Abstract
Influenza A virus (IAV) causes annual epidemics and occasional pandemics, and is one of the best-characterized human RNA viral pathogens1. However, a physiologically relevant role for the RNA interference (RNAi) suppressor activity of the IAV non-structural protein 1 (NS1), reported over a decade ago2, remains unknown3. Plant and insect viruses have evolved diverse virulence proteins to suppress RNAi as their hosts produce virus-derived small interfering RNAs (siRNAs) that direct specific antiviral defence4–7 by an RNAi mechanism dependent on the slicing activity of Argonaute proteins (AGOs)8,9. Recent studies have documented induction and suppression of antiviral RNAi in mouse embryonic stem cells and suckling mice10,11. However, it is still under debate whether infection by IAV or any other RNA virus that infects humans induces and/or suppresses antiviral RNAi in mature mammalian somatic cells12–21. Here, we demonstrate that mature human somatic cells produce abundant virus-derived siRNAs co-immunoprecipitated with AGOs in response to IAV infection. We show that the biogenesis of viral siRNAs from IAV double-stranded RNA (dsRNA) precursors in infected cells is mediated by wild-type human Dicer and potently suppressed by both NS1 of IAV as well as virion protein 35 (VP35) of Ebola and Marburg filoviruses. We further demonstrate that the slicing catalytic activity of AGO2 inhibits IAV and other RNA viruses in mature mammalian cells, in an interferon-independent fashion. Altogether, our work shows that IAV infection induces and suppresses antiviral RNAi in differentiated mammalian somatic cells.
Virus-derived siRNAs (vsiRNAs) serve as both the molecular marker for the induction of antiviral RNAi and the specificity determinants of the defence mechanism5–7. As Dicer products, vsiRNAs are 21–24 nucleotides (nt) long and form short, perfect, base-paired RNA duplexes with 2 nt 3′ overhangs5–7. However, sequencing of total small RNAs from a range of mature mammalian cells infected with diverse human RNA viruses over the past decade has failed to detect the production of a significant amount of vsiRNAs12–16,18. It is known that AGOs selectively bind to Dicer products, and several virus-encoded suppressors of RNAi (VSRs) act to inhibit the biogenesis or Argonaute loading of vsiRNAs5–7. Thus, mammalian vsiRNAs may become readily detectable by deep sequencing when small RNAs are enriched first by Argonaute co-immunoprecipitation (co-IP) from mammalian cells infected with a mutant RNA virus deleted of its VSR gene. Here, we use this strategy to search for vsiRNAs from IAV-infected human somatic cells. NS1 of IAV suppresses both antiviral RNAi in Drosophila cells and engineered RNAi in plant and mammalian cells2,22–25, and shares strong structural similarity3 in dsRNA binding with the nodaviral VSR B2 proteins, known to inhibit the biogenesis of vsiRNAs in animal cells10,11,26. IAV contains a negative-strand RNA genome divided into eight segments, and NS1, encoded by the smallest genome segment, is multifunctional and essential for in vivo infection and virulence1,3. Accordingly, we sequenced small RNAs co-immunoprecipitated by an antibody specific to the four AGOs from human 293T cells infected with PR8/delNS1 (Fig. 1a, left), an NS1-deletion mutant of IAV strain A/Puerto Rico/8/1934(H1N1) characterized previously27.
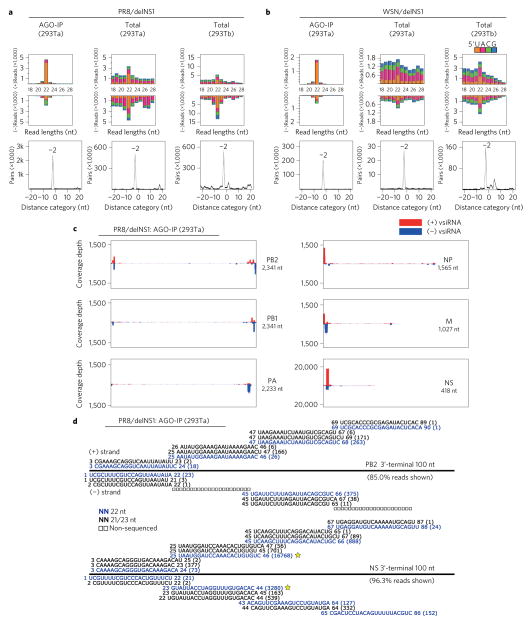
Figure 1. Production of viral siRNAs in mature human somatic cells.
a,b, Size distribution and abundance (per million total mature miRNAs) of total virus-derived small RNAs (vsRNAs) sequenced either directly from two human 293T cell lines (293Ta and 293Tb) 24 h after infection with delNS1 mutants of PR8 and WSN strains (total), or after AGO co-IP from the infected 293Ta cells (AGO-IP). Strong enrichment of total and AGO-bound 22 nt RNAs of both PR8/delNS1 and WSN/delNS1 for pairs (−2 peak) of canonical vsiRNAs with 2 nt 3′ overhangs was detected by computing total pairs of 22 nt vsRNAs with different lengths of base-pairing10. The 5′-terminal nt of vsRNAs is indicated by colour. c, Relative abundance of 21–23 nt vsiRNA hotspots mapped to PR8/delNS1 genomic RNAs, presented from the 3′ end (left) to the 5′ end (right). The genome segments haemagglutinin (HA) and NA, which are targeted by an extremely low density of vsiRNAs, are shown in Supplementary Fig. 1. d, Read sequences along the 3′-terminal 100 nt of PR8/delNS1 mutant genome segments PB2 and NS. Read counts (in brackets), read length, non-sequenced reads, genomic position and percentage of total reads mapped to the region are indicated. The RNAs complementary to the positive (+) or negative (−)-strand vsiRNAs marked by a star were used subsequently as the probes for northern detection of the influenza vsiRNAs.
We found that 93.6% of the 41,324 virus reads, cloned by a protocol requiring the presence of monophosphates at the 5′ termini, were in the 21 to 23 nt size range of Dicer products, with 22 nt as the most dominant size for both positive and negative strands (Fig. 1a, left; Supplementary Table 1). The 22 nt RNAs of IAV exhibited a strong preference for uracil as the 5′-terminal nucleotide (1U) (71.5%, or 62.9% for 21–23 nt vsiRNAs, Supplementary Table 1) and were highly enriched for 20 nt perfect base-paired duplexes with 2 nt 3′ overhangs (Fig. 1a, left). The influenza vsiRNAs were abundant, representing 0.34% of the total sequenced reads and equal to 0.81% of the total mature miRNA content in the library (Supplementary Table 1). Moreover, 91.8% of the virus reads were derived from the terminal 100 nt regions of the eight virion RNA segments (Fig. 1c, Supplementary Table 1 and Supplementary Fig. 1), and these terminal virus reads formed successive (or phased) complementary pairs of vsiRNAs (Fig. 1d and Supplementary Fig. 2). These results suggest that the terminal viral dsRNA replicative intermediates served as the predominant precursors of the influenza vsiRNAs, similar to those terminal vsiRNAs produced by mouse embryonic stem cells to target encephalomyocarditis virus (EMCV)11. Approximately 65% of the vsiRNA reads were mapped to the shortest genome segment NS, which is 418 nt in length after the deletion of the NS1 gene. In particular, we measured 20,048 reads for the second pair of 22 nt vsiRNAs from the 3′ terminus, a number similar to those detected for microRNA 221-3p (20,206), the 22nd most abundant miRNA in the AGO co-IP library. Nevertheless, the total reads mapped to the remaining seven genome segments also exhibited the canonical properties of vsiRNAs (Supplementary Table 2 and Supplementary Figs 3 and 4).
We also sequenced an independent library of small RNAs co-immunoprecipitated with AGOs from 293T cells infected with WSN/delNS1, a NS1-deletion mutant of the human IAV A/WSN/1933(H1N1). Our analysis revealed a highly similar population of vsiRNAs co-immunoprecipitated with AGOs from WSN/delNS1-infected human cells (Fig. 1b, left; Supplementary Figs 1b,2b,3,4b and Supplementary Tables 1 and 2). Therefore, the influenza vsiRNAs from two independent libraries closely resemble previously described vsiRNAs generated in mice10,11 and differ drastically from the virus-derived small RNAs detected previously during wild-type IAV infection, which are almost exclusively from negative strands with a random size distribution and 5′ co-terminal with virion RNAs14,15.
The 22 nt vsiRNAs were more abundant than other sizes, and were highly enriched for canonical pairs of vsiRNAs in the libraries of total small RNAs constructed without AGO co-immunoprecipitated from two different sources of 293T cells infected with either PR8/delNS1 or WSN/delNS1 (Fig. 1a,b and Supplementary Figs 3,4c–f,4h). However, there were also abundant positive- and negative-strand virus reads outside the size range of Dicer products and the peak at 22 nt was weaker in these libraries than in those sequenced after AGO co-IP. These findings together indicate that combining VSR deletion and AGO co-IP improves the detection of human vsiRNAs.
The influenza vsiRNAs of either polarity were readily detectable as discrete bands in the 21 to 22 nt size range by northern blotting from 293T cells infected with PR8/delNS1 (Fig. 2a), providing an independent verification of the production and size of the sequenced influenza vsiRNAs. We found that the influenza vsiRNAs became undetectable in a characterized human 293T cell line28 deficient in human Dicer (hDicer-knockout (KO), Figs 2a,c). Importantly, production of the influenza vsiRNAs was rescued by ectopic expression of wild-type hDicer (Figs 2b,c and Supplementary Fig. 5). Together, our findings show that wild-type hDicer mediates the production of the influenza vsiRNAs, revealing a new activity of hDicer to process virus-specific dsRNA precursors into siRNAs in mature somatic cells, in addition to its role in cellular miRNA biogenesis29.
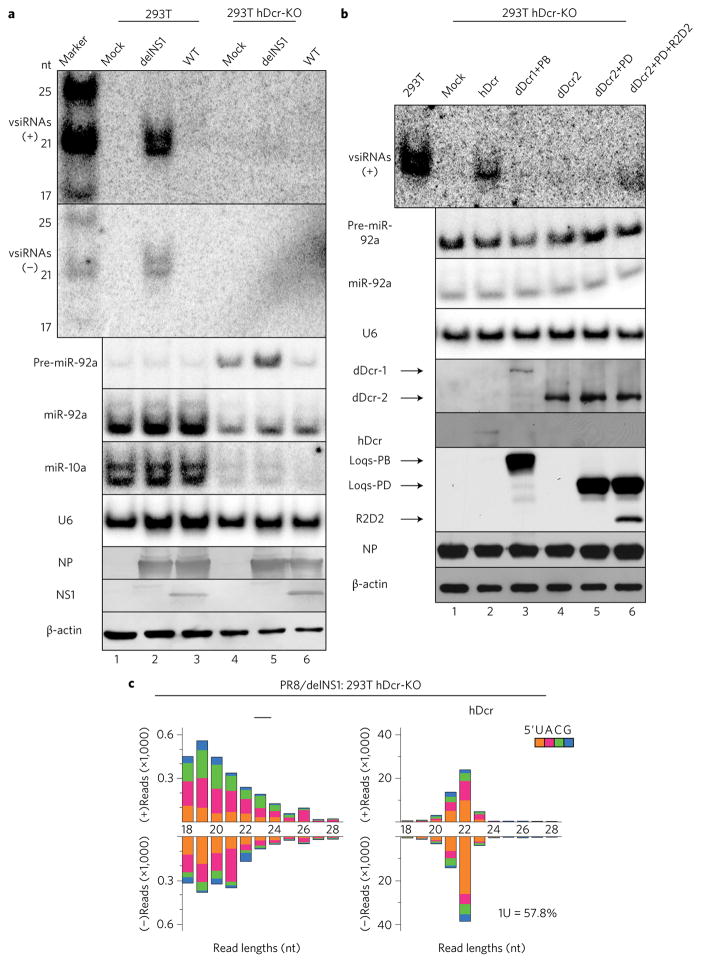
Figure 2. Wild-type (WT) hDicer is necessary and sufficient for the biogenesis of human vsiRNAs in differentiated somatic cells.
a, Northern detection of the influenza vsiRNAs in WT and hDicer-knockout (hDCr-KO) 293T cells 24 h after infection with the WT or mutant (delNS1) viruses of the PR8 strain. b, Production of influenza vsiRNAs in PR8/delNS1-infected hDcr-KO 293T cells ectopically expressing hDcr, dDicer-2 (dDcr2), dDcr1+Loquacious (Loqs) isoform-PB (PB), dDcr2+Loqs-PD (PD) or dDcr2+PD+R2D2, as indicated. The same sets of RNA and protein samples were used for northern or western blot detection of the positive (+) or negative (−)-strand influenza vsiRNAs (RNA sequences marked by a star in Fig. 1d), precursor microRNA-92a (pre-miR-92a), miR-92a, miR-10a, U6 RNA, viral NP and NS1 proteins, hDcr, the tagged Drosophila proteins or β-actin. Each experiment was repeated twice, with reproducible results. c, Properties of the influenza vsiRNAs (per million total reads) sequenced from PR8/delNS1-infected hDcr-KO 293T cells ectopically expressing hDcr (right). The 5′-terminal nt of the vsiRNAs is indicated by colour and the percentage of 1U vsiRNAs is shown. The sRNAs mapped to the viral genome in PR8/delNS1-infected hDcr-KO 293T cells were low in abundance and exhibited a random size distribution (left).
hDicer shares the same domain architecture with Drosophila Dicer-2 (dDicer2), which is known to process viral dsRNA into predominantly 21 nt vsiRNAs in fruit fly somatic tissues29–32. We found that expression of dDicer2 rescued production of influenza vsiRNAs in hDicer-KO cells (Supplementary Fig. 5). This was further enhanced by co-expression with its dsRNA-binding protein partners29 to levels detectable by northern blot hybridization (Fig. 2b and Supplementary Fig. 5), consistent with the recent characterization of dDicer2 expressed in wild-type human 293 cells33. In contrast, the influenza vsiRNAs remained undetectable in the hDicer-KO cells co-expressing dDicer1 with its partner (Fig. 2b), which produces miRNAs in Drosophila29,32. Notably, a dominant size shift of the influenza vsiRNAs to 21 nt was detected by deep sequencing in the hDicer-KO cells ectopically expressing dDicer2 when compared to the 22 nt vsiRNAs in the cells ectopically expressing hDicer (Fig. 2c and Supplementary Fig. 5). By comparison, the 1U preference was less pronounced for the dDicer2-dependent vsiRNAs than for those made by hDicer (Supplementary Fig. 5). However, the distribution pattern of hotspot vsiRNAs over the viral genomic RNAs was highly similar for the 21 and 22 nt vsiRNAs made by dDicer2 and hDicer (Supplementary Figs 6 and 7), respectively, suggesting that a similar set of dsRNA precursors were recognized and processed by the two Dicer nucleases. These findings further illustrate the accessibility of the influenza vsiRNA precursors to Dicer in human somatic cells.
We found that the production of influenza vsiRNAs was also efficiently induced in both human lung epithelial A549 cells and African green monkey epithelial Vero cells by infection with PR8/delNS1 (Fig. 3a). The canonical properties of the influenza vsiRNAs were more clearly visible from the total small RNAs sequenced directly from PR8/delNS1-infected A549 cells without AGO co-IP (Fig. 3b) than in those sequenced without AGO co-IP from 293T cells (Fig. 1a,b and Supplementary Figs 3 and 4). Our results together illustrate that the production of abundant vsiRNAs is a conserved immune response to IAV infection in distinct human and monkey somatic cells that do not encode the rodent-specific Dicer isoform expressed only in oocytes34. A mutational analysis indicated that although a mutant hDicer with an inactive ATPase domain remained active in the production of the influenza vsiRNAs, the influenza vsiRNAs became undetectable when mutations were introduced to disrupt the PAZ, RNase IIIa or RNase IIIb domain of hDicer (Fig. 3c).
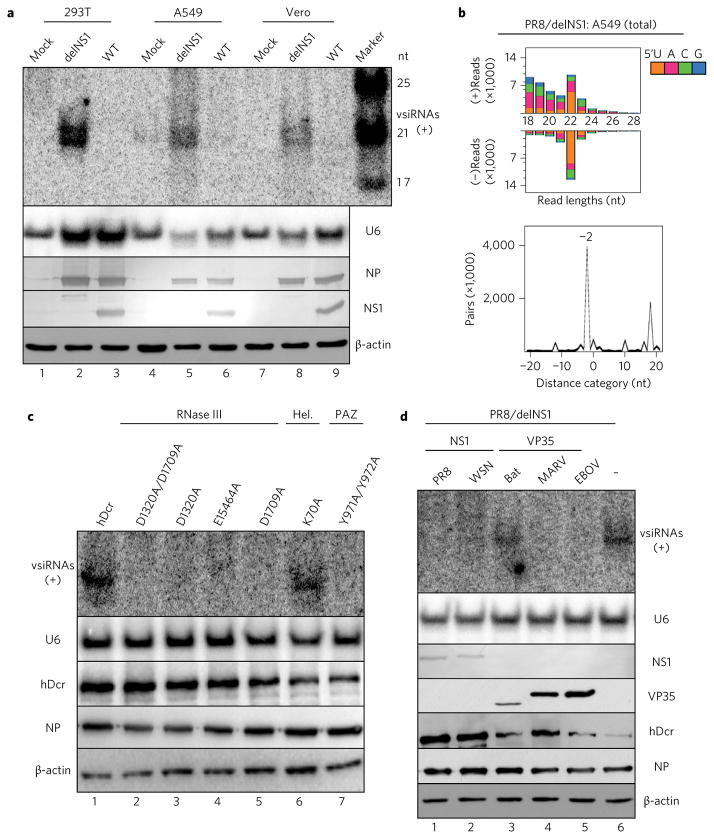
Figure 3. Induction and suppression of influenza vsiRNA biogenesis in distinct human and monkey somatic cells.
a, Northern detection of the influenza vsiRNAs in human (293T and A549) and monkey (Vero) cells 24 h after infection with WT or mutant (delNS1) viruses of the PR8 strain. b, Properties of influenza vsiRNAs (per million total mature miRNAs) sequenced directly without AGO co-IP from PR8/delNS1-infected A549 cells. c, Production of influenza vsiRNAs in PR8/delNS1-infected hDcr-KO 293T cells ectopically expressing WT hDcr, or hDcr mutants carrying the point amino acid mutations29 known to disrupt the function of the helicase (Hel.; K70A), PAZ (Y971A and Y972A), RNase IIIA (D1320A and E1564A) or RNase IIIB (D1709A) domain of hDcr. d, Suppression of influenza vsiRNA biogenesis in PR8/delNS1-infected hDcr-KO 293T cells ectopically expressing hDcr by NS1 of IAV strain PR8 or WSN, FLAG-tagged VP35 of Ebola virus (EBOV), Marburg virus (MARV) or bat, as indicated. The same sets of RNA and protein samples were used for northern or western blot detection of the positive (+)-strand influenza vsiRNAs, U6 RNA, viral NP and NS1 proteins, FLAG-tagged VP35 variants, hDcr or β-actin as in Fig. 2. Each experiment was repeated twice, with reproducible results.
The influenza vsiRNAs were not detected by northern blot analysis in 293T, A549 and Vero cells infected with wild-type PR8 (Figs 2a and 3a), suggesting suppression of the biogenesis of the cognate vsiRNAs by NS1 expressed in cis from the viral genome during IAV infection. Moreover, we found that ectopic expression of NS1 of either PR8 or WSN actively inhibited the production of the influenza vsiRNAs induced by PR8/delNS1 infection (Fig. 3d). These findings define a new activity of NS1 in inhibiting the production of its cognate vsiRNAs induced by an authentic virus infection of human somatic cells, and explains why canonical vsiRNAs are largely undetectable during wild-type IAV infection14,15,25. Next, we investigated if this assay could be used to identify VSRs capable of suppressing the biogenesis of human vsiRNAs. We observed strong suppression of the influenza vsiRNA biogenesis by virion protein 35 (VP35) of the Ebola virus (Fig. 3d), shown previously to exhibit VSR activity10,35. Production of the influenza vsiRNAs was also suppressed by VP35 encoded by the Marburg virus from the same Filoviridae as Ebola virus, but not by the distantly related VP35 encoded by the bat genome (Fig. 3d). These findings show that human viruses have evolved VSRs to inhibit Dicer activity towards viral RNA substrates.
We next determined whether antiviral RNAi restricted IAV infection in mature mammalian cells capable of also eliciting a type I IFN antiviral response. To avoid the pleiotropic effects of the loss of cellular miRNAs on virus infection in Dicer knockout cells28,36, we examined IAV infection in RNAi-defective primary mouse embryonic fibroblasts (MEFs) carrying a genetic mutation abolishing the activity of the RNA-induced silencing complex (RISC) to slice target RNA guided by siRNAs. The Argonaute (AGO) proteins4–7 are essential core components of antiviral RNAi in fungi, plants, insects and nematodes, and are highly conserved in eukaryotes29. Mammals possess four AGOs (AGO 1–4, also known as Eif2c1–4), only one of which, AGO2, exhibits cleavage activity towards target RNA in experimental RNAi37. Furthermore, the catalytic triad in the PIWI domain of AGO2 is deeply conserved, despite the fact that animal miRNAs modulate their targets without AGO-mediated cleavage, suggestive of a function in mammals beyond the processing of a single Dicer-independent miRNA38. Human AGO2 is necessary and sufficient to mediate antiviral RNAi against Nodamura virus infection in mouse embryonic stem cells11, but it is unknown whether this function involves the slicer activity of AGO2, or whether AGO2 is antiviral in differentiated mammalian cells with an intact interferon (IFN) system17,38. We verified efficient loading of the influenza vsiRNAs onto human AGO2 during infection of 293T cells (Supplementary Fig. 8). We took the approach of directly examining the contribution of AGO2 catalytic activity to mammalian antiviral defence in mature, IFN-producing cells. Similar mutational analyses have certainly demonstrated an essential role for the slicer activity of both Drosophila and Arabidopsis AGO2 in antiviral RNAi against wild-type RNA virus infection in plants and fruit flies8,9. We infected two independent primary MEF lines38 where AGO2 is expressed but is catalytically inactive due to substitution of the first aspartic acid in the DDH triad with an alanine (Ago2D597A)38. Importantly, unlike cells deficient in AGO2 protein39, which exhibit a marked loss of host miRNAs, virtually all miRNAs are present in these Ago2D597A cells38. We observed a significant increase in virus levels in cells carrying one allele (Ago2D597A/+), and a further increase in cells carrying two alleles of catalytic-inactive AGO2 (Ago2D597A/D597A) compared to wild-type cells following infection with wild-type PR8 IAV, as measured by plaque assay of cell supernatants and qPCR for virus RNA (Fig. 4a–c). Increased IAV-induced cytopathy was also observed in Ago2D597A MEFs, as demonstrated both by light microscopy images and an increased percentage of Annexin V and 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI)-positive cells following infection (Supplementary Fig. 9). Hence, AGO2 catalytic activity restricts wild-type IAV in mature, IFN-capable mammalian cells. However, the levels of type I IFN and induction of IFN-stimulated genes (ISGs) were similar in wild-type and Ago2D597A MEFs following IAV infection (Fig. 4d) or following infection or stimulation with multiple viruses or viral ligands (Supplementary Fig. 9). Moreover, the expression levels of RNAi components Drosha, Dicer and Ago1–4 (Fig. 4d), growth rate and entry of IAV were equal in wild-type and Ago2D597A MEFs (Supplementary Fig 9). Thus, the slicing activity of AGO2 suppresses IAV infection by RNAi, but not by altering the known IFN-dependent antiviral responses or affecting entry of IAV.
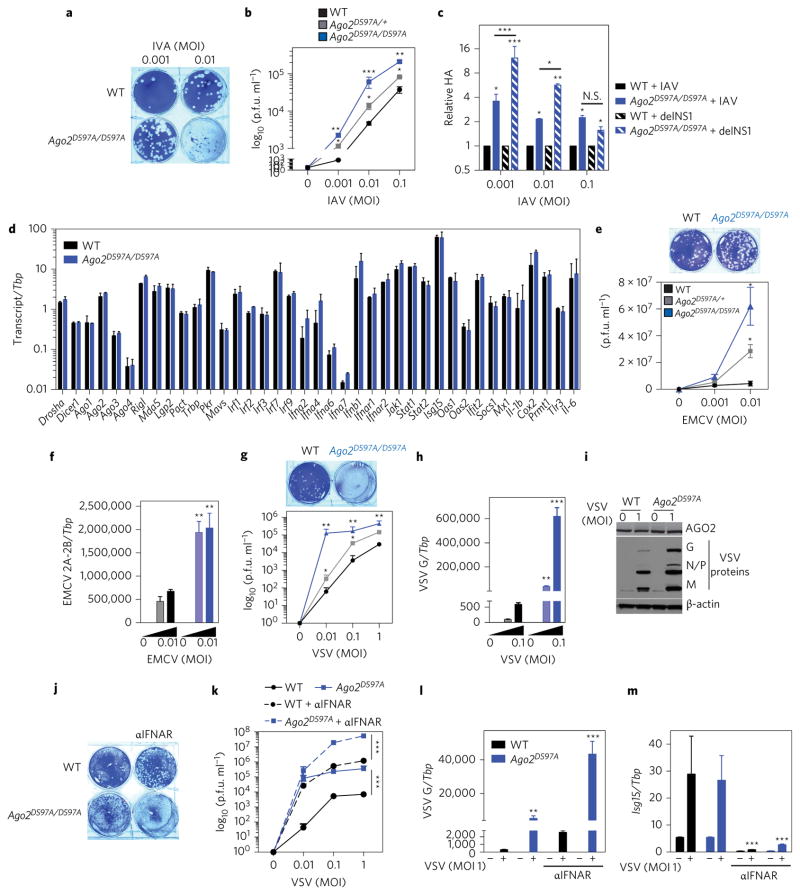
Figure 4. AGO2 slicing activity restricts IAV, EMCV and VSV in mammalian somatic cells.
a,b, Representative plaque assay image of IAV (PR8)-infected (indicated MOIs for 16 h) MEFs from WT mice (black) or mice carrying one (Ago2D597A/+, grey) or two (Ago2D597A/D597A, blue) alleles of catalytic-inactive AGO2. Virus titres quantified as plaque-forming units (p.f.u.) per ml for the indicated MOIs are shown. c, Relative influenza HA RNA as quantified by qPCR in WT or Ago2D597A/D597A MEFs infected with IAV (PR8) or IAV delNS1 mutant. d, Expression of RNAi components, type I IFNs and IFN-stimulated genes (ISGs) in WT (black) or Ago2D597A/D597A (blue) MEFs following influenza infection. e,f, Virus levels in WT (black), Ago2D597A/+ (grey) and Ago2D597A/D597A (blue) MEFs infected with EMCV at indicated MOIs for 16 h as measured by plaque assays of cell supernatants (e) or qPCR of EMCV 2A-2B RNA relative to the housekeeping gene TATA box protein (Tbp) (f). g–i, Virus levels in WT (black), Ago2D597A/+ (grey) and Ago2D597A/D597A (blue) MEFs infected with VSV, at the indicated MOIs, for 16 h as measured by plaque assays of cell supernatants (g) or qPCR of VSV-G RNA relative to Tbp (h) or immunoblot of VSV G, N, P and M proteins (i). j–l, Virus levels in WT (black) and Ago2D597A/D597A (blue) MEFs infected with VSV in the presence of 10 μg ml−1 anti-IFNAR antibody (aIFNAR; MAR1-5A3, dotted lines) as quantified by plaque assays (j,k) or qPCR of VSV-G RNA (l). m, Expression of IFN-stimulated gene 15 (Isg15) relative to Tbp as measured by qPCR-confirmed IFNAR blocking. All data are two separate lines of MEFs performed two to three independent times in triplicate, combined. All error bars are s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, measured by an unpaired t-test or two-way ANOVA (Prism). N.S., not significant.
We next tested the ability of AGO2 catalytic activity to suppress PR8/delNS1 lacking the multifunctional protein NS1. Infection of Ago2D597A MEFs with PR8/delNS1 resulted in significantly enhanced virus levels, as measured by qPCR for haemagglutinin (HA) (Fig. 4c). The enhancement was also significantly more than that observed for wild-type influenza with intact NS1 in Ago2D597A MEFs (Fig. 4c), demonstrating some genetic rescue of PR8/delNS1 in AGO2 catalytic inactive cells. To assess whether AGO2 catalytic activity is a broad antiviral mechanism for a range of virus types, we infected wild-type and Ago2D597A MEFs with the positive-strand RNA virus EMCV or the negative-strand RNA virus vesicular stomatitis virus (VSV). We observed a significant increase in the levels of both viruses in cells carrying one (Ago2D597A/+) or two (Ago2D597A/D597A) alleles compared to wild-type cells following infection, as measured by plaque assay of cell supernatants, qPCR for virus RNA or immunoblots for virus protein (Fig. 4e–i). Hence, AGO2 catalytic activity restricts multiple virus infections in mature mammalian cells. Importantly, we also observed an increase in virus levels following infection of AGO2 catalytic dead MEFs in the absence of type I IFN signalling (Fig. 4j–m), indicating an IFN-independent antiviral function of Ago2 catalytic activity.
Here, we have demonstrated a specific VSR activity of NS1 to inhibit the processing of the cognate viral dsRNA molecules into siRNAs during authentic infection of distinct mammalian cell lines, in addition to the known antagonizing activities of NS1 against IFN-regulated antiviral responses. In the absence of NS1 suppression, we found that wild-type human Dicer mediates the production of highly abundant vsiRNAs in response to IAV infection, revealing innate immune detection of human RNA virus infection for the first time in mature human somatic cells by the RNAi pathway. Our findings illustrate the importance of using VSR-deficient mutant viruses to probe antiviral RNAi and provide the first experimental system to characterize the Dicer-mediated biogenesis of siRNAs from naturally occurring precursors in mammalian cells. It also explains why previous attempts failed to detect significant levels of canonical vsiRNAs in wild-type mammalian cells challenged by wild-type viruses. Our observation that the influenza viral siRNA biogenesis is potently suppressed by VP35 suggests a strategy to characterize the production of Ebola and Marburg viral siRNAs in mammalian cells using mutant viruses expressing VP35 variants defective in RNAi suppression. Moreover, it is likely that the influenza vsiRNAs produced by human Dicer are biologically active, because they exhibit a strong 1U preference and are loaded in AGOs in abundances similar to cellular miRNAs. Consistently, we show that antiviral defence against IAV infection in mature mammalian cells specifically requires the slicing activity of AGO2, known to be essential for RNAi directed by synthetic siRNAs. Notably, antiviral RNAi mediated by the slicing activity of AGO2 functions without altering known IFN-dependent antiviral responses, is effective against both EMCV and VSV, and remains functional in the absence of type I IFN signalling. Although further in vivo characterization is necessary, these findings suggest that antiviral RNAi in cultured mammalian cells can function in an IFN-independent manner. The results presented in this study and in recent publications10,11 together show that infection with distinct families of RNA viruses triggers the production of highly abundant vsiRNAs in mammalian cells. In turn, mammalian RNA viruses have evolved to suppress biogenesis of their cognate vsiRNAs during infection, consistent with a proposed role for VSR activity in human cell infection. We propose that the RNAi pathway has the capacity to confer widespread antiviral function in mammals against RNA viruses.
Methods
Cell culture
Human lung epithelial cells (A549) were maintained in F-12K medium, and human embryonic kidney cells (293T) and African green monkey kidney epithelial cells (Vero) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. hDicer knockout 293T cell line 4–25 and its parental 293T cell line28 were gifts from B. Cullen. Two separate lines38 of MEFs from Ago2 catalytic inactive mice, where the endogenous Ago2 allele was replaced with one carrying an ADH rather than a DDH triad (Ago2D597A), were provided by S. Cheloufi (Massachusetts General Hospital) and G. Hannon (Cold Spring Harbor Laboratories). MEFs were cultured in DMEM with 15% FBS, L-glutamine, penicillin-streptomycin and β-mercaptoethanol. All cell lines were authenticated and tested for mycoplasma contamination.
Virus infections
Influenza A/WSN/1/33 (H1N1) and A/Puerto Rico/8/34 (H1N1) viruses, designated PR8-wild type (WT) and WSN-WT, respectively, and their NS1 deletion mutants, designated PR8/delNS1 and WSN/delNS1, as well as PR8-NS1/RFP, were gifts from A. Garcıa-Sastre and P. Palese27,40. EMCV was purchased from ATCC. VSV (Indiana strain) was provided by C. Rice and M. MacDonald. MEFs were infected in 12-well plates with 100 μl virus for indicated multiplicities of infection (MOIs) diluted in phosphate buffered saline (PBS) supplemented with 1% FBS, calcium and magnesium for 1 h at 37 °C, with rocking every 15 min. Virus was removed, cells were washed once with PBS, and this was replaced with appropriate medium. This was considered time 0. To determine influenza titres in cell supernatants, plaque assays were performed on canine kidney (MDCK) cells using 0.6% BSA, 2% oxoid agarose, 1 mg ml−1 Trypsin-TPCK, 0.6% NaHCO3, 2% sodium pyruvate, 1% penicillin/streptomycin, 50 mM HEPES, 4 mM L-glutamine in 2× DMEM for plaque formation and plaques counted 2–3 days post-infection (d.p.i.). For quantification of EMCV titres, plaque assays were performed on Vero cells with 1.5% low electroendosmosis (LE) agarose and plaques were counted at 24 h.p.i. For quantification of VSV titres, plaque assays were performed on baby hamster kidney (BHK-21) cells with 1.5% LE agarose and plaques were counted at 16 h.p.i., when cells were fixed with 7% formaldehyde followed by crystal violet staining. To assess viral entry, cells were infected with A/Puerto Rico/8/34 at MOI = 50 for 2 h in the presence of 30 μg ml−1 of cylcoheximide. Following infection, cells were incubated in 0.025% Trypsin-EDTA at room temperature for 7 min, to remove any virus that had adsorbed to the cell surface but had not yet been internalized, as previously described41. Incorporated virus was assayed by qPCR. Cell death following virus infection was determined by flow cytometric analysis of Annexin V and DAPI-positive cells. Data analysis was performed with FlowJo (TreeStar).
Plasmids and molecular cloning
The coding sequences for the NS1 of PR8-WT and WSN-WT (ref. 2) were obtained by reverse transcription-polymerase chain reaction (RT-PCR) from infected cells and cloned into pcDNA3.1 vector to generate pcDNA-PR8-NS1 and pcDNA-WSN-NS1. The expression plasmids in pCAGGS for the VP35 of Ebola virus (EBOV), Marburg virus (MARV) and bat with an N-terminal Flag tag42 were gifts from C.F. Basler. The expression plasmids for human AGO2, Dicer and Dicer mutants were purchased from Addgene (cat. nos. 10822, 19873, 41584, 41585, 41586, 41587, 41588, 41589 and 41590). The cDNA clones for Drosophila Loquacious isoforms PB and PD (Loqs-PB and Loqs PD) were from Addgene (nos. 41094 and 42095) whereas the plasmids encoding Drosophila proteins dDicer-1, dDicer-2 and R2D2 were gifts from Q. Liu43,44. The open reading frames (ORFs) of Loqs-PB, Loqs PD and R2D2 were cloned with an N-terminal Flag tag into pcDNAFlag45 to generate pcDNA-Flag-PB, pcDNA-Flag-PD and pcDNA-Flag-R2D2, respectively. The ORFs of dDicer-1 and dDicer-2 were cloned with an N-terminal His tag into pcDNA4HisMax45 to generate pcDNA-His-dDcr1 and pcDNA-His-dDcr2, respectively.
Cell culture infection, transfection and the construction of small RNA libraries
293T and hDcrKO 293T cells were seeded in a 6 cm plate at a density of 2.5 × 106 per plate one day before infection. At 24 h after inoculation with serum-free DMEM (mock), PR8-WT, WSN-WT, PR8/delNS1 or WSN/delNS1 (MOI = 1) as previously described45, infected cells were collected for the extraction of total protein and RNA using TRIzol (Invitrogen) according to the manufacturer’s protocol.
To determine the role of Dicer in the biogenesis of viral siRNAs (vsiRNAs), hDicer-KO 293T cells seeded in a 6 cm plate at a density of 2.5 × 106 per plate were transfected with 8 μg of the plasmid encoding hDicer or a hDicer mutant carrying the following amino acid mutation(s) in key residues (as described in ref. 36): K70A, Y971A/Y972A, D1320A, E1564A, D1709A, D1320A/D1709A or Drosophila Dicer-2 (dDcr2). Alternatively they were co-transfected with (1) 8 μg pcDNA-His-dDcr1 and 4 μg pcDNA-Flag-PB, (2) 8 μg pcDNA-His-dDcr2 and 4 μg pcDNA-Flag-Loqs-PD or (3) 8 μg pcDNA-His-dDcr2, 4 μg pcDNA-Flag-Loqs-PD and 4 μg pcDNA-Flag-R2D2 using TransIT-LT1 transfection reagent (Mirus) following the supplier’s recommended protocol. Six hours after transfection, hDcr-KO 293T cells were infected by PR8/delNS1 (MOI = 1) and the infected cells were collected for the extraction of total protein and RNA using TRIzol 24 h after infection.
To determine the activity of viral suppressors of RNAi (VSRs), hDcr-KO 293T cells seeded in a six-well plate at a density of 1.3 × 106 per well were co-transfected with 4 μg of the hDicer expression plasmid with one (2 μg) of the following VSR-expressing plasmids: pcDNA-PR8-NS1, pcDNA-WSN-NS1, pCAGGS-EBOV-VP35, pCAGGS-MARV-VP35 or pCAGGS-bat-VP35. At 6 h after co-transfection, the hDcr-KO 293T cells were infected by PR8/delNS1 (MOI = 1), and the infected cells were collected for the extraction of total protein and RNA using TRIzol 24 h after infection.
Libraries of small RNAs were constructed from total RNA extracted 24 h after infection of 293T cells with PR8/delNS1 or WSN/delNS1 either without or with co-IP by Anti-pan Ago antibody (Millipore), by a method depending on the 5′ monophosphate of small RNAs (described previously10), with a TruSeq Small RNA Sample Preparation Kit (Illumina). Libraries of small RNAs were also constructed from (1) total RNA from A549 cells 24 h after infection with PR8/delNS1, (2) total RNA from PR8/delNS1-infected hDcr-KO 293T cells without or with ectopic expression of hDicer, dDicer-2 or dDicer-2 plus Loqs-PD and R2D2, and (3) total RNA co-immunoprecipitated from 293T cells over-expressing FLAG-tagged human AGO2 24 h after infection with PR8/delNS1.
Western and northern blot analyses
Western blot analysis was performed as described previously45. Antibodies to NS1, NP and His tag antibodies were gifts of Y. Zhou45. Antibodies to NoV B2 have been described previously10. Antibodies to hDicer (Santa Cruz Biotechnology), β-actin (Cell Signaling Technology) and Flag M2 (Sigma-Aldrich) were sourced from commercial suppliers. For northern blot analysis of low-molecular-weight RNAs, 15 μg total RNA extracted from cells 24 h after infection and a chemical crosslinking protocol46 were used, essentially as described previously, but with one modification10: instead of using locked nucleic acid (LNA) oligonucleotides as probes10, the negative- and positive-strand influenza viral RNAs were detected by the 32P-labelled synthetic RNA oligo, 5′-CAUAAUGG AUCCAAACACUGUG-3′ and 5′-GACACAGUGUUUGGAUCCAUUA-3′, respectively. Each experiment presented in Fig. 2 was repeated twice, with reproducible results.
Deep sequencing and bioinformatic analysis of small RNAs
Libraries of small RNAs cloned from cultured human cells were sequenced by an Illumina HiSeq 2000/2500 at the Core Facility of the Institute for Integrative Genome Biology on campus. Small RNA reads were mapped to the virus and host genome references or compared to mature miRNAs. Mapping was done by Bowtie 1.0.0 with either a perfect match or one mismatch only in Supplementary Fig. 5a. All of the references used were downloaded from web sources as listed below. Subsequent bioinformatics analysis of virus-derived small RNAs was carried out using in-house Perl scripts as described previously10. Pairs of complementary 22 nt vsiRNAs in each library with different base-pairing lengths were computed using a previously described algorithm10, which calculates the total counts of pairs in each nucleotide distance between the 5′ and 3′ ends of complementary 22 nt vsiRNAs. The following reference sequences were used in this study:
PR8/delNS1: Obtained from A/Puerto Rico/8/34 (H1N1) (PR8-WT) by deleting nucleotides 57 to 528 in the NS segment. The sequence of PR8-WT was downloaded from NCBI: AF389115.1, AF389116.1, AF389117.1, AF389118.1, AF389119.1, AF389120.1, AF389121.1 and AF389122.1.
-
WSN/delNS1: Obtained from A/WSN/1/33 (H1N1) (WSN-WT) by deleting nucleotides 57 to 528 in the NS segment. The sequence of PR8-WT was downloaded from NCBI: J02179.1, J02178.1, CY034137.1, J02176.1, CY034135.1, L25817.1, L25818.1 and M12597.1, with the missing terminal sequences from the following segments completed by sequencing (underlined nucleotides are the original terminals from NCBI):
Segment PA (segment 3) 3′ terminal: 3′-AGCGAAAGCAGGTACTGATT…5′
Segment PA (segment 3) 5′ terminal: 3′…AAAAAAGTACCTTGTTTCTACT-5′
SegmentNP(segment5)3′ terminal: 3′-AGCAAAAGCAGGGTAGATAATCACTC…-5′
Segment NP (segment 5) 5′ terminal: 3′-…AAAGAAAAATACCCTTGTTTCTACT-5′
Segment M (segment 7) 5′ terminal: 3′…AAACTACCTTGTTTCTACT-5′
Mature miRNAs and miRNA precursors: miRBase 19 (http://www.mirbase.org/).
Non-coding RNAs: fRNAdb 3.0 (http://www.ncrna.org/frnadb/).
Human mRNAs: Mammalian Gene Collection (MGC).
Human whole genome: GRCh38 project from NCBI, released on 24th December 2013 (GCA_000001405.15).
Correlation analysis of small RNA populations
The correlation analysis of small RNA populations was carried out using the following formulae:
n indicates the total number of nucleotide positions on each strand of the PR8-delNS1 genome in the analysis for correlation between 21 and 22 nt influenza vsiRNAs made by hDicer and dDicer2.
i indicates a specific nucleotide position, as referred to above, that is covered or not covered by either 22 or 21 nt vsiRNAs from the hDicer and dDicer2 libraries, respectively.
yi indicates the relative coverage at position i by 22 nt vsiRNAs made by hDicer with a value between 0 and 1 after normalization by the maximum 22 nt vsiRNA coverage of each virion RNA segment in either polarity. ȳ indicates the average coverage by 22 nt vsiRNAs at all nucleotide positions n.
xi indicates the relative coverage at position i by 21 nt vsiRNAs made by dDicer2 with a value between 0 and 1 after normalization by the maximum 21 nt vsiRNA coverage of each virion RNA segment in either polarity. x̄ indicates the average coverage by 21 nt vsiRNAs at all nucleotide positions n.
Any given nucleotide position (i) with a value change (yi − xi) beyond the interval [μ − 3σ, μ + 3σ] was considered to display significant variation between the two small RNA libraries. Otherwise it was considered to display no significant variation.
Other reagents and cell stimulations
LPS (Escherichia coli 0111:B4, used at 100 ng ml−1) and polyinosinic:polycytidylic acid (PolyI:C, used at 10 μg ml−1) were purchased from Sigma. 5′ Triphosphate double-stranded RNA (5′ ppp-dsRNA) was purchased from InvivoGen. Monoclonal antibody to Interferon A Receptor (anti-IFNAR, MAR103, used at 10 μg ml−1) was purchased from eBiosciences. Sendai virus was purchased from ATCC.
RNA isolation, reverse transcription and qPCR
RNA was isolated with the RNeasy mini kit (Qiagen). RNA was reverse transcribed using Reverse Transcription Supermix (Biorad). qPCR was performed using SYBR green (BioRad), and PCR products were quantified with a standard curve. Gene expression was displayed relative to the housekeeping gene, TATA box protein Tbp. The following primer sequences were used: Flu HA: forward CTGCTCGAAGACAGCCACAA, reverse GAGCCACCGGCGATCTTAC; Flu NP forward ACGGCTGGTCTGACTCACAT, reverse TCCATTCCGGTGCGAACAAG; Ifnb1 forward CTGCGTTCCTGCTGTG CTTCTCCA, reverse TTCTCCGTCATCTCCATAGGGATC Isg15: forward TGAGAGCAAGCAGCCAGAAG, reverse ACGGACACCAGGAAATCGTT, Tbp forward TGATCAAACCCAGAATTGTTCT, reverse TGGTCTTCCTGAAT CCCTTTA. A custom-ordered PrimePCR Mouse Assay panel (BioRad) containing Ifn pathway genes and RNAi machinery components were also used.
Statistical analysis
GraphPad Prism was used for all statistical analysis. All experiments were repeated at least three times. Statistical analysis was carried out by two-way or one-way analysis of variance (ANOVA), or by an unpaired t-test of biological replicates. A P value of <0.05 was considered statistically significant.
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information and are also available from the corresponding authors upon request. The data sets generated during the current study are available from the Gene Expression Omnibus (GEO) database under accession number GSE89514.
Acknowledgments
The authors thank L.A. Ball, C. Basler, B.R. Cullen, A. Garcia-Sastre, C. Rice, M. McDonald, K.L. Johnson, Q. Liu and P. Palese for providing materials, A. Tarakhovsky for scientific discussions and support, and F. Uhl and A.E. Handte-Reinecker for technical assistance. G. Hannon provided Ago2D587A MEF lines. This study was supported by NIH grants R01AI107087 (to K.L.J.), MGH Executive Committee on Research (ECOR) funds (to K.L.J.), R01AI52447 (to S.W.D.) and R56AI110579 (to S.W.D.), CNAS of UC Riverside (to S.W.D.), AI113333 and DK068181 (to H.C.R.), a Department of Defense PRCRP fellowship CA120212 (to S.C.) and NHMRC grants 1027020 and 1083596 (to P.H.).
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
Y.L. and S.D. performed all virus infection experiments in 293T, A549 and Vero cells. M.B. performed and analysed all virus infection experiments in Ago2D597A cells. J.L. performed all bioinformatic analyses of small RNA libraries. Y.H., W.-X.L. and F.V.K. assisted with cloning of small RNAs. D.A.C. and J.T.P. assisted with viral infections. H.C.R and P.H. provided reagents and interpreted results. S.C. provided Ago2D597A MEFs. S.W.D. and K.L.J conceived of the study, designed experiments, interpreted results and wrote the final manuscript.
Additional information
Supplementary information is available for this paper.
References
- 1.Medina RA, García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li WX, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marc D. Influenza virus non-structural protein NS1: interferon antagonism and beyond. J Gen Virol. 2014;95:2594–2611. doi: 10.1099/vir.0.069542-0. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Rice CM. RNAi targeting an animal virus: news from the front. Mol Cell. 2002;9:925–927. doi: 10.1016/s1097-2765(02)00539-7. [DOI] [PubMed] [Google Scholar]
- 5.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 7.Csorba T, Kontra L, Burgyán J. Viral silencing suppressors: tools forged to fine-tune host–pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Carbonell A, et al. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques JT, et al. Functional specialization of the small interfering RNA pathway in response to virus infection. PLoS Pathog. 2013;9:e1003579. doi: 10.1371/journal.ppat.1003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 13.Parameswaran P, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio. 2010;1:e00204–10. doi: 10.1128/mBio.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez JT, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girardi E, Chane-Woon-Ming B, Messmer M, Kaukinen P, Pfeffer S. Identification of RNase L-dependent, 3′-end-modified, viral small RNAs in Sindbis virus-infected mammalian cells. mBio. 2013;4:e00698–13. doi: 10.1128/mBio.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo GJ, et al. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogerd HP, et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J Virol. 2014;88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Backes S, et al. The mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014;8:114–125. doi: 10.1016/j.celrep.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanguy M, Miska EA. Antiviral RNA interference in animals: piecing together the evidence. Nat Struct Mol Biol. 2013;20:1239–1241. doi: 10.1038/nsmb.2708. [DOI] [PubMed] [Google Scholar]
- 21.Sagan SM, Sarnow P. Molecular biology. RNAi, antiviral after all. Science. 2013;342:207–208. doi: 10.1126/science.1245475. [DOI] [PubMed] [Google Scholar]
- 22.Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- 23.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 24.de Vries W, Haasnoot J, Fouchier R, de Haan P, Berkhout B. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J Gen Virol. 2009;90:1916–1922. doi: 10.1099/vir.0.008284-0. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy EM, et al. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human Dicer. Proc Natl Acad Sci USA. 2015;112:E6945–E6954. doi: 10.1073/pnas.1513421112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 28.Bogerd HP, Whisnant AW, Kennedy EM, Flores O, Cullen BR. Derivation and characterization of Dicer- and microRNA-deficient human cells. RNA. 2014;20:923–937. doi: 10.1261/rna.044545.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 30.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 31.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YS, et al. Distinct roles for Drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 33.Girardi E, et al. Cross-species comparative analysis of dicer proteins during Sindbis virus infection. Sci Rep. 2015;5:10693. doi: 10.1038/srep10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flemr M, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Haasnoot J, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurtan AM, Lu V, Bhutkar A, Sharp PA. In vivo structure–function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA. 2012;18:1116–1122. doi: 10.1261/rna.032680.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 38.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A Dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Carroll D, et al. A slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlivan M, et al. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Prins KC, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, et al. Dicer-1, but not Loquacious, is critical for assembly of miRNA-induced silencing complexes. RNA. 2007;13:2324–2329. doi: 10.1261/rna.723707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Anderson DH, Liu Q, Zhou Y. Mechanism of influenza A virus NS1 protein interaction with the p85β, but not the p85α, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J Biol Chem. 2008;283:23397–23409. doi: 10.1074/jbc.M802737200. [DOI] [PubMed] [Google Scholar]
- 46.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available within the paper and its Supplementary Information and are also available from the corresponding authors upon request. The data sets generated during the current study are available from the Gene Expression Omnibus (GEO) database under accession number GSE89514.