Abstract
Elastomeric, robust, and biocompatible hydrogels are rare, while the need for these types of biomaterials in biomedical-related uses remains high. Here, a new family of genetically engineered silk-elastin copolymers (SELPs) with encoded enzymatic crosslinking sites is developed for a new generation of stimuli-responsive yet robust hydrogels. Input into the designs is guided by simulation, and realized via genetic engineering strategies. The avoidance of gamma irradiation or chemical crosslinking during gel fabrication, in lieu of an enzymatic process, expands the versatility of these new gels for the incorporation of labile proteins and cells. In the present study, the new SELP hydrogels offers sequence dependent, reversible stimuli-responsive features. Their stiffness covers almost the full range of the elasticity of soft tissues. Further, physical modification of the silk domains provided a secondary control point to fine-tune mechanical stiffness while preserving stimuli-responsive features, with implications for a variety of biomedical materials and device needs.
Keywords: silk, elastin, stimuli responsive, biomaterials, silk-elastin-like proteins, bioengineering, REMD simulations
1. Introduction
Hydrogels are three-dimensional (3D) insoluble cross-linked polymer networks with an ability to retain a significant amount of aqueous medium within their porous structure. Historically, the designs have consisted primarily of relatively inert matrices, with tremendous impact in medicine, from contact lenses[1] to the field of regenerative medicine.[2] These materials are particularly attractive for tissue engineering,[3, 4] due to the high water content and the ability to mimic aspects of the 3D micro-environment of native tissue extracellular matrices (ECM). However, these conventional hydrogels, once in the equilibrium swelling state, are limited to mimicking static environments and lack responses to endogenous environmental stimuli or natural signals from cells and tissues. These features therefore restrict their applications in cell-mediated delivery of growth factors, swelling-controlled release of drugs in naturally low pH environment such as the gastro-intestinal tract, or detection of symptoms of disease as biological sensors.
Recent trends in hydrogel designs have evolved from static to dynamic, bio-responsive systems, to address broader biomedical needs for higher-performance materials. These hydrogels include stimuli-responsive hydrogels that can dramatically change their volume and other properties in response to environmental stimuli such as temperature, pH and certain biological signals. The most extensively studied stimuli-responsive hydrogels are composed of synthetic polymers, such as temperature sensitive poly(N-isopropyl acrylamide),[5] pH sensitive poly(acrylic acid),[6] and their derivatives.[7] While these studies demonstrated the potential of using stimuli-responsive hydrogels for artificial muscles,[8] controlled drug delivery,[9] smart diagnostics,[10] optical systems,[11] as well as actuated micro-patterned materials and sensors,[12] the polymers utilized to date are non-degradable in vivo and offer limited options in terms of responsive stimuli (e.g., temperature, pH).
In contrast, protein polymers provide a unique opportunity for the tunable design of such dynamic polymer systems, due to the genetic basis of sequence, molecular weight and chirality control. Harnessing proteins to generate stimuli-responsive hydrogels based on protein folding-unfolding for biomedical applications offers tremendous opportunities for fine tuning control, responses and utility, such as for tissue regeneration scaffolds and controlled drug delivery devices.[13] Further, compared to synthetic polymers, protein polymers are generally biocompatible and fully degradable in vivo, which is critical for many biomedical needs. The protein systems are also relatively easy to functionalize through diverse amino acid chemistries, or via the formation of well-defined supramolecular structures, such as β-sheets, α-helices and β-turns. Moreover, with advances in genetic and protein engineering, these protein polymers can be designed de novo by permutation, combination and with the addition of constituent modules to introduce functions, exploiting the modular nature of fibrous proteins. Therefore, protein polymers are ideal for designing new stimuli-responsive biomaterials which can undergo substantial changes in volume, shape, mesh size, mechanical stiffness and optical transparency in response to specific target stimuli. As a starting point for these types of dynamic protein systems, the pioneering studies by Urry et al. on the coacervation phenomena of elastin-like proteins in solution provide a foundation for peptide sequence chemistry and stimuli-responses.[14-16] These designs have been extended over the years by many groups. Self-assembled elastin copolymer micelles, nanoparticles and micellar hydrogels have been used for targeted drug delivery and sensing.[17, 18] While these studies demonstrated the potential of using elastin or elastin block copolymers as dynamic delivery systems, the stimuli-responsive properties of these proteins were mostly appreciated when elastin like peptides were in solution. A few examples demonstrated that elastin coacervate can be chemically cross-linked via (hydroxymethyl)phosphines or physically cross-linked by gamma irradiation for temperature sensitive soft elastomeric matrix. However, the mechanical strength of those biomaterials are weak (less than 0.5 kPa and can’t hold their shape), and thus, refrains their uses for tissue engineering and bio-devices.[19, 20] Therefore, forming solid state, robust protein hydrogels with dynamic mechanical properties and fully translating the molecular level folding-unfolding of these proteins into macroscopic reversibly tunable physical properties remains challenging.
Here we report a new system for the design, synthesis and fabrication of stimuli-responsive, robust and tunable hydrogels, exploiting silk and elastin peptide motifs for the building blocks combined with enzymatic crosslinking to generate the solid state materials. Sequence features of our silk-elastin-like proteins (SELPs) include the elastin domain, GXGVP, as the soft domain providing elasticity and dynamic features, and the silk domain, GAGAGS, as the hard domain regulating the stimuli-responsive features and providing tunable mechanical stiffness by varying the degree of crystallinity (Figure S1a). These new systems provide a number of novel features. (a) The generation of solid state responsive hydrogels from the SELPs were realized by horseradish peroxidase (HRP) catalyzed reactions from the tyrosine crosslinking sites encoded in the proteins (e.g., at the X position in the elastin domain). These elastin crosslinking networks played critical roles to translate molecular level protein folding-unfolding into macroscopic reversibly tunable physical properties (Figure S1b). (b) The incorporation of modeling guided rationale design of stimuli-responsive sequences. Here, replica exchange molecular dynamics simulations (REMD) were integrated with the genetic engineering approaches for the rational design of elastin domains for hierarchical materials with predictable responses to specific environmental stimuli, such as temperature, pH, ionic strength and biological triggers. In particular, sequence variants in the position “X” of the elastin domains that dominate further variants in material features were simulated to address the origin of dynamic material functions. (c) The combination of genetic engineering and molecular modeling provided a toolkit upon which to generate a broad range of stimuli-responsive systems to exploit towards new families of dynamic protein hydrogels. (d) The ability to fine tune the mechanical properties of the hydrogels via the silk domains modulated mechanical properties while preserving the stimuli-responsive features (Figure S1c), in combination with the elastin domains related to sequence-specific responses.
The resultant SELP hydrogels exhibited significant swelling ratios as well as significant reversible changes in optical transparency, mechanical properties and hydrogel pore size upon exposure to designed target stimuli. The development of these dynamic protein hydrogels based on SELPs suggests up new possibilities for these hydrogels as biological structures in tissue regeneration and as vehicles for delivery. The insight gained will also allow enhanced understanding of structure-function relationships with protein designs, thus guiding genetic designs of dynamic hydrogels that respond predictably to virtually any desired environmental input to trigger changes in the material.
2. Results
2.1. Dynamic Design of Stimuli-Responsive SELPs Library
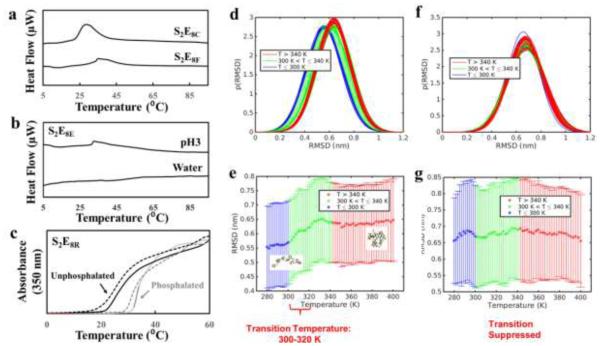
A library of SELPs consisting of 64 different SELP constructs with 12 different monomers and 3 different silk to elastin ratios was biosynthesized via seamless cloning strategies as described previously[18, 21] and purified via inverse temperature transition cycling (Figure S2) with an average yield of purified protein of about 1 g/L. The purity and molecular weight of the proteins were checked by SDS-PAGE (Figure S3). Each individual SELP construct was named as SnEmX, based on both the silk to elastin ratio n: m and its characteristic amino acid “X” in the elastin blocks. The silk and elastin peptide motifs used in our SELP library are shown in Figure S4, with silk blocks serving as the mechanical reinforcement motifs and elastin blocks serving as the stimuli-responsive motifs. Different amino acids (C, F, G, Y, I, E, K, RGYSLG) were employed in the elastin domains at the second amino acid position of the fifth elastin block in each SELP monomer to expand the diversity of the library. This position “X” amino acid in elastin domains was designed to control chain dynamics and the intermolecular or intramolecular interactions of SELP molecules in aqueous environments, in order to control the dynamic functions of the SELPs, via altering the hydrophobicity and surface charge of the “X” residue.[15, 16] Stimuli that were selected to trigger the reversible conformational transitions in the elastin domains were classified into three categories: (1) physical stimuli, such as temperature, electric field, light; (2) chemical stimuli, such as pH, redox state, ionic strength; and (3) biological stimuli, such as enzymatic triggers, glucose. By varying position “X” amino acid in the elastin blocks, SELPs can be designed to predictably respond to a single stimulus in each category, or exhibit multi stimuli-responsive properties that simultaneously respond to more than one stimulus (Figure 1 a-c). The SELPs with the hydrophobic residues at position “X”, such as C and F, responded to temperature changes and exhibited inverse temperature transitions as the temperature was raised through the transition temperature (Figure 2a). The SELPs with the negatively charged residues at position “X”, such as E, exhibited inverse temperature transitions at 34°C in pH4 phosphate-citrate buffer, but no response in water (Figure 2b), suggesting that this SELP responds to pH changes in the temperature window above 34°C. The SELPs with the biological recognition site as position “X” residues, such as RGYSLG that can be recognized by protein kinase (PKA) for phosphorylation[22], exhibited elevated inverse transition temperature once the sequences were phosphorylated (Figure 2c). These results demonstrate that SELPs can be rationally designed to be responsive to a specific stimulus. The insight gained provided a starting base for utilizing the SELP library for the fabrication of dynamic hydrogels.
Figure 1.
SELP dynamic hydrogel designs inspired by the nature of the environmental stimulus. (a) SELPs respond to temperature, a physical stimulus, exemplified by S2E8C and S2E8F. (b) SELPs respond to pH, a chemical stimulus, exemplified by S2E8E. (c) SELPs respond to phosphorylation, a biological stimulus, exemplified by S2E8R. (d) Gaussian fits to the distribution of structures characterized by root mean square deviation (RMSD) from extended linear conformation at a range of temperatures for the X=V sequence. (e) Mean RMSD values for the X=V sequence indicating a transition from extended to folded conformations. (f) Gaussian fits to the distribution of structures characterized by root mean square deviation (RMSD) from extended linear conformation at a range of temperatures for the X=K sequence. (g) Mean RMSD values for the X=K sequence suggesting a suppressed transition.
Figure 2.
SELP dynamic hydrogel schematics and structural characterization. (a) Mechanism of HRP-mediated SELP gelation: the formation of inter- and intramolecular covalent crosslinks (black lines) between elastin blocks allows for chain extension for stimuli responsive hydrogels. (b) Real-time FTIR spectra of S2E8R during gelation suggest the formation of the SELP hydrogel is not based on β-sheet crosslinks. Fluorescence excitation-emission spectra of (c) S2E8Y solution and (d) S2E8Y hydrogel confirmed the formation of dityrosines between the elastin blocks. (e) The resultant S2E8Y hydrogels are optically clear and exhibit blue fluorescence when irradiated with UV, a feature not present in the precursor protein solution.
To study the molecular mechanisms underlying the stimuli-responsive features of the elastin domains, molecular dynamics simulations were used to identify temperature transitions associated with a single peptide, employing the Replica Exchange Molecular Dynamics paradigm for enhanced sampling. We considered two tri-block peptides, with amino acid sequence (GVGVP)(GXGVP)(GVGVP), where the X residue was chosen to be valine (V) or lysine (K) (Figure 2d-g). We observed a transition in the X = V sequence which was characterized by structural folding of the peptide, associated with a hydrophobic collapse thought to be responsible for elastin’s inverse temperature transition (Figure 2d,e). By substituting the lysine residue (X = K), we showed that this structural transition could be suppressed (Figure 2f,g), demonstrating proof of concept of the tunability of elastin domains at the molecular scale.
2.2. Fabrication of SELP Smart Hydrogels and Structural Characterization
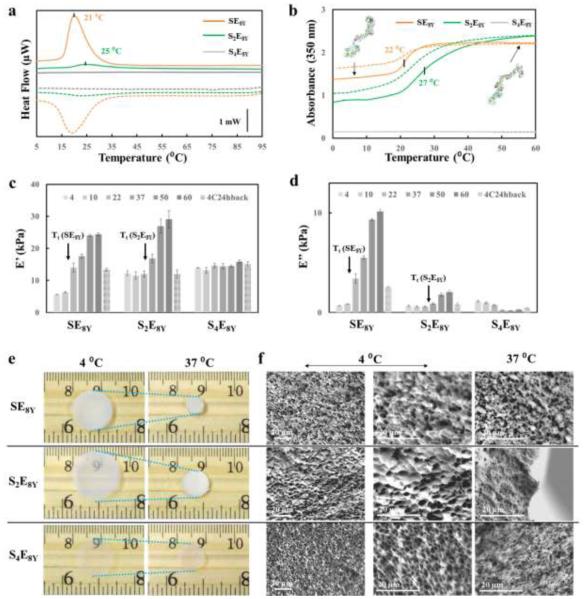
In order to translate protein chain folding-unfolding at the molecular level into reversible macroscopic, solid-state material physical properties changes, the elastin domains were cross-linked to fabricate stimuli-responsive SELP hydrogels (Figure 2). The SELPs that were designed with a tyrosine residue in the “X” position of the elastin blocks, named SnE8Y and S2E8R, were selected as proof-of-concept examples and cross-linked to form hydrogels via an aqueous, enzymatic process based on horseradish peroxidase (HRP)-mediated redox reaction[23] (Figure 2a). In brief, SnE8Y or S2E8R were dissolved in deionized water at 4°C overnight to obtain a homogenous solution; then the addition of H2O2 in the presence of HRP resulted in the formation of inter- and intramolecular covalent crosslinks between tyrosine residues, leading to the formation of a stable, highly elastic hydrogel. These elastin networks, represented by the black solid lines in Figure 2a, are essential for dynamic SELP hydrogels.
The formation of elastin networks based on dityrosine bonds and hydrogel secondary structure during gelation was assessed by real-time Fourier transform infrared spectroscopy (FTIR) and by fluorescence spectroscopy. Real-time FTIR spectra collected during S2E8R gelation (Figure 2b) showed no significant structural changes during the gelation process, especially in the 1624 cm−1 absorbance region for the presence of anti-parallel β-sheets[24]. This result suggested that the enzymatic crosslinking gelation mechanism was not based on β-sheet formation, which would have constrained long range protein chain displacements due to the crystal formation.[25] Fluorescence spectroscopy confirmed dityrosine formation by comparison of the fluorescence excitation-emission matrix (EEM) for the SELP solutions (Figure 2c) and the fully formed hydrogels (Figure 2d). These data show a distinct shift in the fluorescence maxima from the solution with an excitation of 290 nm and an emission of 350 nm to the hydrogel with a peak excitation of 300 nm with an emission at 425 nm. The shift in fluorescence indicates the formation of dityrosine covalent crosslinks during gelation.[4] Furthermore, enzymatic crosslinking of S2E8Y resulted in the formation of optically clear hydrogels (Figure 2e). The resultant hydrogel also emitted blue fluorescence when irradiated under UV due to the presence of the dityrosines, a feature not found in the precursor solution (Figure 2f,g).
2.3. Tunable Responsive Properties of SELP Dynamic Hydrogels
In order to harness the stimuli-responsive properties of the SELP hydrogels, SE8Y, S2E8Y, S4E8Y and S2E8R with concentrations from 2 to 20% were studied upon exposure to specific stimuli. Prior to the stimuli-response studies, all the SELP hydrogels were extensively extracted with deionized water to remove residual reactants and then equilibrated in deionized water, 1M NaCl or PBS buffer for 24 h at 4°C. The equilibrium swelling ratio of the hydrogels in deionized water was between 100% and 200% (Figure S5), which decreased on increasing the hydrogel concentration or the silk-to-elastin ratio, due to increased crosslinking or structural support from the silk domains. The deswelling behavior of the hydrogels in PBS buffer varied with the amino acid in the “X” position: SnE8Y hydrogels maintained constant weight with a degree of swelling about 100% in PBS, while S2E8R significantly contracted to 50% of the original weight due to osmotic pressure induced by electrostatic shielding around charged arginine residues (Figure S6). The physical stimuli responsive properties of SELP hydrogels were investigated via thermodynamic, optical, mechanical and morphological approaches. The thermo-responsive features of SE8Y, S2E8Y and S4E8Y hydrogels with different silk-to-elastin ratios were characterized by differential scanning calorimetry (DSC), UV−vis spectrophotometry, dynamic mechanical analysis (DMA), optical imaging and scanning electron microscopy (SEM) (Figure 3). DSC heat flow curves showed that 2% SE8Y and S2E8Y hydrogels exhibited LCSTs at 21 and 25°C, respectively, while the S4E8Y hydrogel did not display a thermo-response (Figure 3a). The exothermic transition during cooling and the sharp endothermic transitions in the DSC rescans during heating (Figure S7) suggested a reversible temperature transition in SE8Y and S2E8Y hydrogels. Turbidity profiles of 2% SE8Y, S2E8Y and S4E8Y hydrogels confirmed the reversible thermo-response of the SE8Y and S2E8Y hydrogels, and the static nature of the S4E8Y hydrogel (Figure 3b). The optical absorbance from 300 to 800 nm at 4 and 60°C also suggested that these SELP hydrogels changed in optical transparency upon exposure to thermal triggers: the optical profiles of SELP hydrogels showed that the 2% SELP hydrogels were almost clear with negligible absorbance above 500 nm at 4°C, except for SE8Y, and when they were exposed to an environment with a temperature above their LCSTs the optical absorbance increased and became more opaque (Figure S8). Viscoelastic properties, including the storage modulus (Figure 3c) and the loss modulus (Figure 3d) of 10% SELP hydrogels measured in water at 4, 10, 22, 37, 50 and 60°C by DMA showed an increase of hydrogel stiffness and viscosity when the temperature was above the LCST for the SE8Y and S2E8Y hydrogels. No significant change in storage modulus, but a decrease of loss modulus around 37°C, was observed for the S4E8Y hydrogels, suggesting that hydrophobic hydration[15] of the elastin domain still occurred in S4E8Y which led to a less viscose hydrogel matrix at higher temperatures. When the hydrogels were equilibrated back to 4°C after exposure to thermal triggers, the storage and loss modulus of SE8Y was higher than its original values suggesting a semi-reversible response, while the storage and loss modulus of S2E8Y and S2E8Y changed back to their original values suggesting the silk served as a structural support and facilitate the reversible mechanical response. Optical images (Figure 3e) showed that the 2% SELP hydrogels had a large thermo-responsive equilibrium deswelling ratio, except for S4E8Y. Upon exposure to thermal triggers above the LCST, 10% SELP hydrogels contracted to 13%, 64% and 84% of their original weight for SE8Y, S2E8Y and S4E8Y, respectively, depending on the silk-to-elastin ratio (Figure S9). The SEM images (Figure 3f) showed that freeze-dried SELP hydrogels were porous; in swollen states at 4°C (<LCST) the average pore sizes were about 4 µm, and in the contracted states at 37°C (>LCST) the average pore size was about 1 µm in deionized water. In summary, SE8Y and S2E8Y hydrogels showed large responsive deswelling ratios as well as significant reversible changes in optical transparency, viscoelastic properties and micro-morphology in response to environmental temperature changes across the LCST; while S4E8Y exhibited no significant thermo-response. These results suggested that the stimuli responsive properties of the SELP hydrogels were tunable by adjusting the silk-to-elastin ratios.
Figure 3.
Thermo-response of SELP dynamic hydrogels. LCST of 2% SELP hydrogels determined by (a) DSC heat flow vs. temperature curves and (b) turbidity, suggesting a silk-to-elastin ratio governed thermo-responses. Keys: SE8Y (orange curves), S2E8Y (green curves) and S4E8Y hydrogel (grey curves). The heating (solid lines) and cooling (dashed lines) rate was 2°C/min. Viscoelastic properties, including (c) storage modulus and (d) loss modulus of 10% SELP hydrogels measured in water at 4, 10, 22, 37, 50 and 60°C by DMA suggesting an increase of hydrogel stiffness and viscosity when the temperature was raised above the LCST for the SE8Y and S2E8Y hydrogels. (e) Optical and (f) SEM images showing the size and micro-morphology differences of 2% SELP hydrogels in swollen states at 4°C (<LCST) and in the contracted states at 37°C (>LCST) in deionized water.
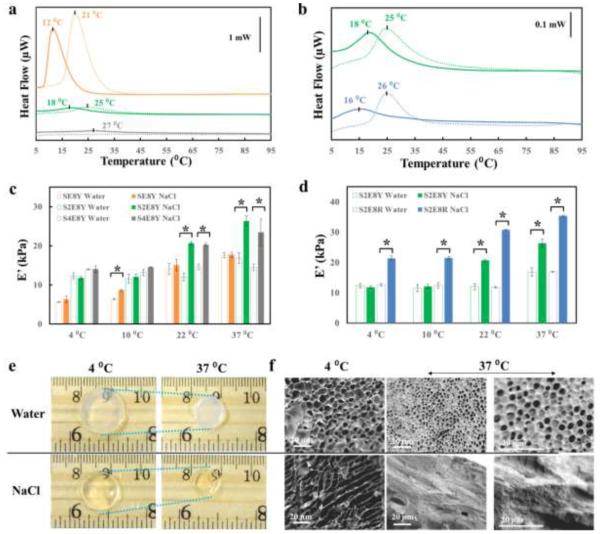
The chemical stimuli responsive properties of SELP hydrogels were also investigated via thermodynamic, optical, mechanical and morphological approaches. In particular, the ionic strength responsive features of SE8Y, S2E8Y, S4E8Y and S2E8R hydrogels with various silk-to-elastin ratios or different “X” position amino acids were characterized (Figure 4). DSC heat flow vs. temperature curves showed that changes in solvent chemical potential caused a shift in the temperature of the inverse temperature transition[20] (Figure 4a,b); when submerged in 1M NaCl, the LCST of 2% SE8Y, S2E8Y, S4E8Y and S2E8R hydrogels were 12, 18, 27 and 16°C, respectively; while when submerged in deionized water, the LCST of 2% SE8Y, S2E8Y and S2E8R were 21, 25 and 26°C, respectively, with no LCST observed for the S4E8Y. These results suggested that the SELP hydrogels exhibited significantly decreased LCST in a solvent with high ionic strength, and thus responded to the changes of solvent chemical potential in a well-defined temperature window. Particularly, SE8Y, S2E8Y, S4E8Y and S2E8R, responded to the changes in solvent from deionized water to 1M NaCl in the temperature ranges 12 - 25, 18 - 25, above 27 and 16 - 27°C, respectively. The storage modulus of 10% SELP hydrogels measured in water and 1M NaCl at 4, 10, 22 and 37°C by DMA further demonstrated that the hydrogels responded to the changes in solvent based on two mechanisms, (a) shifting the LCST or (b) deswelling, depending on the silk-to-elastin ratio and the “X” residue in the elastin block (Figure 4c,d). The DMA showed an increase stiffness for the 10% SE8Y hydrogel at 10 °C, and an increase stiffness of the 10% S2E8Y and S4E8Y hydrogels at 22 °C and above; while the 10% S2E8R hydrogel responded to solvent changes at all temperatures because of its deswelling properties in 1M NaCl. Optical images (Figure 4e) showed that upon exposure to chemical or physical triggers, the S2E8R hydrogels exhibited significant contraction. The 10% S2E8R hydrogels contracted to about 34% of the original weight when exposed to a temperature above the LCST, and 65% when submerged in 1M NaCl (Figure S10). SEM images (Figure 4f) showed that freeze-dried S2E8R hydrogels were porous in deionized water with the average pore size was about 6 µm in the swollen state at 4°C (<LCST) and about 2 µm in contracted state at 37°C (>LCST); when submerged in 1M NaCl the micro-morphology of S2E8R hydrogels changed to a layered structure with the average layer-to-layer distance of about 10 µm in the swollen state at 4°C (<LCST) and less than 1 µm in the contracted state at 37°C (>LCST). In summary, the SE8Y, S2E8Y, S4E8Y and S2E8R hydrogels showed large responsive deswelling ratios as well as significate reversible changes in viscoelastic properties and micro-morphology in response to changes in ionic strength in well-defined temperature windows, depending on silk-to-elastin ratio or the “X” residue in the elastin block.
Figure 4.
Ionic strength responsive properties of SELP dynamic hydrogels. LCST was determined by DSC heat flow vs. temperature curves for 2% SELPs, suggesting (a) a silk-to-elastin ratio and (b) the position of the “X” residue governed the chemical response. Keys: SE8Y (orange curves), S2E8Y (green curves), S4E8Y hydrogel (grey curves) and S2E8R (blue curves) in 1M NaCl (solid curves) or deionized water (dashed curves). Storage modulus of 10% SELP hydrogels was determined by DMA at 4, 10, 22 and 37°C, suggesting the response to ionic strength depended on (c) silk-to-elastin ratio and (d) position “X” residue in elastin block. (e) Optical and (f) SEM images showing the size and micro-morphology of 2% S2E8R hydrogel samples in swollen states at 4°C (<LCST) and in contracted states at 37°C (>LCST) in deionized water and 1M NaCl.
2.4. Tunable Mechanical Properties of SELP Hydrogels
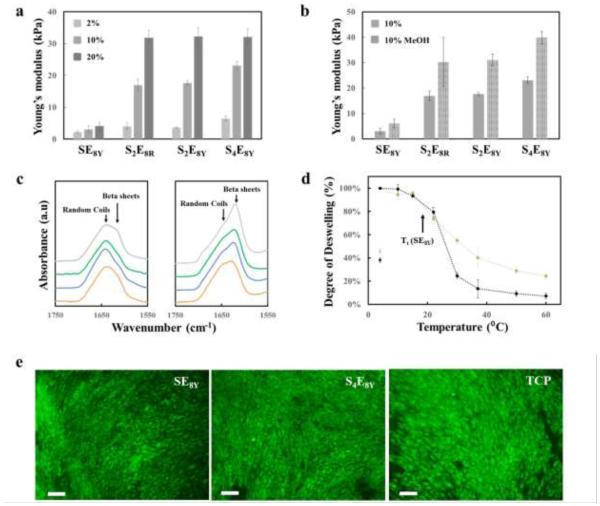
Matching mechanical properties of biomaterials to native tissue extracellular matrix is an important consideration during the selection of biomaterials for tissue engineering and regenerative medicine.[4, 26] In order to fine tune the stiffness of the SELP hydrogels, SE8Y, S2E8Y, S4E8Y and S2E8R from 2 to 20% were studied (Figure 5). The Young’s moduli in aqueous environments demonstrated a wide range of stiffnesses were achievable by adjusting the silk-to-elastin ratio or the hydrogel concentration (Figure 5a). The hydrogel stiffness could also be further reinforced by the formation of β-sheets in the silk blocks via treatment with methanol (Figure 5b). Based on the concentrations and conditions studied, stiffness values between 1 to 64 kPa were obtained, where 2% SE8Y hydrogels had the lowest stiffness and the 20% S4E8Y methanol-treated hydrogel had the highest stiffness. These values cover almost the full range of the elasticity of soft tissues.[27] FTIR absorbance at 1624 cm−1 confirmed that β-sheets were formed in the SELP hydrogels after treatment with methanol (Figure 5c), and these β-sheet crystals served as structural support to increase the stiffness of the hydrogels. The stimuli-responsive properties of the SELP hydrogels before and after methanol treatment were also investigated. In particular, the 10% SE8Y hydrogels with and without β-sheets showed similar thermo-responsive properties; the treated hydrogels had almost the same LCST as the untreated (noncrystalline) hydrogel, while a smaller degree of contraction was found for the treated (crystalline) hydrogel upon exposure to temperature triggers due to the constraints of long range protein chain displacements induced by the β-sheet crystals (e.g., physical crosslinks) (Figure 5d).
Figure 5.
Tunable mechanical properties and biocompatibility of SELP hydrogels. (a) Young’s moduli of SELP hydrogels as a function of silk-to-elastin ratio and hydrogel concentration measured in water at 4°C, demonstrating the wide range of stiffnesses achievable. (b) Young’s moduli of 10% SELP hydrogels before and after methanol treatment measured in water at 4°C, demonstrating tunable stiffness achieved by varying β-sheet content. (c) FTIR spectra of SELP hydrogels before (left) and after (right) methanol treatment, showing the increase of hydrogel stiffness originated from β-sheet formation and the degree of crystallinity induced was positively related to the silk-to-elastin ratio. (d) The responsive feature of SELP hydrogels preserved upon β-sheet formation, exemplified by SE8Y. (e) Human mesenchymal stem cell interactions on SELP hydrogel surfaces: live (green) and dead (red) cell staining on SE8Y (left), S4E8Y hydrogels (middle) and TCP (right) over a 14-day period. Scale bars are 100 µm.
2.5. Biocompatibility of SELP Hydrogels
Apart from desirable material properties, low cytotoxicity and good mammalian cell adhesion for biomaterials are important features, thus human mesenchymal stem cell (hMSCs) were grown on hydrogel surfaces and assessed by live/dead staining. Fluorescent imaging of live/dead staining confirmed that the hMSCs seeded on the SELP hydrogels showed similar adhesion as the control TCP surface and supported cell growth and proliferation for 2 weeks (Figure 5e). An elongated morphology of hMSCs were observed on all tested surfaces after 14 days suggesting a health growth of hMSCs and minimal cytotoxicity of the SELP hydrogels.
3. Discussion
Protein polymers provide a uniquely tunable family of functional biomaterials for biomedical engineering, yet creating stimuli-responsive protein hydrogels with dynamic physical properties remains challenging. This limitation is due to a limited understanding of sequence-function relationships for the rational design of protein polymers, as well as the limited methods to translate the protein folding-unfolding upon triggers at molecular level into the reversible and tunable physical property changes at macroscopic scale. To overcome these gaps between dynamic protein molecules and dynamic actuating biomaterials, a protein material system to systematic study and develop dynamic protein hydrogels is needed, for both fundamental insights and for potential biomedical applications.
The present work demonstrated the design and fabrication of a new family of biocompatible, biodegradable, elastomeric, stimuli-responsive protein hydrogels for use in tissue engineering and regenerative medicine, exploiting silk and elastin peptide motifs for the building blocks. Genetic engineering strategies were used to design a SELP library consisting of 64 different SELP constructs with 12 different monomers designs and 3 different silk to elastin ratios, inspired by prior solution studies of the coacervation phenomena of ELP and ELP copolymers. Replica Exchange Molecular Dynamics simulations (REMD) were also integrated with the experimental studies to guide the rational design of elastin domains for new complex hierarchical materials with predictable responses to specific environmental stimuli, such as temperature, pH, ionic strength and biological triggers. To fabricate dynamic hydrogels, the SELPs were designed with a tyrosine residue in the “X” position of the elastin blocks to form hydrogels via an enzymatic crosslinking reaction based on HRP. The fabrication procedure for SELP dynamic hydrogels is an all aqueous method and the resulting hydrogels SE8Y and S2E8Y showed large responsive deswelling ratios as well as significant reversible changes in optical transparency, viscoelastic properties and micro-morphology in responding to environmental temperature change across the LCST. In contrast, S4E8Y exhibited no significant thermo-response. These results suggested that the stimuli responsive properties of SELP hydrogels were tunable by adjusting the silk-to-elastin ratio of SELP molecules. SE8Y, S2E8Y, S4E8Y and S2E8R hydrogels also showed large responsive deswelling ratios as well as significant reversible changes in viscoelastic properties and micro-morphology in response to changes in ionic strength over well-defined temperature windows, depending on silk-to-elastin ratio or the “X” residue in the elastin block. These results demonstrated the importance of the guest amino acid residue in elastin blocks and silk-to-elastin ratio for the rational design of responsive SELP dynamic hydrogels.
The mechanical properties of hydrogels play important roles in modulating cell-matrix interactions, as well as regulating a variety of biological processes, such as cell proliferation and differentiation. In order to fine tune the stiffness of these new SELP hydrogels, SE8Y, S2E8Y, S4E8Y and S2E8R at various concentrations from 2 to 20% were subjected to methanol treatment to induce β-sheet formation for reinforced structures. Stiffness values between 1 to 64 kPa, which cover almost the full range of the elasticity of soft tissues, were obtained by adjusting the silk-to-elastin ratio or the hydrogel concentration. Apart from these desirable material properties, the SELP hydrogels supported hMSC survival and adhesion, suggesting minimal cytotoxicity as potential biomaterials. Additionally, the ability to trigger the responsive properties of SELP hydrogels suggests utility for delivery and release of bioactive molecules for dynamic biomaterial needs.
Molecular dynamics simulations provided insights into the mechanisms behind the transition capabilities of the elastin domains and confirmed a molecular-scale dependence on sequence, illustrated in a suppressed temperature transition upon changing the X residue within the elastin block. These results suggest that variable intramolecular structural folding at least partially contributes to the tunability of elastin-based protein systems. The effect is scaled up in large protein polymer systems such as the SELPs presented in this work. Sequence change may force a molecular-level suppression of this temperature-driven folding behavior, effectively quelling or modifying the macroscale transitions.
4. Conclusion
In conclusion, a generic design approach for dynamic, fully degradable and biocompatible protein hydrogels was developed and used for the construction of a variety of dynamic SELP hydrogels. A number of studies have demonstrated the potential of using elastin or elastin block copolymers as dynamic drug delivery systems, but the stimuli-responsive properties of these proteins were exploited only in the solution state. In the present study dynamic properties of SELPs were fully translated from molecular level folding-unfolding informed by both modeling and experimental inputs, into macroscopic reversibly tunable solid state materials. The hydrogel fabrication process included a facile approach to obtain functional elastomeric dynamic protein hydrogels via an all aqueous, enzymatic crosslinking method. Select SELP hydrogels exhibited a range of dynamic physical properties including large responsive swelling ratios as well as significant reversible changes in optical transparency, mechanical properties and micro-morphology upon exposure to designed target stimuli, dependent on the silk-to-elastin ratio or the guest amino acid residue designed in the elastin domain. Further, physical modification of the silk domains via β-sheet formation provided a secondary control point to fine-tune mechanical stiffness to match ECM while preserving stimuli-responsive features. Overall, this work systematically studies the sequence-structure-process-property relationships for producing dynamic biomaterials. These results will be useful to guide future library designs with specific targeted properties, leading to new opportunities in biomimetics, biomaterials and tissue engineering.
4. Experimental Section
Synthesis of Polymers
SELP expression plasmids were constructed using our previously established procedures.[18, 21] Briefly, DNA sequences were designed to encode the SELP sequences: SE8Y [(GAGAGS)(GVGVP)4(GYGVP)(GVGVP)3], S2E8X [(GAGAGS)2(GVGVP)4(GXGVP)(GVGVP)3] and S4E8Y [(GAGAGS)4(GVGVP)4(GYGVP)(GVGVP)3], where X was Y, or RGYSLG. The monomer DNA sequences were cloned into Eco-RV site of the vector pUC57 from GenScript (Piscataway, NJ). The BanII restriction sites were designed to flank each of the monomer oligonucleotide sequences. The monomer DNA sequences were liberated by digesting the pUC57 derivatives with BanII (New England Biolabs, Beverly, MA), isolated by preparative gel electrophoresis, and purified using the QIAquick Gel Extraction kit (Qiagen, Valencia, CA). The purified monomer DNA was then self-ligated with T4 DNA ligase (New England Biolabs, Beverly, MA) for 8 h at 16°C to yield DNA multimers. The SELP multimer genes were inserted in the tailor-made expression vector, pET-19b3, and expressed under the T7 promoter in E. coli strain BL21Star (DE3) (Invitrogen, Carlsbad, CA). The recombinant strains were grown at 37°C in 500 mL flasks containing 100 mL of Luria-Bertani medium for overnight culture in a shaking incubator at 250 rpm. A 100 mL seeding culture was transferred to 2 L of yeast extract medium and cultured at 37°C pH 6.8 using a New Brunswick BioFlo 3000 bioreactor (New Brunswick Scientific, NJ). Cells were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Sigma-Aldrich, St. Louis, MO) when the optical density at 600 nm reached approximately 10. At 6 h after induction cells were harvested by centrifugation at 8000 rpm for 15 min at 4°C. SELPs were purified using the inverse temperature cycling method (Figure S2a). Briefly, the bacterial pellet was resuspended in phosphate buffered saline (PBS) with lysozyme, and the cells were disrupted by sonication on ice. The cell lysate was cleared by centrifugation at 8,000 rpm for 15 min at 4°C, and then the supernatant containing SELP was diluted by 2X TN buffer, incubated at 65°C for 2 h, and warm centrifuged at 5,000 rpm for 3 min at 40°C. The supernatant was then discarded, and the pellet containing SELP was recovered by deionized water at 4°C followed by another cold spin at 8000 rpm for 15 min at 4°C. The supernatant containing the purified SELPs were dialyzed (MWCO 3.5 kDa) against deionized water for 2 days. The yield of SELP was approximately 1g of purified protein from per liter of culture medium. The purity of the proteins was monitored via SDS-PAGE (Figure S1b), and the molecular weights of the proteins were determined by MALDI-TOF (Bruker Corporation, Billerica, MA).
Preparation of Enzymatically Cross-linked SELP Hydrogels
The lyophilized SELP powder was dissolved in deionized water at 4°C for 4 h to form a SELP stock solution. Horseradish peroxidase (HRP) type VI lyophilized powder (Sigma-Aldrich, St. Louis, MO) was mixed with deionized water to form a 40 mg/mL HRP stock solution with a concentration of 10,000 U/mL. To fabricate a 10% SELP hydrogel, 6 µL of HRP stock solution was added to 100 µL 10% SELP stock solution, and then the crosslinking reaction of SELP was initiated by adding 0.2 µL of 30 wt% H2O2 solution to the SELP and HRP mixture with a final H2O2 concentration of 18 mM. The reaction mixture was mixed by gentle pipetting prior to gelation. The enzymatically cross-linked SELP hydrogels were formed by incubation at 4°C overnight. To fabricate 2% and 20% SELP hydrogels, the same SELP: HRP: H2O2 ratio and crosslinking reaction protocol was used for SELP gelation. An inversion test was used to qualitatively characterize the gelation time for each SELP hydrogel.
Fluorescence Spectroscopy
The excitation-emission spectra of 2% SELP solution and hydrogel was recorded using a Hitachi F4500 Spectrofluorometer (Hitachi, Schaumburg, IL). The 2% SELP solution was pipetted into a 2 mm pathlength fluorescence cuvette (Hellma USA, Plainview, NY), and the excitation-emission spectra were collected from 260 nm to 600 nm in 10 nm increments at an intensity of 700 V using a 5 nm slit width to avoid saturation. To measure the excitation-emission spectra of SELP hydrogel, 2% SELP solution with HRP and H2O2 was mixed and pipetted into the fluorescence cuvette, allowed to gel at 4°C overnight, and the excitation-emission spectrum of the hydrogel was collected by the spectrofluorimeter. The excitation-emission spectra were then processed to subtract background fluorescence from the solvent and cuvette. Fluorescence images of SELP solution and hydrogel were acquired on a Leica DM IL fluorescence microscope (Leica Microsystems Inc., Buffalo Grove, IL) with an A4S DAPI filter. Images were processed using both Leica Application Suite V3 and ImageJ.
Fourier Transform Infrared Spectroscopy
Fourier transform infrared spectroscopy (FTIR) was carried out on a Jasco (Japan) FT/IR-6200 spectrometer with a deuterated triglycine sulfate detector. To capture the secondary structure changes during SELP sol-gel transition, real-time FTIR was performed in transmission mode with a demountable FTIR liquid cell with BaF2 windows (Pike Tech., Madison, WI). The 2% SELP D2O solution with HRP and H2O2 was mixed and pipetted into the FTIR liquid cell, and the real-time FTIR absorbance spectra were obtained every 5 min till the gelation reaction completed. To access the secondary structure of SELP hydrogels, ATR-FTIR was performed in ATR mode with a horizontal MIRacle ATR attachment (Pike Tech., Madison, WI). The 10% SELP hydrogels were fabricated in D2O, and FTIR absorbance spectra of the SELP hydrogels before and after methanol treatment were obtained. For each measurement, 32 scans were co-added with a resolution of 4.0 cm−1, and the wavenumbers ranged from 400 to 4000 cm−1. The background spectra were taken under the same conditions and subtracted from the sample scans.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) measurements were performed with Nano DSC II Model 6100 (Calorimetry Sciences Corp., Lindon, UT). The 2% SELP solution with HRP and H2O2 was mixed and pipetted into the DSC sample chamber and allowed to gel at 4°C before measurements. To capture the inverse transition temperature of SELP hydrogels, the 2% SELP hydrogels were equilibrated at the initial temperature for 10 min, and then heated in the sample chamber from 0 to 100°C at a rate of 2°C/min and cooled to 0°C at the same rate. The same volume of solvent was placed in the reference chamber during each scan. The baseline scans were taken with the solvent under the same condition and subtracted from the sample scans.
UV–vis Spectrophotometry
The UV-vis spectra of SELP hydrogels were obtained by an Aviv 14DS UV-vis spectrophotometer equipped with a Peltier temperature controller (Aviv Biomedical, Lakewood, NJ). The 2% SELP solution with HRP and H2O2 was mixed and pipetted into a 1 mm pathlength absorption cuvettes (Hellma USA, Plainview, NY) and allowed to gel at 4 °C before measurements. To capture the optical profiles of the 2% SELP hydrogels, wavelength scans were performed at 4 and 60°C from 200 nm to 850 nm in 1 nm increments with an averaging time of 4 s per step. To characterize the inverse temperature transition of SELP hydrogel, temperature scans were performed at 350 nm from 0 to 60°C at a rate of 2°C/min and then cooling to 0°C at the same rate. Absorbance readings were taken after equilibrating SELP hydrogels at the desired temperature for 30 s. The averaging time of each measurement is 4 s per step. The baseline scans were taken with the solvent and cuvette under the same condition and subtracted from the sample scans.
Mechanical Tests
Dynamic mechanical analysis (DMA) studies were performed on a TA Instruments RSA3 dynamic mechanical analyzer (TA Instruments, New Castle, DE) with RSA-G2 immersion system. Cylindrical SELP hydrogels, approximately 9 mm in diameter and 4mm in thickness, were submerged in deionized water or buffers in the RSA-G2 solvent cup during DMA measurements. To assess the viscoelastic properties of the SELP hydrogels, the dynamic single point tests at a strain of 5%, with 3 different frequencies, 0.1, 1.0, and 10 Hz, were performed at 4, 10, 15, 22, 37, 50 and 60°C with an equilibrium time of 30 min per step. The storage modulus, E', which is related to the elastic deformation, and the loss modulus, E", which is related to the viscous deformation and energy absorption were measured. To measure the Young’s modulus of the SELP hydrogels, multiple extension mode tests were performed at a crosshead displacement rates of 0.01 mm/s in compression direction. The Young’s moduli were calculated by fitting the stress–strain curves in the linear response region of the SELP hydrogels (Figure S11). Each sample type was run at least three times, and average values are reported.
Swelling and Deswelling properties
The swelling ratio of the SELP hydrogels at 4°C was determined by the ratio of the weight of the hydrogel equilibrated in deionized water to the weight of corresponding as-prepared gel. For deswelling studies, the swollen samples equilibrated in deionized water at 4°C were transferred to 4°C buffer solution, and the deswelling ratio was determined by the ratio of the weight of the hydrogel in buffer to the weight of the hydrogel equilibrated in deionized water. To investigate the stimuli responsive properties, the hydrogels were equilibrated at 4, 10, 15, 22, 37, 50 and 60°C in deionized water and buffers. The equilibrium deswelling ratio upon environmental stimulus is defined as the ratio of the weight of the hydrogel under the specific stimuli to the weight of the hydrogel equilibrated in deionized water at 4°C.
Scanning Electron Microscopy
The microstructure of SELP hydrogels were imaged using a Zeiss Ultra55 SEM in the Center for Nanoscale Systems at Harvard University. SELP hydrogels were fast frozen by liquid nitrogen, freeze-dried, fractured to expose the cross-sections, and sputter coated with gold for SEM observation. Images were taken using SE2 detectors at 3.00 kV.
Cell Survival and Proliferation
Human mesenchymal stem cells (hMSC's) were isolated from fresh bone marrow aspirates (Lonza, Basel, Switzerland), cultured in Dulbecco’s Modified Eagle Medium (supplemented with 10% fetal bovine serum, 0.1 mM non-essential amino acids, 1 ng/mL bFGF, 1% antibiotic/antimycotic) and seeded at passage 2, as previously described (Altman et al., 2002). For seeding, 2% SELP hydrogels of roughly 400 μm thickness were prepared as described above and allowed to cure overnight at 4°C in fridge. Prior to seeding, hydrogels were extensively extracted with deionized water to remove residual reactants and then incubated in cell culture media for 4 h at 37°C and 5% CO2. Cells were seeded at a density of 200 cells per mm2, and allowed to adhere for 150 min prior to flooding with media. All cell culture was performed in an incubator maintained at 37°C and 5% CO2.
Cell Adherence and viability
Cell adherence, confluence of growth and viability was determined 14 days posterior to seeding, using LIVE/DEAD Viability/Cytotoxicity Kit (Life Technologies, Grand Island, NY) following the protocol recommended by manufacturer. Briefly, cells were incubated with calcein AM and ethidium homodimer-1 (EthD-1) for 60 min to stain live (green) and dead cells (red) respectively. After staining the gels were washed three times with PBS and imaged using a fluorescence microscope (Keyence BZ-X700) with excitation at 488 nm and emission at 499–537 nm for live cells and excitation at 543 and emission at 620–650 nm for dead cells.
Replica Exchange Molecular Dynamics Simulations
Protein structure identification was performed with Replica Exchange Molecular Dynamics (REMD) simulations, using the CHARMM27 all-atom energy function [28]. Replica exchange setup and analysis is carried out using Gromacs simulation software version 5.0.1 [29], 44. Visual Molecular Dynamics (VMD) [30] was used for visualization of protein structures. REMD enhances molecular dynamics (MD) simulation by combining classical MD equilibration runs with the Monte Carlo method. This allows for wide conformational space sampling, avoiding protein entrapment in local-minima free energy states via thermal stimulation through high temperature replicas. Each REMD cycle includes an equilibration phase and an exchange event. 60 replicas are used in simulation, with an initial input structure defined by an extended straight chain of amino acids. Temperatures for replicas are exponentially distributed between 280 K and 400 K. The equilibration phase is 2 ps. 5,000 exchanges, based on the Metropolis criterion, are made per replica, for a total of 600 ns simulations for all replicas. Acceptance ratios for each molecule are between 30-45%, signifying sufficient sampling. Ensemble sampling is done for the last 8 ns for each temperature replica.
Supplementary Material
Acknowledgements
We acknowledge the financial support of the NIH (U01 EB014976) and the Tissue Engineering Resource Center (NIH P41 EB002520) and the Air Force Office of Scientific Research. This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation under NSF award no. ECS-0335765. CNS is part of Harvard University.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Author Contributions: D.L.K., M.J.B. and J.Y.W. designed the research. W.H., Q.W. and X.X designed the SELP library and synthesized SELP protein-polymers. W.H fabricated the SELP hydrogel and performed the stimuli response experiments. A.T. implemented the REMD model and analysis tools, carried out the simulations and collected the data. N.D performed the cytotoxicity experiments. Y.C collected the fluorescence images of SELP hydrogels. All authors contributed to the manuscript writing.
Contributor Information
Dr. Wenwen Huang, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA
Anna Tarakanova, Laboratory for Atomistic and Molecular Mechanics (LAMM), Department of Civil and Environmental Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Dr. Nina Dinjaski, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA
Prof. Qin Wang, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA Hubei Collaborative Innovation Center for Green Transformation of Bio-Resource, College of Life Science, Hubei University, Wuhan 430062, China.
Prof. Xiaoxia Xia, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology Shanghai Jiao Tong University, 800 Dong-Chuan Road, Shanghai 200240, China.
Dr. Ying Chen, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA
Prof. Joyce Y. Wong, Departments of Biomedical Engineering and Materials Science and Engineering, Boston University, Boston, MA 02215, USA
Prof. Markus J. Buehler, Laboratory for Atomistic and Molecular Mechanics (LAMM), Department of Civil and Environmental Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Prof. David L. Kaplan, Department of Biomedical Engineering, Tufts University, 4 Colby Street, Medford, MA 02155, USA.
References
- [1].Wichterle O, Lim D. Nature. 1960;185:117. [Google Scholar]
- [2].Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Advanced Materials. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee KY, Mooney DJ. Chemical Reviews. 2001;101:1869. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- [4].Partlow BP, Hanna CW, Kovacina J. Rnjak, Moreau JE, Applegate MB, Burke KA, Marelli B, Mitropoulos AN, Omenetto FG, Kaplan DL. Advanced Functional Materials. 2014;24:4615. doi: 10.1002/adfm.201400526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schild HG. Progress in Polymer Science. 1992;17:163. [Google Scholar]
- [6].Philippova OE, Hourdet D, Audebert R, Khokhlov AR. Macromolecules. 1997;30:8278. [Google Scholar]
- [7].Schilli CM, Zhang MF, Rizzardo E, Thang SH, Chong YK, Edwards K, Karlsson G, Muller AHE. Macromolecules. 2004;37:7861. [Google Scholar]; Ren LL, Liu TX, Guo JA, Guo SZ, Wang XY, Wang WZ. Nanotechnology. 2010:21. [Google Scholar]; Xia LW, Xie R, Ju XJ, Wang W, Chen QM, Chu LY. Nature Communications. 2013:4. doi: 10.1038/ncomms3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Takashima Y, Hatanaka S, Otsubo M, Nakahata M, Kakuta T, Hashidzume A, Yamaguchi H, Harada A. Nature Communications. 2012:3. doi: 10.1038/ncomms2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stuart MAC, Huck WTS, Genzer J, Muller M, Ober C, Stamm M, Sukhorukov GB, Szleifer I, Tsukruk VV, Urban M, Winnik F, Zauscher S, Luzinov I, Minko S. Nature Materials. 2010;9:101. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- [10].Geraths C, Christen EH, Weber W. Macromolecular Rapid Communications. 2012;33:2103. doi: 10.1002/marc.201200563. [DOI] [PubMed] [Google Scholar]
- [11].Dong L, Agarwal AK, Beebe DJ, Jiang HR. Nature. 2006;442:551. doi: 10.1038/nature05024. [DOI] [PubMed] [Google Scholar]
- [12].Sidorenko A, Krupenkin T, Taylor A, Fratzl P, Aizenberg J. Science. 2007;315:487. doi: 10.1126/science.1135516. [DOI] [PubMed] [Google Scholar]; He X, Aizenberg M, Kuksenok O, Zarzar LD, Shastri A, Balazs AC, Aizenberg J. Nature. 2012;487:214. doi: 10.1038/nature11223. [DOI] [PubMed] [Google Scholar]
- [13].Ehrick JD, Deo SK, Browning TW, Bachas LG, Madou MJ, Daunert S. Nature Materials. 2005;4:298. doi: 10.1038/nmat1352. [DOI] [PubMed] [Google Scholar]; Kong N, Peng Q, Li H. Advanced Functional Materials. 2014;24:4615. doi: 10.1002/adfm.201400526. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yuan W, Yang J, Kopečková P, Kopeček J. Journal of the American Chemical Society. 2008;130:15760. doi: 10.1021/ja805634x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang W, Rollett A, Kaplan DL. Expert Opinion on Drug Delivery. 2014;12:779. doi: 10.1517/17425247.2015.989830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Urry DW, Trapane TL, Prasad KU. Biopolymers. 1985;24:2345. doi: 10.1002/bip.360241212. [DOI] [PubMed] [Google Scholar]; Urry DW. Journal of Protein Chemistry. 1988;7:1. doi: 10.1007/BF01025411. [DOI] [PubMed] [Google Scholar]; Urry DW. Journal of Protein Chemistry. 1988;7:81. doi: 10.1007/BF01025240. [DOI] [PubMed] [Google Scholar]
- [15].Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, Safavy A. Journal of the American Chemical Society. 1991;113:4346. [Google Scholar]
- [16].Urry DW, Peng SQ, Parker TM. Biopolymers. 1992;32:373. doi: 10.1002/bip.360320413. [DOI] [PubMed] [Google Scholar]
- [17].Meyer DE, Kong GA, Dewhirst MW, Zalutsky MR, Chilkoti A. Cancer Research. 2001;61:1548. [PubMed] [Google Scholar]; McDaniel JR, Weitzhandler I, Prevost S, Vargo KB, Appavou M-S, Hammer DA, Gradzielski M, Chilkoti A. Nano Letters. 2014;14:6590. doi: 10.1021/nl503221p. [DOI] [PMC free article] [PubMed] [Google Scholar]; MacEwan SR, Chilkoti A. Journal of Controlled Release. 2014;190:314. doi: 10.1016/j.jconrel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xia X-X, Wang M, Lin Y, Xu Q, Kaplan DL. Biomacromolecules. 2014;15:908. doi: 10.1021/bm4017594. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lin Y, Xia X, Wang M, Wang Q, An B, Tao H, Xu Q, Omenetto F, Kaplan DL. Langmuir. 2014;30:4406. doi: 10.1021/la403559t. [DOI] [PMC free article] [PubMed] [Google Scholar]; Megeed Z, Haider M, Li DQ, O'Malley BW, Cappello J, Ghandehari H. Journal of Controlled Release. 2004;94:433. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]; Dandu R, Megeed Z, Haider M, Cappello J, Ghandehari H. Svenson S, editor. Polymeric Drug Delivery II: Polymeric Matrices and Drug Particle Engineering. 2006;924:150. [Google Scholar]
- [18].Xia XX, Xu QB, Hu X, Qin GK, Kaplan DL. Biomacromolecules. 2011;12:3844. doi: 10.1021/bm201165h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Urry DW, Harris RD, Prasad KU. Journal of the American Chemical Society. 1988;110:3303. [Google Scholar]
- [21].Wang Q, Xia X, Huang W, Lin Y, Xu Q, Kaplan DL. Advanced Functional Materials. 2014;24:4303. doi: 10.1002/adfm.201304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Winkler S, Wilson D, Kaplan DL. Biochemistry. 2000;39:12739. doi: 10.1021/bi001335w. [DOI] [PubMed] [Google Scholar]
- [23].Lopez J. N. Rodriguez, Lowe DJ, Ruiz J. Hernandez, Hiner ANP, Canovas F. Garcia, Thorneley RNF. Journal of the American Chemical Society. 2001;123:11838. doi: 10.1021/ja011853+. [DOI] [PubMed] [Google Scholar]
- [24].Huang W, Krishnaji S, Tokareva OR, Kaplan D, Cebe P. Macromolecules. 2014;47:8107. [Google Scholar]
- [25].Matsumoto A, Chen J, Collette AL, Kim UJ, Altman GH, Cebe P, Kaplan DL. Journal of Physical Chemistry B. 2006;110:21630. doi: 10.1021/jp056350v. [DOI] [PubMed] [Google Scholar]
- [26].An B, Tang-Schomer MD, Huang WW, He JY, Jones JA, Lewis RV, Kaplan DL. Biomaterials. 2015;48:137. doi: 10.1016/j.biomaterials.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hu X, Schomer M. D. Tang, Huang W, Xia X-X, Weiss AS, Kaplan DL. Advanced Functional Materials. 2013;23:3875. doi: 10.1002/adfm.201202685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sartori S, Chiono V, Turo C. Tonda, Mattu C, Gianluca C. Journal of Materials Chemistry B. 2014;2:5128. doi: 10.1039/c4tb00525b. [DOI] [PubMed] [Google Scholar]
- [28].Lazaridis T, Karplus M. Proteins-Structure Function and Genetics. 1999;35:133. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- [29].Feig M, Karanicolas J, Brooks CL. Journal of Molecular Graphics & Modelling. 2004;22:377. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- [30].Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics & Modelling. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.