Abstract
Gametes of the unicellular green alga Chlamydomonas reinhardtii undergo sexual adhesion via enormous chimeric Hyp-rich glycoproteins (HRGPs), the plus and minus sexual agglutinins, that are displayed on their flagellar membrane surfaces. We have previously purified the agglutinins and analyzed their structural organization using electron microscopy. We report here the cloning and sequencing of the Sag1 and Sad1 genes that encode the two agglutinins and relate their derived amino acid sequences and predicted secondary structure to the morphology of the purified proteins. Both agglutinin proteins are organized into three distinct domains: a head, a shaft in a polyproline II configuration, and an N-terminal domain. The plus and minus heads are related in overall organization but poorly conserved in sequence except for two regions of predicted hydrophobic α-helix. The shafts contain numerous repeats of the PPSPX motif previously identified in Gp1, a cell wall HRGP. We propose that the head domains engage in autolectin associations with the distal termini of their own shafts and suggest ways that adhesion may involve head–head interactions, exolectin interactions between the heads and shafts of opposite type, and antiparallel shaft–shaft interactions mediated by carbohydrates displayed in polyproline II configurations.
INTRODUCTION
Sexual adhesion between gametes of the green alga Chlamydomonas reinhardtii is one of the earliest cell–cell recognition systems to have been subjected to experimental analysis. In pioneering studies, Wiese documented that the mating-type-specific components that induce mating-type plus gametic flagella to adhere to mating-type minus gametic flagella could be recovered from the culture medium (Förster and Wiese, 1954; Wiese, 1965). Subsequent analyses of such gamone preparations revealed them to contain flagellar membrane vesicles (Bergman et al., 1975; Snell, 1976; Musgrave et al., 1981), and further studies eventually led to the purification and extensive characterization of their component agglutinin proteins in both C. reinhardtii and the distantly related C. eugametos (Adair et al., 1983; Cooper et al., 1983; Collin-Osdoby et al., 1984; Adair, 1985; Goodenough et al., 1985; Collin-Osdoby and Adair, 1985; Samson et al., 1987; Homan et al., 1988; Goodenough and Adair, 1989).
The agglutinins are displayed by nitrogen-starved gametes but not by mitotic vegetative cells. The plus and minus versions from C. reinhardtii are encoded by different genes and possess complementary adhesive properties, but they also share common features: (1) they are both huge monomeric glycoproteins (native molecular mass > 1000 kD; Adair et al., 1983); (2) their association with the flagellar surface is disrupted by EDTA (Adair et al., 1982), indicating that they are extrinsic membrane proteins; (3) they are both chimeric (Kieliszewski and Lamport, 1994), possessing a large globular head and a fibrous shaft (Goodenough et al., 1985); (4) they are members of the Hyp-rich glycoprotein (HRGP) family (Cooper et al., 1983) found as well in the cell walls of most green organisms (reviewed in Cassab, 1998; Serpe and Nothnagel, 1999) and implicated in sexual interactions in higher plants (reviewed in Wu et al., 2001).
The HRGPs of the Chlamydomonas cell wall self-assemble into dense fibrous meshworks, portions of which are subsequently stabilized by covalent cross-linking and portions of which are chaotrope-soluble (Hills et al., 1975; Homer and Roberts, 1979; Goodenough et al., 1986; Goodenough and Heuser, 1988; Waffenschmidt et al., 1993, 1999; Ferris et al., 2001). Agglutinin adhesion in Chlamydomonas also results in the formation of salt-sensitive plus/minus agglutinin meshworks (Goodenough, 1986; Goodenough and Heuser, 1999). These migrate in the plane of the membrane to the flagellar tips (Goodenough, 1983) in conjunction with an adhesion-associated rise in intracellular levels of cAMP (Pasquale and Goodenough, 1987; reviewed in Pan and Snell, 2000). cAMP elevation also rapidly elicits the downstream events of the mating reaction (cell wall disassembly and mating-structure activation) that culminate in fusion between pairs of plus and minus gametes to form diploid zygotes (reviewed in Snell, 1985; Goodenough, 1991; Beck and Haring, 1996).
Genetic screens of C. reinhardtii have yielded nonagglutinating mutants in both mating types (reviewed in Goodenough et al., 1995). The three mutations that generate nonagglutinating minus strains (imp10, imp12, and agl) all map to the Sad1 (sexual adhesion) gene that resides in the mating-type minus (mt−) locus in linkage group VI (Hwang et al., 1981; Matsuda et al., 1988; Ferris et al., 2002) and are hereafter called sad1-1 to sad1-3. (A functional allele of the Sad1 gene is also located in the mt+ locus [Ferris et al., 2002] but is expressed only when plus cells are induced to differentiate as minus [Galloway and Goodenough, 1985; Ferris and Goodenough, 1997] and is not considered further in this report.) Most mutations affecting plus agglutination map to the Sag1 (sexual agglutination) locus (Goodenough et al., 1978) that does not reside in the mt locus and has recently been mapped near the centromere of linkage group VIII (S.K. Dutcher, personal communication; see Bowers et al., 2003). These mutations, formerly known as imp2, imp5, imp6, imp7, and imp9, are herein designated sag1-1 to sag1-5. An insertional allele of the gene sag1-6 is described in this article.
We report here the cloning and predicted amino acid sequences of the Sad1 and the Sag1 genes from C. reinhardtii and suggest relationships between their predicted primary and secondary structures and the morphology of purified plus and minus agglutinin proteins.
RESULTS
Identification of the Sag1 Gene Encoding Plus Agglutinin
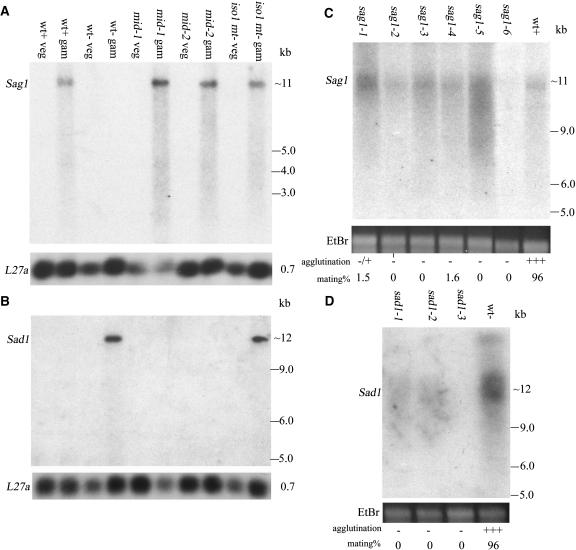
As detailed in Methods, an insertional mutation generating a nonadhesive phenotype was genetically mapped to the Sag1 locus, and DNA flanking the insertion was used to identify the wild-type Sag1 gene in genomic and cDNA libraries. By RNA gel blot analysis (Figure 1A), Sag1 probes hybridize to an ∼11-kb transcript that is absent from vegetative cells and expressed exclusively in gametes that agglutinate as plus. Included in the blot are two mt− strains, mid-1 and mid-2, that carry mutations in the mt−-localized Mid gene necessary for minus gametic differentiation (Ferris and Goodenough, 1997); both strains agglutinate as plus and express the Sag1 gene as gametes. Included also is the iso1 (isoagglutination) mt− strain wherein some cells differentiate and agglutinate as plus and some as minus (Campbell et al., 1995); Sag1 expression is again observed. No transcript is detected in the sag1-6 insertional mutant (Figure 1C), whereas the five other UV-induced mutants (sag1-1 to sag1-5) that mark the Sag1 locus all express transcripts at varying levels (Figure 1C); DNA gel blot analysis indicates that none of these strains has incurred an indel > 0.5 kb (data not shown). Because several of the sag1 mutants have been shown to lack agglutinin on their flagellar surfaces (Adair et al., 1983; Goodenough et al., 1985), they presumably carry either nonsense mutations or missense mutations that disallow proper folding or flagellar targeting.
Figure 1.
RNA Gel Blot Analysis of Wild-Type Sag1 and Sad1 Agglutinin Gene Expression.
(A) and (B) Probes are shown in Figure 2. Sag1 (A) and Sad1 (B). wt, wild type; veg, vegetative (nongametic); gam, gametic; mid-1 and mid-2, mutants in the minus sex determination gene Mid; iso1 mt−, mutant that agglutinates both as plus and minus. Rehybridization of the blots with a probe made from the L27a 60S ribosomal protein gene served as a loading control.
(C) and (D) RNA gel blot analysis of gametes carrying mutations in the Sag1 (C) and Sad1 (D) genes. The sag1-1 to sag1-5 mutants show varying levels of gene expression; the sag1-6 insertional mutant shows no signal. Sad1 expression is undetectable in the sad1-3 mutant and faint in sad1-1 and sad1-2. Loading levels are indicated by ethidium bromide (EtBr) staining of rRNA.
Identification of the Sad1 Gene Encoding Minus Agglutinin
We have cloned both the mt+ and the mt− loci and have recently subjected ∼600 kb of each to RNA gel blot analysis (Ferris et al., 2002). Probes from the −70 to −90 region of the C domain of the mt− locus (Ferris and Goodenough, 1994; Ferris et al., 2002) hybridize to an ∼12-kb message that is expressed exclusively in minus gametes (Figure 1B). As expected (see above), the iso1 mt− gametes also express the gene (Figure 1B). The mid-1 and mid-2 mutants fail to express the gene, documenting that Sad1, like other minus-specific genes, is dependent on the Mid protein for expression (Ferris and Goodenough, 1997). The sad1-1, sad1-2, and sad1-3 mutants express little if any transcript (Figure 1D); sad1-1 carries a 506-bp deletion in the proximal region of the coding sequence (Figure 2B); no indel >0.5 kb is detectable in sad1-2 and sad1-3 coding sequences (data not shown).
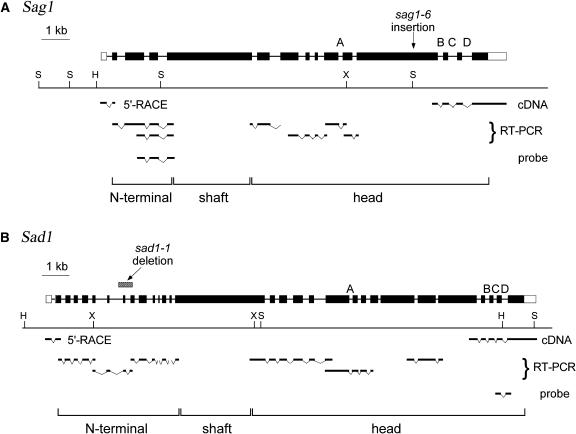
Figure 2.
Structure of the Sag1 and Sad1 Agglutinin Genes.
The center of each diagram shows a restriction map (H, HindIII; X, XhoI; S, SalI). Above the map the agglutinin gene is shown; solid boxes represent coding sequences, open boxes 5′ and 3′ untranslated regions, and thin lines introns. A to D indicate four intron positions conserved in the two genes. The location of the deletion in the sad1-1 mutant is shown, as is the location of the insertion in the sag1-6 mutant. The position of the longest cDNA, the 5′ RACE product, and the RT-PCR products that were used to determine the mRNA structure are indicated below the restriction maps, as are sequences used as probes for Figure 1. The bottom of each diagram indicates which parts of the gene encode the N-terminal, shaft, and head domains (Figure 3).
Cloning and Sequencing the Sag1 and Sad1 Genes
As detailed in Methods, sequencing the enormous Sag1 and Sad1 genes entailed sequencing genomic DNA, identifying putative open reading frames (ORFs) and introns by codon usage, sequencing available cDNAs derived from 3′ ends of the genes, and the use of RT-PCR to verify intron positions and 5′ rapid amplification of cDNA ends (RACE) to obtain N-terminal sequences (Figure 2).
Figure 2A shows the structure of the Sag1 gene, 14,776 bp in length with 15 exons, where the position of the Cry1 plasmid insertion (sag1-6) that identified the gene is indicated. The predicted mRNA encodes a protein of 3409 amino acids; with the predicted signal peptide removed, the protein would be 3349 amino acids, with a molecular mass of 330 kD. An estimate of 400 kD was proposed for the hydrogen fluoride–deglycosylated plus agglutinin (Adair et al., 1983), where the discrepancy is likely attributable both to incomplete deglycosylation and to the reduced ability of (hydroxy)proline-rich peptide domains to bind SDS (Godl et al., 1997).
Figure 2B shows the structure of the Sad1 gene, 17,843 bp in length with 30 exons; introns in similar locations to introns in the Sag1 gene are labeled A to D. The predicted mRNA encodes a protein of 3889 amino acids, or 3853 amino acids with the signal peptide removed, with a molecular mass of 385 kD. The position of the 506-bp deletion carried by the sad1-1 mutant is indicated.
General Relationships between the Plus and Minus Agglutinin Sequences and Agglutinin Protein Morphology
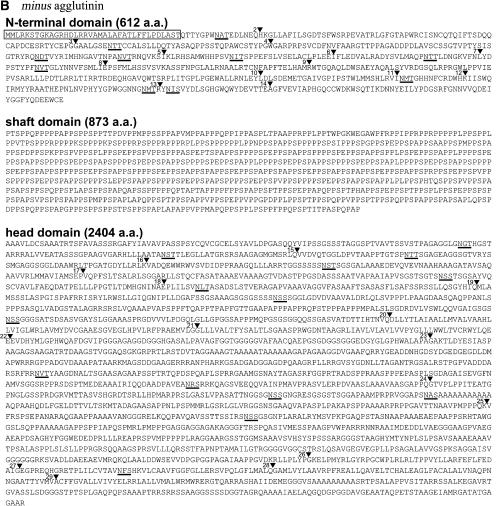
Figures 3A and 3B show the derived amino acid sequences of the plus and minus agglutinin proteins from C. reinhardtii. Three domains are evident: (1) a large C-terminal domain (2006 amino acids in plus, pI 9.71; 2404 amino acids in minus, pI 7.50) with scattered putative N-glycosylation sites (12 in plus, 14 in minus); (2) a central domain (934 amino acids in plus, pI 3.84; 873 amino acids in minus, pI 4.12) rich in P residues (60% P in both plus and minus); (3) a smaller N-terminal domain (409 amino acids in plus, pI 4.03; 576 amino acids in minus, pI 5.60, with signal peptides removed) with putative N-glycosylation sites (three in plus, 10 in minus). The locations of these domains are also indicated in Figure 2.
Figure 3.
Predicted Amino Acid Sequences of the Sag1 and Sad1 Agglutinin Proteins.
(A) Sag1.
(B) Sad1.
Numbered arrowheads indicate the locations of introns (the first intron in both sequences is in the 5′ untranslated region); putative N-glycosylation sites are underlined, and predicted signal sequences are boxed. N-terminal, shaft, and head domains are described in text. a.a., amino acids.
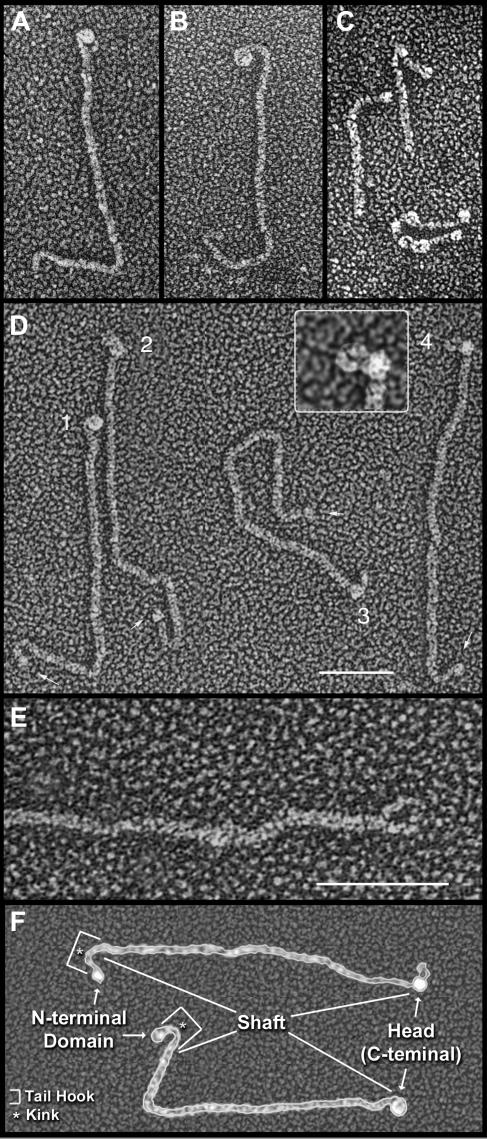
Purified agglutinin proteins from C. reinhardtii, when visualized by transmission electron microscopy (TEM) after adsorption to mica, quick-freeze deep etching (QFDE), and rotary replication with platinum (Heuser, 1983; Goodenough et al., 1985), are shown in Figures 4A (plus) and 4B (minus) and are diagrammed in Figure 4F. Each displays distinct domains: a large globular head and a long fibrous shaft that terminates in a pronounced tail hook. Images of intact flagella show that the tail-hook ends associate with the membrane surface and the heads extend outward (Goodenough et al., 1985; Goodenough and Heuser, 1999).
Figure 4.
Morphology of Agglutinins and Related Cell Wall Proteins.
(A) Plus agglutinin. Curved fibril protrudes beneath the large globular head.
(B) Minus agglutinin.
(C) Gp1 (upper) and Gp2 (lower) cell wall HRGPs. The kink in Gp1 is curved (left) or acute (right).
(D) Plus agglutinins. Arrows indicate globular domains at the proximal shaft termini. Head 1 is globular; heads 2 and 3 are bilobed; head 4 displays a protruding curved fibril, shown at higher magnification in the inset.
(E) Plus shaft showing head loop free of its globular head domain.
(F) Diagram of the agglutinins. All images generated by QFDE-TEM. Bar = 50 nm.
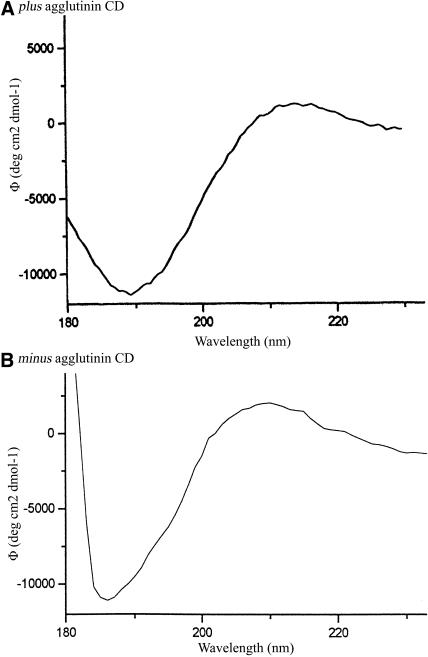
Circular dichroism (CD) spectra of purified agglutinins display the characteristics of polyproline II (PII) helices (Figure 5), with diagnostic extrema at 187 and 217 nm and molar ellipticities of −10,800 and 2000 and −11,000 and 2100 deg cm2 × dmol−1. Polypeptides adopt the PII configuration when they are rich in Pro or its posttranslational derivative, Hyp (reviewed in Creamer, 1998). Therefore, the central P-rich domains are proposed to adopt PII configurations, undergo hydroxylation and glycosylation, and self-organize as fibrous shafts.
Figure 5.
Far-UV CD Spectra of Plus and Minus Agglutinin at pH 7.
Protein concentration was 0.1 mg/mL.
The globular head of the minus agglutinin measures 11.5 ± 1.1 nm in QFDE replicas (Goodenough et al., 1985); for comparison, the globular head of the γ-heavy chain of outer-arm dynein, which possesses a hollow central core, measures 12.6 ± 0.9 nm using the same technique (Goodenough et al., 1987). The minus C-terminal domain contains 2404 amino acids; the γ-heavy chain contains 2405 amino acids. These correspondences indicate that the C-terminal domains correspond to the agglutinin heads. No significant matches to the head sequences are found in the GenBank database.
The smaller (∼500 amino acids) N-terminal domains, not recognized in our previous study (Goodenough et al., 1985; see below), are adjacent to the tail hooks and positioned to mediate the binding of agglutinins to the flagellar membrane. These sequences also lack significant GenBank matches.
Comparison of the Heads of the Plus and Minus Agglutinins
By QFDE-TEM, the heads of the minus agglutinins are invariably globular or somewhat oblate after adsorption to mica (Figure 4B; Goodenough et al., 1985). By contrast, the plus heads, although often also globular (Figures 4A and 4D, head 1), frequently appear as bilobed structures (Figure 4D, heads 2 and 3), apparently as the consequence of denaturation at the time of mica adsorption. The lobe proximal to the shaft is usually globular, whereas the distal lobe tends to splatter (Figure 4D, heads 2 and 3).
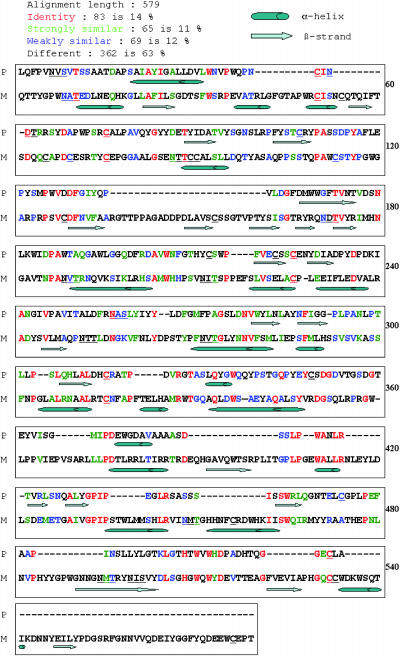
Figure 6 presents a computer-generated alignment of the plus and minus head amino acid sequences (alignment length = 2423 amino acids), wherein 23% (557 positions) are scored as identical, 13% (312 positions) strongly similar, 17% (428 positions) weakly similar, and 46% (1126 positions, many generated via indels) without similarity. Except for the α1 and α2 subdomains described below, the limited sequence similarity between the two heads is patchy and of dubious significance. Of the putative N-glycosylation sites (12 in plus, 14 in minus; see Figure 3), six are in similar locations.
Figure 6.
Sequences of the Plus and Minus Agglutinins Heads Aligned by Computer Algorithms and Marked with Predicted Regions of Secondary Structure (See Icon Key at Top Left).
Aligned amino acids are color coded (key at top left) to indicate their level of similarity. The αβ subdomains are shaded pink, followed by white P-rich subdomains, gray α1 subdomains, white long-unstructured subdomains, yellow α2 subdomains, and then white C-terminal subdomains. P, plus agglutinin heads; M, minus agglutinin heads.
Figure 6 also displays computer-predicted secondary structures of the two head domains as determined by two algorithms. Despite their low sequence similarity, the heads are organized into six comparable subdomains. (1) The αβ subdomain (aligned residues 1 to 599; see legend to Figure 6 for color coding of subdomains), adjacent to the C-terminal end of the shaft, carries scattered predicted hydrophilic α-helices and β-strands. Four C residues mark the first 50 amino acids in the shaft/αβ transition (two additional Cs are found in the minus sequence, residues 342 and 404), and half of the predicted N-glycosylation sites in the head (6/12 for plus, 7/14 for minus) are found in this subdomain. A predicted hydrophobic α-helix is found in a similar location around aligned residue 480 (asterisks). (2) The P-rich subdomain (residues 600 to 780), with strings of up to five contiguous P residues, carries two predicted N-glycosylation sites in minus and lacks C residues in both mating types. (3) The α1 subdomain (residues 781 to 905) includes long tracts of predicted hydrophobic α-helix, with two Cs in each sequence and no predicted N-glycosylation sites. (4) The long unstructured subdomain (residues 906 to 2097) displays little predicted secondary structure, carries two Cs in plus and three in minus, and contains most of the remaining predicted N-glycosylation sites (four in plus and four in minus). (5) The α2 subdomain (residues 2098 to 2265) carries long tracts of predicted hydrophobic α-helix. Most of the remaining C residues (three in plus and five in minus) also localize in α2, and there is one conserved predicted N-glycosylation site. (6) The short C-terminal subdomain (residues 2266 to 2423) is poor in predicted secondary structure and carries one predicted N-glycosylation site and one C residue in plus.
The α1 and α2 subdomains are of particular interest in that their predicted hydrophobic α-helixes are the only head sequences that display strong plus/minus sequence homology (Figure 6). Of the 80 amino acids in aligned α-helixes in α1, 38 (46%) are identical and 38 (25%) are scored as strongly similar; of the 102 amino acids in aligned α-helixes in α2, 54 (53%) are identical and 15 (15%) strongly similar. Many of the identical amino acids are found in blocks of four to eight residues.
Shafts of the Plus and Minus Agglutinins: General Considerations
The 934–amino acid shaft of the plus agglutinin contains 135 iterations of the PPSPX motif; the 873–amino acid shaft of the minus agglutinin contains 122 iterations of the PPSPX motif (Figure 3). These features place the agglutinins in the PPSPX family first defined by the cell wall protein Gp1 (Figure 4C; Ferris et al., 2001) and a gamete-specific protein of unknown function called Mta2 (Ferris et al., 2001). ORFs encoding (PPSPK)20 have also been identified in the genome of a large Chlorella virus (Eriksson et al., 1999; Van Etten, 2003).
In repetitive extensin-type HRGPs in higher plants, most of the shaft Pro are posttranslationally converted to Hyp (Kieliszewski and Lamport, 1994), and when Pro-rich peptides derived from two Volvox HRGPs were directly sequenced, the Pro residues were all found to be hydroxylated (Ertl et al., 1989, 1992). The following considerations indicate that Pro residues in the agglutinin shafts are largely if not completely hydroxylated as well. (1) Expressed as percentage of total amino acids, the purified plus agglutinin protein has been estimated to contain 12.3% Hyp and 4.4% Pro (Cooper et al., 1983). Of the 3349 amino acids in the deduced plus sequence (signal peptide removed), 560 of the 761 Pro residues reside in the shaft; hence, 16.7% of the total amino acids are predicted to be shaft-Pro and 6% non-shaft-Pro. The correspondence between these percentiles indicates that the Pro residues in the shafts are converted to Hyp. (2) By the same rubric, the purified minus agglutinin contains 12% Hyp and 6% Pro (Collin-Osdoby and Adair, 1985), and the minus agglutinin (3853 amino acids), with 526 of its 788 Pro residues in the shaft, carries 13.7% shaft-Pro and 6.8% non-shaft-Pro. The correspondences again indicate that the shaft Pro residues are converted to Hyp. (3) These correspondences are also seen in the cell wall protein Gp1, where amino acid analysis gives 14.7% Hyp/7.7% Pro (Goodenough et al., 1986), and sequence analysis gives 13.2% shaft-Pro and 7.4% non-shaft-Pro.
Although the two agglutinin shafts are different in primary sequence, five similar subdomains can be recognized (Figures 7A and 7B); these are called 2A to 2E (where the shaft is the second of the three agglutinin protein domains). Each of the central 2C subdomains contains repeating motifs of a specific PPSPX motif (as detailed below). The flanking 2B and 2D subdomains are also largely made up of PPSPX motifs, but these are not reiterated in repetitive units; each 2D subdomain also carries two runs of non-PPSPX units (brackets). The 2A subdomains, contiguous to the N-terminal domains, lack PPSPX motifs, are dominated by PPX units, and contain a high density of basic amino acids. The 2E subdomains, contiguous to the head domains, are a mix of PPX, PPSPX, and nonreiterative sequences.
Figure 7.
Subdomain Organization of the Plus and Minus Agglutinin Shafts.
(A) Plus agglutinin shaft.
(B) Minus agglutinin shaft.
Repeating PPSPX motifs are in upper-case letters; PPX motifs are in upper-case letters in subdomains 2A and 2E. Nonrepeating residues and nonconforming positions in PPSPX units are in lower-case letters. Positions judged to be missing based on the nucleotide-level analysis of minus 2C are denoted with an asterisk. Block interruptions in subdomains 2A are indicated with angle brackets (< >); long non-PPSPX sequences in subdomains 2D are indicated with brackets. Numbered rows in the 2C domains indicate endoduplicated regions based on nucleotide alignments: two duplicated regions in plus and one region in minus.
H. Tran and R. Pappu (personal communication) have analyzed the propensity of non-P residues to destabilize a PII helical configuration. By their algorithms, the block interruptions that are set off in brackets in each 2A subdomain (Figure 7) would fail to adopt the PII configuration. Their algorithms further predict that whereas the occasional presence of G, I, L, T, and V residues in the remaining shaft sequences would introduce local interruptions in helical integrity, the dominant amino acids in the sequences (A, E, K, P, Q, R, and S) are all fully compatible with PII helix formation.
The shaft domains of the agglutinin genes share two features with the shaft-encoding sequences of other HRGP-encoding genes in C. reinhardtii listed in Supplemental Table 1 online. First, they are devoid of introns, in contrast with the globular domains of these genes, which invariably contain at least one and often numerous introns. Second, the usual strong bias in C. reinhardtii toward the use of CCC or CCG as the P codon (Naya et al., 2001) is retained in the globular domains but relaxed in the shafts (see Supplemental Table 1 online). A possible interpretation of this observation is that the presence of the CCA and CCT codons renders the DNA less strongly hydrogen bonded and, hence, more readily opened up for replication and transcription; this feature may be under stronger selection in P-rich sequences than is selection for codon preference at the translational level.
The Shaft of the Plus Agglutinin: Specific Features
In our original measurements (Goodenough et al., 1985), the purified plus agglutinin was reported to be 228 ± 7 nm in length; current measurements of digitized images with computer-based measuring programs indicate that the visible plus shaft is 244.5 ± 6.2 nm (n = 17) in length. If the full predicted shaft sequence, subtracting the block interruption, were to adopt a PII helix (3.34 amino acids/nm) it would be 275 nm in length. A possible explanation for the 30-nm discrepancy is offered in a later section.
The plus shaft displays a consistent overall morphology after mica adsorption (Figures 4A and 4D): a tail hook at the nonhead terminus is followed by a more flexible region, followed by a straight region that extends to the head. In images where the head has dissociated from the shaft, a head loop can also be visualized. These regions are related to the shaft subdomains in the sections below.
The Tail Hook (Subdomain 2A)
By QFDE-TEM, the plus (and minus) shafts display a prominent kink at a conserved position, 12 ± 2 nm (n = 23) from the N terminus, whose angle ranges from acute to curved to nearly straight (Figures 4A, 4B, 4D, and 4F; Goodenough et al., 1985). As noted earlier (Figure 7), the predicted amino acid sequence of the plus shaft terminus carries a block interruption, TPVARCIQVGGICD, located at a position corresponding to 19 nm from the terminus. We therefore propose that the block interruption adopts a structural conformation that confers the shaft with the capacity to bend. In the shaft of the Gp1 cell wall protein, a sequence discontinuity, PRPPFPANTPM, also colocalizes with a morphological kink (Ferris et al., 2001), shown in Figure 4C, that has been posited to participate in cell wall assembly (Goodenough and Heuser, 1988).
The kink defines the midpoint of a terminal bend, 24.3 ± 3.7 nm (n = 23) in length, that we call the tail hook (Figure 4F), and we propose that subdomain 2A forms the tail hook, where the block interruption is flanked by a proximal (19 nm predicted) and a distal (14 nm predicted) segment. The plus 2A subdomain lacks PPSPX motifs, is dominated by PPX blocks (where X is often P), and is in general S-poor.
The Medial Region (Subdomains 2B to 2D)
The long medial region of the plus shaft is dominated by PPSPX repeats (Figure 7A). Of these, a central 2C subdomain carries iterated versions of particular repeats, whereas the flanking 2B and 2D subdomains are not reiterated.
The 2C reiterations can in some cases be recognized by repetitive use of particular X amino acids, but they are more reliably identified by the nucleotide sequences of codons. Using nucleotide-based analyses, the plus 2C sequence is found to have been generated by two sets of endoduplication events, indicated by the parentheses in Figure 7A, the first iterated three times and the second twice. Because these analyses have been conducted in the context of an ongoing comparative study of the shafts of C. reinhardtii and its sibling species C. incerta, the data generating these conclusions will be presented in a future publication.
The Head Loop (2E)
In our previous publication (Goodenough et al., 1985), we noted that the plus head occasionally denatures upon mica adsorption to reveal an underlying curved fibril (Figures 4A and 4D, head 4, magnified in the inset). When the heads are absent, a condition stimulated by protease digestion or disulfide reduction/alkylation, the fibril is seen to form a loop (Figure 4E). These head loops measure 26.3 ± 3.3 nm (n = 14) in length.
The 2E subdomain (Figure 7A) is predicted to be 25 nm in length and carries a high density of amino acids (G, L, V, and T) that are predicted to destabilize the PII helix (H. Tran and R. Pappu, personal communication) and hence might generate the propensity to curve. We therefore propose that the head loop corresponds to the 2E subdomain and suggest that the head normally assembles around the 2E sequences such that the head loop is masked from view. This could explain the 30-nm discrepancy, noted earlier, between the measured and predicted shaft length.
In platinum replicas, the agglutinin shaft proper is twice as thick (6 nm) as the head-loop shaft (3 nm) (Figure 4E). As detailed in the Discussion, these observations are consistent with the hypothesis that 2E is glycosylated differently from the bulk of the shaft.
The Shaft of the Minus Agglutinin: Specific Features
Measurements of native minus agglutinins yield a mean length of 224.9 ± 14.1 nm (n = 30) (predicted length 258 nm), with a wider range of values (202 to 248 nm) than the plus agglutinins (232 to 250 nm). Whereas, as noted above, the plus shafts display a consistent morphology, the minus shaft exits the head in a variety of configurations, and the lengths and positions of straight versus flexible segments are varied. A possible contributor to this variability is the presence of more numerous helix-disrupting amino acids (G, I, L, T, and V) in the minus (60 residues) than the plus (48 residues) sequences and their high density in the 2D subdomain of the minus sequence where the shaft emerges from the head.
The central 2C subdomain initiates with 42 PPSPX units that reiterate the sequence PPSPE PPSPA PPSPP, with eight imperfect units (Figure 7B). The next 30 PPSPX units no longer display the serial E, A, and P reiterations. However, nucleotide-based alignments, to be presented in a future publication, indicate that this region is related to the reiterated repeat, with overlapping codon usage for PPSP but relaxed usage at the X position. Although the usage of A and P residues is no longer constrained, an E residue appears in every third motif, meaning that the minus 2C sequence, like the plus 2C sequence, is strongly negatively charged.
The head of the minus agglutinin has not been observed to denature on mica, and we have not subjected minus proteins to proteolysis or reduction/alkylation. We therefore have no direct evidence that the minus 2E subdomain (predicted length 25 nm) forms a head loop like plus 2E, but propose, by virtue of the presumed common ancestry of the two proteins, that this is the case and that the minus head associates with, and covers, the 2E end of the minus shaft.
Although different in primary sequence, the 2A subdomains of the plus and minus agglutinins are similar in predicted length (33 and 42 nm), lack PPSPX motifs, are S-poor and enriched in basic amino acids, and are dominated by blocks of PPX. They both also carry block interruptions in comparable positions, but whereas the plus version (TPVARCIQVGGICD) carries two Cys and no aromatic amino acids, the minus version (TWPGKWEGAWPFR) lacks Cys and has four aromatic amino acids. As argued for the plus protein, we propose that the minus block interruption functions to generate the kink in the minus tail hook (total length of minus tail hook = 22.9 ± 2.2 nm [n = 13]).
The N-Terminal Domains of the Plus and Minus Agglutinins
In our previous study (Goodenough et al., 1985), QFDE-TEM images showed the shafts to end abruptly at the termini of the tail hooks. We were therefore surprised to encounter, in each amino acid sequence, a non-P-rich domain extending from the end of the 2A sequence to the predicted signal sequence (Figures 3A, 3B, and 8). After cleaving the signal sequences, these domains would contain 409 and 576 amino acids, respectively. Both contain putative N-glycosylation sites (three in plus and 10 in minus, with one site conserved); each is enriched in C residues (10 in plus and 16 in minus, with six conserved); their intron profiles are strikingly different (Figure 2); neither displays significant sequence homologies to one another (Figure 8), to the head domains, nor to other proteins in the GenBank database; and neither carries predicted transmembrane α-helices nor putative N-myristylation sequences.
Figure 8.
Sequences of the Plus and Minus Agglutinin N-Terminal Domains Aligned by Computer Algorithms and Marked with Predicted Regions of Secondary Structure (See Icon Key at Top Left).
Aligned amino acids are color coded (key at top left) to indicate their level of similarity. P, plus agglutinin N-terminal domains; M, minus agglutinin N-terminal domains.
Reexamination of the QFDE images reveals globular domains at the termini of tail hooks in some agglutinin preparations (Figure 4D, arrows) but not others (Figure 4A). Possible explanations for this variability include a vulnerability of this domain to proteolysis during protein purification or the possibility that the domain may be capable of forming a sheath-like association with the tail hook so that it does not appear globular.
The N-terminal domains are positioned to mediate the association of the agglutinins with the flagellar membrane, an EDTA-sensitive interaction (Adair et al., 1983) that is presumably mediated by trans-flagellar membrane agglutinin-anchoring proteins.
DISCUSSION
The Plus and Minus Agglutinin Proteins: General Considerations
We have cloned and sequenced the genes encoding the plus and minus sexual agglutinins of C. reinhardtii, the former identified by insertional mutagenesis at a site allelic to mutations that generate nonagglutinating plus sag1 mutants, the latter by RNA gel blot analysis of the cloned mt− locus, where nonagglutinating minus sad1 mutants have been mapped. Gene expression is restricted to gametes of one mating type and is disrupted in some but not all of the nonagglutinating mutants: it is undetectable in the insertional mutant sag1-6 and in the sad1-3 mutant, faint in the sad1-1 and sad1-2 mutants, and detectable to various levels in the sag1-1 to sag1-5 strains, indicating that they carry nonsense or missense mutations.
Sag1 and Sad1 transcripts continue to be present in zygotes 30 min after plus and minus gametes are mixed, but they are undetectable 1.5 h later (data not shown). This same pattern of expression has also been observed for two other gamete-specific genes, Fus1 (Ferris et al., 1996) and Mid (Ferris and Goodenough, 1997). Therefore, transcription of gamete-specific genes appears to terminate rapidly in the zygote, and the transcripts are apparently short-lived.
The sequences of the plus and minus agglutinin genes put to rest earlier speculations that the agglutinin polypeptides might be assembled posttranslationally from smaller units (Goodenough et al., 1985): each ∼12-kb transcript encodes a protein whose predicted secondary structure conforms to the morphology of the molecule visualized by electron microscopy.
Available evidence indicates that sexual adhesion in Chlamydomonas entails direct agglutinin–agglutinin interactions. If it were instead the case, for example, that the plus agglutinin recognized a second agglutinin receptor protein on the minus flagellum, one would expect that the sad1 mutants would continue to express this receptor and hence display at least weak adhesive interactions with plus gametes, which is not the case. Antibody inhibition studies with C. eugametos also indicate that the agglutinins alone participate in adhesion (Homan et al., 1988). That sexual adhesion is effected by similar proteins distinguishes Chlamydomonas and yeasts (Shen et al., 2001) from organisms wherein the sexual (glyco)proteins displayed by the eggs are unrelated to those displayed by the sperm/pollen (reviewed in Vacquier, 1998; Swanson and Vacquier, 2002).
QFDE-TEM images of sexual adhesion in Chlamydomonas show complex meshworks of fibers that disallow identification of specific protein domains (Goodenough and Heuser, 1999). However, images of HRGP assemblages in Chlamydomonas cell walls (Goodenough and Heuser, 1988) and in hammock mastigonemes (Goodenough and Heuser, 1985) have documented that shaft termini, shafts proper, shaft kinks, and globular heads all have the capacity to participate in intermolecular interactions between algal HRGPs.
Given that sexual adhesion in Chlamydomonas is a challenging interaction—flagella that are beating ∼50 times per second must establish sufficient adhesiveness to arrest flagellar movement and bring the cell bodies sufficiently close to achieve cell fusion—it follows that agglutination may entail several modes of adhesive interactions that collectively achieve this outcome. Therefore, we consider below possible ways that protein–protein, protein–carbohydrate, and carbohydrate–carbohydrate interactions may participate in sex-specific and/or species-specific recognition/adhesion events, where it is now possible to test these ideas by expressing partial gene constructs (head-only and shaft-only) and analyzing the adhesive properties of their protein products.
The Head Domains
The agglutinin heads are the most outward-facing domains of the agglutinins on the flagellar surface (Goodenough et al., 1985; Goodenough and Heuser, 1999), and they are therefore positioned to initiate the adhesion reaction. Adhesive proteins often carry internal repeats of putative adhesive motifs (Bierman, 1998; Swanson and Vacquier, 1998; Gao and Garbers, 2001; Swanson et al., 2001), but the agglutinin heads carry no discernable repeats, nor do they carry sequences with suggestive homologies to known adhesive domains.
Confounding any analysis of the heads is their remarkable, and unexplained, size. Even if the head proves to carry several functional subdomains that participate in distinct facets of a multistep adhesion process, it is difficult to imagine a role for ∼2400 amino acids, particularly because the head of its sister cell wall protein, Gp1, binds to several ligands using a mere 180 amino acids (Goodenough and Heuser, 1988; Ferris et al., 2001).
With the exception of the hydrophobic α-helix–rich regions considered below, the plus and minus heads display no significant sequence homology to one another, and whereas the minus head is invariably globular in QFDE-TEM images, the plus head is often bilobed (Figure 4D). Despite these differences, the agglutinin head domains share four intron positions (Figure 2), and their overall topological organization is homologous: six subdomains each display similar predicted secondary structures and similar endowments of C residues and N-glycosylation sites (Figure 6), indicating that both heads derive from a common, albeit distant, ancestral protein domain. Of the six head subdomains, the most N-terminal (αβ) carries the majority of the predicted N-glycosylation sites and hydrophilic α-helical domains and is predicted to be surface localized, whereas the α1 and α2 subdomains, each carrying long tracts of hydrophobic α-helix, are predicted to localize within the head interior. More than half of the amino acids reside in the long unstructured subdomains in the central portions of the sequences.
The α1 and α2 subdomains include predicted hydrophobic α-helices that are ∼50% identical in sequence between the plus and minus proteins of C. reinhardtii. These observations suggest that the α1 and α2 regions in general, and the conserved α-helical residues in particular, confer important head domain properties that are independent of, albeit perhaps necessary for, sex-specific adhesion. One possibility is that they participate in the postulated association of the heads with the head-loop subdomains of the shafts (see below), either directly or by generating a conformation that allows such associations to occur. A second possibility is that helix–helix interactions within each subdomain, and perhaps between α1 and α2, create folds necessary to carry the long unstructured subdomains.
Head–Shaft Interactions
While head–head interactions may initiate adhesion, images of fully adhered flagellar membranes show that they can be far closer than the >500 nm predicted if head–head interactions alone are involved (Goodenough and Heuser, 1999). A second possible agglutination modality, therefore, is that the heads make adhesive contacts with opposite-type shafts.
In Solanaceous plants, chimeric HRGPs (the solanaceous lectins) have been shown to recognize sugar residues in chitin (Kieliszewski et al., 1994; Van Damme et al., 2004), and the Cys-rich extensin-like proteins expressed in the flowers of Nicotiana tabacum have also been suggested to function as lectins (Wu et al., 2001). By analogy, the agglutinin heads may prove to include surface-localized lectin motifs that recognize distinctive carbohydrate moieties carried by the shafts of the opposite mating type, perhaps displayed in a recognized pattern. If these moieties are repetitive, then the heads may ratchet along the shafts during the adhesion process, abetting the apposition of interacting flagella.
Shaft Glycosylation and Shaft–Shaft Interactions
The plus and minus shafts, and the shaft of the cell wall protein Gp1, adopt the PII helical configuration (Ferris et al., 2001; Figure 5) and are dominated by the PPSPX motif over most of their lengths. The similarly wide caliber of the Gp1 and agglutinin shafts (Figures 4A to 4C) suggests that they carry sugar endowments that trap the platinum applied during rotary shadowing in a similar fashion, but given that the three shaft sequences are different from one another at the amino acid level, notably at their X positions, they may acquire distinctive patterns of sugar residues that participate in wall assembly and in sexual adhesion.
Motifs found in Hyp-rich sequences of plant proteins have been proposed to serve as glycomodules (Shpak et al., 1999; Kieliszewski, 2001), directing Hyp-glycosyltransferases to add specific sugar residues to specific Hyp residues in the polypeptide chain (Kieliszewski and Lamport, 1994). Studies identifying the sugars added to the products of synthetic glycogenes suggest two features of such a glycosylation code in higher plants: long branching sugars are found to be attached to alternating Hyp residues via O-galactosyl linkages, whereas short unbranched sugars are attached to contiguous Hyp residues via O-arabinosyl linkages (Shpak et al., 2001; Zhao et al., 2002; Tan et al., 2003, 2004).
Candidate glycomodules have not yet been identified experimentally in Chlamydomonas, but available data indicate that glycosylation patterns will also prove to be specified by the motifs found in HRGP shafts. For example, the PPSPX-rich Gp1 protein carries a complex mixture of sugar side chains, 37% of which are long and branching (Ferris et al., 2001), whereas Gp2, a second cell wall protein that lacks PPSPX motifs and carries numerous blocks of PPP and PPPP (P.J. Ferris, unpublished data), has a very different carbohydrate profile wherein only 11% of the sugars are long and branching (S. Waffenschmidt, unpublished data). Perhaps reflecting this difference, the shaft width of Gp2 (Figure 4C) is distinctly narrower than that of Gp1 (Figure 4C) or the agglutinins (Figures 4A and 4B). It follows that the unique plus and minus agglutinin shaft sequences (Figure 7) may also specify distinctive patterns of sugar addition.
Once sugars are added to such HRGP shafts, their surface topology is expected to be driven by the PII conformation. A PII helix carries three amino acids per helical gyre, meaning that a given amino acid is separated from its two neighbors by 120°; therefore, when viewed en face, each helix displays three longitudinal faces separated from one another by 120°. The amino acid sequence along each face will in most cases be different from the primary amino acid sequence of the polypeptide: for example, PPSPX repeats will generate PPPXS repeats along each face. When such a helix is hydroxylated and glycosylated, the resultant shaft is expected to carry three longitudinal rows of sugar residues, and glycosylation codes may generate distinctive complements of sugars along each face.
Glycosylation defines the interactive molecular surface of an HRGP shaft (Shpak et al., 1999), and glutaraldehyde-fixed gametic flagella continue to display mating-type-specific adhesiveness when mixed together (Goodenough, 1986). Hence, the agglutinin shafts may well associate with one another, and perhaps ratchet along one another, by making transient carbohydrate–carbohydrate interactions along their three faces. These interactions may be individually weak but additively serve to stabilize the whole, a proposal supported by images of adherent flagella that are dominated by meshworks of overlapping shafts (Goodenough and Heuser, 1999). The agglutinin shafts possess an inherent chirality because of the asymmetry of their PPPXS iterations, and they interact with one another in an antiparallel orientation during the mating reaction (Goodenough and Heuser, 1999), features that may also contribute to shaft–shaft interactions.
The shaft of the Gp1 cell wall protein carries discrete subdomains: its dominant PPSPX repeat is interrupted by a tract of poly-P and by a tract of PS repeats (Ferris et al., 2001). The agglutinin shafts are similarly heterogeneous: the terminal 2A and 2E subdomains carry distinctly different motifs from the rest of the shaft (Figure 7) and are predicted to form long faces of poly-P in the PII configuration. Hence, the 2A and 2E sequences may dictate unique glycosylation patterns and hence unique modes of protein–sugar or sugar–sugar interactions.
The 2C subdomain is of particular interest in that it is composed of repeating sets of PPSPX motifs, with the plus sets totally different from the minus. In studies to be published elsewhere, we have established that these repeats have been generated by endoduplication events. We have also shown that the plus 2C sequences from C. reinhardtii are totally different from the plus 2C sequences of its sibling species C. incerta and that the minus 2C sequences are totally different in the two species. These findings point to the 2C subdomain as a candidate carrier of species-specific adhesive information.
The N-Terminal Domains
Unmated gametes display their agglutinins with the heads facing outwards and the shaft ends associated with the flagellar membranes (Goodenough et al., 1985; Goodenough and Heuser, 1999). Membrane association is therefore presumably mediated by the globular N-terminal domains that are found at the shaft termini (Figures 4D and 8). These domains carry no predicted transmembrane nor N-myristilation sequences (nor sites for glycosylphosphatidylinositol anchors, which are added to C termini). Therefore, they presumably associate with the flagellar surface by binding to transmembrane agglutinin-anchoring proteins, a conclusion reached earlier by biochemical analysis (Adair et al., 1982).
In contrast with the heads, the plus and minus N-terminal domains are not obviously homologous to one another in intron structure, sequence, or predicted secondary structure (Figures 2, 3, and 8), indicating that if they share a common ancestral domain at all, it has undergone extensive evolutionary divergence, and suggesting that they bind to different (i.e., plus-specific and minus-specific) agglutinin-anchoring proteins.
If agglutinin heads and/or shafts indeed ratchet along opposing shafts during the adhesion reaction, the heads will eventually reach the opposite ends of their partner agglutinins. Therefore, adhesive interactions between heads and N-terminal domains may also figure in the agglutination process.
Near each N-terminal domain is a block interruption sequence in subdomain 2A of the shaft (brackets in Figure 7) that is proposed to generate the kink in the shaft tail hook (Figure 4F); the plus and minus interruption sequences are totally different from one another. Kink domains in Gp1 (Figure 4C) are positioned to participate in cell wall assembly (Goodenough and Heuser, 1988), and the block interruption sequences may play some homologous role in sexual associations.
Organization of Chimeric HRGPs
The algal HRGPs that have been characterized to date all prove to be head-shaft chimeras (Kieliszewski and Lamport, 1994). Several higher plant HRGPs display this same organization, notably proteins that participate in cell–cell interactions (lectins, AGPs, and proteins found in reproductive tissues). Because the P-rich and non-P-rich tracts in the algal and plant sequences are sharply demarcated, it has been assumed that the two domains form independent modules, and this is demonstrably the case for most regions of the agglutinins and Gp1 (Figure 4).
In addition, however, our analysis of the plus agglutinins indicates that the globular domains also have the capacity to assemble around adjacent shaft domains, analogous to a lollipop surrounding the end of its stick. When the plus head is disrupted before or upon adsorption to mica, a fibrillar structure is visible (Figure 4D, inset), which, in our previous study (Goodenough et al., 1985), we supposed to represent remnants of the denatured head. Reanalysis of such images with the protein sequence in mind suggests that the fibrillar structure is in fact the distal terminus of the shaft, corresponding to the 2E subdomain, around which the head ordinarily assembles. This structure curves back on itself to form a loop (Figure 4E), perhaps as a consequence of its endowment of amino acids that break the PII conformation (H. Tran and R. Pappu, personal communication).
The caliber of the head loop is distinctly narrower (3 nm) than the shaft proper (6 nm) (Figure 4E), but it is not as narrow as an unglycosylated PII helix (<1 nm), which would be barely visible against the mica surface (Heckman et al., 1988; Stafstrom and Staehelin, 1986). We therefore assume that the 2E sequences of the head loops are hydroxylated and glycosylated, albeit differently from the shaft proper, and that the heads then bind to these glycosylated residues, presumably in the Golgi cisternae. It follows that the plus agglutinin can in this sense be considered an autolectin, its head binding to sugars displayed by its own glycopolypeptide backbone. As posited earlier, the head may also function as an exolectin, binding to sugars displayed on opposite-type shafts.
If the globular domains of chimeric HRGPs prove to have both autolectin and exolectin capabilities, then these activities may alternate in different biological contexts. Thus, the head of an agglutinin may bind to the sugars of its own head loop and then, during the mating reaction, be induced to change conformation so as to bind to the sugars displayed on opposite-type shafts.
A more general concept emerges from these considerations. Whereas many chimeric HRGPs in algae and higher plants resemble the agglutinins and Gp1 in having a single long P-rich sequence interconnecting, and perhaps penetrating, globular N-terminal and/or C-terminal domains (Baldwin et al., 1992; Ertl et al., 1992; Rubinstein et al., 1995; Woessner and Goodenough, 1989; Waffenschmidt et al., 1993; Wu et al., 1993; Woessner et al., 1994; Godl et al., 1997; Schultz et al., 1997; Ender et al., 1999, 2002; Rodriguez et al., 1999; Bosch et al., 2001; Hallmann et al., 2001), other chimeric HRGPs have short P-rich segments that would at best form very short shafts (L. Song and W.L. Dentler, unpublished data, GenBank AF508983; Sumper and Hallmann, 1998; R.A. Bloodgood, unpublished data, GenBank AAO25117; Rodriguez et al., 1999), and others have several short P-rich sequences interspersed with several globular sequences (Cheung et al., 1993; Amon et al., 1998; Suzuki et al., 2000; Kubo et al., 2001; P. Ferris, GP2 sequence, unpublished data). Perhaps in many of these proteins, (portions of) the P-rich segments function not as protruding shafts but rather as organizational modules around which fold the globular domains. Such arrangements might also undergo biologically relevant conformational changes. In this regard, it is of interest that very short (5 to 12 residues) PII helices are present in ligands that interact with SH3 domains, WW domains, profilin, and Class II MHC proteins in animals (reviewed in Stapley and Creamer, 1999).
Sexual adhesion between flagellar membranes, although widespread in the algae (Pickett-Heaps, 1975), is by definition a strategy absent from the flagellaless higher plants. It is, however, intriguing that most of the chimeric HRGPs that have been identified in higher plants are either AGPs, implicated in numerous kinds of cell–cell interactions (reviewed in Zhao et al., 2002), or lectins (Kieliszewski et al., 1994) or are preferentially expressed in reproductive tissues (Baumberger et al., 2003). Possibly the modes of interaction displayed by present-day algal chimeras originated early in the green lineage and continue to participate in conserved facets of plant biology.
METHODS
Identification of the Sag1 Gene
Strain 21gr (mt+) was mutagenized by transformation (Kindle, 1990) with the EcoRI-linearized plasmid pJN4 carrying a dominant gene (Cry1) for emetine resistance (emeR) (Nelson et al., 1994). Emetine-resistant colonies were initially screened for their ability to form viable zygotes after mating with an mt− tester strain. Those that failed were examined microscopically to identify mutants that are flagellated, fail to show flagellar agglutination as gametes, and are capable of forming zygotes when treated with cAMP (indicating that they are not defective in differentiation into gametes; Pasquale and Goodenough, 1987).
The 99E6 mutant, which met these criteria, was crossed with an mt− strain using cAMP, and random progeny were analyzed. The mutation was shown to be linked to emeR, not linked to mt, and expressed in a mt+ background only (mt− progeny agglutinated normally), the profile expected for a sex-limited mutation (Goodenough et al., 1978). Then, 99E6 was crossed with mt− strains carrying mutations in Sag1 (sag1-1) and Sag2 (imp8), a gene that influences O-glycosylation of the plus agglutinin (Vallon and Wollman, 1995). Normally agglutinating mt+ progeny (14 out of 96 random progeny tested, where 1/8 or 12/96 are predicted by independent assortment) were recovered in the imp8 cross, whereas none (0 out of 96 progeny tested) were recovered in the cross to sag1-1, documenting Sag1 allelism.
A phage library from strain 99E6 (hereafter sag1-6) was screened with pJN4 sequences to identify the insertion-tagged locus. DNA flanking the insertion was used to identify the wild-type Sag1 gene from a second phage library. Analysis of mutant DNA identified the insertional site (Figure 2B).
Identification of the Sad1 Gene
The Sad1 gene was included in the mt− chromosome walk (Ferris and Goodenough, 1994) and identified by RNA gel blot analysis of the mt− locus (Ferris et al., 2002).
Isolation of cDNA Clones
cDNA clones for the Sag1 and Sad1 genes were identified by screening plaque lifts of a cDNA library in Uni-ZAPXR (Stratagene, La Jolla, CA) prepared from 1 h zygotic poly(A)+ RNA (Armbrust et al., 1993) and by hybridization with appropriate radiolabeled genomic probes. A library prepared from mt+ gamete poly(A)+ RNA (Kurvari et al., 1998), kindly provided by W.J. Snell, provided additional Sag1 cDNAs. Inserts from positive clones were excised as pBluescript II SK− plasmids using Stratagene's rapid excision kit.
DNA Sequencing and Analysis
DNA sequencing entailed subcloning, construction of gene-specific primers, and use of the GPS-1 genome priming system (New England Biolabs, Beverly, MA). Sequence reactions were performed with the ABI PRISM Dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) using double-stranded plasmid DNA and subsequent analysis on an ABI DNA sequencer. Addition of 5% DMSO or 1 M betaine to the sequencing reactions was often necessary. dGTP BigDye terminator cycle sequencing ready reaction mix (Applied Biosystems) was occasionally used to promote extension through particularly difficult regions. Sequence data were compiled and analyzed using the Genetics Computer Group (GCG) sequence analysis software package for VAX/VMS computers (Devereux et al., 1984). Sequences were further investigated using the National Center for Biotechnology Information BLAST program and Tmpred.
ORF Identification
Two approaches were used to identify coding regions in genomic sequences not covered by cDNA clones. (1) Sequences were analyzed using the Codon preference program from the GCG package. Because Chlamydomonas reinhardtii has highly biased codon usage (Naya et al., 2001), candidate ORFs are often identified. (2) Sequences were compared with those obtained from the Sag1 and Sad1 genes of the related species C. incerta (P. Ferris, unpublished data). Because intron sequences are far less conserved than exons, their positions can usually be readily identified. All inferred intron/exon boundaries were confirmed by RT-PCR.
RT-PCR
Approximately 108 gametes were lysed in 300 μL of lysis buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM EDTA, and 2% SDS) at room temperature for 10 min with continuous shaking. The lysate was centrifuged at 13,500 rpm for 5 min, and 200 μL of the supernatant was added to an equal amount of 2× binding buffer containing preequilibrated Dynabeads oligo(dT)25 (Dynal Biotech, Oslo, Norway). After 10 min of incubation at room temperature with continuous shaking, Dynabeads were washed twice with 100 μL of wash buffer (10 mM Tris-HCl, pH 8.0, 150 mM LiCl, 1 mM EDTA, pH 8.0, and 0.05% Triton X-100) and then washed three times with 100 μL of reverse transcription buffer (50 mM Tris acetate, pH 8.4, 75 mM potassium acetate, and 3 mM magnesium acetate). The Dynabeads were resuspended in 10 μL of DEPC-treated water containing 50 ng of random hexamer. The suspension was heated at 70° for 10 min and then immediately cooled on ice. Reverse transcription was performed in 25 μL of the reverse transcription buffer supplemented with the poly(A)+ RNA-attached Dynabeads, 10 mM DTT, 0.5 mM dNTP, 1 M betaine (Sigma-Aldrich, St. Louis, MO), and 40 units of RNaseOUT (Invitrogen, Carlsbad, CA). This reaction was incubated at 25° for 5 min, 15 units of Thermoscript RNase H− reverse transcriptase (Invitrogen) was then added, and the reaction was incubated at 25° for 10 min, 55° for 40 min, and 60° for 30 min. The reaction was terminated by incubating at 85° for 5 min. Then, 2 units of RNase H (Invitrogen) was added and incubated at 37° for 30 min. One microliter of the supernatant was used for each 50-μL PCR reaction. PCR reactions were performed by the standard protocol (Sambrook and Russell, 2001). The amplified fragments were cloned in pGEM-T vector (Promega, Madison, WI) and sequenced.
5′ RACE
Isolation of poly(A)+ RNA and reverse transcription were performed as for RT-PCR. After the RNase H treatment, cDNA was purified with a Prep-A-Gene DNA purification kit (Bio-Rad, Hercules, CA). A polyC linker was added to the 3′ end of cDNA by the terminal transferase reaction. The cDNA was resuspended in 50 μL of the buffer containing 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM DTT, 0.25 mM CaCl2, 2 mM dCTP, and 30 units of terminal transferase and incubated at 37° for 30 min. The reaction was terminated by incubating at 70° for 10 min. One microliter of resultant reaction solution was used for a 50-μL PCR reaction. The first round of PCR was performed with the primer 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′, which binds the polyC tail and a gene-specific primer predicted to be near the 5′ end of the Sad1 or Sag1 mRNA. A second nested PCR was then performed with the primer 5′-GGCCACGCGTCGACTAGTAC-3′ and then another gene-specific primer 5′ to the first primer. The resulting PCR fragments were cloned in pGEM-T vector and sequenced.
Sequence and Secondary Structure Analysis and Image Processing
Signal sequences were predicted in accordance with Nielsen et al. (1997).
Sequences were aligned using the ClustalW program (Thompson et al., 1994) located at http://npsa-pbil.ibcp.fr/ with scoring of strongly similar, weakly similar, and different amino acids according to the Gonnet matrix (Gonnet et al., 1992).
Secondary structure was predicted using two algorithms: the HNN secondary prediction method at http://npsa-pbil.ibcp.fr (Guermeur et al., 1999) and the PSIPRED method (Jones, 1999) at http://bioinf.cs.ucl.ac.uk/psipred/ (McGuffin et al., 2000). Secondary structures predicted by both methods (with a probability of five and higher by the PSIPRED method) were considered to be significant and included in the drawings and calculations.
Using the Adobe Photoshop 5.0 image processing tool kit (Mountain View, CA), the lengths of digitized images of agglutinin molecules were traced with a white line, and the length of the line was measured by the program.
CD Spectra
CD spectra were recorded on a Jasco Model J-175 (Jasco, Gross-Umstadt, Germany), calibrated using ammonium d10-camphorsulfonic acid. All measurements were performed in a heat-controlled 0.1-cm path length cylindrical cuvette at 180 to 260 nm at 25°C. Typically, 10 spectra were recorded at a scan speed of 50 nm/min with a step resolution of 0.1 nm and a nitrogen stream of 8 mL/min. All spectra were corrected for a protein-free spectrum obtained under identical conditions; noise reduction was applied according to the Jasco software.
Sequence data from this article have been deposited with EMBL/GenBank data libraries under accession numbers AY450930 and AY450929.
Supplementary Material
Acknowledgments
We thank Linda Small for expert DNA cloning/sequencing, John Heuser for the QFDE-TEM images, and Rohit Pappu for stimulating discussions of PII helices. This work was supported by grants GM-26150 from the National Institutes of Health and MCB-9904887 from the National Science Foundation to U.W.G. and P.J.F. and by Wa 659/8-1 from Deutsche Forschungsgemeinschaft to S.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ursula W. Goodenough (ursula@biology.wustl.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028035.
References
- Adair, W.S. (1985). Characterization of Chlamydomonas sexual agglutinins. J. Cell Sci. 2 (suppl.), 233–260. [DOI] [PubMed] [Google Scholar]
- Adair, W.S., Hwang, C., and Goodenough, U.W. (1983). Identification and visualization of the sexual agglutinin from mating-type plus flagellar membranes of Chlamydomonas. Cell 33, 183–193. [DOI] [PubMed] [Google Scholar]
- Adair, W.S., Monk, B.C., Cohen, R., Hwang, C., and Goodenough, U.W. (1982). Sexual agglutinins from the Chlamydomonas flagellar membrane: Partial purification and characterization. J. Biol. Chem. 257, 4593–4602. [PubMed] [Google Scholar]
- Amon, P., Haas, E., and Sumper, M. (1998). The sex-inducing pheromone and wounding trigger the same set of genes in the multicellular green alga Volvox. Plant Cell 10, 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust, E.V., Ferris, P.J., and Goodenough, U.W. (1993). A mating type-linked gene cluster expressed in Chlamydomonas participates in the uniparental inheritance of the chloroplast genome. Cell 74, 801–811. [DOI] [PubMed] [Google Scholar]
- Baldwin, T.C., Coen, E.S., and Dickinson, H.G. (1992). The ptl1 gene expressed in the transmitting tissue of Antirrhinum encodes an extensin-like protein. Plant J. 2, 733–739. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., Doesseger, B., Guyot, R., Diet, A., Parsons, R.L., Clark, M.A., Simmons, M.P., Bedinger, P., Goff, S.A., Ringli, C., and Keller, B. (2003). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and Rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, C.F., and Haring, M.A. (1996). Gametic differentiation of Chlamydomonas. Int. Rev. Cytol. 168, 259–302. [Google Scholar]
- Bergman, K., Goodenough, U.W., Goodenough, D.A., Jawitz, J., and Martin, H. (1975). Gametic differentiation in Chlamydomonas reinhardi. II. Flagellar membranes and the agglutination reaction. J. Cell Biol. 67, 606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierman, C.H. (1998). The molecular evolution of sperm bindin in six species of sea urchins (Echinoida: Strongylocentrotidae). Mol. Biol. Evol. 15, 1761–1771. [DOI] [PubMed] [Google Scholar]
- Bosch, M., Knudsen, J.S., Derksen, J., and Mariani, C. (2001). Class III pistil-specific extensin-like proteins from tobacco have characteristics of arabinogalactan proteins. Plant Physiol. 125, 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, A.K., Keller, J.A., and Dutcher, S.K. (2003). Molecular markers for rapidly identifying candidate genes in Chlamydomonas reinhardtii: ERY1 and ERY2 encode chloroplast ribosomal proteins. Genetics 164, 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, A.M., Rayala, H.J., and Goodenough, U.W. (1995). The isol gene of Chlamydomonas is involved in sex determination. Mol. Biol. Cell 6, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 281–309. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., May, B., Kawata, E.E., Gu, Q., and Wu, H. (1993). Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J. 3, 151–160. [PubMed] [Google Scholar]
- Collin-Osdoby, P., and Adair, W.S. (1985). Characterization of the purified Chlamydomonas minus agglutinin. J. Cell Biol. 101, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin-Osdoby, P., Adair, W.S., and Goodenough, U.W. (1984). Chlamydomonas agglutinin conjugated to agarose beads as an in vitro probe of adhesion. Exp. Cell Res. 150, 282–291. [DOI] [PubMed] [Google Scholar]
- Cooper, J.B., Adair, W.S., Mecham, R.P., Heuser, J.E., and Goodenough, U.W. (1983). The Chlamydomonas agglutinin is a hydroxyproline-rich glycoprotein. Proc. Natl. Acad. Sci. USA 80, 5898–5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer, T.P. (1998). Left-handed polyproline II helix formation is (very) locally driven. Proteins 33, 218–226. [PubMed] [Google Scholar]
- Devereux, J., Haeberli, P., and Smithies, O. (1984). A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender, F., Godl, K., Wenzel, S., and Sumper, M. (2002). Evidence for autocatalytic cross-linking of hydroxyproline-rich glycoproteins during extracellular matrix assembly in Volvox. Plant Cell 14, 1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender, F., Hallmann, A., Amon, P., and Sumper, M. (1999). Response to the sexual pheromone and wounding in the green alga Volvox: Induction of an extracellular glycoprotein consisting almost exclusively of hydroxyproline. J. Biol. Chem. 274, 35023–35028. [DOI] [PubMed] [Google Scholar]
- Eriksson, M., Myllyharju, J., Tu, H., Hellman, M., and Kivirikko, K.I. (1999). Evidence for 4-hydroxyproline in viral proteins. Characterization of a viral prolyl 4-hydroxylase and its peptide substrates. J. Biol. Chem. 274, 22131–22134. [DOI] [PubMed] [Google Scholar]
- Ertl, H., Hallmann, A., Wenzl, S., and Sumper, M. (1992). A novel extensin that may organize extracellular matrix biogenesis in Volvox carteri. EMBO J. 11, 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl, H., Mengele, R., Wenzl, S., Engel, J., and Sumper, M. (1989). The extracellular matrix of Volvox carteri: Molecular structure of the cellular compartment. J. Cell Biol. 109, 3493–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., Armbrust, E.V., and Goodenough, U.W. (2002). Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., and Goodenough, U.W. (1994). The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Ferris, P.J., and Goodenough, U.W. (1997). Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146, 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., Woessner, J.P., and Goodenough, U.W. (1996). A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7, 1235–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., Woessner, J.P., Waffenschmidt, S., Kilz, S., Drees, J., and Goodenough, U.W. (2001). Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40, 2978–2987. [DOI] [PubMed] [Google Scholar]
- Förster, H., and Wiese, L. (1954). Gamonewirkungen bei Chlamydomonas eugametos. Z. Naturforsch. [B] 9B, 548–550. [Google Scholar]
- Galloway, R.E., and Goodenough, U.W. (1985). Genetic analysis of mating locus-linked mutations in Chlamydomonas reinhardi. Genetics 111, 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., and Garbers, D.L. (2001). Species diversity in the structure of zonadhesin, a sperm-specific membrane protein containing multiple cell adhesion molecule-like domains. J. Biol. Chem. 273, 3415–3421. [DOI] [PubMed] [Google Scholar]
- Godl, K., Hallmann, A., Wenzl, S., and Sumper, M. (1997). Differential targeting of closely related ECM glycoproteins: The pherophorin family from Volvox. EMBO J. 16, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnet, G.H., Cohen, M.A., and Benner, S.A. (1992). Exhaustive matching of the entire protein sequence database. Science 256, 1443–1445. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W. (1983). Tipping of flagellar agglutinins by gametes of Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 25, 179–189. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W. (1986). Experimental analysis of the adhesion reaction between isolated Chlamydomonas flagella. Exp. Cell Res. 166, 237–246. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W. (1991). Chlamydomonas mating interactions. In Microbial Cell-Cell Interactions, M. Dworkin, ed (Washington, D.C.: American Society for Microbiology), pp. 71–112.
- Goodenough, U.W., and Adair, W.S. (1989). Recognition proteins of Chlamydomonas reinhardtii (Chlorophyceae). In Algae as Experimental Systems, A.W. Coleman, L.J. Goff, and J.R. Stein-Taylor, eds (New York: Alan R. Liss), pp. 171–185.
- Goodenough, U.W., Adair, W.S., Collin-Osdoby, P., and Heuser, J.E. (1985). Structure of Chlamydomonas agglutinin and related flagellar surface proteins in situ and in vitro. J. Cell Biol. 101, 924–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., Armbrust, E.V., Campbell, A.M., and Ferris, P.J. (1995). Molecular genetics of sexuality in Chlamydomonas. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 21–44. [Google Scholar]
- Goodenough, U.W., Gebhart, B., Mecham, R., and Heuser, J.E. (1986). Crystals of the Chlamydomonas reinhardi cell wall: Polymerization, depolymerization, and purification of glycoprotein monomers. J. Cell Biol. 103, 408–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., Gebhart, B., Mermall, V., Mitchell, D., and Heuser, J.E. (1987). HPLC fractionation of Chlamydomonas dynein extracts and characterization of three inner-arm dyneins. J. Mol. Biol. 194, 481–494. [DOI] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1985). The Chlamydomonas cell wall and its constituent glycoproteins analyzed by the quick-freeze deep-etch technique. J. Cell Biol. 101, 1550–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1988). Molecular organization of cell-wall crystals from Chlamydomonas reinhardtii and Volvox carteri. J. Cell Sci. 90, 717–733. [Google Scholar]
- Goodenough, U.W., and Heuser, J.E. (1999). Deep-etch analysis of adhesion complexes between gametic flagellar membranes of Chlamydomonas reinhardtii (Chlorophyceae). J. Phycol. 35, 756–767. [Google Scholar]
- Goodenough, U.W., Hwang, C., and Warren, A.J. (1978). Sex-limited expression of gene loci controlling flagellar membrane agglutination in the Chlamydomonas mating reaction. Genetics 89, 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermeur, Y., Geourjon, C., Gallinari, P., and Deleage, G. (1999). Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15, 413–421. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., Amon, P., Godl, K., Heitzer, K., and Sumper, M. (2001). Transcriptional activation by the sexual pheromone and wounding: A new gene family from Volvox encoding modular proteins with (hydroxy)proline-rich and metalloproteinase homology domains. Plant J. 26, 583–593. [DOI] [PubMed] [Google Scholar]
- Heckman, J.W., Terhune, R.T., and Lamport, D.T.A. (1988). Characterization of native and modified extensin monomers and oligomers by electron microscopy and gel filtration. Plant Physiol. 86, 848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser, J.E. (1983). Procedure for freeze-drying molecules adsorbed to mica flakes. J. Mol. Biol. 84, 560–583. [DOI] [PubMed] [Google Scholar]
- Hills, G.J., Phillips, M., Gay, M.R., and Roberts, K. (1975). Self-assembly of a plant cell wall in vitro. J. Mol. Biol. 96, 431–441. [DOI] [PubMed] [Google Scholar]
- Homan, W., Musgrave, A., de Nobel, H., Wagter, R., de Wit, D., Kolk, A., and van den Ende, H. (1988). Monoclonal antibodies directed against the sexual binding site of Chlamydomonas eugametos gametes. J. Cell Biol. 107, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer, R.B., and Roberts, K. (1979). Glycoprotein conformation in plant cell walls. Planta 146, 217–222. [DOI] [PubMed] [Google Scholar]
- Hwang, C., Monk, B.C., and Goodenough, U.W. (1981). Linkage of mutations affecting minus flagellar membrane agglutinability to the mt- mating type locus of Chlamydomonas. Genetics 99, 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J. (2001). The latest hype on Hyp-O-glycosylation. Phytochemistry 57, 319–323. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J., and Lamport, D.T.A. (1994). Extensin: Repetitive motifs, functional sites, posttranslational codes and phylogeny. Plant J. 5, 157–172. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J., Showalter, A.M., and Leykam, J.F. (1994). Potato lectin: A modular protein sharing sequence similarities with the extensin family, the hevein lectin family and snake venom disintegrins (platelet aggregation inhibitor). Plant J. 5, 849–861. [DOI] [PubMed] [Google Scholar]
- Kindle, K.L. (1990). High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, T., Saito, T., Fukuzawa, H., and Matsuda, Y. (2001). Two tandemly-located matrix metalloprotease genes with different expression patterns in the Chlamydomonas sexual cell cycle. Curr. Genet. 40, 136–143. [DOI] [PubMed] [Google Scholar]
- Kurvari, V., Grishin, N.V., and Snell, W.J. (1998). A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J. Cell Biol. 143, 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, Y., Saito, T., Umemoto, T., and Tsubo, T. (1988). Transmission patterns of chloroplast genes after polyethylene glycol-induced fusion of gametes in non-mating mutants of Chlamydomonas reinhardtii. Curr. Genet. 14, 53–58. [Google Scholar]
- McGuffin, L.J., Bryson, K., and Jones, D.T. (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Musgrave, A., van Eijk, E., te Welscher, R., Broekman, R., Lens, P., Homan, W., and van den Ende, H. (1981). Sexual agglutination factor from Chlamydomonas eugametos. Planta 153, 362–369. [DOI] [PubMed] [Google Scholar]
- Naya, H., Romaro, H., Carels, N., Zavala, A., and Musto, H. (2001). Translational selection shapes codon usage in the GC-rich genome of Chlamydomonas reinhardtii. FEBS Lett. 501, 127–130. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A.E., Savereide, P.B., and Lefebvre, P.A. (1994). The CRY1 gene in Chlamydomonas reinhardtii: Structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14, 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Pan, J., and Snell, W.J. (2000). Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 3, 596–602. [DOI] [PubMed] [Google Scholar]
- Pasquale, S.M., and Goodenough, U.W. (1987). Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 105, 2279–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps, J.D. (1975). Green Algae: Structure, Reproduction and Evolution in Selected Genera. (Sunderland, MA: Sinauer).
- Rodriguez, H., Haring, M.A., and Beck, C.F. (1999). Molecular characterization of two light-induced, gamete-specific genes from Chlamydomonas reinhardtii that encode hydroxyproline-rich glycoproteins. Mol. Gen. Genet. 261, 267–274. [DOI] [PubMed] [Google Scholar]
- Rubinstein, A.L., Broadwater, A.H., Lowrey, K.B., and Bedinger, P.A. (1995). Pex1, a pollen-specific gene with an extensin-like domain. Proc. Natl. Acad. Sci. USA 92, 3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Samson, M.R., Klis, F.M., Homan, W.L., van Egmond, P., Musgrave, A., and van den Ende, H. (1987). Composition and properties of the sexual agglutinins of the flagellated green alga Chlamydomonas eugametos. Planta 170, 314–321. [DOI] [PubMed] [Google Scholar]
- Schultz, C.J., Hauser, K., Lind, J.L., Atkinson, A.H., Pu, Z., Anderson, M.A., and Clarke, A.E. (1997). Molecular characterisation of a cDNA sequence encoding the backbone of a style-specific 120 kDa glycoprotein which has features of both extensins and arabinogalactan proteins. Plant Mol. Biol. 35, 833–845. [DOI] [PubMed] [Google Scholar]
- Serpe, M.D., and Nothnagel, E.A. (1999). Arabinogalactan-proteins in the multiple domains of the plant cell surface. Adv. Bot. Res. 30, 207–289. [Google Scholar]
- Shen, Z.M., Wang, L., Pike, J., Jue, C.K., Zhao, H., de Nobel, H., Kurjan, J., and Lipke, P.N. (2001). Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule α-agglutinin. J. Biol. Chem. 276, 15768–15775. [DOI] [PubMed] [Google Scholar]
- Shpak, E., Elisar, B., Leykan, J.F., and Kieliszewski, M.J. (2001). Contiguous hyrdoxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278. [DOI] [PubMed] [Google Scholar]
- Shpak, E., Leykam, J.F., and Kieliszewski, M.J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation sites. Proc. Natl. Acad. Sci. USA 96, 14736–14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, W.J. (1976). Mating in Chlamydomonas: A system for the study of specific cell adhesion. I. Ultrastructural and electrophoretic analyses of flagellar surface components involved in adhesion. J. Cell Biol. 68, 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, W.J. (1985). Cell-cell interactions in Chlamydomonas. Annu. Rev. Plant Physiol. 36, 287–315. [Google Scholar]
- Stafstrom, J.P., and Staehelin, L.A. (1986). The role of carbohydrate in maintaining extensin in an extended conformation. Plant Physiol. 81, 242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley, B.J., and Creamer, T.P. (1999). A survey of left-handed polyproline II helices. Protein Sci. 8, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumper, M., and Hallmann, A. (1998). Biochemistry of the extracellular matrix of Volvox. Int. Rev. Cytol. 180, 51–85. [DOI] [PubMed] [Google Scholar]
- Suzuki, L., Woessner, J.P., Uchida, H., Kuroiwa, H., Yuasa, Y., Waffenschmidt, S., Goodenough, U.W., and Kuroiwa, T. (2000). A zygote-specific protein with hydroxyproline-rich glycoprotin domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 36, 571–583. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., and Vacquier, V.D. (1998). Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281, 710–712. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., and Vacquier, V.D. (2002). The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W.J., Yang, Z., Wolfner, M.F., and Aquadro, C.F. (2001). Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 98, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., Leykam, J.F., and Kieliszewski, M.J. (2003). Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 132, 1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., Qiu, F., Lamport, D.T.A., and Kieliszewski, M.J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier, V. (1998). Evolution of gamete recognition proteins. Science 281, 1995–1998. [DOI] [PubMed] [Google Scholar]
- Vallon, O., and Wollman, F.-A. (1995). Mutations affecting O-glycosylation in Chlamydomonas reinhardtii cause delayed cell wall degradation and sex-limited sterility. Plant Physiol. 108, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, E.J.M., Barre, A., Rouge, P., and Peumans, W.J. (2004). Potato lectin: An updated model of a unique chimeric plant protein. Plant J. 37, 34–45. [DOI] [PubMed] [Google Scholar]
- Van Etten, J.L. (2003). Unusual life style of giant Chlorella viruses. Annu. Rev. Genet. 37, 153–195. [DOI] [PubMed] [Google Scholar]
- Waffenschmidt, S., Kusch, T., and Woessner, J.P. (1999). A transglutaminase immunologically related to tissue transglutaminase catalyzes cross-linking of cell wall proteins in Chlamydomonas reinhardtii. Plant Physiol. 121, 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waffenschmidt, S., Woessner, J.P., Beer, K., and Goodenough, U.W. (1993). Evidence that isodityrosine crosslinking mediates the insolubilization of cell-wall HRGPs in Chlamydomonas. Plant Cell 5, 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, L. (1965). On sexual agglutination and mating-type substances (gamones) in isogamous heterothallic Chlamydomonads. I. Evidence of the identity of the gamones with the surface components responsible for sexual flagellar contact. J. Physiol. 1, 46–54. [Google Scholar]
- Woessner, J.P., and Goodenough, U.W. (1989). Molecular characterization of a zygote wall protein: An extensin-like molecule in Chlamydomonas reinhardtii. Plant Cell 1, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner, J.P., Molendijk, A.J., van Egmond, P., Klis, F.M., Goodenough, U.W., and Haring, M.A. (1994). Domain conservation in several volvocalean cell wall proteins. Plant Mol. Biol. 26, 947–960. [DOI] [PubMed] [Google Scholar]
- Wu, H., de Graaf, B., Mariani, C., and Cheung, A.Y. (2001). Hydroxyproline-rich glycoproteins in plant reproductive tissues: Structure, functions and regulation. Cell. Mol. Life Sci. 58, 1418–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Zou, J., May, B., Gu, Q., and Cheung, A.Y. (1993). A tobacco gene family for flower cell wall proteins with a proline-rich domain and a cysteine-rich domain. Proc. Natl. Acad. Sci. USA 90, 6829–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z.D., Tan, L., Showalter, A.M., Lamport, D.T.A., and Keiliszewski, M.J. (2002). Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J. 31, 431–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.