Abstract
We previously developed a Deterministic Lateral Displacement (DLD) microfluidic method in silicon to separate cells of various sizes from blood (Davis et al., Proc Natl Acad Sci 2006;103:14779-14784; Huang et al., Science 2004;304:987-990). Here, we present the reduction-to-practice of this technology with a commercially produced, high precision plastic microfluidic chip-based device designed for automated preparation of human leukocytes (white blood cells; WBCs) for flow cytometry, without centrifugation or manual handling of samples. After a human blood sample was incubated with fluorochrome-conjugated monoclonal antibodies (mAbs), the mixture was input to a DLD microfluidic chip (microchip) where it was driven through a micropost array designed to deflect WBCs via DLD on the basis of cell size from the Input flow stream into a buffer stream, thus separating WBCs and any larger cells from smaller cells and particles and washing them simultaneously. We developed a microfluidic cell processing protocol that recovered 88% (average) of input WBCs and removed 99.985% (average) of Input erythrocytes (red blood cells) and >99% of unbound mAb in 18 min (average). Flow cytometric evaluation of the microchip Product, with no further processing, lysis or centrifugation, revealed excellent forward and side light scattering and fluorescence characteristics of immunolabeled WBCs. These results indicate that cost-effective plastic DLD microchips can speed and automate leukocyte processing for high quality flow cytometry analysis, and suggest their utility for multiple other research and clinical applications involving enrichment or depletion of common or rare cell types from blood or tissue samples.
Key terms: microfluidic, cell processing, white blood cells, leukocytes, blood, red blood cells, deterministic lateral displacement, sample preparation, cell sorting
As multiparameter flow cytometry becomes increasingly more valuable and widely used in research and clinical diagnostic testing for many diseases (1-8), the need for new methods to improve the efficiency of sample preparation becomes increasingly critical. Currently, cell membrane and intracellular labeling of cells for multiparameter flow cytometry is a labor- and time-intensive process that involves substantial cell losses, especially using protocols that require samples to be washed multiple times by centrifugation to maximize analytic quality (9). Since each cell wash step may result in a loss of ∼10–15% of the cells (10,11), multiple cell washes during processing for flow cytometry are not only time-consuming, requiring 30 min or more, but also obligate significant overall cell loss and sometimes preferential loss of specific cell types (12). Cell losses during processing necessitate larger starting sample volumes, an especially critical problem in clinical testing of small children and patients who need many blood tests (13–15) and in laboratory research experiments where samples are often unique or of limited volume, such as in small animal studies. In addition, the manual nature of these steps may lead to significant intraoperator and interoperator variability. Finally, improved, automated methods of sample preparation would be valuable for multiple types of analysis beyond flow cytometry, such as genetic and genomic testing, and thus several approaches to take advantage of microfluidic processes for cell sample preparation have been reported (16–18).
Our previous research using silicon devices produced in small numbers in our research laboratories has shown that Deterministic Lateral Displacement (DLD) microfluidic chip (microchip)-processing technology harvests cells from a flow of fluid purely on the basis of cell size (19,20). The DLD approach involves pumping blood through a microfluidic device containing a specifically designed array of microposts that is tilted at a small angle from the direction of the fluid flow. Cells larger than the target size of the micropost array were gently deflected (“bumped”) by the microposts into a stream of buffer, a process that was non-injurious to the cells. This study presents the reduction-to-practice of this approach for clinical applications requiring inexpensive, commercially manufactured, high precision, disposable plastic DLD microchips. These DLD microchips provided rapid (<20 min), automatable, noncentrifugal washing of leukocytes (white blood cells; WBCs) together with depletion of erythrocytes (red blood cells; RBCs) and unbound monoclonal antibodies (mAbs) from small samples of unprocessed human blood, with high viability and recovery of all of the major WBC subsets for high-quality flow cytometry analyses.
Materials and Methods
Microchip Design
In the single channel plastic microchip used in this study, the DLD array consisted of three subarrays of increasingly smaller posts and gaps, and thus critical sizes (Fig. 1A). The cells first enter zone 1 containing 24 μm-diameter posts separated by 18 μm gaps (∼8 μm critical diameter), followed by zone 2 (16 μm posts, 12 μm gaps, ∼5.5 μm critical diameter), and then zone 3 (10 μm posts, 9 μm gaps, ∼4 μm critical diameter). To increase throughput, two micropost arrays were “mirrored” together (Fig. 1B); that is, two micropost arrays were fabricated next to each other, with their tilt axes reversed, so that each micropost array would bump WBCs toward the central bypass channel containing the stream of run buffer terminating in a single Output port. The overall mirror array length and width were 37 and 1.8 mm, respectively.
Figure 1.
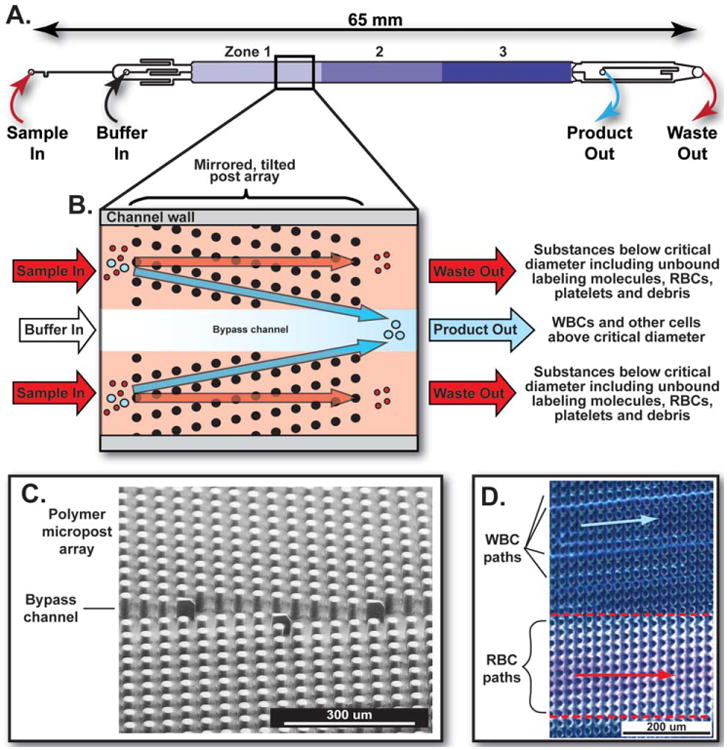
Design of the DLD microfluidic chips used in this project. A: Enlarged view of the DLD microchip array. The full length of the array and microchannel is 65 mm and the width is 1.8 mm. The sample enters the device through the “Sample In” Port, and is directed via a split channel to the outside edges of the array. The run buffer enters the “Buffer In” port and flows down the center of the array. “Product Out” and “Waste Out” ports are used to collect the Product and Waste, respectively. B: Schematic diagram of the mirrored-design DLD microchip to wash and enrich immunostained WBCs: Run buffer enters the left of the microchip via the middle input channel (Buffer In) between the two sample input channels (Sample In) and flows down the central bypass channel toward the middle output channel on the right (Product Out). All fluid streams flow parallel to the channel walls, without turbulent mixing. The blood cell sample, previously incubated with labeled mAbs in buffer, enters the micropost array from the left via duplicate Input channels (Sample In) on the left top and bottom of the micropost array and flows toward the DLD micropost array. The micropost array gently bumps the large cells (in this case, WBCs) from the two Input streams into the central buffer stream and toward the Product Out channel. The smaller cells (in this case, including RBCs and platelets) are not bumped and continue to follow the Input fluid flow direction toward the Waste Out channel. C: Scanning electron microscopic image of an actual plastic mirrored DLD micropost array chip fabricated for this project. Note that the pillars have straight walls without taper. Aspect ratios (diameter of pillars divided by the height) were as high as 5 for this microchip design. D: Enlarged view of the indicated portion of the DLD microchip during use. In this time-lapse image, the path of each immunostained WBC appears as a light blue streak that proceeds along the tilt angle of the micropost array toward the central bypass channel and Product Out port. The pinkish color of the Input stream results from multiple hemoglobin-containing RBCs following the same paths, due to the excess of RBCs over WBCs in blood.
Microchip Fabrication
A silicon master for the plastic DLD microchip was made using standard photolithographic and deep reactive ion etching techniques (21) (A. M. Fitzgerald, Burlingame, CA). The silicon master was then transferred to a soft elastomeric mold (Edge Embossing, Medford, MA). The elastomer was peeled off to create a negative imprint of the silicon master; this soft elastomeric mold was then used to emboss ≥100 plastic chips as follows: using a combination of pressure and temperature, the plastic was extruded into the features (wells) of the soft-elastomer negative mold, replicating the features and depth of the original silicon master. The soft tool was then peeled off from the plastic device, producing a flat piece of plastic surface-embossed to a depth of 60 μm with a pattern of flow channels and trenches around an array of microposts (Fig. 1C). Ports were created for fluidic access to the Input and Output ends of the microchip. After batch cleaning by sonication, the devices were batch lidded with a heat-sensitive, hydrophilic adhesive (ARFlow Adhesives Research, Glen Rock, PA).
Microchip Operation
The microfluidic device was assembled inside a manifold with fluidic connections. Fluids were driven through the DLD microchip using pneumatic pressure (MFCS-EZ, Fluigent, Lowell, MA). The flow path for the buffer line included an in-line degasser (Biotech DEGASi, Minneapolis, MN). The microchip was first primed at low pressure with run buffer [Ca2+- and Mg2+-free phosphate buffered saline (PBS), Mediatech, Manassas, VA] containing 5 μM EDTA (Sigma Aldrich, St. Louis, MO) and either 1% bovine serum albumin (BSA; MP Biomedicals, Santa Ana, CA) or 1% poloxamer (Kolliphor P-188, Sigma Aldrich), as noted. The system was then brought to standard operating pressure and flushed with run buffer for 15 min. The system was depressurized, and the sample loaded into the sample Input port. The sample and buffer Input containers were repressurized to drive samples through the microchip, and the Product and Waste were collected at their Output ports. After the complete sample volume was processed (<20 min), either an air plug or additional buffer was used to flush the chip, as noted. The Product Output volume was similar to the Sample Input volume.
Blood Samples
Anticoagulated normal human donor blood samples for all experiments were obtained with informed consent following FDA guidelines, one day prior to all experiments except that blood was obtained on the day of Experiment 6 of Table 3.
Table 3.
DLD microchip processing of immunostained unlysed blood using optimized buffer resulted in high recovery of washed WBCs with almost 10,000-fold depletion of RBCs.
| Experimenta | Fraction Analyzed |
Volume (μL)b | RBC Count (× 106 Cells/μL) | %RBC Depleted |
%RBC Depleted Flow Cytometryc | WBC Count (× 103 Cells/μL)d | Total % WBC Recoverye | % Granulocytesf | % Monocytesg | % Lymphocytesh | % B-Cellsi | % T-Cellsj | Run Time (Mins) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| With Optimized Buffer | |||||||||||||
| 1 | Input | 200 | 2.66 | – | – | 4.0 | 87 +/− 5% | 54.0 | 5.3 | 40.7 | 9.3 | 75.5 | 21.0 |
| Product | 289 | 0.01 | 99.62% | 99.993% | 2.4 | 52.6 | 14.5 | 32.9 | 11.9 | 77.1 | |||
| 2 | Input | 200 | 2.01 | – | – | 3.2 | 88 +/− 6% | 47.6 | 9.8 | 42.8 | 9.0 | 85.9 | 16.0 |
| Product | 255 | 0.00 | 100.00% | 99.974% | 2.2 | 52.2 | 10.4 | 38.2 | 10.3 | 87.3 | |||
| 3 | Input | 200 | 2.45 | – | – | 2.6 | 78 +/− 7% | 60.3 | 8.6 | 30.0 | 15.7 | 67.3 | 17.0 |
| Product | 240 | 0.00 | 100.00% | 99.997% | 1.7 | 64.4 | 7.4 | 29.1 | 17.0 | 63.8 | |||
| 4 | Input | 200 | 2.23 | – | – | 4.7 | 96 +/− 4% | 60.3 | 9.3 | 30.3 | 13.0 | 79.3 | 18.0 |
| Product | 281 | 0.01 | 99.55% | 99.993% | 3.2 | 61.8 | 8.2 | 28.4 | 14.6 | 74.9 | |||
| 5 | Input | 200 | 2.06 | – | – | 2.0 | 81 +/− 9% | 42.3 | 7.9 | 46.9 | 9.2 | 77.0 | 18.0 |
| Product | 248 | 0.00 | 100.00% | 99.961% | 1.3 | 40.0 | 10.5 | 47.4 | 8.8 | 77.8 | |||
| 6 | Input | 200 | 2.19 | – | – | 2.1 | 98 +/− 8% | 53.6 | 7.3 | 38.4 | 9.4 | 64.3 | 20.0 |
| Product | 241 | 0.00 | 100.00% | 99.991% | 1.7 | 51.2 | 8.0 | 39.8 | 9.0 | 65.6 | |||
| Mean Depletion or Recovery or Run Time | 99.8 + /− 0.09% | 99.985+/− 0.006% | 88 +/− 3% | 18.3+/-0.8 | |||||||||
| Mean Difference {Product vs. Input) | 0.7+/− 1.3% | 1.8 +/− 1.6% | −2.2+/− 1.4% | 1.0 +/− 0.5% | −0.5 +/− 1.1% | ||||||||
| Control with BSA Buffer | |||||||||||||
| 1 | Input | 200 | 2.68 | – | – | 3.8 | 77 +/− 5% | 54.0 | 5.3 | 40.7 | 9.3 | 75.5 | 22.0 |
| Product | 278 | 0.01 | 99.63% | 99.991% | 2.1 | 53.0 | 11.9 | 35.1 | 5.5 | 82.0 | |||
“Control with BSA Buffer” Experiment 1 used BSA buffer and the same donor blood sample, and was run at the same time as Experiment 1 “with Optimized Buffer”. Experiments 1 – 6 with “Optimized Buffer” used the buffer containing Polyoxamer instead of BSA.
%RBC depletion was determined using relative ratios of RBC/WBC for both the Coulter Counter and flow cytometry approaches.
Granulocytes were identified by flow cytometry as DRAQ5+ signals with characteristic high FSC and SSC.
Monocytes were identified as DRAQ5+ with characteristic intermediate FSC and SSC.
Lymphocytes were identified as DRAQ5+ with characteristic low FSC and SSC.
B cells were identified as DRAQ5+/CD19+ with FSC and SSC of lymphocytes.
T cells were identified as DRAQ5+/CD3+ with FSC and SSC of lymphocytes.
Immunostaining and Flow Cytometry
For most experiments, diluted whole blood samples (i.e., 125 μL blood filtered through a 20 μm pore size filter plus 125 μL run buffer) or RBC-lysed preparations of WBCs were incubated (15 min, RT, in the dark) with 40 μL Tritest reagent (containing fluorochrome-conjugated mAbs against CD3, CD19, and CD45; Becton Dickinson, San Jose, CA). In most experiments, an aliquot of the incubation mixture was analyzed directly as the Input sample and 200 μL was processed on a DLD microchip. Immediately prior to analysis on a FACSCanto II flow cytometer (BD), 100 μL reference bead suspension (Flow-Count Fluorospheres, Beckman Coulter, Brea, CA, or BD TruCount Becton Dickinson, San Jose, CA) was added to each sample tube per manufacturer's instructions. Samples were processed until ∼ 10,000 beads were acquired for each tube. Five data parameters were collected simultaneously for each experiment: linear forward-angle light scatter (FSC), linear right-angle light scatter (SSC), log FITC, log PE, and log PerCP immunofluorescence. Data was acquired using DIVA software (BD) and analyzed using DIVA or FlowLogic software (Inivai, Melbourne Australia). WBC subtypes were identified as per Table and Figure legends. The absolute numbers of cells were corrected for dilution and numbers of beads collected.
For the experiments of Figure 2, CD4-FITC, CD56-PE, CD14-PC7, CD41-APC, CD3-APC-AF750, CD19-PB, and CD45-Krome Orange (Beckman Coulter, Brea, CA) were used. Following incubation, 200 μL of the stained sample was processed using the DLD microchip, while two additional 100 μL aliquots were lysed for 10 min with 900 μL of VersaLyse (Beckman Coulter), supplemented with 0.2% formaldehyde (i.e., “VersaFix”). One of the two VersaFixed samples was washed twice by centrifugation with PBS containing 1% BSA. Both the washed VersaFixed sample and 100 μL of the microchip Product were resuspended in PBS containing 0.5% paraformaldehyde to the same 1 mL final volume as the unwashed VersaFixed sample. Samples were analyzed using a 10-color Gallios flow cytometer (Beckman Coulter).
Figure 2.
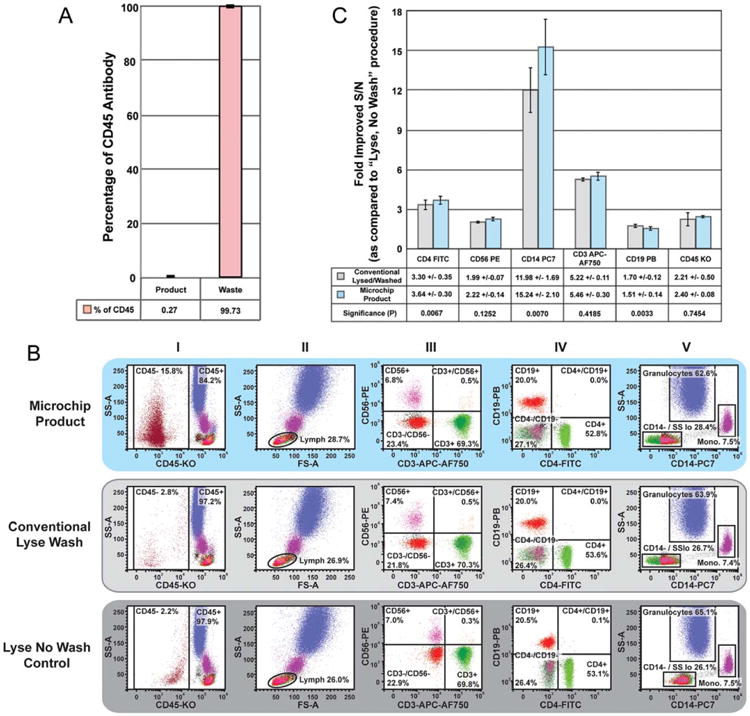
DLD microchip processing of immunostained whole blood resulted in effectively immunostained WBCs. A. 50 μg/mL solution of CD45-Krome Orange was prepared by diluting 150 μL mAb with an equal volume of run buffer. Following priming, this 1:1 mAb solution was pumped through the microchip for 3 min, so that ∼100 μL was equilibrated within the chip prior to collecting the remainder. Subsequently, the chip was processed for 6 min and Product and Waste samples were collected. Sample volume, fluorescence, and absorbance were recorded for both samples. Spectral measurements were compared to independently created standard curves for absorbance and fluorescence using serial dilutions of the CD45-Krome Orange mAb. B: Upper panels are flow cytograms of immunostained blood cells processed via DLD microchip after immunostaining of unlysed blood. Middle panels are comparison flow cytograms of the same donor's starting blood sample processed via a traditional protocol of RBC lysis followed by centrifugal washing after immunostaining. Lower panels are comparison flow cytograms of the same donor's immunostained starting blood sample processed via a Lyse No Wash protocol. Granulocytes were identified as CD45+/FSChi/SSChi, monocytes as CD45+/FSCmed/SSCmed/CD14+, lymphocytes as CD45 ++/FSClow/SSClow/ CD14-, and a Boolean gate for those parameters (i.e., within all three separate lymphocyte gates: FSClow/SSClow, CD45++/SSClow, and CD14-/SSClow) was prepared to eliminate any basophils or monocytes that fell into any of the gates (Column I, II, and V). The following cell types were identified from the lymphocyte Boolean gate for signal-to-noise (S/N) analyses (Column III and IV): CD3 T cells (CD3+/CD56−), NK cells (CD3−/CD56+), NKT cells (CD3+/CD56+), CD4 T cells (CD4+), and B cells (CD19+). CD3−/CD56− lymphocytes were used as the noise population for CD3 and CD56; CD4−/CD19− lymphocytes as the noise population for CD4 and CD19; the lymphocyte population as the noise population for CD14; and platelets as the noise population for CD45. C: The S/N ratio for each cell type was calculated based on flow cytometry profiles in panel B. S/N values for either the lysed and centrifugally washed or the DLD microchip-processed blood samples were divided by the S/N values of Lyse No Wash samples, from four independent experiments (mean ± SEM). Paired t-tests were performed on S/N change between each processing approach for each antigen tested.
For the experiments of Table 3 and Figure 3, cells were first processed via DLD and collected into tubes and stained as above using CD45 PerCP-Cy5.5 (eBioscience, San Diego, CA), then DRAQ5 nucleic acid-binding dye (8.3μM final concentration; eBioscience) and 123count Beads (eBioscience) were added prior to analysis. Fluorescent signals were collected in the Log APC-Cy7 channel (780/60 nm), and data was acquired using a FSC threshold, set to include all bead events, until ∼20,000 bead events were acquired.
Figure 3.
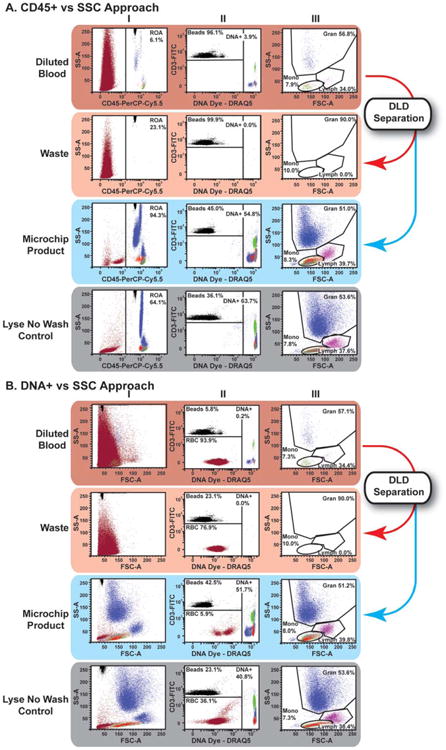
DLD microchip processing of immunostained unlysed blood using optimized buffer and a buffer flush resulted in WBC preparations suitable for standard flow cytometric data analysis approaches. Flow cytometric data from Experiment 6 of Table 3: The top rows of Panels A and B are flow cytograms of immunostained unlysed, unprocessed blood after simple dilution (1:200). The middle two rows of Panels A and B are flow cytograms of the same blood sample after DLD microchip processing into Waste and Product samples. The bottom rows of Panels A and B are flow cytograms of the same blood sample after processing by the Lyse No Wash protocol. Samples were acquired after setting a threshold on FSC signals to include all counting beads. Then, data was analyzed by either the CD45/SSC approach (Panel A) or the DNA/SSC approach (Panel B), as described: A. CD45/SSC approach: To enrich signals from CD45+ WBCs, a gate was set to include all channels in SSC and all CD45+ signals (Column I), including counting beads that registered as fluorescent events. In the CD45+ region of analysis, counting beads discriminated from DNA positive events as shown. CD45+/DNA+ positive events were displayed and gated in forward vs. side scatter to discriminate primary leukocyte subsets suitable for further analysis (Column III). B. DNA/ SSC approach: Using a similar reagent cocktail as panel A, data was acquired with FSC for the purposes of demonstration (Column I), the use of a FITC vs. DNA dye fluorescence plot (Column II) was used to identify nucleated cells (DNA+), counting beads (FITC+/DNA−) and any remaining RBCs (FITC−/DNA−). The Nucleated cell (DNA+) were displayed and gated in forward vs. side scatter to discriminate primary leukocyte subsets suitable for further analysis, similar to the approach in Panel A (Column III).
mAb Quantitation in Product versus Waste Output Samples
A 50 μg/mL solution of CD45-Krome Orange was prepared by diluting 150 μL mAb with an equal volume of run buffer. After priming the DLD microchip, this mAb solution was pumped through the microchip for 3 min, in order that ∼one-third of the solution equilibrated within the chip prior to collecting the remainder of the mAb solution. Subsequently, the Product and Waste samples were collected for an additional 6 min, then the sample volumes were measured, and aliquots were dispensed into a 384-well plate for analysis of fluorescence (405 nm excitation/550 nm emission; Molecular Devices microplate reader, Sunnyvale, CA). The concentrations of mAb in the Product and Waste samples were calculated based on a standard curve prepared using serial dilutions of the CD45-Krome Orange mAb. mAb concentrations used for immunostaining are typically ∼20-fold lower than used in these experiments; this higher concentration of mAb was used to enable the detection of fluorescent mAb in the Product, as smaller quantities are near the lower threshold of detection and thus difficult to quantify.
Results
Application of the DLD Principle for WBC Enrichment and Washing
The central part of the plastic microchip used in this report contains an embossed array of microposts, with the array axis slightly tilted from the horizontal axis that the fluid flow direction follows (Fig. 1). Cells and particles smaller than the critical size (i.e., undesired smaller suspended particles including RBCs, platelets, debris, etc. that are below the specified critical diameter designed to displace WBCs and any larger cells) travel through the micropost array with the average fluid flow, and hence exit the microchip at the same relative horizontal axis at which they enter. However, cells larger than this critical size are bumped by microposts of each row in a direction perpendicular to the flow direction, always following the array axis. The critical size, which is less than the width of the gap between the posts, is determined by the gap and the tilt angle of the array (22). The main stream of Input blood moves horizontally to the Waste Output, while the WBCs separate from the main stream to follow the angle defined by the geometry of the micropost array and exit via the Product Output. Unlike many fractionation methods that rely on a random effect, such as diffusion, the DLD method is deterministic, that is, the path of any cell in the microchip can be predicted precisely, based only on its size and where it enters the chip (19,20). It is also important to note that the DLD micropost array in our current microchip is designed to divert any rare cells of size equal to or larger than WBCs into the Product.
DLD Microchip Processing of RBC-Lysed Human Blood
To simply assess WBC recovery after DLD microchip processing, 200 μL RBC-lysed blood was processed via DLD microchip. WBC recoveries were ≥95% in both these experiments (Table 1).
Table 1. Washing of RBC-lysed blood using DLD microchips resulted in high recovery of total WBCs.
| Experimenta | Fraction Analyzed | Volume (μL)b | WBC Count (×103Cells/μL)c | Total %WBC Recoveryd |
|---|---|---|---|---|
| 1 | Input | 200 | 1.5 | 97%+/−9% |
| Product | 181 | 1.6 | ||
| 2 | Input | 200 | 1.2 | 95%+/−11% |
| Product | 163 | 1.4 |
One RBC-lysed and centrifugally washed human donor blood sample was processed in duplicate experiments, performed on the same day using two separate DLD microchips.
200 μL input volume was pipetted using a hand micropipettor; standard deviation (SD), as determined by weighing, was +/−1.1 in 3 off-line measurements. Output volumes were measured using the same hand micropipettor (SD +/−1.5 in 5 off-line measurements).
Coulter Counter WBC count (SD +/−0.1 in n = 4 off-line measurements).
Calculated from Coulter Counter WBC counts and measured volumes of Input and Output (mean +/− SD; SD propagated as the square root of the sum of the squares of relative SD). The main source of error in these estimates was the number of significant digits provided by the Coulter Counter
DLD Microchip Processing of Whole Blood
In a series of six experiments, diluted unlysed human blood was incubated with a cocktail of fluorochrome-conjugated mAbs against CD3, CD19 and CD45, without prior RBC lysis. In these experiments, the microchips were emptied at the end of each run by following the blood sample with an air plug as a “flush” to clear the microchip and tubing of cells. WBC recovery averaged 102%, with a low of 97% (Table 2); 2.2% (average) of Input WBC were present in the Waste, with a high of 3.7%. When a buffer flush was used, WBCs were essentially undetectable in the Waste by flow cytometry (Fig. 3). The concentration of RBCs in the Product was reduced by 99% (average) compared to the Input sample, based on Coulter counts. The average difference in relative frequency of each immunophenotypic WBC subpopulation found in the Input versus the Product was <0.7%.
Table 2. DLD microchip processing of immunostained whole blood resulted in high recovery of WBCs including major WBC subsets, with >99% depletion of RBCs.
| Experiment | Fraction Analyzed | Volume (μL)a | RBC Count (× 106 Cells/μL) | %RBC Depletedb | WBC Count (× 103 Cells/μL)c | Total % WBC recoveryd | % Granulocytesg | % Monocytesh | % Lymphocytesi | % B-Cellsj | % T-Cellsk | Run Time (Mins) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Input | 200 | 1.99 | – | 2.9 | 97+/−5% | 59.9 | 5.4 | 34.7 | 15.0 | 74.4 | 14.3 |
| Product | 181 | 0.01 | 99.50% | 3.1 | 61.0 | 5.5 | 33.5 | 13.6 | 76.3 | |||
| 2 | Input | 200 | 1.75 | – | 3.6 | 105+/−4% | 68.0 | 4.9 | 27.1 | 18.5 | 75.3 | 12.7 |
| Product | 193 | 0.01 | 99.43% | 3.9 | 67.5 | 5.0 | 27.6 | 17.9 | 76.3 | |||
| 3 | Input | 200 | 1.92 | – | 2.5 | 98+/−7% | 67.9 | 6.7 | 25.4 | 19.5 | 71.9 | 22.0 |
| Product | 258 | 0.02 | 98.96% | 1.9 | 68.7 | 7.0 | 24.2 | 17.7 | 71.7 | |||
| 4 | Input | 200 | 1.85 | – | 2.7 | 101+/−5% | 71.2 | 4.9 | 23.9 | 26.4 | 58.4 | 14.3 |
| Product | 176 | 0.01 | 99.46% | 3.1 | 71.3 | 4.0 | 24.8 | 28.2 | 57.2 | |||
| 5 | Input | 200 | 2.13 | – | 2.4 | 103+/−6% | 70.7 | 6.2 | 23.1 | 10.4 | 83.2 | 18.5 |
| Product | 215 | 0.01 | 99.53% | 2.3 | 72.0 | 5.6 | 22.4 | 9.1 | 84.2 | |||
| 6 | Input | 200 | 2.09 | – | 2.3 | 107+/−6% | 71.2 | 5.3 | 23.6 | 10.2 | 83.8 | 11.7 |
| Product | 223 | 0.02 | 99.04% | 2.2 | 71.9 | 5.9 | 22.2 | 9.5 | 84.0 | |||
| Mean Depletion or Recovery or Run Timee | 99.32+/−0.10% | 102+/−2% | 15.6+/−1.6 | |||||||||
| Mean Difference (Product vs. input)f | 0.6+/−0.3% | -0.1+/−0.2% | −0.5+/−0.4% | −0.7+/−0.5% | 0.4+/−0.4% | |||||||
lnput and Output volumes as in Table 1.
Difference of Coulter Counter RBC count in Input vs Product (column 4) divided by Coulter Counter RBC count in Input. For the Products, the RBC counts were at the lower detection limit of the Coulter Counter and hence are imprecise.
As in Table 1.
Mean +/−standard error of the mean (SEM).
Product value minus Input value (mean +/−SEM).
Granulocytes were identified by flow cytometry as CD45low7CD3−/CD19− signals with characteristic high FSC and SSC.
Monocytes were identified as CD45high/CD3−/CD19− with characteristic intermediate FSC and SSC.
Lymphocytes were identified as CD45high/CD3+ or −/CD19+ or − with characteristic low FSC and SSC.
B cells were identified as CD45high/CD3−/CD19+ with FSC and SSC of lymphocytes.
T cells were identified as CD45high/CD3+/CD19− with FSC and SSC of lymphocytes.
Percent cells of each subtype was calculated using the number of events for that cell population divided by the total number of viable cell events, as determined by flow cytometric FSC/SSC-gating.
Blood from five different donors was analyzed in six replicate experiments. All experiments performed over four weeks, using a fresh microchip for each experiment. The microchips were emptied by following the blood sample with an air plug as a “flush” to clear the microchip and tubing of cells. Prior to flow cytometric analysis, all Input and Output samples were subjected to RBC lysis, to provide comparably treated cells for flow cytometry. For RBC lysis, 100 μL filtered blood was combined with 900 μL 1 × FACS lysing solution (Becton Dickinson, San Jose, CA), vortexed and incubated (RT, 15 min). Following centrifugation (1,000 × g, 10 min), the supernatant was removed and the WBC-enriched pellet resuspended in 1 mL run buffer. After an additional centrifugation, the supernatant was removed and the WBC-enriched pellet resuspended in 250 μL run buffer. This procedure required ∼45 min.
Removal of Unbound mAb by DLD Microchip Processing
To assess the removal of unbound mAb from the microchip Product, 50 μg/mL free CD45-Krome Orange mAb was processed through the microchip, and the Product and Waste Outputs were analyzed for fluorescence. This high concentration of mAb, ∼20-fold higher than used routinely for immunostaining, was used to enable the detection of fluorescent mAb in the Product, as smaller quantities are near the lower threshold of instrument detection. Only 0.27% (SEM 5 0.045%) of the initially added CD45-Krome Orange mAb was detected in the Product Output (Fig. 2A).
To confirm the efficiency of DLD microchip-mediated removal of unbound mAbs from the Product, we compared the Signal:Noise (S/N) ratios of cells that had been immunostained using a panel of mAbs (23). For these experiments, the starting diluted whole blood sample was first stained with a mAb cocktail and then processed by either 1) DLD microchip without RBC lysis, 2) RBC lysis and two subsequent traditional centrifugal washes, or 3) RBC lysis without any washing (24). All WBC subpopulations in the DLD Product had reduced background fluorescence intensities, as compared to sample prepared by a standard “Lyse No Wash” procedure (Fig. 2B Columns III, IV). The resulting improved S/N ratio allowed clear resolution of discrete cell populations in both scatter/fluorescence and fluorescence-only cytograms (Fig. 2B, Panels I and III–V, respectively). The S/N ratios for the DLD Product WBC subsets were comparable to those obtained by traditional post-immunostaining processing via two centrifugal washes after RBC lysis (Fig. 2C). Washing by either DLD microchip or the traditional procedure reduced the background noise from 1.5 to 15 fold, depending on the mAb. In each case, the S/N improvements were equivalent or better in DLD microchip versus centrifugal washing (Fig. 2C).
DLD Microchip Processing of Unlysed Blood Using a Buffer Flush and an Optimized Buffer
In the experiments of Tables (1–3), use of >24 h-old blood samples simulated sample age in routine clinical testing (except in Experiment 6 of Table 3, where blood was drawn on the day of the experiment). Since there are reports indicating that the presence of a membrane-intercalating amphiphilic molecule with surfactant-like properties might reduce cell clumping and adhesion to tubing, connections, and microfluidic chip surfaces (25–27), another series of experiments was performed using a run buffer containing an amphiphilic poloxamer (Optimized Buffer) rather than BSA. In addition, since we suspected and observed anecdotally that the air flush might cause some cell clumping, we replaced the air flush with a buffer flush procedure. In this buffer flush procedure, once the sample reservoir was depleted, the Sample Input port was depressurized, fresh buffer added and the system re-pressurized; by this method, the buffer flushed out any residual cells in the system. As compared with the results of Table 2, the WBC recoveries in Table 3, with optimized buffer and with a buffer flush at the end of the run, were somewhat lower, averaging 88%. In a side-by-side comparison of presence of the amphiphilic poloxamer versus BSA, use of the amphiphilic poloxamer-containing run buffer (Optimized Buffer) resulted in higher WBC recovery (Table 3: 87% for Experiment 1 with Optimized Buffer vs. 77% for Experiment 1 with BSA Buffer), suggesting that the somewhat reduced WBC recovery in the Table 3 vs. Table 2 results was most likely due to the buffer flush being slightly less efficient than the air flush.
In the experiments of Table 3 and Figure 3, we used flow cytometry to more accurately quantitate low numbers of RBCs that are at the limits of or below Coulter Counter resolution. With the optimized buffer and a buffer flush, DLD microchip processing of unlysed blood consistently removed 99.98% (average) of the input RBCs (Table 3). The RBCWBC ratio was reduced from ∼800:1 in the input sample to 0.19:1 in the DLD Product (visualized in Figure 3, blue panels). DLD microchip processing with the optimized buffer and protocol removed RBCs more effectively than the Lyse No Wash control (Fig. 3, gray panel). The microchip-processed sample had (1) fewer remaining RBCs than the Lyse No Wash processed sample, and (2) comparable percentages of immunophenotypic cell types to the Lyse No Wash sample. Furthermore, cell processing time using the DLD microchip was ∼18 min, compared with ∼10 min for Lyse No Wash and >30 min for RBC Lysis with traditional centrifugal washing.
Discussion
Experiments using silicon-based microchips indicated that WBCs can be enriched from blood samples utilizing the DLD principle (19,20). In this project, our initial goal was to reduce DLD microfluidic technology to practice for WBC enrichment and washing by using disposable plastic microchips. In transitioning from silicon to plastic microchips, a major challenge was precision manufacturing of μm-sized microposts. Moving to a soft-embossed plastic device allowed us to achieve reproducible features that faithfully replicated those on the silicon master, while now in a cost-effective, single-use format. An additional challenge of changing materials was the modification in surface properties, which play an important part in the functionality of a microfluidic device. Our current process (manufacturing, cleaning and surface treatment with buffers) evolved to increase reproducibility in processing a range of donor samples, while avoiding cell loss and cell clumping and clogging. Over the course of development, >100 blood samples were run on single-use plastic microchips, including fresh samples, 24 h-old samples, and samples shipped overnight during winter months; >95% of these samples were processed successfully (data not shown).
A second goal of the project was to evaluate DLD microchip-processed human WBCs quantitatively and qualitatively, as compared with WBCs processed by conventional means, utilizing established flow cytometry analytic techniques. A challenge in developing a new technology for WBC enrichment involves the correct control cell processing method against which to compare the new technology. Traditional approaches utilizing centrifugal cell washing yield lower numbers of WBCs in comparison with the blood Input sample, and can bias the WBC subtype yield (12). Therefore, in our first evaluation of the DLD microchip technology (Table 1), we used an enriched WBC population as the Input sample and simply compared it to the microchip Product. By Coulter cell counting, >95% of the Input WBCs were recovered in two experiments, indicating that the DLD microchip was capable of generating a Product with minimal loss of WBCs.
We then progressed to use unlysed blood, the desired starting material, as the Input sample, and assessed numbers of both WBCs and RBCs in the microchip Product vs an Input sample that had been processed via a Lyse No Wash protocol. Microchip processing time averaged 16 min. By Coulter counting, ≥97% (minimum) of starting WBCs were recovered and 99% (average) of starting RBCs were depleted in six independent experiments (Table 2). In these experiments, we also analyzed WBC subtypes by flow cytometry, finding ≤0.7% difference in the frequencies of the major WBC subtypes between Input and Product (Table 2). Since analysis of the Input sample required use of RBC lysing agents (via a Lyse No Wash protocol), we similarly RBC-lysed the DLD microchip Product in these experiments, in order to eliminate any biasing of cell subtypes that might be caused by the RBC lysis procedure.
Next, experiments were done to evaluate the ability of the DLD microchip device to wash unbound mAbs from the microchip Product. When soluble mAb was processed via the DLD microchip, only 0.27% the starting mAb was found in the microchip Product. In addition, microchip-processed WBCs from immunostained unlysed blood samples were analyzed by flow cytometry and compared to two conventional WBC preparations, one in which the input sample was only lysed (Lyse No Wash) and one in which the sample was lysed and washed by traditional centrifugation. The microchip Product had reduced background fluorescence for each mAb-defined WBC subtype, as compared to WBCs prepared by the Lyse No Wash protocol, ranging from 1.5 to 15-fold improvement in S/N (Fig. 2C). This is consistent with the above finding that the DLD microchip efficiently removed free mAbs from the Product (Fig. 2A). This improvement was similar to or higher than the improvement in S/N found via a conventional centrifugal washing procedure, and the DLD processing required <20 min of processing time compared with >30 min required for conventional processing.
In the above experiments, RBC removal as assessed by Coulter counting indicated ≥99% removal (at the Coulter Counter's limit of detection). However, in experiments similar to those of Table 2, close inspection of the tubing connections to the microchip revealed that small clumps of RBCs remained in an ∼2.5 μL dead volume that was air-flushed into the Product, accounting for the small numbers of RBCs that we noted in flow cytometry light scattering analyses performed with the Table 2 experiments. To generate a microchip Product with fewer RBCs, we therefore modified our procedure, replacing the air flush with a buffer flush. In addition, since published reports indicated that use of a membrane-intercalating amphiphilic molecule with surfactant-like properties might reduce cell clumping or adhesion to tubing, connections, or microchip surfaces, we replaced the BSA in our run buffer with an amphiphilic poloxamer (named as the Optimized Buffer). To optimally quantify the number of RBCs remaining in the microchip Product, the microchip Product was not RBC-lysed prior to flow cytometry analysis. As shown in Table 3 and Figure 3 using this modified procedure, the % RBC removal was 99.985% (average) with 88% (average) WBC yield, resulting in a Product with a WBCRBC ratio of 12:1 (average) and WBCs enriched by 9,000 fold (average). In Figure 3B Column II, compared with the Lyse No Wash control sample, the microchip Product had far fewer remaining RBCs–only 6% of the total counts in the microchip Product sample versus 36% RBCs in the Lyse No Wash control. Indeed, the microchip Products had so few RBCs and other debris that they could be analyzed by FSS/SSC alone (without the need for CD45- or DNA-based staining and gates), as is evident in Figure 3B Column I for the microchip Product. The frequencies of gated “lymphocytes” in the microchip Product were similar to those in the Lyse No Wash control sample, indicating that the DLD microchip generated a superior Product with fewer background events of debris and RBCs and with an unchanged subset distribution, as compared with conventional processing.
Multicolor flow cytometric immunophenotyping of processed cells can be readily extended to evaluate numerous potentially interesting cell types that might be assessed, including rare cells such as immune cell subsets, activated cells, stem cells, and circulating tumor cells (CTCs). Because cell recovery is high, detection of extremely rare CTCs for “liquid tumor biopsies” is an attractive area for investigation. In our own ongoing research, addition of an immunomagnetic module downstream of the DLD-based size separation enriched CTCs (28). Assessment of intracellular antigens is another attractive extension of DLD microchip-based cell processing that we are investigating. Our results suggest that plastic DLD microchips can speed and automate cell processing for many other important research and clinical applications involving enrichment or depletion of common or rare cell types, not only from blood but also from tissue samples (e.g. tumor-infiltrating lymphocytes from solid cancers).
Conclusion
These results using a single channel microfluidic design demonstrate that inexpensive plastic DLD microchips can speed and automate the cell separation processing for high quality flow cytometry analysis. DLD processing using an optimized buffer system removed >99.9% of Input RBCs. The low variability in run-to-run performance addressed not only donor-to-donor and day-to-day differences, but also variability of manufacturing μm-sized objects in plastic, thus indicating that these plastic, single-use DLD microchips provide reproducible performance across batches. Comparing the performance to conventional methods of preparation, DLD microchip processing generated a Product that contains fewer RBCs and particulate debris than a Lyse No Wash preparation, and the Product could be analyzed by FSC/SSC alone, without any special staining or gating required. Similar to traditional WBC processing protocols involving RBC lysis followed by centrifugal washing, the DLD microchip delivered Product cells that provide high quality flow cytometry results, including higher S/N ratios than a Lyse No Wash preparation, but with the advantages of short (<20 min) processing time and high (∼90%) WBC recoveries with no biasing of WBC subtypes. DLD microchip processing using single or highly parallel designs should enable effective cell processing at a variety of scales and throughput rates that might be required to support multiple applications beyond flow cytometry.
Acknowledgments
The authors thank Julie Farnsworth, Flow Core Manager at Virginia Commonwealth University, for her technical support with flow cytometry operation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Beckman Coulter and the Beckman Coulter product and service marks mentioned herein are trademarks or registered trademarks of Beckman Coulter, Inc. in the United States and other countries. All other trademarks are the property of their respective owners.
Grant sponsor: National Foundation for Cancer Research; National Institutes of Health, Grant number: R42CA174121.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Contribution: AMS, TW, ZPL, CRD, KG, LA, MJK, GCB, SG, YC, JLD, XC, JCS, and CIC conceived and designed experiments; ZPL, CRD, MJK, YC, JLD, GCB, XC, and JM performed experiments; AMS, ZPL, CRD, provided reagents and materials; AMS, TW, ZPL, CRD, KG, MJK, GCB, and CIC analyzed and interpreted results; CIC, JCS, TW, AMS, KG, LA, GCB, and SG wrote the article.
Disclosure of conflicts of Interest: CIC chairs the Scientific Advisory Board, received consulting payments, and holds equity in GPB Scientific, LLC. These arrangements are being managed by the University of Maryland Baltimore in accordance with its conflict of interest policies. No other conflicts of interest are present.
Literature cited
- 1.Bendall SC, Simonds EF, Qiu P, Amir ED, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler's guide to cytometry Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandy F, Varro R, Recktenwald D. Flow Cytometry Principles. In: Vo-Dinh T, editor. Biomedical Photonics Handbook. Boca Raton, FL: CRC Press; 2003. p. 120. [Google Scholar]
- 4.Maus MV, Fraietta JA, Levine BL, Kalos M, Zhao Y, June CH. Adoptive immunotherapy for cancer or viruses. Annu Rev Immunol. 2014;32:189–225. doi: 10.1146/annurev-immunol-032713-120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Bodovitz S. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Lopez J, Lahuerta JJ, Pepin F, Gonzalez M, Barrio S, Ayala R, Puig N, Montalban MA, Paiva B, Weng L, et al. Prognostic value of deep sequencing method for minimal residual disease detection in multiple myeloma. Blood. 2014;123:3073–3079. doi: 10.1182/blood-2014-01-550020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore J, Yvon P. High dimensional flow cytometry comes of age. Eur Pharm Rev. 2012;17:20–24. [Google Scholar]
- 8.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JKA, Rao PE, Calvelli T, Stetler-Stevenson M, Browning SW, Yeung L, Marti GE. Artifactual staining of monoclonal antibodies in two-color combinations is due to an immunoglobulin in the serum and plasma. Cytometry. 1994;18:140–146. doi: 10.1002/cyto.990180305. [DOI] [PubMed] [Google Scholar]
- 10.Terstappen LW, Meiners H, Loken MR. A rapid sample preparation technique for flow cytometric analysis of immunofluorescence allowing absolute enumeration of cell subpopulations. J Immunol Methods. 1989;123:103–112. doi: 10.1016/0022-1759(89)90034-3. [DOI] [PubMed] [Google Scholar]
- 11.Gratama JW, Menendez P, Kraan J, Orfao A. Loss of CD34(+) hematopoietic progenitor cells due to washing can be reduced by the use of fixative-free erythrocyte lysing reagents. J Immunol Methods. 2000;239:13–23. doi: 10.1016/s0022-1759(00)00154-x. [DOI] [PubMed] [Google Scholar]
- 12.Tamul KR, Schmitz JL, Kane K, Folds JD. Comparison of the effects of Ficoll-Hypaque separation and whole blood lysis on results of immunophenotypic analysis of blood and bone marrow samples from patients with hematologic malignancies. Clin Diagn Lab Immunol. 1995;2:337–342. doi: 10.1128/cdli.2.3.337-342.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madsen LP, Rasmussen MK, Bjerregaard LL, Nøhr SB, Ebbesen F. Impact of blood sampling in very preterm infants. Scand J Clin Lab Invest. 2000;60:125–132. doi: 10.1080/00365510050184949. [DOI] [PubMed] [Google Scholar]
- 14.Testa M, Birocchi F, Carta P, Fanos V. Causes of anaemia in very low birth weight infants. Phlebotomy losses are not the first accused. Minerva Pediatr. 2006;58:263–267. [PubMed] [Google Scholar]
- 15.Broder-Fingert S, Crowley WF, Boepple PA. Safety of frequent venous blood sampling in a pediatric research population. J Pediatr. 2009;154:578–581. doi: 10.1016/j.jpeds.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosic S, Murthy SK, Koppes AN. Microfluidic Sample Preparation for Single Cell Analysis. Anal Chem. 2016;88:354–380. doi: 10.1021/acs.analchem.5b04077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui F, Rhee M, Singh A, Tripathi A. Microfluidic Sample Preparation for Medical Diagnostics. Annu Rev Biomed Eng. 2015;17:267–286. doi: 10.1146/annurev-bioeng-071114-040538. [DOI] [PubMed] [Google Scholar]
- 18.Mach AJ, Adeyiga OB, Di Carlo D. Microfluidic Sample Preparation for Diagnostic Cytopathology. Lab Chip. 2013;13:1011–1026. doi: 10.1039/c2lc41104k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Deterministic hydrodynamics: Taking blood apart. Proc Natl Acad Sci USA. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 21.Madou MJ. Fundamentals of Micro fabrication: The Science of Miniaturization. 2nd. Boca Raton, FL: CRC Press; 2002. pp. 104–107. [Google Scholar]
- 22.Inglis DW, Davis JA, Austin RH, Sturm JC. Critical particle size for fractionation by deterministic lateral displacement. Lab Chip. 2006;6:655–658. doi: 10.1039/b515371a. [DOI] [PubMed] [Google Scholar]
- 23.Donenberg AD, Donnenberg VS. Rare Event Analysis in Flow Cytometry. Clin Lab Med. 2007;27:627–652. doi: 10.1016/j.cll.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson JK, Stein D, Mui T, Mack R, Hubbard M, Denny T. Evaluation of a method for counting absolute numbers of cells with a flow cytometer. Clin Diagn Lab Immunol. 1997;4:309–313. doi: 10.1128/cdli.4.3.309-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB, Rothberg JM, Link DR, Perrimon N, Samuels ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci U S A. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maskarinec SA, Hannig J, Lee RC, Lee KYC. Direct Observation of Poloxamer 188 Insertion into Lipid Monolayers. Biophys J. 2002;82:1453–1459. doi: 10.1016/S0006-3495(02)75499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tharmalingam T, Ghebeh H, Wuerz T, Butler M. Pluronic enhances the robustness and reduces the cell attachment of mammalian cells. Mol Biotechnol. 2008;39:167–177. doi: 10.1007/s12033-008-9045-8. [DOI] [PubMed] [Google Scholar]
- 28.Koesdjojo M, Lee Z, Dosier C, Saini T, Gandhi K, Skelley A, Aurich L, Yang G, Wyer K, Civin C, et al. DLD microfluidic purification and characterization of intact and viable circulating tumor cells in peripheral blood. Cancer Res. 2016;76(Supplement):3956. [Google Scholar]


