Abstract
Mussel adhesion to mineral surfaces is widely attributed to 3,4-dihydroxyphenylalanine (Dopa) functionalities in the mussel foot proteins (mfps). Several mfps, however, show a broad range (30–100%) of Tyrosine (Tyr) to Dopa conversion suggesting that Dopa is not the only desirable outcome for adhesion. Here, we used a partial recombinant construct of mussel foot protein-1 (rmfp-1) and short decapeptide dimers with and without Dopa and assessed both their cohesive and adhesive properties on mica using a surface forces apparatus (SFA). Our results demonstrate that at low pH, both the unmodified and Dopa-containing rmfp-1s show similar energies for adhesion to mica and self-self interaction. Cohesion between two Dopa-containing rmfp-1 surfaces can be doubled by Fe3+ chelation, but remains unchanged with unmodified rmfp-1. At the same low pH, the Dopa modified short decapeptide dimer did not show any change in cohesive interactions even with Fe3+. Our results suggest that the most probable intermolecular interactions are those arising from electrostatic (i.e., cation-π) and hydrophobic interactions. We also show that Dopa in a peptide sequence does not by itself mediate Fe3+ bridging interactions between peptide films: peptide length is a crucial enabling factor.
Keywords: Mussel Foot Protein, bio-adhesion, Dopa, mfp-1 peptide, iron chelation, coating proteins
Introduction
Mussels assemble a battery of proteins known as mussel foot proteins (mfps) into a byssus (plaque and the thread) to adhere to solid surfaces in the high-energy intertidal zone. Dopa (3,4-dihydroxyphenylalanine), a post-translational modification from tyrosine (Tyr), features prominently in mfps, ranging from less than 5 mol % in mfp-4 to 30 mol % in mfp-5.[1] Single molecule tensile tests using an atomic force microscope (AFM) where Dopa was tethered to a cantilever tip showed Dopa contributes to nano-Newton adhesion on iron oxide, titania, and amine-functionalized surfaces.[2] Moreover, several studies with Dopa functionalized polymers have demonstrated a strong positive linear correlation between Dopa content and adhesion to different surfaces.[3] Notwithstanding these trends, much debate persists regarding two critical issues of mfp-mediated adhesion: (1) the actual interfacial chemistry of Dopa side-chains on model surfaces, and (2) the contribution of residues other than Dopa to adhesion. The first issue has seen significant progress by the application of resonance Raman microscopy to detect the pH-dependent formation of bidentate binuclear TiIV coordination complexes between Dopa-containing mfp-1[4] and mfp-3[5] on titania surfaces. The second issue, that is, contribution of other residues, is more challenging because the sequences flanking Dopa in most Mfps are so variable that selecting a representative or relevant sequence to test is difficult. Mfp-1 is a rare exception in this regard in that it contains more than 70 tandem high fidelity decapeptide consensus repeats, e.g. AKP*SY*P*P*TY*K, where * denotes optional hydroxylation sites. That is to say, peptides can be found in the native protein with none, all, or some combination of hydroxylations present.[6]
A significant challenge to assessing the adhesive contributions of other amino acids is the complexity of most native mfp sequences, which are polar with high charge density and little to no 2° structure in solution.[7] The sequences are further complicated by the highly variable post-translational modification by enzymes. In purified native mfp-1, for example, overall Tyr→Dopa and Pro→Hyp conversion can range from 50 to 80%. To reduce sequence complexity, we used a recombinant mfp-1 (rmfp-1) analog that contains 12 tandem repeats of the decapeptide sequence AKPSYPPTYK. This is less than a sixth of the 75 decapeptide repeats in native mfp-1 from Mytilus edulis,[6a] has no post-translational modifications, and limits Tyr to a simple repeating consensus sequence P-T/S-Y-X, where X is P or K. For purposes of the present study, we propose that the decapeptide repeats are not uniformly converted to the fully hydroxylated version because there is some adaptive advantage in not doing so. The present investigation examines the consequences of including/excluding hydroxylation on adhesion, cohesion and Fe3+ -binding.
In a previous study, the sequence differences of mfp-1 from 2 related species (Mc- Mytilus californianus and Me- Mytilus edulis) were investigated with regard to Fe-mediated cross-linking of mfp-1 films.[8] The interaction between Fe3+ and mfp-1 using surface sensitive and solution phase techniques showed that the mfp-1 homologs bind Fe3+ differently: mfp-1 (Mc) Dopa groups interact with Fe3+ to form intramolecular complexes, whereas mfp-1 (Me) Dopa groups form intermolecular complexes.[8] Similarly, the adhesive and cohesive contributions of residues other than Dopa in other mfps are the topic of recent studies[8–9] and will be discussed later.
An important assumption in this study is that a recombinant mfp-1 (rmfp-1) analog with only 12 tandem repeats of the unmodified decapeptide sequence (the native mfp-1 sequence from Mytilus edulis[6a] has 75 decapeptide repeats) retains some attributes of unmodified decapeptides in native protein. More than 80% of the Tyr in rmfp-1 can be converted to Dopa by tyrosinase,[10] enabling a separate assessment of contributions by Dopa. Accordingly, rmfp-1 with and without Dopa was tested for adhesion and cohesion on mica using a surface forces apparatus (SFA). We also tested shorter decapeptide dimers (two repeats of the decapeptide sequence, monomer = AKPSYPPTYK) with and without the hydroxylation of Tyr (Y) to Dopa (Y*) and Pro (P) to Hydroxyproline (P*) for cohesion in metal ion (Fe3+) environments to assess the role of peptide length in the formation of metal-protein complexes.
Our results are remarkable in showing that rmfp-1 without Dopa achieves adhesion comparable to Dopa-modified rmfp-1 on mica. Cohesive interactions are also comparable except when Fe3+ is added to symmetric surfaces of rmfp-1 with Dopa. However, the cohesive interactions between short decapeptide dimers remained the same regardless of presence or absence of Dopa, thus stressing the importance of understanding the molecular parameters beyond Dopa that contribute to mussel adhesion.
Results and Discussion
Cohesion (self-interaction) between the protein films and interaction with mica
The cohesive force of interaction between two symmetric rmfp-1 films, Dopa modified and unmodified, was measured in a SFA (Fig. 1A) at two different pH values, pH 3.7 and 7.5 (Fig. 2). The effect of Fe3+ on the cohesive force between the protein films was also investigated (Fig. 3). The protein film studies were conducted under low pH environment because it was recently shown that mussels dramatically acidify (pH~2–4) the local environment at the substrate-plaque interface during plaque formation.[11]
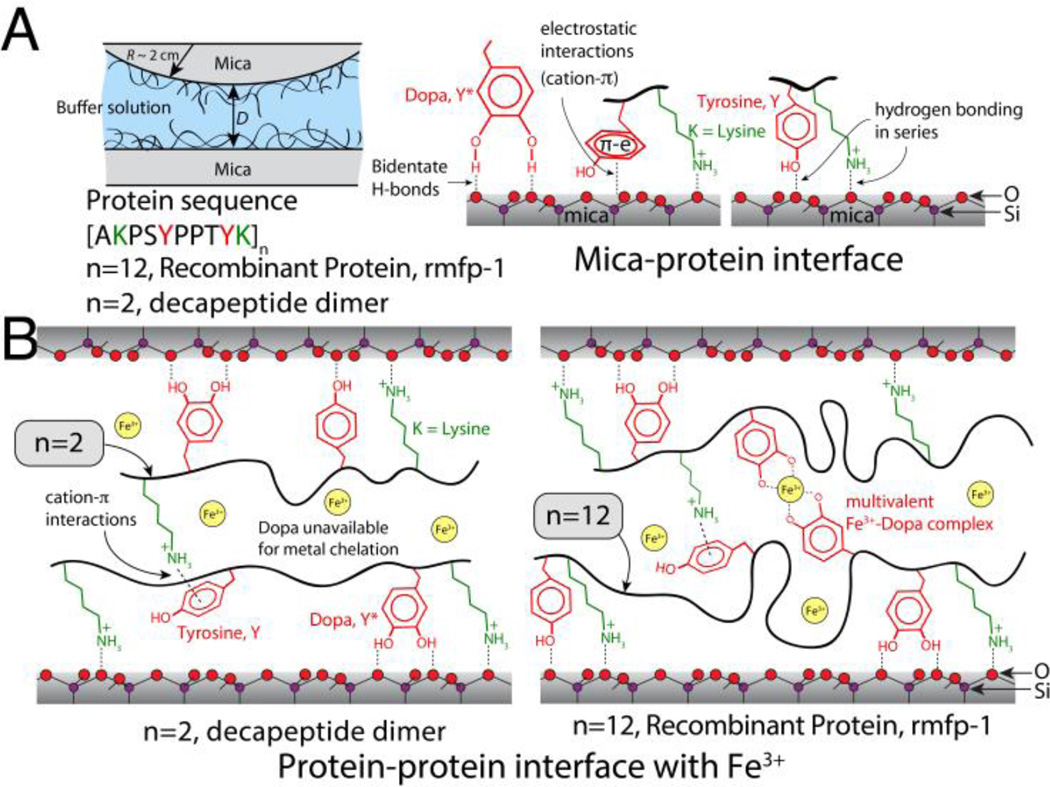
Fig. 1.
Scheme of the surfaces analyzed by the surface forces apparatus. (A) rmfp-1 and short peptides with or without Dopa are adsorbed as thin films onto one or both mica surfaces; Schematics of the bidentate H-bonds, electrostatic and π-cation interactions between the protein and K+ ions adsorbed (not shown for the sake of clarity) to the mica surface (see supplementary Fig. S1). Our results suggest that electrostatic, π-cation and hydrophobic interactions between aromatic residues and mica are more probable than bidentate H-bonding interactions. (B) Schematics showing the effect of peptide length on the adhesive interactions between the protein films. Metal mediated cross-links across the films are possible for proteins containing Dopa residues only when the number of decapeptide monomers is greater than a critical number (n between 2 and 12). For the short decapeptide dimers, most Dopa residues get recruited to the substrate whereas for the decapeptide 12-mer, free Dopa residues remaining at the protein-solution interface are available to bridge with exposed Dopa on the opposing surface through Fe3+ mediated chelation.
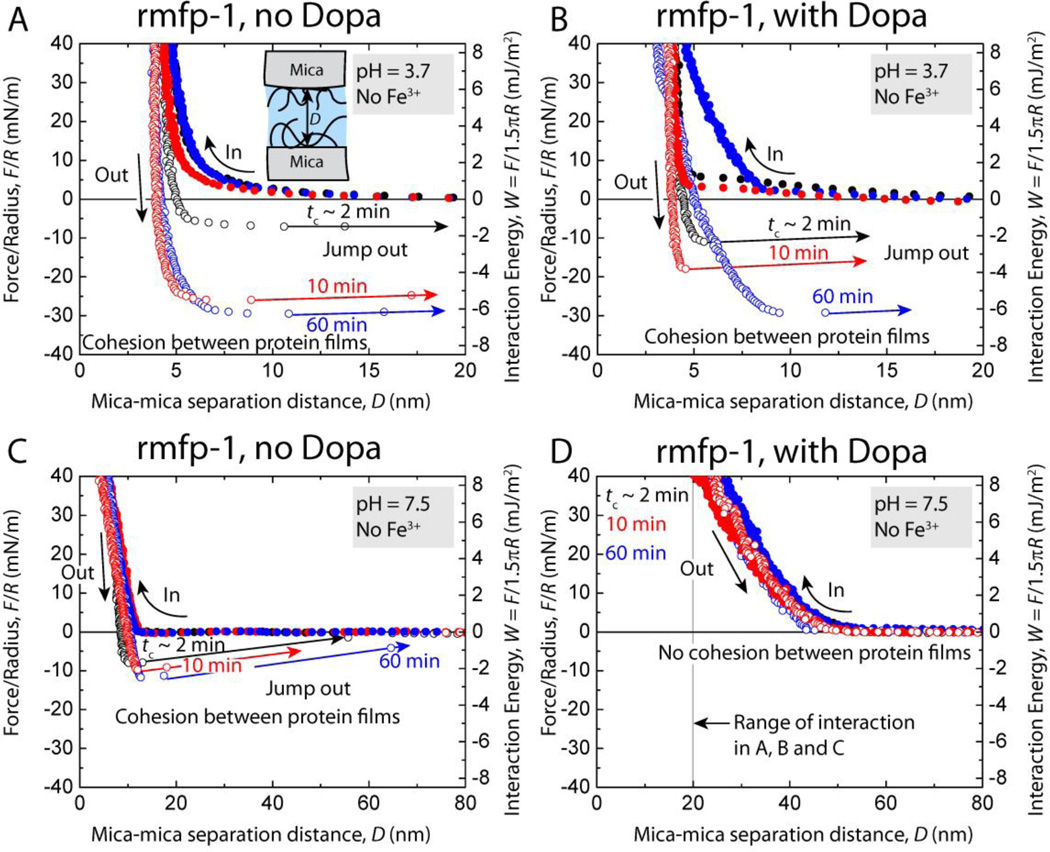
Fig. 2.
Representative force vs. distance plots showing the effect of contact time, tc, on the cohesion between two symmetric rmfp-1 films without Dopa (A, C) as well as two Dopa-containing rmfp-1 films (B, D) at pH 3.7 and, pH 7.5, respectively.
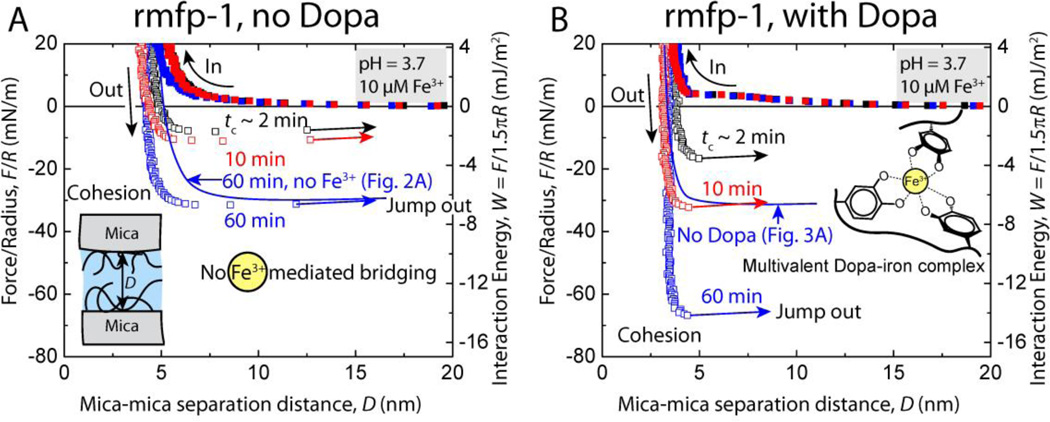
Fig. 3.
Representative force vs. distance plots showing the effect of contact time, tc, on the cohesion between two symmetric (A) unmodified rmfp-1 and (B) Dopa-containing rmfp-1 films at pH 3.7 with 10 µM Fe3+ between the surfaces.
At pH 3.7, similar cohesive interactions were measured for Dopa-containing and unmodified rmfp-1 (no Dopa) when surfaces were kept under compressive contact at t ≥ 10 min (Wc = 4.9 ± 0.6 mJ/m2) (Fig. 2A, B). For short contact times, tc ~ 2 min, the Dopa modified rmfp-1 showed almost 60 % higher cohesion (Wc = 2.40 ± 0.6 mJ/m2) compared to the unmodified protein film (Wc = 1.5 ± 0.8 mJ/m2). This suggests that Dopa may accelerate the development of cohesion between the protein films; however, given enough interaction time, Dopa adds little to the magnitude of cohesive strength between the protein films at equilibrium. The kinetics of bonding interactions during the contact between the films remains complex and somewhat beyond the reach of experiment, however, we observe that Dopa expedites cohesion between the films at short contact times.
At pH 7.5, the Dopa-containing rmfp-1 ceased to cohere and instead showed long-range steric repulsion (Fig. 2D). This is similar to the trend reported for the native mfp-1 and attributed to Dopaquinone formation and the conformational consequences of the tautomerization of Dopaquinone to Δ–Dopa.[12] Interestingly, the unmodified rmfp-1 showed significant cohesion (Wc = 2.0 ± 0.5 mJ/m2), perhaps because there was no Dopa to oxidize. However, unlike native mfp-1 the range of interaction between the rmfp-1 films was not altered (Fig. 1C). The cohesion measured in the unmodified rmfp-1 is contrary to previous observation where the protein did not show cohesion at similar salt concentrations and at a lower pH 5.5.[13] This could possibly be due to the dimerization of the protein since the authors had observed a thicker hardwall (DH = 20 – 25 nm compared 3 – 5 nm in our work) in their experiments and suggested aggregation of the proteins during its synthesis. Recent results suggest that the starting concentration of solutions used for bulk deposition plays a crucial role in determining the adhesive and cohesive properties of a protein film.[8] Hence, the disparity in the results could also be attributed to the lower protein deposition concentrations (20 µg/ml compared with 50 µg/ml in this work) used in the earlier work.
The cohesion between the unmodified rmfp-1 films was completely recovered when the pH of the buffer was switched from 3.7 to 7.5 and back to 3.7 unlike the Dopa modified rmfp-1 where the protein underwent pH-induced irreversible structural changes and cohesion could not be recovered. At low pH and low salt concentrations, π–cation[14] and hydrophobic[15] interactions are strong and these interactions tend to get weaker at higher pH and high salt conditions. Thus, the reversible cohesive behavior of the unmodified rmfp-1 film demonstrates that cohesion in rmfp-1 films could be due to electrostatic (e.g., π–cation)[16], hydrophobic interactions[17] and π–π stacking[18] and that Dopa is not essential for cohesion as has been repeatedly argued in the literature.[3a, 5, 19]
Another intriguing finding was related to the adhesion of the unmodified (no Dopa) and the Dopa modified rmfp-1 film to mica. Both the proteins showed similar time dependence and adhesion energies to mica. Unmodified rmfp-1 adhered to mica with Wad = 8.0 ± 0.1 mJ/m2 whereas, the Dopa modified rmfp-1 showed similar adhesion energy of Wad = 9.8 ± 1.2 mJ/m2 at tc = 60 min (Fig. S1). Protein adsorption experiments in a Quartz Crystal Microbalance (QCM) further established that presence of Dopa in the protein does not change the mass of protein (m ~ 80 ng/cm2) adsorbed to a TiO2 surface (Fig. S2). The negligible change in the dissipation of the quartz crystal (Fig. S2) upon the adsorption of the protein at pH 3.7 indicates that rmfp-1, both with and without Dopa, forms a stiff film on TiO2, and bidentate coordination bond[3a, 19a] of the Dopa to the crystalline TiO2 is not the dominant mechanism that binds the protein to the surface at these solution conditions. It was previously demonstrated that hydrophobicity in the mfps mediates dehydration at substrate protein interface to allow force-free adhesion of the protein to a substrate[20] and that the adsorption of the proteins to a surfaces depends on the Dopa content for small decapeptide monomers or dimers.[21] However, present results argue that for a decapeptide 12-mer, the force-free adsorption of the protein (as measured in the QCM-D) is surprisingly independent of the presence of the Dopa residue. It should be noted that the thickness of the rmfp-1 film with Dopa was about 4 – 5 nm compared to 0.7 – 1.5 nm for the rmfp-1 film without Dopa as measured in the SFA (Fig. S1). The presence of Dopa might affect the structure of the adsorbed rmfp-1 film on the surface, however, both films showed similar adhesive/cohesive properties (SFA studies) and stiffness (QCM-D measurements).
The similar adhesion energies of Dopa modified and unmodified protein to mica also suggest that the primary interaction between the protein film and mica could be due to specific coulombic interactions between the lysine and negatively charged mica or mono-dentate hydrogen bonding in series with lysine-mica interactions (Fig. 1B). Hydrophobic interactions between the aromatic residues and the hydrophobic domains in the mica crystal[15] could also cause a strong adhesion between protein and the surface. π–cation interaction between the aromatic residues of the peptides in the protein and the K+ in the mica crystal lattice could also possibly cause enhanced interaction between the protein and the surface, and bidentate bonds between Dopa and the polysiloxane lattice of mica might play a minor role in the adhesion. Similar π–cation interactions were previously proposed between lignin and gold[22] and lipid bilayers and proteins.[23] The work of adhesion between the mica and rmfp-1 was approximately Wad = 7.8 ± 0.6 mJ/m2 for both Dopa modified and unmodified rmfp-1 (bidentate H-bonds not possible) at short contact times tc ~ 2 min (Fig. S1) which suggests that bidentate Dopa bond to mica cannot be the primary mode of binding to mica surfaces by rmfp-1. It should be noted that the true adhesion energy of the protein to the substrate is likely to be greater than the value measured in the SFA. On pre-adsorbing the protein to mica, most residues endowed with surface-binding reactivity get recruited to the substrate thus become unavailable to bind the opposing interface. Hence, our measurements show that the binding strength of the decapeptide 12-mer to a mica surface is > 7.8 mJ/m2.
There was no material transfer between the surfaces during the force measurements because the approach force-run profiles for the very first contact between the surfaces were similar to the successive runs repeated at least 6 times at the same contact point. The measured cohesive force also didn’t change significantly (< 1 %) for the successive force measurements at a given contact point. The failure during the separation of the protein films was determined to be the protein-protein interface and not the mica-protein interface as the adhesion measured between rmfp-1 (unmodified or Dopa-containing rmfp-1) and mica was significantly higher (Wad = 8.4 ± 0.8 mJ/m2) than the cohesive energies (Wc = 3.9 ± 1.7 mJ/m2) of symmetric rmfp-1 films at tc = 2 to 60 min (Fig. 2 and S1).
Introduction of 10 µM Fe3+ into the gap between rmfp-1 surfaces did not change the cohesion between the unmodified rmfp-1 films (Wc = 5.9 ± 0.8 mJ/m2 for tc = 60 mins with and without Fe3+). However, Fe3+ doubled the cohesion energy between the Dopa – containing rmfp-1 after similar contact times (Fig. 3) and the forces measured were reversible. Contact time tc, between the surfaces significantly changed the cohesive energy from Wc = 3.3 ± 0.4 mJ/m2 for t = 2 min to Wc = 10.0 ± 2.8 mJ/m2 at 60 min for the Dopa-containing rmfp-1 surfaces apparently due to Fe3+ bridging coordination or previously observed Fe3+ mediated covalent cross-linking at low pH.[24] To determine the mechanism of Fe3+ mediated cohesion between the Dopa modified rmfp-1 films, the force measurements were repeated several times (N = 6, see methods) at a given contact point. There was no material transfer between the surfaces during the force measurements because the approach force-run profiles for the very first contact between the surfaces were similar to subsequent force runs and reversible. This observation argues against the covalent cross-linking (irreversible process) of the peptide films by Fe3+ in acidic pH and suggests that Fe3+ bridging between the Dopa modified rmfp-1 films is limited to coordination complexes (Fig. 1B and 3B).
The temporal increase in the Fe3+ mediated cohesive forces (or energies, Wc increases for contact time, tc = 2 min to 60 min) indicates that it takes time for the Fe3+ to recruit two or more Dopa and bridge them across the surfaces. These results also show that Fe3+ is involved in chelating only the Dopa moieties in the rmfp-1 films by forming multivalent catecholate-Fe complexes across the surfaces; however, other hard Lewis acid donors such as the –OH of the Tyrosine or the –NH2 of Lysine between rmfp-1 surfaces are not coordinated. The ligand number of the Fe3+-Dopa complex depends on the pH and the ratio of Dopa to Fe3+,[24b] and the bridging of rmfp-1 surfaces is by bis- and tris-catecholato-Fe3+ complex formation. The local pH within the protein film can be different from the bulk pH[25] (rmfp-1 has a pI of ~10); hence determining the ratio of bis to tris complexes at an interface is challenging and beyond the scope of this work. The magnitude of Fe3+ mediated cohesion between the Dopa modified rmfp-1 films measured in this work is comparable with biotin-avidin interfacial bond energy (Wad ~ 10 mJ/m2),[26] the strongest known non-covalent interaction between a protein and ligand. Two to three Dopa residues of mfp-1 in the cuticle of the marine mussels complex with a single Fe3+,[27] thereby creating a stable complex that can, in principle, be translated to cross-link other structural proteins. These iron-protein complexes have a breaking force nearly half that of covalent bonds (as measured in our experiments), but unlike covalent bonds they can form and break reversibly, making them ideal for creating sacrificial cross-links to prevent catastrophic failure of a material.
Cohesive interactions between mfp-1 short peptide dimers with Dopa
Cohesive interactions between short decapeptide dimers (Pro-pep, [AKPSYPPTYK]2) of the consensus decapeptide repeat unit of mfp-1 were measured to determine the effect of peptide length on the energy of interaction between the protein films uniformly deposited on mica surfaces. We investigated the effect of Fe3+ on the change in cohesive energy between the short peptide films. Another short decapeptide dimer (Hyp-Pep, [AKP*SYP*P*TYK]2, P* = trans-4-hydroxyproline) with hydroxyproline modification was also tested for cohesion. Hyp-pep dimer is a closer mimic of the consensus decapeptide repeat unit of mfp-1 which has trans-4-hydroxyproline modification at P-3, P-6 and P-7 of the decapeptide (additional trans-3 modification occurs at P-6, but was not tested here). We also assessed if hydroxylation of proline has an effect on the cohesive and metal chelating properties between the protein films.
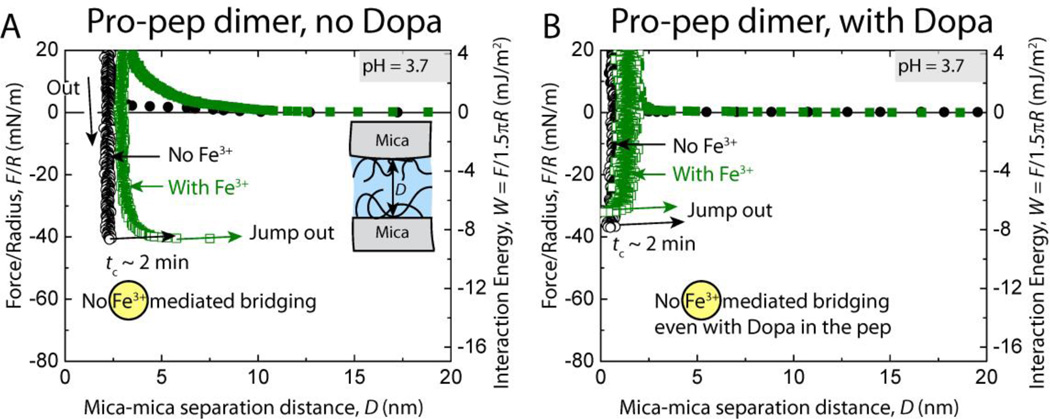
At pH 3.7, the cohesive energy of interaction between unmodified mfp-1 Pro-pep (proline containing dimer) film was Wc = 8.1 ± 1.1 mJ/m2 at short contact times, tc = 2 min (Fig. 4A), and did not change when the surfaces were kept under compressive contact for t = 10–60 min unlike rmfp-1 (Fig. 2A, B). Dopa-modified Pro-pep dimer showed cohesion energy similar to the unmodified dimer. The forces measured between unmodified mfp-1 Pro-pep dimer films on approach were purely repulsive due to steric and hydration forces[28] (Fig. 4A).
Fig. 4.
Representative force vs. distance plots of cohesion between two symmetric (A) unmodified (no Dopa) and (B) Dopa-containing mfp-1 peptide dimer (with proline, Pro-pep) films at pH 3.7 with (green points) and without (black points) 10 µM Fe3+ between the surfaces.
The cohesion energy between the mfp-1 peptide films did not change on introducing 10 µM Fe3+ between the surfaces regardless of the Dopa modification of the decapeptide dimers (Fig. 4) for upto tc = 60 min. In a separate experiment, the Dopa modified decapeptide dimers were given longer times (up to tc = 24 h) to interact cohesively in the presence of Fe3+; however, the cohesive energy of interaction did not change significantly (Wc = 7.7 ± 0.9 mJ/m2, ntrials = 4). This is contrary to the commonly observed property of ferric ions to chelate Dopa containing protein films across surfaces as shown in our rmfp-1 films experiments and previously seen in natural mussel foot protein films.[8, 13] Perhaps the Dopa needed to coordinate and form Fe3+-mediated bridges between the films is unavailable by virtue of interacting with the mica surface through various interactions as shown in Fig. 1B.
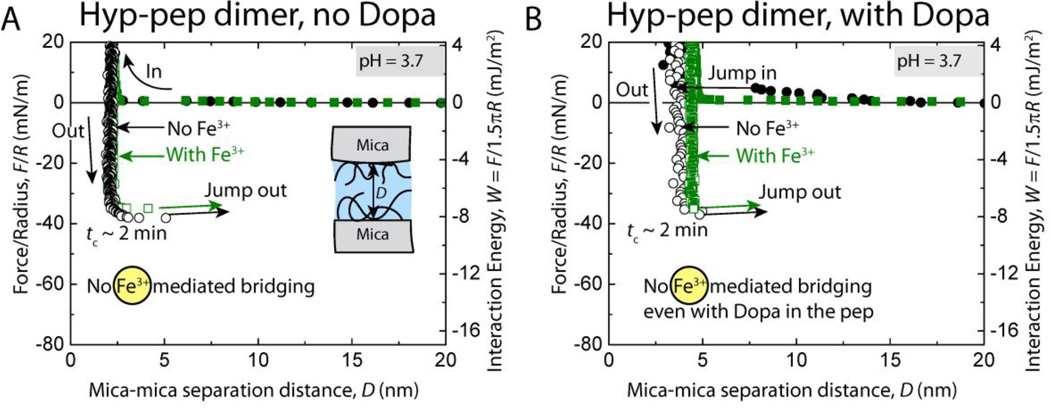
Interestingly, the peptide dimers with hydroxyproline (Hyp-pep) showed cohesion energies similar to the Pro-pep dimers (Wc = 9.4 ± 1.2 mJ/m2) and Dopa did not have an effect on the interaction energies between the films (Fig. 5). Fe3+ was also unable to enhance the cohesive interactions between the Hyp-pep films. These results suggest that peptide length is a critical design parameter for Fe3+-mediated cohesive bridging. We showed that there is a critical number for the repeating decapeptide unit of the monomer between 2 and 12 necessary to trigger metal chelation (Fig. 1B) between the peptide films and that incorporating Dopa into a peptide sequence does not necessarily guarantee the formation of metal mediated cross-links between the peptide films.
Fig. 5.
Representative force vs. distance plots of cohesion between two symmetric (A) unmodified (no Dopa) and (B) Dopa-containing mfp-1 peptide dimer (with trans-4-hydroxyproline, Hyp-pep) films at pH 3.7 with (black points) and without (green points) 10 µM Fe3+ between the surfaces.
Conclusions
In this work, we demonstrate that bidentate hydrogen bonding by Dopa plays only a minor role in the adhesion of mfp-1 to mica (or adsorption to titania surface). The adhesion of the proteins or peptides to a mica surface is more due to specific coulombic interactions between lysine and the negative mica surface or mono-dentate hydrogen bonding in series with Lysine-mica interactions. Hydrophobic interaction between the aromatic residues and the hydrophobic domains in the mica crystal lattice or π–cation between the aromatic rings in the protein and the ions adsorbed to the mica interface are possibly responsible for the adhesion.
Since the catechol group did not influence the cohesive strength between the protein films, π–π stacking, hydrophobic and π–cation interactions are more likely to contribute to the strong cohesion at pH 3.7. Dopa residues tend to accelerate bond formation between the peptide films, however, given enough time, the equilibrium cohesive energy between the films is independent of the Dopa residues in the protein film. The cohesion energy between the protein films was similar for a decapeptide dimer and a 12-mer suggesting that entanglement-entrapment mechanisms[29] are not responsible for the bonding between the mussel inspired peptide films. Cohesion between Dopa-containing rmfp-1 surfaces can be doubled through Fe3+ mediated chelation resulting in an interfacial energy of Wc ~ 10 mJ/m2 which is equivalent to biotin-avidin interfacial adhesion energy, the strongest known non-covalent interaction; but unlike the protein and ligand interaction, the iron mediated cohesive bond can be broken and formed reversibly.[30] This interaction is absent without Dopa in the protein.
Incorporating Dopa into a peptide sequence does not guarantee the formation of metal mediated cross-links between peptide films and the length of the peptide is a very crucial parameter that determines the performance of the materials that involve coordination chemistry. Hence, Dopa containing proteins and peptides with appropriate length could be used as tunable systems for applications in strain resistant coatings, drug delivery and bio-adhesives.
Methods
Modification of rmfp-1
Rmfp-1 used in this work is a shorter synthetic analogue of the natural mussel foot protein mfp-1 from Mytius edulis with 12 tandem repeat units of the mefp-1 consensus decapeptide AKPSYPPTYK. The protein had a M+H+ of 13,619 Da by MALDI TOF mass spectrometry. Tyr in rmfp-1 was converted to Dopa by mushroom tyrosinase (Sigma-Aldrich) using the borate capture method[10] and then purified by C-18 reverse phase HPLC column, eluted with a linear gradient of aqueous acetonitrile. Eluent was monitored continuously at 230 and 280 nm, and 0.33 ml fractions containing peptides were pooled and freeze-dried. Sample purity and hydroxylation were assessed by MALDI-TOF. M+H+ was 13,939 Da with > 83% conversion efficiency. The short peptide dimers ([AKPSYPPTYK]2 and [AKP*SYP*P*TYK]2, P*= trans-4-hydroxyproline) used in these experiments were obtained from GenScript USA Inc. and tyrosine was modified to Dopa by similar methods described above.
Measuring the adhesive/cohesive interactions
The surface forces apparatus (SFA, SurForce LLC) was used to measure the normal forces between two mica surfaces in a cross-cylindrical geometry as a function of the separation distance, D, between them and has been described elsewhere.[28, 31] The protein films were made by adsorbing 50 µL of the protein from a 50 µg/ml in a buffer solution (10 mM sodium acetate buffer, pH 3.7) onto the mica surfaces for 15 minutes, then rinsing the excess protein with the same buffer. It should be noted that the protein deposition concentration was set at 50 µg/ml as previously optimized for mfp1 for achieving maximum cohesive interactions.[8] During the protein adsorption, the discs were kept in a saturated Petri dish to minimize evaporation of the water from the surfaces. The discs were then mounted in the SFA in one of two configurations. In a symmetric configuration (Fig. 1A), the mussel protein film was deposited on both surfaces in order to measure cohesion between the protein films. Cohesion was tested with and without iron. To test the effect of Fe3+, a 10 µM FeCl3 in acetate buffer (as above) was freshly made and added to the reservoir between the symmetrically deposited protein films on mica.
The protein films were always hydrated (i.e. never allowed to dry) and a droplet of the acetate buffer was injected between the surfaces immediately after loading in the SFA. During a typical approach-separation force measurement cycle, the surfaces were first moved towards each other (approach) until reaching a "hardwall" and then separated. The hardwall distance, DH, is the separation distance between the two mica surfaces upon compression that does not change with increased compression. There was no material transfer between the surfaces during the force measurements because the approach force profiles for the initial contact between the surfaces were similar to the successive runs repeated at least 6 times at the same contact point. All the experiments were repeated 3 times. The energy of interaction between two crossed-cylinder geometry, roughly corresponds to a sphere of radius R approaching a flat surface based on the Derjaguin approximation, W(D) = F(D)/2πR where, W(D) is the energy of interaction per unit area between two flat surfaces and F(D) is the measured force of interaction in the SFA.[29] The measured adhesion (or cohesion) force Fad (or Fc) is related to the adhesion (or cohesion) energy per unit area by Wad = Fad/2πR for rigid surfaces with weak adhesive interactions, and by Wad = Fad/1.5πR (used in this study) for soft deformable surfaces with strong adhesion or cohesion.[29, 32]
Protein adsorption experiments
Quartz Crystal Microbalance with Dissipation (QCM-D) experiments were done with a Q-Sense E4 open module to characterize the adsorption of rmfp-1 (Dopa modified and unmodified) to TiO2 surfaces independently of the SFA experiments. The QCM crystals were cleaned in 3% SDS solution, rinsed in distilled water, cleaned with ethanol and then treated with UV-Ozone for 10 min. Frequency and dissipation baselines were established in 100 µL of acetate buffer solution on the crystal followed by injection of 25 µL of 50 µg/ml rmfp-1. The QCM experiments were repeated 3 times on each surface for each protein.
Supplementary Material
Acknowledgments
This research was supported by grants from NIH (R01 DE018468). QCM measurements were done at the MRL shared experimental facility. The MRL Shared Experimental Facilities are supported by the MRSEC Program of the NSF under Award No. DMR 1121053; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). S.D., J.H.W., and J.N.I. designed research; S.D. and N.R.M.R. performed research; W.W. contributed new reagents/analytic tools; S.D., N.R.M.R, J.H.W., and J.N.I. analyzed data; and S.D. and J.H.W. wrote the paper. S.D and N.R.M.R. contributed equally to this work.
Footnotes
The authors declare no conflict of interest.
Contributor Information
Dr. Saurabh Das, Department of Chemical Engineering, University of California, Santa Barbara, California 93106, USA; Materials Research Laboratory, University of California, Santa Barbara, California 93106, USA.
Dr. Nadine R. Martinez Rodriguez, Department of Molecular, Cell & Developmental Biology, University of California, Santa Barbara, California 93106, USA Materials Research Laboratory, University of California, Santa Barbara, California 93106, USA.
Dr. Wei Wei, Materials Research Laboratory, University of California, Santa Barbara, California 93106, USA
Prof. J. Herbert Waite, Department of Molecular, Cell & Developmental Biology, University of California, Santa Barbara, California 93106, USA; Materials Research Laboratory, University of California, Santa Barbara, California 93106, USA; Department of Chemistry and Biochemistry, University of California, Santa Barbara, California 93106, USA.
Prof. Jacob N. Israelachvili, Department of Chemical Engineering, University of California, Santa Barbara, California 93106, USA; Materials Research Laboratory, University of California, Santa Barbara, California 93106, USA.
References
- 1.a) Taylor SW, Luther GW, Waite JH. Inorg. Chem. 1994;33:5819. [Google Scholar]; b) Papov VV, Diamond TV, Biemann K, Waite JH. J. Biol. Chem. 1995;270:20183. doi: 10.1074/jbc.270.34.20183. [DOI] [PubMed] [Google Scholar]; c) Rzepecki LM, Waite JH. Mol. Mar. Biol. Biotech. 1995;4:313. [PubMed] [Google Scholar]; d) Sun CJ, Waite JH. J. Biol. Chem. 2005;280:39332. doi: 10.1074/jbc.M508674200. [DOI] [PubMed] [Google Scholar]; e) Vreeland V, Waite JH, Epstein L. J. Phycol. 1998;34:1. [Google Scholar]; f) Zhao H, Waite JH. J. Biol. Chem. 2006;281:26150. doi: 10.1074/jbc.M604357200. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Scherer NF, Messersmith PB. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12999. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Anderson TH, Yu J, Estrada A, Hammer MU, Waite JH, Israelachvili JN. Adv. Funct. Mater. 2010;20:4196. doi: 10.1002/adfm.201000932. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heo J, Kang T, Jang SG, Hwang DS, Spruell JM, Killops KL, Waite JH, Hawker CJ. J. Am. Chem. Soc. 2012;134:20139. doi: 10.1021/ja309044z. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chung HY, Grubbs RH. Macromolecules. 2012;45:9666. doi: 10.1021/ma301666x. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Liu B, Burdine L, Kodadek T. J. Am. Chem. Soc. 2006;128:15228. doi: 10.1021/ja065794h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Saxer S, Portmann C, Tosatti S, Gademann K, Zurcher S, Textor M. Macromolecules. 2010;43:1050. [Google Scholar]
- 4.Hwang DS, Harrington MJ, Lu Q, Masic A, Zeng H, Waite JH. J. Mater. Chem. 2012;22:15530. doi: 10.1039/C2JM32439C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu J, Wei W, Menyo MS, Masic A, Waite JH, Israelachvili JN. Biomacromolecules. 2013;14:1072. doi: 10.1021/bm301908y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Laursen R. In: Structure, Cellular Synthesis and Assembly of Biopolymers. Case S, editor. Vol. 19. Berlin Heidelberg: Springer; 1992. p. 55. [Google Scholar]; b) Waite JH, Housley TJ, Tanzer ML. Biochemistry. 1985;24:5010. doi: 10.1021/bi00340a008. [DOI] [PubMed] [Google Scholar]
- 7.Hwang DS, Waite JH. Protein Sci. 2012;21:1689. doi: 10.1002/pro.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das S, Miller DR, Kaufman Y, Martinez Rodriguez NR, Pallaoro A, Harrington MJ, Gylys M, Israelachvili JN, Waite JH. Biomacromolecules. 2015;16:1002. doi: 10.1021/bm501893y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Martinez Rodriguez NR, Das S, Kaufman Y, Wei W, Israelachvili J, Waite JH. Biomaterials. 2015;51:51. doi: 10.1016/j.biomaterials.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wei W, Yu J, Gebbie M, Tan Y, Martinez Rodriguez NR, Israelachvili J, Waite J. Langmuir. 2015;31:1105. doi: 10.1021/la504316q. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Seo S, Das S, Zalicki P, Mirshafian R, Eisenbach CD, Israelachvili JN, Waite JH, Ahn BK. J. Am. Chem. Soc. 2015 doi: 10.1021/jacs.5b03827. [DOI] [PubMed] [Google Scholar]
- 10.Taylor SW. Anal. Biochem. 2002;302:70. doi: 10.1006/abio.2001.5522. [DOI] [PubMed] [Google Scholar]
- 11.Martinez Rodriguez NR, Das S, Kaufman Y, Israelachvili J, Waite JH. Biofouling. 2015;31:221. doi: 10.1080/08927014.2015.1026337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rzepecki LM, Waite JH. Arch. Biochem. Biophys. 1991;285:27. doi: 10.1016/0003-9861(91)90324-c. [DOI] [PubMed] [Google Scholar]
- 13.Zeng HB, Hwang DS, Israelachvili JN, Waite JH. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12850. doi: 10.1073/pnas.1007416107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kearney PC, Mizoue LS, Kumpf RA, Forman JE, Mccurdy A, Dougherty DA. J. Am. Chem. Soc. 1993;115:9907. [Google Scholar]
- 15.Donaldson SH, Das S, Gebbie MA, Rapp M, Jones LC, Roiter Y, Koenig PH, Gizaw Y, Israelachvili JN. ACS Nano. 2013;7:10094. doi: 10.1021/nn4050112. [DOI] [PubMed] [Google Scholar]
- 16.Mecozzi S, West AP, Dougherty DA. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10566. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson SH, Jr, Røyne A, Kristiansen K, Rapp MV, Das S, Gebbie MA, Lee DW, Stock P, Valtiner M, Israelachvili J. Langmuir. 2014;31:2051. doi: 10.1021/la502115g. [DOI] [PubMed] [Google Scholar]
- 18.Hunter CA, Sanders JK. J. Am. Chem. Soc. 1990;112:5525. [Google Scholar]
- 19.a) Lin Q, Gourdon D, Sun C, Holten-Andersen N, Anderson TH, Waite JH, Israelachvili JN. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3782. doi: 10.1073/pnas.0607852104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nicklisch SCT, Das S, Martinez Rodriguez NR, Waite JH, Israelachvili JN. Biotechnol. Progr. 2013;29:1587. doi: 10.1002/btpr.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akdogan Y, Wei W, Huang K-Y, Kageyama Y, Danner EW, Miller DR, Martinez Rodriguez NR, Waite JH, Han S. Angew. Chem. 2014;126:11435. doi: 10.1002/anie.201406858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter K, Diaconu G, Rischka K, Amkreutz M, Müller FA, Hartwig A. Bioinspired, Biomimetic Nanobiomater. 2012;2:45. [Google Scholar]
- 22.Pillai KV, Renneckar S. Biomacromolecules. 2009;10:798. doi: 10.1021/bm801284y. [DOI] [PubMed] [Google Scholar]
- 23.Grauffel C, Yang BQ, He T, Roberts MF, Gershenson A, Reuter N. J. Am. Chem. Soc. 2013;135:5740. doi: 10.1021/ja312656v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Sever MJ, Weisser JT, Monahan J, Srinivasan S, Wilker JJ. Angew. Chem. 2004;116:454. doi: 10.1002/anie.200352759. [DOI] [PubMed] [Google Scholar]; b) Holten-Andersen N, Harrington MJ, Birkedal H, Lee BP, Messersmith PB, Lee KYC, Waite JH. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2651. doi: 10.1073/pnas.1015862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longo GS, de la Cruz MO, Szleifer I. Soft Matter. 2012;8:1344. [Google Scholar]
- 26.Helm CA, Knoll W, Israelachvili JN. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8169. doi: 10.1073/pnas.88.18.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington MJ, Masic A, Holten-Andersen N, Waite JH, Fratzl P. Science. 2010;328:216. doi: 10.1126/science.1181044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S, Donaldson SH, Kaufman Y, Israelachvili JN. RSC Adv. 2013;3:20405. [Google Scholar]
- 29.Israelachvili JN. Burlington, MA: Academic press; 2011. [Google Scholar]
- 30.a) Helm CA, Knoll W, Israelachvili JN. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8169. doi: 10.1073/pnas.88.18.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wong JY, Kuhl TL, Israelachvili JN, Mullah N, Zalipsky S. Science. 1997;275:820. doi: 10.1126/science.275.5301.820. [DOI] [PubMed] [Google Scholar]
- 31.a) Israelachvili J, Min Y, Akbulut M, Alig A, Carver G, Greene W, Kristiansen K, Meyer E, Pesika N, Rosenberg K, Zeng H. Rep. Prog. Phys. 2010;73 [Google Scholar]; b) Das S, Banquy X, Zappone B, Greene GW, Jay GD, Israelachvili JN. Biomacromolecules. 2013;14:1669. doi: 10.1021/bm400327a. [DOI] [PubMed] [Google Scholar]
- 32.Das S, Chary S, Yu J, Tamelier J, Turner KL, Israelachvili JN. Langmuir. 2013;29:15006. doi: 10.1021/la403420f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.