (Möller and Cronk, 1997). Like most Gesneriaceae, caulescent Streptocarpus species favor warm, humid, and aseasonal habitats (Burtt, 1998). Clade II species, corresponding largely to subgenus Streptocarpus, lack a vegetative SAM and therefore have no erect vegetative stem. Some acaulescent species, termed unifoliate, have a single leafy organ (a phyllomorph) that arises by continued growth of one cotyledon after germination (Figure 1A; Jong, 1970, 1978; Jong and Burtt, 1975; Burtt, 1994). In other species, additional phyllomorphs are produced from existing phyllomorphs without involving a conventional SAM. Such plants are rosette like and are called rosulate (Figure 1C). Acaulescent species are able to abscise the distal part of the phyllomorph under drought conditions and hence colonize drier and more seasonal habitats than caulescent species (Burtt, 1998). Although lacking a conventional vegetative SAM, all acaulescent species form inflorescence meristems that arise from phyllomorph midribs and produce bracts and flowers conventionally (Figures 1A and 1C). Whereas unifoliate species are monocarpic and die after flowering, rosulate plants are perennial.
Figure 1.
Streptocarpus Growth Forms.
(A) Unifoliate S. dunnii.
(B) Caulescent S. saxorum.
(C) Rosulate S. rexii.
(D) S. dunnii (left) and S. rexii (right). The S. dunnii × S. rexii F1 hybrid (center) has a rosulate arrangement of multiple, large phyllomorphs.
Species with different growth forms within Clade II can form fertile hybrids. In crosses between rosulate and unifoliate species, inheritance of the rosulate character was suggested to involve dominant alleles at two loci (Oehlkers, 1938, 1942, 1964). In some acaulescent species, application of gibberellic acid (GA) resulted in either the formation of extra phyllomorphs or induction of a vegetative shoot (Rosenblum and Basile, 1984).
Studies in model species have demonstrated the importance of Class I KNOTTED-like homeobox (KNOX) genes in SAM function and leaf development. Several of the effects of altered KNOX activity resemble the morphological differences among Streptocarpus species. For example, Arabidopsis thaliana mutants lacking activity of the KNOX gene SHOOT MERISTEMLESS (STM) do not form a SAM during embryogenesis (Barton and Poethig, 1993), as in acaulescent Streptocarpus species. In weaker stm mutants, leaves are formed from the base of cotyledons or existing leaves, apparently without the involvement of a SAM (Clark et al., 1996), resembling rosulate acaulescent Streptocarpus. Gain-of-function KNOX mutations that cause misexpression in leaves can have the opposite effect of conferring meristematic properties on the leaf. These properties are manifest as ectopic leaf outgrowths, and occasionally, SAMs (reviewed in Tsiantis and Hay, 2003), and therefore mimic the morphologies of acaulescent Streptocarpus. A further suggestion that altered KNOX activity might be involved in the novel morphology of Streptocarpus is that KNOX activity is able to repress GA biosynthesis (Sakamoto et al., 2001; Hay et al., 2002), and unifoliate Streptocarpus can phenocopy caulescent Streptocarpus when they are treated with GA, suggesting that acaulescent morphology might involve reduction of GA levels by KNOX activity (Rosenblum and Basile, 1984).
To further investigate the role of KNOX activity in Streptocarpus evolution, we isolated paralogous STM-like genes, SSTM1 and SSTM2, from a range of Streptocarpus species. We found SSTM1 expression in leaves of rosulate but not unifoliate species, suggesting that changes in SSTM1 expression have been involved in evolution of novel morphologies. We were able to attribute rosulate and unifoliate morphologies to genetic differences at two loci, although neither locus was found to correspond to SSTM1. This suggested that mutations in SSTM1 itself have not been responsible for altered gene expression or morphology.
RESULTS
Isolation of SSTM1 Genes
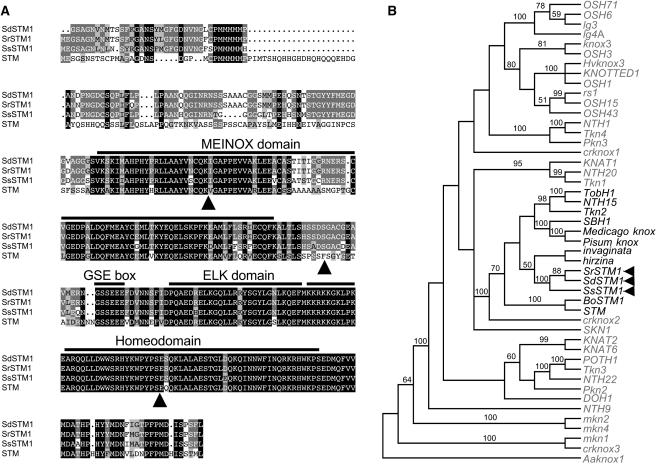
To investigate the role of KNOX genes in morphological evolution, we isolated cDNA sequences of STM orthologs from three Streptocarpus species representing the main growth forms: caulescent S. saxorum, unifoliate S. dunnii, and rosulate S. rexii. Primers were designed to exon sequences conserved between STM and its orthologs from other species and used to amplify Streptocarpus STM-like cDNA sequences by rapid amplification of cDNA ends (RACE). Alignment of the inferred amino acid sequences of the Streptocarpus products (SSTM1 genes) with other KNOX proteins showed that SSTM1 had all typical features of a Class I KNOX gene, including regions encoding the MEINOX, ELK, and homeodomains (Figure 2A; Bürglin, 1997, 1998). Phylogenetic comparison of the SSTM1 sequences with those of other KNOX genes placed them together in a well-supported clade with STM but no other Arabidopsis gene (Figure 2B). The SSTM1 genes were therefore likely to be orthologs of Arabidopsis STM.
Figure 2.
Structure and Phylogeny of SSTM1 Genes.
(A) Alignment of inferred SSTM1 protein sequences from S. dunnii (SdSTM1), S. rexii (SrSTM1), and S. saxorum (SsSTM1) to Arabidopsis STM, showing shared conserved domains and intron positions (arrowheads). Black boxes highlight invariant amino acids, dark gray boxes amino acids conserved between Streptocarpus proteins, and light gray boxes conservative substitutions. Sequences were aligned using ClustalW.
(B) Strict consensus of the four most parsimonious KNOX trees using homeobox cDNA sequences. Dicot STM-like genes are highlighted in black and Streptocarpus genes with arrowheads. Aaknox1, mkn1, and Crknox3 were included as outgroups and trees rooted on the algal sequence, Aaknox1. Bootstrap values are given above branches.
Streptocarpus Contains Two STM-Like Genes That Are Part of a Small Gene Family
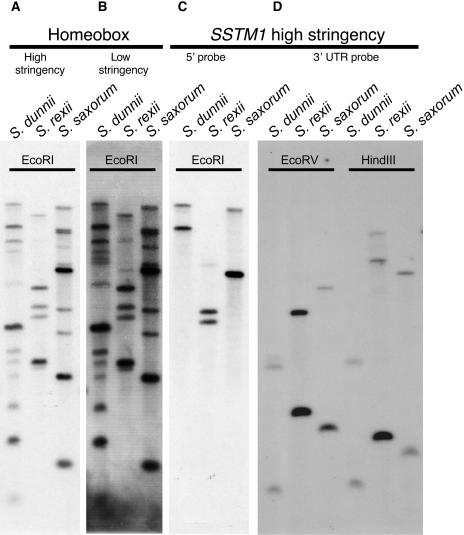
DNA gel blot hybridization was used to determine the number of STM-like KNOX genes in Streptocarpus and to estimate the size of the Streptocarpus KNOX gene family. Genomic DNA from S. dunnii, S. rexii, and S. saxorum was digested with different restriction enzymes and probed either with the highly conserved SSTM1 homeobox sequence (which is potentially able to detect all KNOX genes) or with fragments from the poorly conserved 5′ and 3′ regions of SSTM1 to detect only STM-like genes.
High stringency hybridization with the homeobox probe detected two to three strongly hybridizing fragments in each species and 4 to 10 additional fragments that hybridized weakly (Figure 3A). At low stringency, two additional fragments were detected in S. dunnii and S. rexii (Figure 3B). This suggested that the KNOX gene family comprises 6 to 14 members in Streptocarpus. High stringency hybridization with either the 5′ region of SSTM1 or its 3′ region detected a minimum of two fragments in each species, supporting the existence of two STM-like genes in Streptocarpus (Figures 3C and 3D).
Figure 3.
DNA Gel Blot Analysis of Streptocarpus KNOX Genes.
DNA gel blots of genomic DNA were probed at both high and low stringency with an SSTM1 fragment that includes the homeobox (left) or at high stringency with the poorly conserved 5′ region of SSTM1 or its 3′ untranslated region (right).
Streptocarpus STM1 Genes Are Orthologous
To confirm that Streptocarpus STM1 genes were likely orthologs of each other, we did a phylogenetic analysis. STM-like sequences were amplified from the genomic DNA of 26 species representing the variation within the genus. The amplified region contained parts of the last two exons, encoding the highly conserved homeodomain, and the divergent intervening third intron.
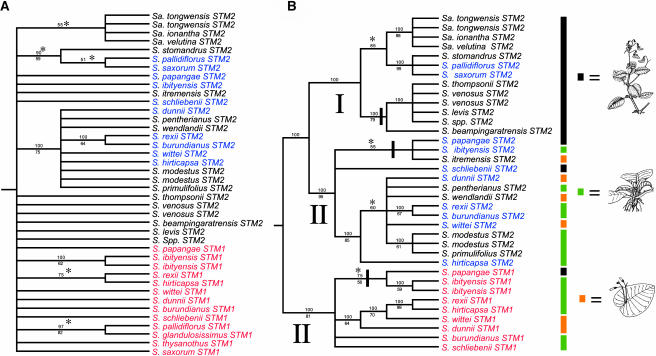
More than one sequence was amplified from many species, supporting the presence of two STM-like genes. Parsimony analysis of the highly conserved exon sequences did not fully resolve gene relationships (Figure 4A). Intron sequences from the SSTM1 genes of three species—S. saxorum, S. pallidiflorus, and S. glandulosissimus—could not be included in the intron phylogeny because they contained direct imperfect repeats of ∼100 nucleotides (see Supplemental Figure 1A online). However, analysis of aligned intron sequences from remaining species placed the genes into two clades (Figure 4B). This suggests that STM-like genes duplicated before the subgeneric split between Clades I and II Streptocarpus. One gene clade was identified as SSTM1 (on the basis of exon sequence shared with cDNAs) and the second designated SSTM2.
Figure 4.
SSTM Phylogenies for Streptocarpus.
(A) Relationships between SSTM exon sequences flanking intron 3 derived from a midpoint-rooted majority rule consensus of 2480 most parsimonious trees. Exon sequences for SSTM1 genes with repetitive introns are included. Percentage of the majority is given above branches with bootstrap support below. Branches that collapse in the strict consensus are indicated with an asterisk. Taxa represented more than once are highlighted in red and blue.
(B) Phylogeny of SSTM third intron shown as a midpoint rooted majority rule consensus of 9002 most parsimonious trees. For each paralogue (SSTM1 or SSTM2) Clade I (I) and Clade II (II) species are grouped. Clades of Madagascan species are indicated by black bars across their bases. Morphological types are indicated at the right (illustrations redrawn from Hilliard and Burtt [1971] and Jong [1978]).
We also investigated the phylogenetic utility of the SSTM third intron by comparing our SSTM phylogeny with a phylogeny of published ITS sequences (see Supplemental Figure 1B online). This showed that the SSTM genes provide novel, low copy number markers for determining species level phylogeny in Streptocarpus that are likely to be useful in other plant groups.
KNOX Expression in Streptocarpus
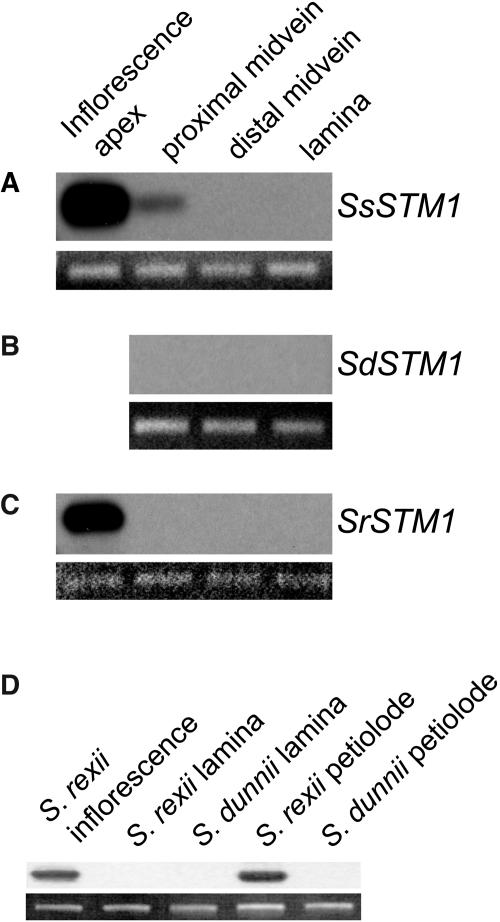
To determine whether changes in KNOX activity may have been involved in the evolution of Streptocarpus, we compared SSTM1 expression patterns by reverse transcription and PCR in different growth forms. In caulescent S. saxorum, expression was detected in apical tissues containing vegetative SAMs or inflorescence meristems and at a low level in the proximal leaf petiole but not in other parts of the leaf (Figure 5A). In unifoliate S. dunnii, no SSTM1 expression was detected in leaves (plants had no inflorescences) or the petiolode (Figures 5B and 5D), although transcripts were detected in inflorescence apices using the same method (data not shown). In the rosulate S. rexii, SSTM1 was expressed in the inflorescence apex but not in the leaf or distal midrib. However, in contrast with unifoliate S. dunnii, SSTM1 expression was also detected in the S. rexii petiolode in the region from which further phyllomorphs were initiating (Figures 5C and 5D).
Figure 5.
SSTM1 Expression Patterns.
(A) to (C) The expression of SSTM1 in inflorescence apices, proximal and distal midvein, and leaf lamina was compared by RT-PCR with a ubiquitously expressed 40S ribosomal subunit gene as a control.
(A) Caulescent S. saxorum.
(B) Unifoliate S. dunnii.
(C) Rosulate S. rexii.
(D) SSTM1 expression was detected under the same conditions in the organogenic petiolode region of rosulate S. rexii but not in the petiolode of unifoliate S. dunnii.
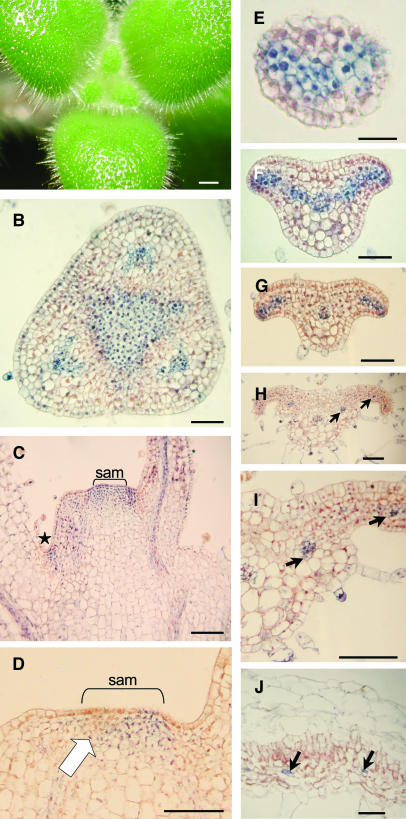
To further locate the sites of KNOX expression, tissue sections were challenged with antibodies raised against STM and able to recognize STM-like KNOX proteins in Arabidopsis (Lynn et al., 1999). The caulescent S. saxorum produces alternate whorls of three leaves from a flat SAM (Figure 6A). KNOX protein was detected in the SAM, visible as a triangular domain between the bases of existing leaf primordia in a transverse section (Figure 6B). In longitudinal sections, protein accumulation was seen in the SAM, axillary meristems, and provasculature extending into leaves but not at the sites of leaf initiation in the SAM (Figure 6D). Signal was detected in leaves at later developmental stages (e.g., Figure 6C), suggesting that the absence of KNOX protein from the site of leaf initiation is transient. In newly initiated leaves, KNOX protein accumulated in two to three layers of cells abaxial to the prospective palisade mesophyll and persisted in these cells at the growing leaf margins and developing vasculature (Figures 6E to 6G) before becoming restricted to the vasculature (Figures 6H to 6J).
Figure 6.
KNOX Immunolocalization in S. saxorum.
(A) A vegetative apex of S. saxorum showing leaves in whorls of three. New leaves are initiated directly opposite a leaf in the preceding whorl (bar = 1 mm).
(B) Transverse section of apex. Nuclear-localized KNOX proteins can be seen in the central, triangular SAM and in leaf veins (bar = 50 μm).
(C) and (D) Longitudinal sections of different S. saxorum apices, showing staining in the central SAMs, SAMs in the axils of leaves (star), and in developing veins. An unstained region was detected at the site of leaf initiation from the SAM (arrow in [D]). Bar = 100 μm.
(E) to (J) Transverse sections through leaves at increasing stages of maturity. Arrows indicate vascular localization of KNOX proteins. Bars = 25 μm in (E), 50 μm in (F) and (G), and 100 μm in (H) to (J).
Unifoliate acaulescent species lack a conspicuous meristem until inflorescences arise from base of the midvein. We were unable to detect SdSTM1 expression in plants without inflorescences (Figure 5B). We were also unable to detect KNOX proteins in juvenile S. dunnii plants (Figure 7C). Once plants started to make inflorescences, we detected KNOX proteins in the meristem (Figures 7A to 7D).
Figure 7.
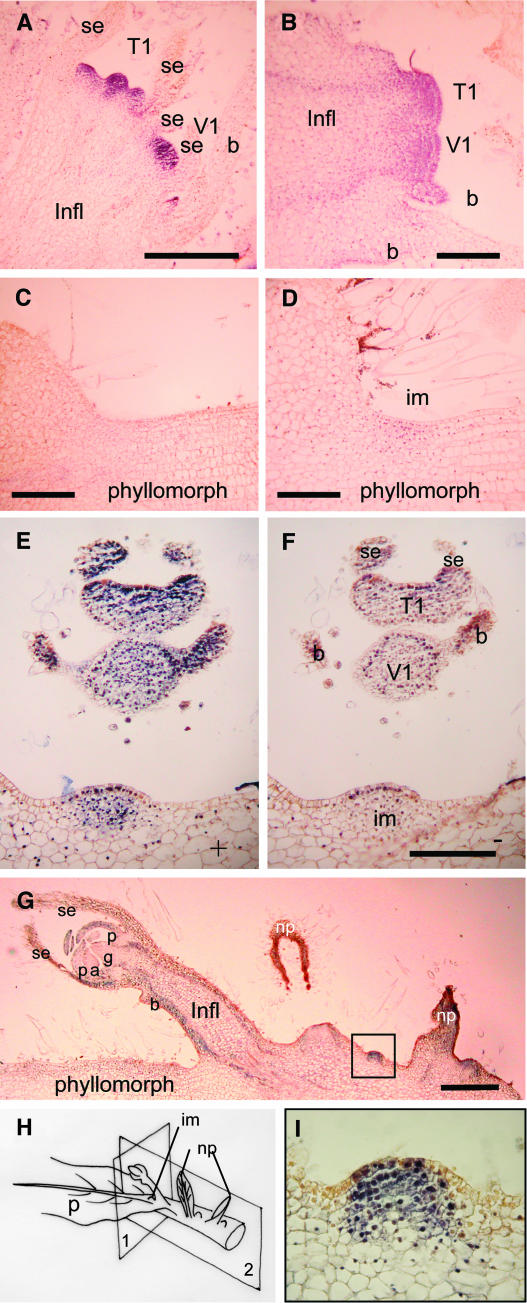
KNOX Immunolocalization in Acaulescent Species.
The inflorescence meristems of S. rexii and S. dunnii are formed in an acropetal sequence from the groove meristem at the proximal midrib. Each inflorescence meristem (im) gives rise to lateral bracts (b), a terminal older flower (T1) proximally, and a younger distal flower (V1) or repetitions of that unit (nomenclature according to Weber [1982]). se, sepal; p, petal; a, anther; g, gynoecium; np, new phyllomorph; Infl, inflorescence. Bars = 0.5 mm.
(A) to (D) S. dunnii.
(A) Two floral meristems in longitudinal section.
(B) A pair of floral meristems at an earlier stage in development, subtended by a bract.
(C) Longitudinal section through a juvenile phyllomorph in which we were unable to detect KNOX expression.
(D) Longitudinal section through a juvenile phyllomorph showing localized KNOX expression at its base.
(E) to (H) S. rexii. New phyllomorphs arise in two ranks from the more proximal petiolode.
(E) KNOX localization in a transverse section of an inflorescence, corresponding to plane 1 in (H), and new inflorescence meristem using antibodies raised against Arabidopsis STM.
(F) An adjacent section to (E) probed only with secondary antibody (bar = 0.1 mm).
(G) A longitudinal section through a flowering phyllomorph corresponding to plane 2 in (H).
(I) The inset region of (G) at higher magnification showing KNOX expression in a mound of cells at the prospective site of phyllomorph initiation.
Rosulate species also lack a shoot, and new phyllomorphs are formed from the base of a mature phyllomorph on the petiolode, characterized by its cylindrical shape. Inflorescence meristems arise from the more distal laminar phyllomorph midrib. Because SSTM1 was expressed in the petiolode of S. rexii, we examined KNOX protein expression in this region to determine whether phyllomorphs are initiated from regions with SAM-like properties. As expected, the STM antibody was able to detect KNOX protein in inflorescence meristems of S. rexii (Figures 7E and 7F). However, KNOX proteins were also observed in mounds of cells in the more proximal petiolode at a position from which new phyllomorphs would initiate (Figures 7G to 7I). The mounds of KNOX-expressing cells therefore resemble transient meristems that, on the basis of their positions, could each give rise to a single phyllomorph.
Two Loci Determine the Differences in Form in Clade II Streptocarpus
To examine the genetic basis for the differences in form in Clade II Streptocarpus, we crossed the rosulate S. rexii with two unifoliate species: S. dunnii or S. wittei. F1 hybrids, which were rosulate in form (Figure 1D), were backcrossed to the unifoliate parent. The forms of the S. wittei backcross progeny (n = 112) were scored in juvenile and adult plants, and the S. dunnii backcross progeny (n = 16) were scored only at maturity (Table 1; see supplemental data online). Plants with one major phyllomorph were classed as unifoliate and those with more than one as rosulate. Previous studies had suggested that the rosulate character was determined by a dominant allele at either of two loci: one promoting the rosulate character early in development, the other acting later (Oehlkers, 1938, 1942, 1964). If this were the case, the F1 plants should have been heterozygous at both loci and therefore rosulate. Approximately half their backcross progeny should have inherited the early acting dominant allele and have developed the rosulate character early. Only 39% of the S. wittei backcross progeny (44/112) were scored as rosulate while juvenile, providing no support for the action of an early-acting dominant allele (P = 0.023 in a χ2 test). If dominant alleles of two unlinked loci were responsible for the rosulate morphology at maturity, ∼75% of the backcross progeny should have inherited at least one dominant allele sufficient to specify rosulate morphology. At maturity, 76% (98/128) of the S. wittei backcross progeny had rosulate morphology, consistent with the action of dominant alleles at two loci that come into effect gradually during development (Table 1; see supplemental data online). The frequency of rosulate plants in the small S. dunnii backcross population was also consistent with the action of two loci.
Table 1.
Data from Backcross Plants
| Backcross Family Number and RBGE Accession Number | Qualifier | Genotype | Form during Juvenile Development | Form at Flowering | Summary of Data for Each Family during Juvenile Development | Summary of Data for Each Family at Flowering |
|---|---|---|---|---|---|---|
| Family 1 (19980107) | A | d/d | – | R | ||
| C | d/r | – | R | |||
| E | d/d | – | R | |||
| F | d/r | – | R | |||
| G | d/d | – | R | |||
| H | d/d | – | U | |||
| J | d/r | – | R | |||
| K | d/r | – | U | |||
| M | d/r | – | R | |||
| P | d/r | – | R | |||
| Q | d/d | – | R | |||
| R | d/r | – | R | |||
| S | d/d | – | R | |||
| T | d/r | – | R | |||
| U | d/d | – | U | |||
| V | d/d | – | U | – | 12R 4U | |
| Family 2 (19982603) | B | w/r | R | R | ||
| C | w/w | U | R | |||
| D | w/w | U | R | |||
| E | w/w | U | R | |||
| F | w/r | U | R | |||
| G | w/r | U | R | |||
| H | w/w | R | R | |||
| I | w/r | R | R | 3R 5U | 8R 0U | |
| Family 3 (19982606) | B | w/r | U | R | ||
| C | w/r | R | R | |||
| D | w/w | R | R | |||
| F | w/r | U | R | |||
| H | w/r | U | R | |||
| I | w/w | R | R | |||
| J | w/r | U | U | |||
| K | w/w | U | R | |||
| L | w/w | U | R | |||
| M | w/w | U | R | 3R 7U | 9R 1U | |
| Family 4 (19982607) | A | w/w | U | R | ||
| B | w/w | U | U | |||
| C | w/r | R | R | |||
| D | w/w | U | R | |||
| E | w/r | R | R | |||
| F | w/r | U | R | |||
| G | w/r | U | U | |||
| H | w/r | R | R | |||
| I | w/w | U | U | |||
| J | w/r | U | U | |||
| K | w/r | U | R | 3R 8U | 7R 4U | |
| Family 5 (19982610) | B | w/w | U | U | ||
| D | w/w | R | R | |||
| E | w/r | U | R | |||
| F | w/r | U | R | |||
| G | w/w | U | R | |||
| J | w/w | R | R | |||
| K | w/w | R | R | |||
| N | w/r | U | R | |||
| O | w/r | R | R | |||
| S | w/r | R | R | 5R 5U | 9R 1U | |
| Family 7 (19981619) | A | w/r | U | R | ||
| D | w/w | R | R | |||
| E | w/r | U | U | |||
| F | w/r | U | U | |||
| H | w/w | U | R | |||
| J | w/w | R | U | |||
| K | w/w | U | R | 2R 5U | 4R 3U | |
| Family 8 (19982620) | A | w/r | R | R | ||
| C | w/w | R | R | |||
| D | w/w | R | R | |||
| E | w/w | R | R | |||
| F | w/w | U | R | |||
| G | w/r | U | R | |||
| H | w/r | R | R | |||
| J | w/r | U | R | |||
| M | w/w | U | U | |||
| N | w/r | U | U | |||
| O | w/r | R | R | |||
| Q | w/r | R | R | 7R 5U | 10R 2U | |
| Family 9 (19982621) | A | w/r | U | U | ||
| B | w/r | R | R | |||
| C | w/r | U | U | |||
| D | w/r | R | R | |||
| F | w/r | U | R | |||
| G | w/w | U | R | |||
| H | w/r | U | U | |||
| I | w/w | R | R | |||
| J | w/r | U | R | |||
| L | w/r | R | R | |||
| M | w/w | R | R | |||
| N | w/w | R | R | |||
| P | w/w | R | R | 7R 6U | 10R 3U | |
| Family 10 (19982627) | A | w/w | U | R | ||
| B | w/r | U | R | |||
| C | w/r | R | R | |||
| D | w/r | U | U | |||
| E | w/r | U | R | |||
| F | w/w | R | R | |||
| H | w/w | U | R | |||
| I | w/r | U | R | |||
| J | w/r | U | U | |||
| K | w/w | R | U | |||
| L | w/r | R | R | |||
| M | w/r | R | U | |||
| N | w/w | R | R | |||
| O | w/w | U | U | |||
| P | w/r | U | R | |||
| Q | w/r | U | R | |||
| R | w/w | R | R | |||
| S | w/w | U | U | 7R 11U | 12R 6U | |
| Family 11 (19982623) | A | w/w | U | R | ||
| B | w/w | U | R | |||
| C | w/r | U | U | |||
| D | w/r | R | R | |||
| E | w/r | U | R | |||
| F | w/r | U | R | |||
| G | w/w | R | R | |||
| H | w/w | U | U | |||
| I | w/w | U | R | |||
| J | w/w | R | R | |||
| K | w/w | U | R | |||
| L | w/r | R | R | 4R 8U | 10R 2U | |
| Family 12 (19982624) | A | w/r | U | U | ||
| B | w/r | U | R | |||
| C | w/w | U | U | |||
| D | w/w | R | R | |||
| E | w/w | U | R | |||
| G | w/w | U | U | |||
| I | w/w | U | R | |||
| J | w/r | R | R | |||
| K | w/w | U | U | |||
| L | w/w | U | R | |||
| M | w/r | R | R | 3R 8U | 7R 4U |
Genotypes of plants are represented as d/w (the SSTM1 allele from the unifoliate recessive parent S. dunnii) and r (the SSTM1 allele from the rosulate parent S. wittei). U denotes plants with unifoliate form, R denotes plants with rosulate form. RBGE, Royal Botanic Garden Edinburgh.
SSTM1 Does Not Specify Rosulate Morphology
To determine whether SSTM1 is one of the two loci directly responsible for morphological differences between unifoliate and rosulate species, and also whether SSTM1 expression is cis-regulated, we followed segregation of SSTM1 alleles in the two backcross populations.
If dominant alleles at two loci, R1 and R2, determine rosulate morphology, the rosulate S. rexii parent will have been homozygous R1/R1 R2/R2 and the S. dunnii or S. wittei parent homozygous for the recessive alleles r1 and r2. The F1 S. rexii × unifoliate hybrids will have been heterozygous at both loci and their backcross progeny composed of four different genotypes (R1/r1 R2/r2, R1/r1 r2/r2, r1/r1 R2/r2, or r1/r1 r2/r2) in equal proportions. Rosulate progeny would carry at least one dominant R1 or R2 allele.
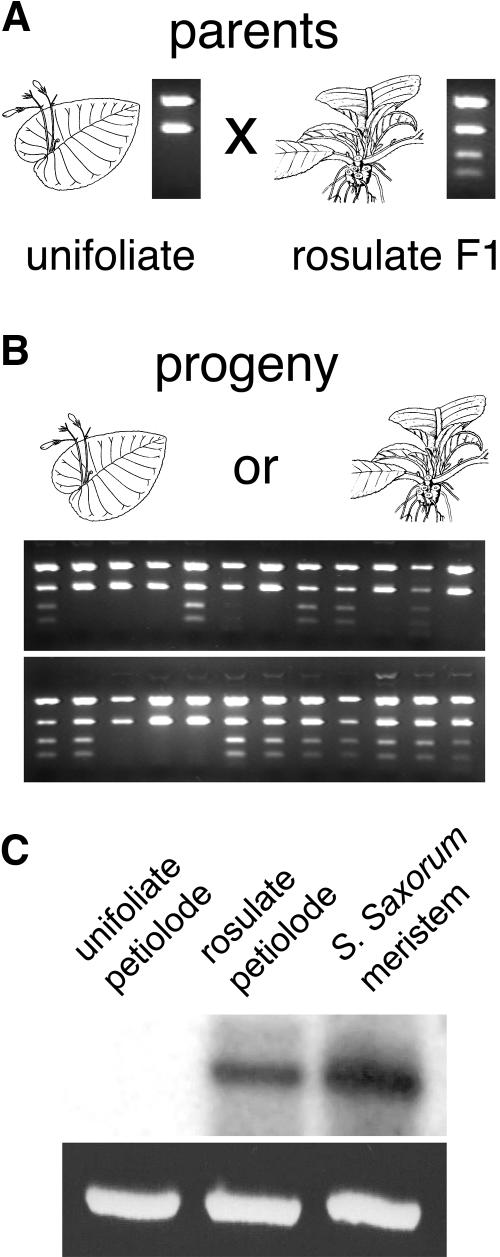
SSTM1 genotypes of the backcross population were determined by amplification of an ∼400-bp region of SSTM1 followed by digestion with a restriction enzyme that cuts the S. dunnii or S. wittei products once and the S. rexii product twice to yield two smaller fragments (Figure 8). As expected, approximately half (62/128) of the backcross progeny were found to be homozygous for the SSTM1 allele from the unifoliate parent, and the remainder were heterozygous (Table 1; see supplemental data online). If SSTM1 is one of the R loci, unifoliate backcross progeny (r1/r1 and r2/r2) should all be homozygous for the SSTM1 allele from the unifoliate parent, and all plants carrying the S. rexii SSTM1 allele should be rosulate. Neither of these criteria were fulfilled in the backcross populations (Figure 8, Table 1; see supplemental data online), suggesting that SSTM1 is not one of the two loci that specifies the morphological differences between unifoliate and rosulate Streptocarpus.
Figure 8.
Inheritance of SSTM1 Alleles and Expression in Different Growth Forms.
(A) and (B) SSTM1 alleles were amplified from the F1 (S. rexii × unifoliate) parent of the backcross, from the unifoliate parent, and from a selection of their progeny and distinguished by restriction site polymorphisms after digestion. No correlation is apparent between SSTM1 genotype and morphology.
(C) SSTM1 expression correlates with plant form in backcross plants.
To further test whether differences in SSTM1 expression correlate with form, we used backcross plants with different forms in RT-PCR. This showed that as in S. dunnii there was no SSTM1 expression in the juvenile phylomorph of a unifoliate plant but that there was SSTM1 expression in the rosulate plant (Figure 8C). Thus, although SSTM1 does not cause morphological differences between forms, its expression does correlate with form.
DISCUSSION
Monophyly of the SSTM1 and SSTM2 Gene Clades
We identified two paralogous Class I KNOX genes, SSTM1 and SSTM2, in Streptocarpus. Their highly conserved homeobox is interrupted by an intron that proved useful for phylogenetic reconstruction, giving comparable resolution to ITS. Our phylogenetic analyses showed that the SSTM1 genes are likely to be orthologs and, thus, that it was appropriate to compare their function between species.
Changes in KNOX Activity Accompanied Evolution of Novel Morphologies
All Old World Gesneriaceae whose early growth has been examined have suppressed plumule development and unequal cotyledon development (anisocotyly). These traits are thought to give the seeds a boost in growth when they germinate, as there is little energy stored (Burtt, 1970, 1994). Taken to its extreme, this pattern of growth results in acaulescence, and the cotyledon gives rise to the first (rosulate species) or only phyllomorph (unifoliate species).
In Arabidopsis, caulescence requires STM to promote SAM activity and repress leaf fate (Barton and Poethig, 1993; Byrne et al., 2000). STM is expressed in cells of the SAM but not in cells that will form leaf primordia (Long et al., 1996)—a pattern of expression that is conserved in diverse seed plants (e.g., Jackson et al., 1994; Sundas-Larsson et al., 1998) and that makes STM expression a marker for SAM cell identity. In Arabidopsis, the domain of STM expression overlaps with those of other Class I KNOX genes that are largely restricted to the SAM (e.g., Lincoln et al., 1994; Chuck et al., 1996).
This pattern of KNOX protein expression was also seen in S. saxorum, suggesting conservation of KNOX function in promoting activity of the SAM in caulescent Streptocarpus. However, KNOX proteins also accumulated in S. saxorum leaf primordia after they had initiated from the SAM. Proteins were present in a layer of cells at the adaxial–abaxial boundary and later restricted to cells of the developing leaf vasculature. A very similar pattern of KNOX expression in leaves has been described for various taxa with compound leaf primordia, which form either compound leaves or derived simple leaves (Bharathan et al., 2002). A causal relationship between KNOX activity and compound leaf development has also been demonstrated in Arabidopsis and tobacco (Nicotiana tabacum), where ectopic KNOX expression in leaves causes compounding (e.g., Chuck et al., 1996; Hay et al., 2003), and in the compound leaves of tomato (Lycopersicon esculentum), where increased KNOX activity increases the degree of compounding (e.g., Hareven et al., 1996). S. saxorum provides an exception to the correlation between KNOX expression and compound leaf morphology because, like all caulescent Streptocarpus, it produces simple leaves from simple primordia. This suggests that downstream targets of KNOX regulation needed for compounding in other taxa were either unresponsive or lost in Streptocarpus. Known targets of KNOX repression include genes needed for synthesis of GA phytohormones (e.g., Sakamoto et al., 2001). GA signaling is needed for compound leaf development in response to ectopic KNOX expression in Arabidopsis and in tomato leaves that normally express KNOX genes (Hay et al., 2002). Uncoupling of GA biosynthesis from KNOX control could explain why S. saxorum leaf development appears insensitive to KNOX expression.
Previous histological studies have suggested that acaulescent Streptocarpus might retain a vegetative SAM, albeit in an unconventional position (Jong and Burtt, 1975). Cells between the embryonic cotyledons of acaulescent species show the layered arrangement of conventional SAMs but do not contribute to growth after germination. Instead, a SAM-like structure subsequently appears in the petiole of the dominant cotyledon and later gives rise to inflorescence meristems as the groove meristem. This lead to the suggestion that the groove meristem represents a vegetative SAM displaced into the cotyledon petiole (termed the petiolode in this case; Jong and Burtt, 1975).
Our expression analyses with S. dunnii, a unifoliate acaulescent species, suggest that this is not the case because we detected no SdSTM1 expression in juvenile plants. Our immunolocalizations corroborated this result in that we did not always detect KNOX protein accumulation in plants lacking conspicuous inflorescences. However, we did detect protein accumulation in inflorescence meristems. Thus, our results suggest that the groove (inflorescence) meristem initiates de novo in S. dunnii and is therefore not equivalent to a conventional SAM. This implies that the vegetative SAM was lost in evolution of the unifoliate growth form. The loss of KNOX expression needed for SAM formation in other angiosperms is consistent with the lack of a vegetative SAM in unifoliate Streptocarpus. SSTM1 expression initiates as the inflorescence meristem initiates, suggesting that a change in the pattern of SSTM1 expression, rather than a complete loss of SSTM1 function, accompanied evolution of the unifoliate growth form.
Although the rosulate S. rexii also appears to lack a vegetative SAM, it expresses SSTM1 and possibly other Class I KNOX genes in the mounds of petiolode cells from which phyllomorphs initiate. These mounds have also been suggested to arise from the groove meristem (Jong, 1978), although we have no direct evidence for this origin, and they may initiate de novo. They therefore resemble conventional SAMs in their structure, expression of SSTM1, and ability to form organs but may be unable to produce organs reiteratively (indeterminacy) and might therefore result from redeployment of only part of the mechanism needed for SAM formation and function. The full mechanism is presumably employed on formation of inflorescence meristems from the groove meristem.
In Streptocarpus, we found that the difference in SSTM1 between a unifoliate and rosulate species correlated with the formation of additional phyllomorphs from SAM-like structures. Crosses between rosulate and unifoliate Streptocarpus indicated that the rosulate character, and hence vegetative SSTM1 expression, was specified by dominant alleles at two loci. Neither locus corresponded to SSTM1. This suggested (1) that vegetative SSTM1 expression was not sufficient to specify the SAM-like structures of rosulate Streptocarpus and (2) that the differences in SSTM1 expression were not the result of differences in SSTM1 alleles themselves but in the genetic prepattern of other factors that regulate SSTM1 expression.
These conclusions are consistent with the findings from Arabidopsis and other model angiosperms that have shown that STM is only one of several genes required for normal SAM formation and function (reviewed in Veit, 2004). Other genes include WUSCHEL (WUS), which is expressed independently of STM and also required for SAM indeterminacy (Mayer et al., 1998). WUS or STM activity alone is insufficient for SAM formation, but WUS and STM together specify at least transient SAMs (Gallois et al., 2002). Expression of cofactors like WUS might therefore be required for the vegetative SAM-like structures of rosulate Streptocarpus. Other genes are known to promote STM and WUS expression, including the CUP-SHAPED COTYLEDON (CUC) genes that are both necessary and sufficient for STM expression and SAM formation in Arabidopsis (Takada et al., 2001). Therefore, differences in regulatory genes like CUC might underlie the evolutionary differences between unifoliate and rosulate Streptocarpus and between caulescent and acaulescent species.
METHODS
Twenty-six different species were used in this study (Table 2; see supplemental data online). Tissue was obtained from glasshouse-grown plants or herbarium specimens at the Royal Botanic Garden Edinburgh. Genomic DNA for PCR was extracted as by Doyle and Doyle (1987) and for DNA gel blot hybridizations using the method of Chen and Dellaporta (1994).
Table 2.
Streptocarpus and Saintpaulia Species Used in This Study and Their RBGE Accession Numbers
| Species | Authority | RBGE Accession Number |
|---|---|---|
| S. beampingaratrensis | Humbert | 19972887 |
| S. burundianus | Hilliard and Burtt | Herbarium S137a |
| S. dunnii | Hooker | 19972909 |
| S. glandulosissimus | Engler | – |
| S. hirticapsa | Burtt | 19932793 |
| S. ibityensis | Humbert | 19932867 |
| S. itremensis | Burtt | 19972889 |
| S. levis | Burtt | 19982242 |
| S. modestus | Britten | 19943058 |
| S. pallidiflorus | Clarke | 19691211 |
| S. papangae | Humbert | 19972886 |
| S. pentherianus | Fritsch | 19972034 |
| S. primulifolius | Gandoger | 19912192 |
| S. rexii | (Hook.) Lindl. | 19870333 |
| S. saxorum | Engler | 19711885 |
| S. schliebenii | Mansfeld | Herbarium S136a |
| Streptocarpus spp | – | 19972893 |
| S. stomandrus | Burtt | 19711392 |
| S. thompsonii | Brown | 19941334 |
| S. thysanotus | Hilliard and Burtt | No. 18 S129a |
| S. venosus | Burtt | 19982247 |
| S. wendlandii | Sprenger | 19982266 |
| S. wittei | Hilliard and Burtt | 19981673 |
| Sa. ionantha | Wendl. | 19970092 |
| Sa. tongwensis | Burtt | 19970096 |
| Sa. velutina | Burtt | 19872179 |
STM-like sequences were amplified from genomic DNA using primers Fc (5′-TGGAGCCGCCACTACAAATG-3′) and GR22 (5′-TGAACCAGTTGTTGATYTGCTT-3′) designed to homeobox sequences conserved between Class I KNOX genes of Arabidopsis thaliana and Antirrhinum majus by Ian Oliver and Michael Möller. These were used in 50 μL of PCR with 20 ng of genomic DNA, 2 mM MgCl2, and 1 mM primers in 35 cycles of 94°C (30 s), 50°C (30 s), and 72°C (1 min). PCR products were gel purified and either sequenced directly or after cloning into a plasmid. Sequences have been deposited in GenBank, and their accession numbers are given in Table 3 (see supplemental data online).
Table 3.
GenBank Accession Numbers of Genes Isolated in This Study
| Species | Gene | Accession Number |
|---|---|---|
| S. beampingaratrensis | S. beampingaratrensis STM2 | AY662116 |
| S. burundianus | S. burundianus STM1 | AY662123 |
| S. burundianus STM2 | AY662108 | |
| S. dunnii | S. dunnii STM1 | AY655752 |
| S. dunnii STM2 | AY662103 | |
| S. glandulosissimus | S. glandulosissimus STM1 | AY662126 |
| S. hirticapsa | S. hirticapsa STM1 | AY662122 |
| S. hirticapsa STM2 | AY662109 | |
| S. ibityensis | S. ibityensis STM1a | AY662120 |
| S. ibityensis STM1b | AY662121 | |
| S. ibityensis STM2 | AY662100 | |
| S. itremensis | S. itremensis STM2 | AY662101 |
| S. levis | S. levis STM2 | AY662117 |
| S. modestus | S. modestus STM2a | AY662110 |
| S. modestus STM2b | AY662111 | |
| S. pallidiflorus | S. pallidiflorus STM1 | AY662125 |
| S. pallidiflorus STM2 | AY662097 | |
| S. papangae | S. papangae STM1 | AY662119 |
| S. papangae STM2 | AY662099 | |
| S. pentherianus | S. pentherianus STM2 | AY662104 |
| S. primulifolius | S. primulifolius STM2 | AY662112 |
| S. rexii | S. rexii STM1 | AY655753 |
| S. rexii STM2 | AY662106 | |
| 40S | AY796341 | |
| S. saxorum | S. saxorum STM1 | AY655754 |
| S. saxorum STM2 | AY662098 | |
| S. schliebenii | S. schliebenii STM1 | AY662124 |
| S. schliebenii STM2 | AY662102 | |
| Streptocarpus spp | Streptocarpus spp STM2 | AY662118 |
| S. stomandrus | S. stomandrus STM2 | AY662096 |
| S. thompsonii | S. thompsonii STM2 | AY662113 |
| S. thysanotus | S. thysanotus STM1 | AY662127 |
| S. venosus | S. venosus STM2a | AY662114 |
| S. venosus STM2b | AY662115 | |
| S. wendlandii | S. wendlandii STM2 | AY662105 |
| S. wittei | S. wittei STM1 | AY655755 |
| S. wittei STM2 | AY662107 | |
| Sa. ionantha | Sa. ionantha STM2 | AY662094 |
| Sa. tongwensis | Sa. tongwensis STM2a | AY662092 |
| Sa. tongwensis STM2b | AY662093 | |
| Sa. velutina | Sa. velutina STM2 | AY662095 |
RNA was extracted using Trizol reagent (Gibco BRL, Paisley, UK). For 3′ RACE, reverse transcription from 1 μg of total DNAase-treated RNA was primed using Qt primer (Frohman, 1995). Nested amplification was performed using primers PHRILL (5′-CCRCACTACCCTCGCCTCTT-3′) and Qo (Frohman, 1995), then MEINOX1 (5′-CGCCTGAAGTKGTGGCAAAG-3′) and Qi (Frohman, 1995). The same PCR conditions were used as for amplification from genomic DNA, except that annealing was performed at 55°C with primers at a final concentration of 0.2 mM.
For 5′ RACE, reverse transcription was primed with ELKup (5′-GCTTGGGGRTCAATGAAACTGT-3′) and cDNA products ligated to an adaptor (Siebert et al., 1995). DNA was amplified using primers MEINOX2 (5′-AAGGGCTTTGAACTGRCAYTC-3′) and LAPCRT3 (Siebert et al., 1995).
To test for the presence of transcripts in different tissues, total RNA was treated with DNase and used in reverse transcription with Qt primer. Multiplex PCR was then performed with two SSTM1 primers, MegsD (5′-CACGCAGTAGTGTGTGTAATGGAG-3′) and ELKup, and two primers for a constitutively expressed Streptocarpus gene, ribosomal up (5′-CCTGCAACTTGGTGGTACGGTA-3′) and ribosomal down (5′-CGACACACCCCTGGTACTTT-3′) as a control. Both primer pairs span introns. SSTM1 products were detected by DNA gel blot hybridization with a DIG-labeled probe from the conserved region of S. rexii SSTM1 (see below).
For DNA gel blot hybridization, 5 μg of genomic DNA was digested with EcoRI, EcoRV, or HindIII. Fragments were separated, blotted, and hybridized as by Langdale et al. (1991). High stringency hybridization was performed in 3× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 65°C and low stringency hybridization in 5× SSC at 58°C.
Immunolocalizations were performed essentially according to Donlin et al. (1995) using paraffin embedded tissue. The anti-STM antibody (a gift from Kathy Barton) was used at 1:500 dilution and detected with an alkaline-phosphatase conjugated secondary antibody. Nonspecific binding was blocked with 10% horse serum in PBS containing 0.1% BSA.
Sequence Alignment and Phylogenetic Analysis
SSTM1 and SSTM2 intron sequences were aligned manually using Se-Al v2.0a11 (Rambaut, 1996). ITS sequences were aligned as discussed previously (Möller and Cronk, 2001), except that gaps no longer necessary in the reduced data set were removed. Sequences for the placement of SSTM1 within the KNOX gene family were aligned using ClustalW and adjusted manually using Se-Al v2.0a11. Positions that were not unambiguously alignable were excluded from analyses. The KNOX phylogeny was rooted on Aaknox1 (an algal KNOX gene that shows features of Class I and Class II KNOX genes). To verify that Class I KNOX genes fell in a separate clade from Class II genes, two Class II genes (mkn1 from a moss and Crknox3 from a fern) were also included. Accession numbers of KNOX sequences are given in Table 4 (see supplemental data online), and alignments have been submitted to TreeBASE (www.treebase.org/treebase/).
Table 4.
Gene Accession Numbers for KNOX Phylogeny
| Species | Gene | Accession Number | Species | Gene | Accession Number |
|---|---|---|---|---|---|
| Oryza sativa | OSH71 | AB028885 | Medicago trunculata | M. trunculata knox | AF308454 |
| OSH6 | AB028883 | A. thaliana | KNAT1 | U14174 | |
| OSH3 | AB028882 | STM | U32344 | ||
| OSH1 | D16507 | KNAT2 | U14175 | ||
| OSH15 | AB016071 | KNAT6 | AB072361 | ||
| OSH43 | AB028884 | Antirrhinum majus | hirzina | AY072735 | |
| Zea mays | lg3 | AF100455 | invaginata | AY072736 | |
| lg4A | AF457121 | Nicotiana tabacum | NTH1 | AB025573 | |
| Knox3 | BQ486722 | NTH20 | AB025714 | ||
| KNOTTED1 | X61308 | TobH1 | AY169493 | ||
| rs1 | L44133 | NTH15 | AB004785 | ||
| Acetabularia acetabulum | Aaknox1 | AF170172 | NTH9 | AB025713 | |
| Ceratopteris richardii | Crknox1 | AB043955 | NTH22 | AB025715 | |
| Crknox2 | AB043956 | Lycopersicon esculentum | Tkn1 | U32247 | |
| Crknox3 | AB043957 | Tkn3 | U76408 | ||
| Physcomitrella patens | mkn1 | AF285148 | Tkn4 | AF533597 | |
| mkn2 | AF285147 | Tkn2 | U76407 | ||
| mkn4 | AF28417 | ||||
| Ipomoea nil | Pkn3 | AB016002 | Picea mariana | SKN1 | U90091 |
| Pkn2 | AB016000 | Solanum tuberosum | POTH1 | U65648 | |
| Dendrobium grex | DOH1 | AJ276389 | |||
| Brassica oleracea | BoSTM1 | AF193813 | Hordeum vulgare | Hvknox3 | X83518 |
| Pisum sativum | P. sativum knox | AF063307 | Glycine max | SBH1 | L13663 |
PAUP version 4.0b10 (Swofford, 1998) was used to generate Streptocarpus phylogenies. For all analyses, heuristic searches were performed using parsimony. Trees were started using stepwise addition, followed by 5000 random addition sequence replicates and then swapping on best trees. Branch swapping used tree bisection and reconnection with the MULTREES option on. For each addition sequence replicate, two trees of score ≥5 were saved. Tree statistics are shown in Table 5.
Table 5.
Tree Statistics for Phylogenies
| KNOX Gene Family Phylogeny | ITS | SSTM Coding Phylogeny | SSTM Intron Phylogeny | |
|---|---|---|---|---|
| Length of aligned sequence (bp) | 564 | 630 | 57 | 169 |
| Number of variable characters | 508 | 276 | 13 | 88 |
| Number of parsimony informative characters | 456 | 156 | 9 | 68 |
| Percentage of informative characters | 80.8 | 24.7 | 22.81 | 40.2 |
| Number of trees | 4 | 32 | 2480 | 9002 |
| Length of trees | 6151 | 556 | 25 | 98 |
| CI | 0.195 | 0.678 | 0.8 | 0.776 |
| RI | 0.498 | 0.648 | 0.932 | 0.924 |
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY655752 to AY655755 and AY662092 to AY662127.
Supplementary Material
Acknowledgments
This work was supported by a Sainsbury PhD studentship and short-term postdoctoral fellowship to J.H. We thank Kathy Barton for her generous gift of the STM antibody, Daniel Barker, Toby Pennington, Hirokazu Tsukaya, and the Oxford Systematics Discussion Group for useful discussions, Miltos Tsiantis, Kathy Barton, and three anonymous reviewers for comments on the manuscript, and Steve Scott for growing our plants.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jill Harrison (jill.harrison@plants.ox.ac.uk).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028936.
References
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOX1 gene expression during development. Science 296, 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bürglin, T.R. (1997). Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25, 4173–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin, T.R. (1998). The PBC domain contains a MEINOX domain: Coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208, 113–116. [DOI] [PubMed] [Google Scholar]
- Burtt, B.L. (1970). Studies in the Gesneriaceae of the Old World XXXI: Some aspects of functional evolution. Notes R. Bot. Gard. (Edinb.) 30, 1–10. [Google Scholar]
- Burtt, B.L. (1994). A commentary on some recurrent forms and changes of form in angiosperms. In Shape and Form in Plants and Fungi, A. Hudson and D.S. Ingram, eds (London: Academic Press), pp. 143–152.
- Burtt, B.L. (1998). Climatic accommodation and phytogeography of the Gesneriaceae of the Old World. In Diversity and Taxonomy of Tropical Flowering Plants, P. Mathew and M. Sivadasan, eds (Calicut, India: Mentor Books), pp. 1–27.
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Carroll, S.B. (2000). Endless forms: The evolution of gene regulation and morphological diversity. Cell 101, 577–580. [DOI] [PubMed] [Google Scholar]
- Chen, J., and Dellaporta, S.L. (1994). Urea-based plant DNA miniprep. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 522–525.
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1998). The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Donlin, M.J., Lisch, D., and Freeling, M. (1995). Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell 7, 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J., and Doyle, J.L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Frohman, M.H. (1995). Rapid amplification of cDNA ends. In PCR Primer, C.W. Diffenbach and G.S. Dveksler, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 381–409.
- Gallois, J.-L., Woodward, C., Reddy, G.V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129, 3207–3217. [DOI] [PubMed] [Google Scholar]
- Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84, 735–744. [DOI] [PubMed] [Google Scholar]
- Hay, A., Jackson, D., Ori, N., and Hake, S. (2003). Analysis of the competence to respond to KNOTTED1 activity in Arabidopsis leaves using a steroid induction system. Plant Physiol. 131, 1671–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hilliard, O.M., and Burtt, B.L. (1971). Streptocarpus: An African Plant Study. (Pietermaritzburg, South Africa: University of Natal Press).
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED 1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jong, K. (1970). Developmental Aspects of Vegetative Morphology of Streptocarpus. PhD dissertation (Edinburgh, UK: University of Edinburgh).
- Jong, K. (1978). Phyllomorphic organization in rosulate Streptocarpus. Notes R. Bot. Gard. (Edinb.) 36, 369–396. [Google Scholar]
- Jong, K., and Burtt, B.L. (1975). The evolution of morphological novelty exemplified in the growth patterns of some Gesneriaceae. New Phytol. 75, 297–311. [Google Scholar]
- Langdale, J.A., Taylor, W.C., and Nelson, T. (1991). Cell-specific accumulation of maize phospoenolpyruvate carboxylase is correlated with demethylation at a specific site >3kb upstream of the gene. Mol. Gen. Genet. 225, 49–55. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has over-lapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Möller, M., and Cronk, Q.C.B. (1997). Origin and relationships of Saintpaulia (Gesneriaceae) based on ribosomal DNA internal transcribed spacer (ITS) sequences. Am. J. Bot. 84, 956–965. [PubMed] [Google Scholar]
- Möller, M., and Cronk, Q.C.B. (2001). Evolution of morphological novelty: A phylogenetic analysis of growth patterns in Streptocarpus (Gesneriaceae). Evolution 55, 918–929. [DOI] [PubMed] [Google Scholar]
- Oehlkers, F. (1938). Bastardierungsversuche in der gattung Streptocarpus Lindl. 1. Plasmatische vererbung und die geschlechtbestimmung von zwitterpflanzen. Zeitschrift f. Botanik 32, 305–393. [Google Scholar]
- Oehlkers, F. (1942). Faktorenanalytische ergebnisse an artbastarden. Biol. Zbl. 62, 280–289. [Google Scholar]
- Oehlkers, F. (1964). Cytoplasmic inheritance in the genus Streptocarpus. Adv. Genet. 12, 329–370. [Google Scholar]
- Rambaut, A. (1996). Se-Al: Sequence Alignment Editor (Se-Al v2.0a11). http://evolve.zoo.ox.ac.uk/.
- Rosenblum, I.M., and Basile, D.V. (1984). Hormonal regulation of morphogenesis in Streptocarpus and its relevance to evolutionary history of the Gesneriaceae. Am. J. Bot. 7, 52–64. [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.L. (1998). A role of Ultrabithorax in morphological differences between Drosophila species. Nature 396, 463–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena, E., Delon, I., Jones, I., Payre, F., and Stern, D.L. (2003). Regulatory evolution of shaven-baby/ovo underlies multiple cases of morphological parallelism. Nature 424, 935–938. [DOI] [PubMed] [Google Scholar]
- Sundas-Larsson, A., Svenson, M., Liao, H., and Engstrom, P. (1998). A homeobox gene with potential developmental control function in the mersitem of the conifer Picea abies. Proc. Natl. Acad. Sci. USA 95, 15118–15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (1998). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- Takada, S., Hibara, K.-I., Tetsuya, I., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., and Hay, A. (2003). Comparative plant development: The time of the leaf? Nat. Rev. Genet. 4, 169–180. [DOI] [PubMed] [Google Scholar]
- Veit, B. (2004). Determination of cell fate in apical meristems. Curr. Opin. Plant Biol. 7, 57–64. [DOI] [PubMed] [Google Scholar]
- Wang, R., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- Weber, A. (1982). Evolution and radiation of the pair-flowered cyme in Gesneriaceae. Newsletter of the Australian Systematic Botany Society 30, 23–41. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.