Abstract
The AUXIN RESPONSE FACTOR (ARF) gene family products, together with the AUXIN/INDOLE-3-ACETIC ACID proteins, regulate auxin-mediated transcriptional activation/repression. The biological function(s) of most ARFs is poorly understood. Here, we report the identification and characterization of T-DNA insertion lines for 18 of the 23 ARF gene family members in Arabidopsis thaliana. Most of the lines fail to show an obvious growth phenotype except of the previously identified arf2/hss, arf3/ett, arf5/mp, and arf7/nph4 mutants, suggesting that there are functional redundancies among the ARF proteins. Subsequently, we generated double mutants. arf7 arf19 has a strong auxin-related phenotype not observed in the arf7 and arf19 single mutants, including severely impaired lateral root formation and abnormal gravitropism in both hypocotyl and root. Global gene expression analysis revealed that auxin-induced gene expression is severely impaired in the arf7 single and arf7 arf19 double mutants. For example, the expression of several genes, such as those encoding members of LATERAL ORGAN BOUNDARIES domain proteins and AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, are disrupted in the double mutant. The data suggest that the ARF7 and ARF19 proteins play essential roles in auxin-mediated plant development by regulating both unique and partially overlapping sets of target genes. These observations provide molecular insight into the unique and overlapping functions of ARF gene family members in Arabidopsis.
INTRODUCTION
The plant hormone auxin, typified by indole-3-acetic acid (IAA), regulates a variety of physiological processes, including apical dominance, tropic responses, lateral root formation, vascular differentiation, embryo patterning, and shoot elongation (Davies, 1995). At the molecular level, auxin rapidly induces various genes (Abel and Theologis, 1996). Several classes of early auxin-responsive genes have been identified including the Aux/IAA, GH3, and SAUR-like genes (Abel and Theologis, 1996; Guilfoyle et al., 1998). The GH3-like genes encode acyl adenylate–forming isozymes (Staswick et al., 2002). Several GH3-like proteins covalently modify IAA, jasmonic acid, or salicylic acid, indicating that they play a global role in various hormone signaling pathways. The function of the SAUR-like genes is still unknown, but it has been suggested that they may encode short-lived nuclear proteins involved in auxin signaling by interacting with calmodulin (Yang and Poovaiah, 2000; Knauss et al., 2003).
The Aux/IAAs have been among the first auxin-regulated genes to be isolated and are the most characterized among early auxin-responsive genes. They are encoded by a large gene family in Arabidopsis thaliana with 29 members (Abel et al., 1995; Reed, 2001; Liscum and Reed, 2002; Remington et al., 2004). They encode short-lived nuclear proteins, and most of them contain four highly conserved domains (I to IV) (Abel et al., 1994; Reed, 2001). Each domain contributes to the functional properties of the protein. Domain II confers instability of the protein (Worley et al., 2000; Ouellet et al., 2001). Domains III and IV serve for homodimerization and heterodimerization with other Aux/IAA gene family members as well as for heterodimerization with the Auxin Response Factors (ARFs) (Kim et al., 1997; Ulmasov et al., 1997, 1999a, 1999b). Domain I is responsible for the transcriptional repressing activity of the proteins (Tiwari et al., 2004).
The ARF proteins are also encoded by a large gene family in Arabidopsis (23 members). A typical ARF protein contains a B3-like DNA binding domain in the N-terminal region, and domains III and IV are similar to those found in the C terminus of Aux/IAAs. An ARF binds to auxin-responsive cis-acting elements (AuxREs) found in the promoter region of auxin-responsive genes through its DNA binding domain (Abel et al., 1996; Ulmasov et al., 1997, 1999a). The amino acid composition of the middle region between the DNA binding domain and domains III/IV determines whether an ARF protein functions as an activator or repressor (Ulmasov et al., 1999b; Tiwari et al., 2003). The Aux/IAA proteins regulate auxin-gene expression through interaction with the ARF proteins. The Aux/IAAs are targets for degradation by the SCFTIR1 complex, and most importantly, auxin mediates their interaction with the proteolytic machinery (Gray et al., 1999, 2001; Ward and Estelle, 2001; Dharmasiri and Estelle, 2004). Aux/IAA protein stability is a central regulator in auxin signaling.
Several gain-of-function Aux/IAA mutants, including shy2/iaa3 (Tian and Reed, 1999), axr2/iaa7 (Nagpal et al., 2000), bdl/iaa12 (Hamann et al., 2002), slr/iaa14 (Fukaki et al., 2002), arx3/iaa17 (Rouse et al., 1998), msg2/iaa19 (Tatematsu et al., 2004), and iaa28-1 (Rogg et al., 2001), have been isolated by forward genetics. These mutants have amino acid substitutions in highly conserved residues of domain II, resulting in enhanced protein stability that causes altered auxin response and dramatic defects in growth and development. Loss-of-function mutations of AUX/IAAs do not show an obvious visible growth phenotype (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000; P.J. Overvoorde and Y. Okushima, unpublished data). Loss-of-function mutants in five ARF genes have been previously isolated. Mutations in the ARF3/ETT affect gynoecium patterning (Sessions et al., 1997; Nemhauser et al., 2000). Loss-of-function mutations of ARF7/NPH4/MSG1/TIR5 result in impaired hypocotyl response to blue light and other differential growth responses associated with changes in auxin sensitivity (Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998; Harper et al., 2000). Mutations in ARF5/MP interfere with the formation of vascular strands and the initiation of the body axis in the early embryo (Hardtke and Berleth, 1998). Mutations in ARF2/HSS have been identified as suppressors of the hookless phenotype (Li et al., 2004). ARF2 acts as a communication link between the ethylene and the auxin signaling pathways for regulating hypocotyl bending. Lastly, ARF8 functions in hypocotyl elongation, and it is involved in auxin homeostasis (Tian et al., 2004). The biological functions, however, of the remaining ARF gene family members are unknown.
Here, we have employed a functional genomic strategy that involves the identification of T-DNA insertion in the ARF gene family members to elucidate some of the biological functions of the ARF transcription factors. Most of the single arf T-DNA insertion mutants fail to show an obvious growth phenotype. However, double mutants, such as arf7 arf19, show a strong auxin phenotype that results in the absence of lateral root formation than neither the arf7 nor arf19 single mutant expresses. The results suggest that there are unique and overlapping functions among related ARF gene family members in Arabidopsis.
RESULTS
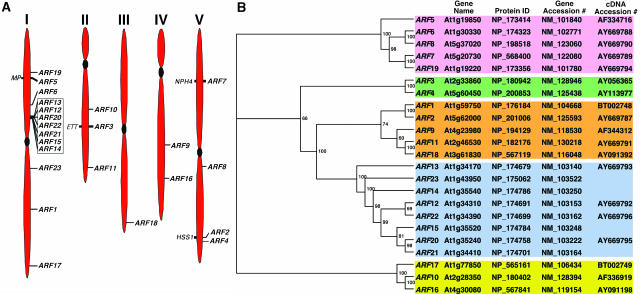
The Arabidopsis ARF Gene Family
The Arabidopsis genome contains 23 ARF genes scattered among the five chromosomes (Arabidopsis Genome Initiative, 2000; annotation version V5.0, Figure 1A). The locations of the four previously described loss-of-function mutations, arf3/ett (Sessions and Zambryski, 1995), arf5/mp (Hardtke and Berleth, 1998), arf7/nph4 (Harper et al., 2000), and arf2/hss (Li et al., 2004), are highlighted in Figure 1A. A cluster of ARF genes, ARF12, 13, 14, 15, 20, 21, and 22, is present in the upper arm of chromosome I (Figure 1A). These genes share a high degree of similarity among their amino acid and nucleotide sequences (see Supplemental Figure 1 and Table 1 online). ARF23 is a pseudogene (see Supplemental Figure 1 online; Guilfoyle and Hagen, 2001). Phylogenetic analysis reveals that the genes fall into three branches (marked with different colors in Figure 1B). Class I has the most members (15) that can be subdivided into three subclasses, Ia (five members, shaded brown), Ib (eight members, shaded blue), and Ic (two members, shaded green). Their middle region is rich in Pro, Ser, Gly, or Leu (Guilfoyle and Hagen, 2001; see Supplemental Figure 1 online), and some of them function as repressors (Ulmasov et al., 1999b; Tiwari et al., 2003). Class II (shaded pink) has five members, and some of them function as activators. Their middle region is rich in Glu (Ulmasov et al., 1999a; Guilfoyle and Hagen, 2001). Class III (shaded yellow) also contains three members that are the most divergent compared with those encoded by the other two classes. ARF3 and ARF17, which are considered to lack the C-terminal domains III and IV (Guilfoyle and Hagen, 2001), may potentially contain highly divergent domains III and IV (see Supplemental Figure 1 online). Furthermore, ARF13 does not have domains III and IV in this new alignment (see Supplemental Figure 1 online). The ARF polypeptides vary in size ranging from ∼57 (ARF13) to ∼129 kD (ARF7) (see Supplemental Table 2 online). This size variation is primarily attributable to the different amino acid content in the middle region (see Supplemental Figure 1 online).
Figure 1.
The ARF Gene Family of Arabidopsis.
(A) Chromosomal location of ARF genes. The locations of 23 putative ARF genes on the Arabidopsis chromosomes (I to V) are shown according to version 5.0 of the Arabidopsis Genome annotation submitted to GenBank. Mutants that have been isolated in the ARF gene are shown on the left side of the chromosomes. The ARF genes clustered on chromosome I are boxed.
(B) Phylogenetic analysis. An unrooted dendogram was generated using ClustalW (Thompson et al., 1994). TreeView was used to generate the graphical output (Page, 1996). The numbers at the branching points indicate the percentage of times that each branch topology was found during bootstrap analysis (n = 1000). The gene names, accession numbers, protein identifier, and the accession numbers of the full-length open reading frames (ORFs) used for this analysis are also shown. Predicted ORFs from the genomic annotation were used for ARF14, ARF15, ARF21, and ARF23 (pseudogene) genes. The full-length ORFs of ARF2, ARF6, ARF7, ARF8, ARF11, ARF12, ARF13, ARF19, ARF20, and ARF22 were constructed during this study. A differential spliced form of ARF13 has been cloned recently (accession number AY680406).
RNA hybridization analysis reveals that ARF1-ARF9, ARF11, ARF16, ARF17, ARF18, and ARF19 are expressed in light-grown seedlings and various plant tissues, including roots, leaves, flowers, and stems (Ulmasov et al., 1999a; data not shown). We were unable to detect expression of the clustered ARF genes in these various RNAs, and there are no ESTs or cDNAs for these genes in public databases. Exploratory RT-PCR analysis using cDNA from various tissues (see Methods) revealed that the clustered genes are expressed during embryogenesis (see Supplemental Figure 2B online). Transgenic plants expressing the β-glucuronidase (GUS) reporter gene from the ARF12 and ARF22 promoters show that the ProARF12:GUS is expressed only in the developing seeds, and its expression is detected in the entire seed, including embryos and the integument surrounding the embryo (see Supplemental Figure 2F online). ProARF22:GUS transgenic plants display an identical GUS expression pattern as the ProARF12:GUS plants (data not shown).
Isolation of ARF T-DNA Insertion Mutants
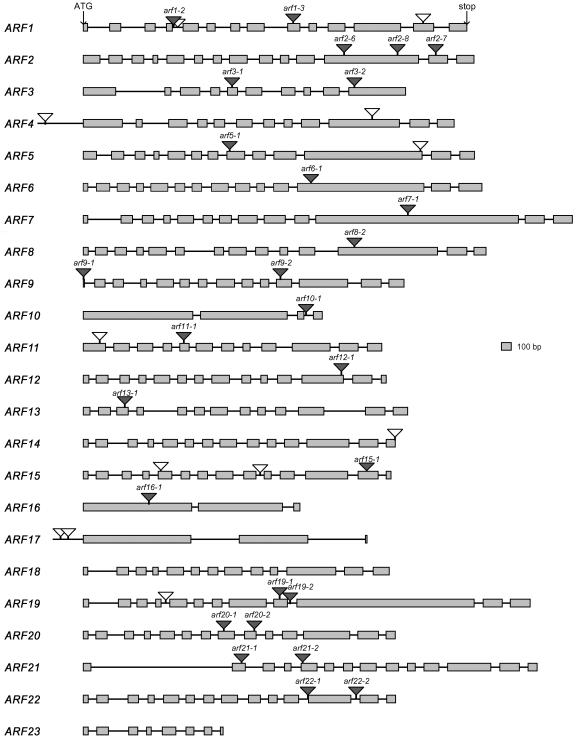
We initiated this project using a PCR-based screening approach to identify T-DNA insertion mutants for a large number of ARF genes. A total of 80,000 T-DNA insertion line populations in the Columbia ecotype were initially screened, and eight lines were identified (Alonso et al., 2003). Subsequently, the laboratory participated in generating the garlic lines in collaboration with the former Torrey Mesa Research Institute, and 10 additional lines were isolated (Sessions et al., 2002). More recently, we obtained another nine T-DNA insertional lines from the Salk T-DNA express line collection (http://signal.salk.edu/cgi-bin/tdnaexpress). Taken together during the last 6 years, we identified 27 T-DNA insertion lines located in the coding region of 18 ARF genes. Figure 2 and Supplemental Table 3 online provide a summary of all the mutants isolated and characterized during the course of this study. All the lines have been backcrossed at least once and partially characterized phenotypically. We plan to deposit all the lines in the Arabidopsis Biological Resource Center (http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/abrchome.htm) for further molecular and phenotypic characterization by the community.
Figure 2.
Location of T-DNA Insertions in the ARF Gene Family Members.
Boxes represent exons. T-DNA insertions with gray triangles denote lines whose characterization has been completed. T-DNA insertions with white triangles denote lines not yet characterized.
Phenotypes of Insertion Mutants
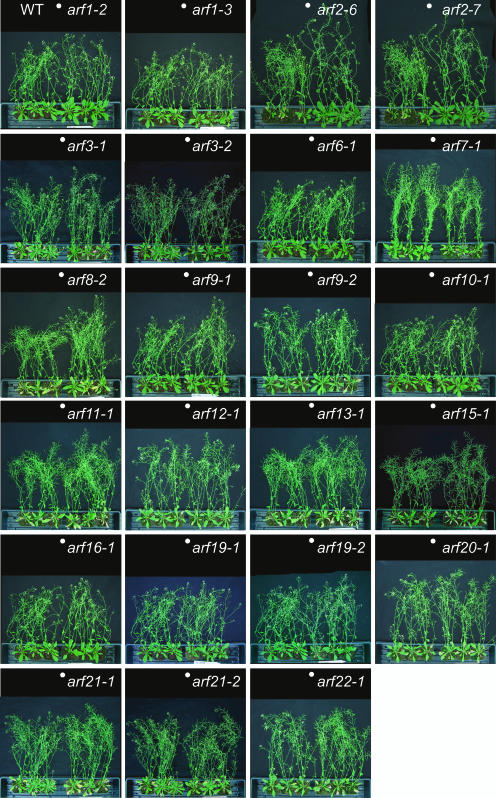
We were able to identify T-DNA insertion lines for arf3/ettin, arf5/mp, arf7/nph4/msg1, and arf2/hss, and their reported phenotypes were confirmed. Two independent arf3 alleles, arf3-1 and arf3-2, have unusual gynoecium and floral patterning defects, including an increased number of sepals and carpals (see Supplemental Figures 3A to 3C online; Sessions et al., 1997). The arf5-1 mutant fails to form root meristem and normal cotyledons (see Supplemental Figure 3D online; Hardtke and Berleth, 1998), and the arf7-1 mutant displays an impaired phototropic response toward blue light (Figure 4F; Harper et al., 2000). The arf2-6, arf2-7, and arf2-8 mutants have a pleiotropic phenotype, including a long, thick, and wavy inflorescence stem, large leaves, abnormal flower morphology, and late flowering under long-day conditions (see Supplemental Figure 3E online; Li et al., 2004; Y. Okushima and A. Theologis, unpublished data). It has been recently reported that arf8 seedlings have long hypocotyls in various light conditions (Tian et al., 2004). We did not examine the light-associated phenotype of arf8, but we saw longer inflorescence stems in the mutant than those in the wild type (Figure 3). The rest of the insertion lines did not show any obvious growth phenotype (Figure 3).
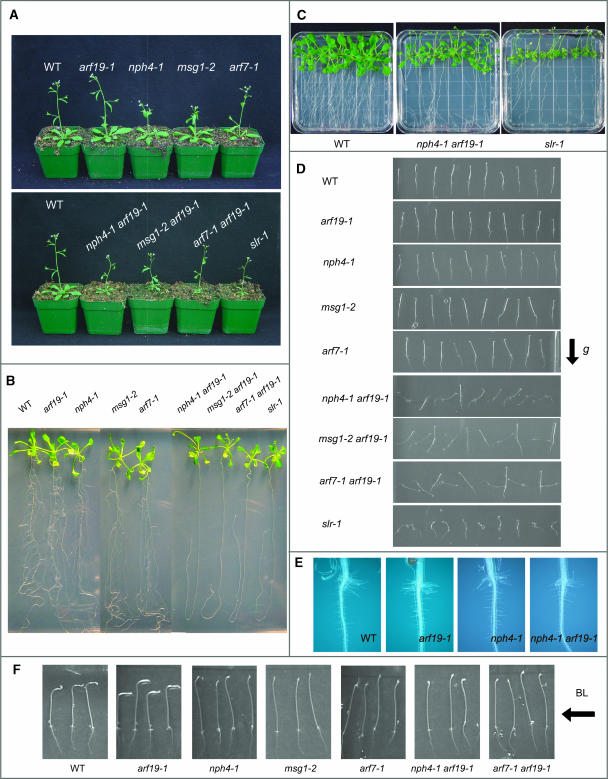
Figure 4.
Phenotypes of the arf7 arf19 Double Mutant.
(A) Four-week-old soil-grown plants of the wild type, arf19-1, nph4-1, msg1-2, and arf7-1 (top) and the wild type, nph4-1 arf19-1, msg1-2 arf19-1, arf7-1 arf19-1, and slr-1 (bottom).
(B) Seventeen-day-old seedlings of wild type, arf19-1, nph4-1, msg1-2, arf7-1, nph4-1 arf19-1, msg1-2 arf19-1, arf7-1 arf19-1, and slr-1.
(C) Twenty-two-day-old seedlings of the wild type, nph4-1 arf19-1, and slr-1 grown on agar plates vertically.
(D) Gravitropic response of 3-d-old dark-grown seedlings of the wild type, arf19-1, nph4-1, msg1-2, arf7-1, nph4-1 arf19-1, msg1-2 arf19-1, arf7-1 arf19-1, and slr-1.
(E) Root hair formations of the wild type, arf19-1, nph4-1, and nph4-1 arf19-1.
(F) Phototropism of 3-d-old dark-grown seedlings of the wild type, arf19-1, nph4-1, msg1-2, arf7-1, nph4-1 arf19-1, and arf7-1 arf19-1. Seedlings were exposed to unilateral blue light from the right for 8 h.
Figure 3.
Phenotype of Mature Mutant Plants.
Three wild-type (left) and three mutant plants (right) are shown. The plants were grown at the same time. White dots indicate the boundaries between the wild-type and the mutant plants.
Because most of the arf T-DNA insertion mutants fail to show an abnormal growth phenotype (Figure 3), we are generating double and higher-order mutants among the various insertion lines. So far, we have generated double mutants among closely related ARF genes, such as arf1 arf2, arf6 arf8, and arf7 arf19 (see Supplemental Figure 4 online). The phenotype of arf1 arf2 is similar but much stronger than that of arf2 (see Supplemental Figure 4A online; Li et al., 2004). ar6 arf8 has dwarfed aerial tissue and exhibits severe defects in flower development (see Supplemental Figure 4C online). The phenotypic and molecular characterization of arf7 arf19 is presented below.
Isolation and Characterization of arf7 arf19 Double Mutants
ARF7 and ARF19 are phylogenetically related (Figure 1B; Liscum and Reed, 2002; Remington et al., 2004). Given the close relationship of ARF7 and ARF19, we tested whether the arf19 mutant had an altered phototropic response similar to that reported for nph4/arf7 (Liscum and Briggs, 1995). We found that the arf19-1 mutant hypocotyl responded to blue light in a wild type–like manner (Figure 4F). Mature arf7 mutant plants (nph4-1, arf7-1, and msg1-2/nph4-102) do not show any gross developmental defects, except that they have epinastic rosette leaves and the length of the inflorescence stems is slightly shorter than that of the wild-type plants (Figure 3; data not shown; Watahiki and Yamamoto, 1997). These characteristics are more pronounced in the arf7 arf19 double mutant. The appearance of mature arf19 plants is identical to that of the wild type (Figures 3 and 4A). The results suggest that the expression of ARF7 functionally compensates for the loss of ARF19 expression responsible for differential hypocotyl growth, but not vice versa.
We initially used the nph4-1 mutant (Liscum and Briggs, 1995) as the arf7 allele for crossing into arf19-1 to generate the arf7 arf19 double mutant. Among the F2 population, approximately one out of 16 plants had short and thin inflorescence stem and small leaves. PCR analysis confirmed that these small plants were double homozygous for both mutations. Because the original nph4-1 line was screened from fast neutron-mutagenized seeds carrying the homozygous recessive glabrous1 (gl1) mutation (Liscum and Briggs, 1995), we backcrossed the nph4-1 and nph4-1 arf19-1 to Columbia (Col) wild-type plants. The nph4-1 and nph4-1 arf19-1 mutant lines without the gl1 mutation were used for further analysis.
The nph4-1 arf19-1 double mutant exhibits much stronger auxin-related phenotypes than those of nph4-1 and arf19-1 single mutants. Adult nph4-1 arf19-1 mutant plants have thin and short inflorescence stems, and their rosette leaves are small and epinastic (Figures 4A to 4C; see Supplemental Figure 4 online; data not shown). In addition, nph4-1 arf19-1 has reduced numbers of inflorescence stems, suggesting enhanced apical dominance. By contrast, the flowers of nph4-1 arf19-1 appear to be normal, and they fertilize normally (data not shown). The phenotype of nph4-1 arf19-1 is the most obvious at its seedling stage, with its most prominent phenotype being severely impaired lateral root formation (Figure 4B, Table 1). The primary roots of arf19-1 produce as many lateral roots as the wild type, whereas the arf7 mutant produces fewer lateral roots compared with the wild type (Figure 4B, Table 1). The primary roots of the nph4-1 arf19-1 seedlings fail to produce lateral roots in 2-week-old seedlings. However, nph4-1 arf19-1 seedlings start to generate several lateral roots after ∼2 weeks of growth, and their morphological appearance is normal (Figure 4C; data not shown). The nph4-1 arf19-1 mutant also displays agravitropic responses in both hypocotyls and roots (Figure 4D). When seedlings are grown vertically under dark conditions, the hypocotyl growth orientation of arf7 is significantly skewed compared with the wild type, whereas the arf19-1 mutant has a normal gravitropic response (Figure 4D; Harper et al., 2000). Interestingly, in the nph4-1 arf19-1 seedlings, regulation of growth orientation is disrupted in both hypocotyls and roots, with the hypocotyls occasionally growing downward and the roots upward (Figure 4D). Also, the roots and hypocotyls of nph4-1 arf19-1 show reduced gravitropic curvatures compared with the wild type when vertically dark-grown seedlings are reoriented by 90° (data not shown). The phototropic response toward blue light in hypocotyls of nph4-1 arf19-1 seedlings is disrupted as in the arf7 single mutants (Figure 4F). We generated additional combinations of arf7 arf19 double mutants using other alleles of arf7 and arf19 to confirm the phenotypes of nph4-1 arf19-1. We used msg1-2/nph4-102 (Watahiki and Yamamoto, 1997) and arf7-1 as the arf7 alleles for crosses with arf19-1 and arf19-2. All five additional arf7 arf19 double mutant alleles, msg1-2 arf19-1, arf7-1 arf19-1, nph4-1 arf19-2, msg1-2 arf19-2, and arf7-1 arf19-2 (Figures 4A, 4B, and 4D, Table 1; data not shown), display the same phenotypes as nph4-1 arf19-1: smaller plant size, impaired lateral root formation, and agravitropic response. These results confirm that the phenotypes of nph4-1 arf19-1 are caused by the loss of ARF7 and ARF19 function.
Table 1.
Lateral Root Formation in arf7, arf19, and arf7 arf19 Seedlings
| Mutant | Number of Lateral Roots |
|---|---|
| Wild type (Col) | 7.6 ± 3.2 |
| nph4-1 | 1.3 ± 1.1 |
| msg1-2 | 0.6 ± 0.7 |
| arf7-1 | 1.7 ± 1.4 |
| arf19-1 | 6.8 ± 1.6 |
| nph4-1 arf19-1 | 0.0 ± 0.0 |
| msg1-2 arf19-1 | 0.0 ± 0.0 |
| arf7-1 arf19-1 | 0.0 ± 0.0 |
The number of lateral roots in 10-d-old seedlings was determined. The numbers represent the average of more than 18 seedlings ± sd.
The phenotypes of the arf7 arf19 mutant are similar to those reported for the solitary root (slr)/iaa14 mutant (Fukaki et al., 2002). The slr mutant also shows strong auxin-related phenotypes, including complete lack of lateral roots, agravitropic roots, and hypocotyls, small plant size, and few root hairs (Figures 4A, 4B, and 4D; data not shown). Whereas the nph4-1 arf19-1 mutant seedlings exhibit severely impaired lateral formation, their primary roots start to produce lateral roots ∼2 weeks from germination (Figure 4C). By contrast, slr-1 seedlings do not produce any lateral roots even after 4 weeks from germination (Figure 4C; data not shown). We also examined the effect of exogenous auxin on lateral root formation in the nph4-1 arf19-1 seedlings. Four-day-old light-grown seedlings of the wild type, nph4-1 arf19-1, and slr-1 were transferred to medium containing 1 μM IAA. After an additional 3 d of incubation, wild-type seedlings started to produce many lateral roots, but nph4-1 arf19-1 and slr-1 fail to produce any lateral roots. However, after 5 d of incubation on IAA, several lateral roots are induced in nph4-1 arf19-1 but not in slr-1 (data not shown). Lower concentrations of IAA (1 to 100 nM) fail to induce lateral root formation in nph4-1 arf19-1 even after 5 d of incubation (data not shown). These results suggest that the auxin- induced lateral root formation is inhibited in nph4-1 arf19-1, but is more severely impaired in slr-1. Also, both slr-1 and arf7 arf19 mutants have smaller size aerial tissues compared with the wild type and single mutants, but slr-1 has smaller rosette leaves and shorter petioles than arf7 arf19 (Figure 4C). The most striking phenotypic difference between the arf7 arf19 and slr-1 mutants is the root hair formation. The slr-1 mutant has very few root hairs (Fukaki et al., 2002), whereas the arf7 arf19 mutant and the arf7 and arf19 single mutants show normal root hair formation (Figure 4E).
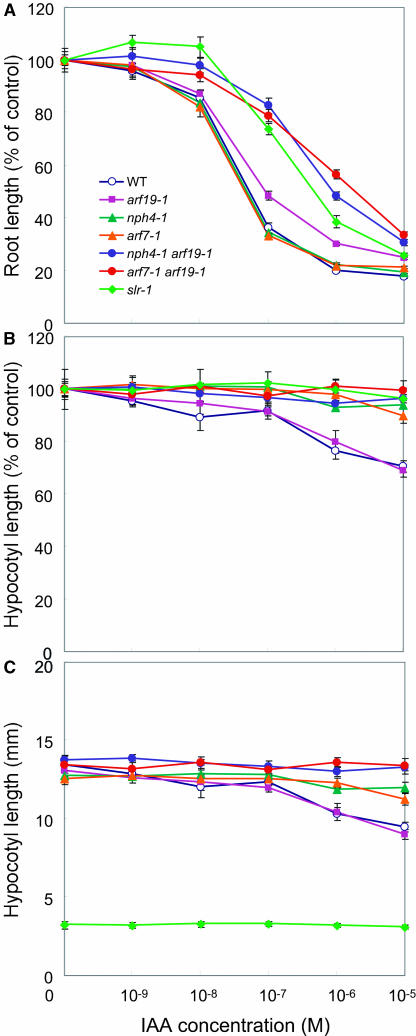
Auxin Sensitivity of arf7 arf19
The arf7 single mutants display reduced auxin sensitivity in hypocotyl growth, whereas they show normal auxin response in the roots (Figures 5A and 5B; Watahiki and Yamamoto, 1997; Stowe-Evans et al., 1998). By contrast, arf19-1 shows normal auxin sensitivity in the hypocotyls and a mild but significant resistance to exogenous auxin in the roots (Figures 5A and 5B). The same level of auxin resistance is also observed in the roots of arf19-2 (data not shown), suggesting that the auxin response is slightly impaired in the roots of the arf19 single mutants. Interestingly, the arf7 arf19 double mutants display severely reduced auxin sensitivity in both roots and hypocotyls (Figures 5A and 5B). The root auxin sensitivity is impaired in arf7 arf19 to the same degree as in slr-1. The data suggest that the hypocotyl auxin sensitivity is impaired in the arf7 single mutants, the root auxin sensitivity is impaired in the arf19 single mutants, and both are severely impaired in the arf7 arf19 double mutant. Surprisingly, the slr-1 hypocotyls fail to elongate after transfer to dark conditions, and exogenous auxin application does not affect their hypocotyl growth (Figures 5B and 5C).
Figure 5.
Auxin Sensitivity of the Wild Type, arf7, arf19, arf7 arf19, and slr Mutants.
(A) Inhibition of root growth by exogenous auxin. Each value represents the average of more than 10 seedlings. Bars represent se of the average.
(B) and (C) Inhibition of hypocotyl elongation by exogenous auxin. Data represent the mean of hypocotyl length as a percent of controls (B) or of actual measurements (C). Bars represent se of the average. See Methods for experimental details.
Expression Patterns of ARF7 and ARF19
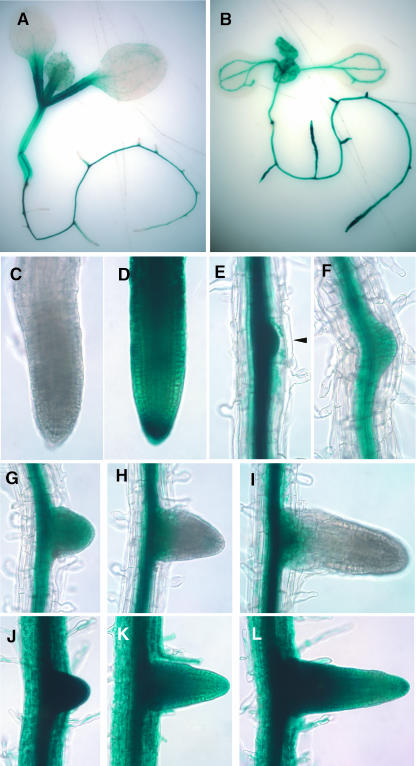
We generated transgenic plants with ProARF7:GUS and ProARF19:GUS to gain a better understanding of the tissue-specific expression of ARF7 and ARF19. The expression patterns of ProARF7:GUS and ProARF19:GUS are distinct, with partial overlap in light-grown seedlings (Figures 6A and 6B). Strong GUS expression is observed in the hypocotyls and petioles of ProARF7:GUS seedlings (Figure 6A), whereas ProARF19:GUS expression is restricted to the vascular tissue in the aerial parts (Figure 6B). In root tissue, unlike the aerial part, ProARF19:GUS is strongly expressed throughout, including vascular tissue, the meristematic region, root cap, root hair, and the sites of newly forming lateral roots (Figures 6B, 6D, and 6J to 6L). By contrast, ProARF7:GUS expression in the primary root is restricted to the vascular tissues and is not detected in the meristematic region, root cap, and root hairs (Figures 6A, 6C, and 6E to 6I). ProARF7:GUS is expressed in the early stages of lateral root primordia (Figure 5E). However, after the root primordia emerge from the parental primary roots, the expression of ProARF7:GUS dissipates from the meristematic region (Figures 6G to 6I). ProARF7:GUS expression is detected in the vascular tissue after the lateral root is elongated (data not shown). The results suggest that both ARF7 and ARF19 are expressed in sites where lateral roots are initiated, consistent with the observation of impaired lateral root formation in the arf7 arf19 double mutants.
Figure 6.
Expression of GUS in ProARF7:GUS and ProARF19:GUS Transgenics.
(A) GUS expression in a 6-d-old light-grown ProARF7:GUS seedling.
(B) GUS expression in a 6-d-old light-grown ProARF19:GUS seedling.
(C) Root apex of a ProARF7:GUS seedling primary root.
(D) Root apex of a ProARF19:GUS seedling primary root.
(E) to (I) ProARF7:GUS expression in the vascular tissue of mature primary root, lateral root primordia ([E] and [F], arrowhead), and developing lateral roots ([G] to [I]).
(J) to (L) ProARF19:GUS expression in entire tissue of primary root and developing lateral roots.
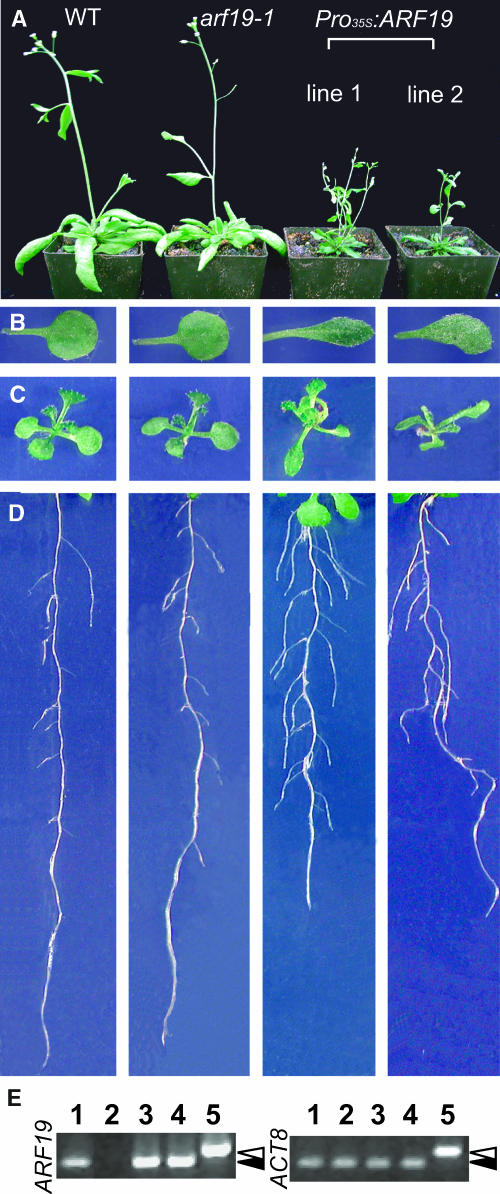
ARF19 Overexpression
Although the loss of ARF19 function does not alter plant development, overexpression of ARF19 has a dramatic effect on plant morphology (Figures 7A to 7D). Overexpression of ARF19 results in alternation of root architecture (Figure 7D). The leaves of Pro35S:ARF19 plants are narrower, elongated, and misshapen (Figures 7B and 7C). The Pro35S:ARF19 plants exhibit strong reduction in apical dominance and have a dwarf phenotype (Figure 7A). They produce a small number of siliques and have lower seed production (data not shown). The phenotype of Pro35S:ARF19 plants is associated with higher levels of the ARF19 transcript (Figure 7E).
Figure 7.
Developmental Defects by ARF19 Overexpression.
(A) to (C) Growth inhibition in 5-week-old plants (A), first true leaves (B), and 12-d-old light-grown seedlings (C).
(D) Alteration of root architecture in 10-d-old seedlings.
(E) Expression of ARF19 in overexpressing lines from 7-d-old light-grown seedlings. ARF gene expression was assessed by RT-PCR as described in Methods. The lanes are as follows: 1, the wild type; 2, arf19-1; 3, Pro35s:ARF19 line 1; 4, Pro35s:ARF19 line 2; 5, genomic DNA. Accumulation of the ACT8 transcript was used as an internal control. White and black arrowheads indicate the size of genomic and cDNA fragments, respectively.
Transcriptional Profiling of the arf7, arf19, and arf7 arf19 Mutants
The auxin-related phenotypes of arf7, arf19, and arf7 arf19 mutants prompted us to perform detailed microarray analysis with these mutants using the Affymetrix whole-genome ATH1 GeneChip. We used the nph4-1, arf19-1, and nph4-1 arf19-1 mutants as representatives for each mutant allele during this experiment. Light-grown seedlings of the wild type, nph4-1, arf19-1, and nph4-1 arf19-1 were treated for 2 h with the carrier solvent ethanol (control sample) or 5 μM IAA (auxin-treated sample). Each experiment was performed in triplicate, and total RNA was independently isolated to generate biotin-labeled cRNA for hybridization (see Methods).
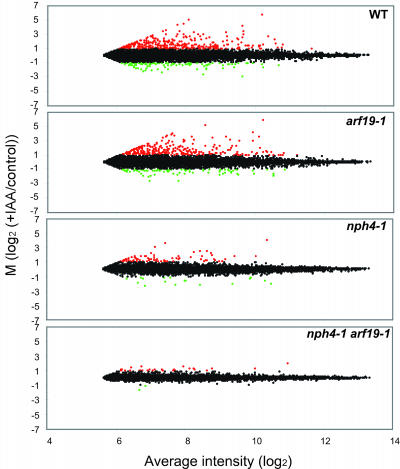
Figure 8 shows the scatter plots representing the auxin-regulated transcriptional profiles of wild-type, arf19-1, nph4-1, and nph4-1 arf19-1 mutants. A cursory examination of these scatter plots demonstrates that the loss of ARF7 and ARF19 causes gross changes in auxin-induced gene expression. The wild-type scatter plot shows that the gene expression profile is globally altered by exogenous auxin treatment. The scatter plot of arf19-1 shows a similar degree of distribution as with the wild type, suggesting that almost normal auxin-regulated gene expression is maintained in the arf19 single mutant (Figure 8). However, the scatter plots of nph4-1 and nph4-1 arf19-1 display a smaller degree of distribution than that of the wild type, indicating that the auxin-mediated transcriptional regulation is globally repressed in these mutants (Figure 8). We extracted the auxin-regulated genes using the log2 expression values from the robust multichip analysis (RMA) output file (Irizarry et al., 2003) and established rigorous statistical criteria based on a variance measurement to generate auxin-regulated gene lists (see Methods). Among the 22,800 genes, only 203 met the criteria for more than twofold auxin induction (I, induced genes), and 68 genes met the criteria for more than twofold repression (R, repressed genes). A complete list of all the auxin-regulated genes and how they are affected by the mutants can be found in the Supplemental Tables 4 and 5 online. These gene lists include various classes of known auxin-regulated genes, such as Aux/IAA, GH3, SAUR, and ACS, consistent with similar studies reported previously (Tian et al., 2002; Ullah et al., 2003; Redman et al., 2004). The genes identified as auxin-regulated (induced or repressed) were functionally categorized to examine the auxin-regulated cellular and metabolic processes affected by either or both loss-of-function mutations of ARF7 and ARF19. Supplemental Figure 6 online shows their functional classification. Approximately 80% of the auxin-regulated genes is currently annotated as encoding proteins of known or putative function.
Figure 8.
Global Gene Expression Profiling.
MA plots (Dudiot et al., 2002) showing changes of auxin-regulated gene expression levels in the wild type, arf19-1, nph4-1, and nph4-1 arf19-1. Each plot represents the log ratio of the average of the auxin-treated samples (I) to the control samples (C) [M = log2 (I/C)] versus overall average intensity [A = log2√(I*C)]. The genes induced by auxin treatment (M > 1) are highlighted in red, and the genes repressed by auxin treatment (M < −1) are highlighted in green. The data were further analyzed for variance to extract statistically valid auxin-regulated genes (see Methods).
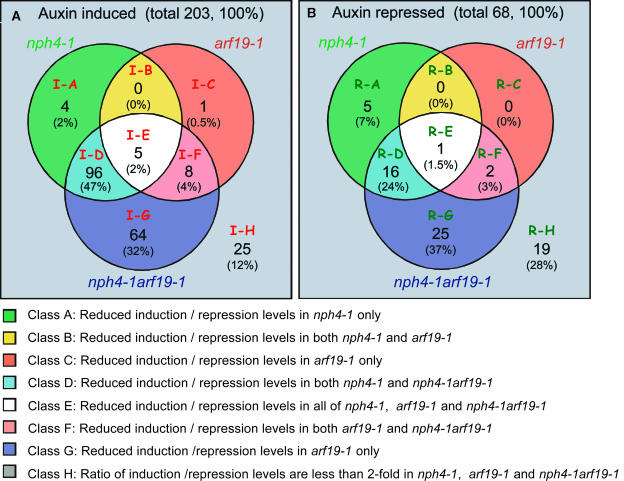
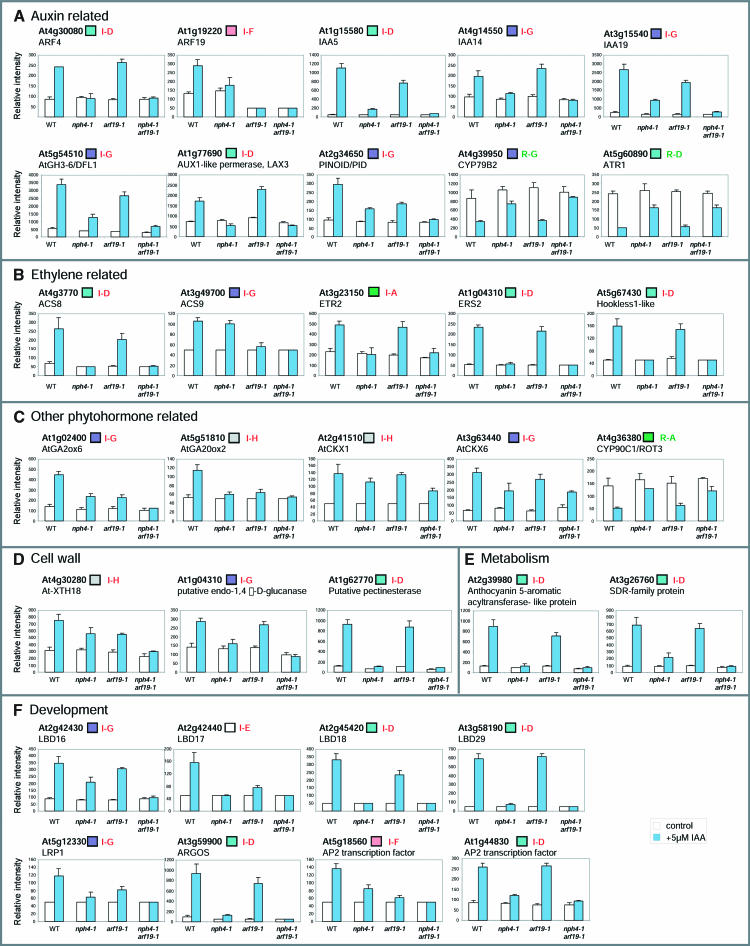
We subsequently extracted the gene sets that were induced or repressed by auxin in the wild type, which do not respond, or were only slightly responsive to auxin in the mutants (see Methods). Among the 203 auxin-induced genes, 105 (51.7%), 14 (6.9%), and 173 (85.2%) were identified as differentially regulated genes by nph4-1, arf19-1, and nph4-1 arf19-1, respectively. Likewise, 22 (32.4%), 3 (4.4%), and 44 (64.7%) among the 68 auxin-repressed genes were identified as differentially regulated genes by nph4-1, arf19-1, and nph4-1 arf19-1, respectively. This comparative analysis of differentially regulated genes among the three mutants revealed overlapping genes among these gene sets (Figure 9). For example, among the 203 auxin-induced genes, 96 were similarly affected by the nph4-1 single and nph4-1 arf19-1 double mutants (Figure 9A, class I-D). The class I-D genes are considered to be preferentially regulated by ARF7. Likewise, eight auxin-induced genes are similarly affected in the arf19-1 single and nph4-1 arf19-1 double mutants (Figure 9A, class I-F). These genes are considered to be preferentially regulated by ARF19. By contrast, 64 auxin-induced genes are differentially regulated only by the nph4-1 arf19-1 double mutant (Figure 9A, class I-G). The genes classified into class I-G are considered to be redundantly regulated by both ARF7 and ARF19. Similar distribution of differentially regulated genes is found among auxin-repressed genes (Figure 9B, class R-D to R-G). Supplemental Figure 5 online shows the expression behavior of individual genes that belong to each class (class I-A to I-H and class R-A to R-H). Figure 10 shows the expression behavior of some representative auxin-regulated genes of various functional categories in these various classes. In addition to classical auxin-regulated genes, such as IAA5, IAA14, and IAA19, various classes of genes involved in ethylene biosynthesis and perception, phytohormone-related, and cell wall biosynthesis and development show defective auxin-regulated gene expression in the mutants, especially in nph4-1 arf19-1. A wide range of auxin-regulated cellular and metabolic processes is affected by the loss of ARF7 and ARF19 gene function.
Figure 9.
Comparative Analysis of Genes Differentially Regulated by Auxin in nph4-1, arf19-1, and nph4-1 arf19-1.
Differentially regulated genes in mutants among auxin-induced (A) and repressed (B) genes are shown. Each circle within the Venn diagram indicates numbers and percentages (in parentheses) of genes with repressed induction or repression levels. Only those genes with greater than twofold fold change ratio (FCR) in nph4-1, arf19-1, and nph4-1 arf19-1 were analyzed (see Methods). We defined each area of the Venn diagram from A to H, and each class was further divided into two subgroups based on their auxin-induced expression profiles in the wild type. The genes classified into class D are considered to be preferentially regulated by ARF7, and those classified into class F are considered to be preferentially regulated by ARF19. The genes classified into classes E and G are considered to be redundantly regulated by ARF7 and ARF19. The class A genes have similar expression profiles to class D genes. Likewise, class C genes have similar expression profiles to class F genes. The expression profiles of the representative genes from each class are shown in Supplemental Figure 5 online.
Figure 10.
The Expression Profiles of Representative Auxin-Regulated Genes in the Wild Type, nph4-1, arf19-1, and nph4-1 arf19-1.
The data represent the average relative intensity expression level of control (open bar) or auxin-treated (blue bar) samples from triplicate experiments. Bars represent sd of the average. Boxes next to gene names indicate classification color codes according to Figure 9.
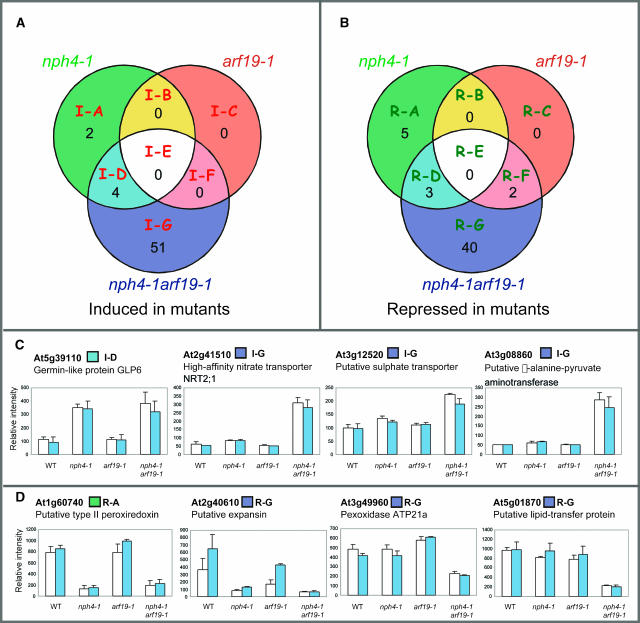
The transcriptional profile of the untreated control seedlings is also altered in the nph4-1 arf19-1 double mutant. Comparison of the transcriptional profiles between the nph4-1 arf19-1 mutant and the wild type in the absence of auxin treatment reveals that 55 and 45 genes are induced and/or repressed twofold or higher in nph4-1 arf19-1, respectively (Figure 11; see Supplemental Tables 6 and 7 online). Interestingly, 20 of the 55 induced genes in nph4-1 arf19-1 are involved in metabolism (see Supplemental Table 6 online). Fewer genes have altered gene expression in untreated nph4-1 and arf19-1 seedlings (Figures 11A and 11B). Only two genes are repressed in arf19-1, and one of them is ARF19 itself, suggesting that the arf19-1 mutation does not affect gene expression in untreated seedlings. Figures 11C and 11D show some representatives of induced or repressed genes in nph4-1 arf19-1 or both nph4-1 and nph4-1 arf19-1.
Figure 11.
Effect of the nph4-1, arf19-1, and nph4-1 arf19-1 Mutations on Global Gene Expression in Untreated Control Samples.
(A) Induced genes in the mutants under control conditions.
(B) Repressed genes in the mutants under control conditions. Each circle within the Venn diagram indicates the number of genes with greater than twofold induction or repression.
(C) Expression profiles of induced classes of genes.
(D) Expression profiles of repressed classes of genes. Data represent the average relative intensity expression levels of control (open bar) or auxin-treated (blue bar) samples from triplicate experiments. Bars represent sd of the average. Boxes next to gene names indicate classification color codes according to (A) and (B).
DISCUSSION
The ARF gene family encodes transcriptional regulators that are involved in auxin signaling. Despite their essential role in auxin-mediated gene regulation, little is known regarding their biological functions, except for very few of them studied by classical molecular genetic analysis. Questions arise, such as why does Arabidopsis have so many ARFs? What is the biological function of each ARF? Which genes do they regulate? To answer these questions, we have attempted to isolate loss-of-function T-DNA insertion mutants for all the ARF gene family members using a reverse-genetics strategy. PCR-based reverse genetic screens provide a systematic strategy for analyzing gene function (Borevitz and Ecker, 2004). We have identified T-DNA insertion alleles for 19 out of 23 ARF genes, and initial characterization has been conducted for 18 ARF T-DNA insertion alleles among the 27 lines isolated. Among the 18 arf single mutants, obvious growth phenotypes were observed only in the previously identified mutants using forward genetics (i.e., arf2/hss, arf3/ett, arf5/mp, and arf7/nph4). The rest of the arf single mutants fail to show an obvious growth phenotype. However, in-depth analysis of these lines regarding their auxin resistance, gravitotropic behavior, and inhibition of root elongation may detect biological phenotypes associated with these lines. These ARFs may act redundantly in auxin-mediated gene regulation and provide compensatory functions during plant development. The expression of at least two clustered ARF genes in a specific stage of embryogenesis reinforces the concept of functional redundancy among the ARF proteins. To query the concept of gene redundancy, we generated several double mutants among closely related ARF members. Their phenotypic analysis indicates that related pairs of ARFs, namely, ARF1/ARF2, ARF6/ARF8, and ARF7/ARF19, act redundantly in a distinct developmental manner. During this study, we focused on the redundant functions of ARF7 and ARF19 using biological and molecular approaches. A similar picture was recently presented with the ARF5/ARF7 pair (Hardtke et al., 2004). The in planta interaction between ARF5 and ARF7 suggested by the experiments of Hardtke et al. (2004) raises the possibility that different combinations of ARF heterodimers may have various selective functions in regulating targeted gene expression. Potential heterodimerization between ARF7 and ARF19 is also suggested by the inhibition of auxin-induced expession of genes such as At2g23060 (Hookless1-like) and At4g22620 (AtSAUR-34) by either the arf7 or the arf19 mutant (see Supplemental Figure 5 online; class I-E). Consequently, the generation of double and higher-order mutants using available arf T-DNA insertion mutants will be beneficial to understand auxin-regulated processes mediated by ARF–ARF and ARF–Aux/IAA interactions. Similar studies using reverse genetics have also revealed unique and overlapping functions among the R2R3-MYB and MADS box transcription factor gene family members (Meissner et al., 1999; Parenicova et al., 2003; Pinyopich et al., 2003).
Unique and Overlapping Developmental Functions of ARF7 and ARF19
Considering the phenotypes of arf7 and arf19 single mutants, ARF7 appears to regulate auxin-dependent differential growth in the hypocotyls, and ARF19 partially mediates auxin signaling in the roots. The severity of their phenotypes is greatly enhanced in the double mutant compared with the single mutations, demonstrating redundant functions between ARF7 and ARF19. The arf7 arf19 mutant exhibits strong auxin-related phenotypes, including severely impaired lateral root formation, agravitropic hypocotyls and roots, and small organs and enhanced apical dominance in aerial portions. These phenotypes are observed only in the arf7 arf19 double mutant, but not in the single mutants, indicating that these developmental events are redundantly regulated by ARF7 and ARF19. Expression of one ARF allows for functional compensation for the loss of the other in arf7 and arf19 single mutants. This may be because of the high similarity of these two proteins. The analysis of promoter-GUS transgenic plants demonstrated that there is a significant agreement between the expression patterns and the developmental defects in the single and double mutants. ProARF7:GUS is strongly expressed in the hypocotyls, whereas ProARF19:GUS is strongly expressed in the roots. Furthermore, expression of ProARF7:GUS is detected throughout the hypocotyl, whereas the expression of ProARF19:GUS is restricted to the vascular tissue of the hypocotyls (Figures 6A and 6B). However, despite the global ProARF19:GUS expression and an altered auxin sensitivity in arf19 root, only the arf7 mutants have slightly reduced numbers of lateral roots (Table 1), suggesting that the ARF7 has a regulatory function in lateral root initiation. The microarray experiments show that the auxin-dependent induction of ARF19 is impaired in the nph4-1 mutant (Figure 10A). Interestingly, the promoter region of ARF19 contains two AuxREs (data not shown), suggesting that ARF7 may directly modulate the expression of ARF19. This may provide an alternative explanation for the apparent phenotype of the arf7 mutants. The inadequate auxin-mediated induction of ARF19 expression may have an additive effect on the loss of ARF7 function, yielding an obvious phenotype. We have not tested yet whether the ARF7 and ARF19 proteins can complement the loss of each other. Promoter-swapping experiments using transgenic arf7 and arf19 single or double mutants harboring ARF7 promoter:ARF19 and ARF19 promoter:ARF7 gene constructs have the potential to clarify this issue.
ARF7 and ARF19 Regulate Both Unique and Partially Overlapping Sets of Target Genes
The microarray data provide clear evidence for the unique and redundant functions of ARF7 and ARF19 on auxin-mediated gene expression. The almost complete lack of auxin-mediated transcriptional regulation in the arf7 arf19 mutant is puzzling (Figure 9). It implies that ARF7 and ARF19 are the only ARF factors that are necessary and sufficient for auxin signaling in 7-d-old light-grown seedlings. Are the rest of the ARFs dispensable? The possibility exists that the majority of auxin-regulated gene expression during this stage of development is mediated by the ARF7/ARF19 pair. It should be noted that the adult arf7 arf19 plants, although smaller in size, have a normal appearance with normal flowers and fertility, suggesting that the ARF7/ARF19 pair may not be critical for auxin-mediated transcriptional regulation during the development of aerial organs. Such a proposition is supported by the phenotypes of two other ARF mutants, arf5/mp and arf3/ett; they control auxin-mediated gene regulation responsible for axial cell and gynoecium patterning during organogenesis, respectively, indicating that ARF5 and ARF3 may also act in a particular developmental window. In addition, several single and double arf mutants, including arf2, arf1 arf2, arf3, and arf6 arf8, have flowers with abnormal morphology and/or poor fertility, suggesting that these ARFs may act redundantly in auxin-mediated gene regulation responsible for flower development. Comparative microarray analysis with different double mutants at different developmental stages has the potential to clarify this view. Alternatively, the remaining ARFs may regulate genes that are not auxin regulated at that particular developmental stage. The current prevailing view that all ARFs regulate auxin-mediated gene expression has not been tested experimentally with vigor. Finally, the remaining ARFs may regulate genes in a cell-specific manner (distinct cell types) that the microarray analysis fails to detect. This last possibility points to the necessity of conducting global expression studies in specific cell types (Birnbaum et al., 2003).
Comparative analysis of the gene sets in which auxin-mediated regulation was suppressed in nph4-1, arf19-1, and nph4-1 arf19-1 mutants allowed us to classify the auxin-regulated genes into gene sets preferentially regulated by ARF7 and ARF19 alone or redundantly regulated by both ARF7 and ARF19 (Figure 9). The data suggest that the ARF7 and ARF19 regulate both distinct and partially overlapping sets of target genes (Figure 9). ARF7 appears to regulate many more auxin-induced genes (47%) than ARF19 (4%), and ∼30% of the auxin-induced genes are redundantly regulated by ARF7 and ARF19. It is of a great interest that 90% of the auxin-induced or -repressed genes contain at least one AuxRE (TGTCnC or GnGACA) in their ∼2-kb promoter region (data not shown), suggesting that they are directly regulated by these ARFs. This suggests that the ARF7 and ARF19 proteins have the capacity to act as transcriptional activators or repressors of various auxin-regulated genes. The current assignment of ARF7 and ARF19 solely as transcriptional activators is not warranted. Although microarray analysis provides useful and a vast amount of information regarding the genes regulated by the ARF7/ARF19 pair, more direct global technologies, such as chromatin immunoprecipitation and DNA CHIP (ChIP:CHIP), have the potential to identify target genes that are regulated by this and other ARF pairs (Ren et al., 2000; Iyer et al., 2001).
The lists of auxin-regulated genes in which expression is inhibited in the mutants contain putative downstream targets of ARF7 and ARF19. LATERAL ROOT PRIMORDIUM1 (LRP1) is one such candidate gene. The expression level of LRP1 is induced by auxin treatment in the wild type (Figure 10F; Ullah et al., 2003), and its auxin-mediated induction is inhibited in nph4-1 arf19-1 (Figure 10F). LRP1 is expressed during the early stage of lateral root primordia (Smith and Fedoroff, 1995), and its inhibition is consistent with impaired lateral root formation in the nph4-1 arf19-1 mutant. Another potential candidate is the AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS) gene, which is inhibited in auxin-treated and -untreated nph4-1 and nph4-1 arf19-1 mutants (Figure 10F). Loss-of-function and gain-of-function mutants of ARGOS result in smaller and larger plant sizes, respectively (Hu et al., 2003). The small plant size of arf7 arf19 may be related to the low expression level of ARGOS. Other potential targets of ARF7 and ARF19 are the genes encoding LATERAL ORGAN BOUNDARIES (LOB) domain (LBD) family members (Iwakawa et al., 2002; Shuai et al., 2002). The current analysis reveals that four LBD genes, LBD16, LBD17, LBD18, and LBD29, are induced by auxin, and their auxin-dependent induction is severely impaired in nph4-1 and nph4-1 arf19-1 mutants (Figure 10F). All four highly similar auxin-inducible LBD genes contain potential AuxREs in their regulatory regions (data not shown). Although the function of these LBD genes is still unclear, LOB is considered to participate in boundary establishment or communication links between the meristems and initiating lateral organs (Shuai et al., 2002). Overexpression of several LBD gene family members results in strong morphological changes (Nakazawa et al., 2003). The root-specific expression of LBD16 and LBD29 (Shuai et al., 2002) suggests that these two LBDs may be involved in lateral root formation. Overexpression of LBD16 rescues the lateral root phenotype of the arf7 arf19 double mutant (Y. Okushima and H. Fukaki, unpublished data). Finally, multiple classes of genes encoding auxin conjugating or auxin synthesis enzymes, cell wall–related proteins, metabolic enzymes, and transcription regulators are potential targets of the ARF7/ARF19 pair (Figure 10; see Supplemental Tables 4 and 5 online).
Regulation of ARF7 and ARF19 by IAA14 and Other Aux/IAAs
The phenotypes of the arf7 arf19 mutants are quite similar to those observed in the iaa14/slr mutant. Enhanced IAA14 protein level and the loss of both ARF7 and ARF19 functions have similar effects, indicating that all three proteins act on the same developmental pathway. Promoter-GUS expression analysis has revealed that the ARF7, ARF19, and IAA14 have overlapping expression patterns at least in the root tissue (Fukaki et al., 2002). This raises the prospect that IAA14 may be a molecular partner of ARF7 and ARF19 by forming heterodimers in planta, thereby repressing the activity of these two ARFs. This interaction may inhibit ARF7- and ARF19-mediated transcriptional activation/repression. Division of pericycle cells is blocked during lateral root initiation in the iaa14/slr-1 mutant (Fukaki et al., 2002). The stronger phenotype of iaa14/slr compared with that observed in arf7 arf19 (i.e., complete lack of lateral roots and few root hairs) may be attributable to the inhibition of other ARFs by the stabilized IAA14 protein. In addition to the iaa14/slr mutant, iaa3/shy2 (Tian and Reed, 1999), iaa19/msg2 (Tatematsu et al., 2004), and iaa28-1 (Rogg et al., 2001) also have reduced numbers of lateral roots, whereas the iaa14 T-DNA insertion mutant (loss of function) has a normal root phenotype (Y. Okushima and A. Theologis, unpublished data). These data suggest that the function of ARF7 and ARF19 may be negatively regulated by multiple Aux/IAA proteins. Similar functional interactions have been proposed between ARF5 and IAA12 (Hamann et al., 2002; Vogler and Kuhlemeier, 2003), IAA19/MSG2 and ARF7 (Tatematsu et al., 2004), and ARF7 and IAA12 (Hardtke et al., 2004). In planta heterodimerization studies using bimolecular fluorescence complementation have the potential to elucidate the heterodimeric interactions among the Aux/IAA and ARF gene family products (Hu et al., 2002; Tsuchisaka and Theologis, 2004).
METHODS
Materials
The pBI101 vector was purchased from Clontech (Palo Alto, CA). All chemicals used for this study were American Chemical Society reagent grade or molecular biology grade. Oligonucleotides were purchased from Operon Technologies (Alameda, CA) or synthesized in house with a Polyplex oligonucleotide synthesizer (GeneMachines, San Carlos, CA).
Molecular Biology
Standard protocols were followed for DNA manipulations described by Sambrook et al. (1989). Standard protocols for DNA sequencing were used to confirm the accuracy of the DNA constructs.
Plant Growth Conditions
Arabidopsis thaliana ecotype Col was used throughout this study. Seeds were surface sterilized for 8 min in 5% sodium hypochlorite + 0.15% Tween-20, excessively rinsed in distilled water and plated on 0.8% agar plates containing 0.5× MS salts (Life Technologies, Rockville, MD) + 0.5 mM Mes, pH 5.7, + 1% sucrose + 1× vitamin B5. The plates were incubated in the dark at 4°C for 2 d and were subsequently transferred to a 16-h-light/8-h-dark cycle at 22°C for light-grown seedlings or in the dark for etiolated seedlings. Mature plants were also grown under the light conditions mentioned above. The root auxin sensitivity assay was performed as follows: 4-d-old light-grown seedlings were transferred to vertically oriented agar plates containing appropriate concentrations of IAA. The root length was determined after an additional 5 d of growth. The auxin sensitivity assay for hypocotyl elongation was performed with 3-d-old seedlings grown on plates lacking auxin and then was transferred to the plates containing various concentrations of IAA and grown for an additional 5 d in the dark. The root and hypocotyl lengths were determined using the NIH Image 1.63 program (http://rsb.info.nih.gov/nih-image/download.html). The phototropic response of etiolated seedlings to blue light was performed as previously described by Liscum and Briggs (1995). Three-day-old etiolated seedlings were exposed to unilateral blue light (1 μmol m−2 s−1) for 8 h and then photographed.
Identification and Characterization of T-DNA Insertion Alleles
Screening for T-DNA Insertions
The identification of insertional mutants was performed using a PCR-based screen. For each gene, a forward (F) primer annealing to 100 to 150 bp 5′ of the ATG and a reverse (R) primer annealing to 100 to 150 bp 3′ of the translation stop codon were designed. The size of the genomic products ranged from 6 to 3.2 kb. Eight sets of DNA template derived from 10,000 plants each (80,000 lines total) were screened. Each set of template contained 40 tubes of DNA (10 each of DNA combined from column, row, plate, and individual superpools). Identification of an individual requires a PCR product in each of the four superpools. Using all combinations of F and R primers with primers annealing to the left border and right border of the T-DNA, PCRs were run (4 × 40 × 8 = 1280 reactions per gene). All operations were adapted to a 384-well format and handling of samples performed with a BioMek robot (Beckman, Palo Alto, CA). The products were analyzed by DNA gel blotting to allow increased sensitivity of detection and assess the specificity of screening. Subsequent to this screen, two large databases containing sequence of DNA flanking T-DNA inserts in 100,000 and 20,000 independent lines have been screened in silico. Data for the 100,000 lines were generated in a collaboration of the University of California, Berkeley, with the Torrey Mesa Research Institute, and the 20,000 lines have been obtained by SIGNAL (http://signal.salk.edu/cgi-bin/tdnaexpress).
Confirmation of T-DNA Lines
The nature and location of the T-DNA insertion is confirmed by sequencing PCR products. Once the location of the T-DNA insertion was confirmed, we designed gene-specific PCR primers that flank the T-DNA for use in a codominant genotyping analysis. By performing two sets of PCR, one using the gene-specific primer pair and the other using a gene-specific primer and the T-DNA border primer, we could determine whether the individual is homozygous for no T-DNA insertion, heterozygous for the T-DNA insertion, or homozygous for the T-DNA insertion.
Molecular Characterization of the T-DNA Lines
To determine the number of T-DNA inserts present in the lines, we compared the DNA gel blot hybridization patterns arising from sibling plants that were either homozygous for the T-DNA insertion or homozygous for no T-DNA. To remove additional T-DNA loci from the lines of interest, backcrosses to wild-type Col were performed, and plants homozygous for the T-DNA insertion were again identified.
Construction of Promoter-GUS Fusions
The following primers were used to amplify the ARF promoter fragments: ARF7, F 5′-CTAAGCTTGTCGACAGTACGTAGATTATTTTCCACAACTCTCTC-3′ and R 5′-GAGGATCCATGATCACTCAACTTTACTTTCTCTGAAG-3′; ARF12, F 5′-GGAGGTCGACACAAACAACATGATTGAATAAG-3′ and R 5′-GATCGGATCCCCAAAATATGTTATCTCAAC-3′; ARF19, F 5′-ACTGAAGCTTTGGGCTAGATTCATCCGTATCTGGGT-3′ and R 5′-CCCGGGAATTCTCATGATGGTTTGGTGCAGGGAAG-3′; ARF22, F 5′-GAAGAAGAGTGAAATCCAGTGACC-3′ and R 5′-AGGATCCATAAGCTCGTATCTAAAGCTCGG-3′.
Promoter fragments (ARF12 and ARF22, 2 kb; ARF7, 2.5 kb; ARF19, 3.2 kb) upstream of the translation initiation codon were synthesized by PCR using wild-type (Col) genomic DNA and the primers listed above. The fragments were sequenced and subcloned into the pBI101.2 (ARF7, ARF12, and ARF22) or pZP121 (ARF19; Hajdukiewicz et al., 1994) vectors as SalI/BamHI (ARF7 and ARF12), HindIII/BamHI (ARF22), and SalI/BspHI (ARF19) fragments. The pZP121 vector was modified by introducing the GUS gene as an NcoI/SacI fragment. Among the four promoter GUS constructs, ProARF12:GUS, ProARF22:GUS, ProARF7:GUS, and ProARF19:GUS, the ProARF19:GUS promoter also contains 889 bp of the 3′ region of the ARF19 gene (from the 41-bp 5′ of the ARF19 translation stop codon to the 848-bp 3′ of the translation stop codon). It was amplified by PCR with the primers, F 5′-ACTGGAGCTCGTACACTATGAAGACACTTCTGCTGCAGCT-3′ and R 5′-TGACGAATTCAAGACGCGATTGAACCAACCCGGTATGA-3′, using BAC T29M8 DNA as a template. It was subcloned as a SacI/EcoRI fragment into a pZP121-ProARF19-GUS construct. With the SacI site present in the forward primer and the EcoRI site located in the reverse primer, the PCR product was cloned into pNcoI-GUS to create pGUS-3A11.
These constructs were introduced into Agrobacterium tumefaciens strain GV3101, and wild-type Col plants were transformed by dipping (Clough and Bent, 1998). Kanamycin-resistant plants in the T2 (ProARF7:GUS) and T3 (ProARF12:GUS, ProARF19:GUS, and ProARF22:GUS) generations were histochemically stained to detect GUS activity by incubating seedlings or tissues in 100 mM sodium phosphate buffer, pH 7.5, containing 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, and 0.1% Triton X-100 for 5 h at 37°C followed by dechlorophylation in 70% ethanol. Several independent lines were examined for GUS staining.
Overexpression of ARF19
Transgenic plants overexpressing the ARF19 protein (Pro35S:ARF19) under the control of the 35S promoter were generated by subcloning the 35S-ARF DNA (pS-A11) as a XhoI fragment into the binary vector pKF111.XL (Ni et al., 1998) and transforming plants as described (Clough and Bent, 1998). Fifty-two T1 transformants were selected in soil based on resistance to Finale (Farnam Companies, Phoenix, AZ) diluted 1:1,000 (final concentration 0.05% glufosinate ammonium) in 0.005% Silwet, and sprayed on the germinating seedlings. Two lines (line 1 and line 2) were examined in detail.
RT-PCR Analysis
Total RNA was isolated from various stages of flower and silique samples using RNAqueous RNA isolation kit with Plant RNA isolation aid (Ambion, Austin, TX). For each sample, 2.5 μg of total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI) to eliminate genomic DNA contamination. First-strand cDNA was synthesized with an oligo(dT)24 primer using a SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA). Then, 1/100th of the resulting cDNA was subjected to 35 cycles of PCR amplification (95°C for 20 s, 62°C for 20 s, 72°C for 45 s). A mixture of ARF12, ARF13, ARF14, ARF15, ARF20, ARF21, and ARF22 cDNA was amplified using primers designed based on the ARF12 coding region: 5′-TCTGGACACTCCTCCGGTGA-3′ and 5′-TGAGAGACTCTTCCTGGACTTCAAA-3′. Because the nucleotide sequences of ARF12, ARF13, ARF14, ARF15, ARF20, ARF21, and ARF22 cDNA are very similar (see Supplemental Table 1 online), the same expression patterns shown in Supplemental Figure 2B online were also observed when we used primer pairs based on the ARF21 and ARF22 coding region (data not shown). The expression level of ARF19 in wild-type, arf19-1, and Pro35S:ARF19 plants was performed using the primers 5′-ACAAAGGTTCAAAAACGAGGGTCA-3′ and 5′-CGATGGCCCTCGAATGATAATGTAA-3′. ACT8 gene-specific primers described by An et al. (1996) were used for control amplification.
Microarray Analysis
Surface-sterile seeds (1.8 mg) were germinated in 40 mL of 0.5× MS medium (Life Technologies) containing 1.5% sucrose and cultured in a 16-h-light/8-h-dark cycle with gentle shaking (100 rpm). After a 7-d culture period, the seedlings were treated with 5 μM IAA (IAA treated) or EtOH (control) for 2 h. Total RNA was prepared using RNAqueous RNA isolation kit with Plant RNA isolation aid (Ambion). After LiCl precipitation, RNA was purified using RNeasy columns (Qiagen, Valencia, CA) and reprecipitated with LiCl. RNA pellets were washed with 70% EtOH (three times) and resuspended in diethyl pyrocarbonate–treated water. Five micrograms of total RNA was used for biotin-labeled cRNA probe synthesis. cRNA probe synthesis, hybridization, washing, and scanning and detection of the array image were performed according to the manufacturer's protocols (Affymetrix, Santa Clara, CA). Twenty-four independent hybridization experiments with three independent biological replicates were performed in this study.
Microarray Data Analysis
Affymetrix GeneChip Microarray Suite version 5.0 software was used to obtain signal values for individual genes. The data files containing the probe level intensities (cell files) were used for background correction and normalization using the log2 scale RMA procedure (Irizarry et al., 2003). The R environment (Ihaka and Gentleman, 1996) was used for running the RMA program. Data analysis and statistical extraction were performed using log2 converted expression intensity data within Microsoft Excel 98 (Microsoft, Redmond, WA). Based on preliminary analysis, a hybridization signal <5.64 (= log2 50) was considered as background; all signals <5.64 were converted to 5.64 before further analysis. The entire data set is provided in the supplemental data online and has been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession numbers GSE627 and GSM9571 to GSM9594.
We used an MA-plot (Dudiot et al., 2002) to represent the difference between two data sets (Figure 10). M = log2 (X/Y) and A = log2 √X*Y (X and Y are the average expression levels for X and Y data sets, respectively). Also, a t value (Dudiot et al., 2002) cutoff was used to identify the statistically valid differentially regulated genes among the two data sets. The t value was calculated using the following formulas; t = M/SE (SE2 = 1/n2 (var1 + var2…+ varn); var is the variance of the expression intensity of the triplicate experiments; n is the number of data sets. A high t value corresponds to low variability (high confidence) data, whereas a low t value corresponds to high variability (low confidence) data. We use 7 as the cutoff t value; data with |t| < 7 were excluded from our differentially regulated gene list.
For example, to extract statistically valid auxin-regulated genes in the wild type, (1) we first calculated the ratio of the average gene expression intensities for the auxin-treated samples to control samples (M). Genes with |M| ≥ 1 (twofold or more induced or repressed; log2 2 = 1) were extracted to generate a preliminary gene list for auxin-regulated genes. At this stage, 294 and 112 genes were identified as auxin induced and repressed genes, respectively. (2) t values for auxin-treated and control samples were calculated, and genes with |t| < 7 were excluded from the list. After this process, 203 of the 294 auxin induced genes in step (1) met this criterion and were extracted as statistically valid auxin-induced genes. Also, 65 genes among 112 repressed genes in step (1) met this criterion and were extracted as statistically valid auxin-repressed genes. The same procedure was employed to identify the genes with induced or repressed expression levels in mutants. Forty-three, 15, and 145 genes were identified as induced genes in nph4-1, arf19-1, and nph4-1 arf19-1 mutants, respectively, in step (1). Among them, 6, 0, and 55 genes passed the step (2) statistical test and then identified as statistically valid induced genes in nph4-1, arf19-1, and nph4-1 arf19-1 mutants, respectively. For identification of repressed genes in the mutants, 28, 11, and 100 genes were extracted as repressed genes in nph4-1, arf19-1, and nph4-1 arf19-1 by step (1), respectively. Among them, 8, 2, and 45 genes passed the step (2) statistical test and then identified as statistically valid repressed genes in nph4-1, arf19-1, and nph4-1 arf19-1 mutants, respectively. To extract the differentially regulated genes in mutants among auxin-regulated genes, we used FCR of induction or repression levels between mutants and the wild type as criteria, with a cutoff FCR value of ≥ 2. Venn diagrams were drawn using GeneSpring software package version 5.1 (Silicon Genetics, Redwood, CA).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY669787 to AY669796 and AY680406.
Supplementary Material
Acknowledgments
We thank E. Liscum, K. Yamamoto, and H. Fukaki for providing nph4-1, msg1-2, and slr-1 seeds, respectively, and T. Speed for helpful discussions regarding microarray data analysis. We also thank D. Hantz for greenhouse work. This research was supported by the National Institutes of Health Grant GM035447 to A.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Athanasios Theologis (theo@nature.berkeley.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028316.
References
- Abel, S., Ballas, N., Wong, L.-M., and Theologis, A. (1996). DNA elements responsive to auxin. Bioessays 18, 647–654. [DOI] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Abel, S., Oeller, P.W., and Theologis, A. (1994). Early auxin-induced genes encode short-lived nuclear proteins. Proc. Natl. Acad. Sci. USA 91, 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M.A., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., and Ecker, J.R. (2004). Plant genomics: The third wave. Annu. Rev. Genomics Hum. Genet. 5, 443–477. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones, Physiology, Biochemistry and Molecular Biology, 2nd ed. (Dordrecht, The Netherlands: Kluwer).
- Dharmasiri, N., and Estelle, M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308. [DOI] [PubMed] [Google Scholar]
- Dudiot, S., Yang, Y.H., Callow, M.J., and Speed, T.P. (2002). Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica 12, 111–139. [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118, 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T.J., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 20, 281–291. [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Benkova, E., Baurle, I., Kientz, M., and Jurgens, G. (2002). The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Harper, R.M., Stowe-Evans, E.L., Luesse, D.R., Muto, H., Tatematsu, K., Watahiki, M.K., Yamamoto, K., and Liscum, E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.-D., Chinenov, Y., and Kerppola, T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Xie, Q., and Chua, N.H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, K., and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Statist. 5, 299–314. [Google Scholar]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Iyer, V.R., Horak, C.E., Scafe, C.S., Botstein, D., Snyder, M., and Brown, P.O. (2001). Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss, S., Rohrmeier, T., and Lehle, L. (2003). The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem. 278, 23936–23943. [DOI] [PubMed] [Google Scholar]
- Li, H., Johnson, P., Stepanova, A., Alonso, J.M., and Ecker, J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7, 1–20. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Reed, J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. [PubMed] [Google Scholar]
- Meissner, R.C., et al. (1999). Function search in a large transcription factor gene family in Arabidopsis: Assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11, 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, M., Ichikawa, T., Ishikawa, A., Kobayashi, H., Tsuhara, Y., Kawashima, M., Suzuki, K., Muto, S., and Matsui, M. (2003). Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34, 741–750. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L., Feldman, L.J., and Zambryski, P.C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Parenicova, L., De Folter, S., Kieffer, M., Horner, D.S., Favalli, C., Busscher, J., Cook, H.E., Ingram, R.M., Kater, M.M., Davies, B., Angenent, G.C., and Colombo, L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Redman, J.C., Haas, B.J., Tanimoto, G., and Town, C.D. (2004). Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J. 38, 545–561. [DOI] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Remington, D.L., Vision, T.J., Guilfoyle, T.J., and Reed, J.W. (2004). Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135, 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, B., et al. (2000). Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- Rogg, L.E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., Nemhauser, J.L., McColl, A., Roe, J.L., Feldmann, K.A., and Zambryski, P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sessions, R.A., and Zambryski, P.C. (1995). Arabidopsis gynoecium structure in the wild and in ettin mutants. Development 121, 1519–1532. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.L., and Fedoroff, N.V. (1995). LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 7, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate Response Locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Harper, R.M., Motchoulski, A.V., and Liscum, E. (1998). NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiol. 118, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C., Muto, H., Higuchi, M., Matamura, T., Tatematsu, K., Koshiba, T., and Yamamoto, K.T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40, 333–343. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka, A., and Theologis, A. (2004). Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 101, 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah, H., Chen, J.G., Temple, B., Boyes, D.C., Alonso, J.M., Davis, K.R., Ecker, J.R., and Jones, A.M. (2003). The β-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell 15, 393–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. a). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999. b). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler, H., and Kuhlemeier, C. (2003). Simple hormones but complex signalling. Curr. Opin. Plant Biol. 6, 51–56. [DOI] [PubMed] [Google Scholar]
- Ward, S.P., and Estelle, M. (2001). Auxin signaling involves regulated protein degradation by the ubiqutin-proteasome pathway. J. Plant Growth Regul. 20, 265–273. [Google Scholar]
- Watahiki, M.K., and Yamamoto, K.T. (1997). The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol. 115, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Yang, T., and Poovaiah, B.W. (2000). Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 275, 3137–3143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.