Abstract
We have identified and characterized a 17- to 18-kD Ser50-phosphorylated form of maize (Zea mays) CENTROMERIC HISTONE H3 (phCENH3-Ser50). Immunostaining in both mitosis and meiosis indicates that CENH3-Ser50 phosphorylation begins in prophase/diplotene, increases to a maximum at prometaphase-metaphase, and drops during anaphase. Dephosphorylation is precipitous (approximately sixfold) at the metaphase–anaphase transition, suggesting a role in the spindle checkpoint. Although phCENH3-Ser50 lies within a region that lacks homology to any other known histone, its closest counterpart is the phospho-Ser28 residue of histone H3 (phH3-Ser28). CENH3-Ser50 and H3-Ser28 are phosphorylated with nearly identical kinetics, but the former is restricted to centromeres and the latter to pericentromeres. Opposing centromeres separate in prometaphase, whereas the phH3-Ser28–marked pericentromeres remain attached and coalesce into a well-defined tether that binds the centromeres together. We propose that a centromere-initiated wave of histone phosphorylation is an early step in defining the two major structural domains required for chromosome segregation: centromere (alignment, motility) and pericentromere (cohesion).
INTRODUCTION
Throughout the eukaryotes, a complex set of interacting posttranslational modifications is known to regulate the interaction of histones with transcription factors and other chromatin-binding proteins. Histone modifications, such as acetylation, methylation, and phosphorylation, are so prevalent that Strahl and Allis (2000) envisioned a combinatorial histone code, which in principle could extend the effective coding capacity of the genome. A strength of the histone code hypothesis is that the major histones are well conserved across all eukaryotes, as are the locations of many known posttranslational modifications. The proposed histone code, however, is confounded by a divergent group of histone variants, many of which have important functions (Malik and Henikoff, 2003). Histone variants typically have strong homology to canonical histones within the C-terminal section that lies within nucleosomes but lack homology in the flexible N-terminal tails that extend outside nucleosomes. Because the N-terminal tails are where the most important posttranslational modifications occur, histone variants may have different but functionally similar codes (Strahl and Allis, 2000), erase previously set histone codes (Ahmad and Henikoff, 2002), or present altogether new codes (Smith, 2002).
On histone H3, four residues on the N-terminal tail are known to be phosphorylated: Ser10, Ser28, Thr3, and Thr11 (Hendzel et al., 1997; Gernand et al., 2003; Preuss et al., 2003; Polioudaki et al., 2004). At all four residues and in all species studied, the level of phosphorylation is low or undetectable at interphase, increases at prophase, and decreases during anaphase and telophase. Among these, phospho-Ser10 residue of histone H3 (phH3-Ser10) is the most extensively characterized (Prigent and Dimitrov, 2003). In mammalian cells, the phosphorylation of H3-Ser10 initiates in pericentric heterochromatin and spreads to chromosome arms during mitotic and meiotic metaphase (Hendzel et al., 1997). These staining patterns and earlier biochemical studies (Gurley et al., 1978; Allis and Gorovsky, 1981) suggest that H3-Ser10 phosphorylation may have a role in chromosome condensation (Wei et al., 1998). However, in the mitotic cells of most plants, phH3-Ser10 phosphorylation never extends beyond pericentric regions. It is only in meiosis I that the entire plant chromosomes stain uniformly with anti-phH3-Ser10 antisera (Houben et al., 1999; Kaszas and Cande, 2000; Manzanero et al., 2000). These staining patterns and the analyses of univalents suggest that phH3-Ser10 is involved in sister chromatid cohesion (Kaszas and Cande, 2000; Gernand et al., 2003). In animals, phH3-Ser10 recruits Aurora B kinase (Crosio et al., 2002), suggesting that the phosphoserine residues may also function as docking sites for proteins involved in cytokinesis (Andrews et al., 2003). Finally, a variety of data indicate that phosphorylation is involved in transcriptional activation (Clayton and Mahadevan, 2003). Given the varied and often conflicting data, the conserved functions of histone phosphorylation remain controversial. The primary function of histone phosphorylation may be to identify different domains of the chromosome and mark their progress through the cell cycle (Prigent and Dimitrov, 2003).
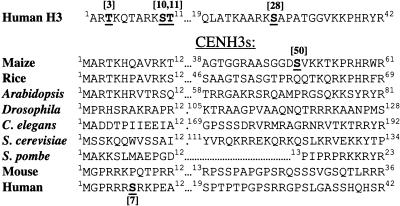
A key histone variant is CENTROMERIC HISTONE H3 (CENH3), the only universally conserved essential inner kinetochore protein (Choo, 2001). CENH3 is characterized (and usually identified) by remarkable sequence and length polymorphism in the N-terminal tail (Henikoff et al., 2000). In humans, there is weak homology between histone H3 and CENH3 over a region containing H3-Ser10. The analogous Ser in human CENH3 is found at position 7 and is phosphorylated in a temporal pattern that is similar to H3-Ser10 (Zeitlin et al., 2001b). Recent data suggest that Ser7-phosphorylated CENP-A (phCENP-A-Ser7) is required for proper chromosome alignment (Kunitoku et al., 2003). However, any model based on phCENP-A-Ser7 is difficult to reconcile with the sequences of other CENH3s. As shown in Figure 1, residues that could be interpreted as Ser7 counterparts are entirely absent in most CENH3s. Although there is no Ser7 counterpart in maize (Zea mays) CENH3, there are several serines in the core-proximal portion of the tail that could in principle correspond to histone H3 phospho-Ser28 (phH3-Ser28), namely CENH3 Ser25, Ser46, and Ser50 (Figure 1). One of these, Ser50, is within a peptide that was previously used to generate antisera for maize CENH3 (Zhong et al., 2002).
Figure 1.
N-Terminal Tails of Human Histone H3 and Nine CENH3s.
Only the CENH3s with documented centromeric localization are listed. The first 12 and (no more than) the last 24 amino acids of the N-terminal tail region are shown (the N terminus of S. pombe SpCENP-A contains only 23 amino acids). The residues known to be phosphorylated on human histone H3, human CENP-A, and maize CENH3 are indicated. The GenBank identification numbers for CENH3 homologs are as follows: maize, AAM74226; rice (Oryza sativa), AAR85315; Arabidopsis, NP_563627; Drosophila, AAF72652; C. elegans, NP_499128; budding yeast, NP_012875; S. pombe, BAA94760; mouse, NP_068686; and human, NP_001800.
Here, we show that maize CENH3-Ser50 is efficiently phosphorylated. The excellent cytology of maize allowed us to extend the description of phCENH3-Ser50 from mitosis to meiosis, to show that phosphorylation is a prophase-to-anaphase event, and that in meiosis II dephosphorylation is rapid and coincident with anaphase onset. The temporal staining pattern matches the pattern we observed for phH3-Ser28, except that the latter is strictly pericentromeric in its spatial distribution. These data suggest that the position of a phosphorylated residue, not necessarily its sequence context, may be a better predictor of which residues are phosphorylated on histone H3 variants. Further, we argue that the two histone modifications define their respective domains in the centromere-active period between prometaphase and anaphase.
RESULTS
Weak CENH3 Staining in Meiotic Metaphase Is Reversed by Phosphatase
Using a well-characterized anti-maize CENH3 polyclonal antibody (Zhong et al., 2002), we immunolocalized CENH3 in both meiotic and mitotic cells. As shown in Figure 2, CENH3 is present at all prophase stages of meiosis I: from premeiotic interphase (data not shown), to the synapsed chromosomes of pachytene (Figure 2A), and further in condensed chromosomes of diplotene and diakinesis (Figures 2D and 2G). Interestingly, CENH3 staining was weak or absent in prometaphase and metaphases I and II (Figures 2J and 2M), but from the onset of anaphase CENH3 staining was again visible (Figure 2P). Similar staining was observed in mitosis: anti-CENH3 staining was apparent in interphase cells, weak staining in prophase, and almost no staining in metaphase (Figure 2S).
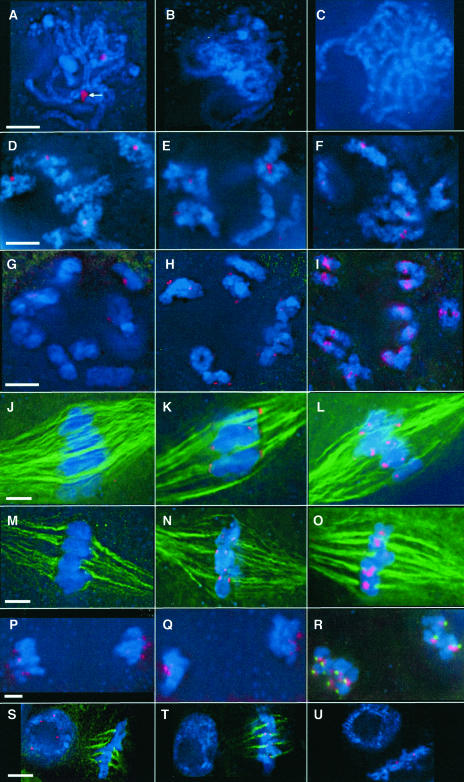
Figure 2.
CENH3, phCENH3-Ser50, and phH3-Ser28 Localization in Meiosis and Mitosis.
All images are partial projections from three-dimensional data sets. CENH3, phCENH3-Ser50, and phH3-Ser28 staining are shown in red, microtubules in green, and chromosomes in blue. Bars (5 μm) indicate the scale for all images in a given row.
(A) CENH3 at pachytene of meiosis I, when kinetochores are paired (arrow).
(B) phCENH3-Ser50 at pachytene.
(C) phH3-Ser28 staining at pachytene.
(D) CENH3 at diplotene.
(E) phCENH3-Ser50 at diplotene.
(F) phH3-Ser28 at diplotene.
(G) CENH3 at diakinesis.
(H) phCENH3-Ser50 at diakinesis.
(I) phH3-Ser28 at diakinesis.
(J) CENH3 at prometaphase I.
(K) phCENH3-Ser50 at prometaphase I.
(L) phH3-Ser28 at prometaphase I.
(M) CENH3 at prometaphase II.
(N) phCENH3-Ser50 at prometaphase II.
(O) phH3-Ser28 at prometaphase II.
(P) CENH3 at anaphase II.
(Q) phCENH3-Ser50 at anaphase II.
(R) phH3-Ser28 at anaphase II. CENPC is shown in green to illustrate that the phH3-Ser28 staining trails kinetochores.
(S) Mitotic CENH3 staining. As in meiosis, CENH3 antisera stain only those cells in interphase, early prophase, and late anaphase. The cell at left is in interphase, and the cell at right is in metaphase.
(T) Mitotic phCENH3-Ser50 staining. As in meiosis, phCENH3-Ser50 antisera stain only those cells in late prophase through early anaphase. The cell at left is in interphase, and the cell at right is in metaphase.
(U) Mitotic phH3-Ser28 staining. The cell at upper left is in interphase, and the cell at lower right is in metaphase.
Blocking of the epitope by posttranslational modification seemed the most attractive explanation for the absence of staining in prometaphase and metaphase. Because histone H3 is phosphorylated during mitosis and meiosis in several organisms, including Tetrahymena, Aspergillus, Caenorhabditis elegans, plants, and vertebrates (e.g., Wei et al., 1999; Hsu et al., 2000; Souza et al., 2000; Crosio et al., 2002; Gernand et al., 2003), we theorized that Ser50, located within the peptide used to prepare antibodies, is phosphorylated.
As an initial test of the idea, we applied calf intestinal phosphatase (CIP) to fixed meiocytes and stained them with anti-CENH3 antisera. CIP preferentially releases phosphate groups from phosphoserine/Thr residues. As shown in Figures 3B and 3C, the typically weak or absent CENH3 staining during meiotic metaphase was reversed by CIP treatment.
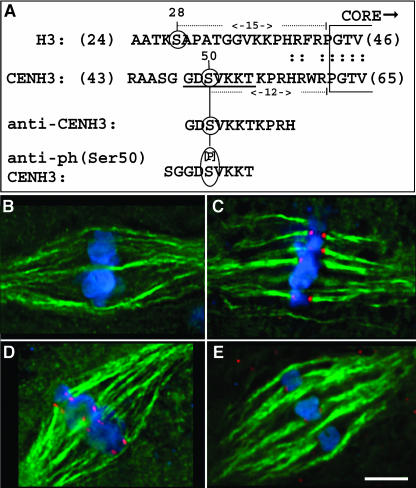
Figure 3.
Descriptions of Peptides Used to Develop Antisera, and the Reactivity of the Antisera with Metaphase Chromosomes after Phosphatase Treatment.
(A) The locations of peptides used to prepare antisera. The top line shows a portion of histone H3, ranging from amino acids 24 to 46. Ser28 is indicated. The second line shows the corresponding region of maize CENH3 with Ser50 indicated. The two peptides used for antibody production are shown below. The CENH3 antiserum (Zhong et al., 2002) recognizes both of the nonphosphorylated peptides, GDSVKKTKPRH and SGGDSVKKT, with similar specificity (Figure 4), indicating that the primary epitope for the CENH3 antibody (or antibodies) is the sequence GDSVKK.
(B) Image showing the characteristic absence of CENH3 staining at metaphase II. CENH3 staining is shown in red, microtubules in green, and chromosomes in blue.
(C) Bright CENH3 staining is revealed after alkaline phosphatase treatment.
(D) Image showing the characteristically strong phCENH3-Ser50 staining in untreated metaphase II cells. phCENH3-Ser50 is shown in red, microtubules in green, and chromosomes in blue.
(E) Alkaline phosphatase treatment reduces phCENH3-Ser50 staining by ∼90%. Bar = 5 μm.
Anti-phCENH3-Ser50 Antibodies Recognize CENH3 on Condensed Chromosomes
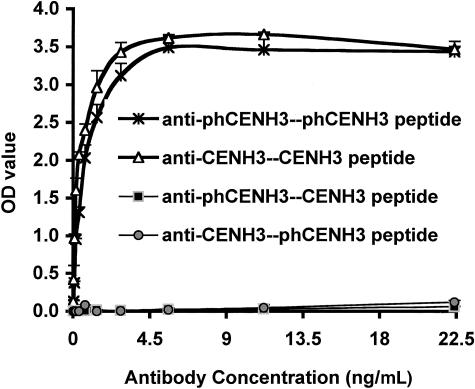
To better understand the phosphorylation of CENH3, we raised rabbit antisera against a synthetic peptide corresponding to amino acids 46 to 54 (SGGDS[p]VKKT) with a phosphorylated Ser at position 50 (Figure 3A). The antisera were analyzed by ELISA, and the resulting data are shown in Figure 4. We found that both our original anti-CENH3 antibodies (Zhong et al., 2002) and the anti-phCENH3-Ser50 antibodies bound specifically to the peptides they were raised against (the CENH3 and phCENH3-Ser50 peptides, respectively) but showed no binding when incubated with the opposite peptides (the phCENH3-Ser50 and CENH3 peptides, respectively).
Figure 4.
ELISA Analyses.
Anti-phCENH3-Ser50 antisera recognize the phCENH3-Ser50 peptide but not CENH3 peptide, whereas anti-CENH3 antisera recognize the CENH3 peptide but not the phCENH3-Ser50 peptide.
Next, we determined the distribution of phosphorylated CENH3 in both meiotic and mitotic cells. During meiosis, phCENH3-Ser50 staining was first detected in diplotene (Figure 2E) and persisted through diakinesis and metaphase (Figures 2H, 2K, and 2N). The staining then became weak in late anaphase (Figure 2Q) and was lost in telophase (data not shown). To confirm that the antisera recognize the phosphate moiety, alkaline phosphatase (CIP) was applied to meiocytes (Figures 3D and 3E). CIP treatment reduced the intensity of anti-phCENH3-Ser50 immunostaining by ∼90% ± 2% (n = 26 before and 8 cells after CIP treatment).
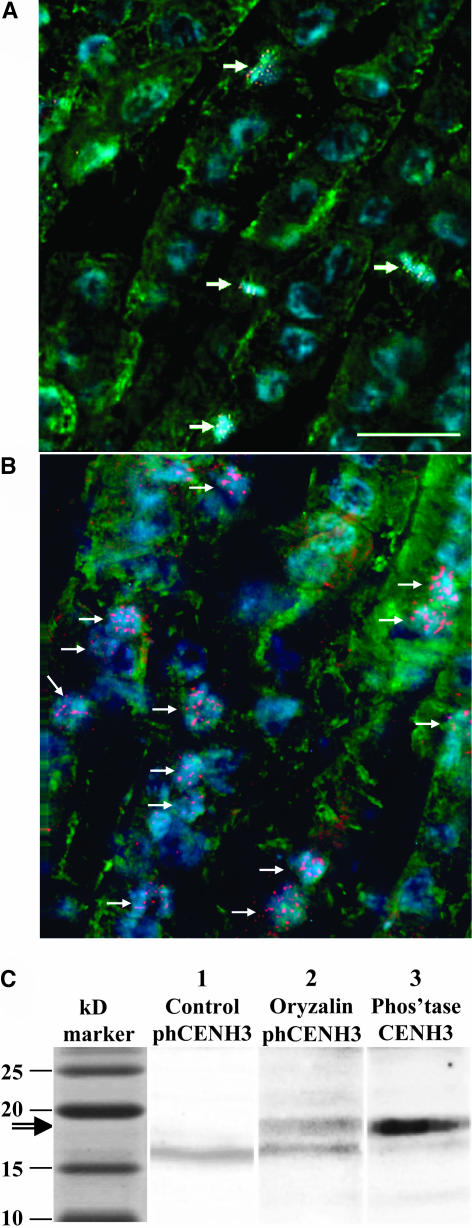
Similar results were obtained from mitotic root tip cells. Although no phCENH3-Ser50 localization was detected in interphase (Figure 2T, left), strong staining was detected in metaphase (Figure 2T, right). When a root tip was viewed in cross-section as shown in Figure 5A, staining was limited to the few cells in prophase, metaphase, and anaphase.
Figure 5.
Effect of the Microtubule-Destabilizing Drug Oryzalin on phCENH3-Ser50 Staining.
(A) An optical section from the meristematic zone of a (untreated) root tip after incubation with anti-phCENH3-Ser50 antisera (red), anti-α-tubulin antisera (green), and the DNA stain 4,6-diamidino-2-phenylindole (blue). Note that phCENH3-Ser50 staining is restricted to cells in prometaphase and metaphase (arrows).
(B) A section from a seedling grown at the same time as the seedling shown in (A), except in the presence 10 μM oryzalin for 8 h. The number of phCENH3-Ser50-positive cells increases dramatically.
(C) Protein blot of extracts derived from root tips. Lane 1 shows anti-phCENH3-Ser50 staining in untreated root tip extracts. Lane 2 shows the results after treating root tips with 10 μM oryzalin for 8 h. Oryzalin induces a phCENH3-Ser50–positive band at 17 to 18 kD. Lane 3 shows the lane 2 membrane after it was stripped, alkaline phosphatase treated, and reprobed with anti-CENH3 antisera.
phCENH3-Ser50 Antibodies Recognize a 17- to 18-kD Protein in Oryzalin-Treated Cells
Given the limited number of phCENH3-Ser50–positive cells even in the most actively dividing tissue (Figure 5A), we expected the quantity of phosphorylated protein to be very low in whole protein extracts. To increase the amount of phosphorylated protein for protein gel blot analysis, root tips were treated with various concentrations of the microtubule-depolymerizing drug oryzalin. A 4- to 8-h treatment with 10 μM oryzalin was most effective, increasing phCENH3-Ser50–positive cells by twofold to fourfold (cf. Figures 5A and 5B). Protein extracts from untreated and oryzalin-treated root tips were processed for protein gel blot analysis and compared side by side (Figure 5C). Although the predicted 17- to 18-kD band was sometimes observed in untreated roots, the intensity of the band was consistently higher in oryzalin-blocked root tissue. When blots were washed, incubated with CIP, and reprobed with anti-CENH3 antisera, a wider band in the same molecular mass range was observed. It is likely that the wide anti-CENH3 band is composed of two bands because both the phosphorylated and nonphosphorylated forms of the protein should be recognized after CIP treatment.
Other bands were also observed to a lesser and variable extent on protein gel blots. Of these, the brightest and most consistently observed band was at ∼16 kD (Figure 5C). We cannot rule out the possibility that it represents a second phosphorylated histone or chromatin protein. However, the fact that anti-phCENH3-Ser50 can be detected in cells only at kinetochores and that CIP removes ∼90% of this signal (Figure 3D) demonstrates that the antiserum binds most effectively to phCENH3-Ser50.
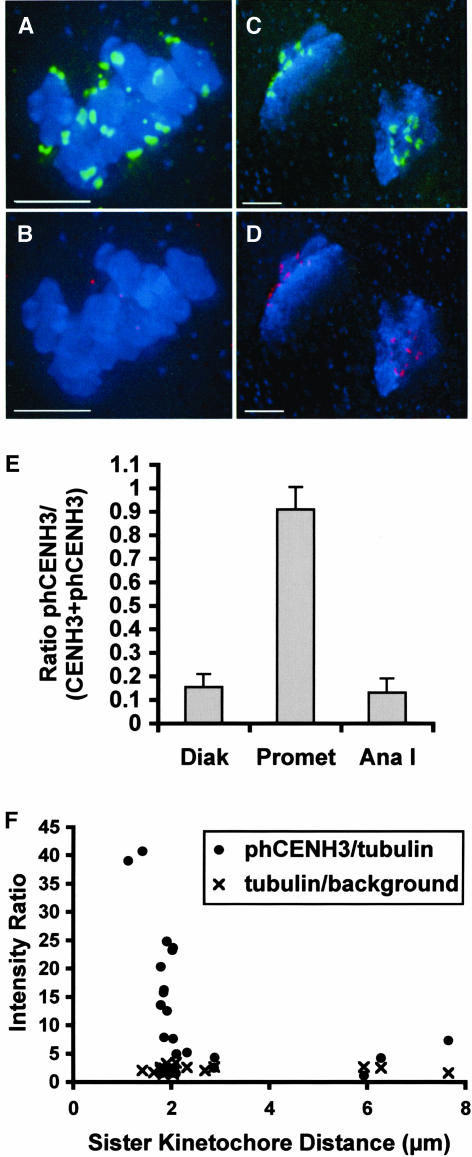
CENH3 Is Rapidly Dephosphorylated at Meiotic Anaphase in a Manner That Is Reminiscent of a Role in the Spindle Checkpoint
In an effort to quantify the onset of CENH3 phosphorylation and dephosphorylation, we performed simultaneous immunodetection of CENH3 and phCENH3-Ser50 at different meiotic stages. Representative microscope images and an analysis of the resulting data are shown in Figure 6. Consistent with the indirect immunolocalization data, only nonphosphorylated CENH3 was observed from the zygotene to pachytene stages of meiosis I, and only phCENH3-Ser50 was observed in metaphase. However, in diakinesis and anaphase I, both proteins were detectable. Over the diakinesis-anaphase I period, the percentage of kinetochore-localized phCENH3-Ser50 changed from 15% in diakinesis to nearly 100% in late metaphase, and back to ∼13% in anaphase I (Figures 6A to 6E).
Figure 6.
Quantitative Analysis of phCENH3-Ser50 Staining at Various Stages of Meiosis.
(A) to (D) Partial projections from three-dimensional data sets, in which phCENH3-Ser50 is shown in green, CENH3 in red, and chromosomes in blue. Bars = 5 μm.
(A) and (B) Colabeling of phCENH3-Ser50 and CENH3 at prometaphase I. Nearly all of the staining is from anti-phCENH3-Ser50 antisera (A); only random background staining is visible in the CENH3 channel (B).
(C) and (D) Colabeling of phCENH3-Ser50 and CENH3 at anaphase I. phCENH3-Ser50 staining is relatively weak (C), scaled up here to make it visible, whereas CENH3 staining is bright (D).
(E) The ratio of phCENH3-Ser50 to CENH3 plus phCENH3-Ser50 staining at three stages of meiosis I. Staining data were averaged from three to five cells at each stage, all from the same slide.
(F) CENH3 is rapidly dephosphorylated at anaphase onset. Data are expressed as tubulin S/N ratios (×) or as phCENH3-Ser50 S/N divided by tubulin S/N (closed circles) plotted against the distance between sister kinetochores. Previous analysis of meiosis II in the W23 inbred (Yu et al., 1999) established that anaphase II commences when the kinetochores are roughly 1.8 to 2.1 μm apart.
To further refine the stage at which CENH3 is dephosphorylated, we analyzed cells in meiosis II, in which the distance between sister kinetochores can be used to predict anaphase with accuracy (Yu et al., 1999). Control experiments established that the total tubulin staining intensity did not change appreciably from prometaphase II to anaphase II (Figure 6F). Based on this observation, phCENH3-Ser50 staining was normalized to tubulin staining and plotted against the distance between sister kinetochores. As shown in Figure 6F, we found that phCENH3-Ser50 staining fell dramatically as the distance between sister kinetochores reached 2.0 to 2.1 μm. As prior data had established that anaphase commences when the kinetochores are 1.8 to 2.1 μm apart (in this same inbred line; Yu et al., 1999), these data indicate that CENH3 is rapidly dephosphorylated at or near the metaphase–anaphase transition.
Antibodies to phH3-Ser28 and phCENH3-Ser50 Stain Chromosomes at the Same Time but in Juxtaposed Domains
Antisera to phH3-Ser28 stain pericentromeric regions of the chromosomes in Arabidopsis thaliana, wheat (Triticum aestivum), barley (Hordeum vulgare), and rye (Secale cereale; Gernand et al., 2003). We confirmed this basic staining pattern in maize and performed a detailed analysis of the early prophase I stages of meiosis. Like phCENH3-Ser50, phH3-Ser28 could not be detected in pachytene (Figure 2C) but was consistently observed in the regions surrounding kinetochores at late diplotene (Figure 2F). Staining increased in the pericentromeric domain at diakinesis (Figure 2I) and was visible trailing the kinetochores at prometaphase I (Figure 2L). By contrast, during prometaphase II, phH3-Ser28 appeared to lie between aligned chromatids at the metaphase plate (Figure 2O). After chromatid separation in anaphase, the staining began to lessen (Figure 2R) and was undetectable in telophase. Similarly, in mitosis, phH3-Ser28 was undetectable in interphase but pronounced at metaphase (Figure 2U).
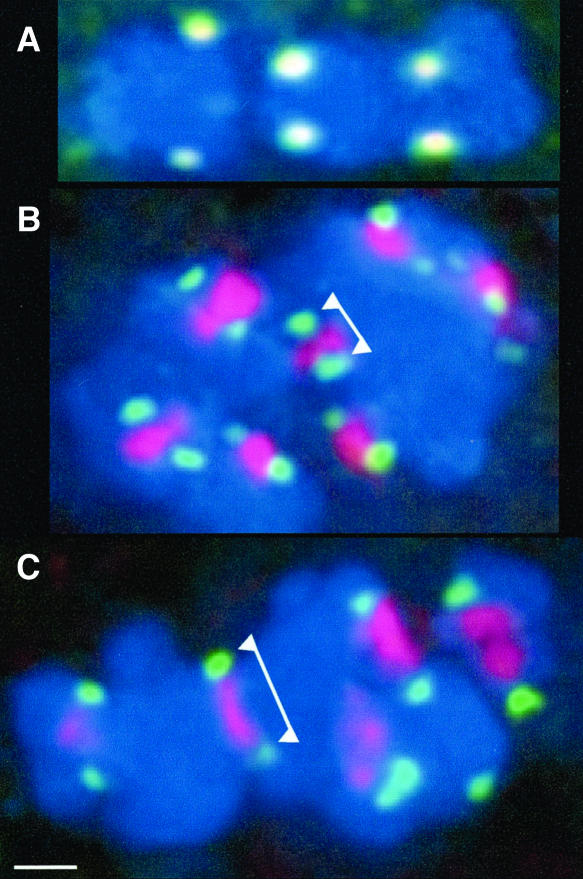
The intriguing between-chromatid localization of phH3-Ser28 at metaphase II (Figure 2O) prompted us to pursue the observation in the double-labeling experiments, as shown in Figure 7. Because Centromere Protein C (CENPC) and phCENH3-Ser50 (like CENH3; Zhong et al., 2002) label the same region of the kinetochore (Figure 7A), anti-CENPC antisera were used to mark the presence of the kinetochores in phH3-Ser28–stained cells. The data confirmed that kinetochores and phH3-Ser28 not only localize to different domains of the chromosome, but also in quite different positions relative to the spindle axis (Figures 7B and 7C). Opposing kinetochores separate early in prometaphase and are never attached by detectable CENPC-positive material. The regions that lie between homologous kinetochores are uniformly stained by anti-phH3-Ser28 antisera (Figure 7C). As kinetochores are drawn farther apart in late metaphase II (to ∼1.8 to 2.1 μm), the phH3-Ser28 domain appears to be under tension, strongly suggesting phH3-Ser28 localizes within the cohesive region that binds chromatids together.
Figure 7.
Differential Localization of phH3-Ser28 and CENPC.
(A) Double labeling of CENPC (green) and phCENH3-Ser50 (red). The two signals overlap to produce a yellow color.
(B) Double labeling of CENPC (green) and phH3-Ser28 (red) at prometaphase (average kinetochore to kinetochore distance in this cell was 1.40 μm, n = 7). Note that the phH3-Ser28 lies between sister chromatids and does not overlap with the CENPC staining (arrowheads).
(C) Double labeling of CENPC (green, arrowheads) and phH3-Ser28 (red) at late metaphase (average kinetochore to kinetochore distance in this cell was 1.93 μm, n = 7). In this case, the phH3-Ser28 domain is stretched between the kinetochores as if it were under tension. Bar = 1 μm.
DISCUSSION
CENH3 is a highly conserved histone H3–like protein that is found uniquely at centromeres. Data from several organisms indicate that CENH3 is at the core of centromere/kinetochore complex, where it helps to establish a specialized chromatin environment and recruits a subset of kinetochore proteins (Hooser et al., 2001; Ando et al., 2002). Although CENH3 is an important histone H3 variant, little is known about its phosphorylation. The only known CENH3 phosphorylation event occurs on Ser7 of human CENP-A (Zeitlin et al., 2001a). Immunolocalization using anti-phCENP-A-Ser7 antisera revealed a pattern similar to what is observed with anti-phH3-Ser10 (Wei et al., 1998), except that phCENP-A-Ser7 is quickly dephosphorylated at anaphase (Zeitlin et al., 2001b). A Ser7-to-Ala7 substitution (Kunitoku et al., 2003) caused chromosome missegregation as well as cytokinesis defects (Zeitlin et al., 2001a), suggesting a dual role in chromosome alignment and cytokinesis.
In this study, we raised antibodies that recognize Ser50-phosphorylated CENH3 and confirmed the specificity of the antibodies by ELISA, protein gel blot analysis, and alkaline phosphatase treatment (Figures 3D, 3E, 4, and 5C). Immunolocalization demonstrated that CENH3 phosphorylation is cell cycle dependent: phosphorylation initiates in diplotene (meiosis) or prophase (mitosis), increases as the chromosomes condense, and at metaphase phosphorylation is almost complete (Figures 2N and 6E). After anaphase onset, CENH3 is dephosphorylated rapidly but incompletely (Figures 2Q and 6E). Remarkably, the temporal staining of phCENH3-Ser50 is nearly identical to the staining of phH3-Ser28 (Figure 2); however, the staining patterns are juxtaposed, with phH3-Ser28 found only in well-demarcated cohesive regions between centromeres.
Histone H3 and CENH3 Are Phosphorylated with Similar Kinetics
In animals, each of four different sites on the N-terminal tail of histone H3—Thr3, Ser10, Thr11, and Ser28—are phosphorylated in a similar prophase-telophase–specific pattern (Hendzel et al., 1997; Gernand et al., 2003; Preuss et al., 2003; Polioudaki et al., 2004). However, none of these residues is found on any known centromeric histone H3 variant (Figure 1A). Indeed, a lack of homology in the N terminus is a key identifier of histone variants (Henikoff et al., 2000), making the question of how the variants might fit into the proposed histone code a matter of speculation. Our data are the first to address this issue in earnest because the previously characterized phCENP-A-Ser7 lies in a relatively well-conserved region of the protein (Zeitlin et al., 2001b; Figure 1A). In the region of the N-terminal tail downstream of the first 12 amino acids (where H3-Ser28 and CENH3-Ser50 lie), there is a near complete absence of homology among the known CENH3s. Here, the sequence data seem incompatible with the idea of a strict combinatorial phosphorylation code because the code would have to be effectively reinvented with the evolution of each new variant. Our data demonstrate that phosphorylation not only occurs in this region, but also that it occurs in a temporal manner that mirrors other histone H3 phosphorylation events. Because both the location and context of the phosphorylation event are novel (relative to H3 and other CENH3s), the data support the view that the presence or absence phosphorylation and/or the combined charges of the residues (Masayoshi and Smith, 2003) may be more important than their relative order or proximity to each other.
An issue of particular interest is whether a maize Aurora kinase is responsible for the phosphorylation of CENH3-Ser50. The Aurora kinases (A and B) belong to a conserved family of Ser/Thr kinases with critical roles in centrosome separation, spindle assembly, chromosome alignment, and cytokinesis (Andrews et al., 2003; Pascreau et al., 2003). Human Aurora B phosphorylates histone H3 at Ser10 and Ser28, and CENP-A at Ser7 (Giet and Glover, 2001; Zeitlin et al., 2001a; Crosio et al., 2002; Goto et al., 2002). More recent data suggest that CENP-A is initially phosphorylated by Aurora A and that the completed reaction is required for the recruitment (i.e., docking) of Aurora B at the inner centromere (Kunitoku et al., 2003).
Numerous Aurora kinase homologs exist in maize and other plants. Although no data are available on the localization or specificity of plant Aurora kinases, human and budding yeast (Saccharomyces cerevisiae) Aurora kinases have known consensus recognition sites. In humans, there is a strong requirement for Arg at the −2 position relative to the phosphorylated Ser and evidence for binding preferences as far away as +4, whereas in budding yeast the consensus site is {RK}X{ST}{LIV} (Cheeseman et al., 2002; Sugiyama et al., 2002). The evident differences between Aurora kinase recognition sites in these species (and the fact that deviations from a consensus generally reduce but do not abolish phosphorylation) make it difficult to predict whether CENH3-Ser50 is an Aurora kinase target. The involvement of Aurora kinase in maize CENH3 phosphorylation is nevertheless quite likely, given the strong conservation of function between the yeast and animal Aurora kinases (Pascreau et al., 2003). Should Aurora kinase prove to regulate maize CENH3-Ser50, the data will provide a strong endorsement of the idea that the presence and function of histone H3 phosphorylation events are broadly conserved (Strahl and Allis, 2000).
The Timing of CENH3 Phosphorylation and Dephosphorylation as It Relates to Anaphase Onset
One of the most important functions of kinetochores is to facilitate the activities of the spindle checkpoint, a surveillance mechanism that regulates the timing and coordination of anaphase. After every kinetochore has attached to the spindle properly, a signal cascade is initiated (Lew and Burke, 2003) that results in the breakdown of cohesin, the protein complex responsible for holding chromatids together (Haering and Nasmyth, 2003). In maize, the spindle checkpoint proteins MAD2 and the 3F/2 antigen mark the progression of metaphase (Yu et al., 1999). Both proteins bind to outer kinetochores in early and mid-metaphase, and are removed/degraded as opposing kinetochores are pulled to roughly 1.8 to 2.1 μm apart and anaphase begins (Yu et al., 1999). Similarly, we show here that phCENH3-Ser50 is rapidly dephosphorylated at the 2.0- to 2.1-μm mark that is indicative of anaphase (Figure 6). Similar timing of phosphorylation and dephosphorylation was reported for phCENP-A-Ser7 (Zeitlin et al., 2001b).
Interestingly, recent data have established that spindle assembly is regulated in part by Aurora kinases (Kallio et al., 2002). Aurora B is a kinetochore passenger protein that localizes to kinetochores only during chromosome alignment (in humans, by docking to phCENP-A-Ser7; Kunitoku et al., 2003) and recruits MAD2 as well as other spindle checkpoint proteins (Ditchfield et al., 2003; Petersen and Hagan, 2003). Further, Aurora kinase is required to correct improper kinetochore microtubule attachments during chromosome alignment (Tanaka et al., 2002; Hauf et al., 2003). Without phCENP-A-Ser7 and Aurora kinase, the accuracy of chromosome segregation drops measurably (Tanaka et al., 2002; Hauf et al., 2003; Kunitoku et al., 2003). Further studies will be required to determine if maize phCENH3-Ser50 recruits a similar kinase that functions in chromosome alignment and segregation.
A Centromere-Initiated Phosphohistone Code for the Centromere and Pericentromere
While the major roles of the centromere/kinetochore complex in chromosome segregation are well known (Nicklas, 1988; Choo, 2001), pericentromeres have remained relatively vague, often identified only by their characteristic deep staining pattern (e.g., Dawe, 2003). Only in Schizosaccharomyces pombe have pericentromeres been molecularly defined (Bannister et al., 2001), and in this species they are the primary cohesive domains that bind chromatids together during chromosome alignment (Appelgren et al., 2003). In plants, phH3-Ser10 (Kaszas and Cande, 2000; Shibata and Murata, 2004) and phH3-Ser28 (Gernand et al., 2003; Figure 7) are the only known molecular markers for the presumed pericentromeric domain. Our phH3-Ser28 localization data and similar phH3-Ser10 staining from Arabidopsis (Shibata and Murata, 2004) appear to confirm the interpretation from S. pombe that chromosomes are held together primarily by their pericentromeres (Appelgren et al., 2003).
It is now well established that cohesin preferentially associates with pericentromeres at metaphase (in mitosis and meiosis II; Haering and Nasmyth, 2003). Recent data demonstrate that the centromere/kinetochore complex has a strong influence on cohesin deposition in these pericentromeric regions (Meluh and Strunnikov, 2002). The most convincing results come from budding yeast, in which cohesin is poorly recruited unless centromeric DNA is present (Megee and Koshland, 1999), and humans, in which mistargeted CENP-A (but not CENP-C) causes the corecruitment of cohesin (Van Hooser et al., 2001). Budding yeast centromeres appear to enhance an existing pattern of cohesin distribution, such that the overall quantity of cohesin on either side of a (existing or newly introduced) centromere is elevated fivefold to sixfold (Weber et al., 2004). Similarly, a cohesin-enhancing role for human centromeres would help to explain how new centromeres, such as neocentromeres (Choo, 2001) or artificial chromosomes (e.g., Mee et al., 2003), are regularly segregated to progeny. Although the available data are compelling, the molecular basis for centromere-mediated cohesin accumulation remains unclear.
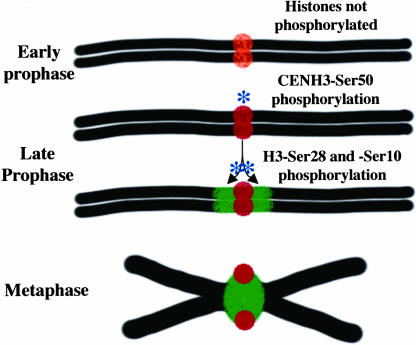
Based on the strict temporal coordination of CENH3 and H3 phosphorylation (Figure 2), we suggest that one signal for centromere-mediated cohesin accumulation is a histone kinase, which binds first at CENH3 and diffuses outward over histone H3 to define the boundaries of the pericentromeric domains. As shown in Figure 8, such CENH3-centered diffusion events would satisfy the need to place the pericentromeres, cell cycle after cell cycle, in discrete domains immediately adjacent to centromeres. Human Aurora B phosphorylates both CENP-A and histone H3, providing a precedent for the idea that a single kinase can regulate the centromere and pericentromere (Zeitlin et al., 2001a; Goto et al., 2002).
Figure 8.
A Kinase Diffusion Model for Pericentromere Determination.
At top is a prediplotene chromosome and its centromere with unphosphorylated CENH3 (orange). At diplotene, a histone kinase phosphorylates CENH3 first (red), then travels outward over the pericentromere and phosphorylates histone H3 (green) in a diffusion-limited manner. The phosphorylated CENH3 interacts with the spindle, whereas phosphorylated histone H3 marks the pericentromere and serves to enhance or stabilize cohesion deposition.
By analogy to the effects of centromeres on cohesin distribution in budding yeast (Weber et al., 2004), we further suggest that histone H3 phosphorylation stabilizes or enhances cohesin distribution within pericentromeres. Previous data support this view. phH3-Ser10 and phH3-Ser28 are entirely absent on maize and rye chromosomes that lack a sister chromatid at meiosis II, suggesting a causal relationship between histone phosphorylation and cohesin deposition (or vice versa; Kaszas and Cande, 2000; Gernand et al., 2003). In addition, H3-Ser10 phosphorylation is one of the few (if only) phosphorylation events that accompanies the dissolution of sister chromatid linkages in Xenopus extracts (Losada et al., 2002).
Whether histone phosphorylation is a cause or consequence (or both) of chromosome alignment and segregation, our data provide compelling correlations between the two events. Foremost among these is the striking temporal coordination between and CENH3-Ser50 and H3-Ser28 phosphorylation (Figure 2) and the fact that phH3-Ser28 defines the cohesive pericentromeric domain with apparent precision (Figure 7; Gernand et al., 2003). Our description of phCENH3-Ser50 also closely parallels the early descriptions of phCENP-A-Ser7 (Zeitlin et al., 2001b), suggesting that CENH3 phosphorylation may have a similar role in regulating anaphase onset (Kunitoku et al., 2003). The availability of well-characterized plant phosphohistone antibodies also has practical implications. The observation that anti-phCENH3-Ser50 antisera identify only segregating chromosomes (Figure 2) opens the door to identifying the DNA in biologically active centromeres (by chromatin immunoprecipitation), an issue that has yet to be addressed in any organism. In addition, our data and the prior data from Gernand et al. (2003) establish anti-phH3-Ser28 antibodies as an excellent reagent for identifying the DNA sequences of plant pericentromeres.
METHODS
Antisera
A peptide was designed to correspond to residues 46 to 54 of maize (Zea mays) CENH3 (Zhong et al., 2002), with a single phosphorylated Ser at position 50 (SGGDS[p]VKKT). Anti-phCENH3-Ser50 antibodies were raised against the peptide conjugated to keyhole limpet hemocyanin. The preparation and affinity purification of antisera were performed by BioSource International (Camarillo, CA). Antibodies to phH3-Ser28 were obtained from Upstate (07-145; Lake Placid, NY).
ELISA Assays
ELISA was performed according to a protocol provided by Biosource International. Plates were coated with 50 mL of phCENH3-Ser50 peptide (10 mg/mL) or CENH3 peptide at 37°C overnight. After washing three times with double-distilled water, the plates were blocked with 50 mL of 0.3% BSA buffer (0.3% BSA, 0.3% Carnation nonfat milk, 0.0002% NaN3, 0.03% Tween 20 in filtered Tris-buffered saline) at room temperature for 1 h. Samples were washed three times in double-distilled water and incubated for 2 h at root temperature in 0.8% BSA buffer. Secondary antibodies were then applied at 1:2500 dilutions for 2 h at room temperature (diluted in 0.83% BSA buffer). After a washing step (three times with double-distilled water), 75 mL of p-nitrophenyl phosphate substrate solution (6 mM p-nitrophenyl phosphate, 0.05M Na2CO3, 0.05 mM MgCl2) was added to each well for 2 h. The data were analyzed using a plate reader set at 405 nm.
Indirect Immunolocalization in Meiotic Cells
Meiocytes were prepared from the W23 inbred line as described by Yu et al. (1999). Fixed samples were incubated with rabbit anti-CENH3 antibodies (1:25), rabbit anti-phCENH3-Ser50 antibodies (1:25), rabbit anti-phH3-Ser28 antibodies (1:25), chicken anti-CENPC antibodies (1:25; Dawe et al., 1999; Zhong et al., 2002), and/or mouse anti-tubulin antibodies (1:500; Asai et al., 1982) as appropriate. Rhodamine-conjugated goat anti-rabbit antibodies (1:25; Jackson Immunoresearch, West Grove, PA) and/or fluorescein isothiocyanate–conjugated goat anti-mouse or donkey anti-chicken (1:25; 14274020; Boehringer Mannheim, Mannheim, Germany) secondary antibodies were then applied for 2 h at room temperature. Procedures for the necessary washing steps, mounting, and 4,6-diamidino-2-phenylindole staining have been described previously (Yu et al., 1997). For alkaline phosphatase treatment, meiocytes were fixed, adhered to cover slips, and incubated with 10 units of CIP (p4252; Sigma-Aldrich, St. Louis, MO) diluted in alkaline phosphatase buffer (100 mM NaCl, 5 mM MgCl2, 100 mM Tris, pH 9.5) at 37°C overnight. Cells were then washed three times in 1× PBS for 5 min each and processed for immunofluorescence.
Direct Immunolocalization in Meiotic Cells
Anti-phCENH3-Ser50 antibodies were directly labeled with Alexa Fluor 488 (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Labeling efficiency was 3.2 mols Alexa Fluor 488/mol phCENH3-Ser50 antibody. For the double staining shown in Figure 6, meiocytes were first incubated with anti-CENH3 antibodies (overnight), then goat anti-rabbit secondary antibodies (3 h), and finally 3.75 μg of direct-labeled phCENH3-Ser50 antisera (overnight). The protocol was finished in the same manner as a standard indirect immunofluorescence experiment.
Indirect Immunolocalization in Root Tips
Seeds from the maize inbred W23 were germinated in a moist incubator at 26°C for 3 d. In some experiments (Figure 5), 3-d-old seedlings were treated for 6 to 8 h with 10 mM oryzalin (Chem Service, West Chester, PA) to depolymerize microtubules. Root tips ∼3 mm in length were fixed, sectioned, and mounted as described previously (Yu et al., 1999). Slides were washed three times in 1× PBS and processed for immunofluorescence as above, except secondary antibodies were applied for 3 h at room temperature.
Image Analysis
Data were acquired and analyzed using a DeltaVision 3D light microscopy workstation and associated software (Applied Precision, Issaquah, WA). Staining intensity measurements were averaged from 4 × 4-pixel boxes centered over 10 different kinetochores or spindle fibers (next to kinetochores), as appropriate. Intensity values were divided by background staining (calculated in the same way, from the cytoplasm) to obtain signal to noise (S/N) ratios. For the data in Figure 6E, the S/N ratios from phCENH3-Ser50 staining were divided by the sum of the S/N ratios from phCENH3-Ser50 and CENH3 staining. For Figures 3E and 6F, phCENH3-Ser50 S/N ratios were divided by the tubulin S/N ratios, in effect normalizing phCENH3-Ser50 staining to tubulin staining.
CENH3 Extraction and Blotting
Root tips ∼3 mm in length were ground in liquid nitrogen and protein extracted as described previously (Pilch et al., 2004). Samples were electrophoresed by SDS-PAGE and blotted to nitrocellulose. Blots were blocked for 1.5 h with 5% Carnation nonfat milk in 0.1% TBST (20 mM Tris, 137 mM NaCl, 0.05% Tween 20, pH 7.6), and incubated for 4 h with phCENH3-Ser50 or CENH3 antibodies at dilutions of 1:2000 (0.56 mg/mL) or 1:5000 (0.4 mg/mL), respectively. After washing three times with TBST, the blots were incubated with horseradish peroxidase–conjugated goat anti-rabbit secondary antibodies (1:3000 dilution; Amersham, Piscataway, NJ) and detected using the ECL protein gel blotting kit (Amersham).
Acknowledgments
This work was supported by the National Science Foundation (Grant 9975827 to R.K.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: R. Kelly Dawe (kelly@plantbio.uga.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028522.
References
- Ahmad, K., and Henikoff, S. (2002). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99, 6477–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis, C.D., and Gorovsky, M.A. (1981). Histone phosphorylation in macro- and micronuclei of Tetrahymena thermophila. Biochemistry 20, 3828–3833. [DOI] [PubMed] [Google Scholar]
- Ando, S., Yang, H., Nozaki, N., Okazaki, T., and Yoda, K. (2002). CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22, 2229–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, P.D., Knatko, E., Moore, W.J., and Swedlow, J.R. (2003). Mitotic mechanics: The auroras come into view. Curr. Opin. Cell Biol. 15, 672–683. [DOI] [PubMed] [Google Scholar]
- Appelgren, H., Kniola, B., and Ekwall, K. (2003). Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116, 4035–4042. [DOI] [PubMed] [Google Scholar]
- Asai, D.J., Brokaw, C.J., Thompson, W.C., and Wilson, L. (1982). Two different monoclonal antibodies to tubulin inhibit the bending of reactivated sea urchin spermatozoa. Cell Motil. 2, 599–614. [DOI] [PubMed] [Google Scholar]
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates III, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. (2001). Domain organization at the centromere and neocentromere. Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]
- Clayton, A.L., and Mahadevan, L.C. (2003). MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 546, 51–58. [DOI] [PubMed] [Google Scholar]
- Crosio, C., Fimia, G.M., Loury, R., Kimura, M., Okano, Y., Zhou, H., Sen, S., Allis, C.D., and Sassone-Corsi, P. (2002). Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K. (2003). RNA interference, transposons, and the centromere. Plant Cell 15, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L., Yu, H.-G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield, C., Johnson, V.L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N., and Taylor, S.S. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernand, D., Demidov, D., and Houben, A. (2003). The temporal and spatial pattern of histone H3 phosphorylation at serine 28 and serine 10 is similar in plants but differs between mono- and polycentric chromosomes. Cytogenet. Genome Res. 101, 172–176. [DOI] [PubMed] [Google Scholar]
- Giet, R., and Glover, D.M. (2001). Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, H., Yasui, Y., Nigg, E.A., and Inagaki, M. (2002). Aurora-B phosphorylates histone H3 at serine 28 with regard to the mitotic chromosome condensation. Genes Cells 7, 11–17. [DOI] [PubMed] [Google Scholar]
- Gurley, L.R., D'Anna, J.A., Barham, S.S., Deaven, L.L., and Tobey, R.A. (1978). Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur. J. Biochem. 84, 1–15. [DOI] [PubMed] [Google Scholar]
- Haering, C.H., and Nasmyth, K. (2003). Building and breaking bridges between sister chromatids. Bioessays 25, 1178–1191. [DOI] [PubMed] [Google Scholar]
- Hauf, S., Cole, R.W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C.L., and Peters, J.M. (2003). The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel, M.J., Wei, Y., Mancini, M.A., Van Hooser, A., Ranalli, T., Brinkley, B.R., Bazett-Jones, D.P., and Allis, C.D. (1997). Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J.S., and Steensel, B.V. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooser, A.V., Ouspenski, I., Gregson, H., Starr, D., Yen, T., Goldberg, M., Yokomori, K., Earnshaw, W., Sullivan, K., and Brinkley, B. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- Houben, A., Wako, T., Furashima-Shimogawara, R., Presting, G., Kunzel, G., Schubert, I., and Fukui, K. (1999). The cell cycle dependent phosphorylation of histone H3 is correlated with the condensation of plant mitotic chromosomes. Plant J. 18, 675–679. [DOI] [PubMed] [Google Scholar]
- Hsu, J.Y., et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Kallio, M.J., McCleland, M.L., Stukenberg, P.T., and Gorbsky, G.J. (2002). Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12, 900–905. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and Cande, W.Z. (2000). Phosphorylation of histone H3 is correlated with changes in the maintenance of sister chromatid cohesion during meiosis in maize, rather than the condensation of the chromatin. J. Cell Sci. 113, 3217–3226. [DOI] [PubMed] [Google Scholar]
- Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Honda, A., Kobayashi, O., Hatakeyama, K., Ushio, Y., Saya, H., and Hirota, T. (2003). CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell 5, 853–864. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and Burke, D.J. (2003). The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37, 251–282. [DOI] [PubMed] [Google Scholar]
- Losada, A., Hirano, M., and Hirano, T. (2002). Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 16, 3004–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, M.S., and Henikoff, S. (2003). Phylogenomics of the nucleosome. Nat. Struct. Biol. 10, 882–891. [DOI] [PubMed] [Google Scholar]
- Manzanero, S., Arana, P., Puertas, M.J., and Houben, A. (2000). The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109, 308–317. [DOI] [PubMed] [Google Scholar]
- Masayoshi, L., and Smith, M.M. (2003). Functional consequences of histone modifications. Curr. Opin. Genet. Dev. 13, 154–160. [DOI] [PubMed] [Google Scholar]
- Mee, P.J., Shen, M.H., Smith, A.G., and Brown, W.R. (2003). An unpaired mouse centromere passes consistently through male meiosis and does not significantly compromise spermatogenesis. Chromosoma 112, 183–189. [DOI] [PubMed] [Google Scholar]
- Megee, P.C., and Koshland, D. (1999). A functional assay for centromere-associated sister chromatid cohesion. Science 285, 254–257. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., and Strunnikov, A.V. (2002). Beyond the ABCs of CKC and SCC. Do centromeres orchestrate sister chromatid cohesion or vice versa? Eur. J. Biochem. 269, 2300–2314. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B. (1988). The forces that move chromosomes in mitosis. Annu. Rev. Biophys. Biophys. Chem. 17, 431–449. [DOI] [PubMed] [Google Scholar]
- Pascreau, G., Arlot-Bonnemains, Y., and Prigent, C. (2003). Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog. Cell Cycle Res. 5, 369–374. [PubMed] [Google Scholar]
- Petersen, J., and Hagan, I.M. (2003). S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr. Biol. 13, 590–597. [DOI] [PubMed] [Google Scholar]
- Pilch, D.R., Redon, C., Sedelnikova, O.A., and Bonner, W.M. (2004). Two-dimensional gel analysis of histones and other H2AX-related methods. Methods Enzymol. 375, 76–88. [DOI] [PubMed] [Google Scholar]
- Polioudaki, H., Markaki, Y., Kourmouli, N., Dialynas, G., Theodoropoulos, P.A., Singh, P.B., and Georgatos, S.D. (2004). Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 560, 39–44. [DOI] [PubMed] [Google Scholar]
- Preuss, U., Landsberg, G., and Scheidtmann, K.H. (2003). Novel mitosis-specific phosphorylation of histone H3 at Thr11 mediated by Dlk/ZIP kinase. Nucleic Acids Res. 31, 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent, C., and Dimitrov, S. (2003). Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116, 3677–3685. [DOI] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117, 2963–2970. [DOI] [PubMed] [Google Scholar]
- Smith, M. (2002). Centromeres and variant histones: What, where, when and why? Curr. Opin. Cell Biol. 14, 279–285. [DOI] [PubMed] [Google Scholar]
- Souza, C.P.D., Osmani, A.H., Wu, L.P., Spotts, J.L., and Osmani, S.A. (2000). Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102, 293–302. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Sugiyama, K., Sugiura, K., Hara, T., Sugimoto, K., Shima, H., Honda, K., Furukawa, K., Yamashita, S., and Urano, T. (2002). Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. (2002). Evidence that the IpI1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome biorientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A.A., Ouspenski, I.I., Gregson, H.C., Starr, D.A., Yen, T.J., Goldberg, M.L., Yokomori, K., Earnshaw, W.C., Sullivan, K.F., and Brinkley, B.R. (2001). Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542. [DOI] [PubMed] [Google Scholar]
- Weber, S.A., Gerton, J.L., Polancic, J.E., DeRisi, J.L., Koshland, D., and Megee, P.C. (2004). The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2, 1340–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Mizzen, C.A., Cook, R.G., Gorovsky, M.A., and Allis, C.D. (1998). Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. USA 95, 7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Yu, L., Bowen, J., Gorovsky, M.A., and Allis, C.D. (1999). Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97, 99–109. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., Chan, A., Sweeney, M., and Dawe, R.K. (1997). Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Muszynski, M.G., and Dawe, R.K. (1999). The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 145, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin, S.G., Barber, C.M., Allis, C.D., and Sullivan, K.F. (2001. b). Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J. Cell Sci. 114, 653–661. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S.G., Shelby, R.D., and Sullivan, K.F. (2001. a). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]