Summary
Plants sense microbial signatures via activation of pattern recognition receptors (PPRs), which trigger a range of cellular defences. One response is the closure of plasmodesmata, which reduces symplastic connectivity and the capacity for direct molecular exchange between host cells.
Plasmodesmal flux is regulated by a variety of environmental cues but the downstream signalling pathways are poorly defined, especially the way in which calcium regulates plasmodesmal closure.
Here, we identify that closure of plasmodesmata in response to bacterial flagellin, but not fungal chitin, is mediated by a plasmodesmal‐localized Ca2+‐binding protein Calmodulin‐like 41 (CML41). CML41 is transcriptionally upregulated by flg22 and facilitates rapid callose deposition at plasmodesmata following flg22 treatment. CML41 acts independently of other defence responses triggered by flg22 perception and reduces bacterial infection.
We propose that CML41 enables Ca2+‐signalling specificity during bacterial pathogen attack and is required for a complete defence response against Pseudomonas syringae.
Keywords: At3g50770, biotic stress, cell‐to‐cell communication, electrophoresis mobility shift, maltose‐binding protein, pathogen‐associated molecular pattern (PAMP)
Introduction
Plasmodesmata are plasma membrane‐lined pores that connect the cytoplasm of adjoining plant cells, allowing the passage of small molecules and ions through the plant symplast (Lucas & Lee, 2004). Our understanding of plasmodesmata has grown in the last decade (Lucas & Lee, 2004; Lee & Lu, 2011; Han & Kim, 2016; Tilsner et al., 2016). For instance, it is now clear that plasmodesmata dynamically regulate cell‐to‐cell connectivity during developmental transitions and in response to environmental change, e.g. the down‐regulation of plasmodesmal flux is an essential defence response following pathogen attack (Lee et al., 2011; Faulkner et al., 2013).
Bacterial flagellin triggers many defence responses via the perception of the immunogenic peptide of flagellin (flg22) by its cognate receptor FLS2 (Gómez‐Gómez & Boller, 2000). In Arabidopsis these responses include both the influx of Ca2+ and plasmodesmal closure; Ca2+ signalling is believed to be a critical component of a successful immune response (Lecourieux et al., 2006; Seybold et al., 2014). Previous studies have shown that changes in cytosolic Ca2+ concentration close plasmodesmata (Tucker & Boss, 1996; Holdaway‐Clarke et al., 2000) – this has led to speculation that Ca2+ signals regulate plasmodesmal flux following pathogen perception (e.g. Han & Kim, 2016; Tilsner et al., 2016). While plasmodesmata‐located calcium responsive proteins and putative calmodulin‐binding sites have been identified (Baluška et al., 1999; Chen et al., 2005; Fernandez‐Calvino et al., 2011; Vaddepalli et al., 2014) none have been characterized for their specific role in plasmodesmal function.
Here, we identify Calmodulin‐like protein 41 (CML41) as a plasmodesmata‐located, Ca2+ responsive protein that mediates flg22‐induced callose‐dependent plasmodesmal closure. CML41 expression is upregulated by flg22 and positively regulates defence against Pseudomonas syringae; therefore, we have identified a novel component of plant defence that links Ca2+ signals with callose deposition and plasmodesmata closure.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Col‐0 and transgenic plants were grown in soil under short day conditions (9 h : 15 h, light : dark, 22°C) for 5–6 wk (Conn et al., 2013), unless indicated otherwise.
Gene cloning and plasmid construction
The coding sequence of CML41 (At3g50770) with or without a stop codon was cloned via PCR (Phusion™ Hot Start High‐Fidelity DNA polymerase; Finnzymes, Vantaa, Finland) with the primers listed in Supporting Information Table S1. To silence CML41, an artificial micro RNA (amiRNA, 5′‐TAAACCGTCATCATTTGACCA‐3′) was designed against the CML41 mRNA sequence using Web Micro RNA Designer (Wmd3, http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) (Schwab et al., 2006). Whilst a 2‐kb sequence upstream from the CML41 ATG start codon was amplified by PCR to represent the CML41 promoter (proCML41). All these PCR products were cloned via the Gateway® system (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturers’ instructions. The CML41 gene with a stop codon was recombined into pDEST566 for protein expression in Escherichia coli and into the binary vector pMDC32 for plant overexpression. CML41 without the stop codon was recombined into the binary vector pMDC83 containing a green fluorescent protein (GFP) tag on the C‐terminus for protein localization and CML41‐amiRNA was recombined into the binary vector pMDC32 for knockdown of CML41 in the plant. The proCML41 was recombined into the binary vector pMDC162 for a GUS histochemical assay (Curtis & Grossniklaus, 2003) and 35S:PDLP1‐mCherry was made by Golden Gate cloning. Stable transformation of Arabidopsis was performed by floral dip and T3 homozygote plants were used for all experiments.
Subcellular localization
Ectopically expressed fluorescent proteins in transgenic Arabidopsis plants were imaged using a confocal laser scanning microscope equipped with a Zeiss Axioskop 2 mot plus LSM5 PASCAL and argon laser (Carl Zeiss, Oberkochen, Germany) or a Leica SP5 confocal microscope (Leica Microsystems, Wexlar, Germany). Sequential scanning and laser excitation was used to capture fluorescence from GFP (excitation = 488 nm, emission = 505–530 nm), aniline blue (excitation = 405 nm, emission = 440–490 nm) and mCherry (excitation = 561 nm, emission = 600–640 nm).
GUS histochemical analysis
Transgenic proCML41::GUS plants were stained in the dark using a buffer containing 50 mM sodium phosphate pH = 7.0, 10 mM EDTA, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 0.1% (v/v) Triton X‐100, 0.1% (w/v) X‐Gluc (5‐bromo‐4‐chloro‐3‐indolyl β‐d‐glucuronide) vacuum infiltrated for 15 min, followed by a 3 h incubation at 37°C. The plants were cleared of chlorophyll in 70% ethanol and imaged using a SMZ800 Stereo Fluorescence microscope (Nikon, Tokyo, Japan).
Quantitative RT‐PCR analysis
RNA was extracted from leaves using TRIzol (Invitrogen) and treated with Turbo DNA‐free kit (Ambion, Thermo Fisher Scientific) before cDNA synthesis using SuperScript® III Reverse Transcriptase (Invitrogen). Quantitative reverse transcription polymerase chain reaction (RT‐PCR) was performed on the cDNA samples with primers listed in Table S1 using the fluorescence output from a QuantStudio™ 12K Flex Real‐Time PCR System. Quantitative RT‐PCR analysis via the 2−ΔCт method to calculate the gene expression level relative to either GAPDH‐A (At3g26650) or UBI10 (At4g05320) as an internal control (Schmittgen & Livak, 2008).
Electrophoresis mobility shift assays
Recombinant CML41 was expressed in T7 Expression lysY/I q E. coli cells (New England Biolabs, Ipswich, MA, USA) using a pDEST566 vector tagged with a 6×His maltose‐binding protein (MBP) to enhance solubility of CML41 (Kapust & Waugh, 1999). The recombinant CML41 was purified using Poly‐Prep® Chromatography Columns (Bio‐Rad, Hercules, CA, USA) and Talon® Metal Affinity Resin (Clontech, Mountain View, CA, USA) and desalted using Zeba™ Spin Desalting Columns (Thermo Fisher Scientific) following the manufacturers’ guide. The electrophoresis mobility shift assay was optimized from methods previously described (Garrigos et al., 1991). Either 1 mM CaCl2 or 10 mM EGTA (ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid) was added to the purified desalted recombinant protein samples. These samples were heated at 95°C for 2 min before electrophoresis separation on an 8% SDS‐PAGE gel containing either 1 mM CaCl2 or 10 mM EGTA. The mobility of proteins was determined by comparison with the Precision Plus Protein™ Standards (Bio Rad).
Plasmodesmal callose staining assay
The eighth rosette leaf was infiltrated with ultrapure water, or ultrapure water containing 100 nM flg22 or 1 mM EGTA for 30 min, followed by an infiltration of aniline blue (0.01% (w/v) in PBS buffer, pH 7.4). Callose deposits were imaged from the abaxial side of the leaf using a SP5 confocal microscope (Leica) or a Nikon A1R laser scanning confocal with excitation = 405 nm and emission = 500–550 nm. Z‐stack images were collected from 24 to 27 technical replicates from two independent biological replicates. Data from different microscopes was not pooled. Callose was quantified using automated image analysis. All annotated images were inspected before inclusion of any data in the statistical analysis. The image analysis pipeline was written in Python; image analysis scripts and further information are available under the open source MIT licence on GitHub (https://github.com/JIC-CSB/find-plasmodesmata).
Macroscopic callose deposition assay
Either ultrapure H2O or ultrapure H2O containing 1 μM flg22 was infiltrated into rosette leaves. After 24 h, the infiltrated leaves were incubated in staining solution (150 mM sodium phosphate, 0.05% (w/v) aniline blue, pH = 8) for an additional 1 h in the dark. The stained leaves were mounted in 50% (v/v) glycerol and imaged with an Axiophot Photomicroscope excitation from a mercury light source and captured with a UV filter (LP = 470 nm) (Carl Zeiss). Callose deposited in leaves was measured by ImageJ, using particle analysis (http://rsbweb.nih.gov/ij/).
Reactive oxygen species (ROS) burst assay
Leaf discs were obtained by using a 4 mm disposable biopsy punch (Kei Medical, Tokyo, Japan) and incubated overnight with 100 μM L‐012 (Wako Chemical, Osaka, Japan) in a Greiner 96 well white plate. Leaf discs were washed once with sterile deionized (DI) water and then triggered with 1 μM flg22 and luminescence was measured for 30 min (Tristar2 LB 942; Berthold Technologies, Bad Wilbad, Germany). The data was replicated three times in independent trials.
Bacterial growth assay
Bacterial growth assays were performed as described previously (Kadota et al., 2014), with slight modification. We assessed the significance of CML41 activity in overall plant resistance by first infiltrating leaves of different Arabidopsis lines with the virulent bacterial pathogen P. syringae pv. tomato (Pst) DC3000 (OD = 0.0002). We next performed infections by surface inoculation with the less virulent, coronatine deficient strain DC3000 (cor −). Briefly, Pst bacterial suspension with OD600 nm = 0.2 in 0.02% Silwet L‐77 were generously sprayed onto leaf abaxial and adaxial surfaces of 5–6‐wk‐old plants. Plants were covered during the course of infection and leaf discs were taken 3 h post‐inoculation (day 0) or 3 d post‐inoculation (day 3) from three leaves per plant, with six plants per genotype per independent trial. Bacterial growth was assessed by colony counting.
GFP bombardment assay
Microprojectile bombardment assays were performed as previously described (Faulkner et al., 2013). Bombardment sites were assessed by epifluorescence microscopy (Leica DM6000).
Results and Discussion
The 50 members of the CML gene family of A. thaliana have been proposed to encode proteins that facilitate specificity during Ca2+ signalling (McCormack et al., 2005; Bender & Snedden, 2013). CML41 expression was previously shown to be upregulated by flg22 (Denoux et al., 2008), so we investigated whether it had a role in pathogen responses. First, we confirmed that CML41 transcription was induced flg22 using quantitative RT‐PCR and expression analysis of the CML41 promoter (Fig. 1a,b). We next investigated whether CML41 could bind Ca2+ and, therefore, has the potential to decode flg22‐induced Ca2+‐signals. CMLs can change conformation upon Ca2+ binding (Bender & Snedden, 2013), to test whether this is the case for CML41 we performed an electrophoresis mobility shift assay (Garrigos et al., 1991). Initial attempts to express and purify CML41 in E. coli were unsuccessful as the purified CML41 was insoluble. Therefore, CML41 was tagged with MBP to enhance its solubility and probed by western blot (Kapust & Waugh, 1999) (Fig. S1). The purified and soluble MBP‐CML41 fraction migrated faster in the presence of Ca2+ relative to EGTA (Fig. 1c), indicating its ability to bind Ca2+. This result is consistent with the recent report that CML41 binds to phenyl sepharose in a Ca2+‐dependent manner (Dell'Aglio et al., 2016). Attempts to obtain the Ca2+‐binding affinity for CML41 using microscale thermophoresis in a range of buffers were unsuccessful due to protein aggregation, even when tagged with MBP. However, the presence in CML41 of EF‐hands, which are conserved in other CML and have already been shown to bind Ca2+ in the nanomolar range, suggests that the Ca2+ responsiveness of CML41 could have a physiological role (Fig. S2).
Figure 1.
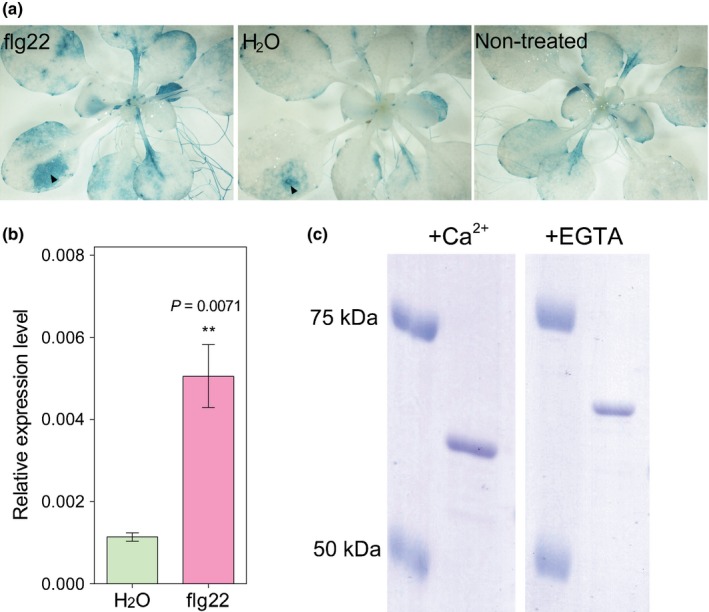
CML41 is induced by flg22 in leaves and binds Ca2+. (a) GUS histochemical staining of 24‐d‐old proCML41::GUS plants treated with either H2O or flg22 infiltration for 4 h, as well as nontreatment control as indicated. Both flg22 and H2O injection at the wound/infiltration site is indicated by arrowheads, there was a localized increase in GUS activity induced by both flg22 and H2O injection at the wound site. (b) Quantitative RT‐PCR analysis of CML41 in the leaves of 5–6‐wk‐old wildtype Arabidopsis Col‐0 plants grown in short‐day conditions (with 9 h : 15 h, light : dark) pre‐infiltrated with either H2O (green) or 1 μM flg22 (magenta) for 12 h. Gene transcript level was relative to GAPDH‐A (At3g26650). Data represent the mean ± standard error of the mean (SEM), n = 3 biological replicates. Primer pairs used for (b) listed in Supporting Information Table S1. Statistical difference was determined by Student's t‐test, asterisks indicate statistical significance, P‐value as indicated. (c) Gel shift Ca2+ binding assay, purified recombinant MBP‐CML41 protein was separated on 8% SDS‐PAGE gel in the presence of 1 mM CaCl2 or 10 mM EGTA, the mobility of proteins was determined by comparison with the Precision Plus Protein™ Standards as indicated.
To investigate the subcellular localization of CML41 we overexpressed CML41 fused with GFP (CML41‐GFP). In leaves, CML41‐GFP localizes to punctate spots at the cell periphery (Fig. 2a–c), patterning that is reminiscent of plasmodesmata (Thomas et al., 2008; Simpson et al., 2009). We confirmed CML41‐GFP was localized to plasmodesmata by co‐staining plasmodesmal callose in Arabidopsis with aniline blue (Fig. 2d–f) or with Plasmodesmata‐located Protein 1 (PDLP1) in Nicotiana benthamiana (Fig. 2g–i) (Thomas et al., 2008). Plasmodesmal‐association has not yet been observed for any other CMLs, suggesting a novel role for CML41 of decoding flg22‐induced Ca2+ signals at plasmodesmata (Bender & Snedden, 2013).
Figure 2.
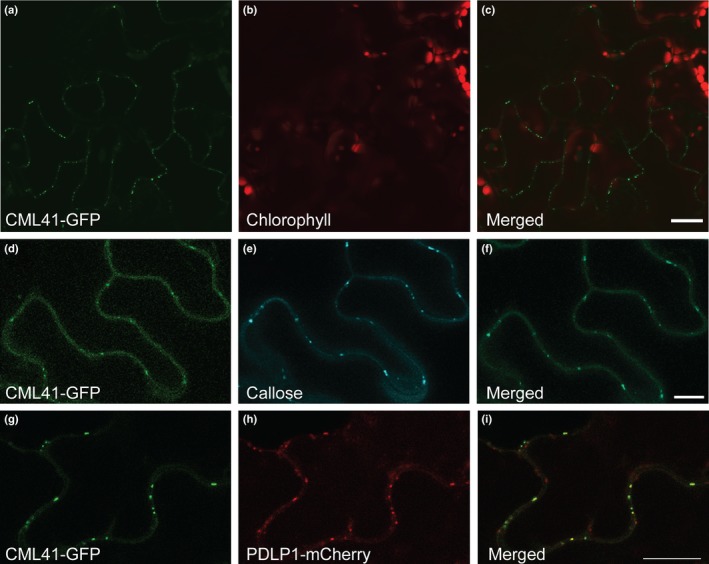
CML41 localizes to plasmodesmata. (a–c) Confocal image of CML41 tagged with GFP in the leaves of 5–6‐wk‐old 35S::CML41‐GFP Arabidopsis plant; bars, 20 μm. (d–f) Co‐localization of CML41‐GFP (d) with callose stained by aniline blue (e) in the leaves of 5–6‐wk‐old 35S::CML41‐GFP Arabidopsis leaf; bars, 10 μm. (g–i) Co‐localization of CML41‐GFP (g) with PDLP1‐mCherry (h), transiently expressed in Nicotiana benthamiana; bars, 10 μm.
To further investigate the role of CML41 at plasmodesmata, we generated CML41 gain‐ and loss‐of‐function transgenic lines using either gene overexpression (OEX) or artificial micro RNA (amiRNA) as no T‐DNA lines were available (Fig. S3). To determine whether CML41 has a role in flg22‐induced plasmodesmal closure we performed intercellular flux assays by measuring GFP diffusion from single cell transformation sites (Faulkner et al., 2013). Whilst flg22 treatment reduced spread of GFP in wildtype plants, indicating plasmodesmal closure, CML41‐amiRNA lines did not close their plasmodesmata in response to flg22 (Fig. 3a). CML41‐OEX plants showed increased basal levels of plasmodesmal closure (reduced spread of GFP), which was unaffected by flg22‐treatment (Fig. 3a).
Figure 3.
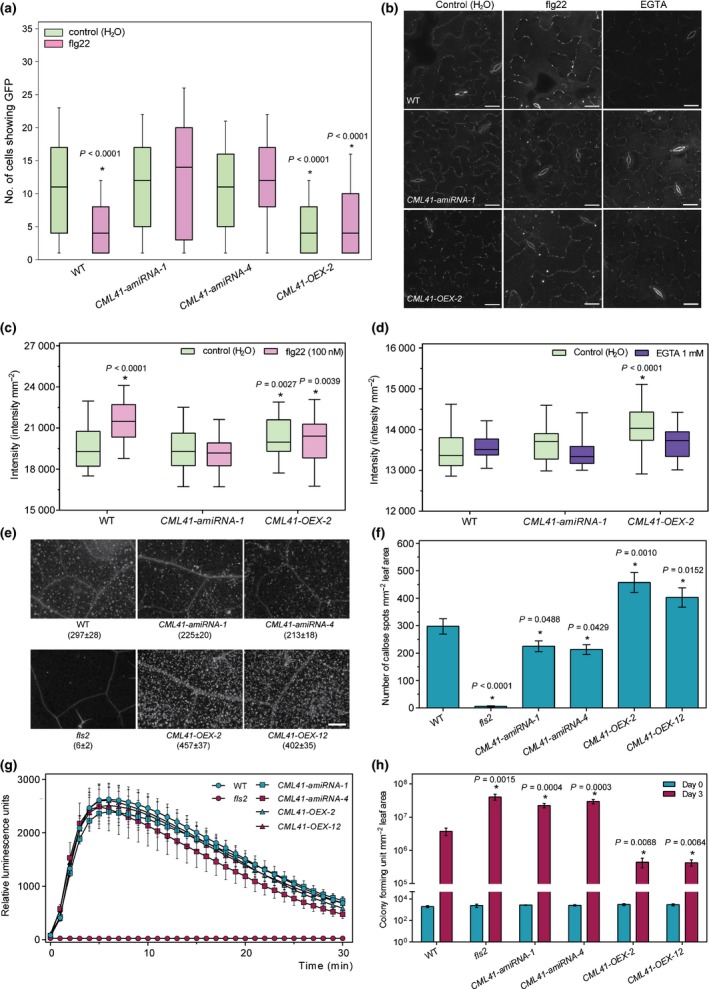
CML41 negatively modulates plasmodesmatal permeability and positively regulates callose production and plant defence. (a) Plasmodesmata permeability of wildtype (WT) Arabidopsis and CML41 transgenic lines (CML41‐amiRNA‐1, ‐4 and CML41‐OEX‐2) in response to 100 nM flg22. Plants were bombarded with constructs capable of producing GFP. Diffusion of GFP to surrounding cells provided a measure of molecular flux through plasmodesmata. Plants were infiltrated with flg22 2 h after bombardment. In each box‐plot, the white line indicates the median value, the shaded area represents the lower and upper quartiles, and the error bars indicate the minimum and maximum values, n = 187 cells for WT (control), 137 for WT (flg22), 85 for CML41‐amiRNA‐1 (control), 103 for CML41‐amiRNA‐1 (flg22), 145 for CML41‐amiRNA‐4 (control), 163 for CML41‐amiRNA‐1 (flg22), 666 for CML41‐OEX‐2 (control) and 198 for CML41‐OEX‐2 (flg22). Statistical difference from the WT control was determined by Student's t‐test, asterisks indicate statistical significance, P‐values as indicated. (b) Confocal images of aniline blue stained plasmodesmal callose in the leaves of 5–6‐wk‐old Arabidopsis WT, CML41‐OEX‐2 and CML41‐amiRNA‐1 lines upon H2O, flg22 and EGTA treatment for 30 min; bars, 20 μm. (c, d) Quantification of PD callose fluorescence intensity in the leaves of 5–6‐wk‐old Arabidopsis WT, CML41‐OEX‐2 and CML41‐amiRNA‐1 lines following flg22 (c) and EGTA treatment (d). Box plots in (c) and (d) are as mentioned earlier, n = 24 (c) and 27 (d). Statistical difference from the WT control was determined by Student's t‐test, asterisks indicate statistical significance, P‐values as indicated. (e) Macroscopy images and (f) quantification of callose deposition upon flg22 or H2O treatments in WT Col‐0, fls2,CML41‐amiRNA‐1, ‐4 and CML41‐OEX‐2, ‐12 lines upon 1 μM flg22 for 24 h, as indicated; bars, 200 μm in (e). Data represent the mean ± SEM, n = 18 leaves. Statistical difference as determined by one‐way analysis of variance (ANOVA), asterisks indicate statistical significance, P‐values as indicated. (g) Reactive oxygen species (ROS) production stimulated by 1 μM flg22 was monitored in Arabidopsis WT, fls2,CML41‐amiRNA‐1, ‐4 and CML41‐OEX‐2, ‐12 leaf discs recorded at every minute using a luminol assay in a microplate reader, fls2 was used as a control. Data are given as relative luminescence units and represent in mean ± SEM, from three independent trials with six technical replicates per biological replicate, n = 6. (h) Evaluation of CML41 transgenic plant susceptability to Pst DC3000 cor −; quantification of bacterial growth in fls2,CML41‐amiRNA‐1, ‐4,CML41‐OEX‐2, ‐12 lines and Arabidopsis WT plants upon 0 and 3 d post‐inoculation of Pst DC3000 cor − suspension. The bacterial colony number was counted in colony‐forming unit (CFU) per cm2. Data represent mean ± SEM, n = 6. Statistical difference as determined by multiple Student's t‐test, asterisks indicate statistical significance from WTcontrol or flg22 treated, P‐values as indicated. The experiments were repeated three times with similar results. See Supporting Information Fig. S7 for the equivalent assays using Pst DC3000.
To test whether CML41‐mediated, flg22‐induced plasmodesmal closure is executed by apoplastic callose deposition adjacent to plasmodesmata (Maule et al., 2012), we used automated image analysis to quantify plasmodesmata‐located aniline blue fluorescence. At 30 min post‐treatment, plasmodesmal callose was significantly increased in wildtype plants (Fig. 3b,c), supporting a model where flg22 induces plasmodesmal closure via rapid callose deposition. CML41‐amiRNA lines showed no difference in callose deposition between H2O and flg22 treated tissue (Fig. 3b,c), which coincided with the loss of the plasmodesmal closure response in these plants (Fig. 3a). CML41‐OEX plants had an increased presence of plasmodesmal callose in the basal state, which was reduced by the Ca2+‐chelator EGTA (Fig. 3b,d). These data indicate that Ca2+‐responsiveness of CML41 is likely to play a critical and physiologically relevant role in flg22‐induced plasmodesmal closure. Furthermore, this result implies that infiltration per se without EGTA (as performed in control experiments in Fig. 3a–f) may be sufficient to raise cytosolic Ca2+ concentration, as could be expected following a manipulation of the leaf apoplastic environment and water relations.
To examine whether CML41 is involved more broadly in pathogen‐associated molecular pattern (PAMP) responses we examined GFP movement in response to chitin, a fungal PAMP that reduces flux via plasmodesmata (Faulkner et al., 2013). CML41‐amiRNA plants showed the wildtype response to chitin (Fig. S4a); furthermore, chitin did not induce CML41 transcription (Fig. S4b). This confirms that plants regulate plasmodesmal closure through different mechanisms following bacterial or fungal infection.
To assess CML41 specificity to plasmodesmal function we examined other flg22‐induced responses. Macroscopic callose deposition in the apoplast in response to flg22 was detectable 24 h post‐treatment (Gómez‐Gómez et al., 1999). In wildtype plants we observed widespread and greater callose deposition following flg22 infiltration compared to water treatment (Figs 3e,f, S5) and the fls2 control (Gómez‐Gómez et al., 1999). The CML41‐amiRNA lines accumulated fewer callose deposits than wildtype plants after flg22 treatment (Fig. 3e,f). CML41‐OEX lines produced more callose deposits in response to flg22 (Fig. 3e,f).
Reactive oxygen species (ROS) accumulate rapidly upon flg22 treatment (Gómez‐Gómez et al., 1999; Boudsocq et al., 2010). While flg22‐induced ROS production is abolished in fls2 (Gómez‐Gómez et al., 1999; Boudsocq et al., 2010), it was observed at similar levels to wildtype in CML41‐amiRNA and CML41‐OEX plants (Fig. 3g) indicating that CML41 does not play a role in flg22‐induced ROS production. The flg22 induces the expression of defence genes such as flg22‐induced Receptor Kinase 1 (FRK1), cytochrome P450 monooxygenase (CYP81F2) and NDR1/HIN1‐like 10 (NHL10) (Boudsocq et al., 2010). All these genes were significantly up‐regulated across all flg22‐infiltrated plants (Fig. S6). Transcript abundance of the salicylic acid (SA) inducible gene, PATHOGENESIS‐RELATED 1 (PR1) (Wildermuth et al., 2001; Zipfel et al., 2004) is also enhanced by flg22 and showed no obvious difference between CML41 transgenic lines and wildtype plants within the same treatments (Fig. S6). This contrasts the action of CML9 in the nucleus, which appears to negatively regulate PR1 expression and the deposition of callose during flg22 responses (Leba et al., 2012). As the same transcriptional responses of these known early innate immunity induced genes are activated in wildtype, CML41 overexpression and knockdown plants, this is strong evidence that CML41 does not work upstream of any of the corresponding response pathways.
In combination, this data establishes that CML41 functions in a specialized signalling pathway, downstream of FLS2 recognition of flg22. The localization of CML41 at plasmodesmata suggests that this signalling pathway directly regulates plasmodesmal function, closing the plasmodesmata within 30 min of pathogen perception (Fig. 3a). CML41 also functions in the production of large deposits of callose over a longer response timescale (Fig. 3e). It is not clear yet whether CML41 directly stimulates deposition via regulation of a callose synthase or inhibits a constitutive process of removal by interfering with β‐1,3‐glucanase activity (Luna et al., 2011), this may become clearer when interacting partners are identified for CML41. It should be noted that we detected CML41 expression in the roots following flg22 treatment (Fig. 1a); therefore, it is likely that CML41 plays a similar role in PD flux regulation when challenged with soil bacterial pathogens. An investigation is also warranted into: the role of CML41 in the other tissues in which it is expressed such as senescent leaves and flowers (McCormack et al., 2005); and, the significance of its transcriptional regulation by RNA‐dependent methylation (Baev et al., 2010).
In this study we focused on assessing the significance of CML41 activity in overall plant resistance by surface inoculation of different Arabidopsis lines with P. syringae (Figs 3h, S6). Three days post‐infection (3 dpi), CML41‐amiRNA lines showed more bacterial growth than wildtype, similar to fls2, indicating they were more susceptible; while the CML41‐OEX lines showed less bacterial growth than wildtype (Fig. 3h). This suggests that flg22‐induced Ca2+ signalling via CML41 is a critical component of callose deposition. A parallel role for Ca2+ in a callose‐independent plasmodesmal closure pathway has also been proposed (Sager & Lee, 2014); however, this does not appear to play a major role in the conditions assayed here.
We have identified CML41 as a Ca2+ responsive component of defensive plasmodesmal closure. Chitin‐induced plasmodesmal closure has previously been shown to be critical to defence and overall resistance against a fungal pathogen and the data presented here identifies that flg22‐induced plasmodesmal closure is similarly critical to defence against bacterial pathogens. Beyond establishing new understanding of the mechanisms of plasmodesmal function via Ca2+, this highlights a role for symplastic connectivity in the full and complete execution of defence responses.
Author contributions
M.G., C.F. and K.S. supervised the research. B.X. performed the majority of experiments except the following. B.H. cloned the amiRNA. B.X. and D.C. purified the protein and performed Ca2+ shift experiments. A.L. performed callose deposition, ROS burst and bacterial growth assays. C.C. performed bombardment assays and plasmodesmal callose quantification. T.S.G.O. wrote the image analysis script. C.C. and C.F. performed co‐localization experiments. B.X., C.C., A.L., K.S., M.G. and C.F. drafted the manuscript. All authors commented on the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 CML41 was produced through heterologous expression in Escherichia coli.
Fig. S2 In silico analysis of CML41 protein sequence and EF domains.
Fig. S3 CML41 transcript abundance was modified in transgenic CML41 misexpression lines.
Fig. S4 CML41 does not have a role in chitin response of plasmodesmata.
Fig. S5 Callose production is not induced by water infiltration in CML41 overexpression or knockdown lines.
Fig. S6 Early immune response gene transcription is not affected by CML41 overexpression or knockdown.
Fig. S7 CML41 overexpression increases bacterial resistance.
Table S1 Primers used for quantitative RT‐PCR analysis and molecular cloning
Acknowledgements
The authors thank Dr Yasuhiro Kadota for critical review of the research, Dr Matthew Hartley for assistance with plasmodesmal image analysis, Roger Leigh, Steve Tyerman, Brent Kaiser, Matt Tucker and Rachel Burton for discussions. Epifluorescence and confocal microscopy were facilitated by access to the JIC Bioimaging and Computational Biology Platforms, respectively, and Adelaide Microscopy. Funding: B.X. was supported by an University of Adelaide Graduate Research Scholarship; research was supported by the Australian Research Council through Centre of Excellence (CE140100008) and Future Fellowship (FT130100709) funding to M.G.; by the Biotechnology and Biological Sciences Research Council (BB/L000466/1) to C.F.; and by KAKENHI (24228008 and 15H05959) to K.S.
Contributor Information
Christine Faulkner, Email: christine.faulkner@jic.ac.uk.
Matthew Gilliham, Email: matthew.gilliham@adelaide.edu.au.
References
- Baev V, Naydenov M, Apostolova E, Ivanova D, Doncheva S, Minkov I, Yahubyan G. 2010. Identification of RNA‐dependent DNA‐methylation regulated promoters in Arabidopsis . Plant Physiology and Biochemistry 48: 393–400. [DOI] [PubMed] [Google Scholar]
- Baluška F, Šamaj J, Napier R, Volkmann D. 1999. Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant Journal 19: 481–488. [DOI] [PubMed] [Google Scholar]
- Bender KW, Snedden WA. 2013. Calmodulin‐related proteins step out from the shadow of their namesake. Plant Physiology 163: 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng S‐H, Sheen J. 2010. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M‐H, Tian G‐W, Gafni Y, Citovsky V. 2005. Effects of calreticulin on viral cell‐to‐cell movement. Plant Physiology 138: 1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, Tyerman SD, Gilliham M. 2013. Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high‐throughput functional analysis of genes in planta . Plant Physiology 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Aglio E, Salvi D, Kraut A, Baudet M, Macherel D, Neveu M, Ferro M, Curien G, Rolland N. 2016. No plastidial calmodulin‐like proteins detected by two targeted mass‐spectrometry approaches and GFP fusion proteins. New Negatives in Plant Science 3: 19–26. [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. 2008. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant 1: 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez‐Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ. 2013. LYM2‐dependent chitin perception limits molecular flux via plasmodesmata. Proceedings of the National Academy of Sciences, USA 110: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez‐Alfonso Y, Maule A. 2011. Arabidopsis plasmodesmal proteome. PLoS ONE 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigos M, Deschamps S, Viel A, Lund S, Champeil P, Møller JV, le Maire M. 1991. Detection of Ca2+‐binding proteins by electrophoretic migration in the presence of Ca2+ combined with 45Ca2+ overlay of protein blots. Analytical Biochemistry 194: 82–88. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez L, Boller T. 2000. FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gómez‐Gómez L, Felix G, Boller T. 1999. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana . Plant Journal 18: 277–284. [DOI] [PubMed] [Google Scholar]
- Han X, Kim J‐Y. 2016. Integrating hormone‐and micromolecule‐mediated signaling with plasmodesmal communication. Molecular Plant 9: 46–56. [DOI] [PubMed] [Google Scholar]
- Holdaway‐Clarke TL, Walker NA, Hepler PK, Overall RL. 2000. Physiological elevations in cytoplasmic free calcium by cold or ion injection result in transient closure of higher plant plasmodesmata. Planta 210: 329–335. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A. 2014. Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Molecular Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Waugh DS. 1999. Escherichia coli maltose‐binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Science 8: 1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leba L‐J, Perochon A, Cheval C, Ranty B, Galaud J‐P, Aldon D. 2012. CML9, a multifunctional Arabidopsis thaliana calmodulin‐like protein involved in stress responses and plant growth? Plant Signaling & Behavior 7: 1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourieux D, Ranjeva R, Pugin A. 2006. Calcium in plant defence‐signalling pathways. New Phytologist 171: 249–269. [DOI] [PubMed] [Google Scholar]
- Lee J‐Y, Lu H. 2011. Plasmodesmata: the battleground against intruders. Trends in Plant Science 16: 201–210. [DOI] [PubMed] [Google Scholar]
- Lee J‐Y, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H. 2011. A plasmodesmata‐localized protein mediates crosstalk between cell‐to‐cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Lee J‐Y. 2004. Plasmodesmata as a supracellular control network in plants. Nature Reviews Molecular Cell Biology 5: 712–726. [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch‐Mani B, Ton J. 2011. Callose deposition: a multifaceted plant defense response. Molecular Plant‐Microbe Interactions 24: 183–193. [DOI] [PubMed] [Google Scholar]
- Maule A, Faulkner C, Benitez‐Alfonso Y. 2012. Plasmodesmata “in communicado”. Frontiers in Plant Science 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Tsai Y‐C, Braam J. 2005. Handling calcium signaling: arabidopsis CaMs and CMLs. Trends in Plant Science 10: 383–389. [DOI] [PubMed] [Google Scholar]
- Sager R, Lee J‐Y. 2014. Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. Journal of Experimental Botany 65: 6337–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real‐time PCR data by the comparative CT method. Nature Protocols 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold H, Trempel F, Ranf S, Scheel D, Romeis T, Lee J. 2014. Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytologist 204: 782–790. [DOI] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ. 2009. An Arabidopsis GPI‐anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell‐to‐cell trafficking. Plant Cell 21: 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez‐Calvino L, Maule AJ. 2008. Specific targeting of a plasmodesmal protein affecting cell‐to‐cell communication. PLoS Biology 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Nicolas W, Rosado A, Bayer EM. 2016. Staying tight: plasmodesmal membrane contact sites and the control of cell‐to‐cell connectivity in plants. Annual Review of Plant Biology 67: 337–364. [DOI] [PubMed] [Google Scholar]
- Tucker EB, Boss WF. 1996. Mastoparan‐induced intracellular Ca2+ fluxes may regulate cell‐to‐cell communication in plants. Plant Physiology 111: 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaddepalli P, Herrmann A, Fulton L, Oelschner M, Hillmer S, Stratil TF, Fastner A, Hammes UZ, Ott T, Robinson DG. 2014. The C2‐domain protein QUIRKY and the receptor‐like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana . Development 141: 4139–4148. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 CML41 was produced through heterologous expression in Escherichia coli.
Fig. S2 In silico analysis of CML41 protein sequence and EF domains.
Fig. S3 CML41 transcript abundance was modified in transgenic CML41 misexpression lines.
Fig. S4 CML41 does not have a role in chitin response of plasmodesmata.
Fig. S5 Callose production is not induced by water infiltration in CML41 overexpression or knockdown lines.
Fig. S6 Early immune response gene transcription is not affected by CML41 overexpression or knockdown.
Fig. S7 CML41 overexpression increases bacterial resistance.
Table S1 Primers used for quantitative RT‐PCR analysis and molecular cloning


