Abstract
Background
The phytopathogenic Xanthomonas campestris pv.campestris is a gram-negative bacterium and the causal agent of black-rot disease of cruciferous crops. Many gram-negative bacteria possess a family of proteins, called Dsbs, which are involved in disulfide bond formation in certain periplasmic proteins. In our preliminary screening of the virulence to the plants we identified that gene XC_0531 which annotated gene dsbD of Xanthomonas campestris pv. campestris (Xcc) is related to the virulence to the host plants.
Results
Here, we found XC_0531 encoded a DsbD like protein. Its deletion is sensitive to DTT and copper, decreased accumulation of free thiols in periplasm. Its deletion also affected heme synthesis, position of Soret band and the production of peak c550. This suggests that XC_0531 is related to c-type cytochromes biogenesis. XC_0531 mutation decreased the utilization of different carbon sources (such as galactose, xylose, maltose, saccharose and glucose), reduced extracellular polysaccharide (EPS) production, decreased extracellular enzyme activities (protease, cellulose and amylase), slowed down growth rate of Xcc and weakened virulence to the plants. These results suggest that these phenotypes caused by XC_0531 mutation is possibly due to deficient biosynthesis of c-type cytochromes in respiration chain and the formation of disulfide bonds. Our work confirmed the function of XC_0531 and provide theory basis for scientists working on molecular mechanisms of cytochrome c biogenesis, pathogenesis of Xcc, development of EPS commercial values and protecting plant from black rot.
Conclusion
We confirmed the function of gene XC_0531, which encodes a DsbD like protein, a protein correlated with c-type cytochrome biogenesis. This gene is related to the virulence to plants by affecting funtion of cytochromes c and probably disulfide bonds modification of proteins in type II secretion system (T2SS).
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-1056-9) contains supplementary material, which is available to authorized users.
Keywords: Cytochrome c, Eps, Xanthomonas campestris, Pathogenesis
Background
The phytopathogenic Xanthomonas campestris pv. campestris is a gram-negative bacterium and the causal agent of black-rot disease of cruciferous crops. Many gram-negative bacteria possess a family of proteins, called Dsbs, which are involved in disulfide bond formation in certain periplasmic proteins [1]. These disulfide bonds, which are structurally modified via disulfide bond formation in the periplasm, are critical for the maturation of virulence factors. The Dsb proteins also have thiol:disulfide oxido-reductase activities [2–7]. DsbA is a strong oxidant with a weak isomerase activity in the periplasmic space for disulfide bond formation [2, 8–10]. DsbB together with DsbA catalyze the formation of cysteine thiols [2]. DsbC has a disulfide isomerase activity catalyzing disulfide formation [10, 11]. DsbD acts as a reductase in the periplasmic space [12]. DsbD also participates in the biosynthesis of cytochromes c [1].
C-type cytochromes act as a partner in the respiration chains and play an important role in the metabolism of bacteria [13] and fungi [14, 15]. C-type cytochromes are distinguished from cytochromes of the other classes by covalent attachment of the heme group to the conserved CXXCH motif of the apocytochromes. The biogenesis of c-type cytochromes has two systems class I and class II. Most gram-negative bacteria harbor the class I system including at least 11 proteins (Dsb ABD and Ccm ABCDEFGH). The heme group attachment to the apoprotein takes place in the oxidative environment of the periplasm [2]. Thus, c-type cytochromes apoproteins need to be reduced prior to heme binding by CcmH, CcmG and DsbD in cytochrome c maturation in Escherichia coli [2, 16].
Our previous studies have shown that DsbB proteins play important roles in pathogenesis of Xcc [17] and in a preliminary screening for pathogenic genes we found that gene dsbD is needed for the virulence of Xcc to plants. Here, we confirmed the function of gene XC_0531, which encodes DsbD, a protein correlated with c-type cytochrome biogenesis and is related to the virulence to plants.
Materials and methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in LB medium at 37 °C. Xcc strains were grown at 28 °C in NYG medium (5 g of peptone, 3 g of yeast extract and 20 g of glycerol per liter), NYGA medium (NYG medium with 1.5% agar), NY medium (NYG medium without glycerol) or non-carbohydrate minimal medium MMX [18]. Antibiotics were added at the following concentrations as required: kanamycin (Kan), 25 μg ml−1; rifampicin (Rif), 50 μg ml−1; ampicillin, 100 μg ml−1; spectinomycin, 50 μg ml−1; tetracycline (Tet), 5 μg ml−1 for Xcc and 15 μg ml−1 for E. coli.
Table 1.
Strains and plasmids used in this study
| Strains and Plasmids | Genotype/Properties | Reference/Resource |
|---|---|---|
| Escherichia coli | ||
| JM109 | RecA1, endA1, gyrA96, thi, supE44, relA1△ (lac-proAB)/F′ [traD36, lacI q, lacZ△M15] | Our lab’s collection |
| ED8767 | RecA, met,containing pRK2073,Spcr | Our lab’s collection |
| M15 | Kmlac ara gal mtl recA1 uvr1 [pREP4 lacI Kanr] | Our lab’s collection |
| Xanthomonos campestris pv. compestris | ||
| 8004 | Wild type; Rif r | Our lab’s collection |
| 015F07 | As 8004,but XC_3579::Tn5gusA5;Rifr, Kmr | This work |
| 045F12 | As 8004, but XC_3579::Tn5gusA5;Rifr, Kmr | This work |
| 142E11 | As 8004, but XC_3579::Tn5gusA5;Rifr, Kmr | This work |
| 0531pk | As 8004, but XC_0531::pK18mob; Rifr, Kmr | This work |
| C0531 | 0531pk harboring pLA0531; Rifr, Kmr, Tetr | This work |
| Plasmids | ||
| pK18mob | pUC18 derivative, lacZα, Kanr, mob site, suicide plasmid in X. campestris pv. campestris. | Our lab’s collection |
| pRK2073 | Helper plasmid,Tra+, Mob+, ColE1, Spcr. | Our lab’s collection |
| pET30a+ | T7 promoter, Kmr | Our lab’s collection |
| pLC0531 | pLAFR3 containing the whole XC_0531 gene;Tcr | This work |
| pLAFR3 | Broad host range cloning vector, Tcr | Our lab’s collection |
DNA manipulations
Methods as described previously [19] were used for preparation of plasmid and chromosomal DNAs, restriction digestion, DNA ligation, agarose-gel electrophoresis and DNA transformation of E. coli. Conjugation between Xcc and E. coli strains was performed as described previously [20]. Restriction endonucleases, T4 DNA ligase and Pfu polymerase were purchased from Promega.
Insertional mutant construction and complementation
An insertional mutant of the ORF XC_0531 was constructed using the suicide plasmid pK18mob [21, 22] as described previously [23]. A 539 bp internal fragment of the XC_0531 ORF sequence was amplified by PCR using the total DNA of wild-type Xcc strain 8004 as a template. The following pairs of oligo nucleotides were used as primers (Additional file 1: Table S1). Primers were modified to provide BamHI-HindIII ends. The amplified DNA fragments were cloned into pK18mob in the same orientation as the lacZ promoter. The resulting recombinant plasmid was introduced from E. coli strain JM109 [24] into Xcc wild-type strain 8004 by triparental conjugation, using pRK2073 as the helper plasmid [25]. The mutant was confirmed by PCR using the primers P18conF and 0531F (Additional file 1: Table S1). The obtained mutant strain was named 0531pk (Table 1). For complementation of mutant 0531pk, a 2786 bp DNA fragment containing the XC_0531 ORF was amplified using the following pair of oligonucleotides as primers: C0531-F and C0531-R (Additional file 1: Table S1). Primers were modified to give BamHI- or HindIII-compatible ends. The amplified DNA fragment was cloned into plasmid pLAFR3 [26]. The obtained recombinant plasmid pLC0531 was transferred into the mutant strain 0531pk by triparental conjugation, resulting in the complemented strain named C0531 (Table 1). The C0531 strain was confirmed by PCR using the primers P18conF and C0531F (Additional file 1: Table S1).
Virulence assay
The virulence of Xcc to the host plant Chinese radish (Raphanus sativus) was tested by the leaf-clipping method [27]. Lesion length was measured 10 days after inoculation.
Sequence analyses
The amino acid sequences of DsbD and DsbC proteins of Xcc, Escherichia coli str. K-12, Shewanella oneidensis MR-1 and Pseudomonas aeruginosa PAO1 were obtained from the database of the National Center for Biotechnology Information (NCBI). Prediction of transmembrane helices was performed with TMHMM Server v.2.0 software [28].
Heme staining
SDS-PAGE was carried out without the addition of DTT. Coomassie brilliant blue staining was carried out as described previously [29]. TMBZ was dissolved in methanol to a final concentration of 6.3 mM. The gel was covered with a solution of 3 parts TMBZ and 7 parts 0.5 M sodium acetate and incubated in the dark for 20 min. H2O2 was added to the final concentration of 30 mM for the visible protein gel bands, as describes [30].
Analysis of c-type cytochromes
Xcc strains were cultured for 14 ~ 16 h to the exponential phase of growth in the NYG and then cellular membranes were prepared from cultures grown for 24 h on minimal medium (MMX) for Xanthomonas campestris with 1% mannitol. The cells were harvested by centrifugation, washed in 0.1 M phosphate buffer (pH 7.4), and resuspended in 0.1 M phosphate buffer (pH 7.4) with 30% (vol/vol) glycerol.
Total soluble protein fractions were prepared by sonication of washed cells in 0.1 M phosphate buffer (pH 7.4). Sonicates were centrifuged to remove unbroken cells at 10,000×g for 15 min at 4 °C. The soluble part and membrane fractions were separated by ultracentrifugation at 100,000×g for 2 h. The soluble proteins were resuspended in 10 mM Tris-HCl (pH 7.4) and membrane fractions were resuspended in 16 mM Tris-HCl (pH 7.4).
Cytochrome spectra were recorded at room temperature using a Beckman DU730 spectrophotometer (Beckman Coulter Inc.). The samples were diluted by PBS and reduced with a few granules of sodium dithionite or oxidized with ammonium persulfate. Reduced minus oxidized spectra were obtained by recording differences between the spectra of the dithionite-reduced sample and the ammonium persulfate-oxidized sample. Protein concentrations were measured with a protein assay kit (Pierce), using BSA as the standard. Proteins were separated by electrophoresis through 15% SDS-polyacrylamide gels by mixing with dithiothreitol (DTT)-free 5 × SDS loading buffer. Samples prepared with buffer containing DTT followed by heating resulted in a loss of staining of c-type cytochromes. Highest levels of heme stain signal were obtained using DTT-free buffer and no heat.Proteins containing covalently bond heme iron were visualized using 3,3′,5,5′- tetramethyl benzidine (TMBZ), as described [30].
Ellman’s assay
Periplasmic protein samples from bacterial cells were prepared using the chloroform method with minor modification [17]. Cells were grown in NYG at 28 °C to the optical density of OD600 = 0.5, 500 ul cultures were centrifuged at 1000 rpm for 10 min to remove the supernatant. The strains were treated with chloroform for 15 min, and then 200 ul of 0.8 mM DTNB (5, 5′-dithiobis-[2-nitrobenzoic acid]) was added into the samples for 5 min. The supernatant containing periplasmic protein was obtained by centrifugation at 6000 rpm for 20 min. The absorbance was determined at 412 nm. The assays were carried out in three independent experiments.
DTT sensitivity tests
For the DTT sensitivity test, 2.5 μl of overnight cultures of each strain OD600 = 1.0 were spotted onto NYG plates supplemented with 4 mM DTT and incubated at 28 °C for 48 h. At least three plates were inoculated for each strain and each experiment was repeated three times.
EPS assay
To estimate EPS production, strains were cultured in 100 ml NY medium supplemented with 2% (w/v) of various sugars at 28°C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried and weighed as described [31, 32].
Motility assay
To test the swarming motility, 3 μl of overnight culture (OD600 of 1.0) of each Xcc strain was inoculated onto NY plates containing 2% glucose and 0.6% agar, and then incubated at 28 °C for 4 days. The diameters of the area occupied by the bacterial cells were measured and these values were used to indicate the motility of the Xcc strains [33]. The experiment was repeated at least three times.
Extracellular enzyme activity analysis
To estimate the activity of the extracellular enzymes endoglucanase (cellulase), amylase and protease, Xcc strains were cultured in NYG medium for 12 h. For protease, 3 μl of overnight cultures (OD600 of 1.0) was spotted onto NYGA plates containing 1% skim milk, after incubation at 28 °C for 24 h, plates were photographed. For cellulose, 3 μl of overnight culture was spotted onto NYGA plates containing 0.5% carboxymethyl cellulose after incubation at 28 °C for 48 h, plates were stained with I2/KI (0.08 M I2, 3.2 M KI) and washed by 70% ethyl alcohol. For amylase, 3 μl of overnight cultures was spotted onto NYGA plates containing 0.1% starch incubation at 28 °C for 24 h. The plates were stained with 0.1% Congo Red and then washed 2 times. At last plates were destained using 1 M NaCl solution. For extracellular protease activity, the method was as described previously [34]. For cellulase (endoglucanase), 10 μl of enzyme-containing extracts was added to 200 μl of indicator buffer containing 1% (wt/vol) carboxymethyl cellulose (CMC, Sangon, Shanghai, China) as the substrate. The reactions were carried out for 30 min at 28 °C. The released reducing sugars were measured as D-glucose equivalents, as described [17]. One unit (U) of the cellulase (endoglucanase) activity was defined as the amount of enzyme releasing 1 μM of reducing sugar per minute. Amylase activity quantification was conducted in the same way as for the cellulase (endoglucanase) measurement, except that the substrate was replaced by 1% (wt/vol) starch solution.
Copper stress response analysis and RT-qPCR analysis
The overnight cultures of Xcc (OD600 of 1.0) were cultured in NYG medium with different concertration of CuSO4 (0.4 mM, 0.8 mM, 1.2 mM and 1.6 mM) for 2 days. The OD600 values were measured and recorded.
Real-time quantitative PCR (RT-qPCR) analysis was carried out and refered to our previous study [32]. Primers were list in Additional file 1: Table S1. All RT-qPCRs were performed in triplicate.
Data analysis
One-way analysis of variance (ANOVA) was used to detect significant differences between the treatments. These analyses were carried out with SPSS 22.0 software.
Results
Disruption of XC_0531 reduces virulence to the host plant
In a preliminary screening, we found that gene XC_0531 of Xcc is related to virulence to plants (unpublished data). To further confirm whether XC_0531 is involved in the pathogenicity, the virulence of Xcc was tested on the host plants cabbage and radish by the leaf-clipping method [27]. As shown in Fig. 1, the wild type (strain 8004) produced a lesion length of 12.27 ± 1.33 mm in cabbage 10 days after inoculation. The lesion length was significantly reduced to 10.35 ± 0.28 mm in the mutant (0531pk) (P < 0.05). The complemented strain (C0531) induced lesion length similar to the wild type (12.05 ± 1.23 mm) in cabbage (Fig. 1a and c). Similar results were obtained in radish leaves (Fig. 1b and c). These results suggest that XC_0531 plays an important role for Xcc virulence to plants.
Fig. 1.
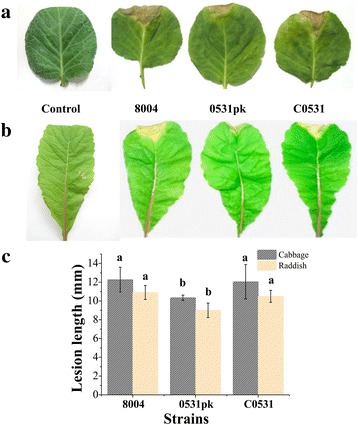
Pathogenic phenotype of the mutant in cabbage and radish. a and b Symptom production on leaves 10 days after inoculation by clipping with strains 8004, 0531pk and C0531. c Mean lesion lengths caused by different Xcc strains in cabbage and radish. Values are calculated from ~30 measurements. Different letters within a plant indicate significant differences at a level of p < 0.05 based on Duncan’s test by one way-ANOVA
The ORF XC_0531 of Xcc encodes a c-type cytochrome biogenesis protein
The protein sequence encoded by XC_0531 of Xcc (strain 8004) (NCBI-Protein ID: AAY47612) was annotated as a c-type cytochrome biogenesis protein (http://xgb.leibniz-fli.de). It had 29% identity and 43% similarity to the thiol:disulfide interchange protein DsbD (NCBI-Protein ID: NP_418559, encoded by the gene b4136 in Escherichia coli str. K-12, and 28% identity and 43% similarity to DsbD of Shewanella oneidensis MR-1 (NCBI-Protein ID: NP_716329) encoded by gene SO_0696. The ORF XC_0531 is located between 626,891 and 629,197 of the Xcc 8004 [35]. The ORFs upstream and downstream of XC_0531 are XC_0529 (encoding an acetyl-CoA carboxylase biotin carboxyl carrier protein subunit), XC_0530 (encoding a 3-dehydroquinate dehydratase) and XC_0532 (encoding a divalent cation tolerance protein) (Fig. 2a). A redox active Cys-X-X-Cys motif, the conserved motif for the Dsb protein family [36], is embedded between the 686th and 683th amino acid residues of the XC_0531 protein (Fig. 2b). Transmembrane helices analysis of XC_0531 showed that the protein spans the membrane nine times with its N terminus facing the cytoplasm and C terminus facing the outside (Fig. 2c), which is in accordance with the identified Dsb proteins [37]. These results suggest that the deduced protein of the ORF XC_0531 may be a DsbD protein. Another Dsb family protein encoded by XC_3579 in Xcc was used in this study to compare the function of XC_0531. XC_3579 is predicted to encode protein DsbC, a disulfide isomerase. It displays a 42% identity and 57% similarity to the thiol:disulfide interchange protein DsbC of Shewanella oneidensis MR-1 (NCBI-Protein ID: NP_716580, encoded by the SO_0951 gene), and 39% identity and 59% similarity to the DsbC protein (NCBI-Protein ID: NP_252426) of Pseudomonas aeruginosa PAO1.
Fig. 2.
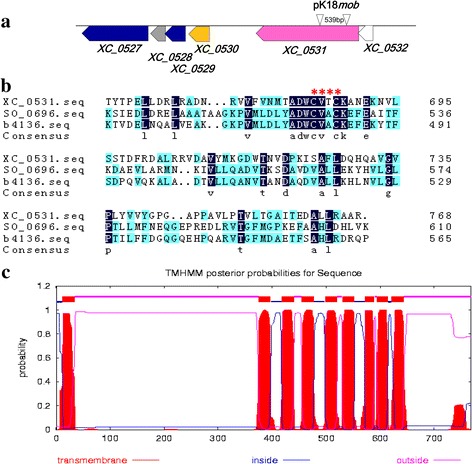
Sequence analyses of XC_0531 of Xanthomonas campestris pv. campestris. a Physical and genetic map of the Xanthomonas campestris pv. campestris dsbD region. The pink arrowhead stands for the dsbD encoding open reading frame XC_0531. The small arrows represent the position of primers used to construct of the 0531pk. b Alignment of amino acid sequence of DsbD homologs from Xanthomonas campestris pv. campestris 8004, Shewanella oneidensis MR-1and Escherichia coli str. K-12. The gene XC_0531 of Xanthomonas campestris pv. campestris, SO_0696 of Shewanella oneidensis MR-1 and b4136 of Escherichia coli str. K-12 were aligned. Navy blue boxes denote identical amino acid residues, whereas baby blue boxes are similar between the two sequences. The starred area denotes the typical redox active -Cys-X-X-Cys- motif of the disulfide bond formation protein family. c Prediction of transmembrane helices of XC_0531 of Xanthomonas campestris pv. campestris. This was analyzed with TMHMM Server v.2.0 software. Red blocks stand for the transmembrane helices in cytoplasmic membrane while pink threads for periplasmic loops and blue for regions inside the cytoplasm
Mutant of XC_0531 is deficient in biosynthesis of c-type cytochromes
XC_0531 encodes a putative, c-type cytochrome biogenesis protein homolog to DsbD (http://xgb.leibniz-fli.de). SDS-PAGE and heme staining were used to confirm this molecular property. Heme staining showed four major bands in the wild type (8004), the complemented strain (C0531) and deletion of XC_3579 (015F07), but no band was observed in the XC_0531 mutant (0531pk) (Fig. 3). Three of the four bands had similar molecular weights of c-type cytochromes, including 30 kD (might be cytochrome c553), 22 kD (might be cytochrome c552) and 14 kD (might be cytochrome c550) [38].
Fig. 3.
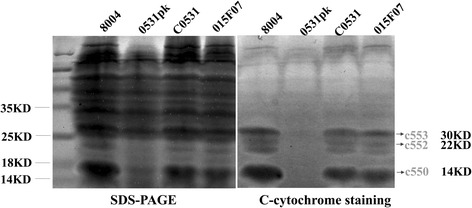
Heme staining of soluble membrane-bound proteins. Soluble membrane-bound proteins in 8004, 0531pk, C0531 and 015F07 were analyzed by heme staining after SDS-PAGE. Equivalent amounts of protein were loaded in each lane, shown stained in panel left. The membrane components were stained (Mrs, 14 kDa, 22 kDa and 30 kDa) in panel right. Lanes contained total soluble membrane-bound heme from 8004, 0531pk, C0531 and 015F07, as indicated. The positions of molecular mass markers are indicated
Redox difference spectroscopy was used to further analyze the nature of the Xcc cytochromes. A peak absorption at ~427 nm was observed in the wild type, C0531 and 015F07, but in the mutant (0531pk) the peak was shifted to 434 nm (Fig. 4). Characteristic of the porphyrin compounds Soret band is the peak absorption at ~420 nm [39]. In addition, the wild type strain (8004) showed a peak plateau at absorption from 553.5 nm to 564 nm, while the mutant (0531pk) had only one peak at the absorption of 564 nm, which is typical of b-type cytochromes [40] (Fig. 4). A previous study has shown that cytochromes c and b have two peaks between 550 nm and 560 nm, when absorption was measured at ultralow temperature [40]. It is likely that the plateau we observed in wild type is composed of two peaks of cytochromes c and b, but the peaks were not resolved at the room temperature we used for the measurement.The complement strain (C0531) and 015F07 had a wide peak similar to wild type with the maximum absorption at 558 nm and 556 nm, respectively. These results further suggest that XC_0531 not XC_3579 encodes a protein related to the biosynthesis of c-type cytochromes.
Fig. 4.
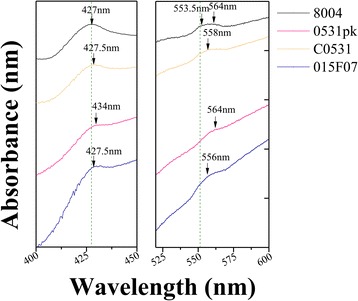
Room-temperature absorption spectra of soluble cytochromes in the membranes of 8004, 0531pk, C0531 and 015F07. The c-type cytochromes showed peaks at about 552 nm and the b-type cytochromes at about 560 nm. Samples were reduced with sodium dithionite or oxidized with ammonium persulfate. The spectra were reduced-minus-oxidized spectra at equal protein concentrations. Absorption maxima were indicated by arrows
XC_0531 affects the disulfide bond formation and copper tolerance
To determine whether disruption of the XC_0531 gene has any effect on the formation of disulfide bonds in the periplasmic proteins, we examined the free thiol groups in the periplasm by measuring the accumulation of proteins with reduced cysteines in periplasm using Ellman’s reaction method [41]. The result showed that the 0531pk had an increased accumulation of proteins with free thiols (Fig. 5a). In addition, the mutant strain 0531pk was more sensitive to the strong reducing agent DTT than the wild type strain (Fig. 5b). These phenotypes of the mutant were restored to wild type in the complement strain. The mutant 015F07 showed an accumulation similar to the wild type. These results suggest that ORF XC_0531 affects the disulfide bond formation, similar to Dsb proteins that catalyze disulfide bond formation in the periplasm.
Fig. 5.
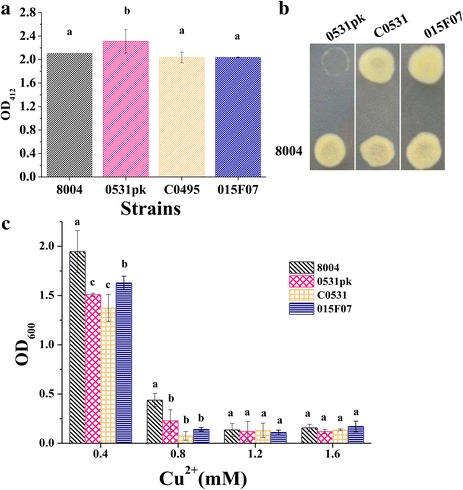
Analysis of the periplasmic disulfide oxidoreductase activity and copper tolerance of Xanthomonas campestris pv. campestris strains. a Accumulation of proteins with reduced cysteines determined by the Ellman’s reaction analysis. Value is the mean ± standard deviation from three repeats; OD412 = optical density at 412 nm. b Dithiothreitol (DTT) sensitivity detection. Overnight culture (2 μl) of each X. campestris pv. campestris strain was spotted onto NYGA supplemented with DTT to a final concentration of 4 mM and incubated at 28 °C. Photographs were taken 2 days after inoculation. Three plates were inoculated in each experiment and each experiment was repeated three times. Data presented were from representative plates and similar results were obtained in all plates of an experiment or in the plates of other independent experiments. c Copper tolerance of wild-type Xcc 8004, 0531pk, C0531 and 015F07. Overnight cultures were cultured in NYG containing various concentrations of CuSO4 for 24 to 48 h at 30 °C. OD600 were test. The values are averages of three independent experiments. Different letters indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA
In Escherichia coli, DsbD protein functions in both cytochromes c and in copper tolerance [12]. The analysis of copper tolerance of Xcc showed after 2 days’ culture in the presence of copper (0.4 mM or 0.8 mM of CuSO4), the OD600 of wide type is higher than strains 0531pk, C0531 and 015F07 (Fig. 5c). This indicated that 0531pk, C0531 and 015F07 were sensitive to copper. XC_0531 and XC_3579 functioned in copper tolerance. We supposed the complement strain might grow slower in double pressures with an introduced plasmid artificially and metal ions that did not compensate the phenotype of XC_0531 mutant in the test of Cu stress.
Deletion of XC_0531 delays cell growth and reduces EPS production
To further understand the physiological role of XC_0531 in Xcc, we compared the growth rates between the wild type (8004) and the mutant (0531pk) in NYG medium. As shown in Fig. 6a, the mutant grew much slower within the first 24 h of culture. The complemented strain showed a similar growth curve as the wild type. Cell growth was further tested on NY agar plates supplied with different carbon sources, including fructose, mannose, arabinose, rhamnose, galactose, xylose, maltose, saccharose and glucose. The mutant and the wild type grew similarly in colony size on fructose, mannose, arabinose and rhamnose. However, the colonies of mutants were smaller than the wild-type strain when growing on plates with galactose, xylose, maltose, saccharose and glucose (Fig. 6b). The complemented strain formed colonies similar to those of the wild-type on plates. These results suggest that the XC_0531 mutant is deficient in EPS production, which is critical to colony growth. To confirm that the XC_0531 is involved in EPS production, strains were cultured in NY liquid medium supplemented with 2% various sugars for 3 days. As summarized in Fig. 6c, the EPS production in the mutant was significantly lower than the wild type (P < 0.05) with galactose, xylose, maltose and saccharose. In addition, the EPS yield of the complemented strain showed no significant difference (P > 0.05) from that of the wild-type when cultured in medium containing these five carbohydrates. These results further confirm that the XC_0531 is involved in EPS production in Xcc. We also compared the colonies of Tn5 insertions in gene XC _ 3579 encoding DsbC (015F07, 045F12 and 142E11) to the wild type in the plates containing glucose. There was no significant difference between mutants and wild type. The EPS level in XC_3579 Tn5 gus mutants also showed similar characteristics to wild type (Additional file 2: Figure S1).
Fig. 6.
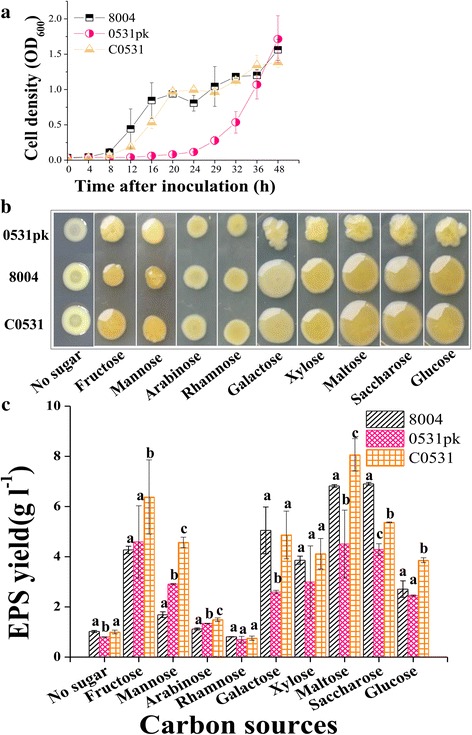
Cell growth and EPS production of Xcc strains at different sugars. a The growth of bacteria in radish leaf tissues was determined at OD600 in NY medium. b Strains were stabled into the plates followed by incubation at 28 °C for 4 days. c Strains in100 ml NY medium supplemented with or without 2% (wt/vol) various sugars at 28 °C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried and weighed. Significant difference (P < 0.05) tested by one way-ANOVA. Different letters within one sugar indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA
Mutant of XC_0531 is deficient in mobility
The cell motility of the XC_0531 mutant was also significantly reduced compared to the wild type using different sugars (P < 0.05) such as galactose, xylose, maltose, saccharose and glucose. The growth zone diameter of mutant 0531pk is 1.48 ± 0.04 cm with glucose as the carbon source and the wild type is 2.28 ± 0.11 cm (Fig. 7a and b). We don’t know why the mobilities of mutant and completment strains formed colonies larger than those of the wild type on plates containing fructose and mannose as sole carbon source (Fig. 7a and b). There was no difference in growth zone diameter between XC_3579 Tn5 mutants’ and wild type (Additional file 3: Figure S2). These results suggest that the XC_0531, but not XC_3579 affects the motility of Xcc.
Fig. 7.
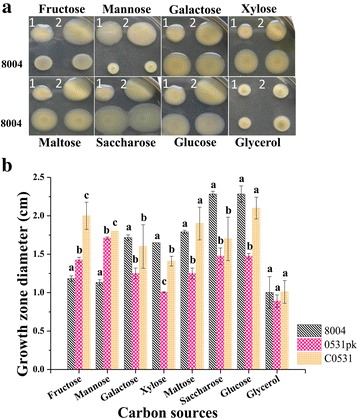
Test of the cell motility. Three μl of overnight culture (OD600 of 1.0) of each Xcc strain was inoculated onto NY plates containing 2% different sugars and 0.6% agar, and then incubated at 28 °C for 4 days to detect swarming motility. a Growth zone of each strain and b Quantitative measurement of growth zone in a. 1, 0531pk; 2, C0531. The experiment was repeated at least three times. Different letters within one sugar indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA
Mutant of XC_0531 reduces extracellular enzyme activities
Spots in the milk plates showed the transparent circle formed by mutant 0531pk is smaller than the wild type (Fig. 8a). The protease activity assay showed that the transparent circle in the mutant 0531pk was significantly smaller than that of the wild type (Fig. 8a and B). The complemented strain C0531 recovered the extracellular protease activity to that in the wild type in the plate assay but not in the liquid activity test (Fig. 8a and b). The Tn5 mutant 015F07 also showed a smaller transparent circle (Fig. 8a and b). This suggests that both XC_0531 and XC_3579 are involved in extracellular protease activity of Xcc.
Fig. 8.
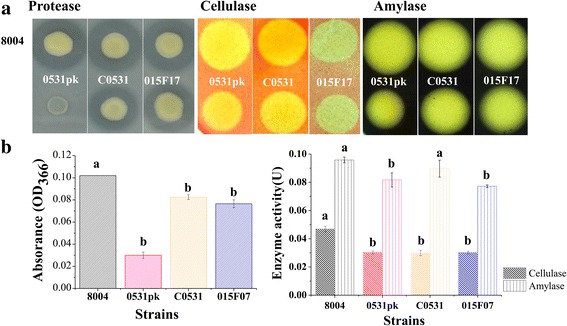
Examination of extracellular enzymes’ activities. a Overnight culture (3 ul) was spotted onto NYGA plates containing 1% skim milk (for protease), after incubation at 28 °C for 24 h, plates were photographed. For cellulose, 3 ul overnight culture was spotted onto NYGA plates containing 0.5% carboxymethyl cellulose after incubation at 28 °C for 48 h, plates were stained by I2/KI (0.08 M I2, 3.2 M KI) and washed by 70% ethyl alcohol. For amylse, 3 ul overnight cultures were spotted onto NYGA plates containing 0.1% starch incubation at 28 °C for 24 h. The plates were stained by 0.1% Congo Red and then washed 2 times. At last plates were destained by 1 M NaCl solution. b For extracellular protease activity, the method described by Swift and associates was used. 10 μl of enzyme-containing extracts were added to 200 μl of indicator buffer containing 1% (wt/vol) carboxymethyl cellulose as the substrate. The reactions were carried out for 30 min at 28 °C. The released reducing sugars were measured as D-glucose equivalents, as described by Miller. One unit (U) of the cellulase (endoglucanase) activity was defined as the amount of enzyme releasing 1 μmol of reducing sugar per minute. Amylase activity quantification was conducted in the same way as for the cellulose (endoglucanase) measurement, except that the substrate 1% (wt/vol) CMC was replaced by 1% (wt/vol) starch solution. Different letters within one enzyme activity assay indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA
In the test of extracellular cellulose and amylase activities, smaller transparent circles of 0531pk and 015F07 were compared to wild type 8004. The Tn5 mutant 015F07 showed a smaller transparent circle and lower protease activity compared to wild type) (P < 0.05) (Fig. 8a and c). The complemented strain C0531, however, did not fully recover the extracellular cellulase activity to the wild type (Fig. 8c).
The relationship between dsbD and dsbA, dsbB, or dsbC
To verify whether the function of gene dsbD in pathogenicity of Xcc is due to affect genes dsbA (XC_0674 and XC_0675), dsbB (XC_3314) or dsbC (XC_3579) expression, we checked transcript levels of dsbA, dsbB and dsbC in dsbD deleted and complemented strains. Real-time quantitative PCR was employed to assay the dsbA, dsbB and dsbC transcripts in dsbD deleted and complemented strains grown in NYG medium. The results showed that the transcription levels of dsbA, dsbB and dsbC between these two strains differed only within 1.1 folds (P > 0.05) (Additional file 4: Figure S3), suggesting that the dsbD expression of Xcc does not affect other dsb genes.
Discussions
Iron ions in the heme act as an activator to catalyze H2O2 to produce O2 and then the O2 oxidize TMBZ to turn the heme into blue [42]. Sequence analysis suggests that XC_0531 likely encodes protein DsbD. dsbD deletion caused deficient in c-type cytochromes, similar to a report in Brassica japonicum cox3, lacking all soluble c-type cytochromes [43]. By complement pL0531, all the deficit phenomena could be reversed to those phenomena in wild type. In the Rhodobacter species protein DsbD provides electrons to apoCyt C [13]. DsbD is also reported in Neisseria meningitidis to provide reducing power to DsbC that shuffles incorrect disulfide bond as well as to the periplasmic enzymes that reduce apo-cytochrome c (CcsX) [44]. In our results, no soluble heme was observed in the XC_0531 mutant, suggesting that XC_0531 deletion might affect the process of electrons to apoCyt C and then the process of heme ligation to the apoCyts C and the maturation of cytochromes C will be disturbed. Further test of cytochromes C using reduced-minus-oxidized spectra showed the characteristics of the porphyrin compounds Soret band in dsbD mutant red shift to 434 nm compared to that in wild type. The phenomena of red shift usually appeared due to the chromophores with unsaturated group such as heme with unsaturated group. The Soret band is a characteristic of porphyrin, therefore, the red shift of Soret band suggests that the proximal ligands with stronger ability of supply electrons (more electron clouds) appeared in the sole compand study [45]. Each strain could be supposed to a compand and the gene deletion could be regard as a replace of proximal ligands. The dsbD deletion could form a larger electrons cloud that could not be transferred smoothly. This also implied the process of electrons to apoCyt C was interrupted.
The c-type cytochromes are needed in the cellular respiratory chain for the electrons transfer to provide energy to the cells. Deletion of dsbD might affect the respiration of Xcc by losing synthesis of c-type cytochromes. This is one of the reasons that the dsbD deletion strain of Xcc grows slower, with poor carbohydrate’ utilization (such as galactose, xylose, maltose, saccharose and glucose), decreased enzymes activity (such asprotease, cellulose and amylase) and less production of EPS (under different sugar, such as, galactose, xylose, maltose, saccharose and glucose). EPS is the important pathogenic factor to the host plant. So the XC_0531 deletion also weakened virulence to plants. On the other hand, Dsb proteins of gram-negative bacteria are a group of proteins that catalyze disulfide bond formation in the periplasm [17]. DsbD has the capacity to reduce disulfide bonds and keeps the DsbC in a reduced state [46, 47]. Virulence factors such as extracellular enzymes and EPS are secreted via type II secretion system (T2SS) in gram-negative bacteria [48, 49]. The proteins of T2SS such as XpsD, XpsM, and XcsN located in periplasm all have cysteine residues and their assembly and cysteine residues modifications were completed in periplasm [35, 48–51]. Disulfide bonds are important for maintaining the structure of proteins. dsbD deletion might decrease the secretion and activities of extracelluar enzymes by affecting synthesis of the contents of T2SS and extracellular enzymes. These all affected the pathogenicity to plants.
Proteins DsbC and DsbA could affect extracellular enzyme formation in Pseudomonas aeruginosa or Erwinia carotovora [52–54]. In our results, we found the extracellular enzyme activities such as protease, cellulose and amylase all decreased in mutant of dsbD but not dsbC. This indicated that the decrease of extracellular enzyme activities in dsbD mutant is not related to DsbC. DsbD protein is also reported in Neisseria meningitidis to provide reducing power to the periplasmic enzymes relative to reduction of apo-cytochrome c [44]. Cytochrome c is a part of respiratory chain. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells [55]. Our qPCR results also showed that in complemented strain C0531 transcription levels of dsbA, dsbB and dsbC were the same as that in mutant 0531pk (Additional file 4: Figure S3). Gene dsbD expression in C0531 did not affect transcription of dsbA, dsbB or dsbC. This further demonstrated dsbD deletion might affect the protein modification process of DsbA-DsbB disulfide bond formation system by deficiency of cytochromes C on the protein level not the mRNA level.
Our results suggest that, similar to DsbB [17], DsbD affects the formation of disulfide bond. Proteins DsbD and DsbC functioned in copper tolerance like those in Escherichia coli [12]. The complement strain did not compensate the phenotype of XC_0531 mutant. We believe that the complement strain might grow slower in double pressures with an introduced plasmid and the presence of copper stress. Deletion of XC_3579 (encodes a DsbC protein) gave no significant differences to the wild type on the EPS production, utilization of carbohydrates, virulence to plant and c-type cytochromes biogenesis. dsbC encoded disulfide isomerase could correct misfolded proteins [12]. Its possible recessive nature (functions only in the presence of misfolded proteins) may explain why its deletion did not affect any phenomena in Xcc.
Conclusions
The phytopathogenic Xanthomonas campestris pv. campestris is a gram-negative bacterium and the causal agent of black-rot disease of cruciferous crops. This study is of important to Xanthomonas pathogenesis. We confirmed the function of gene XC_0531, which encodes a DsbD like protein, a protein correlated with c-type cytochrome biogenesis, polysaccharide production and the virulence to plants. It is also probably related to disulfide bonds modification of proteins in type II secretion system (T2SS). These results will provide data to better understand the molecular mechanism of pathogenesis related to Xanthomonas campestris pv. campestris.
Additional files
Primers used in this study. (TIFF 3416 kb)
EPS production of Xcc strains at glucose plate. Strains were stabled into the plates followed by incubation at 28 °C for 4 days. Strains in 100 ml NY medium supplemented with 2% (wt/vol) various sugars at 28 °C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried and weighed. Different letters within one sugar indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA. (TIFF 5151 kb)
Test of the cell motility. An overnight culture (OD600 of 1.0) of each Xcc strain was inoculatedonto NY plates containing 2% glucose and 0.6% agar using a toothpick, and then incubated at 28 °C for 4 days to detect swarming motility.1, 015F07; 2, 045F12; 3, 142E11. (TIFF 2153 kb)
Real-time quantitative PCR analysis. Real-time quantitative PCR to analyze the expression of the genes dsbA (XC_0674 and XC_0675), B (XC_3314) and C (XC_3579) in strains 0531pk and C0531. RNA was isolated from cultures of Xcc strain 0531pk and C0531 grown in NYG medium alone for 24 h. The relative mRNA levels of dsbA (XC_0674 and XC_0675), B (XC_3314) and C (XC_3579) in C0531 was calculated with levels of the corresponding transcription in cells of 0531pk (values was specified as 1). Values given are the means ± SD of triplicate measurements. (DOCX 13 kb)
Acknowledgements
None.
Funding
This research was supported by the National Natural Science Foundation of China (No.31371263), the Young Scientists Fund (No. 41401285) of the National Natural Science Foundation of China, the Open Project of State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (HC201713) and Yantai Primary Research & Development Plan (2016ZH072).
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
Abbreviations
- ANOVA
One-way analysis of variance
- DTNB
5, 5′-dithiobis-[2-nitrobenzoic acid]
- DTT
dithiothreitol
- EPS
extracellular polysaccharide
- Kan
kanamycin
- NCBI
National Center for Biotechnology Information
- Rif
rifampicin
- Tet
tetracycline
- TMBZ
3,3′,5,5′- tetramethyl benzidine
- Xcc
Xanthomonas campestris pv. campestris
Authors’ contributions
LC performed the experiments, analyzed the data and wrote the paper. MW and LH performed partial experiments. ZZ participated in the design of study, results analsis and revision of manuscript. FL assisted in results analysis and revision of manuscript. GL designed the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-017-1056-9) contains supplementary material, which is available to authorized users.
References
- 1.Bardischewsky F, Friedrich CG. Identification of ccdA in Paracoccus pantotrophus GB17: disruption of ccdA causes complete deficiency in c-type cytochromes. J Bacteriol. 2001;183(1):257–263. doi: 10.1128/JB.183.1.257-263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell JCA, Mcgovern K, Beckwith J. Identification of a protein required for disulfide bond formation Invivo. Cell. 1991;67(3):581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell JCA, Beckwith J. The bonds that tie - catalyzed disulfide bond formation. Cell. 1993;74(5):769–771. doi: 10.1016/0092-8674(93)90455-Y. [DOI] [PubMed] [Google Scholar]
- 4.Dailey FE, Berg HC. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia-Coli. Proc Natl Acad Sci U S A. 1993;90(3):1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamitani S, Akiyama Y, Ito K. Identification and characterization of an Escherichia-Coli Gene required for the formation of correctly folded alkaline-phosphatase, a periplasmic enzyme. EMBO J. 1992;11(1):57–62. doi: 10.1002/j.1460-2075.1992.tb05027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia-Coli Gene Dsbb, whose product is involved in the formation of disulfide bonds in-vivo. Proc Natl Acad Sci U S A. 1993;90(15):7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Missiakas D, Georgopoulos C, Raina S. The Escherichia-Coli Dsbc (Xpra) Gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13(8):2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wunderlich M, Otto A, Seckler R, Glockshuber R. Bacterial protein disulfide-isomerase - efficient catalysis of oxidative protein-folding at acidic Ph. Biochemistry. 1993;32(45):12251–12256. doi: 10.1021/bi00096a039. [DOI] [PubMed] [Google Scholar]
- 9.Zapun A, Bardwell JCA, Creighton TE. The reactive and destabilizing disulfide bond of Dsba, a protein required for protein disulfide bond formation Invivo. Biochemistry. 1993;32(19):5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 10.Zapun A, Creighton TE. Effects of Dsba on the disulfide folding of bovine pancreatic trypsin-inhibitor and alpha-Lactalbumin. Biochemistry. 1994;33(17):5202–5211. doi: 10.1021/bi00183a025. [DOI] [PubMed] [Google Scholar]
- 11.Zapun A, Missiakas D, Raina S, Creighton TE. Structural and functional-characterization of Dsbc, a protein involved in disulfide bond formation in Escherichia-Coli. Biochemistry. 1995;34(15):5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 12.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (Dsbd) in Escherichia-Coli. EMBO J. 1995;14(14):3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders C, Turkarslan S, Onder O, Frawley ER, Kranz RG, Koch HG, et al. Biogenesis of c-type cytochromes and cytochrome complexes. In The purple phototropic bacteria Springers Netherlands. 2009;
- 14.Chen L, Liu LY, Wang MP, Fu JF, Zhang ZJ, Hou J, Bao Xm : Hal2p Functions in Bdf1p-Involved Salt Stress Response in Saccharomyces cerevisiae. PLoS One 2013, 8(4). [DOI] [PMC free article] [PubMed]
- 15.Chen L, Wang MP, Hou J, Liu LY, Fu JF, Shen Y, Zhang ZJ, Bao XM. Regulation of Saccharomyces cerevisiaemef1 by Hda1p affects salt resistance of bdf1 mutant. FEMS Yeast Res. 2014;14(4):575–585. doi: 10.1111/1567-1364.12144. [DOI] [PubMed] [Google Scholar]
- 16.Fabianek RA, Hofer T, Thony-Meyer L. Characterization of the Escherichia Coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171(2):92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 17.Jiang BL, Liu J, Chen LF, Ge YY, Hang XH, He YQ, Tang DJ, Lu GT, Tang JL. DsbB is required for the pathogenesis process of Xanthomonas campestris pv. Campestris. Mol Plant Microbe In. 2008;21(8):1036–1045. doi: 10.1094/MPMI-21-8-1036. [DOI] [PubMed] [Google Scholar]
- 18.Tang DJ, He YQ, Feng JX, He BR, Jiang BL, Lu GT, Chen BS, Tang JL. Xanthomonas campestris pv. Campestris possesses a single gluconeogenic pathway that is required for virulence. J Bacteriol. 2005;187(17):6231–6237. doi: 10.1128/JB.187.17.6231-6237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mamiatis T, Fritsch ET, Sambrook J, Engel J. Molecular cloning–A laboratory manual., 1982 edn: New York. Cold Spring Harbor Laboratory. 1985;
- 20.Turner P, Barber C, Daniels M. Behavior of the transposons Tn5 and Tn7 in Xanthomonas-Campestris Pv Campestris. Mol Gen Genet. 1984;195(1–2):101–107. doi: 10.1007/BF00332731. [DOI] [Google Scholar]
- 21.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia Coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145(1):69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 22.Windgassen M, Urban A, Jaeger KE. Rapid gene inactivation in Pseudomonas Aeruginosa. FEMS Microbiol Lett. 2000;193(2):201–205. doi: 10.1111/j.1574-6968.2000.tb09424.x. [DOI] [PubMed] [Google Scholar]
- 23.Lu GT, Ma ZF, Hu JR, Tang DJ, He YQ, Feng JX, Tang JL. A novel locus involved in extracellular polysaccharide production and virulence of Xanthomonas campestris pathovar campestris. Microbiol-Sgm. 2007;153:737–746. doi: 10.1099/mic.0.2006/001388-0. [DOI] [PubMed] [Google Scholar]
- 24.Yanischperron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains - nucleotide-sequences of the M13mp18 and Puc19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 25.Leong SA, Ditta GS, Helinski DR. Heme-biosynthesis in rhizobium - Identification of a cloned Gene coding for Delta-Aminolevulinic-acid Synthetase from rhizobium-Meliloti. J Biol Chem. 1982;257(15):8724–8730. [PubMed] [Google Scholar]
- 26.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned Avirulence genes from race-0 and race-1 of Pseudomonas-Syringae Pv Glycinea. J Bacteriol. 1987;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, Tang JL. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A. 2003;100(19):10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 29.Dumont ME, Ernst JF, Sherman F. Coupling of Heme attachment to import of cytochrome-C into yeast mitochondria - studies with Heme Lyase-deficient mitochondria and altered Apocytochromes-C. J Biol Chem. 1988;263(31):15928–15937. [PubMed] [Google Scholar]
- 30.Chen L, Wang M, Hou J, Fu J, Shen Y, Liu F, et al. HAL2 overexpression induces iron acquisition in bdf1Delta cells and enhances their salt resistance. Curr Genet. 2016; [DOI] [PubMed]
- 31.Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, Daniels MJ. Genetic and molecular analysis of a cluster of Rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas-Campestris Pathovar Campestris. Mol Gen Genet. 1991;226(3):409–417. doi: 10.1007/BF00260653. [DOI] [PubMed] [Google Scholar]
- 32.Lu GT, Xie JR, Chen L, Hu JR, An SQ, Su HZ, Feng JX, He YQ, Jiang BL, Tang DJ, et al. Glyceraldehyde-3-phosphate dehydrogenase of Xanthomonas campestris pv. campestris is required for extracellular polysaccharide production and full virulence. Microbiol-Sgm. 2009;155:1602–1612. doi: 10.1099/mic.0.023762-0. [DOI] [PubMed] [Google Scholar]
- 33.Su HZ, Wu L, qi YH, Liu GF, Lu GT, Tang JL. characterization of the GntR family regulator HpaR1 of the crucifer black rot pathogen Xanthomonas campestris pathovar campestris. Sci Rep. 2016;6:19862. doi: 10.1038/srep19862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swift S, Lynch MJ, Fish L, Kirke DF, Tomas JM, Stewart GSAB, Williams P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun. 1999;67(10):5192–5199. doi: 10.1128/iai.67.10.5192-5199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian W, Jia Y, Ren SX, He YQ, Feng JX, Lu LF, Sun Q, Ying G, Tang DJ, Tang H, et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. Campestris. Genome Res. 2005;15(6):757–767. doi: 10.1101/gr.3378705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabianek RA, Hennecke H, Thony-Meyer L. Periplasmic protein thiol:disulfide oxidoreductases of Escherichia Coli. FEMS Microbiol Rev. 2000;24(3):303–316. doi: 10.1111/j.1574-6976.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 37.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 38.Page MD, Ferguson SJ. Cloning and sequence-analysis of Cych Gene from Paracoccus-Denitrificans - the Cych Gene-product is required for assembly of all C-type cytochromes, including cytochrome C(1) Mol Microbiol. 1995;15(2):307–318. doi: 10.1111/j.1365-2958.1995.tb02245.x. [DOI] [PubMed] [Google Scholar]
- 39.Loew GH, Harris DL. Role of the heme active site and protein environment in structure, spectra, and function of the cytochrome p450s. Chem Rev. 2000;100(2):407–420. doi: 10.1021/cr980389x. [DOI] [PubMed] [Google Scholar]
- 40.Albouy D, Sturgis JN, Feiler U, Nitschke W, Robert B. Membrane-associated c-type cytochromes from the green sulfur bacterium Chlorobium limicola forma thiosulfatophilum: purification and characterization of cytochrome c(553) Biochemistry. 1997;36(7):1927–1932. doi: 10.1021/bi962624g. [DOI] [PubMed] [Google Scholar]
- 41.Riddles PW, Blakeley RL, Zerner B. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)--a reexamination. Anal Biochem. 1979;94(1):75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- 42.Josephy PD, Eling T, Mason RP. The horseradish peroxidase-catalyzed oxidation of 3,5,3′,5′-Tetramethylbenzidine - free-radical and charge-transfer complex intermediates. J Biol Chem. 1982;257(7):3669–3675. [PubMed] [Google Scholar]
- 43.Ritz D, Bott M, Hennecke H. Formation of several bacterial C-type cytochromes requires a novel membrane-anchored protein that faces the periplasm. Mol Microbiol. 1993;9(4):729–740. doi: 10.1111/j.1365-2958.1993.tb01733.x. [DOI] [PubMed] [Google Scholar]
- 44.Kumar P, Sannigrahi S, Scoullar J, Kahler CM, Tzeng YL. Characterization of DsbD in Neisseria meningitidis. Mol Microbiol. 2011;79(6):1557–1573. doi: 10.1111/j.1365-2958.2011.07546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang MX, Li Q. UV-visible spectra of porphyrin and its derivatives. Chinese Journal of Spectroscopy Laboratory. 2011;3:045. [Google Scholar]
- 46.Stewart EJ, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia Coli. EMBO J. 1999;18(21):5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sone M, Akiyama Y, Ito K. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J Biol Chem. 1997;272(16):10349–10352. doi: 10.1074/jbc.272.16.10349. [DOI] [PubMed] [Google Scholar]
- 48.Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13(12):581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS microbio lett. 2006;255(2):175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 50.Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694(1–3):163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Sandkvist M. Type II secretion and pathogenesis. Infect Immun. 2001;69(6):3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun P, Ockhuijsen C, Eppens E, Koster M, Bitter W, Tommassen J. Maturation of Pseudomonas Aeruginosa elastase. Formation of the disulfide bonds. J Biol Chem. 2001;276(28):26030–26035. doi: 10.1074/jbc.M007122200. [DOI] [PubMed] [Google Scholar]
- 53.Urban A, Leipelt M, Eggert T, Jaeger KE. DsbA and DsbC affect extracellular enzyme formation in Pseudomonas Aeruginosa. J Bacteriol. 2001;183(2):587–596. doi: 10.1128/JB.183.2.587-596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vincent-Sealy LV, Thomas JD, Commander P, Salmond GP. Erwinia carotovora DsbA mutants: evidence for a periplasmic-stress signal transduction system affecting transcription of genes encoding secreted proteins. Microbiology. 1999;145(Pt 8):1945–1958. doi: 10.1099/13500872-145-8-1945. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia Coli cells. Proc Natl Acad Sci U S A. 1997;94(22):11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study. (TIFF 3416 kb)
EPS production of Xcc strains at glucose plate. Strains were stabled into the plates followed by incubation at 28 °C for 4 days. Strains in 100 ml NY medium supplemented with 2% (wt/vol) various sugars at 28 °C with shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, dried and weighed. Different letters within one sugar indicate significant differences at a level of α < 0.05 based on Duncan’s test by one way-ANOVA. (TIFF 5151 kb)
Test of the cell motility. An overnight culture (OD600 of 1.0) of each Xcc strain was inoculatedonto NY plates containing 2% glucose and 0.6% agar using a toothpick, and then incubated at 28 °C for 4 days to detect swarming motility.1, 015F07; 2, 045F12; 3, 142E11. (TIFF 2153 kb)
Real-time quantitative PCR analysis. Real-time quantitative PCR to analyze the expression of the genes dsbA (XC_0674 and XC_0675), B (XC_3314) and C (XC_3579) in strains 0531pk and C0531. RNA was isolated from cultures of Xcc strain 0531pk and C0531 grown in NYG medium alone for 24 h. The relative mRNA levels of dsbA (XC_0674 and XC_0675), B (XC_3314) and C (XC_3579) in C0531 was calculated with levels of the corresponding transcription in cells of 0531pk (values was specified as 1). Values given are the means ± SD of triplicate measurements. (DOCX 13 kb)
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.


