Abstract
We have defined amino acids important for function of the Arabidopsis thaliana Hsp100/ClpB chaperone (AtHsp101) in acquired thermotolerance by isolating recessive, loss-of-function mutations and a novel semidominant, gain-of-function allele [hot1-4 (A499T)]. The hot1-4 allele is unusual in that it not only fails to develop thermotolerance to 45°C after acclimation at 38°C, but also is sensitive to 38°C, which is a permissive temperature for wild-type and loss-of-function mutants. hot1-4 lies between nucleotide binding domain 1 (NBD1) and NBD2 in a coiled-coil domain that is characteristic of the Hsp100/ClpB proteins. We then isolated two classes of intragenic suppressor mutations of hot1-4: loss-of-function mutations (Class 1) that eliminated the 38°C sensitivity, but did not restore thermotolerance function to hot1-4, and Class 2 suppressors that restored acquired thermotolerance function to hot1-4. Location of the hot1-4 Class 2 suppressors supports a functional link between the coiled-coil domain and both NBD1 and the axial channel of the Hsp100/ClpB hexamer. In addition, the strongest Class 2 suppressors restored solubility of aggregated small heat shock proteins (sHsps) after heat stress, revealing genetic interaction of the Hsp100/ClpB and sHsp chaperone systems. These results also demonstrate that quantitative phenotypes can be used for in vivo genetic dissection of protein mechanism in Arabidopsis.
INTRODUCTION
Hsp100/ClpB chaperones are hexameric members of the AAA+ family of proteins, which are ATPases that couple ATP binding and hydrolysis to a variety of protein-remodeling activities (Neuwald et al., 1999; Vale, 2000; Ogura and Wilkinson, 2001; Lupas and Martin, 2002). Hsp100/ClpB proteins have been shown to be essential for the development of thermotolerance to high temperature in bacteria (Goloubinoff et al., 1999), yeast (Glover and Lindquist, 1998), some parasitic protozoa, and higher plants (Hong and Vierling, 2000, 2001), which are the only higher eukaryotes known to express the Hsp100/ClpB subgroup of chaperones. The demonstration that the highly heat-induced Hsp100/ClpB protein in Arabidopsis thaliana, AtHsp101, was essential for acquisition of tolerance to high temperature also represented a direct link of a heat shock protein to heat tolerance in plants. Both in vivo and in vitro evidence indicates that the protective function of these chaperones is a result of their ability to resolubilize protein aggregates in cooperation with the Hsp70/DnaK chaperone system (Goloubinoff et al., 1999; Zolkiewiski, 1999). It remains unknown, however, whether general removal of aggregated proteins or rescue of certain critical substrates defines the essential activity of Hsp100/ClpB in thermotolerance.
This unique subgroup of ATPases contains two AAA modules, each comprised of a nucleotide binding domain (NBD1 or 2) with conserved Walker A, Walker B, and sensor motifs and a smaller C-terminal domain. The AAA modules are flanked by additional N- and C-terminal domains and are separated by a large coiled-coil domain, which is actually an insertion into the C-terminal small domain of AAA module 1 (see Figure 1A). The length of the coiled-coil domain is a major feature that distinguishes Hsp100/ClpB proteins from other Clp proteins with two AAA modules (e.g., ClpA) (Schirmer et al., 1996; Celerin et al., 1998; Nieto-Sotelo et al., 1999). The recent 3.0-Å structure of a complete Thermus thermophilus ClpB subunit, and a cryoelectron microscopy reconstruction of the hexamer (21-Å resolution), has defined the structure of the different ClpB domains and how they are oriented relative to each other (Lee et al., 2003). In each AAA module, NBD1 and 2 have a RecA-like mononucleotide binding fold, and the C-terminal smaller domain has a mixed α/β structure. The two AAA modules are stacked on each other within a subunit such that the hexamer appears as two rings of six ATPase sites, surrounding a pore or axial channel on the order of ∼16 Å in diameter. The coiled-coil domain, which is inserted into the smaller domain of NBD1, is 85-Å long and resembles a two-bladed propeller ringing the outside of the hexamer. Based on the observation of three different conformations in the crystal structure, the N-terminal domain and coiled-coil domain are proposed to be mobile. This is consistent with the fact that deletion of either domain does not disrupt the hexamer (Mogk et al., 2003b).
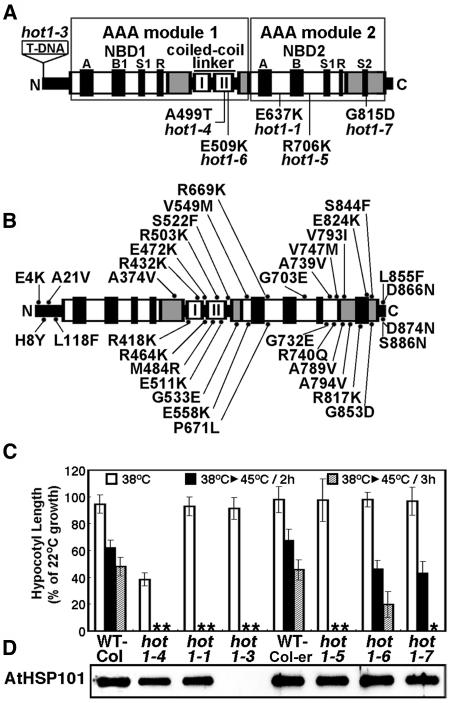
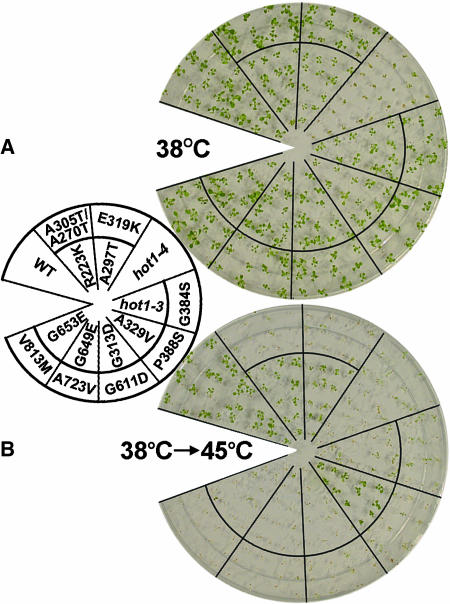
Figure 1.
Location and Phenotype of Mutations in AtHsp101 (See Also Supplemental Figure 1 Online).
(A) Location of hot1 mutations with a thermotolerance phenotype on a schematic diagram of the AtHsp101 protein. The conserved AAA modules consist of two domains, a nucleotide binding domain (NBD1 or 2), containing conserved motifs [Walker A, Walker B, Sensor 1(S1), and Arg finger (R)], and a C-terminal small domain (gray boxes), containing Sensor 2 (S2) in AAA2. The coiled-coil domain contains two signature motifs (Lee et al., 2003), with hot1-4 located in signature motif II.
(B) Location of missense mutations that are wild type for thermotolerance, which were obtained by Tilling analysis (see Methods).
(C) Quantitative assessment of thermotolerance in hot1 mutant seedlings compared with their corresponding wild type (Columbia for hot1-1 and hot1-4, Columbia erecta for all others). After growth for 2.5 d in the dark at 22°C, seedlings were pretreated at 38°C for 90 min and returned to 22°C for 2.5 d (38°C samples), or pretreatment was followed by 2 h at 22°C then 2 or 3 h at 45°C before 2.5 d of recovery (38°C > 45°C samples). hot1-4 seedlings are more sensitive to 38°C treatment than wild-type or null alleles. Asterisks indicate values equal to zero. Mean and standard deviations were derived by measurement from three independent experiments performed with 60 or more seedlings per mutant and values plotted as a percentage of the 22°C value.
(D) Protein gel blot analysis of AtHsp101 protein levels in wild-type and hot1 seedlings after treatment at 38°C.
Biochemical studies using both in vitro assays for ATP hydrolysis and protein disaggregation (chaperone activity), along with in vivo assays for thermotolerance, have also begun to dissect functional domains of Hsp100/ClpB (Smith et al., 1999; Barnett et al., 2000; Clarke and Eriksson, 2000; Schirmer et al., 2001; Beinker et al., 2002; Cashikar et al., 2002; Mogk et al., 2003b; Strub et al., 2003). However, details of the Hsp100/ClpB reaction cycle are unknown. The ATPase activity of both NBDs, as well as an intact coiled-coil domain, are essential for chaperone activity and in vivo thermotolerance (Kim et al., 2000a; Schirmer et al., 2001; Hattendorf and Lindquist, 2002b; Mogk et al., 2003b). A majority of data indicate that deletion of the N-terminal domain does not impair measured activities, and its function remains unknown (Clarke and Eriksson, 2000; Beinker et al., 2002). The C-terminal domain is required for activity, but because deletion of this domain prevents hexamerization, which is required for ATPase activity, its direct role is not revealed by such experiments. Studies with model substrates and effectors of the ATPase activity have suggested that the C-terminal domain is a substrate binding site (Smith et al., 1999), but others conclude this domain modulates ATPase activity only (Strub et al., 2003). The N-terminal domain, NBD1, and the coiled-coil domain have all also been proposed to be involved in substrate interactions (Dougan et al., 2002; Liu et al., 2002; Weibezahn et al., 2003). However, the only data on binding of a natural substrate (TrfA to Escherichia coli ClpB) reveals an interaction only with the surface of NBD1 near the axial pore (Schlieker et al., 2004). Lindquist and coworkers (Cashikar et al., 2002; Hattendorf and Lindquist, 2002a) proposed that in yeast Hsp104, stimulation of NBD2 activity drives a conformational change in the coiled-coil domain, which in turn stimulates ATPase activity in NBD1. Cross-linking studies with ClpB further suggest that movement of the coiled-coil domain is necessary for chaperone activity in vitro (Lee et al., 2003). How these motions are connected to protein disaggregation remains to be defined. It has been suggested that ClpB has a crowbar action in protein disaggregation, perhaps mediated by the coiled-coil domain (Vale, 2000; Maurizi and Xia, 2004; Weibezahn et al., 2004). Alternatively, or in addition, substrate unfolding may be coupled to threading through the axial channel of the ClpB hexamer in a mechanism related to that used by ClpA (Ishikawa et al., 2001) and ClpX (Kim et al., 2000b) for protein unfolding and transfer to the ClpP protease. Recent experiments indicate mutations that affect protein loops in the axial channel eliminate yeast Hsp104 function in protein disaggregation, perhaps because of disruption of protein threading through the channel (Lum et al., 2004).
We have taken a genetic approach to investigating the function and mechanism of Hsp100/ClpB in Arabidopsis. In previous work we showed that AtHsp101, encoded by the HOT1 gene, is a major component controlling acquired thermotolerance, based on the phenotypes of missense and protein null alleles, hot1-1 (E637K; in NBD2) and hot1-3 (a T-DNA insertion in exon 1), respectively (Hong and Vierling, 2000, 2001). Loss of AtHsp101 function, however, does not appear to have major effects on plant growth and development under optimal temperature conditions. To dissect further the mechanism of Hsp101 action in thermotolerance, we sought additional mutant alleles either by direct screening for mutants in a seedling thermotolerance assay (Hong and Vierling, 2000) or by analysis of point mutations in AtHsp101 obtained through the Arabidopsis Tilling Resource (http://tilling.fhcrc.org:9366). We describe four new mutant alleles of Hsp101 that provide insight into structure and function of this class of chaperone ATPase. One mutant allele, hot1-4, which is a missense mutation in the coiled-coil domain (A499T) provides direct in vivo evidence for an essential function of this unique domain. Using this allele, we then screened for suppressor mutations and defined 13 intragenic suppressors. The location of these suppressor mutations defines features essential for in vivo activity of Hsp101 and provides in vivo evidence for a novel mechanistic link between the coiled-coil domain and the axial channel of Hsp100/ClpB proteins. Furthermore, certain suppressors restore the solubility of small heat shock proteins (sHsps) after heat stress in the hot1-4 mutant, providing genetic evidence for interaction of the Hsp100/ClpB and sHsp chaperone systems in higher plants.
RESULTS
AtHsp101 Mutants with Defects in Thermotolerance
Arabidopsis seedlings will continue to grow after a 45°C, 2-h heat treatment if first preconditioned at 38°C for 90 min followed by 1 to 2 h at 22°C. This ability to acclimate to 45°C is lost in the previously characterized missense (hot1-1; E637K) and protein null alleles of AtHsp101 (hot1-3; T-DNA insertion in exon 1) (Hong and Vierling, 2000, 2001). We used this assay to screen for additional AtHsp101 mutants in a population of mutagenized Arabidopsis seedlings and also to assay identified point mutations in AtHsp101 from the Arabidopsis Tilling Resource (http://tilling.fhcrc.org:9366) for loss of thermotolerance. From our screen for additional thermotolerance mutants (Hong et al., 2003), only one other mutant was recovered that failed to complement hot1-1 in allelism tests and that mapped to the Hsp101 chromosomal region (data not shown). Sequencing of the AtHsp101 gene from this mutant identified a single missense mutation resulting in an exchange of Thr for Ala-499 in the coiled-coil domain (Figure 1A; see Supplemental Figure 1 online). Analysis of the AtHsp101 tilling mutants uncovered three additional hot1 alleles that were defective in thermotolerance, R706K (hot1-5), E509K (hot1-6), and R815D (hot1-7), all of which lie in AAA module 2 (Figure 1A). The locations of an additional 35 missense mutations obtained by tilling showed no defect in thermotolerance and are indicated in Figure 1B.
The relative severity of each of the newly identified defective alleles (hot1-4 to hot1-7) was compared with the wild type and the functionally null hot1-1 allele (Hong and Vierling, 2001) in a quantitative assay for seedling thermotolerance (Figure 1C). The hot1-4 and hot1-5 alleles are the most severe, showing no ability to acclimate to treatment at 45°C for 2 h, whereas the hot1-6 and hot1-7 alleles have a less severe phenotype, with hot1-6 showing the mildest phenotype. Differences in severity are not because of differences in protein levels as all the alleles accumulate AtHsp101 protein to wild-type levels (Figure 1D). Therefore, we conclude that relative thermotolerance of the alleles reflects the degree to which each mutation disrupts AtHsp101 function in vivo and defines critical AtHsp101 residues in the coiled-coil domain and NBD2.
hot1-4 Is a Semidominant, Gain-of-Function AtHsp101 Allele
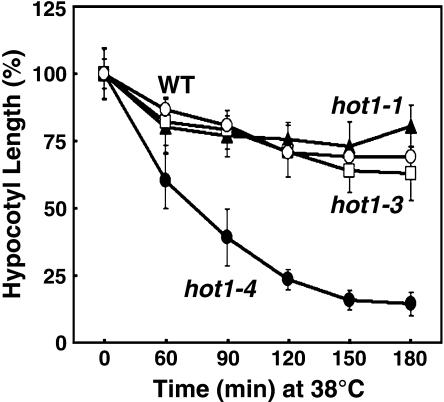
We noted that in contrast with all the other hot1 alleles, hot1-4 was not only defective in acquired thermotolerance to 45°C treatment, but also showed sensitivity to the 38°C acclimation pretreatment (Figure 1C), which is a permissive treatment for the other hot1 alleles. To determine the extent of this sensitivity, elongation of hot1-4 seedlings was compared with the wild type, protein null (hot1-3), and the missense hot1-1 alleles after increasing time at 38°C. The hot1-4 mutant displays dramatic sensitivity to 38°C treatment compared with the wild type and the other loss-of-function alleles (Figure 2). The fact that the phenotype of hot1-4 at 38°C is worse than the absence of the protein (hot1-3 phenotype, Figure 1D) defines hot1-4 as a gain-of-function mutation and indicates that the hot1-4 protein interferes with essential cell functions at 38°C. Furthermore, whereas the hot1-5, hot1-6, and hot1-7 as well as the previously isolated AtHsp101 alleles are recessive, loss-of function mutants, the hot1-4 phenotype proved to be semidominant (Table 1). Compared with wild-type and hot1-4 homozygous seedlings, heterozygous hot1-4 plants showed an intermediate hypocotyl length after a 45°C heat treatment with preconditioning. Despite the sensitivity of hot1-4 to mild heat treatment, the hot1-4 mutation had no obvious effect on growth and development in the absence of stress (data not shown).
Figure 2.
Unusual Sensitivity of hot1-4 to 38°C.
Hypocotyl elongation of hot1 mutant seedlings treated for different times at 38°C, as measured after an additional 2.5 d of growth in the dark at 22°C. Replicated measurements performed as in Figure 1C.
Table 1.
Genetic Analysis of hot1 Mutants
| Cross | Generation | Total | WT | Semi- dominant | hot | χ2 | P |
|---|---|---|---|---|---|---|---|
| hot1-1 | F1 | 23 | 23 | 0 | 0 | ||
| × WT | F2a | 159 | 127 | 0 | 32 | 1.58 | >0.1 |
| hot1-3 | F1 | 31 | 31 | 0 | 0 | ||
| × WT | F2 | 252 | 187 | 0 | 65 | 0.08 | >0.5 |
| hot1-4 | F1 | 30 | 0 | 30 | 0 | ||
| × WT | F2 | 268 | 64 | 134 | 70 | 0.26 | >0.5 |
| hot1-5 | F1 | 7 | 7 | 0 | 0 | ||
| × WT | F2 | 42 | 33 | 0 | 9 | 0.28 | >0.5 |
| hot1-6 | F1 | 8 | 8 | 0 | 0 | ||
| × WT | F2 | 94 | 72 | 0 | 22 | 0.12 | >0.5 |
| hot1-7 | F1 | 4 | 4 | 0 | 0 | ||
| × WT | F2 | 48 | 38 | 0 | 10 | 0.44 | >0.5 |
The phenotypic scoring was based on the hypocotyl elongation phenotypes of the parental lines after 45°C for 2 h with pretreatment (38°C for 90 min). WT, wild type.
The hypocotyl length ranges of the phenotypic classes of F2 progenies were 6.5 to 8 mm for the wild-type phenotype, 2.0 to 4.0 mm for the semidominant phenotype, and 0 mm for hot1-4 mutant phenotype after heat stress.
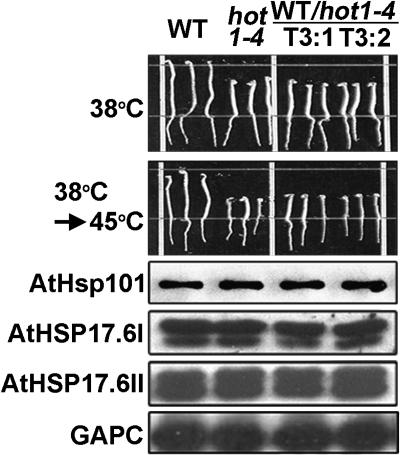
To confirm that the hot1-4 mutant protein causes the observed heat-sensitivity phenotypes, we cloned a hot1-4 mutant genomic fragment, including its own 1.5-kb promoter region, and transformed wild-type Arabidopsis plants with the mutated gene. As shown in Figure 3, the phenotype of hot1-4 can be recapitulated by expression of the hot1-4 mutant protein in wild-type plants. Protein gel blot analysis showed that levels of HSP accumulation were unaffected in the transgenic plants. Thus, the hot1-4 mutant protein functions in a dominant-negative fashion. In total, these results provide evidence that the coiled-coil domain is required for AtHsp101 function during an early step in the development of thermotolerance. In addition, the semidominant, gain-of-function phenotype of hot1-4 suggests that this mutation results in a nonproductive interaction with a limiting substrate or cofactor.
Figure 3.
Expression of Mutant HOT1-4 Protein in Wild-Type Plants Recapitulates the 38°C Sensitivity Phenotype of hot1-4.
The hot1-4 genomic DNA under its own promoter was expressed in transgenic wild-type plants. Two independent transgenic lines (T3:1 and T3:2) are shown compared with the wild type and hot1-4 (three seedlings each). The hypocotyl elongation assay was performed as in Figure 1C. Equal protein samples from heat-stressed seedlings (38°C for 90 min followed by 2 h at 22°C) were analyzed with the indicated antisera against AtHsp101, sHsps, or glyceraldehyde-3-phosphate dehydrogenase (GAPC).
Isolation of Suppressors of hot1-4
The dominant-negative, 38°C-sensitive phenotype of hot1-4 provided an excellent opportunity to screen for suppressor mutations that could restore growth. Suppressor mutations should provide additional insight into structure-function relationships of AtHsp101 and potentially identify important AtHsp101 substrates or cofactors. Approximately 7500 homozygous hot1-4 seeds were mutagenized with ethyl methanesulfonate, and nearly 110,000 dark-grown, 2.5-d-old seedlings from the M2 generation were screened for continued growth after a 2-h treatment at 38°C. In total, 43 lines were isolated that showed a significant reversion of hot1-4 sensitivity to 38°C treatment and that exhibited the same phenotype in the subsequent M3 generation (data not shown).
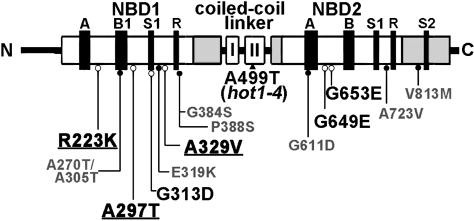
To separate intragenic from extragenic suppressors, we sequenced the AtHsp101 gene from all 43 suppressors. Seven of the suppressors did not carry a second mutation in the AtHsp101 gene and are unlikely to be linked to the original hot1-4 mutation. Presumably, they represent extragenic suppressors of hot1-4. These suppressors could define factors specific to plant stress tolerance and/or identify genes that encode AtHsp101 cofactors or critical substrates. Further genetic and molecular analysis of these mutants is in progress. The other 34 suppressors had a second mutation in the hot1-4 gene, resulting in 13 different amino acid substitutions: eight in the first nucleotide binding domain (NBD1) (R223K, A270T/A305T, A297T, G313D, E319K, A329V, G384S, and P388S), four within NBD2 (G611D, G649E, G653E, and A723V), and one in the C-terminal region (V813M) (Figure 4).
Figure 4.
Location of hot1-4 Suppressor Mutations in AtHsp101.
Class 1 loss-of-function suppressors are labeled in smaller gray font and indicated with closed circles. Restoration-of-function Class 2 suppressors are indicated in larger black font with open circles. The strongest Class 2 suppressors, as discussed in the text, are underlined (see also Supplemental Figure 1 online).
To confirm that the loss of sensitivity to a 38°C treatment in mutants carrying a second site mutation in the Hsp101 gene was because of this mutation and not to another extragenic mutation, a plant carrying each of the individual mutations was crossed to either the wild type or hot1-4. Segregation of the wild type, hot1-4, and suppressed phenotype was then scored in the F1 and F2 generations for each cross. As shown in Table 2 and Supplemental Tables 1 and 2 online, suppression of the 38°C phenotype was recessive and linked to the hot1-4 mutation. These data confirm that the second site mutation in Hsp101 is the cause of the recovery of wild-type growth at 38°C for each of these suppressors.
Table 2.
Genetic Segregation Analysis of Intragenic Suppressors
| No. of plants
|
|||||||
|---|---|---|---|---|---|---|---|
| Genotypes | Total | WT | hot | Sup. | χ2 | P | |
| R223K | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 36 | 36 | 0 | 0 | ||
| R223K | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 36 | 0 | 10 | 26 | 0.14 | >0.9 |
| A270/A305 | F1 | 3 | 3 | 0 | 0 | ||
| × WT | F2 | 38 | 38 | 0 | 0 | ||
| A270/A305 | F1 | 7 | 0 | 0 | 7 | ||
| × hot1-4 | F2 | 40 | 0 | 9 | 31 | 0.13 | >0.9 |
| A297T | F1 | 7 | 7 | 0 | 0 | ||
| × WT | F2 | 36 | 36 | 0 | 0 | ||
| A297T | F1 | 6 | 0 | 0 | 6 | ||
| × hot1-4 | F2 | 29 | 0 | 6 | 23 | 0.73 | >0.5 |
| G313D | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 36 | 36 | 0 | 0 | ||
| G313D | F1 | 6 | 0 | 0 | 6 | ||
| × hot1-4 | F2 | 32 | 0 | 7 | 25 | 0.16 | >0.9 |
| E319K | F1 | 7 | 7 | 0 | 0 | ||
| × WT | F2 | 32 | 32 | 0 | 0 | ||
| E319K | F1 | 2 | 0 | 0 | 2 | ||
| × hot1-4 | F2 | 28 | 0 | 5 | 23 | 0.76 | >0.5 |
| A329V | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 46 | 46 | 0 | 0 | ||
| A329V | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 39 | 0 | 8 | 31 | 0.14 | >0.9 |
| G384S | F1 | 6 | 6 | 0 | 0 | ||
| × WT | F2 | 36 | 36 | 0 | 0 | ||
| G384S | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 32 | 0 | 7 | 25 | 0.16 | >0.9 |
| P388S | F1 | 10 | 10 | 0 | 0 | ||
| × WT | F2 | 40 | 40 | 0 | 0 | ||
| P388S | F1 | 3 | 0 | 0 | 3 | ||
| × hot1-4 | F2 | 40 | 0 | 9 | 31 | 0.13 | >0.9 |
| G611D | F1 | 4 | 4 | 0 | 0 | ||
| × WT | F2 | 46 | 46 | 0 | 0 | ||
| G611D | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 43 | 0 | 10 | 33 | 0.07 | >0.9 |
| G649E | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 45 | 45 | 0 | 0 | ||
| G649E | F1 | 6 | 0 | 0 | 6 | ||
| × hot1-4 | F2 | 44 | 0 | 8 | 36 | 0.36 | >0.5 |
| G653E | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 37 | 37 | 0 | 0 | ||
| G653E | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 34 | 0 | 7 | 27 | 0.35 | >0.5 |
| A723V | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 27 | 27 | 0 | 0 | ||
| A723V | F1 | 8 | 0 | 0 | 8 | ||
| × hot1-4 | F2 | 35 | 0 | 8 | 27 | 0.08 | >0.9 |
| V813M | F1 | 5 | 5 | 0 | 0 | ||
| × WT | F2 | 46 | 46 | 0 | 0 | ||
| V813M | F1 | 5 | 0 | 0 | 5 | ||
| × hot1-4 | F2 | 48 | 0 | 9 | 39 | 1 | >0.5 |
Phenotype scoring was done by comparing the hypocotyl length distribution patterns of the F2 seedlings with those of the parental lines. The hypocotyl length ranges of the phenotypic classes of F2 progenies were 9.5 to 10.5 mm for the wild-type phenotype, 9.0 to 11.0 mm for the suppressor phenotype (Sup.), and 2.0 to 4.0 mm for hot1-4 mutant phenotype after 38°C heat treatment for 2 h. WT, wild type.
The Suppressor Mutations Include Both Loss-of-Function and Restoration-of-Function Alleles
The phenotypes of intragenic suppressor mutants, representing each amino acid substitution, were retested at 38°C to quantify the degree of phenotypic reversion. All the intragenic suppressors showed a strong restoring phenotype of hypocotyl elongation after the same 38°C treatment that was used for their initial identification (Table 3). In addition, when tested for sensitivity to 38°C as 10-d-old seedlings, the suppressors also showed recovery of the wild-type phenotype (Figure 5A). Thus, all of the suppressors clearly eliminated the dominant-negative function of the hot1-4 allele at two distinct growth stages.
Table 3.
Hypocotyl Elongation Phenotype of Intragenic Suppressors
| Hypocotyl Length (%)a
|
|||
|---|---|---|---|
| Genotype | No. of Isolates | 38°C | 38°C/45°C (%) |
| WT | 91.8 ± 9.6 | 68.0 ± 9.2 (100) | |
| hot1-4 (A499T) | 26.2 ± 4.5 | 0 ± 0 | |
| hot1-3 (null) | 90.8 ± 9.6 | 0 ± 0 | |
| Class 1 suppressors | |||
| A270T/A305T | 6 | 89.9 ± 6.4 | 0 ± 0 |
| E319K | 1 | 84.7 ± 6.6 | 0 ± 0 |
| G384S | 3 | 89.7 ± 8.3 | 0 ± 0 |
| P388S | 3 | 87.5 ± 6.8 | 0 ± 0 |
| G611D | 5 | 90.1 ± 6.1 | 0 ± 0 |
| A723V | 3 | 79.4 ± 9.8 | 0 ± 0 |
| V813M | 2 | 91.4 ± 8.3 | 0 ± 0 |
| Class 2 suppressors | |||
| R223K | 2 | 83.6 ± 5.1 | 28.7 ± 5.8 (42) |
| A297T | 2 | 82.1 ± 9.8 | 29.3 ± 6.3 (43) |
| G313D | 3 | 75.3 ± 6.7 | 9.7 ± 4.1 (14) |
| A329V | 1 | 94.1 ± 6.2 | 51.3 ± 6.7 (75) |
| G649E | 1 | 86.7 ± 8.0 | 20.9 ± 6.8 (30) |
| G653E | 2 | 85.2 ± 7.3 | 24.1 ± 7.3 (35) |
Dark-grown, 2.5-d-old seedlings were total hypocotyl elongation after the heat stress was measured as a percentage of the 22°C value. Mean and standard deviations were derived by measurement from three independent experiments performed with 60 or more seedlings per F3 seedling. The numbers in parentheses indicate percentage of elongation compared with that of wild-type plants.
Figure 5.
Thermotolerance Phenotype of 10-d-Old Suppressor Mutants.
After growth for 10 d in the light at 22°C, seedlings were treated either at 38°C for 3 h (A) or at 38°C for 90 min followed by 2 h at 22°C and then 2 h at 45°C (B). Seedlings were photographed 7 d after treatment.
The intragenic suppressor mutants were then tested to determine if the function of AtHsp101 in acquired thermotolerance had been restored by measurement of hypocotyl elongation after 2 h at 45°C of seedlings that had been preconditioned at 38°C (Table 3). The suppressors could clearly be separated into two classes based on their response to this assay. Suppressors in Class 1, which includes A270T/A305T, E319K, G384S, P388S, G611D, A723V, and V813M, were completely arrested in hypocotyl elongation after 2 h at 45°C with preconditioning. Genetic analysis confirmed that this phenotype also segregated with the hot1-4 mutation, again confirming that it arises as a result of the second site mutation in AtHsp101 (see Supplemental Tables 1 and 2 online). This acquired thermotolerance test indicates that the Class 1 suppressors are most likely loss-of-function alleles because they eliminate the dominant-negative phenotype of hot1-4 but do not restore function in acquired thermotolerance. By contrast, the second class of suppressors (Class 2) (R223K, A297T, G313D, A329V, G649E, and G653E) partially restored acquired thermotolerance of hypocotyl elongation, with the best suppression shown by A329V, which elongated to ∼75% of wild-type levels (Table 3).
The same phenotypic behavior was observed when 10-d-old suppressor seedlings were tested for acquired thermotolerance. Class 1 suppressors failed to acquire thermotolerance; a 45°C treatment for 2 h (with preconditioning) completely blocked production of additional leaves, and existing leaves and cotyledons turned white (Figure 5B). Of the Class 2 suppressors, those which restored hypocotyl elongation after 45°C stress to <35% of the wild type (G313D, G649E, and G653E; Table 3) did not exhibit acquired thermotolerance at the 10-d-old seedling stage (Figure 5B). However, suppressors R223K, A297T, and A329V, which showed stronger reversion of the hypocotyl phenotype (>43% of the wild type), also showed acquired thermotolerance at the 10-d-old seedling stage (Figure 5B). Furthermore, the A329V suppressor recovered as much as the wild type, whereas the R223K and A297T suppressors had pale green leaves and were smaller than the A329V suppressor, also consistent with the relative growth of these suppressors in the hypocotyl assay. Thus, the strong suppressors, R223K, A297T, and A329V, clearly compensate for the primary functional defect caused by hot1-4, although with somewhat different effectiveness. These three mutations are all located in NBD1 (Figure 4), supporting a functional interaction of this domain with the coiled-coil domain containing the primary hot1-4 mutation.
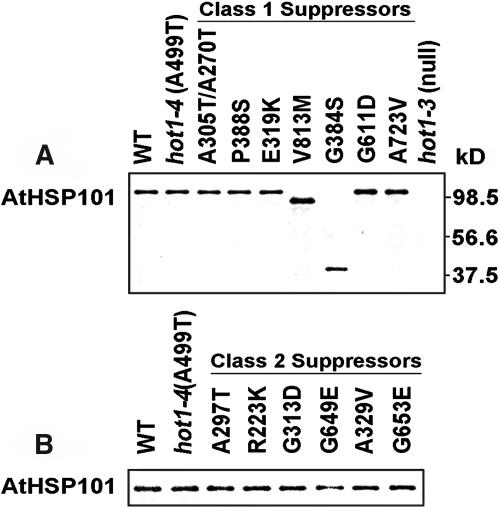
The V813M and G384S Suppressors Accumulate Truncated AtHsp101
To determine whether the restored growth phenotype of the suppressors was because of a dose effect of the mutant protein, protein gel blot analysis was performed on protein samples from heat-treated, 2.5-d-old dark-grown seedlings. Except for V813M and G384S, all the mutant proteins accumulated to similar levels when compared with the wild type and hot1-4 (Figure 6). Although the Class 1 mutants V813M and G384S are obviously missense mutations, ∼90- and 41-kD bands, respectively, were detected with AtHsp101 antibody in these samples (Figure 6). Because the antibody is directed specifically to the N-terminal domain of AtHsp101 (Hong and Vierling, 2001), these bands most likely represent C-terminally truncated proteolytic fragments generated in vivo as a result of protease sensitivity near the mutation site. Thus, these two suppressor mutations clearly produce a nonfunctional protein. This result supports the above conclusion that the defective phenotype in acquired thermotolerance of the Class 1 suppressors is caused by loss of AtHsp101 function.
Figure 6.
Accumulation of AtHsp101 Protein in the Suppressor Mutants.
Protein samples from heat-stressed seedlings (38°C for 90 min followed by 2 h at 22°C) were analyzed with AtHsp101 antiserum.
(A) Class 1 suppressor mutants.
(B) Class 2 suppressor mutants.
Strong Suppressors Restore the Function of AtHsp101 in Protein Resolubilization
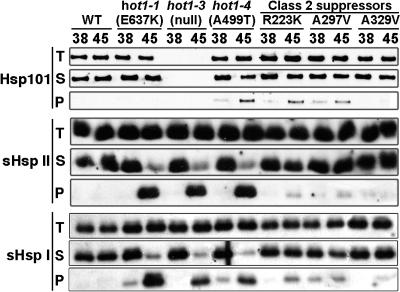
The current model for Hsp100/ClpB function in thermotolerance proposes that these proteins are required for reactivation of aggregated proteins after heat stress (Glover and Lindquist, 1998; Goloubinoff et al., 1999; Motohashi et al., 1999). Therefore, it can be assumed that during recovery from heat stress, heat-aggregated proteins requiring AtHsp101 function should remain in the insoluble cell fraction in hot1 mutants, whereas these proteins should return to the soluble fraction in the wild type. It also follows that if resolubilization is an essential function of AtHsp101 for acquired thermotolerance, the strong hot1-4 suppressor mutations should restore protein solubility after heat stress. As a first step to test this hypothesis, we sought conditions for comparing the insoluble protein fractions from wild-type and mutant plants during recovery from heat stress, along with a suitable protein to assay for solubility. We determined that in wild-type plants, the cytosolic Class I and II sHsps, partitioned during 45°C heat stress into the insoluble cell fraction and almost completely transitioned back to the soluble fraction after 3 h of recovery from the heat stress (data not shown). We therefore compared the solubility of either Class I AtHsp17.6C-I or Class II AtHsp17.6C-II in wild-type, hot1-4, and the strong suppressors, R223K, A297T, and A329V, after 3 h of recovery from a 38°C, 3-h heat stress or from a 45°C, 45-min heat stress with preconditioning. In contrast with the wild type, a majority of the sHsps remained in the pellet fraction during recovery from 45°C stress in the hot1-4, hot1-1, and hot1-3 mutants (Figure 7). The mutants also showed detectably more pelleted sHsps after the 38°C stress. We then examined the solubility of these sHsps in the strong suppressors. sHsp solubility was directly correlated with the strength of the suppressor (Figure 7), with the most effective suppressor, A329V, being almost identical to the wild type and the weaker suppressors, R223K and A297T, showing slightly more sHsp retained in the insoluble fraction. The same samples were also used to examine the solubility of AtHsp101. In contrast with the sHsps, neither the wild type nor the hot1-1 allele of AtHsp101 becomes insoluble during heat stress; similar behavior is reported for E. coli ClpB (Mogk et al., 2003a, 2003c). A fraction of hot1-4 becomes insoluble during heat stress, but insolubility does not correspond to the phenotype, as at 38°C the hot1-4 phenotype is already severe, whereas little protein is insoluble. Only the strongest suppressor reverses the AtHsp101 insolubility. Altogether, these results support the model that one essential thermotolerance function of AtHsp101 involves protein disaggregation and genetically link Hsp101 to the sHsp chaperone system.
Figure 7.
Reversal of sHsp Insolubility in the Suppressor Mutants.
Ten-day-old seedlings were treated either at 38°C for 3 h (38) or at 38°C for 90 min followed by 2 h at 22°C and then 60 min at 45°C (45), and total protein (T) was isolated after further recovery for 3 h at 22°C. The insoluble protein fraction (P) was separated from the soluble fraction (S) by centrifugation and analyzed with AtHsp101, AtHsp17.6C-I (sHsp I), and AtHsp17.6C-II (sHsp II) antiserum.
DISCUSSION
We have been able to identify features critical to Hsp100/ClpB function in vivo using a sensitive, quantitative bioassay for thermotolerance. We identified four missense mutations in the Hsp100/ClpB protein, Arabidopsis AtHsp101, that compromise the function of AtHsp101 in thermotolerance, including an unusual allele, hot1-4 (A499T), which shows a semidominant, gain-of-function sensitivity to a mild, usually nonlethal heat stress. This mutation, which lies in the coiled-coil domain of AtHsp101, supports an essential role for this structural domain in Hsp100/ClpB action in vivo. We also suggest that the gain-of-function phenotype of hot1-4 is consistent with the interpretation that the mutation disrupts AtHsp101 activity such that a critical substrate or cofactor becomes limiting for the cell. Locations of the primary missense mutations hot1-5, -6, and -7 and of suppressors of hot1-4 define other features essential for in vivo function of AtHsp101 and point to protein domain interactions required for Hsp100/ClpB action. Finally, appearance of sHsps in the soluble cell fraction during recovery from heat stress does not occur in hot1-4 but is restored by strong suppressor mutants, in line with the model that disruption of protein aggregates is an essential AtHsp101 function in thermotolerance and supporting the model that sHsps and Hsp100/ClpB proteins interact in a chaperone network in the cell.
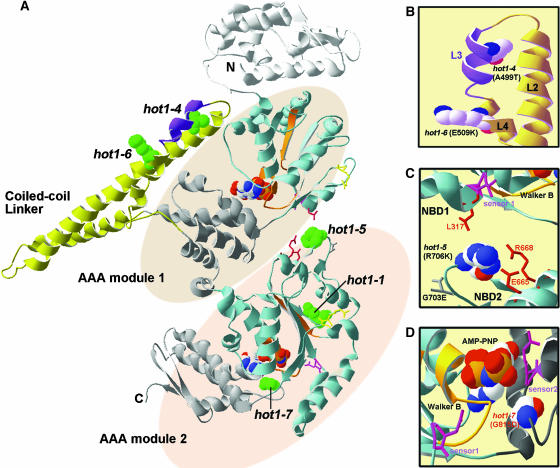
The coiled-coil domain of Hsp100/ClpB is unique to this class of AAA+ proteins (Schirmer et al., 1996; Celerin et al., 1998). Although referred to alternatively as the linker, middle region, or spacer between AAA modules 1 and 2, it is actually an insertion into the C-terminal small domain of AAA module 1 (Figures 1 and 8; see Supplemental Figure 1 online). Deletion of the coiled-coil domain from E. coli ClpB has shown that it is not essential for hexamerization or ATPase activity but is required for thermotolerance in vivo and for disaggregation of firefly luciferase in vitro (Mogk et al., 2003b). The structure of T. thermophilus ClpB reveals that this domain is formed by four helices (L1 to L4) that interact in two coiled-coil motifs comprised of α-helices L1 and L2, and L2 with L3/4 (Figures 8 and 9). Both coiled-coils have features typical of Leu zippers. The hot1-4 mutation (A499T) lies in helix L3, within the conserved sequence segment 494YDLARAADL502 (484YDLNRAAEL492, in T. thermophilus) (Figures 8A and 8B). This sequence motif was previously identified by Schirmer et al. (1996) as characteristic of Hsp100/ClpB chaperones. The weak mutation hot1-6 (E509K) lies in the coiled-coil domain directly outside this conserved motif (Figure 8B). Neither the hot1-4 nor hot1-6 residues are predicted to contact residues in neighboring subunits, which along with the normal accumulation of the proteins and the similar solubility of hot1-4 to the wild type, suggest that these mutations do not significantly disrupt the hexamer structure. In vitro cross-linking experiments also produce the same large, presumably oligomer forms of AtHsp101 in the wild type and the hot1-4 mutants (U. Lee, unpublished data). Eight other mutations in this domain have no effect on AtHsp101 function (Figure 1). Altogether, the hot1-4 and hot1-6 mutations provide direct genetic evidence for the importance of helix L3 and the coiled-coil domain in AtHsp101 function.
Figure 8.
Location of the hot1 Mutations on the T. thermophilus ClpB Structure.
(A) The available structure of the T. thermophilus ClpB monomer (PDB file 1QVR; chain in [A]) is shown with the coiled-coil domain and AAA modules indicated. Within each AAA module, the NBD domain is colored blue, with the Walker A and B motifs in orange, sensor 1 in pink, and the Arg finger in yellow (see also Supplemental Figure 1 online). The small, α-helical domain of each AAA module is shown in gray. Nucleotide (AMP-PNP) is space-filled in CPK coloring (hydrogen, white; oxygen, red; phosphorous, orange; sodium, blue). Motif 1 as defined by Schirmer et al. (1996) is colored purple in the coiled-coil domain. The residue corresponding to each of the hot1 mutation sites is space-filled and colored green.
(B) Alternative view of the motif II region of the coiled-coil domain indicating the L2, L3, and L4 helices and the residue positions of the hot1-4 and hot1-6 mutations (in CPK coloring).
(C) View of interactions of the hot1-5 position (in CPK coloring) with residues within NDB2 and NBD1 as discussed in the text.
(D) Position of hot1-7 (CPK coloring) relative to the NBD2 ATP binding site. The sensor 2 Arg residue, which is part of the GAR motif, as well as sensor 1 are shown in pink. Residue numbering corresponds to AtHsp101. Figure was prepared with Swiss PDB viewer.
Figure 9.
Location of the hot1-4 Suppressor Mutations on the T. thermophilus ClpB Monomer Structure.
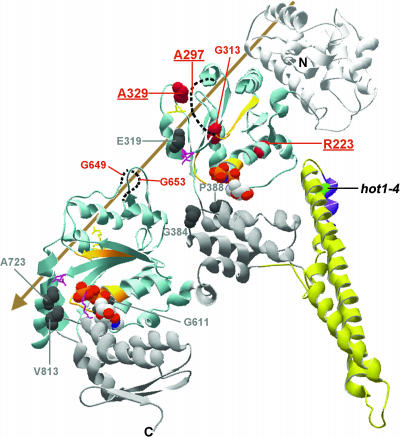
The major domains and motifs of HCP100/ClpB are colored as indicated and described in Figure 8. The sites of Class 1 suppressor mutations are space-filled dark gray, and sites of the Class 2 suppressor mutations are space-filled in red. The three strongest Class 2 suppressors are indicated in bold and are underlined. For A297, G653, and G649, which are in unresolved segments of the structure, dotted lines are used to indicate the relative positions of those segments. The arrow indicates the general position of the axial channel of the hexameric form of the protein. Residue numbering corresponds to AtHsp101. Figure was prepared with Swiss PDB viewer.
Interestingly, in a screen for mutants in Saccharomyces cerevisiae Hsp104, Schirmer et al. (2004) recently reported isolation of a mutation in the same motif, at the adjacent Ala residue (A503V; 497YDTATAADL505 in S. cerevisiae). The Hsp104 A503V mutant was an active ATPase in vitro (Cashikar et al., 2002), exhibiting actually an approximately twofold increase in basal ATPase activity (i.e., activity in the absence of substrate). Therefore, the A503V mutation cannot disrupt the hexameric structure, which is required for ATPase activity. This observation further supports our interpretation that the hot1-4 mutant protein retains its hexameric structure. Furthermore, like hot1-4, the Hsp104 A503V mutation also exhibits a gain-of-function phenotype, leading to cell lethality when induced at the normally permissive temperature of 37°C (Schirmer et al., 2004). The similarity of phenotype of these AtHsp101 and yeast Hsp104 mutations, along with the ability of AtHsp101 to support thermotolerance in yeast (Schirmer et al., 1994), argue that these proteins are true functional homologs.
The other mutations studied also provide new information about interactions within the Hsp100/ClpB proteins. The strong hot1-5 allele alters a residue (R706K) that is conserved in E. coli (R705) and T. thermophilus (R695), and which is in a loop between β-strands 4 and 5 in NBD2, positioned away from the active site (Figures 8A and 8C; see Supplemental Figure 1 online). Normal accumulation and solubility of the hot1-5 protein argues that the protein is not significantly compromised structurally. Based on the T. thermophilus structure, this Arg residue makes intramolecular contact with three other residues conserved in Arabidopsis and E. coli, including a Leu residue directly adjacent to sensor 1 in NBD1, and has side chain interactions with conserved Glu and Arg residues in NBD2 helix D7 (Figure 8C). Notably, a Lys residue is present at the hot1-5 position in yeast Hsp104, and correspondingly the predicted interacting residues are different. The unique importance of hot1-5 interactions, as opposed to variation in the loop itself, is also indirectly supported by the absence of a phenotype for another mutation in the loop connecting β-strands 4 and 5, G703E (Figures 1 and 8C). Hattendorf and Lindquist (2002a) have proposed that ATP hydrolysis at NBD1 depends on the nucleotide bound at NBD2 in yeast Hsp104. We speculate that hot1-5 is in a position involved in communicating nucleotide status between NBD2 and NBD1 and note that the distance between the hot1-5 Arg residue and the conserved Leu in NBD1 varies in the three ClpB molecules in the crystal structure, consistent with a dynamic interaction.
It is difficult to define the possible effects of some of the other AtHsp101 mutations. The hot1-7 mutation represents a dramatic change (G815D) in a conserved motif (GAR) that includes the sensor 2 Arg residue already recognized as important for function (Figure 8D). The residues in the sensor 2 motif are in contact with nucleotides, and the Arg residue in yeast Hsp104 is proposed to contribute binding energy, but not discrimination between ATP and ADP (Hattendorf and Lindquist, 2002a). Mutations in sensor 2, or of all three residues of the GAR motif, do not severely disrupt hexamerization or ATPase activity, but prevent ability to dissociate protein aggregates (Hattendorf and Lindquist, 2002b; Mogk et al., 2003b). In vivo, the moderate phenotype of hot1-7 is consistent with a moderate defect in thermotolerance observed for an R to M mutation of sensor 2 in yeast (Hattendorf and Lindquist, 2002b). The absence of phenotype for 35 other missense mutations analyzed is not surprising, given that many lie in nonconserved residues or represent conservative changes (Figure 1; see Supplemental Figure 1 online). Perhaps the most surprising of the aphenotypic mutations is P671L, which represents a Pro residue conserved not only in NBD2 of all ClpB proteins, but also found in ClpA.
The unusual sensitivity of the hot1-4 allele to 38°C, which is otherwise permissive for wild-type and AtHsp101 null plants, led us to focus further on analysis of this mutation. We performed a screen that identified both loss-of-function (Class 1) and restoration-of-function (Class 2) intragenic suppressor mutations in AtHsp101. Genetic dissection of Hsp100/ClpB function by intragenic suppressor analysis has not been performed in any organism, and we are also unaware of any previous studies using intragenic suppressors for dissection of protein mechanism in higher plants. We argue that the seven Class 1 mutations that eliminated the 38°C heat sensitivity of hot1-4, but did not restore the acquired thermotolerance function, represent loss-of-function AtHsp101 mutants. Consistent with this interpretation, one of these suppressors is in the Walker A motif of NBD 2 (G611D), and three others are predicted to affect the nucleotide binding pocket (E319K, P388S, and A723V) (Figure 9), which should result in defective hexamerization or ATP hydrolysis as demonstrated in vitro for other Hsp100/ClpB proteins (Kim et al., 2000a; Watanabe et al., 2002; Mogk et al., 2003b). These findings suggest that in hot1-4, AtHsp101 retains normal ATP hydrolysis activity and hexamerization and indicates that these properties are essential for the dominant-negative effect of this allele. Two other Class 1 suppressors, G384S and V813M, are both located between the second and third helices in the C-terminal subdomain of NBD1 and NBD2, respectively (Figure 9; see Supplemental Figure 1 online). Both of these mutations lead to accumulation of a truncated AtHsp101 protein with sizes that predict that in vivo cleavage occurs in close proximity to the mutation. Proteolytic removal of the C terminus in V813M supports the requirement of this domain for function, which as stated before, may involve not only hexamerization, but also effector binding (Smith et al., 1999; Strub et al., 2003). Although our screen was not performed to saturation, it is notable that no loss-of-function suppressors were obtained in the N-terminal domain, which has also not been associated with an essential function in other organisms (Clarke and Eriksson, 2000; Beinker et al., 2002; Mogk et al., 2003b).
Whereas the coiled-coil domain is clearly essential for Hsp100/ClpB function, its role in the catalytic cycle is not known. The position of hot1-4 (A499T) (and A503V in S. cerevisiae Hsp104) suggests that this mutation could disrupt interactions of L3 with L2 within a subunit. Lee et al. (2003) have proposed that motion of the coiled-coil domain is required for Hsp100/ClpB chaperone activity. They introduced Cys residues into T. thermophilus ClpB to effect cross-linking of the L2/L3-4 coiled-coil (in which hot1-4 is located) to NBD1 and observed in vitro that ATP hydrolysis continued, but the protein disaggregation activity of ClpB was lost. The three hot1-4 suppressors that restore >43% of acquired thermotolerance of hypocotyl growth and also restore acquired thermotolerance of 10-d-old seedlings are all located in NBD1 (R223K, A297T, and A329V). The strongest of these, A329V, has >70% wild-type activity in the hypocotyl assay and restores solubility of sHsps during recovery from heat stress. Interestingly, A329V lies in close proximity to the NBD1 Arg finger, which is believed to interact with the nucleotide in an adjacent subunit (Figure 9). Another weak suppressor, G313D, is also in NBD1 (Figure 9). Taken together, the location of the Class 2 restoration-of-function suppressors of hot1-4 is consistent with the interpretation that dynamic interaction of the coiled-coil domain with NBD1 is indeed essential in vivo.
How Hsp100/ClpB proteins act to disaggregate proteins has been proposed to involve a crowbar action of the coiled-coil domain and/or threading of substrate through the axial channel of the hexamer, analogous to the mechanism of other hexameric ATPases (Lee et al., 2003; Lum et al., 2004; Schlieker et al., 2004). Location of the Class 2 suppressors provides potential support for the latter mechanism. The strong suppressor A297V is located in a presumably flexible, disordered segment that was not resolved in the TtClpB structure, but which is in the axial channel region. A329V, another strong suppressor, is positioned to potentially alter interactions with another unresolved axial channel loop (TtClpB residues 234 to 246; corresponding to 243 to 255 in AtHsp101), which has recently been proposed to contain residues that bind the E. coli ClpB substrate TrfA (Schlieker et al., 2004). The only restoration-of-function suppressors outside NBD1 are the two weak suppressors, G649E and G653E in NBD 2, which also lie in an unresolved axial channel loop (TtClpB residues 638 to 650; corresponding to AtHsp101 648 to 660) (Figure 9; see Supplemental Figure 1 online). This loop contains the conserved motif GYVG found in other AAA+ proteins (with the first G corresponding to G653 in AtHsp101). Mutation of the Tyr residue in this loop impairs function of E. coli ClpX (Siddiqui et al., 2004) and HslU (Wang et al., 2001b). Structural studies also indicate that this loop gates the axial channel of HslU during the ATPase cycle (Wang et al., 2001a). Recent experiments with yeast Hsp104 also support an essential role for this loop in the protein disaggregation reaction (Lum et al., 2004). In total, we suggest that these intragenic suppressors point to a model in which motions of the coiled-coil domain modulate the ATPase activity of NBD1 and the position of axial channel loops in AtHsp101, which may be involved in threading of substrate through the axial channel.
Analysis of the solubility of plant cytosolic sHsps in the hot1-4 mutant and the strong suppressors also provides data to link these two chaperone systems in a eukaryote. Genetic interactions of sHsps and ClpB have been reported in both E. coli and Synechocystis (Giese and Vierling, 2002; Mogk et al., 2003a) and are supported by in vitro analysis of E. coli ClpB activity in combination with E. coli, Synechocystis, and plant cytosolic class I sHsps (Mogk et al., 2003c). The current model for sHsp function proposes that sHsps bind and maintain denaturing proteins in a form that is accessible to ATP-dependent refolding chaperones. Furthermore, their presence in large protein aggregates makes these aggregates better substrates for the action of Hsp100/ClpB (Mogk et al., 2003c). However, although Hsp100/ClpB disaggregates sHsp containing complexes, there is no evidence for direct physical interaction of Hsp100/ClpB with sHsps (Mogk et al., 2003a, 2003c), and we did not observe association of AtHsp101 with sHsps in the insoluble protein fraction of the cell. In total, the data here indicate that the plant cytosolic Class II, as well as cytosolic Class I sHsps, which diverged on the order of 400 million years ago (Waters and Vierling, 1999), may both facilitate protein disaggregation by Hsp100/ClpB in plants.
At which step the hot1-4 mutation affects catalysis will require additional biochemical studies. It does not appear that the dominant-negative phenotype of hot1-4 is attributable to unregulated ATPase activity of the mutant AtHsp101 protein, leading to depletion of cellular ATP. Measurement of ATP levels in the wild type, hot1-3, and hot1-4 showed no significant differences in ATP levels between the genotypes during heat stress or recovery (U. Lee and E. Vierling, unpublished data). One possibility is that the mutation traps AtHsp101 in a nonfunctional protein complex such that another critical cofactor or substrate becomes limiting for the cell. Such a complex must still be soluble, as our analysis does not indicate that a significant fraction of AtHsp101 becomes insoluble at 38°C where the hot1-4 phenotype is still severe. The Class 1 suppressors that eliminate the dominant-negative phenotype are predicted to disrupt the basic hexameric structure and ATPase activity, consistent with the requirement of these activities to form a trapped complex. By altering structural interactions of the coiled-coil domain with NBD1, the strong Class 2 suppressors may eliminate this trapped intermediate. Weaker Class 2 suppressors may affect the protein's conformation, but not enough to fully disrupt the formation of the nonfunctional complex between hot1-4 and its bound protein(s). Another possibility is that the suppressors directly or indirectly alter a binding site for substrate or functional partner protein, such that a nonfunctional complex no longer forms, but sufficient substrate or cofactor interactions are retained for partial function in vivo. Attempts to coimmunoprecipitate hot1-4 in complex with other proteins has not been successful. Biochemical characterization of hot1-4 along with identity of extragenic suppressors should provide further insight into the nature of the hot1-4 defect and the mechanism of Hsp100/ClpB action.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Columbia accession) plants were grown on plates in the dark or under long-day conditions (16 h light/8 h dark) in a controlled-temperature growth chamber (22°C/18°C). Thermotolerance tests of 2.5-d-old dark-grown or 10-d-old light-grown seedlings were performed basically according to Hong and Vierling (2000).
Isolation and Genetic Analysis of hot1 Mutant Alleles
The hot1-4 mutation was isolated as a thermotolerance-defective mutant in a screen based on hypocotyl elongation of 2.5-d-old dark-grown seedlings (Hong and Vierling, 2000). Allelism tests showed that hot1-4 was tightly linked to the previously described AtHsp101 loss-of-function missense allele, hot1-1 (Hong and Vierling, 2000). For most of the experiments described in this work, the third backcrossed line of hot1-4 to Columbia wild type was used. Tilling analysis (in the Columbia ecotype, carrying the erecta mutation) was performed on three segments of the AtHsp101gene, encompassing approximately amino acid residues 1 to 230, 355 to 595, and 660 to 911 (Arabidopsis Tilling Resource, http://tilling.fhcrc.org:9366) (see Supplemental Figure 1 online). The hot1-5, hot1-6, and hot1-7 mutants were recovered from 37 missense mutations in AtHsp101 obtained from this analysis. Three stop codon mutations (Q409, Q422, and Q704) were also obtained, confirmed to be defective in thermotolerance, and not studied further. Each line was assayed for thermotolerance defects in the homozygous or heterozygous state, and homozygous lines were isolated for lines showing a phenotype (hot1-5, hot1-6, and hot1-7). One homozygous M4 plant from each mutant line was then backcrossed to Columbia erecta wild-type plants, and one homozygous F3 line for each mutation was used for quantitative analysis.
Isolation and Genetic Analysis of hot1-4 Suppressor Mutations
Approximately 7500 homozygous seeds of hot1-4 were mutagenized with ethyl methanesulfonate, and ∼110,000 M2 seed were screened for suppressor mutants as follows. M2 seeds were surface-sterilized and plated on minimal medium containing 0.5% sucrose. Plates were incubated at 4°C for 3 d and then placed in a vertical position at 22°C for 2.5 d in the dark. The dark-grown seedlings were treated at 38°C for 2 h, and then the plates were returned to the dark at 22°C for 1.5 d. Seedlings that showed increased hypocotyl elongation compared with hot1-4 were rescued by growth under light for approximately 1 week before being transplanted to soil. The selected seedlings were retested for tolerance to 38°C in the next generation (M3) as described above.
To distinguish if the suppressor mutations were intragenic or extragenic, the entire AtHsp101 gene from the candidate suppressors was amplified by PCR using specific primers. The amplified DNA products were sequenced on both strands. Thirty-four lines were found to contain the original hot1-4 mutation together with an additional missense mutation within the AtHsp101 coding region. The same suppressor mutation was found in more than two independent lines (R223K, A270/A305T, A297T, G313D, G384S, P388S, G611D, G653E, A723D, and V813M), whereas E319K, A329V, and G649E mutations, which were isolated only once, were confirmed by sequencing from two individual lines (Table 3).
To test genetic linkage between hot1-4 and the suppressors, the homozygous M3 or M4 intragenic suppressors with second site mutations in AtHSP101 were backcrossed to hot1-4. The F1 plants all showed the suppressing phenotype after 38°C heat treatment for 2 h. The subsequent F2 progenies completely segregated in a 3:1 ratio of the suppressor mutant to hot1-4, indicating that the suppressor mutations are tightly linked to the hot1-4 mutation (Table 2; see Supplemental Tables 1 and 2 online).
To obtain F3 homozygous intragenic suppressor lines for phenotypic analysis, the homozygous M3 or M4 intragenic suppressors were outcrossed to wild-type plants. All F1 plants showed the wild-type phenotype, and F2 progenies segregated either the hot1-4 mutant phenotype (Class 1 suppressor) or the suppressor phenotype (Class 2 suppressor) at a ratio of 1:3 to the wild-type phenotype when tested by heating at 45°C for 2 h with pretreatment (38°C for 90 min). Scoring of the hypocotyl length phenotype in F2 progenies was based on the hypocotyl length distribution of the parental lines.
Plant Transformation
A 4.7-kb XhoI/XbaI AtHsp101 genomic region containing the hot1-4 mutation, including 1.5 kb of the promoter region, was cloned into pBin19 and transformed into Columbia wild-type plants by the floral dipping method (Clough and Bent, 1998). A total of 20 lines (T1 generation) were selected on minimal plates with kanamycin (30 μg/mL). The number of T-DNA insertion loci was determined in the T2 generation based on the segregation ratio of both kanamycin resistance and the hot1-4 phenotype. Two independent T3 homozygous lines were used for phenotypic studies.
Fractionation of Heat-Denatured Proteins
Ten-day-old seedlings were pretreated at 38°C followed by 2 h at 22°C, and then heat shocked at 45°C for 60 min (denaturation phase), followed by 3 h of recovery at 22°C (recovery phase). Total protein was extracted before the denaturation phase and after the recovery phase in nondenaturing buffer (25 mM Hepes, pH 7.5, 0.5% Triton X-100, 200 mM NaCl, 0.5 mM EDTA, 5 mM ɛ-amino-N-caproic acid, and 1.0 mM benzamidine). Protein concentration was determined using a Coomassie Brilliant Blue dye binding assay (Ghosh et al., 1988) with BSA as a standard. Total protein solutions were immediately centrifuged at 19,000g for 15 min at 4°C. Supernatants were mixed with the same volume of SDS sample buffer (60 mM Tris-HCl, pH 7.5, 60 mM DTT, 2.0% [w/v] SDS, 15% [w/v] sucrose, 5 mM ɛ-amino-N-caproic acid, and 1.0 mM benzamidine). The pellet was suspended in nondenaturing buffer, washed six times, and then resuspended to the original volume in SDS sample buffer. Proteins were separated by SDS-PAGE on 15% acrylamide gels and processed for protein gel blot analysis (Hong and Vierling, 2001).
SDS-PAGE and Protein Gel Blot Analysis
The 2.5-d-old dark-grown seedlings were treated at 38°C for 90 min, and then total protein was extracted in SDS sample buffer and separated by SDS-PAGE on 7.5% or 15% acrylamide gels and processed for protein gel blot analysis (Hong and Vierling, 2001). Protein blots were probed with rabbit antiserum against AtHsp101 or against AtHsp17.6C-I or -II (Hong and Vierling, 2001). As a loading control, blots were probed for cytosolic glyceraldehyde-3-phosphate dehydrogenase using a GAPC antibody (gift of Ming-Che Shih, University of Iowa) as described (Chan et al., 2002). Blots were incubated with goat anti-rabbit horseradish peroxidase and bands visualized by enhanced chemiluminescence (Amersham International, Piscataway, NJ).
Supplementary Material
Acknowledgments
We thank Ming-Che Shih for the glyceraldehyde-3-phosphate dehydrogenase antibodies, K. Giese, M. Mishkind, and C. Dieckmann for critique of the manuscript, and Joseph T. Carroll for preparation of the structure figures. Supported by Department of Energy Grant DE-FG03-99ER20338 to E.V. S.W.H. was in part supported by the Agricultural Plant Stress Research Center (Kwang Ju, South Korea).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Elizabeth Vierling (vierling@email.arizona.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027540.
References
- Barnett, M.E., Zolkiewska, A., and Zolkiewiski, M. (2000). Structure and activity of ClpB from Escherichia coli. Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 275, 37565–37571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinker, P., Schlee, S., Groemping, Y., Seidel, R., and Reinstein, J. (2002). The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 277, 47160–47166. [DOI] [PubMed] [Google Scholar]
- Cashikar, A.G., Schirmer, E.C., Hattendorf, D.A., Glover, J.R., Ramakrishnan, M.S., Ware, D.M., and Lindquist, S.L. (2002). Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760. [DOI] [PubMed] [Google Scholar]
- Celerin, M., Gilpin, A.A., Schisler, N.J., Ivanov, A.G., Miskiewicz, E., Krol, M., and Laudenbach, D.E. (1998). ClpB in a Cyanobacterium: Predicted structure, phylogenic relationships, and regulation by light and temperature. J. Bacteriol. 180, 5137–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C.S., Peng, H.P., and Shih, M.C. (2002). Mutations affecting light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Plant Physiol. 130, 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A.K., and Eriksson, M.J. (2000). The truncated form of the bacterial heat shock protein ClpB/HSP100 contributes to development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 182, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simple method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dougan, D.A., Mogk, A., Zeth, K., Turgay, K., and Bukau, B. (2002). AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529, 6–10. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., Hepstein, S., Heikkila, J., and Dumbroff, E. (1988). Use of scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissue. Anal. Biochem. 169, 227–233. [DOI] [PubMed] [Google Scholar]
- Giese, K.C., and Vierling, E. (2002). Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J. Biol. Chem. 277, 46310–46318. [DOI] [PubMed] [Google Scholar]
- Glover, J.R., and Lindquist, S.L. (1998). Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82. [DOI] [PubMed] [Google Scholar]
- Goloubinoff, P., Mogk, A., Zvi, A.P., Tomoyasu, T., and Bukau, B. (1999). Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichapherone network. Proc. Natl. Acad. Sci. USA 96, 13732–13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattendorf, D.A., and Lindquist, S.L. (2002. a). Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattendorf, D.A., and Lindquist, S.L. (2002. b). Analysis of the AAA sensor-2 motif in the C-terminal ATPase domain of Hsp104 with a site-specific fluorescent probe of nucleotide binding. Proc. Natl. Acad. Sci. USA 99, 2732–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.W., Lee, U., and Vierling, E. (2003). Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol. 132, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.W., and Vierling, E. (2000). Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc. Natl. Acad. Sci. USA 97, 4392–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.W., and Vierling, E. (2001). Hsp101 is necessary for heat tolerance but dispensable for development and germination in the absence of stress. Plant J. 27, 25–35. [DOI] [PubMed] [Google Scholar]
- Ishikawa, T., Beuron, F., Kessel, M., Wickner, S., Maurizi, M.R., and Steven, A.C. (2001). Translocation pathway of protein substrates in ClpAP protease. Proc. Natl. Acad. Sci. USA 98, 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.I., Cheong, G.W., Park, S.C., Ha, J.S., Woo, K.M., Choi, S.J., and Chung, C.H. (2000. a). Heptameric ring structure of the heat-shock protein ClpB, a protein-activated ATPase in Escherichia coli. J. Mol. Biol. 303, 655–666. [DOI] [PubMed] [Google Scholar]
- Kim, Y.I., Burton, R.E., Burton, B.M., Sauer, R.T., and Baker, T.A. (2000. b). Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5, 639–648. [DOI] [PubMed] [Google Scholar]
- Lee, S., Sowa, M.E., Watanabe, Y., Sigler, P.B., Chiu, W., Yoshida, M., and Tsai, F.T.F. (2003). The structure of ClpB: A molecular chaperone that rescues protein from an aggregated state. Cell 115, 229–240. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Tek, V., Akoev, V., and Zolkiewski, M. (2002). Conserved amino acid residues within the amino-terminal domain of ClpB are essential for the chaperone activity. J. Mol. Biol. 32, 111–120. [DOI] [PubMed] [Google Scholar]
- Lum, R., Tkach, J.M., Vierling, E., and Glover, J.R. (2004). Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279, 29139–29146. [DOI] [PubMed] [Google Scholar]
- Lupas, A.N., and Martin, J. (2002). AAA proteins. Curr. Opin. Struct. Biol. 12, 746–747. [DOI] [PubMed] [Google Scholar]
- Maurizi, M.R., and Xia, D. (2004). Protein binding and disruption by Clp/Hsp100 chaperones. Structure 12, 175–183. [DOI] [PubMed] [Google Scholar]
- Mogk, A., Deuerling, E., Vorderwulbecke, S., Vierling, E., and Bukau, B. (2003. a). Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50, 585–595. [DOI] [PubMed] [Google Scholar]
- Mogk, A., Schlieker, C., Friedrich, K.L., Schonfeld, H.J., Vierling, E., and Bukau, B. (2003. c). Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042. [DOI] [PubMed] [Google Scholar]
- Mogk, A., Schlieker, C., Strub, C., Rist, W., Weiberzahn, J., and Bukau, B. (2003. b). Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrosis, and chaperone activity. J. Biol. Chem. 278, 17615–17624. [DOI] [PubMed] [Google Scholar]
- Motohashi, K., Watanabe, Y., Yohda, M., and Yoshida, M. (1999). Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. USA 96, 7184–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald, A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. (1999). AAA+: A class of chaperone-like ATPases associated with assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43. [PubMed] [Google Scholar]
- Nieto-Sotelo, J., Kannan, K.B., Martínez, L.M., and Segal, C. (1999). Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene 230, 187–195. [DOI] [PubMed] [Google Scholar]
- Ogura, T., and Wilkinson, A.J. (2001). AAA+ superfamily ATPases: Common structure—diverse function. Genes Cells 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Schirmer, E.C., Glover, J.R., Singer, M.A., and Lindquist, S.L. (1996). Hsp100/Clp proteins: A common mechanism explains diverse functions. Trends Biochem. Sci. 21, 289–296. [PubMed] [Google Scholar]
- Schirmer, E.C., Homann, O.R., Kowal, A.S., and Lindquist, S.L. (2004). Dominant gain-of-function mutations in Hsp104p reveal critical roles for the middle region. Mol. Biol. Cell 15, 2061–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, E.C., Lindquist, S., and Vierling, E. (1994). An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell 6, 1899–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer, E.C., Ware, D.M., Queitsch, C., Kowal, A.S., and Lindquist, S.L. (2001). Subunit interactions influence the biochemical and biological properties of Hsp104. Proc. Natl. Acad. Sci. USA 98, 914–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker, C., Weiberzahn, J., Patzelt, H., Tessarz, P., Strub, C., Zeth, K., Erbse, A., Schneider-Mergener, J., Chin, J.W., Schiltz, P.G., Bukau, B., and Mogk, A. (2004). Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 1, 607–615. [DOI] [PubMed] [Google Scholar]
- Siddiqui, S.M., Sauer, R.T., and Baker, T.A. (2004). Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 18, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.K., Baker, T.A., and Sauer, R.T. (1999). Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. USA 96, 6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub, C., Schlieker, C., Bukau, B., and Mogk, A. (2003). Poly-L-lysine enhances the protein disaggregation activity of ClpB. FEBS Lett. 553, 125–130. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. (2000). AAA proteins: Lords of the ring. J. Cell Biol. 150, F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Song, J.J., Franklin, M.C., Kamtekar, S., Im, Y.J., Rho, S.H., Seong, I.S., Lee, C.S., Chung, C.H., and Eom, S.H. (2001. b). Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure 9, 177–184. [DOI] [PubMed] [Google Scholar]
- Wang, J., Song, J.J., Seong, I.S., Franklin, M.C., Kamtekar, S., Eom, S.H., and Chung, C.H. (2001. a). Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure 9, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y.H., Motohashi, K., and Yoshida, M. (2002). Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J. Biol. Chem. 277, 5804–5809. [DOI] [PubMed] [Google Scholar]
- Waters, E.R., and Vierling, E. (1999). The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol. Biol. Evol. 16, 127–139. [DOI] [PubMed] [Google Scholar]
- Weibezahn, J., Bukau, B., and Mogk, A. (2004). Unscrambling an egg: Protein disaggregation by AAA+ proteins. Microb. Cell Fact. 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn, J., Schlieker, C., Bukau, B., and Mogk, A. (2003). Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 278, 32608–32617. [DOI] [PubMed] [Google Scholar]
- Zolkiewiski, M. (1999). ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J. Biol. Chem. 274, 28083–28086. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- Additional published data support threading of the substrate through the axial channel.
- Weibezahn, J., Tessarz, P., Schlieker, C., Zahn, R., Maglica, Z., Lee, S., Zentgraf, H., Weber-Ban, E.U., Dougan, D.A., Tsai, F.T.F., Mogk, A., and Bukau, B. (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.