Abstract
Plants produce diverse isoprenoids, which are synthesized in plastids, mitochondria, endoplasmic reticulum (ER), and the nonorganellar cytoplasm. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) catalyzes the synthesis of mevalonate, a rate-limiting step in the cytoplasmic pathway. Several branches of the pathway lead to the synthesis of structurally and functionally varied, yet essential, isoprenoids. Several HMGR isoforms have been identified in all plants examined. Studies based on gene expression and on fractionation of enzyme activity suggested that subcellular compartmentalization of HMGR is an important intracellular channeling mechanism for the production of the specific classes of isoprenoids. Plant HMGR has been shown previously to insert in vitro into the membrane of microsomal vesicles, but the final in vivo subcellular localization(s) remains controversial. To address the latter in Arabidopsis (Arabidopsis thaliana) cells, we conducted a multipronged microscopy and cell fractionation approach that included imaging of chimeric HMGR green fluorescent protein localizations in transiently transformed cell leaves, immunofluorescence confocal microscopy in wild-type and stably transformed seedlings, immunogold electron microscopy examinations of endogenous HMGR in seedling cotyledons, and sucrose density gradient analyses of HMGR-containing organelles. Taken together, the results reveal that endogenous Arabidopsis HMGR is localized at steady state within ER as expected, but surprisingly also predominantly within spherical, vesicular structures that range from 0.2- to 0.6-μm diameter, located in the cytoplasm and within the central vacuole in differentiated cotyledon cells. The N-terminal region, including the transmembrane domain of HMGR, was found to be necessary and sufficient for directing HMGR to ER and the spherical structures. It is believed, although not directly demonstrated, that these vesicle-like structures are derived from segments of HMGR-ER. Nevertheless, they represent a previously undescribed subcellular compartment likely capable of synthesizing mevalonate, which provides new evidence for multiorganelle compartmentalization of the isoprenoid biosynthetic pathways in plants.
The reaction catalyzed by the enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) is the first committed step in the mevalonate pathway of isoprenoid biosynthesis (Bach, 1995). Plants synthesize an enormous and diverse array of isoprenoids (in excess of 29,000 different compounds), which function in many aspects of growth, development, reproduction, and disease resistance (Chappell, 1995). Consistent with this complexity, all plant species studied so far have several HMGR isoforms that likely are critical in directing the flux of pathway intermediates into specific isoprenoid compounds (Enjuto et al., 1994; Chappell, 1995; Weissenborn et al., 1995). Although synthesis of many of these compounds occurs in the nonorganellar cytoplasm (sterols, sesquiterpenes), several classes of isoprenoids are synthesized in plastids (carotenoids, plastoquinone, and phylloquinone), mitochondria (ubiquinone), or specialized vacuoles (rubber particles; McGarvey and Croteau, 1995). The mevalonate pathway had been considered for many years as the unique source for the synthesis of isoprenoids. However, an alternative mevalonate-independent pathway for isoprenoid biosynthesis, also known as the methylerythritol phosphate pathway, recently has been identified in bacteria, plants, algae, and protozoa. In plants, this novel pathway is confined to the plastidic compartment (Rohdich et al., 2001; Rodríguez-Concepción and Boronat, 2002).
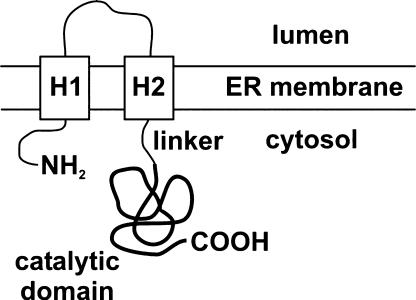
The in vivo subcellular localization of plant HMGRs remains controversial (Bach, 1995), partially due to lack of suitable specific antibodies against the protein. Plant HMGRs have a membrane domain consisting of two separate hydrophobic amino acid stretches that are linked to the highly conserved cytoplasmic domain bearing the catalytic center (Fig. 1). Arabidopsis (Arabidopsis thaliana) contains two differentially expressed HMGR genes (HMG1 and HMG2) that encode three HMGR isoforms: HMGR1S, HMGR1L, and HMGR2. The HMGR1S and HMGR1L isoforms derive from the HMG1 gene (Lumbreras et al., 1995). These proteins are identical in amino acid sequence, except that HMGR1L is extended at its N terminus by an additional region of 50 amino acid residues. The catalytic domains at the C-terminal end are highly conserved, sharing 78% amino acid identity. The different HMGR isoforms exhibit distinct spatial and developmental patterns of expression and respond differently to environmental stimuli (Lumbreras et al., 1995; Weissenborn et al., 1995). Based on protein amounts and expression patterns in both aerial leaves of adult plants and in seedlings, Arabidopsis HMGR1S is believed to function as a housekeeping form of the enzyme (Enjuto et al., 1994). The three Arabidopsis HMGR isoforms insert in vitro into dog pancreas endoplasmic reticulum (ER)-derived vesicles (Enjuto et al., 1994; Lumbreras et al., 1995). The generally accepted hypothesis is that they are first targeted to the ER (Campos and Boronat, 1995), which is in agreement with results showing insertion of tomato HMGR into microsomal membranes (Denbow et al., 1996) and localization of yeast (Saccharomyces cerevisiae) HMGR in ER (Hampton et al., 1996).
Figure 1.
Topological model proposed for plant HMGR in the ER membrane. H1 and H2 represent the transmembrane hydrophobic regions (adapted from Campos and Boronat, 1995). The catalytic domain is represented with a bold line. In the isoform HMGR1S (592 amino acid residues), the catalytic domain starts at position 172.
However, an exclusive ER localization raises the question of how the different isoforms of HMGR, which presumably would all be present in the same endomembrane system with their catalytic domain exposed to the cytoplasm, could channel intermediates into specific families of isoprenoids and discriminate against flux into other isoprenoids. Such functional specializations have been suggested to be facilitated by discrete isoforms within distinct ER regions (Campos and Boronat, 1995; Chappell, 1995; McCaskill and Croteau, 1998). Studies on this topic often have been hampered by the complexity of membrane-trafficking routes in plant cells (Okita and Rogers, 1996; Hawes et al., 1999; Nebenführ, 2002; Neumann et al., 2003). In support of this hypothesis, however, the multifunctional nature of the plant ER has been attributed in several cases to a multiplicity of specialized ER regions other than the three classical subcompartments, rough ER, smooth ER, and the nuclear envelope (Okita and Rogers, 1996; Staehelin, 1997; Mullen et al., 1999; Choi et al., 2000; Naested et al., 2000; Braun, 2001; Sami-Subbu et al., 2001; Hamada et al., 2003).
In this paper, we used a multipronged approach to determine the subcellular site(s) of Arabidopsis HMGR1S, the housekeeping form of the enzyme. Intracellular sorting experiments were performed with transiently expressed green fluorescent protein (GFP) chimeras possessing the N-terminal region of HMGR1S. The main thrust, however, was to determine the site(s) of the endogenous enzyme in wild-type (nontransformed) seedlings, using high-fidelity antibodies raised against the catalytic domain of HMGR1 as a specific probe for in vitro immunoblot analyses of subcellular fractions, confocal immunofluorescence microscopy, and immunogold electron microscopy. The results show that, contrary to the current prevailing hypothesis, the main reservoir of Arabidopsis HMGR in cotyledons is not within the reticular ER, but within novel cytoplasmic and intravacuolar vesicular structures that seem to be derived from subdomains of the ER.
RESULTS
The Amino-Terminal Domain of HMGR1S Is Sufficient for Directing GFP to the ER and to Spherical Structures
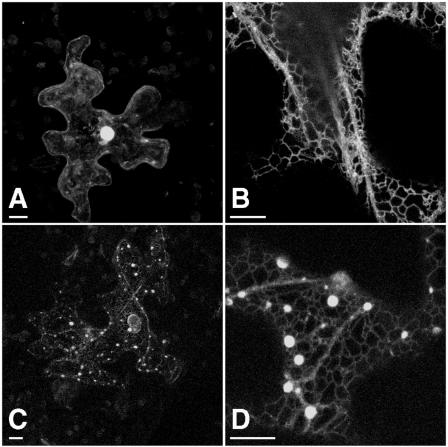
With the objective of investigating the subcellular sorting/targeting of HMGR1S and determining the role in these processes played by its amino-terminal domain, a chimeric protein was constructed such that the catalytic domain encoded by HMG1 was replaced with GFP. Thus, the polypeptide fragment of HMGR1S fused to GFP was amino acid residues 1 to 178 that included the cytoplasm-facing N-terminal region, the membrane domain consisting of both transmembrane sequences (H1 and H2) and the luminal stretch, and the linker region (Fig. 1). The resulting chimeric protein 1S:GFP was transiently expressed under control of the cauliflower mosaic virus 35S promoter in epidermal cells of 5-week-old Arabidopsis leaves after microbombardment. Confocal laser microscopy was used to determine the localization pattern of the expressed proteins (Fig. 2). Figure 2A is a positive control image showing the typical nuclear and nonorganellar cytoplasmic localization of transiently expressed, nonchimeric GFP in a transformed leaf epidermal cell. Figure 2B illustrates the results of another positive control, namely, an ER-targeted construct, pEGFPer (Danon et al., 2004), which clearly sorted to and appears within reticular ER. Figure 2, C and D, shows as expected the localization of chimeric 1S:GFP in reticular ER comparable to that of pEGFPer (Fig. 2B). Of particular interest, however, 1S:GFP also was observed within spherical structures located along the ER strands that ranged in apparent diameter from 0.5 to 2 μm. Measurements of this sort are subject to varying intensities of transiently expressed GFP fusion proteins. In conclusion, the N-terminal region of HMGR1S, with its two transmembrane domains, is sufficient to direct GFP to both reticular ER and the spherical structures.
Figure 2.
Transient expression in Arabidopsis leaves (5-week-old plants) of the chimeric protein composed of the amino-terminal membrane domain of HMGR1S fused to GFP (1S:GFP). Confocal microscopy localization patterns within epidermal cells expressing control GFP (A), the control lumenal ER construct pEGFPer (B), and the fusion 1S:GFP (C and D) at two different magnifications. All images are three-dimensional projections of 10 sections. Bars = 10 μm.
Endogenous Arabidopsis HMGR Is Localized to Spherical Structures in Cotyledon Cells
Overexpressed, membrane-targeted GFP fusion proteins have been reported to form organelle aggregates, which can lead to misinterpretations of sorting pathways for trafficked proteins (Lisenbee et al., 2003b). This concern has been expressed particularly for ER-targeted GFP (e.g. Gunning, 1998; Hawes et al., 2001). Also, elevated levels of yeast HMGR expression can result in its deregulation (Donald et al., 1997) and changes in subcellular localization of its isoforms (Koning et al., 1996). Consideration of these potential problems prompted us to focus on localization of endogenous Arabidopsis HMGR1S.
Rabbit polyclonal antibodies raised against the catalytic domain of HMGR1 (anti-CD1; Manzano et al., 2004) were employed in our confocal immunofluorescence microscopy experiments. Immunoblot analyses of wild-type Arabidopsis extracts (and subcellular fractions; Fig. 6A) revealed a 63-kD polypeptide corresponding to the estimated molecular mass of the HMGR1S isoform. Preincubations of anti-CD1 antiserum with excess recombinant HMGR1 catalytic domain resulted in the absence of immunoblot signals (data not shown), providing further evidence for the specificity of the antiserum. Anti-CD1 antibodies also recognized the catalytic domain of HMGR2 (CD2), as evidenced by immunoblot detections in extracts of transgenic plants transformed with CD2 (González, 2002). The failure to detect endogenous HMGR2 (60 kD) or HMGR1L (69 kD) isoforms in wild-type plant extracts on the same immunoblots as HMGR1S indicates that these isoforms exist at much lower levels than HMGR1S.
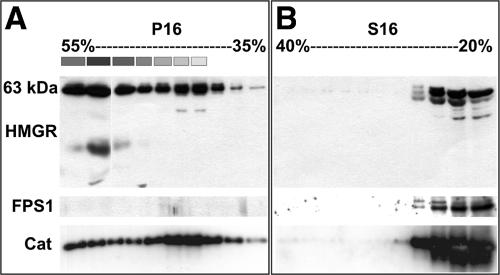
Figure 6.
Immunoblot analyses of HMGR-containing fractions derived from leaf samples (5-week-old plants) separated in Suc density gradients. A, Fractionation of P16 in a 35% to 55% (w/v) Suc density gradient. Thirty microliters of every other fraction (1–19) were loaded in each well of SDS gels. The plastid-containing fractions are indicated by boxes above the corresponding lanes, where the different shades of gray are proportional to the amount of chlorophyll determined spectrophotometrically. B, Fractionation of S16 in a 20% to 40% (w/v) Suc density gradient. Thirty microliters of every other fraction (1–19) were loaded in each well of SDS gels. PVDF membranes representing each gradient were incubated first with anti-CD1 antiserum (top blots), polypeptides were detected, and then the blots were incubated with anti-FPS1 antiserum (middle blots) and polypeptides detected again. For catalase detections (bottom blots), separate gels were loaded, electroblotted, and immunodetected for catalase polypeptides (approximately 57 kD).
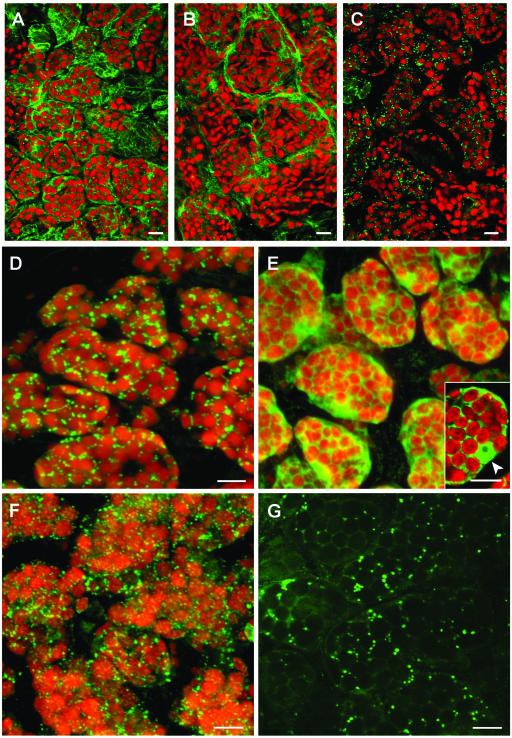
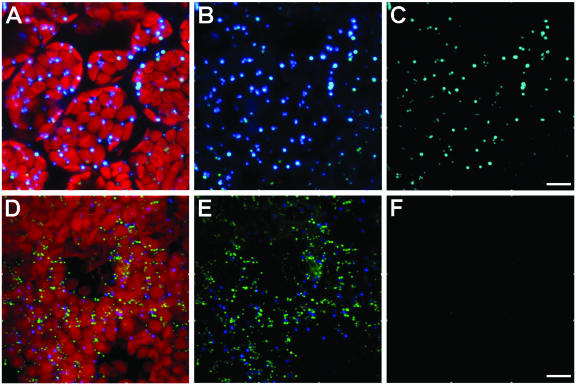
Immunofluorescence experiments (Fig. 3) were done with whole mounts of 6-d-old seedling cotyledons rather than with older plantlets or true leaves because difficulties were encountered with the penetration of the antibodies in the older plant parts, possibly due to the thicker and/or more cross-linked cell walls. Negative controls included omission of primary antibody (data not shown), and positive controls included applications of antiserum raised against the tonoplast membrane protein, α-tonoplast intrinsic protein (α-TIP; Höfte and Chrispeels, 1992; Fig. 3A) or antiserum raised against cytoplasmic farnesyl diphosphate synthase isoform 1 (FPS1; Cunillera et al., 1997; Fig. 3B). Figure 3, C and D, shows that most of the HMGR signal (anti-CD1 antibody) is associated with spherical structures distributed throughout the cell, singly or in small groups, with apparent diameters ranging from 0.4 to 1.3 μm. The presence of HMGR in plastids has been a subject of debate (Brooker and Russell, 1975; Wong et al., 1982). For this reason, the autofluorescence of chlorophyll in Figure 3 is shown to provide evidence for the lack of detectable levels of HMGR in plastids.
Figure 3.
Whole-mount immunofluorescence confocal microscopy of 6-d-old Arabidopsis cotyledons. A to D, Double-fluorescence x-y confocal optical sections of wild-type Arabidopsis cotyledon parenchyma cells showing endogenous plastid fluorescence (red) and subcellular immunodetection (green) of α-TIP (A), FPS1 (B), and HMGR1 (anti-CD1 antibodies; C and D). E and F, Double-fluorescence confocal imaging of the autofluorescence of plastids (red) and anti-CD1 antibodies (green) in cotyledons of plants stably transformed with the catalytic domain of HMGR1 (CD1; E), and with isoform HMGR1S (F). Inset in E is a representative middle section showing the cytoplasmic and nuclear localization of the catalytic domain of HMGR1. The nucleus, containing a nucleolus, is indicated with an arrowhead. G, Single confocal section in a nontransformed cotyledon cell imaged with increased photomultiplier intensity of the green channel revealing HMGR signal in spherical structures as well as in a polygonal, reticular network. A to F, Three-dimensional projections of a series of 40 to 50 sections with a z step of 0.25 μm. Scale bars = 100 μm (A–C); 10 μm (D–G).
To determine whether the amino-terminal domain of HMGR is essential for acquisition of the enzyme into the spherical structures, a transgenic line overexpressing the catalytic domain (CD1) was generated. Figure 3E shows that the truncated protein exhibited an expected cytoplasmic localization, and the inset shows that CD1 also existed in the nucleus of transformed cells. This result provides indirect evidence that the amino-terminal domain of HMGR is necessary for directing the protein to spherical structures. Figure 3F, however, shows that spherical structures acquire HMGR1S in cells of plants stably transformed with this protein (González, 2002), a result that, when coupled with that presented in Figure 3E, provides more direct evidence for the necessity of the HMGR N-terminal region for this targeting.
The confocal immunofluorescence data described above indicate that the spherical structures are the main repository of Arabidopsis HMGR. Figure 3G shows the image of an individual confocal section of nontransformed cotyledon cells when the intensity of the green HMGR signal has been increased significantly over the red channel. This manipulation reveals that the endogenous HMGR immunofluorescence signal emanates also from a reticulated system observed in the spaces between the plastids. This latter image is consistent with a concomitant ER localization, which is similar to the reticular ER localizations observed in our GFP-fusion experiments (Fig. 2).
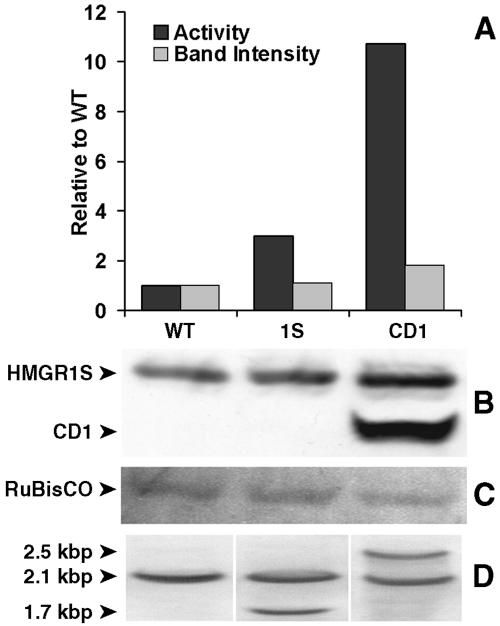
The transgenic line transformed with the sequence coding for isoform 1S had significantly higher HMGR-specific activity than wild-type plants (Fig. 4A), whereas the corresponding increase in protein was relatively modest (Fig. 4B). The CD1-transformed plants showed the highest HMGR-specific activity (Fig. 4A). In these plants, an intense immunoblot signal for CD1 (50 kD) was, intriguingly, accompanied by a clearly increased signal for HMGR1S. Southern blots of the transgenic lines indicated that both contained a single extra DNA copy of the corresponding forms (Fig. 4D).
Figure 4.
HMGR activity assays, immunoblots, and Southern blots of extracts from the wild-type (WT) and transgenic plant lines (1S and CD1) that were analyzed by confocal microscopy (Fig. 3). A, Histograms show the densitometric values for the immunoblot polypeptide bands corresponding to 63-kD HMGR1S (B) revealed by anti-CD1 antiserum (light bars), and the corresponding HMGR activities (dark bars), normalized per total protein content, of extracts from 9-d-old seedlings grown under a continuous-light regime. The specific activity of wild-type plants was 0.26 ± 0.01 nmol mevalonate min−1 mg protein−1. The activities are the mean values of duplicate analyses ±10% variation. Fifteen micrograms of total protein were loaded per lane of SDS gels used for immunoblots. C, Staining of the PVDF membrane with Ponceau to reveal the band corresponding to Rubisco, used as an assessment for equal protein loading. D, Southern blots of the three lines.
The cytoplasmic, punctate pattern revealed by application of anti-CD1 antibodies (Fig. 3, C, D, and G) could be consistent with the known peroxisomal localization of mammalian HMGR (Keller et al., 1985; Appelkvist and Kalen, 1989). Therefore, it was of interest to determine whether these HMGR-containing structures were plant peroxisomes. Lisenbee et al. (2003a) recently showed that the mouse anti-tobacco catalase monoclonal antibodies and the rabbit anti-cottonseed catalase polyclonal antibodies used in our study recognized peroxisomal catalase in Arabidopsis suspension-cultured cells, but not in plant tissue cells. Figure 5 shows the results of dual-labeling immunofluorescence experiments conducted with these antibodies in 6-d-old wild-type Arabidopsis seedling cotyledons. Punctate structures attributable to immunorecognition of the mouse anti-tobacco catalase antibodies and the rabbit anti-cottonseed catalase antibodies are shown among the red plastids (Fig. 5A) and without chlorophyll detection (Fig. 5B). The corresponding binary mask image, where only coincident pixels are shown (Fig. 5C), reveals colocalization of both antibodies in peroxisomes. These results verified the monoclonal antibody recognition of cotyledon peroxisomal catalase and provided a positive control for colocalization.
Figure 5.
Immunofluorescence colocalization evaluations of catalase and HMGR in cotyledons of parenchyma cells of Arabidopsis seedlings. A to C, Colocalization analysis of confocal projections of mouse anti-tobacco catalase and rabbit anti-cottonseed catalase. Punctate mouse anti-catalase and rabbit anti-catalase shown with (A) and without (B) autofluorescent red plastids, and the binary mask result showing the colocalization of both antibodies in punctate peroxisomes (C). D to F, Colocalization evaluations of confocal projections of mouse anti-tobacco catalase (blue) and rabbit anti-CD1 (green). Triple-fluorescence images of both antibodies and plastids of a group of parenchyma cells (D), the combination of the antibody images only (E), and the binary mask colocalization analysis showing the absence of colocalizing pixels (F). Images are three-dimensional projections of a series of 40 to 50 sections with a z step of 0.25 μm. Bars = 10 μm.
Figure 5, D and E, illustrates dual-immunolabeling results with similar cells incubated in the same mouse monoclonal anti-catalase antibodies and rabbit anti-CD1 antiserum. Signals from both antibodies are observed with (Fig. 5D) and without (Fig. 5E) visible plastids. The resulting binary mask image (Fig. 5F) clearly shows that the signals are not colocalized, indicating that, in Arabidopsis seedlings, the HMGR signal is not located within catalase-containing peroxisomes. In confocal images, the mean diameter of HMGR-containing structures is approximately 0.6 μm, whereas peroxisomes are about 1 μm in diameter, in agreement with preexisting data for peroxisome size in Arabidopsis seedlings (Mathur et al., 2002). The data presented above indicate that most of Arabidopsis HMGR resides in as yet unidentified abundant spherical structures somewhat smaller in diameter than cotyledon peroxisomes.
Arabidopsis HMGR1S Localizes to High-Density Subcellular Fractions
Extracts of 9-d-old wild-type Arabidopsis seedlings were cleared of unbroken whole cells and cellular debris with a 200g (5 min) centrifugation. The supernatant (S200) was then centrifuged at 16,000g (20 min), and the resulting pellet (P16) and supernatant (S16) were assayed for their HMGR activity. The results indicated that 58.8% ± 5.7% of total activity was found in P16, and 40.7% ± 5.7% was found in S16. Similar partitioning between the pellet and supernatant was observed with plants grown under long-, short-, and continuous-daylight regimes and with 6-d-old and 5-week-old plants (data not shown). After the 16,000g centrifugation, the sediment contains intact plastids and peroxisomes, while S16 contains the nonorganellar cytoplasmic components and membranes derived from all disrupted organelles, including the ER and Golgi (Leech, 1977; Quail, 1979). We routinely confirmed the reliability of this separation through measurements of chlorophyll absorbance (Gegenheimer, 1990) and analyses of peroxisomal and cytoplasmic protein markers on immunoblots (see below).
With the objective of further characterizing the structures possessing HMGR, Suc density-gradient centrifugations were employed. P16 and S16 prepared from leaves were loaded onto 35% to 55% (w/v) and 20% to 40% (w/v) Suc gradients, respectively. Gradient fractions were analyzed on immunoblots with anti-CD1 antiserum (Fig. 6). The HMGR signal was observed in all the fractions derived from P16 (Fig. 6A, top blot), and in the less dense region of the 20% to 40% gradient derived from S16 (Fig. 6B, top blot). From densitometric analyses of the immunoblots, the partitioning of HMGR between P16 and S16 was approximately 70:30, respectively, in good agreement with activity data (see above text). A main polypeptide band of approximately 63 kD was identified in P16 fractions, consistent with the expected size of HMGR1S, the main isoform in adult leaves (Lumbreras et al., 1995). Additional minor bands of a slightly higher and a lower mobility were observed in S16 fractions. These bands might be HMGR isoforms 2 and 1L, with respective expected molecular masses of 60 and 69 kD. Similar results were obtained when S16 or P16 extracts from plants grown under short- and long-daylight regimes were applied to similar Suc gradients (data not shown).
Subcellular fraction distributions in these gradient experiments were determined with antiserum against cytoplasmic FPS1 and peroxisomal catalase, and from chlorophyll absorbance for chloroplasts. As expected, FPS1 signal was detected only in S16 fractions (Fig. 6B, middle blot; 40-kD polypeptide). Rabbit antiserum against cottonseed catalase recognized the Arabidopsis homolog (57-kD polypeptide) on immunoblots. As was anticipated, catalase released from ruptured peroxisomes was abundant at the top of the S16 20% to 40% gradients (Fig. 6B, bottom blot). Somewhat surprisingly, however, catalase was found in all fractions of the P16 35% to 55% gradient (Fig. 6A, bottom blot). These results suggest that our w/v Suc density gradients separate peroxisomes of varied density.
Immunogold Electron Microscopy of Arabidopsis HMGR in Seedling Cotyledons
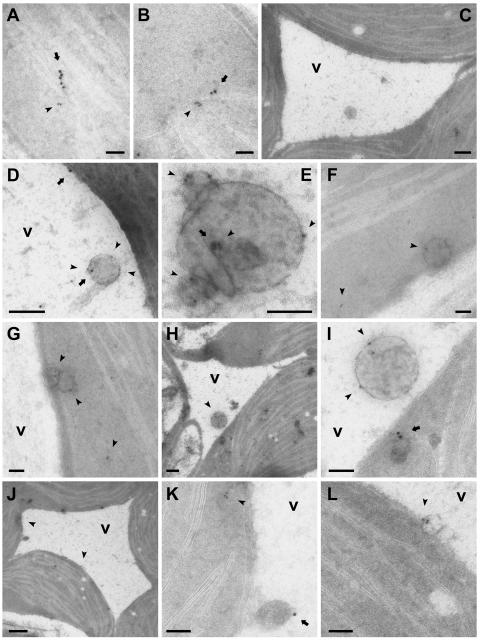
Subcellular localizations and ultrastructure of the HMGR-containing spherical bodies were elucidated through extensive immunogold electron microscopy studies of sections prepared from cotyledons of 6-d-old seedlings, which also were used for confocal immunofluorescence studies (Fig. 3). Figure 7, A and B, illustrates representative images of cell cytoplasm observed following dual immunogold labeling with the ER resident protein BiP (Ig heavy-chain binding protein) marked with 15-nm gold particles and HMGR marked with 10-nm gold particles. Double-labeling experiments for HMGR and BiP were done with two different anti-BiP antibodies: polyclonal rabbit anti-tobacco BiP (data shown) and monoclonal mouse anti-spinach BiP (data not shown), obtaining essentially the same labeling results in both cases. The linear arrangement of both sized gold particles is consistent with the expected colocalization of these proteins in ER. Figure 7C illustrates a representative negative gold-particle control image obtained after application of preadsorbed anti-CD1 antiserum (anti-CD1 antibodies preincubated with excess recombinant HMGR1 catalytic domain) and protein A-conjugated gold particles (10 nm). Gold particles are not observed over chloroplasts or anywhere in the nonorganellar cytoplasm surrounding a portion of the central vacuole containing a spherical structure bounded by a single membrane characteristic of vesicles. Results of dual labeling with anti-α-TIP and anti-HMGR antisera, shown in Figure 7, D and E, revealed HMGR-specific immunogold labeling of spherical, membrane-bounded vesicles within the central vacuole. These structures are similar in appearance to the unlabeled structures in the vacuole observed in the control images (e.g. Fig. 7C). Anti-α-TIP gold labeling was relatively less than HMGR gold labeling. Gold particles attributable to anti-α-TIP were observed over the tonoplast (Fig. 7D) and, less frequently, over intravacuolar vesicles (Fig. 7, D and E), providing additional evidence for the inclusion of the vesicles within the vacuole. The largest diameter of the sectioned vesicles observed in transmission electron microscopy images was approximately 0.4 μm, which is somewhat less than the average 0.6-μm diameter determined from measurements of significantly more spherical structures observed in whole-mount confocal images. This slight difference very likely is due to resolution differences between the two imaging processes.
Figure 7.
Immunogold electron microscopy of HMGR localizations in thin unembedded cryosections of 6-d-old wild-type Arabidopsis cotyledons. A, B, and F to L, Double labeling with anti-CD1 antiserum (10-nm gold particles) and anti-BiP antiserum (15-nm particles). C, Control image of sections incubated first with anti-CD1 antiserum preadsorbed with excess recombinant HMGR1 catalytic domain, then with protein A gold (10-nm particles). D and E, Double labeling with anti-CD1 antiserum (10-nm particles) and anti-α-TIP antiserum (15-nm particles). In all images (except H and J), arrowheads indicate site(s) of HMGR-specific 10-nm gold particles. Arrows indicate site(s) of BiP- or α-TIP-specific 15-nm particles. In images H and J, arrowheads point to regions that are shown at higher magnification in I (H), K, and L (J). v, Vacuole. Bars = 200 nm (C, D, and H), 500 nm (J), and 100 nm (all others).
The HMGR-containing vesicles also were observed in the nonorganellar cytoplasm, appearing as single structures (Fig. 7F) or in small groups (Fig. 7G). Although gold particles attributable to HMGR and BiP often were observed in neighboring areas in the same sections, they were not colocalized within vesicles in the nonorganellar cytoplasm or vacuoles (Fig. 7, H–L). BiP gold particles were observed over structures surrounded by cytoplasm (Fig. 7, H and I) and over structures adjacent to the tonoplast (Fig. 7, J and K) that resemble previously described BiP bodies (Levanony et al., 1992). Similarly, HMGR gold particles were identified in small membrane-bounded vesicles near the tonoplast (Fig. 7, J and L). In most cases, the HMGR-specific gold particles were associated with the membrane of the vesicles (Fig. 7, D–G, I, and L).
DISCUSSION
Several enzymes of the isoprenoid biosynthetic pathway in plants have been identified in more than one subcellular compartment. Such is the case for FPS in the soluble cytoplasm (Feron et al., 1990; Hugueney et al., 1996), mitochondria (Cunillera et al., 1997), and plastids (Sanmiya et al., 1999), and for geranylgeranyl diphosphate synthase in plastids, ER, and mitochondria (Okada et al., 2000). For one of the main regulatory enzymes of this pathway, HMGR, there is a paucity of in vivo data describing its subcellular localization. In vitro, it has been shown that Arabidopsis HMGR1S is inserted into ER microsomal membranes (Enjuto et al., 1994; Campos and Boronat, 1995; Lumbreras et al., 1995). The amino-terminal region of HMGR1S containing the membrane domain (residues 1–178) appears to be necessary (Fig. 3, immunofluorescence data) and sufficient (Fig. 2, GFP data) for targeting to and accumulation in ER and spherical structures. Consistently, the targeting domain of both yeast HMGRs has also been assigned to the amino-terminal transmembrane region of the protein (Hampton et al., 1996), and it was shown that removal of the membrane domain led to increased protein and enzyme activity (Donald et al., 1997). Transgenic plants used in this study carrying an extra CD1 copy showed greatly increased HMGR activity, indicating that when the catalytic domain was removed from its membrane environment, deregulation ensued. This suggests a key role of the endomembrane system in the control of HMGR activity.
Our results show that the main reservoir of Arabidopsis HMGR in cotyledons is not within the reticular ER, but within novel cytoplasmic and intravacuolar vesicular structures that seem to be derived from subdomains of the ER. Given the versatility of the multifunctional plant ER, it seems likely that specialized ER regions exist based on their functional attributes. Papers variously describing such so-called ER domains are plentiful (Okita and Rogers, 1996; Staehelin, 1997; Mullen et al., 1999; Choi et al., 2000; Chrispeels and Herman, 2000; Naested et al., 2000; Braun, 2001; Sami-Subbu et al., 2001; Hamada et al., 2003; Matsushima et al., 2003), although in some cases the suggested domains might have been identified as the result of protein aggregation. The punctate immunofluorescence microscopy pattern revealed by our subcellular localization study of Arabidopsis HMGR is reminiscent of the protein bodies formed in yeast by the ER resident protein BiP upon overproduction of the ER membrane protein Sec12p that inhibits protein transport from the ER to the Golgi apparatus (Nishikawa et al., 1994). In plants, castor bean endosperm BiP has been identified in ricinosomes, an ER-derived organelle that migrates to high-density fractions in Suc density gradients (Schmid et al., 2001). Like BiP, HMGR is an ER resident protein that can leave the reticular ER to be incorporated into high-density vesicles that could be considered a specialized ER subdomain. In angiosperms, it has been suggested that the transport of proteins out of the ER is mediated by membrane-bounded vesicles (Hawes et al., 1999; Pimpl et al., 2000), as is the case in mammalian and yeast cells (Klumperman, 2000), although evidence for the existence of such vesicles in plants is not well documented, and the involvement of vesicles in the ER-to-Golgi transport is being challenged in the plant field (Neumann et al., 2003). The data presented here are consistent with the existence in Arabidopsis cotyledons of trafficking of HMGR out of the ER in vesicular structures. A fraction of the vesicles might be involved in the mobilization of enzymes or isoprenoid products to other parts of the cytoplasm, perhaps representing the physical basis of processes such as the transport of leek seedling sterols from the ER to the plasma membrane (Moreau et al., 1998). However, our data indicate that a significant fraction of HMGR-containing vesicles go to the vacuole.
The transport of HMGR-enriched vesicles to the vacuole could represent, among several other possibilities, the need for storage of HMGR in cotyledons or the entry of ER vesicles into the lytic vacuole by autophagy as part of a continuous or regulated HMGR flux. Vacuoles occupy up to 90% of the volume of a plant cell and are part of the endomembrane system that also includes the secretory pathway. This system comprises several compartments: ER, Golgi apparatus, trans-Golgi network, prevacuolar compartments, the vacuole, and endosomes (Surpin and Raikhel, 2004). In wheat, many storage proteins aggregate within the ER and are subsequently transported as protein bodies to the vacuole through a route independent of the Golgi complex, as is the case for γ-gliadin and BiP in the developing endosperm (Levanony et al., 1992). In developing pumpkin cotyledons, the storage protein 2S albumin is transported from the ER to vacuoles, in vesicles surrounded by an ER-derived membrane (Hara-Nishimura et al., 1998). These data are consistent with the hypothesis that the formation of transport vesicles can be mediated by the ER rather than by the Golgi. Our results provide supporting evidence of a possible role for the ER in vacuolar ontogeny in young seedlings by forming small vesicles that are involved in the mobilization of proteins in the vacuoles (Chrispeels and Herman, 2000).
As yet, no coherent rationale has been developed to explain the presumed regulatory roles played by the different HMGR isoforms. However, trafficking of specific forms of HMGR through the ER to other organelles in the endomembrane system has been suggested (Campos and Boronat, 1995), as has the involvement of metabolite channeling through multienzyme complexes and the presence of specific isoforms of HMGR in discrete regions of the ER (Chappell, 1995; McCaskill and Croteau, 1998). The HMGR-enriched vesicles might have characteristic enzymatic compositions that could represent the physical basis of metabolic channels specialized in the biosynthesis of isoprenoids with a particular structure and/or subcellular distribution. In yeast, there is evidence in favor of the compartmentalization of the two HMGR isoforms HMG1p and HMG2p that are targeted to the perinuclear ER and to the peripheral ER, respectively (Wright et al., 1988). This differential localization has been associated with different responses of each isoform to specific stimuli (Hampton et al., 1996). While HMG1p was relatively stable, HMG2p was quickly degraded in response to an increase in metabolite flow through the sterol pathway. Similarly, it was previously proposed that the various plant HMGR isoforms might act on separate pathways specialized in the synthesis of different isoprenoid products (Chappell et al., 1991; Choi et al., 1992). However, no evidence for a physical association between different enzymes has been reported so far. Our results demonstrating that plant HMGR concentrates in specific regions of the cell might fit the compartmentalization model, but further studies are required to confirm, for instance, that other enzymes of the isoprenoid pathway are also located in the HMGR-containing vesicles.
In some subcellular fractionation studies, plant HMGR activity has been detected not only in ER-derived membranes, but also in high-density fractions containing mitochondria, chloroplasts, and peroxisomes (Brooker and Russell, 1975; Wong et al., 1982; Keller et al., 1985; Appelkvist and Kalen, 1989). These observations led to the suggestion that these organelles contained the various HMGR isoforms. In liver, CHO, and brain cells, HMGR was found in ER and peroxisomes (Keller et al., 1985; Appelkvist and Kalen, 1989; Aboushadi et al., 1999; Kovacs et al., 2001), suggesting an essential role of peroxisomes in isoprenoid biosynthesis (Gupta et al., 1999). Animal peroxisomes apparently are a major site of cholesterol biosynthesis since they contain all of the cholesterogenic enzymes involved in the conversion of mevalonate into farnesyl diphosphate (e.g. Kovacs et al., 2002). However, we show that Arabidopsis cotyledon peroxisomes do not possess HMGR (Fig. 5). Our results presented herein revealing the presence of HMGR in a novel type of dense structure could reconcile those previous data with the current view of HMGR being an enzyme exclusively derived from the ER. Wong et al. (1982) described the existence in pea seedlings of two kinetically distinct HMGR activities. A proposed plastidic subpopulation had a much higher affinity for HMG-CoA and a higher pH optimum than the cytoplasmic reductase. Comparative values for the plastid and cytoplasmic (parentheses) enzymes were: apparent Km for HMG-CoA, 0.77 μm (160 μm); pH optimum, 7.9 (6.9). Although some HMGR activity in plastids cannot be completely ruled out, our results are consistent with the hypothesis that the HMGR-containing vesicles cosediment with plastids, as we observed in our own density gradients shown in Figure 6. Therefore, the bulk of HMGR activity measured in the plastid fraction would be attributable to HMGR in the spherical structures, not in plastids. This is consistent with the proposal that the isoprenoid biosynthetic route in plastids proceeds via a mevalonate-independent pathway (Rohmer et al., 1993; Lichtenthaler et al., 1997).
The dual localization of HMGR in the ER (where it is synthesized and inserted into its membrane) and in cytoplasmic and vacuolar vesicles suggests that, in plants, mevalonate biosynthesis can occur in different cell compartments. This, in turn, could be the molecular basis for a subcellular partitioning of isoprenoid biosynthesis.
MATERIALS AND METHODS
Plant Cell Fractionation
Seeds from Arabidopsis (Arabidopsis thaliana ecotype Columbia) plants were surface sterilized and then sown in petri dishes containing solid (0.8% w/v agar) germination medium (Murashige and Skoog medium supplemented with 10 g L−1 Suc, 0.5 g L−1 MES, pH 5.7). Dishes were incubated at 22°C ± 2°C for the days indicated for long- (16-h light/8-h dark), short- (8/16), or continuous-day (24/0) illumination regimes with 130 μmol m−2 s−1 daylight fluorescent illumination. Transgenic plants transformed with individual HMGR isoforms were grown on plates with germination medium containing 50 μg mL−1 kanamycin. Thereafter, 9-d-old seedlings were transplanted into soil and allowed to grow for up to 5 weeks under the same light/dark conditions. For subcellular fractionation of plant organs, leaves (400 mg) from 5-week-old plants were ground in a mortar on ice with 3 mL of extraction medium containing 0.1 m Tricine, pH 7.5, 10 mm KCl, 1 mm MgCl2, 1 mm EDTA, 1% (w/v) Ficoll, 0.1% (w/v) bovine serum albumin (BSA; Fraction V; Sigma, St. Louis), 0.5 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, and 20% (w/v) Suc. The ground material was cleared of large debris and nuclei in a short centrifugation at 200g (5 min, 4°C). The resulting supernatant was spun in a bench-top centrifuge at 16,000g (20 min, 4°C) to yield a pellet (P16) and supernatant (S16). Protein was determined with the DC Protein Assay from Bio-Rad (Hercules, CA). The protein content was nearly equal in P16 and S16. S16 was spun again as above and 1 mL of the resulting clarified S16, containing approximately 1.5 mg of protein, was loaded directly on top of a 40-mL continuous 20% to 40% (w/v) Suc gradient freshly prepared in Thinwall Ultra-Clear Beckman tubes (Beckman, Fullerton, CA). P16 was washed once with extraction buffer and the resulting cleaned P16, adjusted to contain approximately 1.5 mg of protein, was taken up in 1 mL of extraction buffer containing 35% (w/v) Suc and loaded on top of a 40-mL continuous 35% to 55% (w/v) Suc gradient prepared as described above. The gradients were centrifuged in a Beckman swinging bucket SW 28 rotor at 25,000 rpm (82,700g at rav) for 6 h (4°C). Approximately 20 2-mL fractions were collected immediately from each gradient by puncturing the bottom of tubes with a red-hot hypodermic needle. Fractions were kept frozen at −20°C.
HMGR Activity Assays, Antibody Production, and Immunoblot Analyses
HMGR activity was assayed as described in Masferrer et al. (2002). The mean values and sds were obtained from eight independent assays of the same sample, unless otherwise specified. The cDNA coding for the catalytic domain of Arabidopsis HMGR1 was expressed in Escherichia coli and the protein (CD1) was purified according to Dale et al. (1995). Antibodies against the purified protein (anti-CD1) were generated in rabbits as described in Manzano et al. (2004). For immunoblot analyses, each well in a 12.5% SDS-polyacrylamide gel was loaded with either equal amounts of total protein (Fig. 4; 15 μg/lane) or equal volumes of density gradient fractions (Fig. 6; 30 μL/lane). Electrophoresis was performed as described by Laemmli (1970) and proteins were transferred to a Hybond-P polyvinylidenedifluoride (PVDF) membrane (Amersham, Buckinghamshire, UK) using a mini trans-blot cell (Bio-Rad). Equal protein loading in Figure 4 was further assessed by Ponceau staining of the PVDF membrane blot. Densitometric analysis (see below) of the band corresponding to Rubisco indicated that differences in protein content between lanes were always less than 10%. PVDF membranes were blocked in 0.1 m Tris-HCl, pH 7.4, 0.1 m MgCl2, 0.5% (v/v) Tween 20, 1% BSA (Fraction V; Sigma), and 5% (v/v) fetal calf serum (Reactiva) for 1 h at room temperature. PVDF membranes were incubated with rabbit antiserum raised against either the catalytic domain of Arabidopsis HMGR1 (CD1), Arabidopsis FPS1 (Masferrer et al., 2002), or cottonseed catalase (Lisenbee et al., 2003a), all of them at 1:2,000 dilution in blocking buffer for 16 h at 4°C. The ECL western-blotting detection system (Amersham) was used to visualize the decorated bands. The negatives were scanned at 200 dpi (AGFA Duoscan T2000 XL). Density analyses of each band were made using a computerized densitometer and the IMAT program (Scientific and Technical Services, University of Barcelona, Barcelona).
Genomic DNA Extraction and Southern-Blot Analysis
For extraction of plant genomic DNA, approximately 1 g of fresh Arabidopsis leaves was ground in a mortar in the presence of liquid nitrogen. To the fine powder were added 5 mL of DNA extraction buffer (0.1 m EDTA, 0.25 m NaCl, 100 μg/mL proteinase K, 0.1 m Tris-HCl, pH 8.0), and the grinding was resumed until the complete disappearance of undissolved material. The resulting solution was transferred to a conical tube where N-lauroylsarcosine was added to a final concentration of 1% (w/v). After a 2-h incubation at 55°C, the tube was centrifuged (10 min, 6,000 rpm), and the resulting supernatant centrifuged again several times in the same conditions until the solution was clear. The DNA was then precipitated with 0.6 volumes of isopropyl alcohol, washed twice with 70% ethanol, slightly dried, and finally taken up in Tris EDTA buffer (1 mm EDTA, 10 mm Tris-HCl, pH 7.5). Genomic DNA was further purified by adding 1/10 volume of a NaCl/hexadecyltrimethylammonium bromide solution (5 m NaCl, 10% w/v hexadecyltrimethylammonium bromide). After a 20-min incubation at 65°C, the aqueous phase was extracted several times with 1 volume of CHCl3, and the purified DNA finally precipitated with 0.6 volumes of isopropyl alcohol, washed with 70% ethanol, and taken up in Tris EDTA buffer, pH 7.5. A digoxigenin-labeled DNA probe was amplified by PCR from a region of the HMGR1 catalytic domain cDNA lacking EcoRI sites. Primers SP6 (antisense) and an internal primer located on position 722 of the HMGR1L cDNA (sense) were used with the cloned cDNA as a template to generate an 878-bp probe that stretched until position 1,600 of the coding region. Alkali-labile DIG-dUTP (Boehringer Mannheim, Basel) was included in the PCR reaction mixture with a DIG-dUTP:dTTP ratio of 1:3. For Southern-blot analysis, genomic DNA was digested with EcoRI (Gibco BRL, Cleveland), under the conditions specified by the supplier. The digested DNA was electrophoresed in 1% agarose gels, transferred to a positively charged nylon membrane (Boehringer Mannheim) by capillary transfer, and hybridized with the DIG-labeled DNA probe overnight at 42°C. Hybridization, stringency washes, and detection were performed following the instructions in the DIG DNA-labeling kit (Boehringer Mannheim), with a final wash of 0.1× SSC, 0.1% SDS at 68°C.
Confocal Microscopy
For whole-mount immunofluorescence experiments, 6-d-old Arabidopsis seedlings grown under a short-daylight regime were fixed for 3 h at room temperature under vacuum in 4% (v/v) formaldehyde (prepared from paraformaldehyde) in PMEG buffer (50 mm KPIPES, pH 7.5, 2 mm MgSO4, 5 mm EGTA) and 10% (v/v) dimethyl sulfoxide, washed three times over 30 min in PMEG, and incubated for 30 min at 37°C in a cell wall-digesting solution containing 0.5% (w/v) pectolyase from Aspergillus japonicus (Sigma; 4.5 units/mg solid), 2% (v/v) Triton X-100, and 1% (w/v) BSA (Fraction V; Merck, Rahway, NJ) in PMEG. Seedlings were washed three more times in PMEG and incubated overnight at 4°C with primary antibody (see above) diluted in 0.1 m phosphate-buffered saline (PBS) containing 3% (w/v) BSA (PBS/BSA). Rabbit antiserum against bean α-TIP was kindly provided by Dolors Ludevid (Centre d'Investigació i Desenvolupament-Consejo Superior de Investigaciones Científicas, Barcelona), and originally obtained from H. Höfte (Höfte and Chrispeels, 1992). The working concentration of the primary antibody (generally 1:500 dilution) was empirically determined. After washing with PBS three times (30 min), seedlings were incubated with the secondary antibody (see below) in PBS/BSA for 3 h at 37°C, and washed three more times with PBS. Finally, whole seedlings were mounted on a glass microscope slide with Mowiol (Calbiochem, San Diego), and stored at 4°C in the dark until examination with the confocal microscope. AlexaFluor 488 goat anti-rabbit F(ab′)2 was used as a secondary antibody for the rabbit anti-CD1 and the rabbit anti-cottonseed catalase antiserum (Kunce et al., 1988), and AlexaFluor 350 goat anti-mouse F(ab′)2 was used for the mouse monoclonal anti-tobacco catalase (Chen et al., 1993). Both secondary antibodies were purchased from Molecular Probes (Eugene, OR) and applied at a final concentration of 7 μg/mL. Whole mounts of seedlings were analyzed with a Leica SP2 confocal microscope (Leica, Wetzlar, Germany), using an HCX Plan Apochromatic 63 X/N.A.1.3 oil or an HCX PL APO 100.0 × 1/40 oil objectives at several zoom factors. For double- and triple-fluorescence imaging, sequential excitations of the AlexaFluor 350 (351-nm line of an UV-Ar laser), AlexaFluor 488 (488-nm line of an Ar laser), and the autofluorescence from plastids (633 nm of an He-Ne laser) were combined with a narrow spectral detection to avoid autofluorescence contamination of the AlexaFluor 350 and AlexaFluor 488 detection. For the autofluorescence of plastids, the spectral detection was set. Seedlings also were observed using a fluorescence low-magnification microscope (Leica MZ FLIII) coupled to a CCD (Leica DC camera and Leica ICM 1000 software). Three-dimensional projections and three-dimensional colocalization analyses were made with the Imaris (version 2.7) and Colocalization (version 1.2) Software packages from Bitplane AG (Zurich). Organelle size quantification was made using the Image Morphometric Analysis function of the Metamorph Software package from Universal Imaging (Chester, PA).
Transgenic Plants, Plasmid Construction, and Microbombardment Assays
Plant expression vectors were constructed according to standard molecular biology procedures (Ausubel et al., 1987). Transgenic plants transformed with isoforms HMGR1S, HMGR1L, HMGR2, and with the catalytic domain of isoform HMGR1 were generated by Víctor M. González (González, 2002). The cDNA sequences encoding the different forms were cloned under the control of the cauliflower mosaic virus 35S promoter. Arabidopsis plants were transformed using the in planta vacuum infiltration method (Bechtold et al., 1993) and Agrobacterium tumefaciens. For GFP experiments, pRTL2-sGFP plant expression plasmid containing sGFP(S65T) (Chiu et al., 1996) was modified to generate pGFPau plasmid, by introducing at the NcoI site a polylinker containing the sequence 5′-CATGGGCAATTGGTACCGCCGGCTAGCGGCCGCAGTCGACGG-3′. The polylinker inserted before the ATG start codon of the GFP in pGFPau allowed us to insert DNA codons for in-frame fusion proteins at the amino-terminal site of GFP. To generate the 1S:GFP fusion, a PCR-amplified fragment coding for amino acids 1 to 178 of HMGR1S was previously cloned in the pAS2-1 vector (Clontech Laboratories, Palo Alto, CA) using the NcoI and BamHI sites. It was then recovered with NcoI and SalI and cloned into pGFPau to generate the 1S:GFP vector. All the resulting vectors were verified by sequencing, using the ABI PRISM BigDye DNA sequencing kit (Perkin-Elmer Biosystems, Foster City, CA). For the microbombardment assays, Arabidopsis plants were grown for 4 to 6 weeks under short-daylight conditions. Arabidopsis leaf cells were then microbombarded with the 1S:GFP construct using a Biolistic PDS-1000/He system (Bio-Rad) as described (Lois et al., 2000). After 18 h of expression, subcellular localization of the fusion proteins was determined by analyzing the green fluorescence patterns by confocal laser-scanning microscopy. The pEGFPer vector was kindly provided by Patrick Gallois (University of Manchester, UK). It was created as a positive control for ER localization by adding a basic chitinase secretion peptide and the ER retention signal KDEL to EGFP (Danon et al., 2004).
Immunogold Electron Microscopy
For immunogold labeling of nonembedded cryosections, discs (1-mm diameter) excised from cotyledons of 6-d-old Arabidopsis seedlings grown under a short-daylight regime were chemically fixed overnight at 4°C in a mixture of 2% formaldehyde (prepared from paraformaldehyde; Ted Pella) and 0.1% (v/v) glutaraldehyde (EMS) in 0.1 m sodium phosphate buffer (PB), pH 7.5. After washing with PB containing 50 mm Gly, discs were embedded in 10% (w/v) gelatin (Merck; molecular biology grade) and infused with 2.3 m Suc in PB (Raposo et al., 1997). Mounted gelatin blocks were frozen in liquid propane. Ultrathin sections were cut with an ultracryomicrotome (Leica EM FCS, Vienna). The frozen sections of the discs were collected with a solution containing 1% methylcellulose and 1.15 m Suc (Liou et al., 1996), deposited onto Cu/Pd grids, and then immunogold labeled with anti-CD1 antiserum and protein A coupled to 10-nm gold particles (purchased from Dr. J.W. Slot, Department of Cell Biology, Utrecht University Medical School, Utrecht, The Netherlands). Specifically, fixed cryosections were incubated at 37°C for 20 min in 0.1 m PBS containing 2% (w/v) gelatin, and then washed four times for 3 min each at room temperature in PBS containing 0.4% (w/v) Gly. After a final wash in PBS containing 10% (w/v) fetal bovine serum (FBS; Amresco, Solon, OH; PBS/FBS10), sections were incubated for 30 min with anti-CD1 antiserum diluted 1:50 in PBS/FBS5. After eight 2-min washes in PBS/FBS0.2, the sections were incubated for 20 min with protein A conjugated to 10-nm gold particles diluted 1:60 in PBS/FBS5. Grids were then rinsed with eight 2-min PBS washes and incubated for 5 min in PBS containing 1% glutaraldehyde. At this point, the grids selected for double staining with rabbit anti-tobacco BiP or rabbit anti-bean α-TIP (Höfte and Chrispeels, 1992) were returned to the PBS/gelatin step. The same processing of grids was done as above, except goat anti-rabbit IgGs conjugated to 15-nm gold particles (Aurion-BSA, Science Services, Munich) were applied instead of protein A gold conjugated to 10-nm particles. Thereafter, the PBS washes were the same as above. For double labelings of HMGR and BiP using a monoclonal anti-spinach BiP (mouse anti-HSC70; StressGen Biotechnologies, Victoria, British Columbia, Canada), both primary antibodies were added together; later, protein A gold conjugated to 10-nm beads and goat anti-mouse conjugated to 15-nm gold particles (Biocell British/International, Cardiff, UK) were added also in a single step. Finally, grids were washed in water 8 times, 2 min each. Cryosections were contrasted and embedded in a mixture of methylcellulose and uranyl acetate and observed with a JEOL 1010 transmission electron microscope.
Acknowledgments
We thank D. Ludevid for providing the antisera against BiP and α-TIP; R. Blanvillain and P. Gallois for their gift of the pEGFPer construct; J.D.I. Harper for his helpful comments about immunofluorescence techniques in Arabidopsis; and the Scientific and Technical Services of the University of Barcelona and the Servei de Camps Experimentals (Facultat de Biologia, University of Barcelona) for technical assistance, in particular to Raquel García, Sonia Ruiz, and Josep Matas for their help with the confocal immunofluorescence microscopy, with the preparation of samples for immunogold electron microscopy, and with the maintenance of Arabidopsis plants, respectively.
This work was supported by the Ministerio de Ciencia y Tecnología (MCyT; grant nos. BIO2000–0334 to A.F., BIO2002–00128 to X.F.-B., BMC2003–03450 to N.C., and BMC2003–06833 to A.B.; all grants included Fondo Europeo de Desarrollo Regional funds), and by the National Science Foundation (grant no. MCB–0091826 to R.N.T.). X.F.-B. and N.C. hold Ramón y Cajal tenure-track positions from the MCyT. P.L. acknowledges receipt of a fellowship from the Comissió Interdepartamental de Recerca i Innovació Tecnològica (Generalitat de Catalunya).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.050245.
References
- Aboushadi N, Engfelt WH, Paton VG, Krisans SK (1999) Role of peroxisomes in isoprenoid biosynthesis. J Histochem Cytochem 47: 1127–1132 [DOI] [PubMed] [Google Scholar]
- Appelkvist EL, Kalen A (1989) Biosynthesis of dolichol by rat liver peroxisomes. Eur J Biochem 185: 503–509 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, eds (1987) Current Protocols in Molecular Biology. John Wiley and Sons, New York
- Bach TJ (1995) Some new aspects of isoprenoid biosynthesis in plants: a review. Lipids 30: 191–202 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Braun M (2001) Association of spectrin-like proteins with the actin-organized aggregate of endoplasmic reticulum in the Spitzenkorper of gravitropically tip-growing plant cells. Plant Physiol 125: 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker JD, Russell DW (1975) Subcellular localization of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Pisum sativum seedlings. Arch Biochem Biophys 167: 730–737 [DOI] [PubMed] [Google Scholar]
- Campos N, Boronat A (1995) Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell 7: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J (1995) Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 521–547 [Google Scholar]
- Chappell J, VonLanken C, Vögeli U (1991) Elicitor inducible 3-hydroxy-3-methylglutaryl coenzyme A reductase activity is required for sesquiterpene accumulation in tobacco cell suspension cultures. Plant Physiol 97: 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ricigliano JW, Klessig DF (1993) Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc Natl Acad Sci USA 90: 9533–9537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Choi D, Ward BL, Bostock RM (1992) Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell 4: 1333–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW (2000) Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 407: 765–767 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Herman EM (2000) Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol 123: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388 [DOI] [PubMed] [Google Scholar]
- Dale S, Arró M, Becerra B, Morrice NG, Boronat A, Hardie DG, Ferrer A (1995) Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem 233: 506–513 [DOI] [PubMed] [Google Scholar]
- Danon A, Rotari VI, Gordon A, Mailhac N, Gallois P (2004) Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and Defender against Apoptotic Death. J Biol Chem 279: 779–787 [DOI] [PubMed] [Google Scholar]
- Denbow CJ, Lang S, Cramer CL (1996) The N-terminal domain of tomato 3-hydroxy-3-methylglutaryl-CoA reductases. Sequence, microsomal targeting, and glycosylation. J Biol Chem 271: 9710–9715 [DOI] [PubMed] [Google Scholar]
- Donald KA, Hampton RY, Fritz IB (1997) Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol 63: 3341–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M, Balcells L, Campos N, Caelles C, Arró M, Boronat A (1994) Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA 91: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron G, Clastre M, Ambid C (1990) Prenyltransferase compartmentation in cells of Vitis vinifera cultivated in vitro. FEBS Lett 271: 236–238 [DOI] [PubMed] [Google Scholar]
- Gegenheimer P (1990) Preparation of extracts from plants. Methods Enzymol 182: 174–193 [DOI] [PubMed] [Google Scholar]
- González V (2002) Regulación de la biosíntesis de isoprenoides en plantas: estudio molecular del control de la vía a nivel de la enzima HMG-CoA reductasa. PhD thesis, University of Barcelona, Barcelona
- Gunning BES (1998) The identity of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci 3: 417. [DOI] [PubMed] [Google Scholar]
- Gupta SD, Mehan RS, Tansey TR, Chen HT, Goping G, Goldberg I, Shechter I (1999) Differential binding of proteins to peroxisomes in rat hepatoma cells: unique association of enzymes involved in isoprenoid metabolism. J Lipid Res 40: 1572–1584 [PubMed] [Google Scholar]
- Hamada S, Ishiyama K, Sakulsingharoj C, Choi SB, Wu Y, Wang C, Singh S, Kawai N, Messing J, Okita TW (2003) Dual regulated RNA transport pathways to the cortical region in developing rice endosperm. Plant Cell 15: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Koning A, Wright R, Rine J (1996) In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc Natl Acad Sci USA 93: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura I, Shimada T, Hatano K, Takeuchi Y, Nishimura M (1998) Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C, Saint-Jore C, Martin B, Zheng HQ (2001) ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci 6: 245–246 [DOI] [PubMed] [Google Scholar]
- Hawes CR, Brandizzi F, Andreeva AV (1999) Endomembranes and vesicle trafficking. Curr Opin Plant Biol 2: 454–461 [DOI] [PubMed] [Google Scholar]
- Höfte H, Chrispeels MJ (1992) Protein sorting to the vacuolar membrane. Plant Cell 4: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, Quennemet J, d'Harlingue A, Camara B (1996) Developmental and stress regulation of gene expression for plastid and cytosolic isoprenoid pathways in pepper fruits. Plant Physiol 111: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller GA, Barton MC, Shapiro DJ, Singer SJ (1985) 3-Hydroxy-3-methylglutaryl-coenzyme A reductase is present in peroxisomes in normal rat liver cells. Proc Natl Acad Sci USA 82: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J (2000) Transport between ER and Golgi. Curr Opin Cell Biol 12: 445–449 [DOI] [PubMed] [Google Scholar]
- Koning AJ, Roberts CJ, Wright RL (1996) Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol Biol Cell 7: 769–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs WJ, Faust PL, Keller GA, Krisans SK (2001) Purification of brain peroxisomes and localization of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Eur J Biochem 268: 4850–4859 [DOI] [PubMed] [Google Scholar]
- Kovacs WJ, Olivier LM, Krisans SK (2002) Central role of peroxisomes in isoprenoid biosynthesis. Prog Lipid Res 41: 369–391 [DOI] [PubMed] [Google Scholar]
- Kunce CM, Trelease RN, Turley RB (1988) Purification and biosynthesis of cottonseed (Gossypium hirsutum L.) catalase. Biochem J 251: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leech RM (1977) Subcellular fractionation techniques in enzyme distribution studies. In H Smith, ed, Regulation of Enzyme Synthesis and Activity in Higher Plants. Academic Press, London, pp 289–327
- Levanony H, Rubin R, Altschuler Y, Galili G (1992) Evidence for a novel route of wheat storage proteins to vacuoles. J Cell Biol 119: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Rohmer M, Schwender J (1997) Two independent biochemical pathways for isopentenyl diphosphate and isoprenoid biosynthesis in higher plants. Physiol Plant 101: 643–652 [Google Scholar]
- Liou W, Geuze HJ, Slot JW (1996) Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol 106: 41–58 [DOI] [PubMed] [Google Scholar]
- Lisenbee CS, Heinze M, Trelease RN (2003. a) Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis cells. Plant Physiol 132: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisenbee CS, Karnik SK, Trelease RN (2003. b) Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic 4: 491–501 [DOI] [PubMed] [Google Scholar]
- Lois LM, Rodríguez-Concepción M, Gallego F, Campos N, Boronat A (2000) Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J 22: 503–513 [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Campos N, Boronat A (1995) The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J 8: 541–549 [DOI] [PubMed] [Google Scholar]
- Manzano D, Fernàndez-Busquets X, Schaller H, González V, Boronat A, Arró M, Ferrer A (2004) The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta 219: 982–992 [DOI] [PubMed] [Google Scholar]
- Masferrer A, Arró M, Manzano D, Schaller H, Fernàndez-Busquets X, Moncaleán P, Fernández B, Cunillera N, Boronat A, Ferrer A (2002) Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J 30: 123–132 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Hülskamp M (2002) Simultaneous visualization of peroxisomes and cytoskeletal elements reveals actin and not microtubule-based peroxisome motility in plants. Plant Physiol 128: 1031–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33: 493–502 [DOI] [PubMed] [Google Scholar]
- McCaskill D, Croteau R (1998) Some caveats for bioengineering terpenoid metabolism in plants. Trends Biotechnol 16: 349–355 [Google Scholar]
- McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7: 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Hartmann MA, Perret AM, Sturbois-Balcerzak B, Cassagne C (1998) Transport of sterols to the plasma membrane of leek seedlings. Plant Physiol 117: 931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN (1999) Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. Plant Cell 11: 2167–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Nebenführ A (2002) Vesicle traffic in the endomembrane system: a tale of COPs, Rabs and SNAREs. Curr Opin Plant Biol 5: 507–512 [DOI] [PubMed] [Google Scholar]
- Neumann U, Brandizzi F, Hawes C (2003) Protein transport in plant cells: in and out of the Golgi. Ann Bot (Lond) 92: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Hirata A, Nakano A (1994) Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol Biol Cell 5: 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita TW, Rogers JC (1996) Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol 47: 327–350 [DOI] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG (2000) In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (1979) Plant cell fractionation. Annu Rev Plant Physiol 30: 425–484 [Google Scholar]
- Raposo G, Kleijmeer MJ, Posthuma G, Slot JW, Geuze HJ (1997) Immunolabeling of ultrathin cryosections: application in immunology. In LA Herzenberg, D Weir, C Blackwell, eds, Handbook of Experimental Immunology, Vol 4. Blackwell Science, Cambridge, MA, pp 1–11
- Rodríguez-Concepción M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F, Kis K, Bacher A, Eisenreich W (2001) The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr Opin Chem Biol 5: 535–540 [DOI] [PubMed] [Google Scholar]
- Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sami-Subbu R, Choi SB, Wu Y, Wang C, Okita TW (2001) Identification of a cytoskeleton-associated 120 kDa RNA-binding protein in developing rice seeds. Plant Mol Biol 46: 79–88 [DOI] [PubMed] [Google Scholar]
- Sanmiya K, Ueno O, Matsuoka M, Yamamoto N (1999) Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol 40: 348–354 [DOI] [PubMed] [Google Scholar]
- Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C (2001) The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc Natl Acad Sci USA 98: 5353–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA (1997) The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Surpin M, Raikhel N (2004) Traffic jams affect plant development and signal transduction. Nat Rev Mol Cell Biol 5: 100–110 [DOI] [PubMed] [Google Scholar]
- Weissenborn DL, Denbow CJ, Laine M, Lång SS, Yang Z, Yu X, Cramer CL (1995) HMG-CoA reductase and terpenoid phytoalexins: molecular specialization within a complex pathway. Physiol Plant 93: 393–400 [Google Scholar]
- Wong RJ, McCormack DK, Russell DW (1982) Plastid 3-hydroxy-3-methylglutaryl coenzyme A reductase has distinctive kinetic and regulatory features: properties of the enzyme and positive phytochrome control of activity in pea seedlings. Arch Biochem Biophys 216: 631–638 [DOI] [PubMed] [Google Scholar]
- Wright R, Basson M, D'Ari L, Rine J (1988) Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol 107: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]